Abstract
Peptide macrocycles are widely utilized in the development of high affinity ligands including stapled α-helices. The linear rigidity of a 1,3-diynyl linkage provides an optimal distance (7 Å) between β- carbons of the i, i+4 amino acid side chains, suggesting its utility in stabilizing α-helical structures. Here we report the development of an on-resin strategy for an intramolecular Glaser reaction between two alkyne-terminated side chains using copper chloride, an essential bpy-diol ligand, and diisopropylethylamine, at room temperature. The efficiency of this ligation is illustrated by the synthesis of (i, i+4), (i, i+5), (i, i+6), and (i, i+7)-stapled BCL-9 α-helical peptides using the unnatural amino acid propargyl serine. Overall, this procedurally-simple method relies on inexpensive and widely available reagents to generate low molecular weight 23-, 26-, 29-, and 32-membered peptide macrocycles.
Keywords: peptide stapling, Glaser, copper acetylide, peptide macrocyclization, chemoselective
The alpha-helix is a ubiquitous protein secondary structure motif defined by directional hydrogen bonding between backbone N-H and C=O groups periodically along the amino acid sequence. The most prevalent protein secondary structure, the alpha-helix plays an important functional role in molecular interactions of proteins in their cellular context, including as DNA binding elements, membrane spanning structures, and in mediating protein-protein interactions (PPIs).[1,2] As dynamic protein complexes functioning as intended are critical to cell function, it is no surprise that aberrant PPIs are a major contributor to a truly diverse array of disease states. This fact has stimulated the development of conformationally-constrained stapled peptide ligands that display increased alpha-helicity relative to their unstapled congeners in order to better act as competitive inhibitors to pathogenic PPIs.
While an alpha-helix from a natural protein involved in mediating a disease state via PPIs is an obvious template for designing a therapeutic, the native peptide itself is a poor candidate. Peptides lacking their protein context rarely retain their structure and therefore their binding capability, in addition to being rapidly digested by proteases, general inability to cross cell membranes, and potential triggering of an immune response. Therefore, the development of synthetic methods to stabilize alpha-helical peptide therapeutic candidates represents a viable solution to otherwise ineffective bioactive peptides and has been pursued for decades by a number of research groups.[3-5] The most well-known approach to stabilizing alpha-helices is through side-chain tethering.[3-6] Through the introduction of a synthetic brace along the axis of the helix, the conformational entropy of the peptide is greatly reduced, thereby stabilizing secondary structure and restoring binding affinity, protecting against proteolytic degradation, and improving cell penetration. Typically, amino acids lying on the same face of the helix (i, i+4; i, i+7; etc.) are chosen for substitution to non-native amino acids containing side chains capable of covalent macrocyclization.
Initial efforts to form staples by Felix and coworkers employed the proteinogenic amino acids Lys and Glu/Asp for lactamization of i to i+4 residues.[7] Similarly, the formation of a thioether staple from the i to i+3 positions has also been developed, by the reaction of a Cys thiol and a side chain alpha-bromo amide.[8] As an expansion of two-component disulfide formation stapling,[9-12] Pentelute and coworkers have developed perfluorobenzene as an electrophilic linkage between two thiol side chains.[13] The most well-known methodology for helix stabilization, and from which the stapling field garners its name, is ring-closing metathesis (RCM) performed on-resin between amino acids bearing olefinic side chains.[14,15] Alkene RCM on peptides was first reported by Blackwell and Grubbs, who reported solution-phase metathesis and ensuing hydrogenation of i to i+4 systems to generate parrafinic staples.[16] Stapling between the i to i+7 positions has also been pursued, such as in the stabilization of oxyntomodulin analogues through biaryl crosslinking of two cysteine residues.[17] By introducing azide- and alkyne-terminated amino acids into the sequence, the copper-catalyzed azide-alykne cycloaddition (CuAAC) has also been successfully adapted to peptide stapling by a number of research groups (Figure 1).[18-20]
Figure 1.
