Abstract
cgpPindel is a modified version of Pindel that is optimized for detecting somatic insertions and deletions (indels) in cancer genomes and other samples compared to a reference control. Post-hoc filters remove false positive calls, resulting in a high-quality dataset for downstream analysis. This unit provides concise instructions for both a simple ‘one-shot’ execution of cgpPindel and a more detailed approach suitable for large-scale compute farms.
Keywords: somatic, sequencing, Pindel, cancer
Introduction
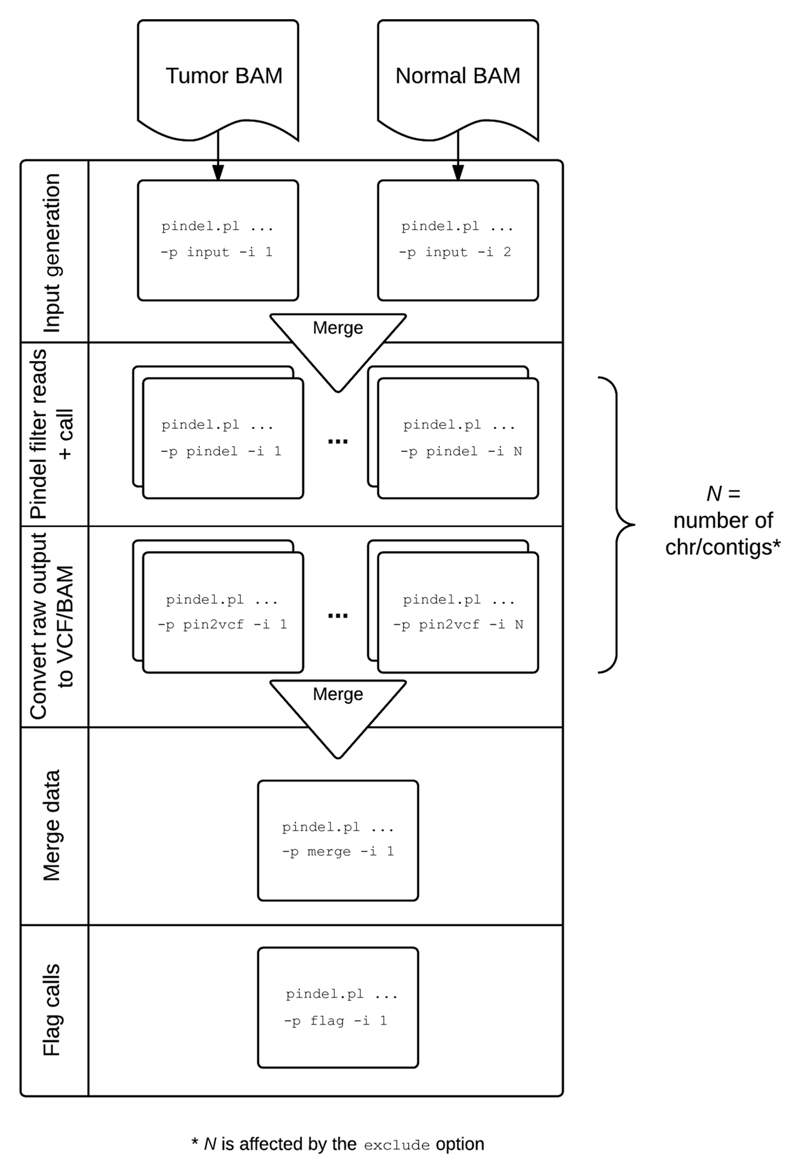
The analysis of next-generation sequencing (NGS) data to identify indels using the Pindel algorithm (Ye et al., 2009) has several steps and requires pre- and post-processing to exclude sequencing artifacts. There are four main steps for processing: input generation, read filtering, detection by Pindel, and flagging (See Fig. 15.7.1). The pre-processing step identifies read pairs where one read has mapped (the anchor) and the read mate is either unmapped or maps with the inclusion of an indel (the query). A modified version of Pindel is subsequently employed to analyze these fragments and performs ‘split read mapping’ on the query read to identify putative indels. Post-processing of the Pindel output involves the application of multiple empirically derived filters aimed at removing false calls and retaining genuine somatic mutations.
