Abstract
Mitochondrial gene expression is largely controlled through post-transcriptional processes including mitochondrial RNA (mt-RNA) processing, modification, decay, and quality control. Defective mitochondrial gene expression results in mitochondrial oxidative phosphorylation (OXPHOS) deficiency and has been implicated in human disease. To fully understand mitochondrial transcription and RNA processing, we performed RNA-seq analyses of mt-RNAs from the fission yeast Schizosaccharomyces pombe. RNA-seq analyses show that the abundance of mt-RNAs vary greatly. Analysis of data also reveals mt-RNA processing sites including an unusual RNA cleavage event by mitochondrial tRNA (mt-tRNA) 5′-end processing enzyme RNase P. Additionally, this analysis reveals previously unknown mitochondrial transcripts including the rnpB-derived fragment, mitochondrial small RNAs (mitosRNAs) such as mt-tRNA-derived fragments (mt-tRFs) and mt-tRNA halves, and mt-tRNAs marked with 3′-CCACCA/CCACC in S. pombe. Finally, RNA-seq reveals that inactivation of trz2 encoding S. pombe mitochondrial tRNA 3′-end processing enzyme globally impairs mt-tRNA 3′-end processing, inhibits mt-mRNA 5′-end processing, and causes accumulation of unprocessed transcripts, demonstrating the feasibility of using RNA-seq to examine the protein known or predicted to be involved in mt-RNA processing in S. pombe. Our work uncovers the complexity of a fungal mitochondrial transcriptome and provides a framework for future studies of mitochondrial gene expression using S. pombe as a model system.
Keywords: tRNase Z, transcriptome, mitochondria, RNA processing, RNA-seq
INTRODUCTION
Mitochondria are eukaryotic energy-producing organelles, which synthesize ATP through oxidative phosphorylation (OXPHOS). In addition, mitochondria are involved in many important cellular processes including amino acid and fatty acid synthesis, reactive oxygen species production, apoptosis and aging (Chan 2006; Suen et al. 2008; Shadel and Horvath 2015). Mitochondrial dysfunction is associated with neurodegenerative diseases including Alzheimer's disease and Parkinson's disease (Wallace 2005; Suzuki et al. 2011) and metabolic disorders such as diabetes (Bhatti et al. 2017).
Mitochondria contain their own DNA (mtDNA) that encodes essential subunits of the OXPHOS complexes. Similar to animal mtDNAs (∼16 kb) and unlike the budding yeast Saccharomyces cerevisiae mtDNA (∼86 kb), the fission yeast Schizosaccharomyces pombe mtDNA is a compact genome (∼19 kb). The two yeast mtDNAs encode seven subunits of the OXPHOS complexes including apocytochrome b (Cob1, also called Cob or Cytb), cytochrome coxidase subunits 1, 2, and 3 (Cox1, Cox2, and Cox3) and three ATP synthase subunits (Atp6, Atp8, and Atp9). Besides, the two mtDNAs encode a ribosomal protein (Var1, also called Rsp3 in S. pombe), 25 tRNAs and two rRNAs (rns and rnl), and the RNA subunit of RNase P (rnpB). In contrast, the human mtDNA encodes 13 OXPHOS subunits including seven subunits of complex I (ND1-ND6 and ND4L) and 22 tRNAs and two rRNAs.
Mitochondrial genes are transcribed into polycistronic RNAs (Supplemental Fig. S1). The two strands of human mtDNA are transcribed into genome-length polycistronic transcripts (Asin-Cayuela and Gustafsson 2007; Scarpulla 2008). In S. pombe, mtDNA-encoded genes are expressed as two primary transcripts (∼19 and ∼11 kb) with the major promoter located before the rnl gene and the minor promoter located just ahead of the cox3 gene (Schäfer et al. 2005). Unlike human and S. pombe mtDNAs, S. cerevisiae mtDNA is transcribed as 11 separate polycistronic units with long noncoding stretches of DNA in between (Foury et al. 1998; Turk et al. 2013).
According to the tRNA punctuation model (Ojala et al. 1981), mitochondrial mRNAs (mt-mRNAs) and rRNAs (mt-rRNAs) are often immediately adjacent to tRNAs (mt-tRNAs) and liberated by mt-tRNA 3′-end endonuclease tRNase ZL and 5′-end processing endonuclease RNase P. In humans, besides these two endonucleases (Daoud et al. 2012; Rossmanith 2012; Salinas-Giége et al. 2015), additional factors such as PTCD1, PTCD2, GRSF1, and the FASTK family proteins are required for processing of human mt-mRNAs that do not have flanking tRNAs at both ends (Xu et al. 2008; Rackham et al. 2009; Sanchez et al. 2011; Jourdain et al. 2013, 2017). In S. cerevisiae, maturation of 3′-ends of mt-mRNAs and mt-rRNAs involves cleavage of two nucleotides downstream from an AU-rich dodecamer sequence by an unknown enzyme. In contrast, mature 5′-ends of mt-mRNAs and mt-rRNAs can be generated by a variety of processes, including transcription (15S rRNA, 21S rRNA, COX1, COX2, and ATP9), removal of the upstream mt-tRNA (COB, COX3, VAR1, and RPM1) and cleavage at the upstream dodecamer site (ATP6/8) (Lipinski et al. 2010). Processing of the 5′-end of COB, ATP6/8, and VAR1 additionally requires a putative 5′- to 3′-exonuclease Pet127 (Lipinski et al. 2010). In Candida albicans, the 5′- and 3′-ends of mitochondrial RNAs (mt-RNAs) are generated by mt-tRNA processing enzymes except for the 3′-ends of NAD1, COX3, NAD3, and RNS that are likely to be generated by transcription termination (Kolondra et al. 2015).
RNase P can be either a ribonucleoprotein (RNP) enzyme (ribozyme) consisting of a catalytic RNA and one or more protein subunits or a protein-only enzyme (PRORP) (Seif et al. 2003; Howard et al. 2013; Lechner et al. 2015). PRORP is widespread in eukaryotes and found in the nuclei and/or organelles whereas RNP-based RNase Ps is identified in bacteria, archaea, the eukaryotic nuclei and the mitochondria of fungi (particularly in yeast) (Lechner et al. 2015; Klemm et al. 2016). Human mitochondrial PRORP contains three protein subunits (MRPP1, MRPP2, and MRPP3) (Holzmann et al. 2008) and is responsible for mt-tRNA 5′-end processing in humans (Brzezniak et al. 2011; Sanchez et al. 2011). In humans, mt-tRNA 5′-end processing precedes 3′-end processing, and is required for mitochondrial ribosome assembly (Rackham et al. 2016). The Arabidopsis organellar PRORP (PRORP1) is a single-subunit enzyme responsible for tRNA 5′-end processing in both mitochondria and plastids (Gobert et al. 2010; Gutmann et al. 2012; Zhou et al. 2015). The S. cerevisiae mt-RNase P contains an mtDNA-encoded RNA (rnpB) and a nuclear-encoded protein (Rpm2) whose close homologs are only found in species of the order Saccharomycetales. Both the S. cerevisiae Rmp2 and catalytic subunit of PRORP are pentatricopeptide repeat (PPR) proteins (Herbert et al. 2013; Pinker et al. 2013).
tRNase Z (also called RNase Z) can be classified into short (tRNase ZS) (Li de la Sierra-Gallay et al. 2005, 2006) and long form (tRNase ZL), which is believed to have arisen by duplication and divergence of the tRNase ZS gene. (Ma et al. 2017a). Bacteria and archaea only have the short form, whereas eukaryotes have either the long form or both forms.
The role of tRNase ZL in mt-RNA processing has been studied in S. cerevisiae (Skowronek et al. 2014), S. pombe (Zhao et al. 2009, 2010; Fan et al. 2011; Wang et al. 2012; Zhang et al. 2013; Zhou et al. 2014), Drosophila (Xie and Dubrovsky 2015) and human (In humans, tRNase ZL is named ELAC2) (Brzezniak et al. 2011; Sanchez et al. 2011). These studies firmly establish that tRNase ZL is responsible for mt-tRNA 3′-end processing.
Transcriptomic studies permit a global view of the expression levels of the RNAs and RNA processing events in living cells. High-throughput RNA sequencing (RNA-seq) has been applied to analyze the mitochondrial transcriptomes of humans (Mercer et al. 2011), S. cerevisiae (Turk et al. 2013), C. albicans (Kolondra et al. 2015), and some plants (Stone and Storchova 2015).
We have previously shown that unlike S. cerevisiae, Drosophila and human, which contain a single tRNase ZL localized to both the nucleus and mitochondria, S. pombe has two tRNase ZLs encoded by two different essential genes which are responsible for nuclear and mitochondrial tRNA 3′-end processing, respectively (Zhao et al. 2009; Gan et al. 2011; Zhang et al. 2013). However, a comprehensive landscape of S. pombe mitochondrial transcriptome is still lacking.
In this study, RNA-seq was used to explore the S. pombe mitochondrial transcriptome. We provided a detailed map of mt-RNA processing sites. Furthermore, we identified previously undescribed mtDNA-encoded transcripts including the rnpB-derived fragment, mt-tRNA-derived fragments, mt-tRNA halves, and mt-tRNAs marked with 3′-CCACCA/CCACC. We also confirmed and extended previous results on mt-tRNA 3′-end processing enzyme trz2 by showing that inactivation of trz2 globally impairs maturation of the 3′-ends of mt-tRNAs and 5′-ends of other mt-RNAs.
