Site-specific ultraviolet “point welding” makes designed DNA objects much more durable.
Abstract
Bottom-up fabrication of custom nanostructures using the methods of DNA nanotechnology has great potential for applications in many areas of science and technology. One obstacle to applications concerns the constrained environmental conditions at which DNA objects retain their structure. We present a general, site-selective, and scalable method for creating additional covalent bonds that increase the structural stability of DNA nanostructures. Placement of thymidines in close proximity within DNA nanostructures allows the rational creation of sites for covalent cyclobutane pyrimidine dimer (CPD) bonds induced via ultraviolet irradiation. The additional covalent bonds may be used in a sequence-programmable fashion to link free strand termini, to bridge strand breaks at crossover sites, and to create additional interhelical connections. Thus designed multilayer DNA origami objects can remain stable at temperatures up to 90°C and in pure double-distilled water with no additional cations present. In addition, these objects show enhanced resistance against nuclease activity. Cryo–electron microscopy (cryo-EM) structural analysis of non–cross-linked and cross-linked objects indicated that the global shape and the internal network of crossovers are preserved after irradiation. A cryo-EM map of a CPD-stabilized multilayer DNA origami object determined at physiological ionic strength reveals a substantial swelling behavior, presumably caused by repulsive electrostatic forces that, without covalent stabilization, would cause disassembly at low ionic strength. Our method opens new avenues for applications of DNA nanostructures in a wider range of conditions.
INTRODUCTION
DNA nanotechnology (1–4) enables the bottom-up self-assembly of discrete three-dimensional (3D) objects with subnanometer precise features and overall dimensions ranging from the nanometer to the micrometer scale (5–16) and with molecular weights up to the gigadalton scale (17, 18). The resulting objects may be site-specifically functionalized and modified with chemical groups and biomolecules (19–21), and objects can be constructed that may also include mechanisms to yield machine-like behavior (22–24). Custom DNA objects have been developed and successfully used in diverse applications in basic research, thereby delivering new scientific insights and underlining the capacity of DNA nanotechnology to yield objects with utility. Examples range from structural biology (25–27) to biophysics (28–33), to photonics (34–37), to plasmonics (38–42), and to molecular electronics (19, 43–45). First steps have also been taken to explore uses of designed DNA objects as programmable agents in medical therapy (24, 46). Scalable biotechnological approaches for producing single-stranded DNA help pave the way to fabricate DNA objects at the quantities required for materials and health care applications (47).
To find use in different contexts, designed DNA objects must remain stable at the target conditions for sufficient time so that the desired application effects can be achieved. Commonly, applications in low ionic strength solutions such as in physiological fluids, in other solvents, in air or vacuum, and at elevated temperatures beyond 50°C are not accessible. Researchers have therefore sought for ways to expand the range of conditions under which designed DNA objects remain stable. Additional covalent bonds have been introduced in exemplary structures between correspondingly modified strand termini via chemical (48, 49) or enzymatic ligation (50). DNA nanostructures may also be further stabilized by the addition of cofactors such as 8-methoxypsoralen (51) or oligolysine and oligolysine–polyethylene glycol (PEG) copolymers (52, 53).
Despite these advances, it remains desirable to establish complementary, generally applicable approaches for the covalent stabilization of DNA nanostructures that do not require costly chemically modified strands or the addition of cofactors. The possibility of creating additional covalent connections at user-defined sites in DNA nanostructures would enable the rational stabilization of entire structures or parts of them for uses in a broader scope of environmental conditions. Moreover, it may enable the stable trapping of conformational states in mechanisms and in higher-order assemblies. Here, we present a general and scalable method for site-selectively introducing additional covalent bonds in DNA nanostructures. The target bonding sites are specified in the sequences of DNA strands alone and do not require the introduction of chemical modifications. Our method is generally applicable to the diverse range of DNA nanostructures, and it functions regardless whether the DNA strands have been produced via solid-phase chemical synthesis or using a biotechnological process (47).
