Abstract
RNA sequencing (RNA-seq) is becoming a prevalent approach to quantify gene expression and is expected to gain better insights into a number of biological and biomedical questions compared to DNA microarrays. Most importantly, RNA-seq allows us to quantify expression at the gene or transcript levels. However, leveraging the RNA-seq data requires development of new data mining and analytics methods. Supervised learning methods are commonly used approaches for biological data analysis that have recently gained attention for their applications to RNA-seq data. Here, we assess the utility of supervised learning methods trained on RNA-seq data for a diverse range of biological classification tasks. We hypothesize that the transcript-level expression data are more informative for biological classification tasks than the gene-level expression data. Our large-scale assessment utilizes multiple data sets, organisms, lab groups, and RNA-seq analysis pipelines. Overall, we performed and assessed 61 biological classification problems that leverage three independent RNA-seq data sets and include over 2000 samples that come from multiple organisms, lab groups, and RNA-seq analyses. These 61 problems include predictions of the tissue type, sex, or age of the sample, healthy or cancerous phenotypes, and pathological tumor stages for the samples from the cancerous tissue. For each problem, the performance of three normalization techniques and six machine learning classifiers was explored. We find that for every single classification problem, the transcript-based classifiers outperform or are comparable with gene expression-based methods. The top-performing techniques reached a near perfect classification accuracy, demonstrating the utility of supervised learning for RNA-seq based data analysis.
Keywords: RNA-seq, alternative splicing, classification, gene expression, machine learning
INTRODUCTION
Ever since the intrinsic role of RNA was proposed by Crick in his Central Dogma (Crick 1970), there has been a desire to accurately annotate and quantify the amount of RNA material in the cell. A decade ago, with the introduction of RNA sequencing (RNA-seq) (Mortazavi et al. 2008), it became possible to quantify the RNA levels on the whole genome scale using a probe-free approach, gaining insights into cellular and disease processes and illuminating the details of many critical molecular events such as alternative splicing, gene fusion, single nucleotide variation, and differential gene expression (Conesa et al. 2016). The basic assessment of RNA-seq is focused on utilizing the data for differential gene expression between the groups of biological importance (Trapnell et al. 2013). However, there are additional patterns that can be elucidated from the same raw sequencing data by extracting the expression levels of the alternatively spliced transcripts (Zhang et al. 2013).
Alternative splicing (AS) of pre-mRNA provides an important means of genetic control (Chen and Manley 2009; Nilsen and Graveley 2010). It is abundant across all eukaryotes and even occurs in some bacteria and archaea (Keren et al. 2010; Barbosa-Morais et al. 2012; Reddy et al. 2013). AS is defined by the rearrangement of exons, introns, and/or untranslated regions that yields multiple transcripts (Kelemen et al. 2013). Furthermore, 86%–95% of multiexon human genes are estimated to undergo alternative splicing (Djebali et al. 2012). Genes tend to express many transcripts simultaneously, 70% of which encode important functional or structural changes for the protein (Djebali et al. 2012). RNA-seq data encompass expression at both gene and transcript levels: the gene-level expression amounts to the combined expression of all transcripts associated with a particular gene. It has been previously demonstrated that the gene-level expression is an excellent indicator of the tissue of origin as well as certain cancer types (Wan et al. 2014; Wei et al. 2014; Achim et al. 2015; Danielsson et al. 2015; Mele et al. 2015). However, transcript-level expression has been shown to provide a more precise measurement of gene product dosage, resulting in the superior performance in predicting the cancer patient prognosis or survival time, and providing further insights into the functional transformations driving cancer (Zhang et al. 2013; Shen et al. 2016; Trincado et al. 2016; Climente-González et al. 2017). Differential AS depends on many factors, including the epigenetic state, genome sequence, RNA sequence specificity, activators and inhibitors from both, proteins and RNAs, as well as post-translational modification (Edwards and Myers 2007; Chen and Manley 2009; Luco et al. 2011; Gamazon and Stranger 2014). These diverse mechanisms control AS to obtain developmental, cell-type, and tissue-specific expression. Furthermore, the patterns driven by AS and specific to cancer and other diseases have been recently identified (Cáceres and Kornblihtt 2002; Sebestyen et al. 2015).
Machine learning tools developed over the last several decades have significantly advanced the analysis of the vast amount of next generation sequencing and microarray expression data by discovering the biologically relevant patterns (Tarca et al. 2007; Liu et al. 2013; Neelima and Babu 2017). Previous studies have utilized unsupervised and supervised machine learning techniques on the microarray gene expression data with variable success rates (Vandesompele et al. 2002; Libbrecht and Noble 2015). Along with the individual approaches (Jagga and Gupta 2014), large-scale comparative studies have been carried out (Costa et al. 2004; Pirooznia et al. 2008). Some studies evaluated both basic and advanced clustering techniques, such as hierarchical clustering, k-means, CLICK, dynamical clustering, and self-organizing maps, to identify the groups of genes that share similar functions or genes that are expressed during the same time point of a mitotic cell cycle (Mudge et al. 2013; GTEx Consortium 2015; Mele et al. 2015). Other studies compared the ability to perform disease/healthy sample classification tasks by state-of-the-art supervised methods, such as support vector machines (SVM), artificial neural nets (ANN), Bayesian networks, decision trees, and random forest classifiers (Pirooznia et al. 2008).
When it comes to the biological classification, the RNA-seq data present an attractive alternative to microarrays, since it is possible to quantify all RNA present in the sample without the need of the a priori knowledge. With RNA-seq rapidly replacing microarrays, it is necessary to assess the potential of the supervised machine learning methodology applied to the RNA-seq data across multiple data sets and biological questions (Byron et al. 2016). Recently, there have been limited studies that have assessed RNA-seq data with supervised and unsupervised machine learning techniques (Thompson et al. 2016). However, these studies utilized RNA-seq data by leveraging only gene-level expression data rather than more detailed transcript-level data available for the alternative splicing transcripts (Chen and Manley 2009). Most recently, a study analyzed the utility of RNA-seq transcript-level data for the disease/nondisease phenotype classification of the samples, showing the advantage of the transcript expression data for the disease phenotype prediction task (Labuzzetta et al. 2016). However, the question of whether or not the utility of transcript-level expression presents a general trend across all main biological and biomedical classification tasks remains open.
This work aims to systematically assess how well state-of-the-art supervised machine learning methods perform in various biological classification tasks when utilizing either gene-level or transcript-level expression data obtained from the RNA-seq experiments. The assessment is done from three different perspectives: (i) by analyzing RNA-seq data from two organisms (rat and human), (ii) by using the increasingly difficult data sets, and (iii) by considering different technical scenarios. The data sets were analyzed using six supervised machine learning techniques, three normalization methods, and two RNA-seq analysis pipelines. Altogether, the method performances on 61 major classification problems that amounted to 2196 individual classification tasks were compared. We define a major classification problem as a combination of the biological class and the data set used. We then define the “individual classification task” as a combination of all machine learning methods, normalization techniques, as well as the major classification problem. The use of multiple data sets allows us to determine if the success of a classification task is due to the discovery of distinct biological patterns by a machine learning algorithm, or if it is due to biologically unrelated patterns such as caused by differences in library preparation and/or the lab source. Finally, we assess whether using the information on alternatively spliced transcripts presented in the form of transcript expression data can provide the higher classification accuracy, compared to the gene expression data.
