Abstract
CaVEMan is an expectation maximization–based somatic substitution-detection algorithm that is written in C. The algorithm analyzes sequence data from a test sample, such as a tumor relative to a reference normal sample from the same patient and the reference genome. It performs a comparative analysis of the tumor and normal sample to derive a probabilistic estimate for putative somatic substitutions. When combined with a set of validated post-hoc filters, CaVEMan generates a set of somatic substitution calls with high recall and positive predictive value. Here we provide instructions for using a wrapper script called cgpCaVEManWrapper, which runs the CaVEMan algorithm and additional downstream post-hoc filters. We describe both a simple one-shot run of cgpCaVEManWrapper and a more in-depth implementation suited to large-scale compute farms.
Keywords: somatic, cancer, sequencing, SNV, substitution
Introduction
CaVEMan is a somatic substitution-detection algorithm written in C, which implements an expectation-maximization (EM) algorithm (Do and Batzoglou, 2008). The EM algorithm is an iterative maximum likelihood estimation algorithm for statistical models that depend on latent (not directly observed/inferred) variables. CaVEMan utilizes BAM (Li et al., 2009) or CRAM (Hsi-Yang Fritz et al., 2011) next-generation sequencing files as input, and uses location-specific copy number information and aberrant cell fraction to calculate the probability of putative somatic genotypes by comparing tumor data against a control sample and the reference genome. If the algorithm determines that the sum of all possible somatic genotype probabilities is above the defined probability cut-off, the position is output to file in VCF (Danecek et al., 2011) format.
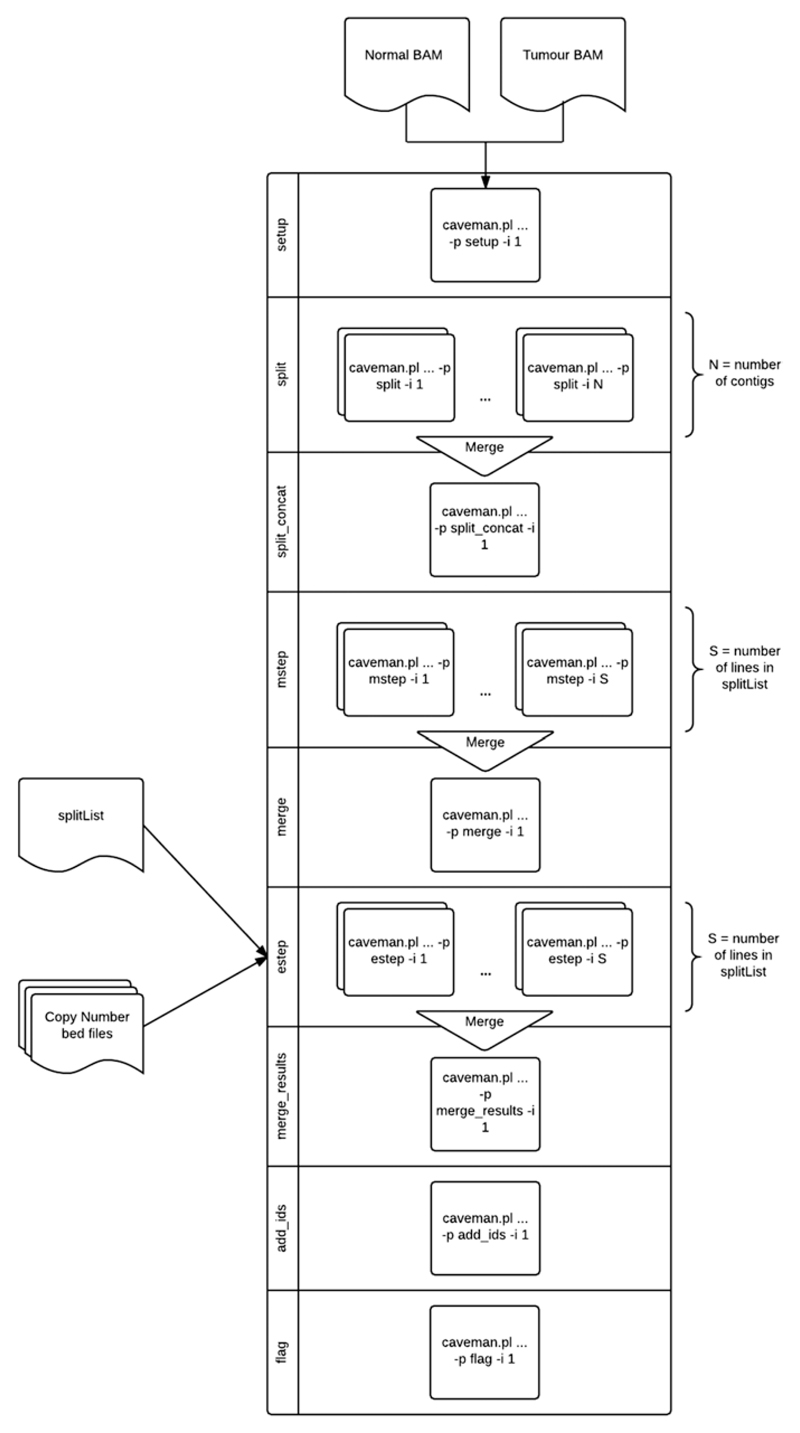
CaVEMan has four main steps: split, mstep, merge, and estep, followed by flagging (post-hoc filtering). The split step divides the genome into regions of roughly equal read count. The number of split regions varies with coverage but may be in the order of 4000 regions for a 30-fold-coverage genome. The split step regions are subsequently used by the mstep and estep. The mstep iterates through each position in a split region, creating a profile of the genomic data using several covariates including base quality, reference base, called base, read position, MAPQ, and mapping strand. Merge combines these per-region profiles into one for the whole genome. The estep uses the genome profile generated by the mstep, location-specific copy number, and the reference base in combination with sequence data to assign a probability to each possible genotype. Finally, flagging applies filters to the somatic calls with the aim of retaining genuine somatic mutations while reducing the number of false positives.