RESULTS
Pyrimidine dimers are molecular lesions produced by photochemical reactions in DNA (54). Ultraviolet (UV) light induces the formation of covalent bonds through reactions at the C=C double bonds in thymine (T) or cytosine (C) bases (Fig. 1, left). Common products are cyclobutane pyrimidine dimers (CPDs), including thymine dimers. Minor by-products, such as (6-4) pyrimidine-pyrimidone and Dewar isomers, may also form upon UV irradiation. These lesions can arrest DNA replication and transcription and thus are cancerogenic and represent targets of the cellular DNA repair machinery (55). In 1982, Lewis and Hanawalt (56) reported that CPDs can also form from adjacent terminal thymines in separate DNA strands that are brought together by a templating complementary DNA strand. However, the potential of this finding for solving the stability problem in DNA nanotechnology has remained unrecognized thus far. The key concept in our work is the user-defined placement of thymidines in close proximity within DNA nanostructures to rationally create sites for introducing covalent CPD bonds via UV irradiation. These additional bonds may be used to link free strand termini, to remove strand breaks that occur at crossover sites, and to create additional interhelical connections (Fig. 1, right).
Fig. 1. Proximal thymidines as sites for cross-linking in DNA nanostructures.
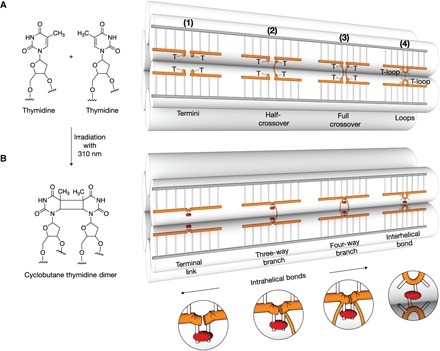
(A) Left: Chemical structures of two proximal thymidines before UV irradiation. Right: Schematic illustration of a six-helix bundle DNA nanostructure featuring single-stranded thymidines at strand termini (1), at half-crossovers (2), at full crossovers (3), and thymidine loops (4) before UV irradiation. (B) As in (A) but after exposure to light with 310-nm wavelength. CPD bonds are indicated as red ellipsoids.
The fundamental building blocks of DNA nanotechnology are double-helical DNA domains. In DNA origami objects (3, 4, 57), these domains form by hybridization of a set of short single-stranded staple oligonucleotides to a long single-stranded scaffold molecule. In other types of DNA nanostructures such as DNA tile-brick objects (16), the double-helical domains are formed only between single-stranded oligonucleotides. DNA origami and tile-brick objects contain hundreds of single-strand breaks, which represent weak points. This is because free ends enable not only the formation but also the dissolution of plectonemic double-helical domains. To create the option for removing the single-strand breaks after self-assembly of a target object, we prepare DNA strands with additional thymidines at both strand termini (Fig. 1A, motif 1). Although the added bases will not be involved in forming Watson-Crick base pairs, the thymidines will come in close proximity at single-strand break sites in the folded object, which allows CPD bonds to form between the two thymidines through irradiation with light of 310-nm wavelength.
In DNA objects, double-helical domains are connected to neighboring double-helical domains by interhelical connections typically formed by antiparallel single-strand crossovers including both half-crossovers and double crossovers (Fig. 1A, motif 2 versus motif 3). For example, DNA tile-brick objects are almost exclusively connected via half-crossovers, while in DNA origami objects, both types of interhelical connections may occur. The crossover positions also represent weak points in DNA nanostructures due to the interrupted backbone bonds in the helical direction. To create the option for closing the weak links after self-assembly, we may add additional unpaired thymidine bases in the staple strands at crossover positions, as indicated in Fig. 1A (motifs 2 and 3). Through proximity, irradiation with 310-nm light again may induce the formation of CPD bonds that covalently connect the strands along the helical direction (Fig. 1B, motifs 2 and 3), thereby creating another topological hindrance for helical unwinding.
When designing DNA objects, strand crossovers are typically placed between neighboring double-helical domains at positions where the helical backbones come close together. Complementary to strand crossovers, we may also exploit the light-induced CPD dimer bonds to create additional interhelical linkages after self-assembly of an object. To this end, we place single-stranded thymidine loops (T loops) at positions where the backbones of neighboring DNA helices roughly align (Fig. 1A, motif 4). Irradiation at 310 nm can then induce the creation of covalent interhelical linkages (Fig. 1B, motif 4).