RESULTS
S. pombe mt-RNAs are differentially expressed
To globally examine S. pombe mitochondrial gene expression levels, we performed RNA-seq on purified mitochondria from the wild-type S. pombe strain yAS56. S. pombe mitochondria were isolated by the spheroplast method and tested for possible nuclear and/or cytosolic contaminants by immunoblotting and qRT-PCR analyses. We did not detect any nuclear and cytosolic markers (Supplemental Fig. S2). To determine the levels of mt-mRNAs and mt-rRNAs, we constructed and sequenced a cDNA library using total RNA isolated from S. pombe mitochondria. The cDNA library was prepared without poly(A) selection, depletion of rRNA and tRNA and size selection. This approach is similar to the strategies used in S. cerevisiae and human. (Mercer et al. 2011; Turk et al. 2013). Totally, ∼30 million mapped reads were unambiguously aligned with the S. pombe mtDNA reference sequence (NC_001326.1) and covered almost the entire mtDNA transcribed. The majority (>85%) of reads that did not map to the reference sequence could be unambiguously mapped to the nuclear genome sequence. As observed in the analyses of human and C. albicans mitochondrial transcriptomes (Mercer et al. 2011; Kolondra et al. 2015), these nuclear match reads possibly resulted from copurification of cytosolic ribosomes with the mitochondrial outer membrane.
To compare the expression levels of different mitochondrial transcripts, we calculated gene expression level as reads per kilobase per million mapped reads (RPKM) (Fig. 1). There appeared to be no significant difference between the two mt-rRNAs levels (rns and rnl). Similar results have been reported for transcriptome analyses of the mitochondria of S. cerevisiae (Turk et al. 2013), C. albicans (Kolondra et al. 2015), and human (Mercer et al. 2011). In contrast, the steady-state levels of different mature mt-mRNAs vary by several orders of magnitude (Fig. 1A). mt-mRNAs showed nearly 800-fold differential expression between cox1 with the highest RPKM values, and atp8 with the lowest RPKM values (Fig. 1A).
FIGURE 1.
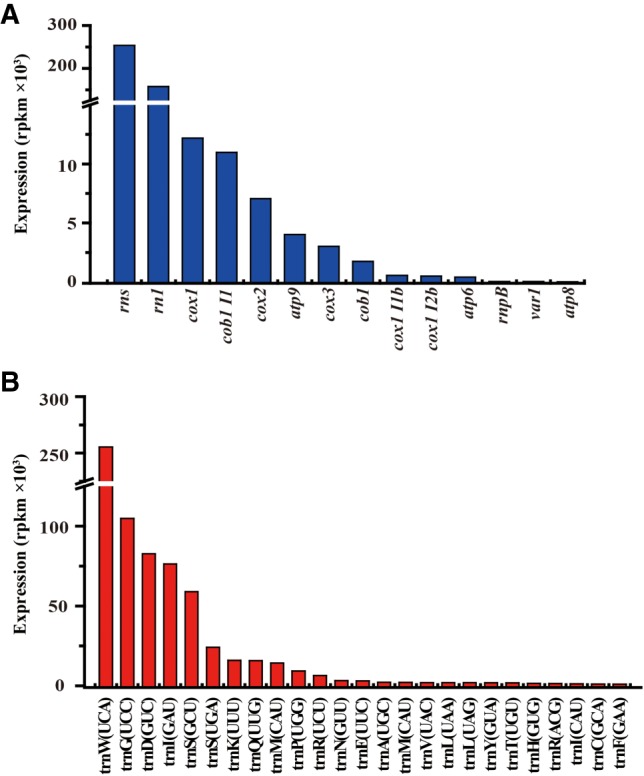
Differential expression of mt-RNAs in S. pombe. (A) Quantification of the levels of mt-rRNAs (rns and rnl), mt-mRNAs, and mtDNA-encoded introns (cob1I1, cox1I1b, and cox1I2b) in S. pombe. (B) Quantification of levels of mature mt-tRNAs in S. pombe. Values are the RNA read counts expressed in RPKM.
To determine mt-tRNA levels and their 5′- and 3′-ends, we constructed and sequenced a cDNA library using mt-tRNA isolated from S. pombe mitochondria. ∼2.5 million reads that mapped uniquely to the mitochondrial genome were obtained. Mature mt-tRNA-derived reads were defined by the presence of 3′-end CCA. The reads that were mapped to the mature mt-tRNA sequences were counted for each unique mature mt-tRNA sequence to calculate RPKM values. Remarkably, the steady state levels of mt-tRNAs appear to vary dramatically (Fig. 1B).
Genome-wide analysis of processing of mt-RNAs
Deep sequencing of the mitochondrial transcriptome allows determination of 5′-and 3′-end processing of all S. pombe mt-RNAs. The 3′-ends of mt-tRNAs were determined by looking for the presence of the 3′-CCA, which is added post-transcriptionally to all tRNAs. The 3′-CCA-containing reads were then used to identify the 5′-ends of mt-tRNAs based on the tRNA secondary structure prediction. This analysis revealed that both ends of mt-tRNAs are generated by a single cleavage event. We also compared the ends of mt-tRNAs determined by RNA-seq and tRNAscan-SE with those of mt-tRNAs in Pombase, which were predicted solely by tRNAscan-SE. Except for mt-tRNAArg(UCU), which has incorrect 5′- and 3′-ends, all predicted mt-tRNAs have correct 5′- and 3′-ends (Supplemental Figs. S3, S4).
Next, we examined processing of mt-mRNAs and mt-rRNAs. We found that 5′-end processing of all mt-mRNAs and mt-rRNAs occurs at a single major site (Fig. 2; Supplemental Fig. S5). The 5′-ends of most mt-mRNAs and mt-rRNAs precisely coincide with the 3′-ends of their upstream mt-tRNAs, indicating that the 5′-ends of most mt-rRNAs and mt-mRNAs are generated by trz2. The positions of 5′-ends of cob1, cox2, and atp6 are one nucleotide further downstream from the 3′-ends of their upstream mt-tRNAs, suggesting that these mt-mRNAs are subsequently subjected to 5′–3′ exonucleolytic trimming for maturation. rnl and cox3 mRNA lack upstream mt-tRNAs. However, the rnl and cox3 genes are adjacent to the major promoter (Pma) and minor promoter (Pmi), respectively. It is most likely that their 5′-ends are generated by transcription initiation sites (Fig. 2). Analyses of 5′-ends of mt-RNAs allow us to define their 5′-UTR sequences (Fig. 3). These 5′-UTR sequences generally matched those reported previously (Schäfer 2005; Schäfer et al. 2005).
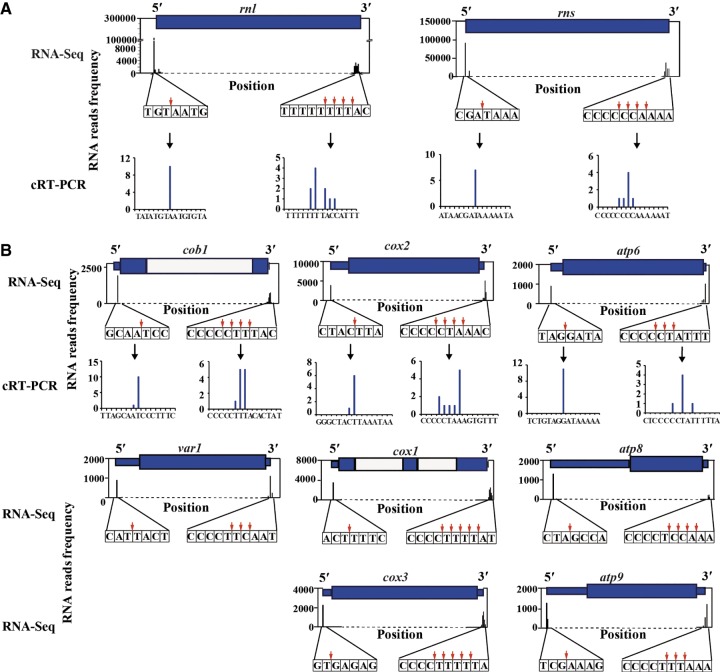
FIGURE 2.
Schematic of the 5′- and 3′-termini of mt-rRNAs and mt-mRNAs in S. pombe. Graphs showing the number of RNA-seq reads that mapped to 5′- and 3′-ends of mt-rRNAs (rns and rnl) (A) and eight mitochondrial encoded genes (cob1, cox1, cox2, cox3, atp6, atp8, atp9, and var1) (B). 5′- and 3′-ends of mt-rRNAs, cox2, atp6, and var1 were also mapped by cRT-PCR, and the data are shown at the bottom. The exons and introns are indicated by the blue boxes and white boxes, respectively. The likely cleavage site is depicted with a down arrow.
FIGURE 3.

Mapping of the 5′- and 3′-termini of mitochondrial transcripts. The number in the parentheses after the name of the gene indicates the size of the gene. The likely cleavage sites are depicted with a down arrow. mt-tRNAs are represented by single-letter code.
Unlike the 5′-processing of mt-mRNAs and mt-RNAs, the 3′-processing involves multiple cleavage sites (Fig. 2; Supplemental Fig. S6). Except for rnl, all mt-mRNAs and mt-rRNAs contain a C-core motif (four or five consecutive C residues) close to the 3′-end of all mt-mRNAs and rns mRNA (Schäfer et al. 2005). 3′-processing of these mt-RNAs occurs primarily at one or a few nucleotides downstream from the C-core motif (Fig. 2). For rnl, which lacks a C-core motif but contains a poly(U) track close to its 3′-end, 3′-processing occurs at the last two Us within a poly(U) track and one or few nucleotides downstream from the poly(U) track (Fig. 2A; Supplemental Fig. S6). This poly(U) track is likely to serve as the signal for 3′-end cleavage. Similar to the situations in S. cerevisiae and unlike humans and consistent with previously reported results (Schäfer et al. 2005), we found that mature mt-mRNAs in S. pombe are not polyadenylated at their 3′-ends.