cgpCaVEManWrapper simplifies the running of the above CaVEMan steps into a single command (see Basic Protocol and Alternate Protocol 1). If the user has access to a large-scale compute farm, additional control is also available (see Alternate Protocol 2). cgpCaVEManPostProcessing provides similar control of the downstream filtering step and can be used in isolation or handled by cgpCaVEManWrapper. While CaVEMan has been optimized for somatic substitution detection in paired tumor and normal samples, it can also detect substitutions in any sample when compared to a ‘normal control’; however, this would result in germline variants in the tumor sample also being output to the somatic calls file unless the control sample also contains these germline variants. CaVEMan also outputs germline substitutions in a separate VCF file where the sum of all possible germline genotype probabilities exceeds the set cut-off.
Support Protocol 1 describes installation instructions for the CaVEMan software suite. Once installed, the following command will list available options:
caveman.pl -h
Together, cgpCaVEManWrapper and cgpCaVEManPostProcessing form a suite of tools that perform the full workflow to identify somatically acquired substitutions (see Fig. 15.10.1). This suite has successfully been used within the International Cancer Genome Consortium (ICGC; Alioto et al., 2015) PanCancer project as well as the Cancer Genome Project (CGP). Perl wrapper scripts have been used in order to simplify usage.
Figure 15.10.1.
cgpCavEManWrapper workflow. If –p/-i options are omitted, individual componenets are automatically executed. On restart, the workflow will automatically recover to the last successful point if killed for any reason.
Basic Protocol 1: Calling Substitutions Via a Single Command for a Tumor/Normal Sample Pair
CaVEMan in combination with post-hoc filters is used to produce a set of high-quality somatic substitution calls in VCF format, including information on coverage of alternative alleles, allowing further filters to be applied by the user. The algorithm compares tumor sample data to normal sample data and reference, then assigns a somatic probability to each position. Those positions above the default (0.8 for somatic genotypes, 0.95 for germline) or user defined cut-off probability are output to VCF. This section describes how to execute cgpCaVEManWrapper with a single command.
Necessary Resources
Hardware
The following resources assume a matched tumor/normal pair of whole genome sequence (WGS) samples with 30- to 40-fold sequence coverage aligned using the Human Genome Reference GRCh37d5. Requirements (for WGS data) include:
Linux cluster with a minimum 4 GB RAM per core
Recommended 32 cores (minimum)
It is of note that samples with greater sequencing depth than 30 to 40-fold or a failure to mask regions of the genome that have a high degree of mis-mapping (repeats regions for example) can increase both run time and hardware requirements (see Support Protocol 2: hiSeqDepth.bed)
Software
See Support Protocol 1 for installation steps:
PCAP-core: https://github.com/ICGC-TCGA-PanCancer/PCAP-core/releases (contains core modules used by CGP algorithm control scripts)
cgpVcf: https://github.com/cancerit/cgpVcf/releases (contains CGP code for VCF manipulation)
cgpCaVEManPostProcessing: https://github.com/cancerit/cgpCaVEManPostProcessing/releases (contains the post processing code and control code for CaVEMan filtering)
cgpCaVEManWrapper: https://github.com/cancerit/cgpCaVEManWrapper/releases (contains the control code for running CaVEMan)
The above packages install other dependencies including:
Biobambam2: https://github.com/gt1/biobambam2 (not used by CaVEMan)
-
libBigWig: https://github.com/dpryan79/libBigWig (not used by CaVEMan)
bwa: https://github.com/lh3/bwa (not used by CaVEMan)
samtools: https://github.com/samtools/samtools
kentUtils: https://github.com/ENCODE-DCC/kentUtils (not used by CaVEMan)
Various perl libraries
Files
Static reference files (see Support Protocol 2):
genome.fa.fai reference genome index (with associated *.fa reference file). Must be the same as the reference genome used during mapping of the input BAMs.
centromeric_repeats.bed: Bed file of centromeric repeats
simple_repeats.bed: Bed file of simple repeats
snps.bed: Bed file of SNP locations
Unmatched normal panel VCF files named unmatchedNormal.[1-numberofcontigs].vcf.gz (or a single bed file named unmatchedNormal.bed.gz) bgzipped and tabix indexed.
ignore_regions.bed: Bed file of regions to exclude from analysis
flag.vcf.config.ini: Config-ini file containing list of flags to apply and related parameters.
Sample Data
<Tumour>.[b|cr]am: aligned tumor sample
<Normal>.[b|cr]am: aligned normal sample
For sample alignment files, both BWA-mem (Li, 2013) and BWA-backtrack (Li and Durbin, 2009) have been tested. Any aligner that makes use of MAPQ (mapping quality) SAM field should be suitable for CaVEMan analysis.