Proof of concept: High temperature and low-salt stability
To test our method, we implemented the design alterations shown in Fig. 1 in several variants of multilayer DNA origami objects. We tested the stability of the resulting objects after irradiation with light of 310-nm wavelength in melting experiments (Fig. 2, left) and in experiments in which we removed cations from solution (Fig. 2, right). We modified a brick-like multilayer DNA origami (58) object in honeycomb-packing geometry (fig. S1) by inserting additional thymidines both at all strand termini and at all strand crossover positions. We found that the nonirradiated control sample disassembled (“unfolded”) around 50°C, as seen in gel electrophoresis by disappearance of the band indicating folded objects, and appearance of free staple strands (Fig. 2A, left gel). By contrast, the irradiated sample preserved its global shape up to 90°C, judging by the fact that the electrophoretic mobility of the folded species remains largely unaltered. A slight smear at high electrophoretic mobilities shows that some strands still separate from the folded objects at high temperatures. However, the strands that separated had much lower electrophoretic mobilities and thus higher mass than the staple strands that emerged from the molten nonirradiated control sample. The high-temperature resistance of the irradiated object and the emergence of higher-mass strands provide evidence for the successful introduction of covalent cross-links at the designed thymidine sites upon UV irradiation.
Fig. 2. Proof of concept of UV cross-linking with multilayer DNA origami.
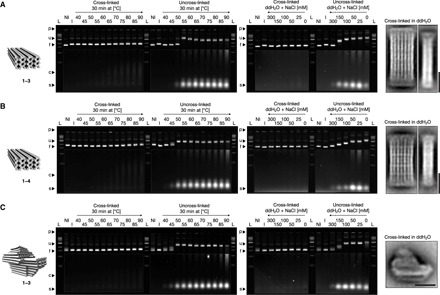
(A) From left to right: Model of the brick-like DNA origami object featuring additional thymidines at all strand termini and at all strand crossover positions; laser-scanned fluorescent images of 2.0% agarose gels stained with ethidium bromide. Irradiated (135 min at 310 nm) and nonirradiated samples were either incubated for 30 min at different temperatures or incubated for 3 hours at room temperature in double-distilled water (ddH2O) containing successively lower concentrations of monovalent sodium chloride, respectively. p, pocket; u, unfolded species; f, folded species; c, cross-linked staple strands; s, uncross-linked staple strands; L, 1kB ladder; NI and I, nonirradiated and irradiated reference samples in folding buffer with 5 mM MgCl2, respectively. The images of the gels were autoleveled, and the highlighted regions were autoleveled twice; average 2D particle micrograph of the irradiated sample in double-distilled water. (B and C) As in (A) but with the brick-like DNA origami object featuring additional thymidines at all strand termini and at all strand crossover positions and with 5-T loops and the pointer object featuring additional thymidines at all strand termini and at all strand crossover positions, respectively. See fig. S9 for globally autoleveled gel images.
We also tested the stability of the irradiated versus nonirradiated sample when removing cations from solution. Using filtration, we exchanged buffers and dissolved the samples in double-distilled water containing successively lower concentrations of monovalent sodium chloride (Fig. 2A, right). The irradiated sample remained folded even in double-distilled water containing zero added cations, whereas the nonirradiated control disassembled between 300 and 150 mM sodium chloride (NaCl), as seen by strong mobility shifts and the emergence of single strands with high mobility. Transmission electron microscopy (TEM) imaging of the irradiated sample dissolved in pure water revealed particles with the expected shape (fig. S2). The degree of heterogeneity of the sample in pure water was higher than at high-salt conditions. Electrophoretic analysis after up to 1 day of storage in pure water showed no changes in electrophoretic mobility, and we could not detect any staple strands that separated from the folded objects (fig. S3). Therefore, simple design alterations and UV irradiation allow the stabilization of the normally quite cation-sensitive multilayer DNA origami for uses under physiological (~150 mM NaCl) and even lower ionic strength conditions. Many other harsh environments may also be accessible after UV stabilization. As a simple demonstration, we dissolved the cross-linked objects in aqueous mixtures of dimethyl sulfoxide (an organic solvent) without added cations (fig. S4).
As a second example, we prepared and tested a variant of the brick-like object in which we also inserted single-stranded T loops (motif 4 in Fig. 1), in addition to extra thymidines at all strand termini and at all crossover positions (fig. S5). The extent of thermal and cationic stabilization after irradiation (Fig. 2B) was similar compared to the design variant lacking the single-stranded T loops. We tested loops containing one, three, and five thymidines. The variant with 5-T loops showed incrementally greater stabilization at high temperatures (80°C), judging by gel electrophoretic band intensity (fig. S6). We note that the irradiation of the brick variant with additional 5-T loops for interhelical bonds also leads to a slight electrophoretic mobility increase compared to the nonirradiated control (Fig. 2B, lane R2 versus lane R1), which suggests that the additional interhelical bonds may lead to some degree of compaction or mechanical stabilization.