RESULTS
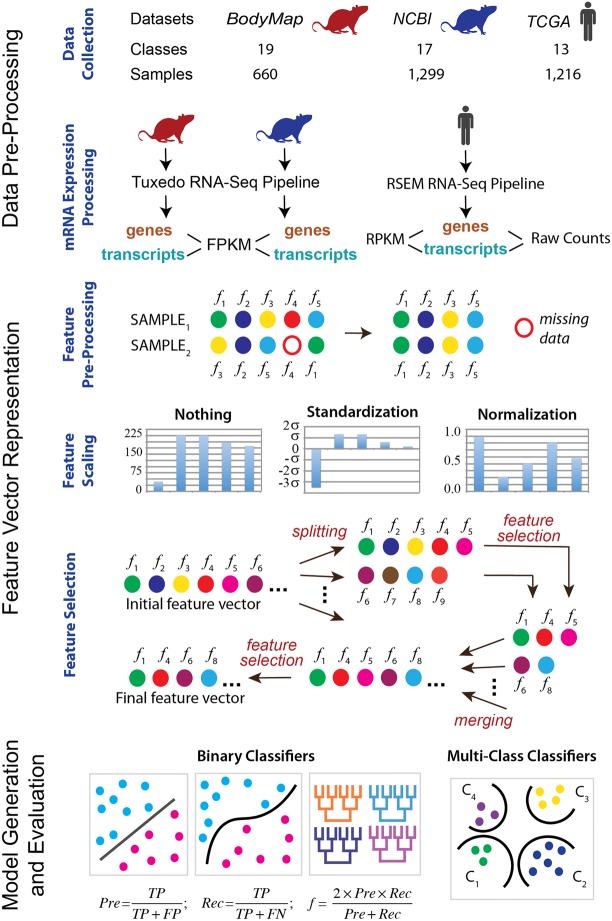
The goal of this work is to examine the capabilities of supervised machine learning methods in performing biological classification based on RNA-seq data. Specifically, we analyzed whether the performance is influenced by (i) the power of the machine learning classifier, and/or (ii) more detailed information extracted from the RNA-seq data. In the first case, we assessed several supervised classifiers, ranging from the very basic methods to the state-of-the-art supervised classifiers, across three different normalization techniques. In the second case, we compared the same classifiers using either gene-level or transcript-level expression data. Together, the study setup utilized three RNA-seq data sets, six supervised machine learning techniques, and three normalization protocols (Fig. 1). Furthermore, each of the 61 classification problems was set to use the numerical features generated either from the gene-level or transcript-level expression data. To the best of our knowledge, this is the largest comparative analysis of biological classification tasks based on RNA-seq data, performed to date.
FIGURE 1.
Overall computational pipeline used in this work. The samples from each of the three data sets are collected. The classification tasks are then defined. The expression data are processed for each sample at the gene and isoform levels using two RNA processing pipelines and three different count measures. Next, feature preprocessing, scaling, and selection are done for each classification task. Finally, the binary as well as multiclass supervised classifiers are trained and tested.
Classification tasks analyzed
Two categories of classification tasks were considered: normal phenotype and disease phenotype. In the first category, we determined whether it was possible to distinguish between age groups, sex, or tissue types in normal rats based on transcriptome analysis. The second category focused on classification tasks associated with breast cancer, with the main goal to differentiate between the pathological tumor stages. Both categories were analyzed using RNA-seq data at the gene and transcript levels. Two types of classification were considered for each category of tasks: binary classification and multiclass, or multinomial, classification. These classification types center around two conceptually different classification problems. The binary classification distinguishes a sample as either belonging to the class or not. The multiclass classification distinguishes which class a specific sample belongs to. For example, for a binary tissue classification task, a sample can be classified as extracted from the brain tissue or not. In the context of a multiclass classification, the same sample is classified as extracted from exactly one of several tissue types.
Data set statistics
Three data sets were used to carry out the classification tasks: two data sets for the normal phenotype classification tasks and one data set for the disease phenotype classification tasks (Fig. 1). The first data set was obtained from the Rat Body Map and is referred to as the RBM data set. It consisted of 660 normal rat samples whose transcriptomes were sequenced from the same rat strain and served as a reference data set for the community (Yu et al. 2014). The transcriptomes were obtained at 40 M reads per sample on average. The data were evenly split between the male and female rats, four age groups, and eleven tissue types (Supplemental Fig. S1). The four age groups included 2, 6, 21, and 101 wk. The eleven tissue types included adrenal gland, brain, heart, thymus, lung, liver, kidney, uterus, testis, muscle, and spleen. All samples used the same library preparation protocol, sequencer, and were prepared by the same laboratory. As a result, the data set was expected to have the least impact from the data inconsistency that arises from the nonbiological sources, such as utilizing different sequencing protocols, instruments, and other parameters.
The second data set, also used in the normal phenotypes classification tasks and referred to as the NCBI data set, included over 1100 samples (Supplemental Fig. S2) with the sequencing depth ranging between 6 and 116 M reads. This data set was prepared by analyzing the collection of rat transcriptomes that were sequenced on an Illumina Hi-Seq 2000 platform and were publicly available from the NCBI SRA database (Kodama et al. 2012). The data set was obtained from the sequencing experiments of 29 research projects (Supplemental Table 1). It contained highly variable transcriptomes due to the differences in library preparation, project goals, and rat strains. The classification tasks for the NCBI data set were the same as for the RBM data set, but with one modification. The age classification was modified from the four age groups into either embryo or adult age groups and is described later in this section.
The last data set included raw RNA-seq data from 1216 human breast cancer patients from the Cancer Genome Atlas (referred to as TCGA data set) and was used in the disease phenotype classification tasks (Weinstein et al. 2013). At the preprocessing stage, two RNA-seq data normalization techniques were implemented and compared. Classification was performed to distinguish between the pathological cancer stages, as defined by the American Joint Committee on Cancer (AJCC) (Edge and Compton 2010). The AJCC breast cancer staging is based on size of tumors present within the breast, presence or absence of detection of metastases that are not within the breast, and the presence, size, and type of metastases within the lymph nodes. The patients were distributed with high variability especially when considering subcancer stages (Supplemental Fig. S3).
Feature selection and analysis
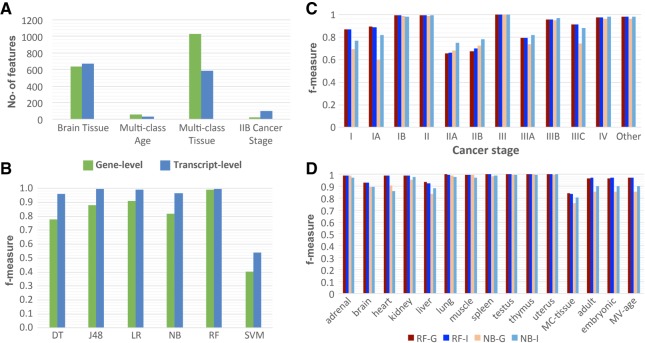
The numerical features for this study represented either gene or transcript expression levels. As a result, the number of features ranged from 10,711 to 73,592, depending on the data set and representation (Supplemental Table S2). Utilizing all features for a classification task greatly increases the computational complexity. Moreover, not all expressed genes or transcripts may be important for a given classification task; using the uninformative features during the training process could potentially decrease the accuracy of the classifier. To reduce the dimensionality of the feature space, a feature selection method (Hall and Smith 1998) was applied in a classification-specific and data set-specific manner, resulting in a significant reduction of features ranging from 107 to 735 folds (Fig. 2A; Supplemental Figs. S4–S7; Supplemental Tables S3).
FIGURE 2.
Overview of feature selection and the performance of classifiers using gene and isoform level expression data. (A) Comparison of the number of features between gene and isoform after feature selection. Each classification task has the same number of features selected for each classifier at the gen-level and isoform-level. The four selected classes represent the four types of patterns seen between gene-level (green) and isoform-level classifiers. The brain tissue class is the most common pattern of feature selection. In general, more features are selected for isoform-level classifiers versus gene-level. (B) Example of the variability of gene and isoform performance determined by f-measure across the six methods ([DT] Decision Table, [J48] J48 Decision Tree, [LR] Linear Regression, [NB] Naïve Bayes, [RF] Random Forest, [SVM] Support Vector Machine). This example is from the RBM data set for the Multi Age class without normalization. While there is a high degree of variability in performance, isoform-level classifiers consistently perform either comparably or better than gene-level classifiers. (C,D) Summary of the performance variability across classes for gene and isoform f-measure for the most frequent top and bottom performance methods ([RF-G] Random Forest Gene, [RF-I] Random Forest Isoform, [NB-G] Naïve Bayes Gene, [NB-I] Naïve Bayes Isoform). The data used in C is TCGA data set and in D is NCBI data set. MC stands for multiclass.