<Tumour>.cn.bed: Tumor copy number bed file from <Tumour>.copynumber.caveman.csv (see UNIT 15.9; Raine et al., 2016)
<Normal>.cn.bed: Normal copy number bed file from <Tumour>.copynumber.caveman.csv (see UNIT 15.9; Raine et al., 2016)
Normal-contamination value from <Tumour>.samplestatistics.txt (see UNIT 15.9; Raine et al., 2016)
The above three files are derived from results generated by ascatNgs. See UNIT 15.9 (Raine et al. 2016) and Support Protocol 3 for further instructions on these files. Should ascatNGS not be available, the Advanced Parameters section provides instructions on using default copy-number values. Copy number information is used by CaVEMan to generate a list of possible genotypes at each position.
<Tumour>_vs_<Normal>.germline.bed.gz[.tbi]: bgzip compressed germline indel bed file and tabix index. Original file generated by cgpPindel (see UNIT 15.7; Raine et al., 2015)
Should germline indel calls not be available from pindel, a bgzipped and tabix indexed bed file of germline indels from another calling algorithm or a blank bed file can be provided. Should a blank file be provided, the germline indel filter will not be applied during flagging.
Example Data
Pre-generated reference files and COLO-829/COLO-829-BL BAM (Pleasance et al., 2010) files aligned with BWA-mem alongside expected results can be found at the ftp site: ftp://ftp.sanger.ac.uk/pub/cancer/support-files/CPIB/caveman/
In all steps, modify locations of files as appropriate
-
Set environmental variables:
For ease of execution set environmental variables to point to the reference area, output directory, results directory, and example data set. To set environmental enter the following in your Linux terminal (modify locations as appropriate for your system).
Location of the reference files (downloaded or otherwise):export REF=/referenceAreaCreate an output directory for results and variable for the output location:export POUT=/outputmkdir $POUT/resultThe output dataset:export CAVE=/exampleData - Build the caveman.pl command (in this example we use 32 cores) by running the CaVEMan algorithm. This single execution solution will run CaVEMan from start to finish:
caveman.pl \ -reference $REF/genome.fa.fai \ -outdir $POUT/result \ -tumour-bam $CAVE/tumour/HCC1143.bam \ -normal -bam $CAVE/normal/HCC1143_BL.bam \ -ignore -file $REF/ignore_regions.bed \ -tumour-cn $CAVE/tumour/HCC1143.cn.bed \ -normal-cn $CAVE/normal/HCC1143_BL.cn.bed \ -species Human \ -species-assembly GRCh37d5 \ -flag-bed-files $REF/flagging \ -germline-indel $CAVE/tumour/HCC1143_vs_HCC1143_BL.germline.bed \ -unmatched-vcf $REF/flagging/unmatched_vcf \ -seqType genomic \ -normal-contamination \ -threads 32 \ -limit 32 \ -flagConfig $REF/flagging/ \ -flagToVcfConfig $REF/flagging/ \ -annot-bed-files $REF/vagrent/ \ >& $POUT/caveman_run.log
Note this is not a quick process. The example can take ∼3500 CPU hours to run to completion. On completion, caveman.pl will exit with a zero exit code and emit no errors. You will find your results in your output directory ($POUT). The following files should be present (where TUMOUR and NORMAL are the names of your tumor/normal samples):
TUMOUR_vs_NORMAL.muts.ids.vcf.gz[.tbi]
TUMOUR_vs_NORMAL.snps.ids.vcf.gz[.tbi]
For explanations of all result files see Guidelines for Understanding Results.
Alternate Protocol 1: Processing Other Sequencing Types
Basic Protocol 1 describes SNV calling in WGS data. This alternate protocol describes how to call somatic substitutions in other types of sequencing experiments [exome (WXS) and targeted pull-down] along with explaining how to choose various post-processing flags.
Necessary Resources
Other DNA sequencing experiment types such as whole exome sequencing (WXS) and targeted pull-down will generally have more modest hardware requirements than WGS analysis (as described in Basic Protocol). However, where extreme sequencing depth is encountered (PCR-based amplicon sequencing), an increase in compute memory requirements may be observed.
Follow steps 1 to 4 of the Basic Protocol.
-
Follow all further steps in the Basic Protocol from step 5 on, modifying –seqType as appropriate.
In post-hoc filtering, the flags applied to different datatypes have different sections in a config file. For details on modifying flags and flagging parameters, refer to the cgpCaVEManPostProcessing wiki:
Support Protocol 1: Installation of cgpCaVEManWrapper and Dependencies
cgpCaVEManWrapper has few dependencies and is packaged with the aim of minimizing complexity of installation. The examples provided below demonstrate the usage of the versions that are available at time of publication. Please see repositories for current software versions. In the following steps please replace the /your/scratcharea and /installbase variables with those that you require for your scratch space and installed executables, respectively.