As a third example, we chose the previously described “pointer” object (13), which is a multilayer DNA origami in square lattice packing geometry, and added additional thymidines at all strand termini and at all crossover positions (fig. S7). As for the brick variants, irradiation with UV light stabilized this object against exposure of temperatures up to 90°C, and the pointer object may now be dissolved in pure water without any cations (Fig. 2C). The nonirradiated control pointer sample already disassembled between 45° and 50°C and required more than 300 mM NaCl in solution to remain folded, as seen by electrophoretic mobility analysis (Fig. 2C) and TEM imaging (fig. S8).
In the course of establishing our method, we tested several parameters such as time of exposure to UV irradiation. In addition, we performed a defect analysis using the de Bruijn assay (59) to evaluate the structural integrity of double-helical domains in DNA objects upon UV irradiation. Approximately 2 hours of exposure at our UV setup lead to most efficient stabilization for all structures tested (figs. S10 and S11) without signs of structural degradation (fig. S12). For shorter irradiation times, the cross-linking was not complete, meaning that structures did not survive exposure to temperatures substantially above the melting temperatures before UV treatment. For irradiation times longer than the optimal, structural radiation damages accumulated, as reflected in successively lower electrophoretic mobilities of the objects. Therefore, the exposure to UV irradiation follows a Goldilocks principle. Because the optimal irradiation time will depend on details of the UV light source and other parameters, our optimal irradiation times will not necessarily hold up in other contexts. However, users can identify the optimum irradiation time by using screens similar to those we performed.
As outlined in Introduction, our method relies on CPDs, which could form between T-T but also between, for example, T-C contacts. We compared the efficiency of cross-linking for brick variants prepared with T-T versus T-C at all strand termini and at all strand crossover positions (fig. S13). On the basis of the amount of structures that survive exposure to high temperatures after UV exposure, T-T bonds form significantly more efficiently than the T-C contacts and lead to complete stabilization. The cross-linking worked successfully through exposure to 310-nm light. Longer wavelengths, such as 365 nm, did not lead to stabilization in our hands (fig. S14), although it has been reported that CPD bonds may also form through exposure to 365-nm UVA light (60).
Stability under physiological conditions
Our UV cross-linking method may be used to substantially enhance the stability of DNA nanostructures, and in particular multilayer DNA origami objects, for applications under physiological conditions. As a demonstration, we dissolved the brick-like multilayer DNA origami object, which contained additional Ts at all staple termini and at all crossover positions in physiological phosphate-buffered saline (PBS) solution, and incubated the objects at the physiological temperature of 37°C. Even after 2 days of storage in PBS at 37°C, there was no detectable degradation of the irradiated and covalently cross-linked sample (Fig. 3A). By contrast, the nonirradiated control disassembled within minutes after exposure to these conditions. Judging by the extent to which irradiated design variants of the brick sample remained stable at physiological temperature and ionic strength, the stabilization appeared complete for the design variant that had additional T bases to all strand termini and at all half-crossover and full crossover positions. Ligating only the free strand termini and a subset of crossovers was not sufficiently effective in preserving the full structure (fig. S15). In 10% fetal bovine serum (FBS) at 37°C, the irradiated brick sample survived for several hours and substantially longer than a nonirradiated control (Fig. 3B). In serum, presumably the loss of folded structures was caused by enzyme activity rather than the low ionic strength of the solution.
Fig. 3. Stability under physiological conditions.
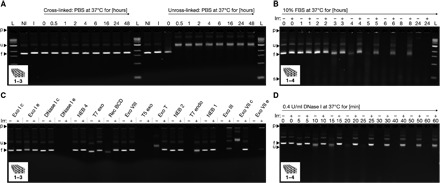
Laser-scanned fluorescent images of 2.0% agarose gels stained with ethidium bromide. Cross-linked samples were irradiated for 135 min at 310 nm. (A) The brick-like DNA origami object featuring additional thymidines at all strand termini and at all strand crossover positions was incubated for different periods of time in PBS solution at 37°C. (B) The brick-like DNA origami object featuring additional thymidines at all strand termini, at all strand crossover positions, and 5-T loops was incubated for different periods of time in 10% FBS at 37°C. (C) The brick-like DNA origami object from (A) was exposed to a set of different nucleases (100 U/ml) for 24 hours at 37°C. Lanes labeled with a “c” indicate controls in which the sample was dissolved in the corresponding buffers in the absence of nuclease. (D) The brick-like DNA origami object from (B) was exposed to DNase I (0.4 U/ml) for different periods of time at 37°C. (B to D) Nonirradiated and irradiated samples were loaded on the gel alternatingly. All images of the gels were globally autoleved.