To verify our RNA-seq data, we performed circularized reverse transcription PCR (cRT-PCR) to map both 5′- and 3′-ends of two mt-rRNAs and three mt-mRNAs. Sequence analysis of the cRT-PCR products revealed that 5′-ends of all mt-rRNAs and mtRNAs examined were precisely cut at a single site, whereas the 3′-ends of all these mt-RNAs were cut at multiple sites. Thus, the cRT-PCR data confirmed our RNA-seq data (Fig. 2).
Examination of the primary sequences and predicted secondary structures of mt-tRNAs revealed that two mt-tRNAs contain an extra 5′-nucleotide. One is mt-tRNAHis(GUG), which has one additional G residue at position −1 at its 5′-end (G−1), and the other is mt-tRNAArg(UCU), which contains an additional A residue at position −1 (A−1) (Fig. 4A). The G−1 residue is a ubiquitous feature of mt-tRNAHis and is required for recognition by the histidyl-tRNA synthetase (Betat et al. 2014), whereas the addition of A−1 at the 5′-end of mt-tRNAArg has not been reported. In addition, the presence of G−1 could potentially extend the acceptor stem by forming an additional base pair with C72 in tRNAHis(GUG). Similarly, the unusual A−1 base pairs with U71 in tRNAArg(UCU), resulting in formation of an unusual acceptor stem (8 base pairs instead of the normal 7).
FIGURE 4.
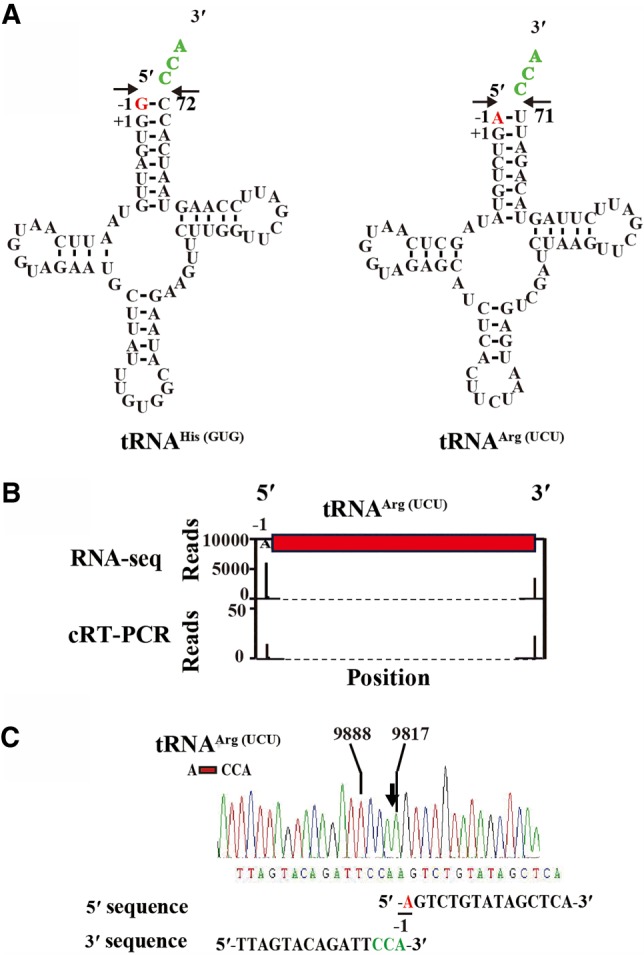
Identification of mt-tRNAs with an extra nt at the 5′-end. (A) The secondary structures of mt-tRNAHis(GUG) and mt-tRNAArg(UCU) each containing an extra nt at the 5′-end. Numbers indicate standard nt numbering positions; −1, the extra nt encoded by the genome; +1, the first nt encoded by the genome. The likely cleavage sites are depicted with an arrow. (B) Abundance of 5′- and 3′-ends sequencing reads at each nt position mapped to mt-tRNAArg(UCU) analyzed by RNA-seq (top) and cRT-PCR (bottom). (C) Sequence of representative cloned RT-PCR products derived from circularized tRNAs. Nucleotide identities are only shown for the predominant sequence. The corresponding sequences up to the 5′- as well as the 3′-end of the cDNA are given. The nonencoded CCA ends are highlighted in green, and the 5′-extra nt of the mature mt-tRNAs are underlined. Numbering is based on the reference sequence NC_001326.1.
To confirm the presence of A−1 in mt-tRNAArg(UCU), we precisely mapped the 5′-end of mt-tRNAArg(UCU) by cRT-PCR. In this method, total mt-tRNA was first treated with T4 RNA ligase to join their 5′- and 3′-ends and then the ligated RNA end of mt-tRNAArg(UCU) was specifically amplified by RT-PCR using specific primers (Supplemental Table S1). The resulting PCR products were cloned into a Sma I-digested plasmid for sequencing. All the 12 sequenced clones contain the A−1 at position 9817 (mt-tRNAArg(UCU) -1 position) and a U71 at position 9888 (numbering based on the reference sequence NC_001326.1), followed by a CCA at the 3′-end (Fig. 4B), except in one clone CC was detected as the 3′-ending nucleotides. Thus, by using cRT-PCR, we were able to detect the presence of the extra A in the mt-tRNAArg(UCU) (Fig. 4B,C).
Trz2 regulates the processing of mt-RNAs
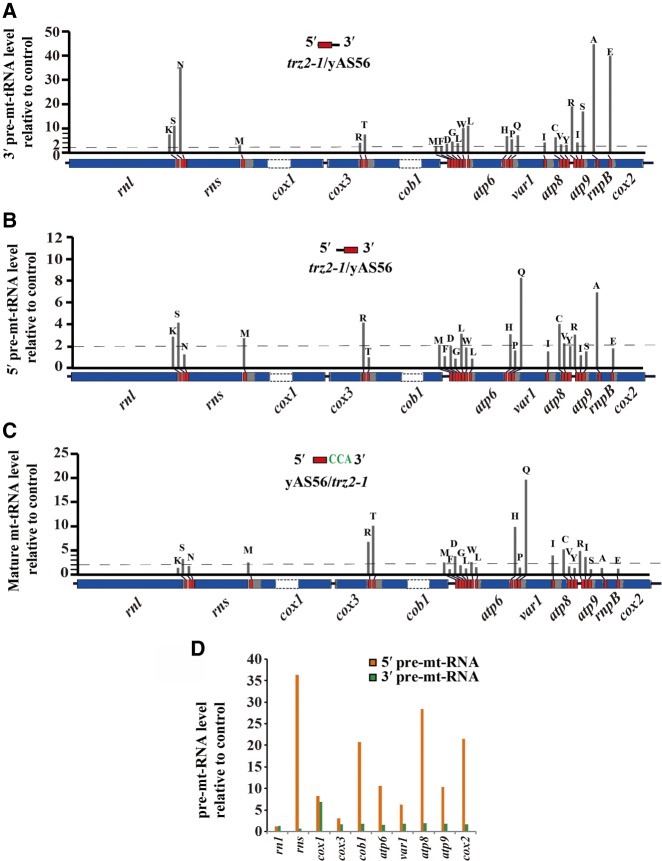
We have previously shown by Northern blot analysis that inactivation of trz2 affects 3′-end processing of mt-tRNALys(UUU), mt-tRNAArg(ACG), and mt-tRNAHis(GUG) (Zhang et al. 2013). To determine whether inactivation of trz2 affected genome-wide mt-tRNA 3′-end processing, we constructed and sequenced cDNA libraries from mt-tRNA of the wild-type strain and trz2-1 mutant, which harbors a temperature-sensitive allele of trz2 (Zhang et al. 2013). The 5′- and 3′-extended precursor mt-tRNAs were defined by the presence of at least 5-nt 5′-flank and at least 5-nt 3′-flank of mature mt-tRNAs, respectively. Reads that were mapped to mature mt-tRNA regions and mt-tRNA flanks were accounted to compute RPKM. RNA-seq revealed that inactivation of trz2 resulted in accumulation of precursor mt-tRNAs with unprocessed 3′-ends (Fig. 5A). The most pronounced increases (∼40- to 50-fold increase) were observed for mt-tRNAAla(GUC) and mt-tRNAGlu(UUC) (Fig. 5A). Since inactivation of tRNase ZL in S. cerevisaie also affects 5′-end processing of pre-tRNAs (Skowronek et al. 2014), we wondered whether trz2 inactivation would affect 5′-end processing of mt-tRNAs. We found that 5′-end processing of nearly half of mt-tRNAs was impaired in the trz2-1 mutant, but 5′-end processing was not impaired as severely as 3′-end processing (Fig. 5B). As expected, we found that the levels of most mature mt-tRNAs were reduced in the trz2-1 mutant strain under nonpermissive temperature (Fig. 5C).
FIGURE 5.
Inactivation of trz2 affects the expression of precursor and mature mt-RNAs. (A,B) Relative expression of precursor mt-tRNAs with unprocessed 3′-end or 5′-end in the trz2-1 mutant compared with the WT control strain (defined value of 1) by RNA-seq. (C) Relative expression of mature mt-tRNAs in the WT strain compared with the trz2-1 mutant (defined value of 1) by RNA-seq. (D) Relative expression of 5′-extended or 3′-extended precursors of mt-rRNAs and mt-mRNAs in the trz2-1 mutant compared with the WT control strain (defined value of 1) by RNA-seq. mt-tRNAs are represented by single-letter code and indicated by red boxes.
Because 3′-end cleavage of mt-tRNAs is also crucial for the maturation of downstream adjacent mt-RNAs, we also analyzed 5′-end processing of mt-mRNAs and mt-rRNAs in the trz2-1 mutant strain under nonpermissive temperature. As expected, we found that the levels of 5′-extened precursors of all mt-mRNAs and mt-rRNAs that are flanked on the 5′-side by an mt-tRNA were increased in the trz2-1 mutant, compared to those from the wild-type strain (Fig. 5D). In contrast, 3′-extended precursors of most mt-mRNAs and mt-rRNAs were much less affected (Fig. 5D). Together, these results suggest that the trz2 inactivation impairs a global mt-tRNA 3′-end processing and 5′-end processing of the adjacent mt-mRNAs and mt-rRNAs.
