Abstract
The extracellular matrix (ECM) plays a crucial role in embryogenesis, serving both as a substrate to which cells attach and as an active regulator of cell behavior. However, little is known about the spatiotemporal expression patterns and 3D structure of ECM proteins during embryonic development. The lack of suitable methods to visualize the embryonic ECM is largely responsible for this gap, posing a major technical challenge for biologists and tissue engineers. Here, we describe a method of viewing the 3D organization of the ECM using a polyacrylamide-based hydrogel to provide a 3D framework within developing murine embryos. After removal of soluble proteins using sodium dodecyl sulfate, confocal microscopy was used to visualize the 3D distribution of independent ECM networks in multiple developing tissues, including the forelimb, eye, and spinal cord. Comparative analysis of E12.5 and E14.5 autopods revealed proteoglycan-rich fibrils maintain connections between the epidermis and the underlying tendon and cartilage, indicating a role for the ECM during musculoskeletal assembly and demonstrating that our method can be a powerful tool for defining the spatiotemporal distribution of the ECM during embryogenesis.
Keywords: Extracellular matrix, morphogenesis, mouse embryo, decellularization
Introduction
The ECM is an intricate network of proteins, proteoglycans, glycosaminoglycans, and other macromolecules. It has several main functions, including providing structure to tissue and regulating cellular behavior during growth, repair, regeneration, and homeostasis (Bonnans et al., 2014; Mouw et al., 2014). During embryogenesis, the ECM provides structural support and mechanical cues to direct global tissue movement and organization (Swinehart and Badylak, 2016). Furthermore, the ECM locally controls cell behavior by defining the stiffness of the cellular microenvironment and activating intracellular signaling via binding with cell surface receptors (Schwartz, 2010). In addition, the ECM is thought to play a key role in paracrine signaling vital to development through growth factor sequestration (de Almeida et al., 2016; Wohl et al., 2016). While it is generally accepted that ECM composition undergoes dramatic remodeling during tissue formation, leading to changes in tissue mechanics and cellular behavior, little is known about how the 3D organization of these ECM components varies as a function of development.
Visualization of the ECM in 3D has been hindered by the inherent difficulty of high resolution imaging within opaque biological tissues. Optical clearing is a powerful approach to enhance the ability to resolve 3D tissue architecture by either reducing the amount and diversity of light scattering elements (e.g. lipids) or minimizing refractive index mismatches (Orlich and Kiefer, 2017; Richardson and Lichtman, 2015). Hydrogel-based clearing techniques have been developed to address the issue of protein loss when using harsh solvents or detergents to remove light scattering biomolecules by first embedding the issue in hydrogel (Richardson and Lichtman, 2015). The tissue is incubated in a monomeric solution followed by polymerization triggered by a thermal initiator (Chung et al., 2013). As polymerization occurs, a hybrid network composed of proteins and the polymer chains of the hydrogel is created that allows for the removal of lipids and other light-scattering biomolecules.
Optical clearing methods based on solutions that match the refractive index of tissue are also becoming more prevalent (Orlich and Kiefer, 2017; Richardson and Lichtman, 2015). One example, SeeDB, is a concentrated aqueous fructose solution with a refractive index close to that of fixed tissue (n~1.49) that greatly enhances the depth at which a variety of tissues can be imaged using confocal microscopy ranging from embryos to dense connective tissues (Calve et al., 2015; Ke et al., 2013). However, not all solvent-based clearing reagents are compatible with immunostaining (Hou et al., 2015), and for those techniques that are, tissue penetration and fluorescent signal attenuation are of concern (Hama et al., 2015; Richardson and Lichtman, 2015).
Previous optical clearing-based approaches have improved the visualization of ECM proteins such as type IV collagen in the rat brain (Lee et al., 2016) and proteoglycans in bovine musculoskeletal tissues (Calve et al., 2015; Neu et al., 2015). Nonetheless, most clearing techniques were designed to resolve cellular structures and there have been few efforts to visualize the organization of the ECM fibrils within intact tissues. To address this need, we developed a method to selectively remove intracellular proteins while leaving behind proteins of the ECM in the native 3D conformation. Typically, decellularization involves using detergents to extract cell membranes and other components (Crapo et al., 2011); however, most decellularization techniques are harsh to the sample and generally only applicable to ECM-rich tissues (Lumpkins et al., 2008; Nakayama et al., 2010). Softer, embryonic tissues are highly cellular and susceptible to damage (Taghiabadi et al., 2015). Therefore, we modified established hydrogel-based clearing methods such that the hydrogel only cross-linked to itself, allowing for removal of soluble, intracellular proteins while providing a 3D framework for the remaining insoluble ECM networks (Chung et al., 2013; Yang et al., 2014). Decellularization significantly enhanced the visualization of ECM proteins and maintained the 3D architecture of independent networks. Using confocal microscopy, we demonstrate that the ECM within intact E12.5 embryos can be visualized at multiple scales. Comparative analysis of the autopod of E12.5 and E14.5 forelimbs revealed continuous proteoglycan-rich fibrils extended between the epidermis and cartilage, which remained present after the formation of the extensor tendons. Overall, we expect our method will be a powerful tool for investigating how the ECM remodels during tissue assembly.