Figure 15.7.1.
cgpPindel processing workflow. Individual components are executed automatically when run without ‘-p/-i’ options. The workflow automatically recovers to the last successful point on restart if killed for any reason. Please see Alternate Protocol 2 for further detail.
In order to run the algorithm on modest hardware, the process requires division into a series of steps. cgpPindel simplifies the running of these steps, allowing a single command to trigger the complete workflow. More granular control is available via the same tool set for users with access to a large-scale compute farm.
cgpPindel has been optimized for detecting somatic mutations in tumor samples, which are paired with a non-neoplastic control from the same individual. In theory, however, it can be used to identify indels present in any sample compared to a reference control.
Strategic Planning
cgpPindel is a suite of tools that performs the full indel calling workflow, which has been used successfully within the Cancer Genome Project (CGP) and the International Cancer Genome Consortium (ICGC) PanCancer project. All components are wrapped in Perl scripts to simplify usage.
Please see Support Protocol 1 for installation instructions.
Once installed, running the following command will list available options:
pindel.pl -hBasic Protocol 1: Calling Indels with a Single Command for a Tumor/Normal Sample Pair
The purpose of cgpPindel is to produce a set of high-confidence somatic indel calls in VCF format (http://vcftools.github.io/specs.html). Additionally, reads that support these calls are aggregated into a pair of BAM files (https://samtools.github.io/hts-specs/SAMv1.pdf) to enable visual inspection if required. The algorithm identifies indels in both the tumor and normal samples and subsequently excludes variants that occur in the normal. This section describes how to execute cgpPindel with a single command.
Necessary Resources
Hardware
The resources listed here assume a dataset of whole genome sequence (WGS) samples with 30- to 40-fold sequence coverage for Human Genome Reference GRCh37d5 from tumor and normal samples. Poor-quality input data (e.g., high indel artifact rates) can greatly increase both the running time and hardware requirements. Requirements include:
A Linux computer with at least 8 GB of RAM
4 to 6 cores (8 GB per core) recommended
Processing storage of 50 GB
Software
PCAP-core: https://github.com/ICGC-TCGA-PanCancer/PCAP-core/releases; this software installs its own dependencies including:
biobambam: https://github.com/gt1/biobambam
bwa: https://github.com/lh3/bwa (not used here)
samtools: https://github.com/samtools/samtools
kentUtils: https://github.com/ENCODE-DCC/kentUtils (not used here)
Various perl libraries
cgpVcf: https://github.com/cancerit/cgpVcf/releases; this software installs its own dependencies including:
Various perl libraries
cgpPindel: https://github.com/cancerit/cgpPindel/releases; this package includes a modified version of the pre 0.2.0 Pindel source with agreement of original author (Ye et al., 2009).
Files
Static reference files (see Support Protocol 2):
genome.fa: reference genome (with associated *.fai index). This must be the same as the reference used during mapping of the input BAMs.
simpleRepeats.bed.gz: tabix-indexed bed file of simple repeats
codingexon_regions.bed.gz: tabix-indexed bed file of coding exons (see unit 15.8)
NormalPanel.gff3.gz: tabix indexed gff3 file of events seen in a panel of normal sequencing
*.lst: List of rules to be applied depending on data type being analyzed
Sample data
<Tumour>.bam: aligned paired-end sequencing for tumor sample
<Normal>.bam: aligned data paired-end sequencing for normal sample
For sample alignments, both BWA-mem (Li, 2013) and BWA-backtrack (Li and Durbin, 2009) have been tested. Any other aligner that makes proper use of the MAPQ (mapping quality) SAM field should be suitable.
Example data
Pre-generated reference files and COLO-829/COLO-829-BL (Pleasance et al., 2010) BAM files aligned with BWA-mem along with expected results can be found at ftp://ftp.sanger.ac.uk/pub/cancer/support-files/cgpPindel
-
Collect mapping statistics for input BAM files:
bam_stats -i tumour.bam -o tumour.bam.bas bam_stats -i normal.bam -o normal.bam.basThis will take several hours (files included for example data). The output will be written into the two .bas files.