Previous approaches for i to i+4 stapling include (a) lactamization, (b) thioether bridging, (c) perflurobenzene SNAr, (d) alkene RCM, and (e) CuAAC “Click” chemistry. Present work, (f) applies the Glaser reaction to form a rigid, linear 7 Å diynyl linker between i to i+4 spaced propargyl serine residues. The reaction is also capable of i to i+7 stapling (h), akin to (g) biaryl stapling of cysteine residues. Model helix taken from the protein monellin, PDB entry 3PXM.38
While each of these stapling methods have been shown to increase peptide helicity, no single method has proven generally applicable as a number of shortcomings manifest after macrocyclization. For example, cis-trans isomerization impacts the monodispersity of RCM-generated stapled peptides, while thioether and amide-cyclized stapled peptides lack both a rigid framework and the functionality to be subsequently manipulated. Triazole, aryl, and biaryl linkages address the problem of rigidity by constraining the peptide macrocyle into a bicyclic architecture, but suffer from decreased aqueous solubility stemming from their large hydrophobic surfaces. To overcome these deficiencies, we sought to develop an on-resin peptide stapling strategy between alkyne-terminated amino acid side chains that delivers a unique 1,3-diynyl peptide macrocycle. The 7 Å length of the employed 1,3-diyne motif is the optimal distance between the β-carbons of the i, i+4 amino acid side chains and thereby represents the most atom-economical approach to forming low molecular weight stapled peptide macrocycles. Optimizing this distance and decreasing the number of paraffinic moieties has the added benefit of decreasing bulk hydrophobicity, relative to similarly rigid triazole, aryl, and biaryl linkages. Overall, diyne-stapled peptides enjoy the benefits of all other staple types including increased rigidity, solubility, monodispersity (no cis/trans isomerization), and potential for functionalization.[21]
Recently, Wang and coworkers exploited CuAAC-stapling for the stabilization of a peptide segment of B-cell CLL/lymphoma 9 protein (BCL9).[22] BCL9 is a transcriptional activator of β-catenin, which is a primary mediator of the Wnt signaling pathway whose runaway activation is a hallmark of a number of human cancers, including colon cancer, prostate cancer and melanoma.[23,24] Inspired by recent work on the adaptation of the classic Glaser reaction for diyne formation in aqueous systems,[25] we employed the BCL9 24-mer peptide as a model system to develop a robust platform for peptide stapling, using inexpensive or easily synthesized amino acid building blocks, cheap catalysts and ligands, and mild reaction conditions capable of robust performance on the solid support.
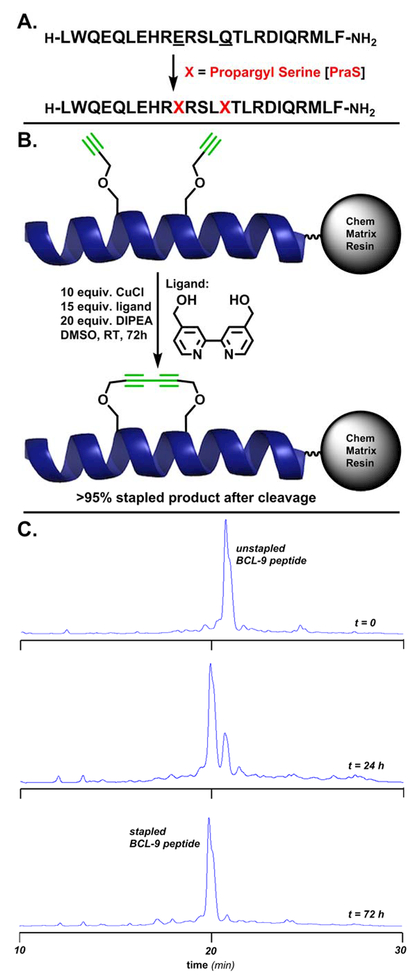
From the wild-type BCL9 24mer sequence, the same residues chosen for substitution by Wang and coworkers for CuAAC stapling were substituted here to propargyl serine residues for on-resin Glaser stapling. In addition to the i, i+4 BCL9 peptide, i, i+5, i, i+6 and i, i+7 peptides were also prepared using propargyl serine as the substitution for both residues. Notably, the i, i+5 and i, i+6 peptides would require the propargyl serine side chain to extend far enough away from the helical core to form the linear staple, while the i, i+7 peptide features both residues on the same face of the helix, as in the i, i+4 molecule. These BCL9 24-mers were synthesized by standard Fmoc solid-phase peptide synthesis (SPPS) on high-swelling Rink Amide ChemMatrix resin (Figure 2A). The N-Fmoc-protected, side-chain-protected peptides were then subjected to the Glaser stapling cocktail.