Results and Discussion
Decellularization increases embryo transparency
To enhance the visibility of the developing matrix, freshly harvested embryonic tissues were decellularized following standard detergent-based methods formulated for adult tissues (Crapo et al., 2011), but we observed that the tissues did not maintain their macroscopic morphology and were easily damaged. Instead, we found that by removing the paraformaldehyde from the hydrogel utilized in the CLARITY method (Chung et al., 2013), a hydrogel framework was formed that did not cross-link to biomolecules and retained the 3D organization of the insoluble matrix (Fig. 1). Hydrogel-based protocols tend to surround samples with an excess of solution, which generates superficial hydrogel after polymerization that needs to be removed prior to imaging (Chung et al., 2013; Yang et al., 2014). Unlike stiffer adult tissues, fragile embryonic samples can be damaged during this process. To address this problem, excess hydrogel solution was removed before polymerization and the embryos were placed in mineral oil, which is immiscible with the aqueous acrylamide-based hydrogel. After polymerization, there was minimal hydrogel on the surface of the specimen that could interfere with confocal imaging. Additionally, the hydrogel increased tissue rigidity, making the samples easier to handle and less susceptible to damage during staining and imaging.
Fig. 1. Decellularization increases transparency of murine embryos.
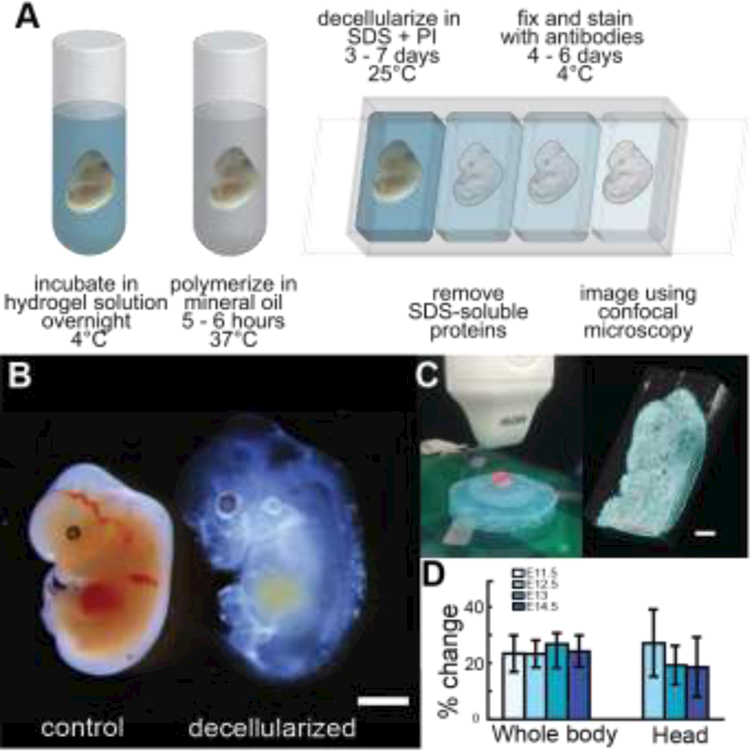
(A) Isolated murine embryos were incubated in hydrogel solution (Table S1) and gently rocked at 4°C overnight. Excess hydrogel was removed, and embryos were submerged in mineral oil at 37°C to induce polymerization. Embryos were decellularized in sodium dodecyl sulfate (SDS) with protease inhibitor (PI) then processed for immunohistochemistry. (B) Comparison of control and decellularized E12.5 embryos from the same litter showed swelling occurred after cell removal; however, overall morphology was retained. Bar = 2 mm. (C) Ultrasound setup used to quantify changes in embryonic volume and segmentations (blue) of a E14.5 embryo before decellularization for whole body 3D rendering. Bar = 2 mm. (D) Volume changes remained consistent over the time periods investigated (n=4; bars=SD) and were largely isotropic (see Fig. S2).
Since the nascent embryonic ECM is likely to be less stable than ECM found in adult tissues, preliminary studies to determine the minimal concentration of sodium dodecyl sulfate (SDS) that increased tissue transparency, while also preserving ECM architecture, were conducted. Network visualization was greatly enhanced after treatment with 0.05% SDS in PBS (Fig. S1). Staining for nucleic acids (DAPI, blue) indicated that tissues were not fully decellularized (Figs. S1, 2A); however, the primary goal of this study was to increase matrix visibility and not to create scaffolds devoid of cellular components for implantation where DNA and proteins can promote an immune response. Since the ability to resolve ECM architecture was greatly enhanced even with the presence of residual DNA, further optimization of the decellularization process was not conducted to minimize disruption of ECM network architecture.
Fig. 2. Decellularization increases matrix visibility and retains independent networks.