- Set an environment variable pointing to the file system location of the reference files (downloaded or otherwise). Modify the path as appropriate:
export REF=/refarea - Set an environment variable to indicate the location for the output data. Modify as appropriate:
export POUT=/workspace - Create the output folder:
mkdir -p $POUT/result - Set an environment variable to indicate the location of the input example data. Modify as appropriate for your system:
export PIN=/exampleData -
Build the pindel.pl command (this example uses 6 cores):
pindel.pl \ -reference $REF/genome.fa \ -exclude NC_007605,hs37d5,GL% \ -simrep $REF/simpleRepeats.bed.gz \ -badloci $REF/hiSeqDepth.bed.gz \ -genes $REF/codingexon_regions.indel.bed.gz \ -unmatched $REF/pindel_np.gff3.gz \ -assembly GRCh37d5 \ -species Human \ -seqtype WGS \ -filter $REF/genomicRules.lst \ -softfil $REF/softRules.lst \ -tumour $PIN/tumour/COLO-829.bam \ -normal $PIN/normal/COLO-829-BL.bam \ -outdir $POUT/result \ -cpus 6 >& $POUT/run.log &This is not a quick process; expect a wall-clock time of around 18 hr when using 6 cores.
Alternate Protocol 1: Processing other Sequencing Types
Basic Protocol 1 describes how to run WGS data to call somatic indels. Here, we address how to call indels in other data types (exome and targeted pulldown) and explain how to pick and choose the post cgpPindel filters. Although it is possible to use cgpPindel to process RNA-seq for indels, it is not considered a primary focus of the tool and consequently is not covered in this unit.
Necessary Resources
In general, analysis of other DNA sequencing types such as whole exome sequence (WXS) and targeted pulldown (TG) have more modest hardware requirements than those described in Basic Protocol 1 Necessary Resources
Follow steps 1 to 4 of Basic Protocol 1.
Modify the parameters of the command from step 5 of Basic Protocol 1 appropriately using Table 15.7.1 as a guide.
Table 15.7.1. Parameters for Alternate Sequencing Types.
| Parameter | Detail | Values |
|---|---|---|
| -seqtype | Correlated with appropriate information in the input BAM headers and will be overridden should the two input BAMs disagree. Primarily provided for population of VCF when not found in BAM header. |
|
| -filter | The list of filtering rules to be applied to the VCF file. This can be an empty file. | Several panels of filtering rule sets are included in the distribution:
|
Alternate Protocol 2: Using cgpPindel with Compute Farm Infrastructure
Running the complete analysis as a single command can be inefficient due to how memory and CPU are utilized in different elements of the analysis. More advanced users may wish to break down execution into subcomponents so that they can be resourced more accurately under a compute farm infrastructure.
Figure 15.7.1 illustrates the different elements of the process flow.
Necessary Resources
Individual steps have different hardware requirements that require tuning on a per species/build basis. This will require experimentation, but the resources described in Basic Protocol 1 can serve as a good starting point.
Follow Basic Protocol 1 steps 1 to 4.