Processing of the S. pombe rnpB RNA
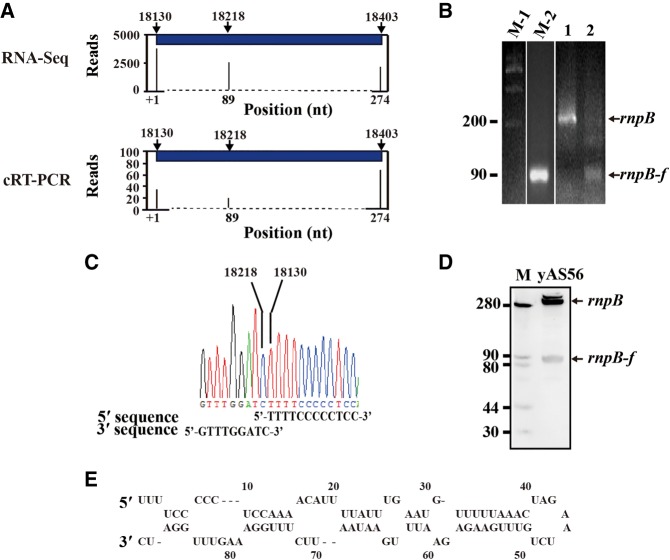
In S. pombe, the rnpB RNA is flanked by mt-tRNAAla(UGC) and mt-tRNAGlu(UUC). RNA-seq analysis revealed that the 5′- and 3′-cleavage sites of the rnpB RNA precisely coincided with the 3′-cleavage site of upstream mt-tRNAAla(UGC) and the 5′-cleavage site of downstream mt-tRNAGlu(UUC), respectively (Figs. 3, 6A), confirming the previous primer extension mapping results (Seif et al. 2003). In our RNA-seq data, we also identified that the rnpB RNA was further cleaved at its 5′-side. The majority of the rnpB-derived fragments (rnpB-f) start with a thymidine at position 18,130 and end with a cytidine at position 18,218 (Fig. 6A). This result was confirmed by cRT-PCR (Fig. 6B,C) and Northern blot analysis using a specific probe for the rnpB-f RNA (Fig. 6D). The rnpB-f RNA is featured with a predicted stem–loop hairpin structure (Fig. 6E). We did not find any fragments with significant coverage from the remaining rnpB RNA.
FIGURE 6.
Mapping of rnpB transcript ends. (A) Analysis of 5′- and 3′-ends of the rnpB and rnpB-f RNAs by RNA-seq (top) and cRT-PCR (bottom). cRT-PCR was performed on total mt-RNA, and the rnpB and rnpB-f products were amplified, cloned, and sequenced. Arrow indicates the cleavage site. (B) An ethidium bromide-stained gel showing detection of the rnpB RNA (lane 1) and the rnpB-f RNA (lane 2) by cRT-PCR analysis. The products were separated on a 3.0% agarose gel. Lanes M-1 and M-2 are DNA size markers in nt. (C) Chromatogram displaying the sequences of the TA cloned cRT-PCR products representing the sense strands of the rnpB-f RNA. The corresponding DNA sequences are aligned below. (D) Northern blot analysis of the rnpB-f RNA. RNA was isolated from S. pombe mitochondria, separated by 9% PAGE-8 M urea, and transferred. The blot was probed with the DIG-labeled oligoDNA probe specific for the rnpB-f RNA. Lane M contains a DNA marker. (E) The secondary structure of the rnpB-f RNA predicted by Mfold (http://unafold.rna.albany.edu/?q=mfold/RNA-Folding-Form).
mt-tRNA halves
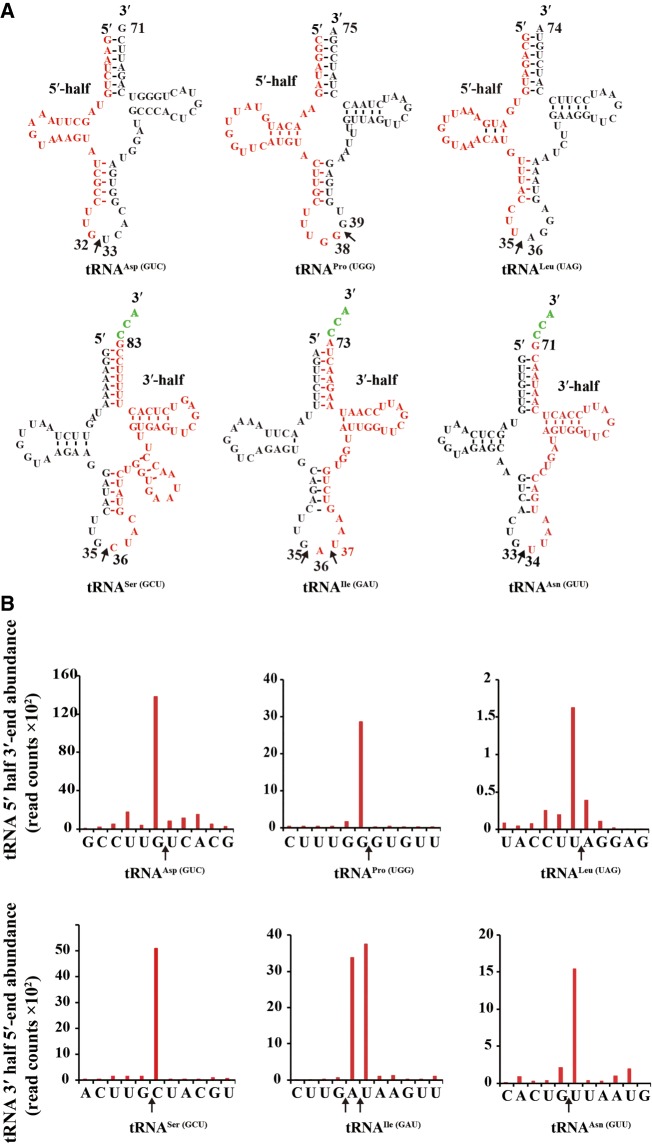
Our deep sequencing of tRNAs extracted from S. pombe mitochondria identified mt-tRNA halves. These identified mt-tRNA halves came from six different mt-tRNAs. The 5′-mt-tRNA halves were derived from mt-tRNAAsp(GUC), mt-tRNAPro(UGG), and mt-tRNALeu(UAG), while the 3′-mt-tRNA halves were derived from mt-tRNASer(GCU), mt-tRNAAsn(GUU) and mt-tRNAIle(GAU) (Fig. 7). The 5′ mt-tRNA halves were 32–38 nt long, and the 3′-mt-tRNA halves were 40–51 nt in length. Both 5′- and 3′-mt-tRNA halves were detected with cleavage sites (Fig. 7). mt-tRNAIle(GAU) was primarily cleaved between 35 and 36 and between 36 and 37 within the anticodon loop, whereas others were primarily cleaved at a single site within the anticodon loop (Fig. 7).
FIGURE 7.
Identification of mt-tRNA halves by RNA-seq analysis. (A) 3′- and 5′- mt-tRNA halves found in S. pombe. Sequences of mt-tRNA halves (in red) were identified by RNA-seq analysis. Arrow indicates the cleavage site. Nt positions are indicted based on the nt numbering system of tRNAs. (B) The distribution of reads that were mapped to the region surrounding the cleavage site (indicated by an arrow).
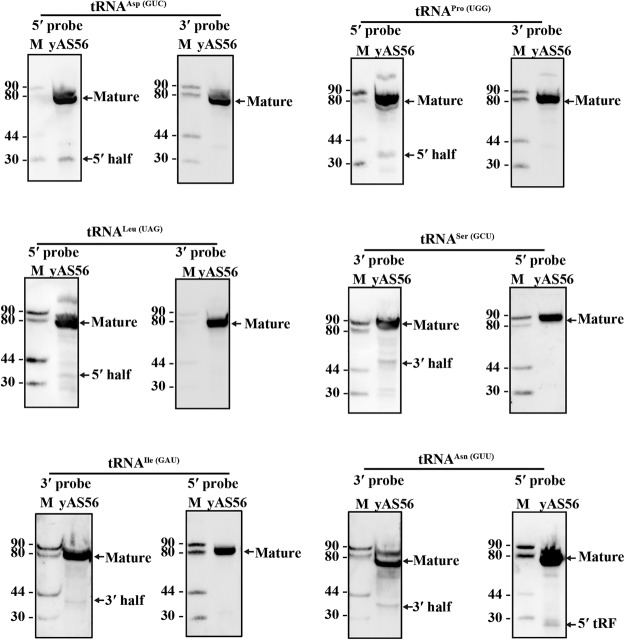
To obtain an independent validation of the RNA-seq results, we used Northern blot analysis to detect mt-tRNA halves in the wild-type S. pombe cells (Fig. 8). We probed total RNA isolated from S. pombe mitochondria with probes specific for the 5′- or 3′-mt-tRNA half. Probes specific for 5′-mt-tRNAAsp(GUC), 5′-mt-tRNAPro(UGG), and 5′-mt-tRNALeu(UAG) detected 5′-mt-tRNA halves (Fig. 8). As controls, probes specific for 3′-halves of these mt-tRNAs did not detect their corresponding 3′-mt-tRNA halves (Fig. 8). Similarly, probes specific for the 3′-mt-tRNASer(GCU), 3′-tRNAIle(GAU), or 3′-tRNAAsn(GUU) detected 3′-mt-tRNA halves. As controls, probes specific for 5′-halves of these mt-tRNAs did not detect their corresponding 5′-mt-tRNA halves (Fig. 8). In the case of the 5′-mt-tRNAAsn(GUU) half probe, a 5′-mt-tRF was detected instead of an mt-tRNA half (Fig. 8). This 5′-mt-tRF was also identified in RNA-seq data (Supplemental Table S2). These results further confirmed the presence of 5′- and 3′-mt-tRNA halves.