Regardless of the classification task or data set, the normalization of the RNA-seq data did not make a significant difference on the choice of the selected features: Variation in the numbers of selected features was <1% (Supplemental Figs. S8–S11). An interesting observation, consistent across different tissue classification tasks, was that the number of features selected for the multiclass classification tasks was significantly greater than for a binary classification task. This observation should not be surprising, because the binary classification task is generally simpler than the multiclass classification task (Supplemental Table S3). However, in our case even if all features of the binary classification tasks related to a single multiclass classification task were combined, it would still not account for all features selected by the feature selection method for the multiclass task.
In many cases, the overall number of features selected for a binary classification task was the same or nearly the same, irrespective of whether the features were gene- or transcript-based (Supplemental Figs. S4–S7). Does it mean that the features from the gene- or transcript-based approaches correspond to the same genes? Not always: The transcripts used for the selected features in a transcript-based, or transcript-based, classifier did not always originate from the genes that were selected for the corresponding gene-based classifier. Indeed, because 70% of transcripts were expected to encode different functional gene products (Djebali et al. 2012), we expected cases where the gene expression features were not as specific as the corresponding transcript features. In general, there was a large portion of 73,592 transcripts from 20,524 genes that corresponded to the same gene set (70%–100%). However, there were several classification tasks, including multiclass tissue classification using the NCBI data set, where there was a lower percentage of such overlap (30%). Furthermore, there were several classification tasks, including multiclass age classification using RBM, multiclass tissue classification using GEO, and stage IIB classification using the TCGA data set, where the numbers of features that used either the gene or transcript level of expression were significantly different, which was usually the case when a multiclass classification task was considered (Fig. 2A; Supplemental Figs. S4–S7; Supplemental Tables S2, S3). Another interesting observation was obtained when comparing the RBM and NCBI Rat data sets: The number of selected features was much smaller for the RBM data set rather than for the NCBI Rat data set (on average 231 versus 588), thus indicating the need for additional features to compensate for the increased data variability found within the NCBI Rat data set.
Overall performance of classifiers trained on gene-based versus transcript-based data
Next, we hypothesized that because of the observed specificity of alternative splicing across tissues, ages, sexes, and between disease/normal phenotypes, training classifiers with the RNA-seq data at the transcript level for the biological classification tasks could increase the classification accuracy (Hall and Smith 1998; Xiong et al. 2015). Consistent with this hypothesis, the supervised learning classifiers that leverage the transcript-based data performed comparably or better than the classifiers trained on the gene data for all classification tasks (Figs. 2B, 3). This observation also held true irrespective of the data sets used, normalization protocols, classification tasks, or supervised classifiers. Furthermore, the differences between the gene- and transcript-based classifiers were consistently less than the standard deviation across all 10-folds, supporting this hypothesis (Supplemental Figs. S12, S13). The most frequently top performing methods were the random forest and logistic regression classifiers, whereas the worst performing method was typically the naïve Bayes classifier (Fig. 2B–D). However, the former approaches were not the most accurate ones for every single classification task, since in some cases the naïve Bayes classifier was capable of outperforming all other methods tested (Stages IIA and IIB, Fig. 2C). In general, the random forest classifier applied to the data without any normalization achieved 83%–100% accuracy (Fig. 2B–D).
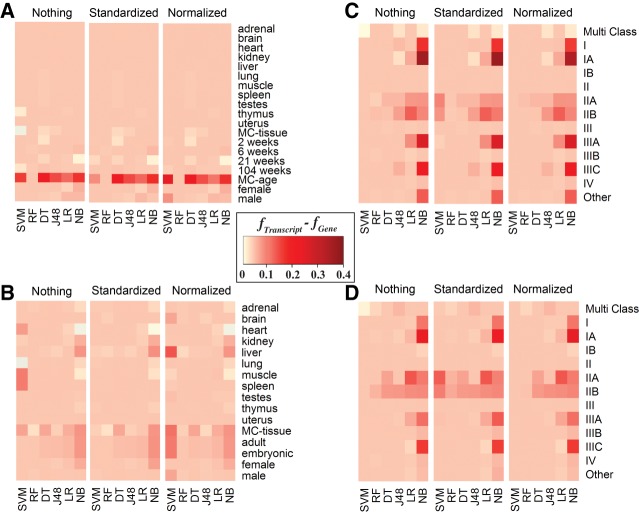
FIGURE 3.
Heat map representation of the difference between Isoform and Gene f-measure across machine learning methods, classes, data sets, and normalization techniques. For the majority of classification tasks, using isoform-level rather than gene-level expression data resulted in a small to substantial increase of the performance accuracy, represented by f-measure values here. The bottom x-axis represents the machine learning techniques ([DT] Decision Table, [J48] J48 Decision Tree, [LR] Linear Regression, [NB] Naïve Bayes, [RF] Random Forest, [SVM] Support Vector Machine). The y-axis represents the classes considered. MC stands for multiclass. The top x-axis represents normalization techniques including Nothing (no normalization), Standardized, and Normalized. Data sets for each panel are (A) RBM, (B) NCBI, (C) TCGA–log2 normalized counts, and (D) TCGA–raw counts.
It was also observed that for 63% of classification tasks, the gene- and transcript-based methods performed with similar accuracy (within 0.2 difference in f-measure value). For 37% of the classification tasks, the transcript-based methods performed better than the gene-based (more than 0.2 gain in f-measure value). The difference between the transcript-based and gene-based classification accuracies was particularly profound when comparing the classification results of naïve Bayes, which was one of the less accurate methods analyzed, while being among the fastest classifiers. However, we did not observe such a drastic difference, and sometimes no difference at all, when considering one of the most accurate classifiers, random forest, across all classification tasks. For instance, when comparing gene- and transcript-based classifiers for stage IA cancer using the raw count expression values and not performing any normalization protocols, the accuracy and f-measure values for the naïve Bayes classifier ranged between 49.5%–76.4% and between 0.60–0.82, respectively, while for random forest the ranges were nearly identical (Fig. 2C,D).
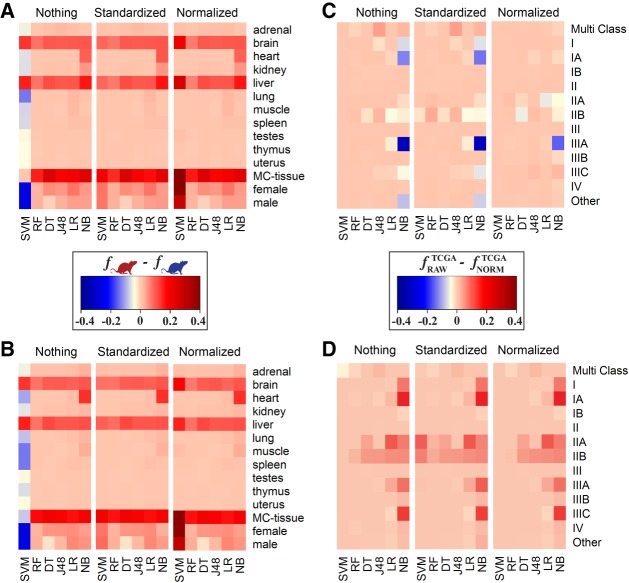
Another potential source of variability in the classifier performance was the difference in the protocols used by different studies. To determine whether the difficulty of classification task increased when using data sets from multiple laboratories rather than from a single one, the classification accuracies between the two rat data sets were compared for each binary or multiclass classification task. Not surprisingly, we found that there was a greater difference in the performance accuracies when relying on the data from one laboratory compared to the data from multiple laboratories (Fig. 4A,B). With the exception of a single worst performing classifier, SVM, the classifiers performed better on the RBM data set, which came from a single study, then on the NCBI data set, which was obtained by merging multiple independent studies. Moreover, this difference held for both the gene- and transcript-based models. Next, we evaluated if the prediction accuracy depended on the transcript counting approach. To do so, TCGA expression values were calculated based on (i) raw counts and (ii) log2 normalized counts with respect to the gene length and sequencing depth. The results showed that there was a strong preference, in terms of accuracy, in raw counts for the gene-based classifiers, but to a lesser extent for the transcript-based models. However, the opposite was observed where transcript-based models were more accurate when using log2 normalized counts (Fig. 4C,D). There was less variability (less than 0.3 in the maximum difference of f-measure values across all methods for each classification task) when considering transcript-based versus gene-based models.