-
Determine the number of contigs/chromosomes that will be processed. This is the number of entries in the *.fa.fai file less any that match the patterns passed to -exclude (these are typically small contigs and other reference entries that you wish to ignore). An example for the provided data:
$ wc -l genome.fa.fai 86Less 61 (NC_007605,hs37d5,GL%), resulting in 25 (1-22+X,Y,MT)
Please use this stub command from this point for all items indicated by ‘pindel.pl …’
pindel.pl \ -reference $REF/genome.fa \ -exclude NC_007605,hs37d5,GL% \ -simrep $REF/simpleRepeats.bed.gz \ -badloci $REF/hiSeqDepth.bed.gz \ -genes $REF/codingexon_regions.indel.bed.gz \ -unmatched $REF/pindel_np.gff3.gz \ -assembly GRCh37d5 \ -species Human \ -seqtype WGS \ -filter $REF/genomicRules.lst \ -softfil $REF/softRules.lst \ -tumour $PIN/tumour/COLO-829.bam \ -normal $PIN/normal/COLO-829-BL.bam \ -outdir $POUT/result - Run the input generation steps, always 2 jobs. The option -cpus N can be included here to reduce run time (max 3):
pindel.pl … -cpus 3 -process input -index 1 pindel.pl … -cpus 3 -p input -index 2 - Run the filtering and calling step once for each of the 25 chromosomes/contigs:
pindel.pl … -process pindel -index 1 … pindel.pl … -process pindel -index 25 - Run the conversion of raw pindel output to VCF and BAM once for each of the 25 chromosomes/contigs:
pindel.pl … -process pin2vcf -index 1 … pindel.pl … -process pin2vcf -index 25 - Merge the per-contig outputs:
pindel.pl … -process merge -index 1 -
Run the flagging and clean up step:
pindel.pl … -process flag -index 1It is possible to use parallel ‘round-robin’ processing on a limited number of cores on a single host for steps 4 and 5. This is achieved by omitting -index, then specifying -cpus and -limit with the number of cores to utilize. A secondary advantage is that this removes the need to determine the number of jobs required.
Support Protocol 1: Installation of cgpPindel and Dependencies
cgpPindel has been packaged to minimize the installation complexity. The examples below use the versions available at the time of publication. Please see the repositories for current versions.
Necessary Resources
Linux-based system with Web access
- Install PCAP-core (which contains the thread framework for cgpPindel). In this and following steps, please replace /your/scratcharea and ~/installBase with the file system paths that you desire to use for scratch space and installed executables, respectively:
$ cd /your/scratcharea $ wget https://github.com/ICGC-TCGA-PanCancer/PCAP-core/archive/v1.9.4.tar.gz $ tar -zxf v1.8.2.tar.gz $ rm v1.8.2.tar.gz $ cd PCAP-core-1.8.1 $./setup.sh ~/installBase - Install cgpVcf (reusable VCF manipulation tools common to many CGP projects):
$ cd /your/scratcharea $ wget https://github.com/cancerit/cgpVcf/archive/v1.2.3.tar.gz $ tar -zxf v1.2.3.tar.gz $ rm v1.2.3.tar.gz $ cd cgpVcf-1.2.3 $./setup.sh ~/installBase -
Install cgpPindel (modified version of Pindel optimized for somatic mutation detection):
$ cd /your/scratcharea $ wget https://github.com/cancerit/cgpPindel/archive/v1.5.2.tar.gz $ tar -zxf v.1.3.2.tar.gz $ rm v1.3.2.tar.gz $ cd cgpPindel-1.3.2 $./setup.sh ~/installBaseAll of the setup scripts above will complete with a message along the lines:'Please add the following to beginning of path …'If any fail to give this message, examine the setup.log file (co-located with setup.sh).
Support Protocol 2: Static Reference Files
The genome reference file is an essential requirement to run the algorithm. The following are recommended for human WGS analysis. Examples of each of these are available on the FTP site indicated in Basic Protocol 1. Note that unlike some other human genome references, these files do not prefix each chromosome name with chr.
genome.fa
This is the reference assembly as used for the mapping of the paired end sequencing data. The fasta index (fai) is also required. This can be generated by executing:
samtools faidx genome.fa
samtools is included in the install detailed in Support Protocol 1.