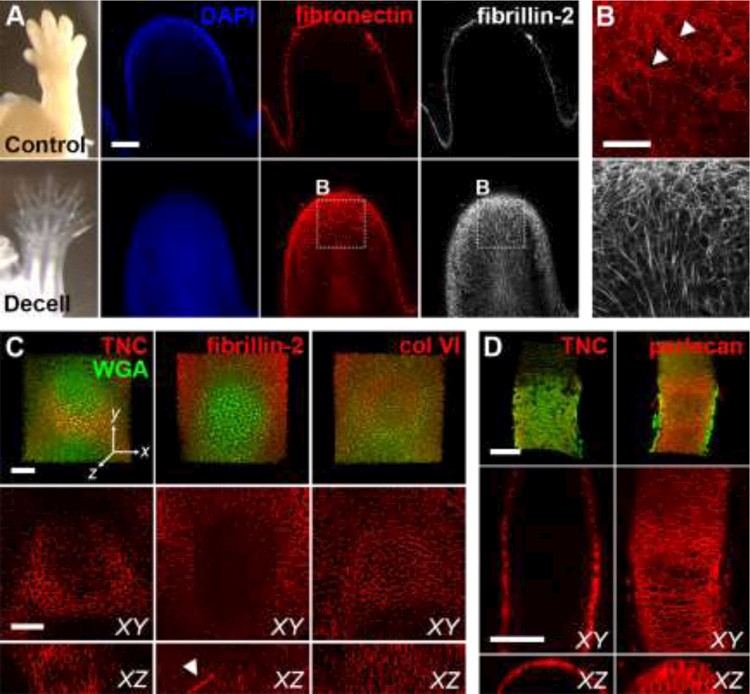
(A) E14.5 control and decellularized forelimbs were stained for fibronectin (FN; red) and fibrillin-2 (FBN2; white) and digit tips were imaged using confocal microscopy (blue = DAPI). Bar = 100µm. (B) Higher magnification view of A reveals FN and FBN2 maintain independent, interpenetrating networks after removal of cells with SDS. Arrowheads show FN+ blood vessels. Bar = 50µm. (C, D) The native distribution of different ECM was maintained in developing cartilage and bone in decellularized E14.5 forelimbs. Tenascin-C (TNC, red) expression was restricted to the periphery of the developing cartilage (left). FBN2 (red) was localized to fibrils within the cartilage (middle). YZ section shows extended FBN2-expressing microfibril (arrowhead). Type VI collagen (col VI, red) was found throughout the developing cartilage (right). Green = WGA. Bars = 50 μm. Dimensions of stacks in C: 237 × 237 × 78 μm (x × y × z). (D) TNC (red, left) was restricted to the perichondrium/periosteum in the developing long bone, whereas perlecan (red, right) was found throughout the tissue. Green = WGA. Bars = 100 μm, axes oriented as in C. Dimensions of stacks in D: 242 × 338 × 69 μm.
Removal of soluble components greatly increased embryo transparency; however, noticeable swelling occurred (Fig. 1B). Swelling was attributed to removal of cellular links between matrices and/or from the inherent swelling properties of polyacrylamide (Tanaka and Fillmore, 1979). To assess if swelling was isotropic, changes in embryo volume were quantified using ultrasound before and after incubation in 0.05% SDS. Swelling caused by decellularization remained constant across all time points investigated and was generally isotropic (Fig. 1B). Expansion in the direction of the sagittal plane tended to be lower than in other directions but was only significant at E14.5 (Fig. S2). Significant differences between changes in area at E14.5 were also observed between coronal, transverse and sagittal planes, which was attributed to swelling constraints brought upon by the formation and connection of cartilage elements in the developing skeleton, particularly the vertebral column (Fig. S2). Sample expansion is not typically considered detrimental to the 3D study of native tissue and has been documented during the implementation of techniques such as CUBIC (Susaki et al., 2014), CLARITY (Chung et al., 2013), and PACT (Yang et al., 2014). Indeed, tissue swelling has been used to enhance the resolution of smaller samples as demonstrated with expansion microscopy in which a hydrogel framework swells to 4.5× the original size and does not negatively influence cellular architecture (Chen et al., 2015).
Independent ECM networks are maintained after cell removal
To demonstrate that our method increases ECM visibility and maintains independent protein networks, control and decellularized E14.5 digit tips were imaged at approximately the same depth. The structure of fibronectin (FN) and fibrillin-2 (FBN2) within control tissue could not be visualized beyond the surface (Fig. 2A). In contrast, independent and interpenetrating networks of FN and FBN2 were clearly resolved in the decellularized samples (Fig. 2B). FN was widely distributed and outlined the blood vessels (arrowheads; Fig. 2B), consistent with the proposed role for FN during vasculogenesis (George et al., 1993; Risau and Lemmon, 1988). FBN2 was arranged in a dense network of microfibrils, similar to what has been shown in mouse hindlimb (Arteaga-Solis et al., 2001).
To confirm SDS treatment did not interfere with the spatial organization of ECM at the tissue level, E14.5 forelimbs were stained for proteins with known differential distributions in developing cartilage and images were collected of the distal phalanx and long bones in the zeugopod. Samples were counterstained with AF488-conjugated wheat germ agglutinin (WGA; green), which binds to proteoglycans containing sialic acid and N-acetylglucosamine and provides a global label for musculoskeletal tissue ECM (Kostrominova, 2011). As expected, tenascin-C expression (TNC; red) was restricted to the periphery of the cartilage condensation (Fig. 2C; (Ros et al., 1995; Swinehart et al., 2013)), which persisted during ossification (Fig. 2D; (Koyama et al., 1996)). FBN2 (red) was distributed as discrete microfibrils within cartilage and was present at a higher density in the surrounding mesenchyme (Fig. 3C; (Zhang et al., 1995)). In contrast, type VI collagen (col VI) and perlecan were found throughout the entire cartilage (Fig. 3C,D). While col VI and perlecan are associated with the pericellular ECM in adult cartilage, both are widely distributed during the early stages of chondrogenesis and gradually become restricted to the periphery of the chondrocytes as development progresses (Xu et al., 2016). In addition, perlecan is highly expressed during the transition from cartilage to bone (Arikawa-Hirasawa et al., 1999).
Fig. 3. 3D visualization of ECM in E12.5 embryo.