FIGURE 8.
Identification of mt-tRNA halves in S. pombe mitochondria by Northern blot analysis. Total mt-RNA was isolated from S. pombe cells grown to mid-log phase and was separated by electrophoresis in a 9% PAGE containing 8 M urea. Blots were probed with oligoDNAs that are specific to 5′- or 3′-mt-tRNA halves. M, single-stranded DNA markers in nt. Mature mt-tRNA, 5′- and 3′-mt-tRNA halves and 5′-tRF are indicated to the right.
CCACCA/CCACC addition in S. pombe mt-tRNAs
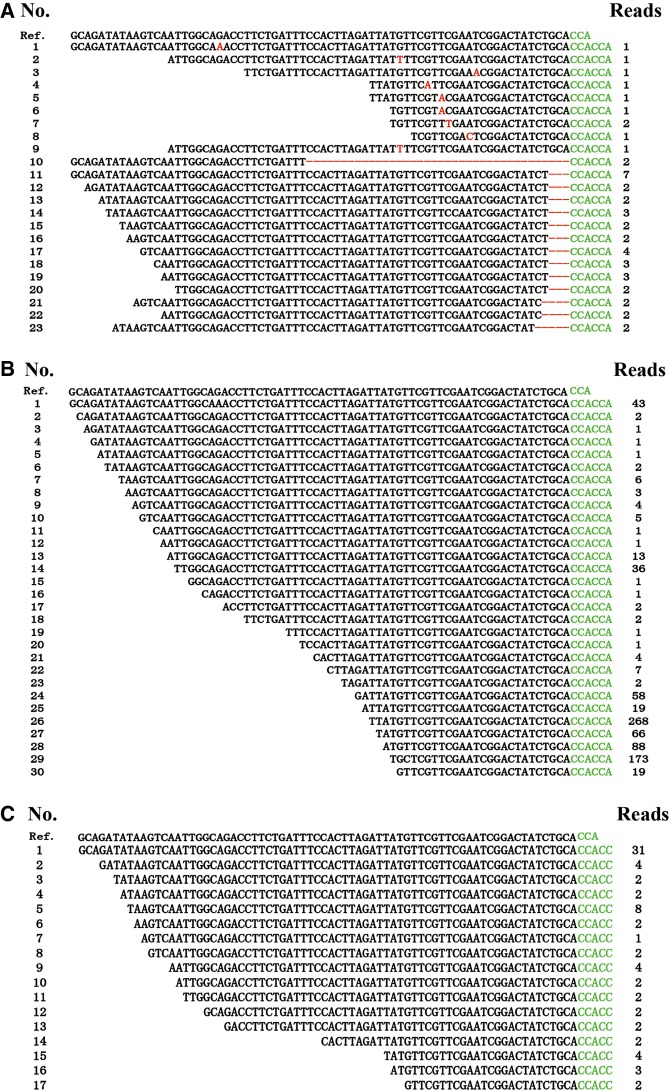
Interestingly, RNA-seq revealed addition of CCACCA/CCACC to the 3′-end of some mt-tRNAs from wild-type S. pombe (Fig. 9; see Supplemental Table S3 for a complete list). As an example, with mt-tRNAGly(UCC), we found that a fraction of the CCACCA/CCACC tagged mt-tRNAGly(UCC) contains point mutations and/or truncations (Fig. 9). Truncations at the 3′-end of mt-tRNAGly(UCC) apparently disrupt the base-pairing of the acceptor stems and cause destabilization of tRNA (Fig. 9A). We also found that a series of mt-tRNAGly(UCC) 3′-fragments with successive 5′-truncations were marked with 3′-CCACCA/CCACC (Fig. 9B,C). These 5′-truncations suggest progressive 5′–3′ exoribonucleolytic degradation of CCACCA/CCACC-containing mt-tRNAsGly(UCC). Similar results were obtained in several other mt-tRNAs such as mt-tRNATrp(UCA), mt-tRNAAsp(GUC), mt-tRNAAsn(GUU), and mt-tRNAIle(GAU) (Supplemental Table S3).
FIGURE 9.
List of mt-tRNAGly(UCC) transcripts containing 3′-CCACCA/CCACC. (A) Alignment of the sequences of mt-tRNAGly(UCC) marked with 3′-CCACCA and containing mutation (shown in red colored character) or 3′-deletion (shown as a hyphen). (B) Alignment of the sequences of the 5′-truncated mt-tRNAGly(UCC) transcripts marked with 3′-CCACCA. (C) Alignment of sequences of the 5′-truncated mt-tRNAGly(UCC) transcripts marked with 3′-CCACC. Ref., the sequence of the S. pombe mt-tRNAGly(UCC) gene.
S. pombe mitosRNAs
To determine whether mitosRNAs exist in S. pombe, we sequenced the 15- to 30-nt small RNA (mitosRNA) library constructed from RNA extracted from S. pombe mitochondria. The numbers of reads mapped to S. pombe mtDNA are ∼1.8 million. To distinguish mitosRNAs from potential random by-products of the degradation of mt-RNAs, we filtered out lowly expressed mitosRNAs (expressed at >850 reads per million, the median level of sRNA expression in S. pombe). As expected, the predominant size of sequences mapping to S. pombe mtDNA was in the size range of 15–30 nt. Totally, 32 unique mitosRNA sequences were mapped to 14 different loci in the S. pombe mtDNA (Supplemental Table S2). More than half of mitosRNA sequences were mapped to mt-tRNA loci and are candidate mt-tRNA-derived fragments (mt-tRFs). Sequences that were derived from the 5′- and 3′-regions of tRNA were categorized as mt-tRF-5s and mt-tRF-3s, respectively. Since mt-tRF-5s and mt-tRF-3s are significantly enriched, these sequences are probably not random by-products of the degradation of tRNAs.
DISCUSSION
In this study, we presented a mitochondrial transcriptome map from S. pombe, which is evolutionarily distant from S. cerevisiae and other well-studied model yeasts. Compared to traditional low-throughput techniques such as Northern blot analysis or reverse transcriptase PCR, which are limited to examine individual RNAs, RNA-seq allows simultaneous high-resolution analysis of all RNAs. Our transcriptome analysis revealed that S. pombe mt-mRNAs as well as mt-tRNAs display significant abundance differences. Because mtDNA-encoded genes are transcribed as polycistronic transcripts, these differences are likely due to differences in mt-RNA stability. Similar observations have been found in S. cerevisiae, C. albicans, and human mitochondria (Krause and Dieckmann 2004; Piechota et al. 2006; Mercer et al. 2011; Turk et al. 2013; Kolondra et al. 2015). These findings suggest the complexity of post-transcriptional regulation in mitochondria. Future experiments will be required to elucidate the mechanism for the post-transcriptional regulation of mt-RNA expression. It should be noted that tRNA modifications and its stable and extensive secondary structure can interfere with cDNA synthesis and adaptor ligation during the cDNA library construction. These problems for the sequencing of tRNA could be overcome in future studies by using engineered tRNA demethylase and highly processive thermostable group II intron reverse transcriptase (TGIRT) (Cozen et al. 2015; Zheng et al. 2015).
Our transcriptome data show that similar to the situation in human mitochondria, 5′-ends of all S. pombe mt-mRNAs and mt-rRNAs are generated by tRNase ZL, consistent with the tRNA punctuation model (Ojala et al. 1981), except for rnl and cox3 RNAs whose 5′-ends are generated directly from transcription. However, unlike the situation in human mitochondria, S. pombe mt-mRNAs and mt-rRNAs have heterogeneous 3′-ends likely resulting from exonucleolytic trimming.
Our analysis reveals that unlike 5′-end maturation, the generation of the 3′-ends of S. pombe mt-mRNAs and rns rRNA involves additional exonucleolytic processing after mt-RNase P cleavage. This is likely responsible for 3′-heterogeneity, which has also been observed previously (Hoffmann et al. 2008). A previous genetic study suggests that the 3′-ends are matured by a complex consisting of a putative exoribonuclease Rpm1 (also called Par1) and a putative DEAD box helicase Rpm2 (also called Pah1), which are homologous to S. cereveisiae exoribonuclease Dss1 and the RNA helicase Suv3, respectively (Hoffmann et al. 2008). It is likely that the final maturation of the 3′-ends requires exonucleolytic trimming by Rpm1. Rpm2 is likely to play a supportive role in this process. However, S. cerevisiae Dss1 and Suv3 do not appear to be involved in the 3′-end formation of mt-RNAs. These two proteins form the mitochondrial degradosome and are involved in degradation of mt-RNAs (Dziembowski et al. 2003).
Our analysis also shows the C-core motif located a few nucleotides close to the 3′-cleavage site appears to serve as the 3′-processing signal for Rpm1 and Rpm2, which is consistent with a previous study (Schäfer et al. 2005). It is unclear whether this motif serves a binding site for a yet-to-be-identified protein, which plays a role in mt-RNA 3′-end formation. The C-core motif appears to be present not only in the fission yeasts but also in Saccharomcotina, Basidiomycota and basal groups of fungi (Supplemental Table S4; Supplemental Fig. S7), suggesting that this may be a basic type of mechanism of mt-RNA 3′-end processing.