simpleRepeats.bed.gz[.tbi]
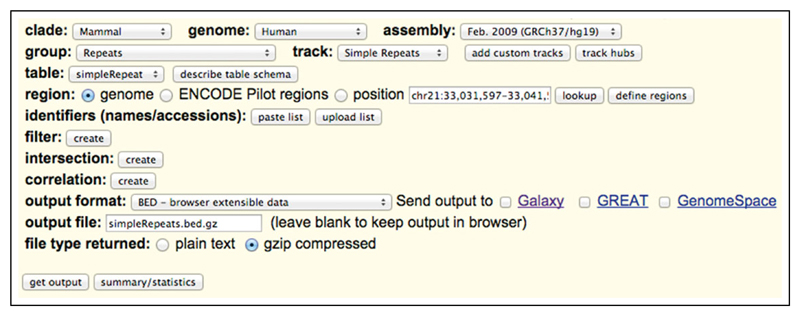
This is a tabix (Li, 2011) indexed bed file of simple repeats, required for filtering of results. This is generated as follows for Human GRCh37/hg19:
Using a Web browser, navigate to https://genome.ucsc.edu/cgi-bin/hgTables.
Set all of the options to match those shown in Figure 15.7.2.
Select the create button for ‘filter:’.
Modify ‘period’ to be ‘<=6’ and submit this form.
Now, back on the original form select ‘get output’.
- Index the resulting file using tabix (i.e., create the *.tbi file):
tabix -p bed simpleRepeats.bed.gz
Figure 15.7.2.
UCSC Table Browser Settings for generation of simpleRepeats.bed.gz file.
In the example data, the chr prefix has been stripped to match the mapping chromosomes.
pindel_np.gff3.gz
This file supports one of the most powerful filters in the cgpPindel workflow. A normal panel is a list of locations found to be aberrant in normal genome sequencing, such as sequencing and mapping artifacts. There are several criteria to be considered when generating these:
Sequencing technology
Sequencing chemistry
Read lengths of paired-end reads
Base aligner used
It is not necessary to have a different panel for each permutation. In practice, the best approach is to ensure that any existing panel is augmented with data from new chemistry/read-length as it becomes available.
It is recommended that a new panel be created if the base alignment tools are changed (e.g., BWA backtrack -> BWA mem).
From our experience, a minimum of 20 normal samples (from different donors) is required to generate an effective panel.
- Create a BAM file with no reads (all relevant tools included in distributions):
(echo '@1/1';echo 'A';echo '+';echo 'B' \ ;echo '@1/2';echo 'A';echo '+';echo 'B') \ | fastqtobam namescheme=generic \ RGID=1 RGLB=X RGPL=ILLUMINA RGPU=X RGSM=FAKE \ | samtools view -h - \ | samtools view -Sbt genome.fa.fai -o FAKE.bam - - Index the BAM.
samtools index FAKE.bam - Generate a *.bas file:
bam_stats -i FAKE.bam -o FAKE.bam.bas Run Basic Protocol 1 for each of the normal samples you want to include using FAKE.bam as the file for -tumour.
- Run pindel_np_from_vcf.pl using the VCF output from all of the data run in step 3:
pindel_np_from_vcf.pl -o normalPanel -samp_id NORMAL results/*.vcf.gz
codingexon_regions.bed.gz
This is a listing of coding exons required for filtering of results. Generation of this file is covered in unit 15.8.
Guidelines for Understanding Results
cgpPindel generates several result files of the format:
<TUMOUR>_vs_<NORMAL>[._]*
The TUMOUR and NORMAL values are taken from the SM field of the BAM read-group headers. Table 15.7.2 details the different extensions:
Table 15.7.2. Files Created by cgpPindel.
| File | Type |
|---|---|
| T_vs_N.flagged.vcf.gz[.tbi] | Variant call format (bgzip compressed) |
| T_vs_N_wt.bam[.bai|.md5] | Pindel-aligned reads from the wild-type/normal sample in BAM format |
| T_vs_N_mt.bam[.bai|.md5] | Pindel-aligned reads from the mutant/tumour sample in BAM format |
| T_vs_N.germline.bed | BED file containing ranges of events highly likely to be germline |
*.germline.bed
This file is primarily used as an input for cgpCaVEManPostprocessing (filtering step for the CaVEMan substitution caller; please see the wrapping project cgpCaVEManWrapper at https://github.com/cancerit/cgpCaVEManWrapper).
The content provides regions where germline indels may cause incorrect primary alignment, which in turn can produce false-positive results in the substitution caller.