FIGURE 4.
Heat map representation showing the influence of different factors on the accuracy performance. Panels A and B represent the difference in performance accuracies, calculated with f-measure, between RBM (single-lab) and NCBI (multi-lab) data sets for gene-based (A) and isoform-based (B) classifications, respectively. Panels C and D represent the difference in f-measure between the classifiers trained on the TCGA expression values, quantified as either raw counts or log2 normalized counts with respect to gene length and sequencing depth. Shown are f-measure differences for gene-based (C) and isoform-based (D) classifications, respectively. The bottom x-axis represents the machine learning techniques ([DT] Decision Table, [J48] J48 Decision Tree, [LR] Linear Regression, [NB] Naïve Bayes, [RF] Random Forest, [SVM] Support Vector Machine). The y-axis represents the classes considered. MC stands for multiclass. The top x-axis represents normalization techniques including Nothing (no normalization), Standardized, and Normalized.
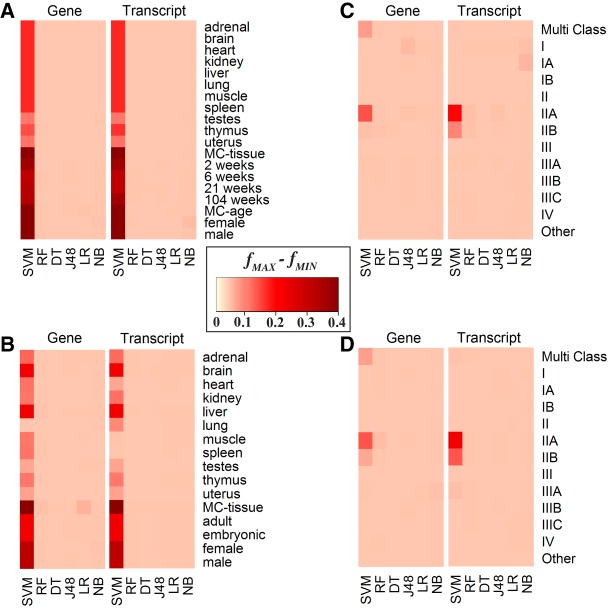
Finally, we considered different normalization techniques across the gene- and transcript-based classifiers. The general trend observed was little to no difference in performance accuracies using either different normalization protocols or no normalization at all. The only exception was the performance of the SVM classifier used by both, the transcript-based and gene-based, approaches: Differences in the accuracy values between the various normalization techniques for some classification tasks were as high as 40.3% and 30.7%, respectively (Fig. 5).
FIGURE 5.
Heat map representation of the difference between maximum f-measure and minimum f-measure across normalization techniques. To demonstrate the variability attributed to the machine learning normalization technique, the intensity of the color represents the difference between the maximum and minimum f-measures achieved for a specific classification task and specific classifier across all three normalization protocols. The upper x-axis reflects if the difference is from gene or isoform expression values. SVM is the only method that has significant changes due to normalization. The lower x-axis represents machine learning techniques ([DT] Decision Table, [J48] J48 Decision Tree, [LR] Linear Regression, [NB] Naïve Bayes, (RF) Random Forest, [SVM] Support Vector Machine). The y-axis represents the classes considered. MC stands for multiclass. Data sets for each panel are (A) RBM, (B) NCBI, (C) TCGA–log2 normalized counts, and (D) TCGA–raw counts.
Normal phenotype classification tasks: age, sex, and tissue classification of rat samples
The Rat Body Map (RBM) represents a data set with the least amount of noise due to nonbiological variation because it comes from a single laboratory, which uses the same sample and library preparation protocols and a fixed sequencing depth (Fig. 3A). From this data set we identified eleven tissue types, four age groups, and both sexes. We then defined 17 “one-against-all” binary classification problems. Additionally, we merged the tissue and age groups and applied a multiclass classifier.
For the tissue classification, including multiclass tissue classification, the models achieved 100% accuracy and 1.0 f-measure based on the assessment protocol and irrespective of the machine learning method. However, when considering the normalization technique, SVM had the accuracy ranged between 75.3% to 99.8% and 0.39 to 1.00 f-measure. The age group classification represented a more challenging task, with the classification accuracy ranging between 40.2% to 100% and f-measure ranging from 0.40 to 1.00. For the 2-wk and 104-wk age groups, the classifiers again achieved nearly 100% accuracy and 1.0 f-measure across all machine learning techniques. The 6-wk and 21-wk age groups were predicted with over 97% accuracy using random forest, j48, and logistic regression classifiers, while naïve Bayes could only achieve 81.1% and SVM with 40.2%. A similar pattern was observed in sex classification, where logistic regression and random forest achieved more than 97.3% in accuracy, but naïve Bayes could reach only 86.1%.
The NCBI data set was expected to result in a greater variation of the feature values, compared with the RBM data set, since it included the data from multiple research laboratories that sequenced different rat samples and even strains using different library preparation protocols (Fig. 3B). The same types of classification tasks were considered, including tissue, age, and sex. Since this data set represents all publicly available data in rat obtained using the same sequencer model, it included more tissue types than the RBM data set. For consistent comparison, only those tissue types that were previously included in the RBM data set were chosen for the NCBI data set for the binary classification. However, for the multiclass tissue classification problem, the labels were determined based on the entire range of organs and tissues that the samples originated from, thus including more tissue types than in the RBM data set. In contrast, the age group classification for the NCBI data set was more limited than the one for the RBM data set, since some samples in the former did not include the detailed age information. Therefore, the age types for the NCBI data sets were reduced to either adult or embryonic types.
The RNA-seq data normalization did not have an effect on the classification results for the NCBI data set: The performance difference when using the normalized and unnormalized data sets was only observed for the SVM classifier, the method that performed the worst out of the six supervised learning methods. The binary tissue-based classification performed well overall, reaching over 99.7% in accuracy and 0.99 in f-measure for the top-performing random forest classifier. Interestingly, the worst performing classifier, SVM, achieved the accuracy of only 21.2% and 0.07 f-measure for the gene-based tissue multiclass. The analysis of method performances for multiclass classification tasks revealed that classification of several tissue types was particularly challenging for some of the less accurate methods. The binary tissue type classification tasks reporting the lowest accuracies included brain and liver tissue classification, with 79.1%–94.3% in accuracies and 0.70–0.94 in f-measure values, depending on the supervised learning method used. For the harder problem of multiclass tissue classification, the performance of the classifiers was highly variable, with the accuracy ranging from 0.7% to 84.3% and f-measure from 0.07 to 0.84, and with the observation that the random forest classifier was, again, the best performing method. Differentiating between embryonic and adult samples as well as between the sexes were easier tasks compared to the tissue origin. The age classification accuracy ranged from 83.2% to 98.3% and f-measure from 0.75 to 0.97 across all six supervised learning methods. The sex classification task had classification accuracy ranging between 71.1% and 97.3% and f-measure between 0.59 and 0.97. Interestingly, the consistently poor performance of the SVM classifier was not dependent on the normalization technique.