Necessary Resources
A Linux-based system with Web access
- Install PCAP-core (contains the thread framework for cgpCaVEManWrapper and cgpCaVEManPostProcessing):
$ cd /your/scratcharea $ wget https://github.com/ICGC-TCGA-PanCancer/PCAP-core/archive/v2.4.1.tar.gz $ tar -zxf v2.4.1.tar.gz $ rm v2.4.1.tar.gz $ cd PCAP-core-2.4.1/ $./setup.sh ∼/installBase Install cgpVcf (reusable VCF manipulation tools common to many CGP projects): $ cd /your/scratcharea $ wget https://github.com/cancerit/cgpVcf/archive/v2.0.4.tar.gz $ tar -zxf v2.0.4.tar.gz $ rm v2.0.4.tar.gz $ cd cgpVcf-2.0.4/ $./setup.sh ∼/installBase/- Install cgpCaVEManPostProcessing (package for applying post hoc flags to CaVEMan substitution calls):
$ cd /your/scratcharea $ wget https://github.com/cancerit/cgpCaVEManPostProcessing/archive/1.7.0.tar.gz $ tar -zxf 1.7.0.tar.gz $ rm -f 1.7.0.tar.gz $ cd cgpCaVEManPostProcessing-1.7.0/ $./setup.sh ∼/installBase/ - Install cgpCaVEManWrapper (including the CaVEMan executable this package wraps all CaVEMan processes in a simple perl script):
$ cd /your/scratcharea $ wget https://github.com/cancerit/cgpCaVEManWrapper/archive/1.9.8.tar.gz $ tar -zxf 1.9.8.tar.gz $ rm 1.9.8.tar.gz $ cd cgpCaVEManWrapper-1.9.8/ $./setup.sh ∼/installBase/
All of the above scripts will complete with a message similar to:
Please add the following to beginning of path …
If any step fails to provide the above message, inspection of the setup.log file (colocated with setup.sh) will show the messages emitted during the install process.
Alternate Protocol 2: Using cgpCaVEManWrapper with Compute Farm Infrastructure
Given how memory and CPU usage varies between sections of the CaVEMan analysis, running cgpCaVEManWrapper as a single process can be inefficient. More advanced users may prefer to break the cgpCaVEManWrapper process into component parts in order to more accurately resource each step.
Necessary Resources
Each individual step will have different hardware requirements and will require tuning on a sequencing type/species basis. Requirements described in Basic Protocol will serve as a good starting point.
Follow steps 1 to 4 of the Basic Protocol.
-
Find the number of contigs/chromosomes that are to be processed. This is the number of entries in the *.fa.fai file. For instance in the example data:
$ wc -l $REF/genome.fa.fai 86From this point please use the following stub to replace caveman.pl …:caveman.pl \ -reference $REF/genome.fa.fai \ -outdir $POUT/result \ -tumour-bam $CAVE/tumour/HCC1143.bam \ -normal-bam $CAVE/normal/HCC1143_BL.bam \ -ignore-file $REF/ignore_regions.bed \ -tumour-cn $CAVE/tumour/HCC1143.cn.bed \ -normal-cn $CAVE/normal/HCC1143_BL.cn.bed \ -species Human \ -species-assembly GRCh37d5 \ -flag-bed-files $REF/flagging/ \ -germline-indel $CAVE/tumour/HCC1143_vs_HCC1143_BL. germline.bed \ -unmatched-vcf $REF/flagging/unmatched_vcf \ -seqType genomic \ -normal-contamination \ -flagConfig $REF/flagging/ \ -flagToVcfConfig $REF/flagging/ \ -annot-bed-files $REF/vagrent/ \ - Run the single job setup step:
caveman.pl … -process setup –index 1 - Run the split step once per contig (86 in the given example) in the *.fa.fai file:
caveman.pl … -process split –index 1 … caveman.pl … -process split –index 86 - Run a single merge split sections job to create a single splitList file:
caveman.pl … -process split_concat –index 1 -
Calculate the number of sections in the split file and run a mstep job per section:
wc –l splitListIn this example we will assume 200 split sections:caveman.pl … -process mstep –index 1 … caveman.pl … -process mstep –index 200It is possible to use a parallel ‘round-robin’ approach to process the mstep and estep on a limited number of cores for steps 6 and 8. Assuming N is the number of cores available; adding – limit N to the command and running jobs with index 1 …N will enable a limited number of cores to be used when processing the mstep and estep.
- Merge the profiles generated in the mstep jobs into a single file:
caveman.pl … -process merge –index 1 -
Run an estep (variant calling step) job per section in the splitList:
caveman.pl … -process estep –index 1 … caveman.pl … -process estep –index 200See step 6 for a note on using a ‘round-robin’ approach to parallelized processing on a reduced number of cores.
- Merge the CaVEMan results into a single VCF result file:
caveman.pl … -process merge_results –index 1 - Add UUIDs to each VCF entry:
caveman.pl … -process add_ids –index 1 - Flag (post process) the VCF results:
caveman.pl … -process flag –index 1
Support Protocol 2: Static Reference File Generation
Alongside the genome reference file, CaVEMan and its associated post-hoc flags require several static files related to the reference. The following files are required or recommended (as marked) for WGS analysis and examples of these can be found on the FTP site provided in Internet Resources.
genome.fa
Required for CaVEMan execution.
The reference assembly as was used for mapping of the sample data. For details of how to generate this file, see UNIT 15.7 (Raine et al., 2015), Support Protocol 2.
simpleRepeats.bed.gz[.tbi]
Recommended for flagging. Enables use of the simple repeat flag.
A tabix (Li, 2011)–indexed bed file of simple repeats used in post-analysis flagging.
For details of how to generate this file, see UNIT 15.7 (Raine et al., 2015), Support Protocol 2.
centromericRepeats.bed.gz[.tbi]
Recommended for flagging. Enables use of the centromeric repeat flag.