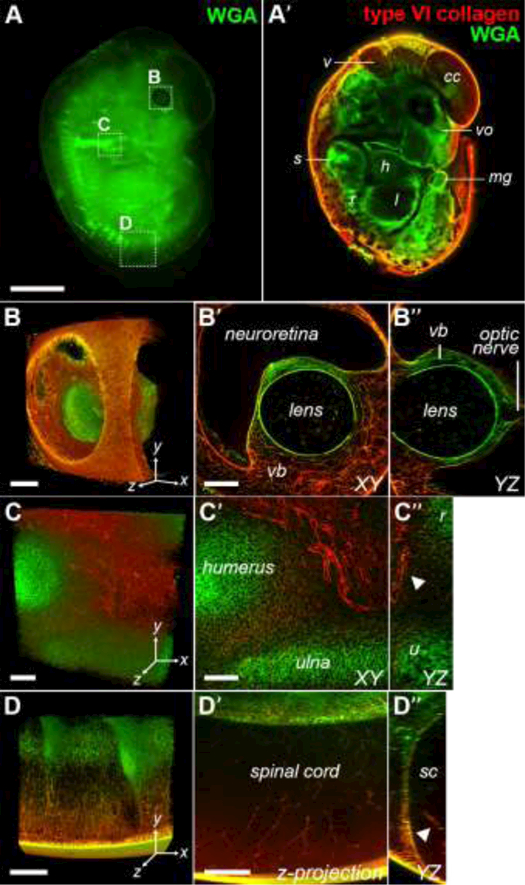
(A) Widefield view of WGA-stained E12.5 decellularized embryo. (A’) Virtual confocal section of E12.5 embryo stained with WGA (green) and type VI collagen (red). Various structures can be identified including the including the 4th ventricle of the brain (v), future cerebral cortex (cc), vomeronasal organ (vo), scapula (s), midgut (m), liver (l), heart (h) and ribs (r). 10×, bar = 2 mm. (B-B”): 3D rendering of the eye at 25× (567 × 567 × 395 μm; x × y × z). The vascularization of the vitreous body (vb) can be visualized (B’) and the lens and optic nerve can be clearly resolved in a YZ projection of the image stack. Bars = 100 μm. (C-C”) 3D rendering reveals dense vascular network around the elbow (567 × 567 × 186 μm). Comparison of XY (C’) and YZ planes (C’’) revealed that blood vessels remained patent. 25×, bars = 100 μm. (D-D”) 3D rendering of developing spinal cord at 10× (850 × 850 × 251 μm). Z-projection of 12 slices (70 μm) shows maintenance of spinal cord vascularization (D’) and intact blood vessel in the YZ projection (arrowhead; D”). Bars = 200 μm.
The ability of our method to resolve matrix architecture was compared with SeeDB. FBN2 (red) and WGA (green) visualization was greatly enhanced in decellularized samples (Fig. S3). The ECM of a tendon within the zeugopod was intensely stained in the decellularized tissue, whereas little to no FBN2 signal was observed in the same tendon in the limb cleared with SeeDB, which was attributed to limited antibody penetration in the denser SeeDB sample.
Intact embryos can be visualized at multiple scales
To demonstrate the macroscopic structure of developing tissue was maintained, E12.5 embryos were stained with WGA (green) and an antibody against col VI (red; Fig. 3). Skeletal elements of the forelimbs and ECM supporting the dorsal root ganglia along the developing spinal cord were easily resolved under a dissecting microscope (Fig. 3A). Sagittal confocal slices enabled the identification of internal organs and tissues within the embryo, including the 4th ventricle of the brain (v), future cerebral cortex (cc), vomeronasal organ (vo), scapula (s), midgut (m), liver (l), heart (h) and ribs (r; Fig. 3B). WGA (MW = 38 kDa) diffused through the entire specimen, whereas the antibody against col VI (MW = 150 kDa) could only be imaged to a depth of 150 – 200 µm. The limited depth of staining can be attributed to a large number of type VI epitopes in the surface tissues rather than a limitation in diffusion as an antibody against perlecan was able to penetrate further into the same sample (Fig. S4). We expect that increasing the concentration of anti-type VI collagen will enhance the depth of labeling.
The developing eye could be clearly visualized at 25x, wherein blood vessels of the vitreous body (vb) were outlined with col VI (Fig. 3B). In contrast, the predominantly cellular neuroretina and lens were devoid of signal. An orthogonal cross-section of the z-stack from Fig. 3B shows that lens architecture and stalk of the optic nerve were maintained (Fig. 3B’B”). Cartilage elements of the elbow were intensely stained with WGA and were surrounded by a col VI+ network of patent blood vessels (arrowhead, Fig. 3C-C”). WGA staining formed an inverse pattern with the dorsal root ganglia along the developing spinal cord (Fig. 3A,D), which is likely made up of proteoglycans such as versican that inhibit neural crest and axon migration (Landolt et al., 1995). Notably, the intricate architecture of the vasculature within the spinal cord was maintained (arrowhead; Fig. 3D’,D”), providing further evidence that our method can maintain the integrity of delicate networks.