We also investigated the role of S. pombe Trz2 in mt-RNA processing using RNA-seq, which allows comprehensive analysis of the 5′- and 3′-ends of all mt-tRNAs. The data derived from RNA-seq have been verified by comparing them with our previous data obtained with Northern blotting (Zhang et al. 2013). We found that while trz2 deletion blocks the 3′-end processing of all mt-tRNAs, it has varying effects on mt-tRNAs. These results indicate that Trz2 is responsible for 3′-processing of all mt-tRNAs. This is consistent with the notion that unlike nuclear tRNA 3′-ends, which can be generated endonucleolytically or exonucleolytically (Blewett and Maraia 2018), mt-tRNA 3′-ends are generated endonucleolytically by tRNase ZL. We also found nearly a half of mt-tRNA 5′-end processing was also affected in the trz2-1 mutant although not as severely as 3′-end processing. Inhibition of 5′-processing when mitochondrial tRNase ZL activity is compromised has also been observed in S. cerevisiae (Skowronek et al. 2014). It is very likely that trz2 inactivation may compromise the mt-RNase P activity since both mt-tRNA 5′- and 3′-end processing enzymes may be integrated into a stable protein supercomplex and the tRNA 5′- and 3′-end processing may be somehow linked (Ma et al. 2017b). Unexpectedly, the levels of some mature mt-tRNAs are not affected upon trz2 inactivation. One possible explanation is that the levels of residual tRNAse ZL activity in the trz2-1 mutant may be sufficient for 3′-end processing of some mt-tRNAs. Another possibility is that these mt-tRNAs may have slower decay rates.
Our high-throughput sequencing analysis of the transcriptome of S. pombe mitochondria also reveals a plethora of mitochondrial sRNAs (mitosRNAs) including mt-tRNA-derived fragments (mt-tRFs) and mt-tRNA halves. mt-tRFs revealed by deep sequencing are in a size range from 17 to 28 nt. The biogenesis and biological functions of S. pombe mt-tRFs remain to be determined. mt-tRFs have been also found in animal mitochondria (Mercer et al. 2011; Ro et al. 2013). The mammalian mt-tRFs have been suggested to play a role in regulation of mtDNA gene expression (Ro et al. 2013). However, their biogenesis is unclear. In humans, DICER and tRNase ZL have been shown to be involved in the biogenesis of nuclear tRFs (Shigematsu et al. 2014).
tRNA halves identified in S. pombe mitochondria are 32–51 nt in size and generated by cleavage of mt-tRNA within the anticodon. Our analysis identified at least six mt-tRNA halves under nonstressed conditions, which were confirmed by Northern blotting (Fig. 8). mt-tRNA halves have previously only been reported in rainbow trout; however, their functions are unknown (Ma et al. 2016). Unlike mt-tRNA halves, tRNA halves derived from cytosolic tRNAs have been widely found in eukaryotes under stress as well as nonstress conditions (Huang and Hopper 2016; Soares and Santos 2017). tRNA halves whose expression is induced in response to a variety of stresses are known as tRNA-derived stress-induced RNAs (Yamasaki et al. 2009).
tRNA halves have been implicated in inhibition of protein synthesis (Soares and Santos 2017), promoting formation of stress granules (Yamasaki et al. 2009; Emara et al. 2010), and protection of cells from apoptosis through interaction with cytchrome c (Saikia et al. 2014). A type of 5′-tRNA halves termed sex hormone-dependent tRNA derivatives (SHOT-RNAs) can enhance cell proliferation in breast and prostate cancer cell lines (Honda et al. 2015). A subset of mouse sperm tRNA-derived small RNAs (mainly from 5′-tRNA halves), whose expression is affected by paternal diet, may contribute to diet-induced metabolic disorders (Chen et al. 2016; Sharma et al. 2016).
So far ribonucleases Rny1 in yeast and angiogenin (ANG) in mammals have been identified to cleave mature cytosolic tRNAs to generate tRNA halves during stress. Rny1 and ANG belong to the RNase T2 family (Thompson and Parker 2009) and the RNase A superfamily, respectively (Yamasaki et al. 2009). The homologs of Rny1 and ANG do not appear to exist in S. pombe, suggesting an alternative mechanism for generation of mt-tRNA halves.
In our RNA-seq data, we also found that a small fraction of the full-length rnpB RNA was cleaved into an 89-nt fragment (rnpB-f). In S. cerevisiae, three fragments of mt-RNase P RNA (RMP1 RNA) (52, 71, and 89 nt) but not full-length RMP1 RNA were detected, suggesting that an intact RMP1 RNA is not required for mt-RNase P activity (Daoud et al. 2012; Turk et al. 2013). Because the rnpB-f RNA contains only conserved region CRI but lacks other conserved regions that are essential for catalytic activity or substrate binding (Supplemental Fig. S8), it cannot fulfill the function of the full-length RNA. Thus, unlike S. cerevisiae mt-RNase P, an intact rnpB RNA is likely to be required for S. pombe mt-RNase P activity. However, elucidation of the biological role and biogenesis of the rnpB-f RNA will require further investigation.
Our study provides the first evidence that mt-tRNAHis(GUG) from a fungal mitochondria contains an extra guanylate residue at its 5′-end (G−1). With very few exceptions, all known mt-tRNAHis contain an extra G−1 residue, which is required for recognition by histidyl-tRNA synthetase (Betat et al. 2014). Various mechanisms used to generate this extra G−1 have been demonstrated. In bacteria and archaea and presumably in fungal mitochondria and plant chloroplasts, the G−1 residue is encoded in the mt-tRNAHis(GUG) gene and generated by an unusual mt-RNase P cleavage. In the cytoplasm of eukaryotes and animal mitochondria, the G−1 residue is gene-encoded and is generated after transcription by histidinyl tRNA guanylyl transferase (Thg1), which is more common in eukaryotes, and also found in a limited number of bacterial and archaeal species (Jackman and Phizicky 2006; Abad et al. 2010). The G−1 residue can also be generated by transcription initiation. In the archaeon Nanoarchaeum equitan, RNase P is lost and all tRNA 5′-ends are generated by transcription initiation. Because the G−1 residue in S. pombe mt-tRNAHis(GUG) is gene-encoded, it is very likely that this addition nucleotide is the result of unusual processing by mt-RNase P. Consistent with this prediction, we found that the mitochondrial Thg1 could be predicted bioinformatically in humans, but not in S. cerevisiae and S. pombe.
Besides mt-tRNAHis(GUG), our analysis also revealed that mt-tRNAArg(UCU) possesses an extra adenylate residue at its 5′-end. This extra nucleotide is gene-encoded and presumably generated because of an unusual cleavage by mt-RNase P. Our finding adds to a growing list of tRNAs, which contain an extra 5′-nucleotide. Several tRNAs other than tRNAHis have also been found to contain an extra 5′-nucleotide. For example, mt-tRNAPhe from the mitochondria of the ciliate protozoon Tetrahymena pyriformis has an extra 5′ U−1. (Schnare et al. 1985). In the archaeon Nanoarchaeum equitans, tRNATyr and tRNAiMet are found to possess an extra 5′ G−1 or 5′ A−1, respectively. These extra 5′-nucleotides are generated by transcription initiation, and the resulting unusual tRNAs are substrates for their cognate amino-tRNA synthatases.
One interesting finding of our deep sequencing analysis is identification of a low but reproducible level of extended CCA motifs (defined as CCACCA or CCACC) on mt-tRNAs, such as mt-tRNAGly(CGA), mt-tRNATrp(UCA), mt-tRNAAsp(GUC), mt-tRNAAsn(GUU), and mt-tRNAIle(GAU) in S. pombe. Some of our detected CCACCA-marked mt-tRNAs are truncated at the 3′-ends. Because these truncations were in the acceptor stems, they apparently disrupt base-pairing and cause instability of mt-tRNA. Most of the CCACCA-marked mt-tRNAs have common 3′-ends and variable 5′-truncations, indicative of exonucleolytic degradation. It is unclear how the 5′-sequences are missing. It has previously been shown that the CCA-adding enzyme, ATP (CTP):tRNA nucleotidyltransferase can add CCACCA to structurally unstable tRNAs and hypomodified tRNAs (Wilusz et al. 2011). Unlike S. cerevisiae and human, which contain a single CCA-adding enzyme, S. pombe contains two candidate genes (cca1 and cca2) encoding the CCA-adding enzymes. Cca1 and Cca2 display 30% identity and 53% similarity. It is unclear which of these proteins can add CCACCA to structurally unstable or hypomodified mt-tRNAs. CCACCA-marked tRNAs are difficult to be detected because they are degraded rapidly and efficiently by 3′-5′ exonuclease RNase R in E. coli and 3′-5′ exonuclease Rrp44 (also called Dis3), in cooperation with 5′-3′ exonucleases Xrn1 in cooperating with Rrp44 in S. cerevisiae (Wilusz et al. 2011). It remains to be determined how CCACCA-marked mt-tRNAs are degraded. Our findings also suggest that the CCACCA-mediated quality control mechanism may also operate in mitochondria.
MATERIALS AND METHODS
Strains and media
S. pombe strains used in this study are yAS56 (wild-type; h+ leu1-32 ura4-D18) and yHD1 (h+ trz2-1 leu1-32 his3-D1 ura4-D18 ade6-210), which exhibits a temperature-sensitive phenotype resulting from mutation of Ala to Val at position 623 of trz2 (Zhang et al. 2013). S. pombe cells were grown in yeast extract supplement (YES) medium with the appropriate supplements (0.5% yeast extract, 3% (w/v) glucose, and 225 mg/liter each adenine, histidine, leucine, and uracil) and the cell growth was monitored at OD600.