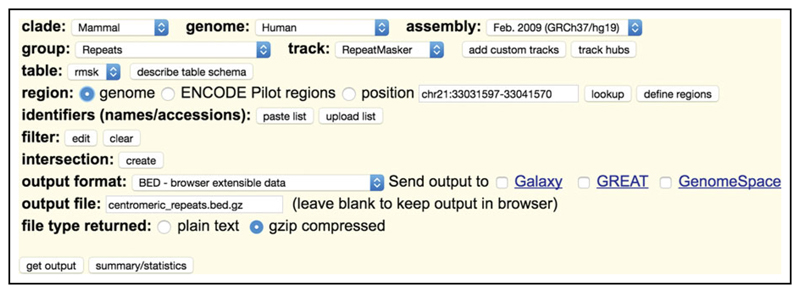
A tabix-indexed bed file of centromeric repeats used in post-analysis flagging. This is generated for GRCh37/hg19 as described in the following steps:
Navigate your Web browser to https://genome.ucsc.edu/cgi-bin/hgTables.
Set all of the options to match those shown in Figure 15.10.2.
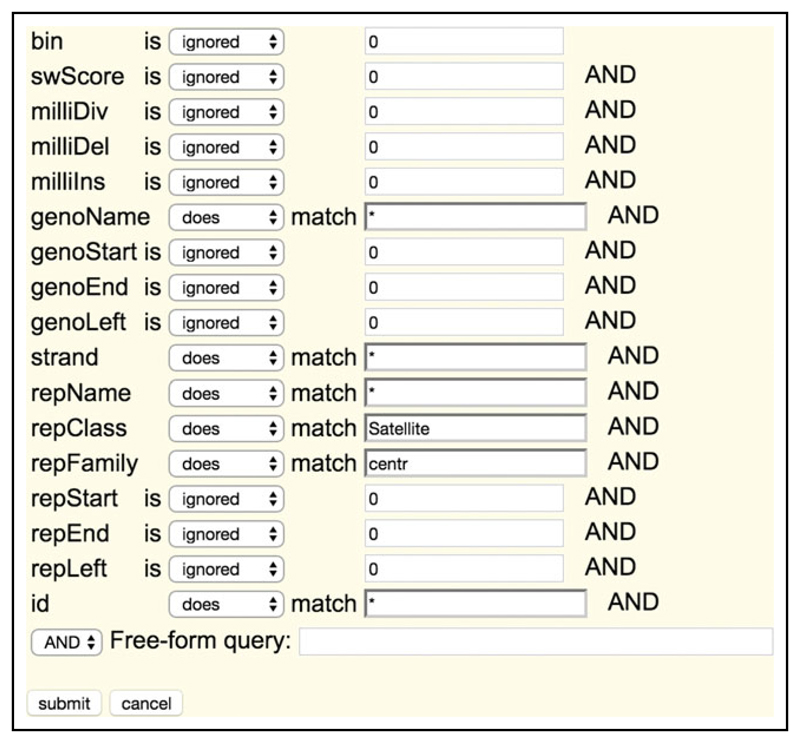
- Select the create button for filter and modify the filters as per Figure 15.10.3.
- Modify repClass to match Satellite.
- Modify repFamily to match centr.
- Click the Submit button.
Select the get output button.
-
Index the resulting output file using tabix:
tabix –p bed centromeric_repeats.bed.gzIn the example data, the chr prefix has been stripped from the contig names to match the reference data.
Figure 15.10.2.
UCSC Table Browser settings for generation of the centromeric_repeats.bed.gz file.
Figure 15.10.3.
UCSC Table Browser filter settings for generation of the centromeric_repeats.bed.gz file.
snps.bed.gz[.tbi]
Recommended for flagging. Enables use of the SNP flag.
A tabix indexed bed file of positions used in post-hoc flagging (when applying the ‘snpFlag’). The example data contains a filtered set of dbSNP confirmed variants.
unmatchedNormal.bed.gz[.tbi]
Highly recommended for flagging. Enables use of the unmatched normal panel flag.
A tabix indexed bed file of positions that show a non-reference allele in a panel of unmatched normal samples. This file is utilized by the unmatchedNormalVcfFlag in the post-processing filters. This flag is one of the most powerful filters applied to CaVEMan data and filters both polymorphisms and locations prone to aberrant mapping or systematic sequencing artifacts in genome sequencing. In order to reduce the likelihood of filtering out driver mutations, normal samples rather than tumor samples are used in this panel. When generating a normal panel, it is important to consider criteria that may influence the artifacts recorded (and therefore to be filtered). Variations in sequencing technology, sequencing chemistry, read length, and aligner could all impact on the data, resulting in systematic errors. Thus, it is recommended that the normal panel be augmented with data from new occurrences of sequencing technology, chemistry, and read length.