Figure 2.
A. The BCL9 24-mer peptide. Underlined residues were chosen for substitution to propargyl serine to form the i, i+4 diyne staple. The peptide was synthesized on Rink Amide ChemMatrix resin. B. Reaction conditions for on-resin Glaser stapling. The displayed helix is the wild-type BCL9 24-mer, PDB entry 2GL7. C. From top to bottom, HPLC traces of crude peptide cleaved in 10% DMS, 2.5% TIPS, 2.5% H2O cocktail (2 hours) after Glaser stapling reaction at t=0, t=24 (76% conversion by peak area), and t=72 hrs (>95% conversion by peak area).
While copper-mediated terminal alkyne couplings have been known for over a century,[26,27] the mechanistic requirement of maintaining catalytically competent Cu(I) has limited its applicability in manipulations involving biomolecules.[28-31] Previous work on the stabilization of a β-turn motif formed by an 8-mer peptide via Glaser-Hay coupling on-resin was met with limited success, as peptide degradation, high amounts of side products and incomplete reactions over long time periods arose from requirements such as mixed solvent systems, heating/cooling cycles and microwave reactors.[32] The recent application of a bpy-diol ligand, specifically 4,4’-bis(hydroxymethyl-2,2’-bipyridine (Figure 2B), which functions to broker Cu(I) towards the formation of Cu(I)-acetylides for the Glaser coupling while simultaneously sequestering harmful Cu(II) species capable of oxidative damage has rendered the reaction useful in aqueous bioconjugation reactions.[25] In contrast to intermolecular chemoselective ligation onto unprotected peptides, the introduction of staples is most advantageous when performed on-resin, as the desired amount of peptide can be exposed to the stapling conditions and yield resin-linked, fully recoverable stapled product, following the removal of excess reagents by flow wash.
Drawing from our previous work on adapting the Glaser reaction to bioconjugation,[25] suitable conditions for on-resin stapling in solvents typically employed during Fmoc-SPPS to limit peptide aggregation (DMF, NMP, DMSO) were developed. Following a screening of common organic bases amenable to Fmoc-SPPS, diisopropylethylamine (DIPEA) was chosen as the optimal base for the reaction given its miscibility in these solvents and well-documented compatibility in Fmoc-SPPS protocols. While the reaction is successful in each of these solvents, the optimal conditions (Figure 2B) utilized DMSO as the solvent to deliver 76% product over 1 day and >95% stapled product over 3 days at room temperature (25°C) from the linear starting material by HPLC peak area (Figure 2C). A summary of the reaction optimization for a canonical i, i+4 staple across a number of organic solvents and bases is presented in Table 1.
Table 1.
Base and solvent screen for stapling of BCL-9 peptide (i, i+4) shown in Figure 2B. t=24 hrs, T=25°C.
| Solvent | Base (20 equiv.) | Conversion[a] (%) |
|---|---|---|
| DMF | DIEA | 63 |
| DMF | DBU | 28 |
| DMSO | DIEA | 76 |
| DMSO | DBU | 38 |
| NMP | DIEA | 16 |
| TFE | DIEA | 9 |
Percent conversion by HPLC peak area.
A summary of percent conversion by HPLC peak area for the i, i+4; i, i+5; i, i+6 and i, i+7 BCL9 24-mer peptides is presented in Table 2. Notably, the stapling of the i, i+5; i, i+6 and i, i+7 peptides proceeded less smoothly than the i, i+4 peptide, most likely due to physical limitations of the propargyl side chains reaching one another to form the diyne product – in anticipation of this result, these reactions were left for 5 days (120 hrs) instead of 3 days (72 hrs). ESI-MS data of the linear starting materials and the stapled products are presented in the Supporting Information. Following the Glaser reaction, the resin was flow-washed, N-Fmoc deprotected by standard procedure.