Disease phenotype classification tasks: breast cancer versus healthy and stage classification of human samples
Based on the promising results for the normal phenotype classification tasks, we further increased the difficulty of classification task by predicting different pathological stages of breast cancer using gene-based and transcript-based data. To evaluate if this classification task could benefit from additional information, we assessed the method performances based on the RNA-seq data with log2 normalization in addition to the three types of normalization used in the two previous classification problems. The classification performance was heavily dependent on the supervised learning method with accuracies ranging from 20.2% to 99.8% and f-measure ranging from 0.21 to 1.00, and with naïve Bayes and SVM classifier being the worst performing classifiers (Fig. 3C,D). Furthermore, when considering all classes and log2 normalization, the accuracies decreased by as much as 60%, and the only method that benefited from the normalization was the poorly performing SVM classifier.
For each stage of breast cancer, we were able to achieve at least 78.3% in accuracy and 0.77 in f-measure. However, there is a significant variability within all parameters tested (Figs. 2C, 3C,D). Similar to the analysis for the RBM and NCBI data sets, random forest had the highest performance across all stages of breast cancer based on 71.3% to 99.8% accuracy and 0.64 to 1.00 f-measure. The most difficult stages to classify were stages IIA and IIB, with the average difference in accuracy between 21.3% accuracy and 0.29 f-measure. Unlike the RBM and NCBI data sets, there were classes, such as Stage IIB, where naïve Bayes and SVM outperformed random forest by 5% in accuracy and 0.11 in f-measure. The easiest stages to classify were stages II and III with 99.8% accuracy and 1.00 f-measure.
In contrast to the RBM and NCBI data sets, the worst performing models for each class were highly variable, depending on the parameters chosen. For example, for stage IIA, the logistic regression classifier was the best performing model at 78.2% accuracy and 0.77 f-measure. However, the worst performing model was J48 at 60.1% accuracy and 0.60 f-measure. Similarly, for stage I the worst performing classifier was naïve Bayes with 53.7% accuracy and 0.63 f-measure, while the best performing classifier was random forest with 91.4% accuracy and 0.84 f-measure. On the other hand, for stage III binary classification the performance was 99.5%–99.8% accuracy and 0.996%–0.998 f-measure across all classifiers and parameter sets. These results demonstrated that no single method and parameter set was able to always outperform all others.
DISCUSSION
This work achieves two aims. The first aim is to broadly assess how well the supervised machine learning methods perform in various biological classifications by utilizing exclusively the RNA-seq data. This aim is supported by our rationale that the key biological patterns should be recoverable from the transcriptomics data. Our second aim is to investigate whether relying on the transcript-level expression, which provides details on the alternatively spliced transcripts, can increase the accuracy of biological classification compared to the gene-level expression. Since the data patterns detected by the machine learning techniques during their training stage are highly dependent on the type of biological classification problem, we wanted our assessment to cover multiple aspects. Specifically, we evaluated the performance of six widely used supervised classifiers across different RNA-seq data sets, organisms, and normalization protocols, totaling in 61 classification problems and 2196 individual classification tasks. The different RNA-seq data sets were selected based on the increasing difficulty of classification tasks due to the background noise. The RBM data set represented the “easiest” data set as the level of background noise was expected to be low due to using a single data source and well-defined biological classification problems: tissue-, age-, and gender-based. The assumption of a single data source implies a well-defined animal model, with the genetically identical specimina, and the same RNA-seq library protocol. The NCBI data set increases the background noise by including multiple RNA-seq protocols and different genetic backgrounds, but keeping the classification the same, to allow for comparison with the RBM data set. The TCGA data set further increases the background noise due to increasing genetic and environmental variability by switching from a model organism to human, from the normal to disease-specific phenotype, and by relying on a potentially biased definition of the biological classes (breast cancer pathological stages are not defined from the molecular perspective, but by a pathologist). Each task separately utilized the gene-level and transcript-level expression data sets. The main purpose behind our study was to demonstrate the importance of enriching RNA-seq data with the differentially expressed transcripts for the biological classification tasks, suggesting that limiting the RNA-seq analysis to the differentially expressed genes would, in turn, limit the capabilities of machine learning algorithms. As a result, several important conclusions were made.
First, we found that the accuracy of machine learning classifiers depended on how much data variation associated with the type of sequencers, library preparation, or sample preparation was introduced. Our rat data sets were specifically selected to compare the differences in data variation and in classification accuracies. The first data set (RBM) was chosen because it included samples representing multiple age groups, tissue types, as well as sex (Yu et al. 2014), while these data were generated by only one research group and using the same sequencer. Thus, possible variation due to the type of sequencers or preparation protocols was expected to be minimal. Furthermore, we downloaded and processed the raw RNA-seq reads using our in-house protocol and thus excluding possible variation due to different RNA-seq analysis techniques. Our second data set (NCBI) incorporated all publicly available RNA-seq data for rat using the same sequencer model, thus minimizing possible sequencer-based bias, a well-documented source of variation (Schirmer et al. 2016). The NCBI data set included 29 studies from multiple laboratories and represented the same classes as in the RBM except for the age groups. As expected, higher variation negatively affected the accuracy across predominantly all machine learning methods, normalization protocols and classification tasks. On the other hand, even for the NCBI data set, the accuracies for all top-performing binary classifiers were never below 90% either for gene-level or for transcript-level expression data, suggesting minimal influence of the batch effect on the supervised classifiers.
Second, our study suggested that the standard data normalization techniques were not needed for RNA-seq data, except when using the poor-performing SVM classifiers. Random forest and logistic regression classifiers performed consistently well with each of the normalization techniques but also without them, regardless of the classification task. However, there are several normalization techniques specific to RNA-seq data, including RPKM (reads per kilobase per million reads), FPKM (fragments per kilobase per million reads), and TPM (transcripts per kilobase per million reads) (Conesa et al. 2016). Assessing whether these normalization techniques have an effect on classification accuracy should be considered for future studies.
Third, we found that the overall performance of the most accurate machine learning classifiers was very strong, with a few exceptions. In fact, for several classification tasks including all tissue classes, 2 wk, and 104 wk from the RBM data set, stage I from the TCGA data set, and the top-performing classifiers achieved a perfect 1.0 f-score, while for the majority of other tasks, the accuracy and f-measure were no less than 0.9 and often achieved by more than one classifier. From the biological perspective, it was surprising to see how well the classifiers performed on the normal phenotype data sets, in spite of significant variations in the sample and library preparation by different labs as well as the difference in rat strains. Intuitively, the expression values should have high variability due to these differences. The few exceptions in excellent performance were the multiclass age group classification for the normal phenotype data sets and classifications of clinical stages I, IIA, IIB, and IIIA for the disease phenotype data set, with stages IIA and IIB performing significantly worse. The clinical definition of IIA and IIB are based on the size of the tumor as well as evidence of cancer movement, and the reduced performance on each of these stages suggests that while there is a phenotype difference there may not be a strong molecular expression difference, which would cause a higher error rate by a classifier. The results also suggested that, from the diagnostic perspective, a more accurate AJCC classification methodology to distinguish those two phenotypes might be required to improve the stage prediction accuracy. The most consistent in the overall performance across all tasks were the random forest classifiers, which had been previously shown to perform exceptionally well for a number of bioinformatics tasks (Boulesteix et al. 2012) and can be suggested as a reliable first choice for a biological classification task. Overall, our findings provided strong evidence that the supervised learning approach is readily available for the majority of the biological classification tasks.
Finally, we found that the classifiers that leveraged the transcript-level expression never performed worse and often outperformed the classifiers that used the gene-level expression data. This observation was consistent across data sets, normalization techniques, RNA-seq pipelines, and classification tasks. For the normal phenotype tasks, the most profound difference was when considering the most challenging classification task—the multiclass classification of age groups. For the disease phenotype tasks, the most significant difference in performances of the classifiers that used gene-based and transcript-based expression data was again for the most challenging classes, the clinical stages IIA and II B of breast cancer. The better performance for the classifiers on the transcript-level data seems to be the expected result because the methods are trained on the enriched data, from the biological point of view. However, we note that the transcript-level data provide a significantly higher number of initial features, which could result in adding more noise to or potential overfitting of a classifier. Hence the importance of the feature selection and thorough model evaluation, which in this work suggests that the transcript-level information is a better choice when developing a biological classifier. Given that the transcript extraction methods continue to improve (Alamancos et al. 2015; Goldstein et al. 2016), we expect further improvement in the accuracy of transcript-level based classifiers.