The sample data unmatchedNormal.bed.gz was generated from a panel of 100 samples:
- Create a bed file of split sections to analyze named contigs.bed. These should be one per contig, in bed format (0 based start), and listing start and end position of each contig in the reference genome. It is possible to divide the workload further, but that would require a merge into a per-contig vcf file after this step. For example, the entry for chromosome 1 would look as follows:
1 0 249250621 -
For each line in contigs.bed, run the following command:
let $index = line number in contigs.bed/your/scratcharea/caveman/bin/generateCavemanUMNormVCF\ --index $index \ --out-file /your/reference-data/unmatchedNormal.$index.vcf\ --reference /your/reference-data/genome.fa \ --split-sections contigs.bed \ --species human \ --spp-vers GRCh37d5 \ --bam-files <comma separated paths to normal panel sample bam files> \ --sample-names <comma separated names of samples (in the same order as --bam-files> - Compress the produced VCF files using bgzip:
bgzip /your/reference-data/unmatchedNormal.$index.vcf tabix --p vcf /your/reference-data/unmatchedNormal.$index.vcf.gz - Convert each of the VCF files to bed format:
convertVCFUnmatchedToBed.pl \ -infile /your/reference-data/unmatchedNormal.$index.vcf.gz \ -output /your/reference-data/unmatchedNormal.$index.bed \ -vcfUnmatchedMinMutAlleleCvg 3 \ -vcfUnmatchedMinSamplePct 1 - Concatenate the generated bed files and sort:
head -2 /your/reference-data/unmatchedNormal.1.bed > /your/reference-data/unmatchedNormal.bed cat /your/reference-data/unmatchedNormal.*.bed | grep -ve 'ˆ#' | sort -k 1,1 -k2,2n >> /your/reference-data/unmatchedNormal.bed
- Compress and tabix index the single normal panel bed file:
bgzip /your/reference-data/unmatchedNormal.bed tabix –p bed/your/reference-data/unmatchedNormal.bed.gz -
Clean up the working area:
rm /your/reference-data/unmatchedNormal.*.bedIf you choose not to retain the VCF files generated you can also remove those:rm /your/reference-data/unmatchedNormal.*.vcf.gz*
hiSeqDepth.bed
Recommended for flagging. Enables use of the high seq depth flag.
This bed file contains a list of regions that should be ignored throughout cgpCaVEManWrapper analysis. The file is useful for excluding regions where depth is abnormally high and where there is therefore a higher likelihood of false-positive calls. A description of how to generate this can be found in the PCAP-core wiki (https://github.com/ICGC-TCGA-PanCancer/PCAP-core/wiki/Hi-seq-depth-regions).
codingexon_regions.bed.gz
Optional. Enables use of the coding flag.
A listing of coding exons used in post analysis flags (when applying codingFlag). For details of how to generating this file see UNIT 15.8 (Menzies et al., 2015).
gene_regions.bed.gz
Optional. Enables use of the ‘is annotatable’ flag.
A listing of annotatable regions used in post analysis flags (when applying annotationFlag). For details of how to generating this file see UNIT 15.8 (Menzies et al., 2015).
Support Protocol 3: ASCAT and PINDEL Output File Manipulation
The copy number and indel bed files can be generated using ASCAT and PINDEL, respectively, using the following perl one liners:
<Normal>.cn.bed:
perl -ne '@F=(split q{,}, $_)[1,2,3,4];$F[1]-1; print
join(''\\t'',@F).''\\n'';' < ascatResults/<Tumour>
.copynumber.caveman.csv > <Normal>.cn.bed
<Tumour>.cn.bed:
perl -ne '@F=(split q{,}, $_)[1,2,3,6]; $F[1]-1; print
join(''\\t'',@F).''\\n'';' < ascatResults/<Tumour>
.copynumber.caveman.csv > <Tumour.>cn.bed
Normal contamination value:
awk '($1==''NormalContamination'') {print $2}'
<Tumour>.samplestatistics.txt
<Tumour>_vs_<Normal>.germline.bed.gz:
bgzip pindelResults/<Tumour>_vs_<Normal>.germline.bed
tabix –p bed <Tumour>_vs_<Normal>.germline.bed.gzGuidelines for Understanding Results
cgpCaVEManWrapper generates several results files with the following naming convention:
<TUMOUR>_vs_<NORMAL>[._]*
The values TUMOUR and NORMAL are taken from the sample (SM) field of the BAM read-group header lines. All VCF format outputs are currently VCF version v4.1 (http://vcftools.github.io/specs.html). VCF files are also compressed with bgzip and tabix indexed. The various output files are described below. Refer to Table 15.10.1 for a brief description of VCF output and to Tables 15.10.2 and 15.10.3 for descriptions of the CaVEMan specific INFO fields and FORMAT fields.
Table 15.10.1. Description of Standard VCF Output Fields.
| VCF field | Description | Example data |
|---|---|---|
| CHROM | Chromosome/contig name | 1 |
| POS | Position | 10010 |
| ID | ID assigned to this variant (UUID for CaVEMan data) | 81b61f98-f614-11e5-8014-d8d2535b31b0 |
| REF | Reference allele at this position | A |
| ALT | Mutant allele at this position | T |
| QUAL | Quality score associated with this variant (“.” if blank) | . |
| FILTER | List of filter flags this variant fails (“PASS” if none fail) | PASS |
| INFO | List of key value pairs separated by a semicolon describing the variation. Keys and associated values described in the VCF header. See Table 15.10.2. for CaVEMan INFO field entries. | DP = 90;MP = 1.0e + 00;GP = 5.8e-08;TG = TT/CT;TP = 9.9e-01; SG = TT/CC;SP = 7.1e-03 |
| FORMAT | Colon-separated list of fields that constitute the entries under the NORMAL and TUMOUR headings. Keys and associated values are described in the VCF header. See Table 15.10.3. for CaVEMan FORMAT field entries. | GT:FAZ:FCZ:FGZ:FTZ:RAZ:RCZ:RGZ:RTZ:PM |
| NORMAL | Values described in the FORMAT field for the normal sample. | 0|0:0:0:1:20:0:1:0:10:3.1e-02 |
| TUMOUR | Values described in the FORMAT field for the tumor sample. | 0|1:0:7:1:32:0:1:0:17:1.4e-01 |
Table 15.10.2. Explanation of VCF Fields Controlled by INFO Convention.