Biological fluids, such as serum, contain a variety of exo- and endonucleases for digesting DNA molecules. To elucidate the activity of various nucleases, we exposed the brick-like multilayer DNA origami object featuring Ts at strand termini, at all crossover positions, and T loops to a panel of such enzymes (Fig. 3C). Some enzymes, such as Exo VIII and T7 Exo, by default, appear inactive on the brick sample whether it was irradiated or not. However, for others (such as Exo I e, Exo T, T7 Endo, and Exo III), introduction of the additional covalent bonds through irradiation substantially enhanced the lifetime of the cross-linked object compared to the nonirradiated control sample. The most active DNA-degrading enzyme was deoxyribonuclease I (DNase I). A kinetic analysis of the digestion of the brick-like object using DNase I at blood plasma activity levels (61) revealed that the irradiated and thus stabilized brick sample was digested much more slowly than the nonirradiated control. Analysis of the band intensities reveals an approximately five- to sixfold life-span expansion from 10 to 60 min through cross-linking under the conditions tested (Fig. 3D). The brick variant featuring only extra Ts at strand termini and at all crossover positions (lacking interhelical T loops) showed somewhat weaker resilience against DNase I digestion (fig. S16).
Cryo-EM structural analysis of UV–cross-linked multilayer DNA origami objects
To elucidate the effects of UV irradiation and CPD bond formation on the structure of a DNA object, we exemplarily determined five electron density maps using single-particle cryo–electron microscopy (cryo-EM) (Fig. 4, A to C). First, we collected single-particle cryo-EM data on a nonirradiated control multilayer brick sample containing additional Ts at all strand termini and all strand crossover positions (fig. S17). The reconstructed 3D EM density map revealed the expected global rectangular brick-like shape (Fig. 4A). However, the object showed also a global twist deformation (Fig. 4D), whose extent was more pronounced than what was expected on the basis of a previous analysis of a similar brick-like object lacking the additional Ts (17). Presumably, the added Ts increase the flexibility of crossover sites, which may affect the packing geometry of helices. We determined the chirality of the twist deformation to be right-handed using a tomographic tilt series (movie S1).
Fig. 4. Cryo-EM structural analysis before and after UV irradiation.
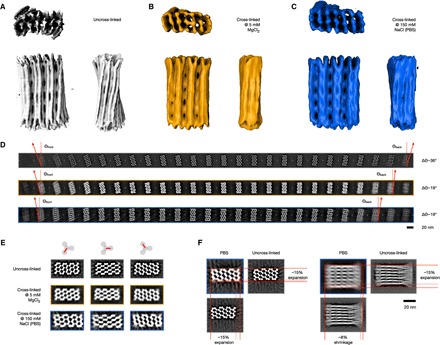
(A) Cryo-EM density map of the nonirradiated brick-like object with TT motifs 1 to 3 (Electron Microscopy Data Bank identifier EMD-4354). (B and C) Cryo-EM density map of the irradiated (135 min at 310 nm) brick-like object with TT motifs 1 to 3 in buffer containing 5 mM MgCl2 or in PBS buffer, respectively. The electron density thresholds are chosen in such a way that all crossovers in the top layer are visible, as seen in the side view (Electron Microscopy Data Bank identifier EMD-0027 and EMD-0028, respectively). (D) Slices along the z direction obtained from the three density maps shown in (A) to (C) from top to bottom. To determine the twist angle delta Theta, the first and last slices were chosen. (E) Slices showing the three crossover layers in the reconstructions shown in (A) to (C). (F) Comparison of the global dimensions of the uncross-linked variant in 5 mM MgCl2 buffer and the cross-linked variant in PBS buffer. Color code as shown in (A) to (C).