The epidermis is linked to the underlying musculoskeletal tissues via proteoglycan–rich fibrils
Comparative analysis of the developing forelimbs between E12.5 and E14.5 revealed that FBN2 expression decreased between the time points (Fig. 4A), consistent with a decrease in FBN2 transcripts as a function of limb development (Arteaga-Solis et al., 2001). The increase in the number of phalanges and articulation between the joints as development progresses was resolved by WGA labeling (Fig. 4B,E, Movies 1,2). Higher resolution imaging of the ECM in the digit revealed continuous proteoglycan-based fibrils extending from the epidermis (epi) to the developing cartilage (c), even after the formation of the extensor tendons (t; Fig. 4C,F; Movies 3,4). Cryosections of control E14.5 limbs show FBN2+ fibrils extending between the ventral epidermis and internal tissues (Fig. S5), indicating that the architecture visualized in Figs. 4C and 4F is likely not an artifact of the decellularization process. Recent genetic knockdown studies indicated that signals secreted by cartilage and epidermis, such as Wnts and Noggin, can influence the induction of autopod tendons (Huang et al., 2015; Yamamoto-Shiraishi and Kuroiwa, 2013). The persistence of proteoglycan-rich connections between the epidermis and cartilage during tendon specification from E12.5 to E14.5, in combination with previous studies showing that elastic fibrils appear to anchor developing tendons to the perichondrium in the developing chick limb (Hurle et al., 1994; Hurle et al., 1989), suggest that the matrix also plays a role in regulating extensor tendon development in the autopod.
Fig. 4. Multiscale imaging of the musculoskeletal system in the developing forelimb.
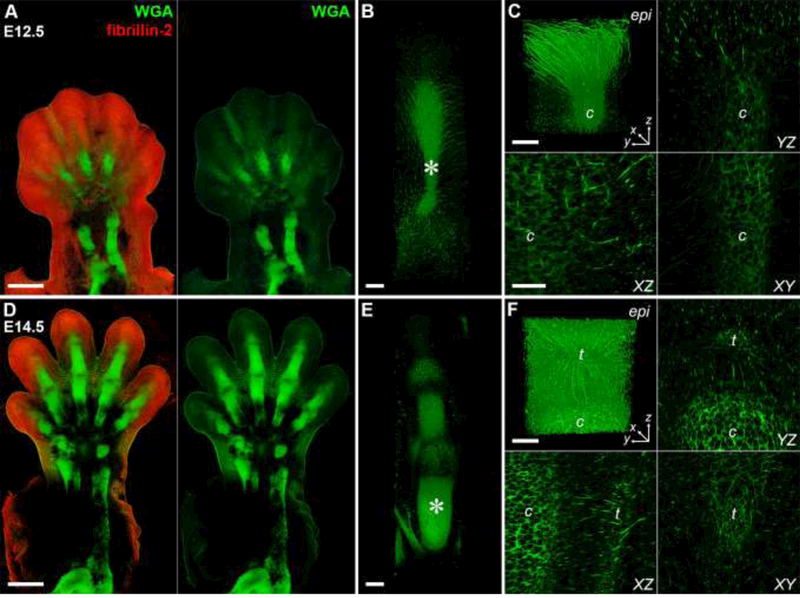
E12.5 (A – C) and E14.5 (D – F) forelimbs were decellularized and stained with WGA (green) and an antibody against fibrillin-2 (FBN2, red). Dorsal view. (A, D) Cartilage elements of the autopod and zeugopod were easily resolved with WGA. FBN2 became more restricted with time. 10×, bar = 500 μm. (A: 2471 × 3599 × 600 μm; D: 2799 × 4733 × 450 μm; x × y × z) (B, E) 3D rendering of digits showed a network of WGA+ fibrils radiating from the cartilage as well as staining of joint tissues and the extensor tendons. 25×, bar = 100 μm. (381 × 1364 × 289 μm). (C, F) Higher magnification of digits in B, E (*) revealed WGA+ fibrils extended from dorsal epidermis (epi) to cartilage (c) at E12.5 and at E14.5 fibrils appeared to connect the extensor tendons (t) to the epidermis and cartilage. 40×, bars = 50 μm, (213 × 213 × 227 μm). See Movies 1 – 4.
The goal of this study was to establish a method that would enable us to visualize the ECM during the early stages of musculoskeletal development (E11.5 – E14.5). Our method will have limited application for intact embryos after E15.5 when the barrier function of the skin is acquired (Hardman et al., 1998), preventing the diffusion of the hydrogel solution into the tissues. Nevertheless, we expect that it should be feasible to process isolated tissues from most developmental stages. It may be necessary to increase the SDS concentration as the tissues become more dense; however, by those stages, the stabilization of the geometry using the hydrogel will likely be unnecessary. Our future studies aim to investigate more finely resolved time points so that a clearer picture will emerge regarding the role the ECM plays during forelimb musculoskeletal assembly as well as all other organ systems within the developing embryo.
Materials and Methods
Unless otherwise specified, all reagents were of chemical grade from Sigma-Aldrich.
Embryo harvest
Tissues from E11.5-E14.5 mice were generated by the timed matings of wild type C57Bl/6 mice. All murine experiments were approved by the Purdue Animal Care and Use Committee (PACUC; protocol 1209000723). PACUC ensures that all animal programs, procedures, and facilities at Purdue University adhere to the policies, recommendations, guidelines, and regulations of the USDA and the United States Public Health Service in accordance with the Animal Welfare Act and Purdue’s Animal Welfare Assurance. Dams were euthanized via CO2 inhalation, confirmed by cervical dislocation. Embryos were removed from the uterine horns and rinsed in 1× PBS. Dissection of the yolk sac and amnion was performed under magnification to avoid damaging the embryos.