*.bam
These files contain a BAM representation of all events called by Pindel. They can be integrated in to a genome browser and used with the primary alignment BAM for further analysis employing scripting tools such as Bio::DB::Sam.
*.vcf.gz
The VCF file is the final result, currently v4.1 (http://samtools.github.io/hts-specs/VCFv4.1.pdf). Any variant not marked as PASS in the filter field can be ignored with high confidence.
For details of the individual flags applied please see Advanced Parameters, below.
The formatted Tumour/Normal fields require some explanation. Table 15.7.3 details the ‘FORMAT’ elements with basic background.
Table 15.7.3. Explanation of VCF Fields Controlled by FORMAT Convention.
| ID | Description (as VCF header) | Detail |
|---|---|---|
| GT | Genotype | ‘./.’, see VCF specification |
| PP | Pindel calls on the positive strand | Number of reads mapped by Pindel to positive strand |
| PN | Pindel calls on the negative strand | Number of reads mapped by Pindel to negative strand |
| PB | BWA calls on the positive strand | Number of reads mapped by the primary aligner to positive strand showing a similar indel event. Other aligners can be used. |
| NB | BWA calls on the negative strand | Number of reads mapped by the primary aligner to negative strand showing a similar indel event. Other aligners can be used. |
| PD | BWA mapped reads on the positive strand | Count of positive strand mapped reads from primary aligner (with or without indel) |
| ND | BWA mapped reads on the negative strand | Count of negative strand mapped reads from primary aligner (with or without indel) |
| PR | Total mapped reads on the positive strand | Unique union of PP and PD |
| NR | Total mapped reads on the negative strand | Unique union of NP and ND |
| PU | Unique calls on the positive strand | Unique union of PP and PB |
| NU | Unique calls on the negative strand | Unique union of NP and NB |
| TG | Total distinct contributing read groups | Number of read groups represented in PR/NR values |
| VG | Variant distinct contributing read groups | Number of read groups represented in PU/NU values |
The values described are used in the filtering/flagging process; exceptions are TG and VG, which are for information only. These two values should be used with care, as increases in sequencing depth from single lanes has resulted in it being common for only one readgroup to be necessary to generate sufficient coverage for many applications.
Commentary
Background Information
The major drive to package cgpPindel was the ICGC/TCGA PanCancer project. This large-scale project involves the systematic analysis of 2500 WGS Tumour/Normal sample pairs (http://icgc.org). To standardize the primary mutation dataset for this endeavor, one mapping workflow was used and the resulting data processed through three calling pipelines: one each from the Broad Institute, DKFZ, and Sanger Institute. To participate in this project, we needed to ensure that all tools could function on any Unix system. This was used as an opportunity to revisit code, remove aligner-specific dependencies, and improve performance.
The Cancer Genome Project originally worked closely with the author of Pindel (around 2010) to improve the sensitivity of the core caller. At this time, longer-read paired-end sequencing was becoming available, resulting in increased tolerance of indels within mapped reads. Pindel itself is a split-read mapper based on a pattern growth model originally designed to capture break points of large deletions and medium-size insertions. Changing the candidate read selection to include mapped reads exhibiting indel events found by the primary aligner was the key to allowing Pindel to detect small events. An early version of this code is included in the Pindel release; however, this is tightly coupled with BWA-backtrack (aln + sampe) using several aligner specific fields.