Table 2.
Stapled product conversion by HPLC peak area.
| BCL9 Peptide | Conversion[a] (%) | Time (hrs) |
|---|---|---|
| i, i+4 | >95 | 72 |
| i, i+5 | 7 | 120 |
| i, i+6 | 10 | 120 |
| i, i+7 | 16 | 120 |
Percent conversion by HPLC peak area after standard conditions (Figure 2B) for the given period of time.
The BCL9 peptides contain methionine, which is susceptible to oxidation to the sulfoxide during routine Fmoc SPPS and downstream TFA cleavage from resin. While the starting material crude peptide contained this species as a minor side product before the stapling reaction, we observed this species as the primary product following exposure to the stapling cocktail in DMSO. Inspired by related work on controlling methionine oxidation in synthetic peptides[33-35] and by classic biochemistry procedures for handling and redox control of methionine-containing proteins,[36] we developed a TFA cleavage cocktail containing 10% dimethyl sulfide (DMS) in addition to the standard 2.5% triisopropylsilane (TIPS) and 2.5% H2O additives. In strong acid such as HCl or TFA, methionine sulfoxide is rapidly reduced to methionine when an excess of reducing agent such as DMS is present. Beyond its application in this setting for repairing methionine oxidation inflicted by the Glaser cocktail, we envision this cleavage cocktail having broad utility in the cleavage of any Fmoc-SPPS peptide containing unwanted methionine oxidation side products.
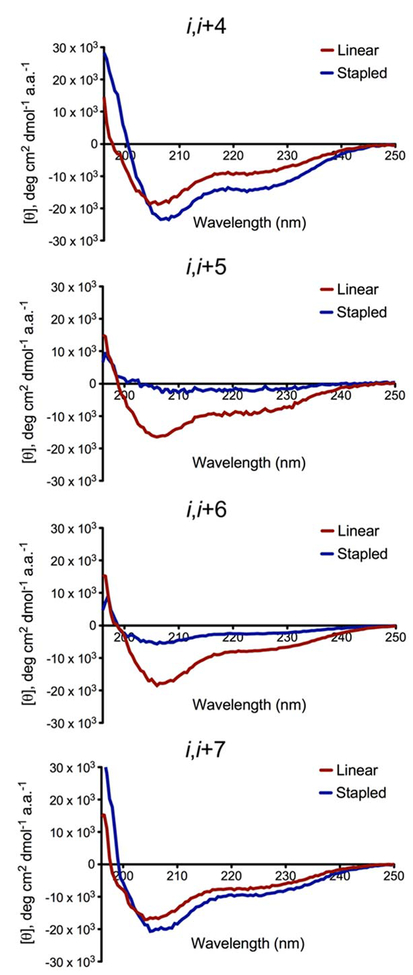
With diyne-stapled products in hand, our next aim was to determine their helical character in aqueous solution versus the unstapled, linear peptides. Circular dichroism (CD) spectroscopy confirmed the stapled products arising from the i, i+4 and i, i+7 peptides as more helical than their linear precursors, while the i, i+5 and i, i+6 peptides displayed more disordered structures versus their unstapled starting materials, Figure 3. Each peptide was purified by preparatory HPLC, lyophilized, and dissolved in phosphate-buffered saline (PBS), pH 7.4, at a concentration of 40 μM at T=25°C. Exact peptide concentrations were determined by UV-Vis spectroscopy prior to the CD assays. Mean residue ellipticity at 222 nm (θ222) is known to be linearly related to mean helix content.[37] From this inference, percent helicities for all peptides were calculated, quantifying the i, i+4 peptide as having a 56% increase in helical content and the i, i+7 peptide having a 40% increase. Conversely, the i, i+5 and i, i+6 peptides had their
Figure 3.