| ID | Description (as in the VCF header) | Detailed description |
|---|---|---|
| DP | Total depth | Total depth of alleles seen by CaVEMan |
| MP | Sum of CaVEMan somatic genotype probabilities | Sum of probabilities for all possible somatic genotypes at this position |
| GP | Sum of CaVEMan germline genotype probabilities | Sum of probabilities for all possible germline substitution genotypes at this position |
| TG | Most probable genotype as called by CaVEMan | Genotype with the highest probability. Genotypes take the form <Normal-Genotype>/<Tumour-Genotype> and are affected by copy number and reference base. |
| TP | Probability of most probable genotype as called by CaVEMan | Probability value of the most likely genotype (TG) |
| SG | 2nd most probable genotype as called by CaVEMan | Genotype with the second highest probability |
| SP | Probability of 2nd most probable genotype as called by CaVEMan | Probability value of the second most likely genotype (SG) |
Table 15.10.3. Explanation of VCF Fields Controlled by FORMAT Convention.
| ID | Description (as in the VCF header) | Detailed description |
|---|---|---|
| GT | Genotype | See VCF specification |
| FAZ | Reads presenting a A for this position, forward strand | Reads seen by CaVEMan and mapping to the forward strand presenting an A at this position |
| FCZ | Reads presenting a C for this position, forward strand | Reads seen by CaVEMan and mapping to the forward strand presenting an C at this position |
| FGZ | Reads presenting a G for this position, forward strand | Reads seen by CaVEMan and mapping to the forward strand presenting an G at this position |
| FTZ | Reads presenting a T for this position, forward strand | Reads seen by CaVEMan and mapping to the forward strand presenting an T at this position |
| RAZ | Reads presenting a A for this position, reverse strand | Reads seen by CaVEMan and mapping to the reverse strand presenting an A at this position |
| RCZ | Reads presenting a C for this position, reverse strand | Reads seen by CaVEMan and mapping to the reverse strand presenting an C at this position |
| RGZ | Reads presenting a G for this position, reverse strand | Reads seen by CaVEMan and mapping to the reverse strand presenting an G at this position |
| RTZ | Reads presenting a T for this position, reverse strand | Reads seen by CaVEMan and mapping to the reverse strand presenting an T at this position |
| PM | Proportion of mut allele | Proportion of mutant allele presenting reads (ALT field) seen by CaVEMan |
TUMOUR_vs_NORMAL.muts.ids.vcf.gz[.tbi]
VCF file containing the raw CaVEMan somatic substitution calls with a UUID assigned to each variant. No post-processing filters have been applied to the data in this file.
TUMOUR_vs_NORMAL.snps.ids.vcf.gz[.tbi]
VCF file containing the raw CaVEMan germline substitution calls with a UUID assigned to each variant. No post-processing filters have been applied to the data in this file.
It should be noted that CaVEMan has been tuned to detect somatic variants, and no work has been done on flags for germline substitution calling.
TUMOUR_vs_NORMAL.flagged.muts.vcf.gz[.tbi]
The somatic mutation VCF output file with post-processing flags applied. Variants containing anything but PASS in the filter field can be excluded with high confidence. Details of flags applied can be found in the in the Advanced Parameters section.
TUMOUR_vs_NORMAL.no_analysis.bed
A bed file of regions not analyzed by CaVEMan. There are several reasons for missing analysis: lack of coverage in normal or tumor sequence data, zero copy number in normal or tumor, or the fact that the position was in the ignore regions bed file. If it is believed CaVEMan has missed mutations, this file can be useful to check whether the position was analyzed before further investigation.
On average, a whole-genome normal/tumor pair with 30× coverage in each pair will produce ∼150,000 mutations, of which an average of 5000 will pass flagging (calculated from 200 breast cancers). This total mutation figure may increase greatly with hyper-mutated cancers or noisy sequencing data. Pull-down and targeted sequencing data will result in fewer somatic mutations, since they target smaller areas than the whole genome. Mutations passing flagging are expected to be between 90% and 95% specific (true positives).
Commentary
Background Information
CaVEMan was developed soon after the advent of next-generation sequencing with the aim of producing a substitution-detection algorithm capable of excluding sequencing artifacts while also handling the large amounts of sequencing data produced by the high-throughput sequencing methods. It was originally written in Java before being reimplemented in C to improve efficiency.
cgpCaVEManWrapper was developed in order to make CaVEMan suitable for use in the ICGC/TCGA Pancancer project, a systematic screen of 2,500 WGS Tumor/Normal sample pairs (http://icgc.org) and to enable ease of use as a stand-alone analysis tool.
Critical Parameters
It is important to correctly set -seqType as this parameter is used during flagging. Flags (and their parameters) applied to CaVEMan calls differ between sequencing types.
Failure to mask regions of much higher than expected sequencing depth via the -ignore-file may result in much higher hardware requirements.
Troubleshooting
See Table 15.10.4 for solutions to common problems.