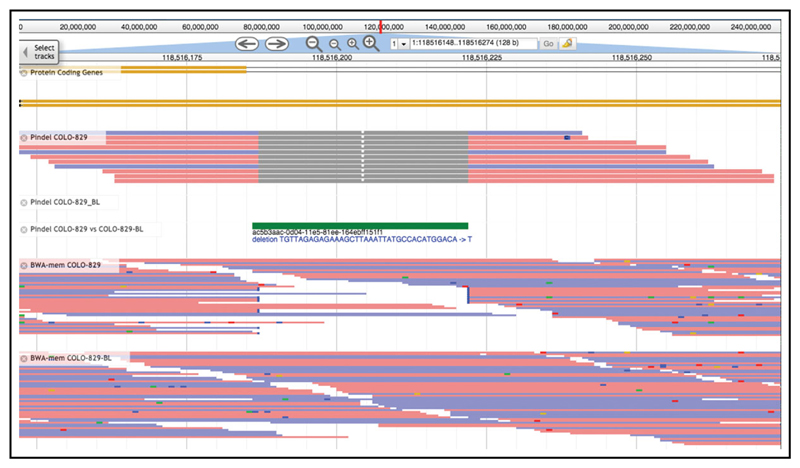
To facilitate analysis and visualization, the conversion of raw Pindel text alignments to BAM format were implemented internally; this feature was subsequently added to the core Pindel codebase. Figure 15.7.3 shows these minimal BAM files viewed in JBrowse (Skinner et al., 2009).
Figure 15.7.3.
A JBrowse view of a medium-size deletion called in the example dataset. The first track shows the structure of a localized protein-coding gene (SPAG17). The next three tracks are generated by cgpPindel, BAM of reads supporting the event from the tumor and normal, and the VCF file data, respectively. The final two tracks show the data from the original BWA-mem mapping for tumor and normal.
The version of Pindel included in cgpPindel has been tuned to our specifications and is divergent from the core algorithm available from the original authors, but any issues detected have been freely communicated.
Critical Parameters
It is essential that the type of sequencing being processed be correctly set for the -filter option. Incorrect values here have a significant effect on specificity and sensitivity.
For targeted screens, you may wish to omit the -badloci and -simrep options.
Troubleshooting
See Table 15.7.4 for solutions to common problems.
Table 15.7.4. Common Problems and Resolutions with cgpPindel.
| Problem | Cause | Solution |
|---|---|---|
| No results | Reference files and inputs have different naming convention | Check BAM is mapped with same reference build and or prefixes e.g., chr1 vs. 1. |
| Known event not reported | Event fails filtering | Search file including all results, not just PASS. If found, see the filter definitions. |
| Known event not reported | Region excluded by -exclude, -simrep, or -badloci options |
Check for overlap with the expected event. -badloci will exclude any reads where the mapped mate is overlapping. |
Advanced Parameters
The filtering step uses a panel of filters that can be switched on and off. As these are likely to be enhanced or augmented over time, please see the cgpPindel wiki for further details (https://github.com/cancerit/cgpPindel/wiki/VcfFilters).
New filters can be added by augmenting the perl module Sanger::CGP:: PindelPostProcessing::Filter Rules with a new subroutine, listing it in the dispatch table and adding the key to the relevant rules file (cgpPindel/perl/rules).
The subroutine uses the VcfTools perl module (http://vcftools.sourceforge.net/perl_module.html#Vcf.pm).
Please feel free to submit new filters for consideration as a pull request.
Suggestions for Further Analysis
CGP has created a variant annotation tool VAGrENT, which can be used to annotate indels and substitutions at the cDNA and protein level in VCF format. Please see unit 15.8 for further details on this tool.
Acknowledgement
The authors wish to thank Kai Ye (The Genome Institute at Washington University in St. Louis), the original author of Pindel for continued support and enhancement of the CGP branch.
This work was supported by the Wellcome Trust grant [098051].
Literature Cited
- Li H. Tabix: Fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2011;27:718–719. doi: 10.1093/bioinformatics/btq671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Quant Biol. 2013 arXiv:1303.3997 [q-bio]. Available at http://arxiv.org/abs/1303.3997. [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin M-L, Ordóñez GR, Bignell GR, Ye K, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: A next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics (Oxford, England) 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Internet Resources
- https://github.com/canceritRepository for Wellcome Trust Sanger Institute Cancer Genome Project public projects.
- http://gmt.genome.wustl.edu/packages/pindelCore Pindel site.
- https://genome.ucsc.edu/cgi-bin/hgTablesUCSC Genome Browser Table Browser.
- http://vcftools.github.io/specs.htmlVCF file format specification.
- https://samtools.github.io/hts-specs/SAMv1.pdfSAM format specification.