Embryo decellularization
Murine embryos were decellularized and imaged as depicted in Fig. 1. After dissection of yolk sac and amnion, embryos were rinsed in 1× PBS, submerged in 1 – 1.5 mL hydrogel solution (Table S1) in a 2.0 mL microcentrifuge tube, and gently rocked overnight at 4°C. Excess hydrogel solution was removed and embryos were immersed in mineral oil in a round bottom 14 mL round bottom polypropylene tube (Falcon 352059). Tubes were sealed with Parafilm before being placed in a 37°C water bath for 5–6 hours to polymerize the hydrogel. Embryos were removed from mineral oil and rinsed three times in 1× PBS to remove excess oil. Each embryo was gently rocked in 1mL 0.05% SDS with 1× HaltTM protease inhibitor (PI, Thermo Scientific, 78430) in 1× PBS in a dish suitable for confocal imaging (Ibidi, 80421) at 25°C. Preliminary experiments showed that 0.05% SDS was the best compromise between cell removal and maintenance of networks (Fig. S1). SDS-PI solution was replaced every 24 hours until decellularization was complete (3–7 days). Completion of decellularization was determined by subjective visual inspection of samples for presence of cells with a light microscope, with variation in timing depending on the age and natural biological variability among samples being decellularized. Upon decellularization, samples were rinsed in an excess of 1× PBS for 1 hour, fixed in 4% paraformaldehyde (PFA) for 1 hour and then rinsed again in 1× PBS for 1 hour at 25°C with gentle rocking.
To confirm decellularization enhanced visualization of the ECM and demonstrate that network structure was maintained, littermates serving as whole-mount and cleared controls were rinsed in 1× PBS immediately after harvest and then fixed in 4% PFA overnight at 4°C. After PFA fixation, control embryos were transferred to 1× PBS for storage at 4°C until processed for imaging as described below.
Fluorescent labeling of ECM
After PFA fixation, decellularized and control samples were incubated in blocking buffer [10% donkey serum diluted in 1× PBS with 1% Triton X-100 (PBST) and 0.02% sodium azide] for 16 hours at 4°C to increase the ability of antibody stains to permeate through the sample and to block non-specific binding. Samples were then incubated with primary antibodies (Table S2) diluted in blocking buffer, and gently rocked at 4°C for 48 hours. Samples were rinsed 3×30 minutes with 1% PBST at 25°C, and then incubated with secondary staining reagents (Table S2) diluted in blocking buffer, placed in a lightproof container, and gently rocked at 4°C for 48 hours. Samples were rinsed 3×30 minutes with 1% PBST at 25°C and stored in 1× PBS at 4°C until imaged.
Clearing of control tissues
To confirm that ECM structure was maintained after decellularization, tissues were cleared following (Ke et al., 2013). D-(−)-fructose (JT Baker, Center Valley, PA) was dissolved in milliQ water with 0.5% α - thioglycerol (Sigma-Aldrich St. Louis, MO) to generate fructose solutions of varying concentrations (20%, 40%, 60%, 80%, 100% and 115% wt/vol). Tissues were equilibrated to increasing concentrations of fructose by incubating in each formulation under gentle rocking at room temperature, following the timeline defined in (Ke et al., 2013).
Cryosectioning of control tissues
To visualize the fibrillar structure of ECM within intact tissues, freshly harvested E14.5 forelimbs were embedded in Optimal Cutting Temperature compound (Sakura Finetek), frozen with dry ice-cooled isopentane and stored at 80°C. 17 µm-thick cryosections were collected on His-bond glass slides (VWR) and processed following (Calve et al., 2010) using the antibody concentrations defined in Table 2. Sections were imaged at 25× using a Zeiss LSM 880 confocal.
Imaging
Decellularized and control samples were imaged using one of the following confocal microscopes: 1) an inverted Zeiss LSM 710 confocal using either a 10× Plan-Neofluar (NA = 0.3) or a 25× multi-immersion LD LCI Plan-Apochromat (NA = 0.8) objective (Figs. 2A,B; S1), 2) an upright Zeiss LSM 800 confocal using either the 10× Plan-Neofluar (NA = 0.3), a 20× water immersion Plan-Apochromat (NA = 1.0) objective or a 63× water immersion C-Apochromat (NA = 1.2) (Figs. 4A,D; S3) or 3) an inverted Zeiss LSM 880 confocal using either the 10× EC-Plan NeoFluar (NA = 0.3), 25× multi-immersion LD LCI Plan-Apochromat (NA = 0.8) or a 40× water immersion C-Apochromat (NA = 1.2) objective (Figs. 2C,D; 3; 4B,C,E,F). Widefield images were acquired using a Leica M80 stereo microscope.
Image processing
Confocal stacks were rendered in 3D using FIJI (NIH; (Schindelin et al., 2012). Background antibody staining in some images was reduced using the despeckle algorithm in FIJI (Figs. 2C,D; 3; 4), which uses a 3×3 median filter to decrease noise. Due to the attenuation of signal when imaging deeper within tissues, some images were processed using the Enhance Local Contrast (CLAHE) plugin to equalize large variations in intensity across tiled image stacks (Figs. 3A’; 4A,D). For cryosections, image stacks were processed into maximum z-projections using FIJI (Fig. S5). Figures were assembled by taking snapshots of 3D-rendered image volumes or selecting 2D slices from stacks and were arranged using Adobe Photoshop and Illustrator.