In summary, this study demonstrates that a supervised learning method leveraging transcript-level RNA-seq data is a reliable approach for many biological classification tasks. We conclude that an appropriate general purpose pipeline for building a RNA-seq based classifier should use (i) transcript-based expression data, (ii) feature selection preprocessing, (iii) Random Forest classification method, and (iv) do not use normalization. The proposed pipeline is computationally fast and can be fully automated for the projects that involve massive volumes of sequencing data and/or high number of samples. However, it is important to note that (i) there are some cases where Random Forest can be outperformed and (ii) the protocols and methods used for data gathering may have an effect on the classifier. With the rapid advancements of RNA sequencing technologies as well as with continuous improvement of the transcript prediction methods, the accuracy of the machine learning approaches will only increase. We also expect for these methods to tackle more challenging tasks such as cell type classification, disease phenotype classification of common and rare complex diseases, and clinical stage classification across all major cancer types. Finally, we expect for advanced machine learning approaches, such as semisupervised learning (Zhu 2005), deep learning (LeCun et al. 2015), and learning under privileged information (Vapnik and Vashist 2009) to step in.
MATERIALS AND METHODS
The methodology used in this study compares three RNA-seq data sets, six supervised machine learning methods, three normalization techniques, two RNA-seq analysis pipelines, and 61 classification problems in order to assess if the features derived from the expression data at the alternative splicing level (i.e., transcript-based) can result in a higher classification accuracy than the features derived from the gene-based expression levels. Our approach attempts to systematically evaluate the classifiers that relied on these features from multiple perspectives, with a goal to provide a comprehensive analysis. We use the increasingly difficult biological classification tasks to assess the performances of classifiers in the presence of noise due to the difference in the biological sources, sequencers, and preparation protocols. The analysis is based on three RNA-seq data sets, two from rat and one from human. The six supervised machine learning methods tested in this work include support vector machines (SVM), random forest (RF), decision table, J48 decision tree, logistic regression, and naïve Bayes. The three normalization protocols used include (i) pipeline-specific RNA-seq count with no post-normalization, (ii) pipeline-specific RNA-seq count with normalization from 0 to 1, and (iii) pipeline-specific RNA-seq count protocol with standardization with respect to standard deviation. The two RNA-seq analysis pipelines in this work, each using different RNA-seq count methods were the standard Tuxedo suite and RSEM. The 61 classification problems include binary and multiclass classifications of tissue types, age groups, sex, as well as clinical stages of breast cancer.
Data sources
Three data sets are used to demonstrate the usability of the transcript-level expression data for the supervised classification. The first two data sets are from rat samples of normal phenotype; the raw RNA-seq data for both data sets is processed using our in-house protocol. The last data set consists of already processed RNA-seq data from human breast cancer samples (Zhu et al. 2014). The first, RBM, data set is obtained from the Rat Body Map and includes 660 samples from 12 different rats from the F344 rat strain (Yu et al. 2014) covering four different age groups, 11 tissues, and both male and female rats. Publicly available raw mRNA RNA-seq data from the Rat Body Map (http://pgx.fudan.edu.cn/ratbodymap/) is downloaded and processed for the gene and transcript levels of expression. The second data set, NCBI, includes all publicly available raw RNA-seq data from rat samples that are sequenced using Illumina Hi-Seq 2000 and available from the NCBI GEO Data sets collection (http://www.ncbi.nlm.nih.gov/gds, Supplemental Table S3). In total, 1308 samples are used, which represents 29 different projects. In contrast to the processing of the data for the first data set, these 29 projects used a variety of library preparation protocols and adapters to process their samples. The third, TCGA, data set is obtained from The Cancer Genome Atlas data repository (Zhu et al. 2014) and includes 1216 breast cancer patients diagnosed with different pathological cancer stages (as defined by the American Joint Committee on Cancer, AJCC [Edge and Compton 2010]). The class distributions for all data sets are shown in Supplemental Figures S9–S11.
RNA-seq pipeline
RNA-seq analysis encompasses three main stages: preprocessing, alignment, and quantification. There are a number of methods to perform each of these three basic steps, while the debate on the most appropriate methodology continues (Conesa et al. 2016). In this work, we expect for the variation due to data processing to be minimal, since the same processing pipeline is used for each data set. Two different RNA-seq pipelines are implemented and applied to each data set for both gene and transcript levels of expression. These two pipelines leverage different algorithms and different metrics (Engström et al. 2013). For the RBM and NCBI data set, all raw RNA-seq data are downloaded from the SRA repository (https://www.ncbi.nlm.nih.gov/sra) using unique project IDs (Supplemental Table S3). SRA file formats are then converted into fastq format. These files are used as input for the preprocessing stage. The preprocessing is done using Fastx Tools with the settings that removed reads shorter than 20 bp. All nucleotides with quality scores of less than 20 are converted into N's (http://hannonlab.cshl.edu/fastx_toolkit/index.html). The alignment is done against the rat genome version rn5 (NCBI Resource Coordinators 2013) using Tophat v2 and its default settings (Kim et al. 2013). Quantification for both gene- and transcript-based expression levels is performed using Cufflinks v2 (Trapnell et al. 2012) and Ensembl transcript annotation v75 (Flicek et al. 2012). The Cufflinks is set to use the transcript annotation for quantification with other settings being default. For the TCGA data set, MapSlice (Wang et al. 2010) is used for alignment and RSEM (Li and Dewey 2011) for quantification. The final output includes expression levels for each sample at both gene and transcript levels. We note that the gene-based expression values are the summation of all transcripts determined to be associated with the corresponding gene.
Supervised learning classifiers
The quantified expression values obtained from Cufflinks are then used to train and assess six supervised classifiers for each task. Two types of classification tasks are considered: one-against-all and multiclass. Our classification approach leverages feature-based supervised learning methods. Each post-processed RNA-seq sample is represented as a feature vector, where each feature represents the transcript- or gene-level expression value for a specific gene or AS transcript corresponding to this gene. Expression samples may vary in length, thus to generate feature vectors of the same length, we compute the intersection of all samples in terms of the feature set that represents each sample. We next rank the importance of each feature and select subsets of the features that best describe their respective classes using the Best First (BF) feature selection method (Hall and Smith 1998). The BF method is driven by the property that the subsets of important features are highly correlated with a specific class and are not correlated with each other. The method is described as a greedy hill climbing algorithm augmented with a backtracking step, where the importance of features is estimated through one-by-one feature removal. All machine learning methods are implemented using the Weka package version 3.7.13 (Hall et al. 2009).
Due to a large number of gene-based and an even greater number of transcript-based features (∼20,000–73,000) using the base classification or even BF method was not computationally feasible, and the original methodology was modified allowing to reduce the processing time. The modifications include introducing multiple splits of the features followed by two rounds of feature selection. Specifically, we split the data into 1000 subsets and performed feature selection on each subset independently. After feature selection is performed on all splits, the selected features are merged, and another round of feature selection is performed. Our solution reduces the time needed to compute from several weeks to hours and still able to successfully select a reduced feature set that allows for accurate classification.
Machine learning technique rationale
A broad range of supervised learning approaches was implemented to test whether performance could be improved, depending on the method tested. The machine learning methods have different assumptions on how the data are structured; the methods also vary in their treatment of the class outliers and convergence w.r.t number of training examples.