CD spectra of diyne-stapled BCL9 24-mers versus unstapled linear BCL9 24-mers. All peptides were dissolved in PBS, pH 7.4, to a final concentration of 40 μM. 3 baseline scans of buffer were taken and averaged at T=25°C, followed by 5 scans of each peptide. These spectra were averaged, baseline-subtracted from the averaged blank scans, and converted to mean residue ellipticity to plot against wavelength. The percent helicity of each peptide was calculated based on [θ]222/[θ]max. [θ]max was calculated according to the formula: [θ]max=−39000(1–3/n), where n is the number of amide bonds.
baseline helical content (~25%) ablated by the installation of the diyne crosslink, Table 3. Together, these data confirm the Glaser stapling method as successful in enhancing the helical conformation of BCL9 peptides when the stapled residues are present on the same face of the helix.
Table 3.
Percent helicity of stapled and linear BCL9 variants.
| i, i+4 | i, i+5 | i, i+6 | i, i+7 | |
|---|---|---|---|---|
| Stapled | 42% | 6% | 8% | 35% |
| Linear | 27% | 27% | 26% | 25% |
In summary, we have adapted the aqueous Glaser bioconjugation conditions published by Silvestri and coworkers as a robust on-resin reaction for helical peptide stapling. By employing a polar solvent (DMSO) that effeciently swells peptide-loaded resins, mild and chemoselective conditions were developed to intramolecularly ligate alkyne-terminated amino acids at positions i, i+4, i, i+5, i, i+6, and i, i+7 of BCL-9 α-helical peptides. This furnished low molecular weight 23-, 26-, 29-, and 32-membered peptide macrocycles cleanly after SPPS, stapling, and cleavage. The procedural simplicity of this method in addition to the ubiquity of the reagents required are expected to facilitate the use of this method in the development of bioactive helical peptide ligands and inhibitors.
Supplementary Material
Acknowledgements
This work was funded by the National Institutes of Health (NIH R01 AI113867). Author J. C. J. Hintzen acknowledges support from the Stichting Fundatie van de Vrijvrouwe van Renswoude (AV 20170242). The authors wish to thank Dr. J. S. Chen and the Automated Synthesis Facility at TSRI for assistance with peptide analysis and purification.
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- [1].Bergey CM, Watkins AM, and Arora PS, Bioinformatics, 2013, 29(21), 2806–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bullock BN, Jochim AL and Arora PS, J. Am. Chem. Soc, 2011, 133, 14220–14223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Walensky LD and Bird GH, J. Med. Chem, 2014, 57, 6275–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lau YH, de Andrade P, Wu Y and Spring DR, Chem. Soc. Rev, 2015, 44, 91–102. [DOI] [PubMed] [Google Scholar]
- [5].White CJ and Yudin AK, Nature Chemistry, 2011, 3, 509–524. [DOI] [PubMed] [Google Scholar]
- [6].Noisier AFM, García Jesús, Ionut IA, and Albericio F, Angew. Chem., Int. Ed 2017, 56, 314–318. [DOI] [PubMed] [Google Scholar]
- [7].Felix AM, Heimer EP, Wang C-T, Lambros TJ, Fournier A, Mowles TF, Maines S, Campbell RM, Wegrzynski BB, Toome V, Fry D and Madison VS, Int. J. Pept. Protein Res, 1988, 32, 441–454. [DOI] [PubMed] [Google Scholar]
- [8].Brunel FM and Dawson PE, Chem. Commun, 2005, 2552–2554. [DOI] [PubMed] [Google Scholar]
- [9].Jackson DY, King DS, Chmielewski J, Singh S and Schultz PG, J. Am. Chem. Soc, 1991, 113, 9391–9392. [Google Scholar]
- [10].Tan WL, Wong KH and Tam JP, FASEB J, 2017, 31, 115. [Google Scholar]