Isolation of mt-RNA
S. pombe cells were grown in YES medium at 30°C overnight, reinoculated in fresh YES medium to an OD600 of 0.2, and grown to an OD600 of 0.6–0.8. Then cells were shifted to 37°C and continued to grow for another 6 h before being harvested. The crude mitochondria were prepared by the spheroplast method and further purified by sucrose gradient centrifugation as previously described (Wang et al. 2017). The sucrose gradient purified mitochondria were treated with DNase I (Qiagen) to reduce cytoplasmic nucleic acid contamination. Total mt-RNA was purified using the miRNeasy Micro Kit (Qiagen) with additional on-column DNase I treatment to remove mt-DNA. RNA purity and integrity was assessed by checking its A260/A280 ratio using BioAnalyser (Agilent) and the enrichment of mt-RNAs (cox2 mRNA) relative to nuclear (uba2 mRNA) and cytoplasmic RNAs (18S rRNA) by Quantitative real-time RT-PCR (qRT-PCR).
RNA sequencing (RNA-seq)
To determine mt-mRNA and mt-rRNA expression levels, we constructed a cDNA library using the Illumina TruSeq Stranded Total RNA protocol with modifications. Because S. pombe mt-mRNAs lack poly(A) tails, mt-RNA samples were used directly for fragmentation without poly(A) selection. In addition, the library was constructed without ribosomal or tRNA subtraction. Approximately 200-nt fragmented mt-RNAs were selected for library construction. The first-strand cDNA was synthesized with random hexamer primers. Adapters were ligated to the 3′-ends and 5′-ends of cDNA. The ligated fragments were amplified by PCR and sequenced with the Illumina HiSeq 2000.
To determine mt-tRNA expression levels and mt-tRNA cleavage sites, the second library was prepared and constructed using the Illumina Small RNA Sample Preparation Kit. Briefly, the mt-RNA fraction (30–120 nt) from total mt-RNA was gel-purified on denaturing urea-PAGE. Sequencing of RNA libraries was performed on an Illumina HiSeq 2000 sequencer. Reads were checked for quality with the program FastQC and trimmed to remove low quality bases and the adaptor sequences with Trimmomatic. The reference genome sequence for mapping the RNA-seq data was retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/nuccore/NC_001326.1) with Bowtie (Langmead et al. 2009; Langmead and Salzberg 2012). Quantification of gene expression was conducted using Cufflinks v2.1.1 to calculate RPKM for each gene.
To identify sRNA candidates in S. pombe mitochondria, a sRNA library was generated using the Illumina Small RNA Sample Preparation Kit. Briefly, total mt-RNA was fractionated on denaturing PAGE. sRNA (15- to 30-nt) was isolated from the gel, ligated to adapters. PCR was performed with primers complementary to the RNA adapters to yield a library. The library was sequenced on an Illumina HiSeq 2000. The raw reads were processed as described above.
Circular reverse transcription-PCR (cRT-PCR)
cRT-PCR was used to determine the termini of mt-RNAs as previously described (Supplemental Fig. S9; Yokobori and Paabo 1995; Forner et al. 2007). Briefly, 5 µg of mt-RNA from one sample was allowed to self-ligate in a total volume of 100 µL containing 100 units of T4 RNA ligase (Takara) at 16°C for 16–18 h. The circular RNAs were purified using the PureLink RNA Mini Kit (Invitrogen). RT-PCR was performed as previously described (Wang et al. 2017). Primer sequences are provided in Supplemental Table S1. For all RT-PCR experiments, a negative control without template RNA was included. PCR products were cloned into the TA vector using the TAcloning kit (Takara) according to the manufacturer's protocol and sequenced (Sangon Technology).
Northern blot analysis
mt-RNA was separated on a 9% polyacrylamide-urea gel and transferred to a nylon membrane (Amersham). The blots were hybridized with digoxigenin (DIG)-labeled oligonucleotide probes complementary to precursors or mature forms of mt-tRNAs. The sequence of the probes is listed in Supplemental Table S5. DIG-labeled probes were synthesized by Sangon Biotech, and immunological detection was performed using the DIG Luminescent Detection Kit (Roche) according to manufacturer's instructions and as described.
qRT-PCR
qRT-PCR was performed as described before (Su et al. 2015). Primer sets are listed in Supplemental Table S1. cDNA was prepared by reverse transcription of RNA using random hexamers and the RevertAidTM First Strand cDNA Sythesis Kit (Fermentas). The SYBR Select Master Mix (Applied Biosystems) and a StepOne real-time PCR system (Applied Biosystems) machine were used for all RT-qPCR experiments. All reactions were performed in three technical replicates and two biological replicates. Data analysis was performed by StepOne software.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. John Mattick, Pawel Golik, and Kai Xu for helpful discussion. This work was supported in part by grants from the National Natural Science Foundation of China (31400032 to J.S., and 31470778 and 31770810 to Y.H.); and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD to Y.H.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.064477.117.
REFERENCES
- Abad MG, Rao BS, Jackman JE. 2010. Template-dependent 3′-5′ nucleotide addition is a shared feature of tRNAHis guanylyltransferase enzymes from multiple domains of life. Proc Natl Acad Sci 107: 674–679. [Google Scholar]
- Asin-Cayuela J, Gustafsson CM. 2007. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem Sci 32: 111–117. [DOI] [PubMed] [Google Scholar]
- Betat H, Long Y, Jackman JE, Mörl M. 2014. From end to end: tRNA editing at 5′- and 3′-terminal positions. Int J Mol Sci 15: 23975–23998. [Google Scholar]
- Bhatti JS, Bhatti GK, Reddy PH. 2017. Mitochondrial dysfunction and oxidative stress in metabolic disorders—a step towards mitochondria based therapeutic strategies. Biochim Biophys Acta 1863: 1066–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewett NH, Maraia RJ. 2018. La involvement in tRNA and other RNA processing events including differences among yeast and other eukaryotes. Biochim Biophys Acta 1861: 361–372. [DOI] [PubMed] [Google Scholar]
- Brzezniak LK, Bijata M, Szczesny RJ, Stepien PP. 2011. Involvement of human ELAC2 gene product in 3′ end processing of mitochondrial tRNAs. RNA Biol 8: 616–626. [DOI] [PubMed] [Google Scholar]
- Chan DC. 2006. Mitochondria: dynamic organelles in disease, aging, and development. Cell 125: 1241–1252. [DOI] [PubMed] [Google Scholar]
- Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, et al. 2016. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351: 397–400. [DOI] [PubMed] [Google Scholar]
- Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. 2015. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods 12: 879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daoud R, Forget L, Lang BF. 2012. Yeast mitochondrial RNase P, RNase Z and the RNA degradosome are part of a stable supercomplex. Nucleic Acids Res 40: 1728–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Piwowarski J, Hoser R, Minczuk M, Dmochowska A, Siep M, van der Spek H, Grivell L, Stepien PP. 2003. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J Biol Chem 278: 1603–1611. [DOI] [PubMed] [Google Scholar]
- Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. 2010. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 285: 10959–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Wang Z, Liu J, Guo W, Yan J, Huang Y. 2011. A survey of green plant tRNA 3′-end processing enzyme tRNase Zs, homologs of the candidate prostate cancer susceptibility protein ELAC2. BMC Evol Biol 11: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner J, Weber B, Thuss S, Wildum S, Binder S. 2007. Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res 35: 3676–3692. [Google Scholar]
- Foury F, Roganti T, Lecrenier N, Purnelle B. 1998. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett 440: 325–331. [DOI] [PubMed] [Google Scholar]
- Gan X, Yang J, Li J, Yu H, Dai H, Liu J, Huang Y. 2011. The fission yeast Schizosaccharomyces pombe has two distinct tRNase ZLs encoded by two different genes and differentially targeted to the nucleus and mitochondria. Biochem J 435: 103–111. [DOI] [PubMed] [Google Scholar]
- Gobert A, Gutmann B, Taschner A, Gössringer M, Holzmann J, Hartmann RK, Rossmanith W, Giegé P. 2010. A single Arabidopsis organellar protein has RNase P activity. Nat Struct Mol Biol 17: 740–744. [DOI] [PubMed] [Google Scholar]
- Gutmann B, Gobert A, Giegé P. 2012. PRORP proteins support RNase P activity in both organelles and the nucleus in Arabidopsis. Genes Dev 26: 1022–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert CJ, Golik P, Bonnefoy N. 2013. Yeast PPR proteins, watchdogs of mitochondrial gene expression. RNA Biol 10: 1477–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B, Nickel J, Speer F, Schäfer B. 2008. The 3′ ends of mature transcripts are generated by a processosome complex in fission yeast mitochondria. J Mol Biol 377: 1024–1037. [DOI] [PubMed] [Google Scholar]
- Holzmann J, Frank P, Löffler E, Bennett KL, Gerner C, Rossmanith W. 2008. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 135: 462–474. [DOI] [PubMed] [Google Scholar]