The first two classifiers, naïve Bayes and logistic regression, are often regarded as the baseline methods due to their simplicity and robustness. Naïve Bayes classifier is a probabilistic method that has been used in many applications including bioinformatics and text mining (McCallum and Nigam 1998; Rish 2001; Liu et al. 2002; Li et al. 2004; Kim et al. 2006). It is a simple model that leverages the Bayes rule and describes a class of Bayesian networks with assumed conditional independence between the numerical features. The use of this “naïve” assumption makes the method computationally efficient during both the training and classification stages. Furthermore, while the probability estimation by naïve Bayes is reported to be not very accurate (Niculescu-Mizil and Caruana 2005), a threshold-based classification performance is typically very robust. In our implementation, the numeric estimator precision values are chosen based on analysis of the training data and is set to 0.1 The batchSize parameter that specifies the preferred number of instances to process during training if batch prediction is being performed is set to 100. “Logistic regression” is another type of a simple machine learning classifier that has been compared with naïve Bayes in terms of accuracy and performance (Ng and Jordan 2002). Different versions of logistic regression models are often used in bioinformatics applications (Shevade and Keerthi 2003; Liao and Chin 2007; Sartor et al. 2009; Wang et al. 2013). In this work, we implemented a boosting linear logistic regression method without regularization and with the optimal number of boosting iterations based on cross validation.
The next three classifiers, decision tables, J48, and random forest, are the decision tree based algorithms. A “decision table” is a rule-based classifier commonly used for the attribute visualization and less commonly for classification. The rules are represented in a tabular format using only an optimal subset of features that are included into the table during training. The decision table is a less popular approach for bioinformatics and genomics classification tasks, however it has showed a superior performance in some bioinformatics applications (Asur et al. 2006), and therefore is included into the pool of methods. The decision table model is implemented as a simple majority classifier using the Best-First method for searching. J48 is an open source implementation of perhaps the most well-known decision tree algorithm, C4.5 (Quinlan 1993), which is, in turn, an extension of the Iterative Dichotomiser 3 (ID3) algorithm (Quinlan 1979). C4.5 uses the information-theoretic principles to build decision trees from the training data. Specifically, it leverages the information gain and gain ratio for a heuristic splitting criterion with a greedy search that maximizes the criterion. Furthermore, the algorithm includes a tree pruning step to reduce the size of the tree and avoid the overfitting. In this work, the implementation of J48 was done with the default confidence threshold of 0.25 and minimum number of instances per leaf set to 2. “Random forest” is an ensemble learning approach, where many decision trees are generated during the training stage, with each tree based on a different subset of features and trained on a different part of the same training set (Breiman 2001). During the classification of unseen examples, the predictions of the individually trained trees are then agglomerated using the majority vote. This bootstrapping procedure is found to efficiently reduce the high variance that an individual decision tree is likely to suffer from. The random forest methods have been widely used in bioinformatics and genomics applications due to their versatility and high accuracy (Breiman 2001). In this work, due to a large but highly variable number of features the number of attributes, K, randomly selected for each tree is dependent on the classification task and is defined as K = [log 2 n + 1], where n is the total number of features. The number of sampled trees per each classifier is set to 100.
The last method, SVM represents yet another family of the supervised classifiers, the kernel methods (Vapnik 1998). It is among the most well-established and popular machine learning approaches in bioinformatics and genomics (Hirose et al. 2007; Zhao et al. 2011; Dou et al. 2014; Libbrecht and Noble 2015). SVM classifiers range from a simple linear, or maximum margin, classifier where one needs to find a decision boundary separating two classes and represented as a hyperplane, in case of a multidimensional feature space, to a more complex classifier represented by a nonlinear decision boundary through introducing a nonlinear kernel function. For our SVM model training, radial basis function (RBF) was used, a commonly used kernel. The two parameters, Gamma and C, were set to 0.01 and 1, respectively.
Training, testing, and assessment of classifiers
To evaluate each of the classifiers, a basic supervised learning assessment protocol is implemented. Specifically, the training/testing stages are assessed as a 10-fold stratified cross validation to eliminate the sampling bias. This protocol is implemented using Weka (Hall et al. 2009). The reported result of assessment is based on the average f-measure for the 10-folds for the testing data set. f-measure incorporates recall (Rec, also called sensitivity) and precision (Pre) into one reported metric:
where TP is the number of true positives (correctly classified as class members for a specified class), TN is the number of true negatives (correctly classified as not class members), FP is the number of false positives (incorrectly classified as class members), and FN is the number of false negatives (incorrectly classified as not class members). While each of the above four measures are commonly used to evaluate the overall performance of a method, we primarily focus on the most balanced metric, f-measure, due to a high number of classification tasks to be reported.
DATA DEPOSITION
The supervised machine learning methods were implemented using the Weka platform (http://www.cs.waikato.ac.nz/ml/weka/). Data used are publicly available from the Rat Body Map (http://pgx.fudan.edu.cn/ratbodymap/), Geo Datasets (http://www.ncbi.nlm.nih.gov/gds), and the Cancer Genome Atlas (https://portal.gdc.cancer.gov/).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
The computations were performed on the HPC resources at the University of Missouri Bioinformatics Consortium (UMBC) and Worcester Polytechnic Institute (WPI). Additionally, we would like to thank Dr. Lane Harrison for useful suggestions about the biological data visualization. This work has been supported by the National Science Foundation (DBI-0845196 to D.K.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.062802.117.
REFERENCES
- Achim K, Pettit JB, Saraiva LR, Gavriouchkina D, Larsson T, Arendt D, Marioni JC. 2015. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat Biotechnol 33: 503–509. [DOI] [PubMed] [Google Scholar]
- Alamancos GP, Pagès A, Trincado JL, Bellora N, Eyras E. 2015. Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA 21: 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asur S, Raman P, Otey ME, Parthasarathy S. 2006. A model-based approach for mining membrane protein crystallization trials. Bioinformatics 22: e40–e48. [DOI] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. 2012. The evolutionary landscape of alternative splicing in vertebrate species. Science 338: 1587–1593. [DOI] [PubMed] [Google Scholar]
- Boulesteix AL, Janitza S, Kruppa J, König IR. 2012. Overview of random forest methodology and practical guidance with emphasis on computational biology and bioinformatics. Wiley Interdiscip Rev Data Min Knowl Discov 2: 493–507. [Google Scholar]
- Breiman L. 2001. Random forests. Mach Learn 45: 5–32. [Google Scholar]
- Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. 2016. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet 17: 257–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Kornblihtt AR. 2002. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet 18: 186–193. [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL. 2009. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10: 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Climente-González H, Porta-Pardo E, Godzik A, Eyras E. 2017. The functional impact of alternative splicing in cancer. Cell Rep 20: 2215–2226. [DOI] [PubMed] [Google Scholar]
- Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szczesniak MW, Gaffney DJ, Elo LL, Zhang X, et al. 2016. A survey of best practices for RNA-seq data analysis. Genome Biol 17: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa IG, de Carvalho FA, de Souto MC. 2004. Comparative analysis of clustering methods for gene expression time course data. Genet Mol Biol 27: 623–631. [Google Scholar]
- Crick F. 1970. Central dogma of molecular biology. Nature 227: 561–563. [DOI] [PubMed] [Google Scholar]
- Danielsson F, James T, Gomez-Cabrero D, Huss M. 2015. Assessing the consistency of public human tissue RNA-seq data sets. Brief Bioinform 16: 941–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Yao B, Zhang C. 2014. PhosphoSVM: prediction of phosphorylation sites by integrating various protein sequence attributes with a support vector machine. Amino Acids 46: 1459–1469. [DOI] [PubMed] [Google Scholar]
- Edge SB, Compton CC. 2010. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol 17: 1471–1474. [DOI] [PubMed] [Google Scholar]