Ultrasound quantification of embryo swelling
A high frequency small animal ultrasound system (Vevo2100; FUJIFILM VisualSonics, Toronto, ON, Canada) was used to quantify the change in embryo dimensions during decellularization. Embryos were imaged immediately after polymerization and again after completion of decellularization (3–7 days). Embryos were placed in an agarose bed and covered with ultrasound transmission gel. A 50 MHz center frequency transducer with an axiolateral resolution of 30×75 µm2 and an elevation resolution of 0.154 mm was positioned perpendicular to the embryo (Fig. 1C). Similar to previous work (Phillips et al., 2015), sequential transaxial brightness mode (B-mode) images were combined to create ultrasound volumes (0.19 mm step size). Images were analyzed with Vevo LAB software (FUJIFILM VisualSonics) to obtain area and volume measurements by manually segmenting the embryonic head and whole body (Fig. 1C). Area measurements in the sagittal, transverse, and coronal planes were used to determine if tissue expansion was isotropic (Fig. S2).
Statistical analyses
Percent change during decellularization was calculated by subtracting the area and volume after decellularization from the initial measurements. Tukey-Kramer HSD was performed on the percentage area change in the transverse, coronal, and sagittal planes (Fig. S2) at E12.5, E13, and E14.5 (n=4) for statistical significance using JMP. Similarly, the percentage change in whole embryo and head volumes at these embryonic stages (n=4) were analyzed with Tukey-Kramer HSD for statistical significance. Confidence quantile alpha was set at 0.05.
Supplementary Material
Highlights.
3D organization of ECM can be resolved in hydrogel-embedded, decellularized embryos
Independent, interpenetrating ECM networks are maintained
Proteoglycan-rich fibrils connect epidermis to underlying tissues in the autopod
Acknowledgments
The authors would like to thank Dr. Robert Mecham for kindly providing the fibrillin-2 antibody and Dr. David Umulis for the use of his Zeiss 800 confocal microscope.
Funding
This work was supported by the National Institutes of Health [R03 AR065201, R21 AR069248, R01 AR071359 and DP2 AT009833 to S.C.].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare no competing interests.
Contributor Information
Michael A. Drakopoulos, Weldon School of Biomedical Engineering, Purdue University, 206 South Martin Jischke Drive, West Lafayette, IN 47907
Yue Leng, Weldon School of Biomedical Engineering, Purdue University, 206 South Martin Jischke Drive, West Lafayette, IN 47907.
Craig J. Goergen, Weldon School of Biomedical Engineering, Purdue University, 206 South Martin Jischke Drive, West Lafayette, IN 47907
References
- Arikawa-Hirasawa E, Watanabe H, Takami H, Hassell JR, Yamada Y, 1999. Perlecan is essential for cartilage and cephalic development. Nat Genet 23, 354–358. [DOI] [PubMed] [Google Scholar]
- Arteaga-Solis E, Gayraud B, Lee SY, Shum L, Sakai L, Ramirez F, 2001. Regulation of limb patterning by extracellular microfibrils. J Cell Biol 154, 275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnans C, Chou J, Werb Z, 2014. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol 15, 786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calve S, Odelberg SJ, Simon HG, 2010. A transitional extracellular matrix instructs cell behavior during muscle regeneration. Dev Biol 344, 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calve S, Ready A, Huppenbauer C, Main R, Neu CP, 2015. Optical clearing in dense connective tissues to visualize cellular connectivity in situ. PLoS One 10, e0116662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tillberg PW, Boyden ES, 2015. Expansion microscopy. Science 347, 543 -- 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, Pak S, Bernstein H, Ramakrishnan C, Grosenick L, Gradinaru V, Deisseroth K, 2013. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crapo PM, Gilbert TW, Badylak SF, 2011. An overview of tissue and whole organ decellularization processes. Biomaterials 32, 3233–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida PG, Pinheiro GG, Nunes AM, Goncalves AB, Thorsteinsdottir S, 2016. Fibronectin assembly during early embryo development: A versatile communication system between cells and tissues. Dev Dyn 245, 520–535. [DOI] [PubMed] [Google Scholar]
- George EL, Georges-Labouesse EN, Patel-King RS, Rayburn H, Hynes RO, 1993. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development 119, 1079–1091. [DOI] [PubMed] [Google Scholar]
- Hama H, Hioki H, Namiki K, Hoshida T, Kurokawa H, Ishidate F, Kaneko T, Akagi T, Saito T, Saido T, Miyawaki A, 2015. ScaleS: an optical clearing palette for biological imaging. Nat Neurosci 18, 1518–1529. [DOI] [PubMed] [Google Scholar]
- Hardman MJ, Sisi P, Banbury DN, Byrne C, 1998. Patterned acquisition of skin barrier function during development. Development 125, 1541–1552. [DOI] [PubMed] [Google Scholar]
- Hou B, Zhang D, Zhao S, Wei M, Yang Z, Wang S, Wang J, Zhang X, Liu B, Fan L, Li Y, Qiu Z, Zhang C, Jiang T, 2015. Scalable and DiI-compatible optical clearance of the mammalian brain. Front Neuroanat 9, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AH, Riordan TJ, Pryce B, Weibel JL, Watson SS, Long F, Lefebvre V, Harfe BD, Stadler HS, Akiyama H, Tufa SF, Keene DR, Schweitzer R, 2015. Musculoskeletal integration at the wrist underlies the modular development of limb tendons. Development 142, 2431–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurle JM, Corson G, Daniels K, Reiter RS, Sakai LY, Solursh M, 1994. Elastin exhibits a distinctive temporal and spatial pattern of distribution in the developing chick limb in association with the establishment of the cartilaginous skeleton. J Cell Sci 107 ( Pt 9), 2623–2634. [DOI] [PubMed] [Google Scholar]