Table 15.10.4. Common Problems and Solutions in cgpCaVEManWrapper.
| Problem | Cause | Solution |
|---|---|---|
| Known mutation not reported | Flagged | Search the results file T_vs_N.flagged.muts.vcf.gz including all mutations rather than just those marked as PASS in the filter column |
| Known mutation not reported | SNP | Search the germline output file T_vs_N.snps.ids.vcf.gz to ensure the position has not been reported as germline. |
| Known mutation not reported | Ignored regions/not analyzed | Check the ignored regions file to see if it covers the expected position. Also check T_vs_N.no_analysis. bed to ensure position was analyzed. Other possible reasons for no analysis are lack of depth or zero copy number in one or both normal and tumor samples. |
| Allele counts in output VCF don’t match the raw BAM file | CaVEMan excludes lower quality bases | Bases at a position with base quality ≤ 10 will be ignored by CaVEMan. |
Advanced Parameters
Probability cut-offs:
-mut_probability_cutoff
<float>
The above is the minimum total probability of all somatic genotypes before a position is output to the muts VCF file.
-snp_probability_cutoff
<float>
The above is the minimum total probability of all germline (SNP) genotypes before a position is output to the snp VCF file.
Prior probabilities:
-prior-mut-probability
<float>
The above is the prior probability of a somatic mutation. It can be useful to increase this value where sensitivity is preferred to specificity.
-prior-snp-probability
<float>
The above is the prior probability of a germline mutation.
Flagging parameters:
Which flags are applied to CaVEMan calls and the parameters behind them can be modified in the flag.vcf.cfg.ini files provided. For detailed information on flags and modification of the ini file, see the cgpCaVEManPostProcessing wiki at http://cancerit.github.io/cgpCaVEManPostProcessing/.
Providing copy number:
Copy number calls from other algorithms can be provided in the form of a bed-like file for tumor and a bed-like file for normal with the format:
<contig> <1-based start> <1 based stop> <CN>
These can be passed using the commandline options:
-tum-cn-default
-norm-cn-default
It is recommended to use the following combination of parameters where no copy number or normal contamination is known:
-tum-cn_default 5
-norm-cn-default 2
-normal-contamination 0.1Suggestions for Further Analysis
CGP has created a variant annotation tool VAGrENT, which can be used to annotate indels and substitutions at the cDNA and protein level in VCF format. Please see UNIT 15.8 (Menzies et al., 2015) for further details on this tool.
Acknowledgements
The authors wish to thank Peter Camp-bell for the mathematical details of the algorithm and Keiran M Raine for his work on adding CaVEMan to the IGCG/TCGA Pan-cancer analysis pipeline. Both are from the Wellcome Trust Sanger Institute. Thanks also go to Pascal Costanza of ExaScience Lab Belgium, Intel Health & Life Sciences, for contributions to the CaVEMan C code.
This work was supported by the Wellcome Trust grant [098051].
Literature Cited
- Alioto TS, Buchhalter I, Derdak S, Hutter B, Eldridge MD, Hovig E, Heisler LE, Beck TA, Simpson JT, Tonon L, Sertier AS, et al. A comprehensive assessment of somatic mutation detection in cancer using whole-genome sequencing. Nat Commun. 2015;6:10001. doi: 10.1038/ncomms10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do CB, Batzoglou S. What is the expectation maximization algorithm? Nat Biotechno. 2008;26:897–899. doi: 10.1038/nbt1406. [DOI] [PubMed] [Google Scholar]
- Hsi-Yang Fritz M, Leinonen R, Cochrane G, Birney E. Efficient storage of high throughput DNA sequencing data using reference-based compression. Genome Res. 2011;21:734–740. doi: 10.1101/gr.114819.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Tabix: Fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2011;27:718–719. doi: 10.1093/bioinformatics/btq671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. [Accessed June 8, 2016];arXiv:1303.3997. 2013 [q-bio]. Available at: http://arxiv.org/abs/1303.3997. [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies A, Teague JW, Butler AP, Davies H, Tarpey P, Nik-Zainal S, Campbell PJ. VAGrENT: Variation annotation generator. Curr Protoc Bioinform. 2015;52:15.8.1–15.8.11. doi: 10.1002/0471250953.bi1508s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin M-L, Ordóñez GR, Bignell GR, Ye K, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raine KM, Van Loo P, Wedge DC, Jones D, Menzies A, Butler AP, Teague JW, Tarpey P, Nik-Zainal S, Campbell PJ. ascatNgs: Identifying somatically acquired copy-number alterations from whole-genome sequencing data. Curr Protoc Bioinform. 2016;56:1. doi: 10.1002/cpbi.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raine KM, Hinton J, Butler AP, Teague JW, Davies H, Tarpey P, Nik-Zainal S, Campbell PJ. cgpPindel: Identifying somatically acquired insertion and deletion events from paired end sequencing. Curr Protoc Bioinform. 2015;52:15.7.1–15.7.12. doi: 10.1002/0471250953.bi1507s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Internet Resources
- https://github.com/canceritRepository for Wellcome Trust Sanger Institute Cancer Genome Project public projects.
- ftp://ftp.sanger.ac.uk/pub/cancer/support-files/CPIB/FTP site for reference and example data listed in this unit.
- https://genome.ucsc.edu/cgi-bin/hgTablesUCSC Genome Browser Table Browser
- http://icgc.orgICGC/TCGA Pancancer project site.
- http://vcftools.github.io/specs.htmlVCF format.
- https//samtools.github.io/hts-specs/SAMv1.pdfSAM format.
- https://github.com/ICGC-TCGA-PanCancer/PCAP-core/wikiPCAP-core wiki describes generation of high sequence depth file.