- [11].Assem N, Ferreira DJ, Wolan DW, Dawson PE, Angew. Chem., Int. Ed. Engl, 2015, 54, 8665–8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jo H, Meinhardt N, Wu Y, Kulkarni S, Hu X, Low KE, Davies PL, DeGrado WF, and Greenbaum DC, J. Am. Chem. Soc 2015, 134, 17704–17713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Spokoyny AM, Zou Y, Ling JJ, Yu H, Lin Y-S and Pentelute BL, J. Am. Chem. Soc, 2013, 135, 5946–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Schafmeister CE, Po J and Verdine GL, J. Am. Chem. Soc, 2000, 122, 5891–5892. [Google Scholar]
- [15].Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL and Korsmeyer SJ, Science, 2004, 305, 1466–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Blackwell HE and Grubbs RH, Angew. Chem., Int. Ed, 1998, 37, 3281–3284. [DOI] [PubMed] [Google Scholar]
- [17].Muppidi A, Zou H, Yang PY, Chao E, Sherwood L, Nunez V, Woods AK, Schultz PG, Lin Q and Shen W, ACS Chem. Biol 2016, 11, 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kolb HC and Sharpless KB, Drug Discov. Today, 2003, 8, 1128–1137. [DOI] [PubMed] [Google Scholar]
- [19].Ingale S and Dawson PE, Org. Lett 2011, 13, 2822–2825. [DOI] [PubMed] [Google Scholar]
- [20].Cantel S, Isaad Ale C, Scrima M, Levy JJ, DiMarchi RD, Rovero P, Halperin JA, D’Ursi AM, Papini AM and Chorev MJ, Org. Chem 2008, 73, 5663–5674. [DOI] [PubMed] [Google Scholar]
- [21].Verlinden S, Ballet S and Verniest G, Eur. J. Org. Chem, 2016, 35, 5807–5812. [Google Scholar]
- [22].Kawamoto SA, Coleska A, Ran X, Yi H, Yang C-Y and Wang S, J. Med. Chem, 2012, 55, 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Polakis P, Genes Dev, 2000, 14, 1837–1851. [PubMed] [Google Scholar]
- [24].Sampietro J, Dahlberg CL, Cho US, Hinds TR, Kimelman D and Xu W, Mol. Cell, 2006, 24, 293–300. [DOI] [PubMed] [Google Scholar]
- [25].Silvestri AP, Cistrone PA and Dawson PE, Angew. Chem., Int. Ed, 2017, 56, 10438–10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Glaser C, Ber. Dtsch. Chem. Ges, 1869, 2, 422–424. [Google Scholar]
- [27].Glaser C, Justus Liebigs Ann. Chem, 1870, 154, 137–171. [Google Scholar]
- [28].Brauer MCN, Neves Filho RAW, Westermann B, Heinke R and Wessjohann LA, Beilstein J. Org. Chem 2015, 11, 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Verlinden S, Geudens N, Martins JC, Tourwe D, Ballet S and Verniest G, Org. Biomol. Chem, 2015, 13, 9398–9404. [DOI] [PubMed] [Google Scholar]
- [30].Lampkowski JS, Villa JK, Young TS and Young DD, Angew. Chem. Int. Ed, 2015, 54, 9343–9346. [DOI] [PubMed] [Google Scholar]
- [31].Minakawa N, Ono Y and Matsuda A, J. Am. Chem. Soc 2003, 125, 11545–11552. [DOI] [PubMed] [Google Scholar]
- [32].Auberger N, Di Pisa M, Larregola M, Chassaing G, Peroni E, Lavielle S, Papini A-M, Lequin O and Mallet J-M, Bioorg. Med. Chem, 2014, 22, 6924–6932. [DOI] [PubMed] [Google Scholar]
- [33].Hackenberger CPR, Org. Biomol. Chem, 2006, 4, 2291–2295. [DOI] [PubMed] [Google Scholar]
- [34].Vilaseca M, Nicolás E, Capdevila F and Giralt E, Tetrahedron, 1998, 54, 15273–15286. [Google Scholar]
- [35].Kukowska M, Kukowska-Kaszuba M and Dzierzbicka K, Tet. Lett, 2015, 56, 525–528. [Google Scholar]
- [36].Shechter Y, J. Biol. Chem, 1986, 261, 66–70. [PubMed] [Google Scholar]
- [37].Rohl CA and Baldwin RL, Biochemistry, 1997, 36, 8435–8442. [DOI] [PubMed] [Google Scholar]
- [38].Templeton CM, Ostovar Pour S, Hobbs JR, Blanch EW, Munger SD and Conn GL, Chem. Senses 2011, 36, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.