- Hurle JM, Hinchliffe JR, Ros MA, Critchlow MA, Genis-Galvez JM, 1989. The extracellular matrix architecture relating to myotendinous pattern formation in the distal part of the developing chick limb: an ultrastructural, histochemical and immunocytochemical analysis. Cell Differ Dev 27, 103–120. [DOI] [PubMed] [Google Scholar]
- Ke MT, Fujimoto S, Imai T, 2013. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci 16, 1154–1161. [DOI] [PubMed] [Google Scholar]
- Kostrominova TY, 2011. Application of WGA lectin staining for visualization of the connective tissue in skeletal muscle, bone, and ligament/tendon studies. Microsc Res Tech 74, 18–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama E, Shimazu A, Leatherman JL, Golden EB, Nah HD, Pacifici M, 1996. Expression of syndecan-3 and tenascin-C: possible involvement in periosteum development. J Orthop Res 14, 403–412. [DOI] [PubMed] [Google Scholar]
- Landolt RM, Vaughan L, Winterhalter KH, Zimmermann DR, 1995. Versican is selectively expressed in embryonic tissues that act as barriers to neural crest cell migration and axon outgrowth. Development 121, 2303–2312. [DOI] [PubMed] [Google Scholar]
- Lee E, Choi J, Jo Y, Kim JY, Jang YJ, Lee HM, Kim SY, Lee HJ, Cho K, Jung N, Hur EM, Jeong SJ, Moon C, Choe Y, Rhyu IJ, Kim H, Sun W, 2016. ACT-PRESTO: Rapid and consistent tissue clearing and labeling method for 3-dimensional (3D) imaging. Sci Rep 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumpkins SB, Pierre N, McFetridge PS, 2008. A mechanical evaluation of three decellularization methods in the design of a xenogeneic scaffold for tissue engineering the temporomandibular joint disc. Acta Biomater 4, 808–816. [DOI] [PubMed] [Google Scholar]
- Mouw JK, Ou G, Weaver VM, 2014. Extracellular matrix assembly: a multiscale deconstruction. Nat Rev Mol Cell Biol 15, 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama KH, Batchelder CA, Lee CI, Tarantal AF, 2010. Decellularized Rhesus Monkey Kidney as a Three-Dimensional Scaffold for Renal Tissue Engineering. Tissue Eng Pt A 16, 2207–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu CP, Novak T, Gilliland KF, Marshall P, Calve S, 2015. Optical clearing in collagen- and proteoglycan-rich osteochondral tissues. Osteoarthritis Cartilage 23, 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlich M, Kiefer F, 2017. A qualitative comparison of ten tissue clearing techniques. Histol Histopathol 10. [DOI] [PubMed] [Google Scholar]
- Phillips EH, Yrineo AA, Schroeder HD, Wilson KE, Cheng JX, Goergen CJ, 2015. Morphological and Biomechanical Differences in the Elastase and AngII apoE(−/−) Rodent Models of Abdominal Aortic Aneurysms. Biomed Res Int 2015, 413189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson DS, Lichtman JW, 2015. Clarifying Tissue Clearing. Cell 162, 246–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risau W, Lemmon V, 1988. Changes in the vascular extracellular matrix during embryonic vasculogenesis and angiogenesis. Dev Biol 125, 441–450. [DOI] [PubMed] [Google Scholar]
- Ros MA, Rivero FB, Hinchliffe JR, Hurle JM, 1995. Immunohistological and ultrastructural study of the developing tendons of the avian foot. Anat Embryol (Berl) 192, 483–496. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MA, 2010. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb Perspect Biol 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, Abe T, Kiyonari H, Shimizu Y, Miyawaki A, Yokota H, Ueda HR, 2014. Whole-Brain Imaging with Single-Cell Resolution Using Chemical Cocktails and Computational Analysis. Cell 157, 726–739. [DOI] [PubMed] [Google Scholar]
- Swinehart IT, Badylak SF, 2016. Extracellular matrix bioscaffolds in tissue remodeling and morphogenesis. Dev Dyn 245, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinehart IT, Schlientz AJ, Quintanilla CA, Mortlock DP, Wellik DM, 2013. Hox11 genes are required for regional patterning and integration of muscle, tendon and bone. Development 140, 4574–4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghiabadi E, Nasri S, Shafieyan S, Jalili Firoozinezhad S, Aghdami N, 2015. Fabrication and characterization of spongy denuded amniotic membrane based scaffold for tissue engineering. Cell J 16, 476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Fillmore DJ, 1979. Kinetics of swelling of gels. The Journal of Chemical Physics 70, 1214–1218. [Google Scholar]
- Wohl AP, Troilo H, Collins RF, Baldock C, Sengle G, 2016. Extracellular Regulation of Bone Morphogenetic Protein Activity by the Microfibril Component Fibrillin-1. J Biol Chem 291, 12732–12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Li Z, Leng Y, Neu CP, Calve S, 2016. Knockdown of the pericellular matrix molecule perlecan lowers in situ cell and matrix stiffness in developing cartilage. Dev Biol 418, 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto-Shiraishi Y, Kuroiwa A, 2013. Wnt and BMP signaling cooperate with Hox in the control of Six2 expression in limb tendon precursor. Dev Biol 377, 363–374. [DOI] [PubMed] [Google Scholar]
- Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, Shah S, Cai L, Gradinaru V, 2014. Single-Cell Phenotyping within Transparent Intact Tissue through Whole-Body Clearing. Cell 158, 945–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hu W, Ramirez F, 1995. Developmental expression of fibrillin genes suggests heterogeneity of extracellular microfibrils. J Cell Biol 129, 1165–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