- Edwards TM, Myers JP. 2007. Environmental exposures and gene regulation in disease etiology. Environ Health Perspect 115: 1264–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström PG, Steijger T, Sipos B, Grant GR, Kahles A, Rätsch G, Goldman N, Hubbard TJ, Harrow J, Guigó R, et al. 2013. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat Methods 10: 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. 2012. Ensembl 2012. Nucleic Acids Res 40: D84–D90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Stranger BE. 2014. Genomics of alternative splicing: evolution, development and pathophysiology. Hum Genet 133: 679–687. [DOI] [PubMed] [Google Scholar]
- Goldstein LD, Cao Y, Pau G, Lawrence M, Wu TD, Seshagiri S, Gentleman R. 2016. Prediction and quantification of splice events from RNA-seq data. PLoS One 11: e0156132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. 2015. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348: 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MA, Smith LA. 1998. Practical feature subset selection for machine learning. In Computer Science '98, Proceedings of the 21st Australasian Computer Science Conference ACSC'98, Perth, 4–6 February, 1998 (ed. McDonald C), pp. 181–191, Springer, Berlin. [Google Scholar]
- Hall MA, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. 2009. The WEKA data mining software: an update. SIGKDD Explorations 11: 10–18. [Google Scholar]
- Hirose S, Shimizu K, Kanai S, Kuroda Y, Noguchi T. 2007. POODLE-L: a two-level SVM prediction system for reliably predicting long disordered regions. Bioinformatics 23: 2046–2053. [DOI] [PubMed] [Google Scholar]
- Jagga Z, Gupta D. 2014. Classification models for clear cell renal carcinoma stage progression, based on tumor RNAseq expression trained supervised machine learning algorithms. BMC Proc 8: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S. 2013. Function of alternative splicing. Gene 514: 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G. 2010. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11: 345–355. [DOI] [PubMed] [Google Scholar]
- Kim SB, Han KS, Rim HC, Myaeng SH. 2006. Some effective techniques for naive Bayes text classification. IEEE Trans Knowl Data Eng 18: 1457–1466. [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y, Shumway M, Leinonen R. 2012. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res 40: D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuzzetta CJ, Antonio ML, Watson PM, Wilson RC, Laboissonniere LA, Trimarchi JM, Genc B, Ozdinler PH, Watson DK, Anderson PE. 2016. Complementary feature selection from alternative splicing events and gene expression for phenotype prediction. Bioinformatics 32: i421–i429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y, Bengio Y, Hinton G. 2015. Deep learning. Nature 521: 436–444. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Zhang C, Ogihara M. 2004. A comparative study of feature selection and multiclass classification methods for tissue classification based on gene expression. Bioinformatics 20: 2429–2437. [DOI] [PubMed] [Google Scholar]
- Liao J, Chin KV. 2007. Logistic regression for disease classification using microarray data: model selection in a large p and small n case. Bioinformatics 23: 1945–1951. [DOI] [PubMed] [Google Scholar]
- Libbrecht MW, Noble WS. 2015. Machine learning applications in genetics and genomics. Nat Rev Genet 16: 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Li J, Wong L. 2002. A comparative study on feature selection and classification methods using gene expression profiles and proteomic patterns. Genome Informatics 13: 51–60. [PubMed] [Google Scholar]
- Liu C, Che D, Liu X, Song Y. 2013. Applications of machine learning in genomics and systems biology. Comput Math Methods Med 2013: 587492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. 2011. Epigenetics in alternative pre-mRNA splicing. Cell 144: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum A, Nigam K. 1998. A comparison of event models for naive Bayes text classification. AAAI-98 workshop on learning for text categorization, Madison, WI. [Google Scholar]
- Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. 2015. Human genomics. The human transcriptome across tissues and individuals. Science 348: 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Mudge JM, Frankish A, Harrow J. 2013. Functional transcriptomics in the post-ENCODE era. Genome Res 23: 1961–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators. 2013. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 41: D8–D20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelima E, Babu MP. 2017. A comparative study of machine learning classifiers over gene expressions towards cardio vascular diseases prediction. Int J Comput Intell Res 13: 403–424. [Google Scholar]
- Ng AY, Jordan MI. 2002. On discriminative vs. generative classifiers: a comparison of logistic regression and naive Bayes. Adv Neural Inform Process Syst 2: 841–848. [Google Scholar]
- Niculescu-Mizil A, Caruana R. 2005. Predicting good probabilities with supervised learning. Proceedings of the 22nd international conference on machine learning ACM, Bonn, Germany. [Google Scholar]
- Nilsen TW, Graveley BR. 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirooznia M, Yang JY, Yang MQ, Deng Y. 2008. A comparative study of different machine learning methods on microarray gene expression data. BMC Genomics 9: S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan JR. 1979. Discovering rules by induction from large collections of examples. Expert systems in the micro electronic age. Edinburgh University Press, UK. [Google Scholar]
- Quinlan J. 1993. C4.5: Programs for machine learning by J. Ross Quinlan. Morgan Kaufmann Publishers, San Mateo, CA. [Google Scholar]
- Reddy AS, Marquez Y, Kalyna M, Barta A. 2013. Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rish I. 2001. An empirical study of the naive Bayes classifier. IJCAI 2001 workshop on empirical methods in artificial intelligence IBM, New York. [Google Scholar]
- Sartor MA, Leikauf GD, Medvedovic M. 2009. LRpath: a logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics 25: 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, D'Amore R, Ijaz UZ, Hall N, Quince C. 2016. Illumina error profiles: resolving fine-scale variation in metagenomic sequencing data. BMC Bioinformatics 17: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebestyen E, Zawisza M, Eyras E. 2015. Detection of recurrent alternative splicing switches in tumor samples reveals novel signatures of cancer. Nucleic Acids Res 43: 1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Wang Y, Wang C, Wu YN, Xing Y. 2016. SURVIV for survival analysis of mRNA isoform variation. Nat Commun 7: 11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevade SK, Keerthi SS. 2003. A simple and efficient algorithm for gene selection using sparse logistic regression. Bioinformatics 19: 2246–2253. [DOI] [PubMed] [Google Scholar]
- Tarca AL, Carey VJ, Chen XW, Romero R, Draghici S. 2007. Machine learning and its applications to biology. PLoS Comput Biol 3: e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JA, Tan J, Greene CS. 2016. Cross-platform normalization of microarray and RNA-seq data for machine learning applications. PeerJ 4: e1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trincado JL, Sebestyen E, Pages A, Eyras E. 2016. The prognostic potential of alternative transcript isoforms across human tumors. Genome Med 8: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik VN. 1998. Statistical learning theory. Wiley, New York. [DOI] [PubMed] [Google Scholar]
- Vapnik V, Vashist A. 2009. A new learning paradigm: learning using privileged information. Neural Netw 22: 544–557. [DOI] [PubMed] [Google Scholar]
- Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, et al. 2014. Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505: 706–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. 2010. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 38: e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. 2013. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res 41: e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei IH, Shi Y, Jiang H, Kumar-Sinha C, Chinnaiyan AM. 2014. RNA-seq accurately identifies cancer biomarker signatures to distinguish tissue of origin. Neoplasia 16: 918–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, Cancer Genome Atlas Research Network. 2013. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45: 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RK, Hua Y, Gueroussov S, Najafabadi HS, Hughes TR. 2015. The human splicing code reveals new insights into the genetic determinants of disease. Science 347: 1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Fuscoe JC, Zhao C, Guo C, Jia M, Qing T, Bannon DI, Lancashire L, Bao W, Du T, et al. 2014. A rat RNA-seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun 5: 3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Pal S, Bi Y, Tchou J, Davuluri RV. 2013. Isoform level expression profiles provide better cancer signatures than gene level expression profiles. Genome Med 5: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Pang B, Shyu CR, Korkin D. 2011. Feature-based classification of native and non-native protein–protein interactions: comparing supervised and semi-supervised learning approaches. Proteomics 11: 4321–4330. [DOI] [PubMed] [Google Scholar]
- Zhu X. 2005. Semi-supervised learning literature survey (1530). Computer Sciences, University of Wisconsin–Madison. [Google Scholar]
- Zhu Y, Qiu P, Ji Y. 2014. TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat Methods 11: 599–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.