- Honda S, Loher P, Shigematsu M, Palazzo JP, Suzuki R, Imoto I, Rigoutsos I, Kirino Y. 2015. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci 112: E3816–E3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MJ, Liu X, Lim WH, Klemm BP, Fierke CA, Koutmos M, Engelke DR. 2013. RNase P enzymes: divergent scaffolds for a conserved biological reaction. RNA Biol 10: 909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, Hopper AK. 2016. Multiple layers of stress-induced regulation in tRNA biology. Life (Basel) 6: E16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Phizicky EM. 2006. tRNAHis guanylyltransferase adds G−1 to the 5′ end of tRNAHis by recognition of the anticodon, one of several features unexpectedly shared with tRNA synthetases. RNA 12: 1007–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain AA, Koppen M, Wydro M, Rodley CD, Lightowlers RN, Chrzanowska-Lightowlers ZM, Martinou JC. 2013. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab 17: 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain AA, Popow J, de la Fuente MA, Martinou JC, Anderson P, Simarro M. 2017. The FASTK family of proteins: emerging regulators of mitochondrial RNA biology. Nucleic Acids Res 45: 10941–10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm BP, Wu N, Chen Y, Liu X, Kaitany KJ, Howard MJ, Fierke CA. 2016. The diversity of ribonuclease P: protein and RNA catalysts with analogous biological functions. Biomolecules 6: E27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolondra A, Labedzka-Dmoch K, Wenda JM, Drzewicka K, Golik P. 2015. The transcriptome of Candida albicans mitochondria and the evolution of organellar transcription units in yeasts. BMC Genomics 16: 827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K, Dieckmann CL. 2004. Analysis of transcription asymmetries along the tRNAE-COB operon: evidence for transcription attenuation and rapid RNA degradation between coding sequences. Nucleic Acids Res 32: 6276–6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M, Rossmanith W, Hartmann RK, Thölken C, Gutmann B, Giegé P, Gobert A. 2015. Distribution of ribonucleoprotein and protein-only RNase P in eukarya. Mol Biol Evol 32: 3186–3193. [DOI] [PubMed] [Google Scholar]
- Li de la Sierra-Gallay I, Pellegrini O, Condon C. 2005. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature 433: 657–661. [DOI] [PubMed] [Google Scholar]
- Li de la Sierra-Gallay I, Mathy N, Pellegrini O, Condon C. 2006. Structure of the ubiquitous 3′ processing enzyme RNase Z bound to transfer RNA. Nat Struct Mol Biol 13: 376–377. [DOI] [PubMed] [Google Scholar]
- Lipinski KA, Kaniak-Golik A, Golik P. 2010. Maintenance and expression of the S. cerevisiae mitochondrial genome—from genetics to evolution and systems biology. Biochim Biophys Acta 1797: 1086–1098. [DOI] [PubMed] [Google Scholar]
- Ma H, Weber GM, Wei H, Yao J. 2016. Identification of mitochondrial genome-encoded small RNAs related to egg deterioration caused by postovulatory aging in rainbow trout. Mar Biotechnol (NY) 18: 584–597. [DOI] [PubMed] [Google Scholar]
- Ma M, Li de la Sierra-Gallay I, Lazar N, Pellegrini O, Durand D, Marchfelder A, Condon C, van Tilbeurgh H. 2017a. The crystal structure of Trz1, the long form RNase Z from yeast. Nucleic Acids Res 45: 6209–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Li de la Sierra-Gallay I, Lazar N, Pellegrini O, Lepault J, Condon C, Durand D, van Tilbeurgh H. 2017b. Trz1, the long form RNase Z from yeast, forms a stable heterohexamer with endonuclease Nuc1 and mutarotase. Biochem J 474: 3599–3613. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, et al. 2011. The human mitochondrial transcriptome. Cell 146: 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature 290: 470–474. [DOI] [PubMed] [Google Scholar]
- Piechota J, Tomecki R, Gewartowski K, Szczesny R, Dmochowska A, Kudla M, Dybczynska L, Stepien PP, Bartnik E. 2006. Differential stability of mitochondrial mRNA in HeLa cells. Acta Biochim Pol 53: 157–168. [PubMed] [Google Scholar]
- Pinker F, Bonnard G, Gobert A, Gutmann B, Hammani K, Sauter C, Gegenheimer PA, Giegé P. 2013. PPR proteins shed a new light on RNase P biology. RNA Biol 10: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham O, Davies SM, Shearwood AM, Hamilton KL, Whelan J, Filipovska A. 2009. Pentatricopeptide repeat domain protein 1 lowers the levels of mitochondrial leucine tRNAs in cells. Nucleic Acids Res 37: 5859–5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham O, Busch JD, Matic S, Siira SJ, Kuznetsova I, Atanassov I, Ermer JA, Shearwood AM, Richman TR, Stewart JB, et al. 2016. Hierarchical RNA processing is required for mitochondrial ribosome assembly. Cell Rep 16: 1874–1890. [DOI] [PubMed] [Google Scholar]
- Ro S, Ma HY, Park C, Ortogero N, Song R, Hennig GW, Zheng H, Lin YM, Moro L, Hsieh JT, et al. 2013. The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res 23: 759–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmanith W. 2012. Of P and Z: mitochondrial tRNA processing enzymes. Biochim Biophys Acta 1819: 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M, Jobava R, Parisien M, Putnam A, Krokowski D, Gao XH, Guan BJ, Yuan Y, Jankowsky E, Feng Z, et al. 2014. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol 34: 2450–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas-Giegé T, Giegé R, Giegé P. 2015. tRNA biology in mitochondria. Int J Mol Sci 16: 4518–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MI, Mercer TR, Davies SM, Shearwood AM, Nygård KK, Richman TR, Mattick JS, Rackham O, Filipovska A. 2011. RNA processing in human mitochondria. Cell Cycle 10: 2904–2916. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. 2008. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88: 611–638. [DOI] [PubMed] [Google Scholar]
- Schäfer B. 2005. RNA maturation in mitochondria of S. cerevisiae and S. pombe. Gene 354: 80–85. [DOI] [PubMed] [Google Scholar]
- Schäfer B, Hansen M, Lang BF. 2005. Transcription and RNA-processing in fission yeast mitochondria. RNA 11: 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnare MN, Heinonen TY, Young PG, Gray MW. 1985. Phenylalanine and tyrosine transfer RNAs encoded by Tetrahymena pyriformis mitochondrial DNA: primary sequence, post-transcriptional modifications, and gene localization. Curr Genet 9: 389–393. [DOI] [PubMed] [Google Scholar]
- Seif ER, Forget L, Martin NC, Lang BF. 2003. Mitochondrial RNase P RNAs in ascomycete fungi: lineage-specific variations in RNA secondary structure. RNA 9: 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS, Horvath TL. 2015. Mitochondrial ROS signaling in organismal homeostasis. Cell 163: 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, et al. 2016. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351: 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu M, Honda S, Kirino Y. 2014. Transfer RNA as a source of small functional RNA. J Mol Biol Mol Imaging 1: 8. [PMC free article] [PubMed] [Google Scholar]
- Skowronek E, Grzechnik P, Späth B, Marchfelder A, Kufel J. 2014. tRNA 3′ processing in yeast involves tRNase Z, Rex1, and Rrp6. RNA 20: 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares AR, Santos M. 2017. Discovery and function of transfer RNA-derived fragments and their role in disease. Wiley Interdiscip Rev RNA 8 10.1002/wrna.1423. [DOI] [PubMed] [Google Scholar]
- Stone JD, Storchova H. 2015. The application of RNA-seq to the comprehensive analysis of plant mitochondrial transcriptomes. Mol Genet Genomics 290: 1–9. [DOI] [PubMed] [Google Scholar]
- Su Y, Chen C, Huang L, Yan J, Huang Y. 2015. Schizosaccharomyces pombe homologs of human DJ-1 are stationary phase-associated proteins that are involved in autophagy and oxidative stress resistance. PLoS One 10: e0143888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen DF, Norris KL, Youle RJ. 2008. Mitochondrial dynamics and apoptosis. Genes Dev 22: 1577–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Nagao A, Suzuki T. 2011. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet 45: 299–329. [DOI] [PubMed] [Google Scholar]
- Thompson DM, Parker R. 2009. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 185: 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk EM, Das V, Seibert RD, Andrulis ED. 2013. The mitochondrial RNA landscape of Saccharomyces cerevisiae. PLoS One 8: e78105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet 39: 359–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zheng J, Zhang X, Peng J, Liu J, Huang Y. 2012. Identification and sequence analysis of metazoan tRNA 3′-end processing enzymes tRNase Zs. PLoS One 7: e44264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yan J, Zhang Q, Ma X, Zhang J, Su M, Wang X, Huang Y. 2017. The Schizosaccharomyces pombe PPR protein Ppr10 associates with a novel protein Mpa1 and acts as a mitochondrial translational activator. Nucleic Acids Res 45: 3323–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Whipple JM, Phizicky EM, Sharp PA. 2011. tRNAs marked with CCACCA are targeted for degradation. Science 334: 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Dubrovsky EB. 2015. Knockout of Drosophila RNase ZL impairs mitochondrial transcript processing, respiration and cell cycle progression. Nucleic Acids Res 43: 10364–10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Ackerley C, Maj MC, Addis JB, Levandovskiy V, Lee J, Mackay N, Cameron JM, Robinson BH. 2008. Disruption of a mitochondrial RNA-binding protein gene results in decreased cytochrome b expression and a marked reduction in ubiquinol-cytochrome c reductase activity in mouse heart mitochondria. Biochem J 416: 15–26. [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P. 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori S, Paabo S. 1995. Transfer RNA editing in land snail mitochondria. Proc Natl Acad Sci 92: 10432–10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhao Q, Huang Y. 2013. Partitioning of the nuclear and mitochondrial tRNA 3′-end processing activities between two different proteins in Schizosaccharomyces pombe. J Biol Chem 288: 27415–27422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Su W, Yuan S, Huang Y. 2009. Functional conservation of tRNase ZL among Saccharomyces cerevisiae, Schizosaccharomyces pombe and humans. Biochem J 422: 483–492. [DOI] [PubMed] [Google Scholar]
- Zhao W, Yu H, Li S, Huang Y. 2010. Identification and analysis of candidate fungal tRNA 3′-end processing endonucleases tRNase Zs, homologs of the putative prostate cancer susceptibility protein ELAC2. BMC Evol Biol 10: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. 2015. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods 12: 835–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zhou M, Yang Y, Li J, Zhu L, Jiang D, Dong J, Liu Q, Gu L, Zhou L, et al. 2014. RNase ZS1 processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice. Nat Commun 5: 4884. [DOI] [PubMed] [Google Scholar]
- Zhou W, Karcher D, Fischer A, Maximova E, Walther D, Bock R. 2015. Multiple RNA processing defects and impaired chloroplast function in plants deficient in the organellar protein-only RNase P enzyme. PLoS One 10: e0120533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.