Second, we collected single-particle cryo-EM data of the brick-like object after exposure to UV irradiation (fig. S18). The reconstructed 3D EM density map again revealed the global rectangular brick-like shape (Fig. 4B). After irradiation, the right-handed global twist was significantly diminished (Fig. 4D). We attribute the twist reduction to the creation of the additional covalent bonds at the crossover sites, which reduces the junction flexibility and aligns the helices again in a geometry closer to the default honeycomb-packing design. Previously, Chen and coworkers (62) UV-irradiated single-layer DNA origami rectangles to investigate radiation damages and observed a twist-reducing flattening effect. However, because the samples of Chen and coworkers were not specifically designed to contain thymidine-thymidine cross-linking sites, the mechanism leading to twist removal may be different from our samples. Third, we collected single-particle cryo-EM data of the irradiated brick-like object once it had been dissolved in physiological ionic strength PBS buffer (fig. S19). The resulting 3D EM density map again revealed a global rectangular brick-like shape (Fig. 4C). A slice-by-slice comparison of the three cryo-EM maps indicates that the internal network of crossovers has been preserved after irradiation and exposure to low ionic strength conditions (Fig. 4E, fig. S20, and movies S2 to S4).
The overall aspect ratio of the cryo-EM density map determined for the cross-linked sample at low (physiological) ionic strength was different compared to the cryo-EM density maps, which were determined at higher ionic strength in the presence of magnesium (Fig. 4, A to C and F). The object’s cross section expanded under physiological conditions by approximately 15% and shrank in the helical direction by approximately 8%. The deformation presumably is a consequence of the strong electrostatic repulsion in PBS buffer, which pushes the helices away from each other. Without UV irradiation, these forces would normally lead to the disassembly of the object. However, the additional covalent CPD bonds after UV light exposure prevent the double-helical DNA domains from dissociating.
Finally, we also collected single-particle cryo-EM data of the brick-like variant designed with additional T loops for interhelical bonds before and after exposure to UV irradiation, respectively. The resulting 3D EM density maps again revealed the expected global rectangular brick-like shape (figs. S21 and S22, respectively). However, the internal crossover lattice was less well resolved than in the design variants that lacked the additional T loops, which we attribute to the more pronounced molecular heterogeneity in these samples that is caused by the presence of additional flexible T loops.
Covalently bonding conformational states and higher-order assemblies across interfaces
The targeted introduction of base-paired thymidines also enables us to covalently cross-link DNA-based mechanisms and higher-order assemblies across binding interfaces. We demonstrate here the possibility of locking conformational states with a previously described two-state switch (fig. S23) (11). The closed state of the switch is stabilized by base pair–stacking contacts when the shape-complementary surfaces of its two beams come in direct contact (Fig. 5A). The object may be switched between the two states by raising and lowering the temperature or through addition of cations such as magnesium chloride. We hypothesized that in the closed state, terminal thymidines positioned directly at the blunt-ended base pair–stacking contacts may be in sufficient proximity to allow the formation of CPD dimer bonds upon UV irradiation. The switch design already contained several such TT-stacking contacts. A time-resolved analysis of the effects of UV irradiation on the switch in the presence of 30 mM MgCl2 (which stabilizes the closed state) reveals that, after 30-min exposure, about 80% of the particles were irreversibly trapped in the closed state. We conclude this from the band pattern in gel electrophoresis under the low ionic strength conditions, which normally lead to the opening of the switch at 5 mM MgCl2 (Fig. 5, B and C). Hence, the CPD bonds may also be formed between fully separate double-helical DNA domains that are held in proximity.
Fig. 5. Covalent bonding of conformational states and higher-order assemblies.
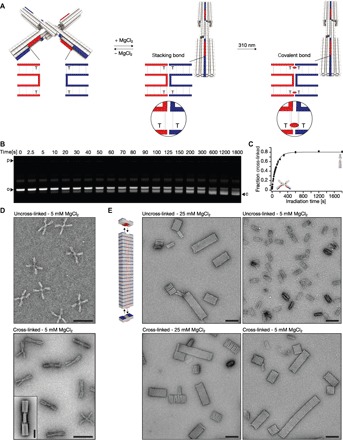
(A) Schematics of the two-state switch that consists of two rigid beams flexibly connected in the middle by an immobile Holliday junction. Cylinders in the models represent double-helical DNA domains, and shape-complementary surface features are highlighted in red and blue. Insets show blow-ups of the blunt-ended interfaces of protruding (red) and recessive (blue) surface features. Thymidines directly located at the blunt-end site can be cross-linked upon UV light irradiation. The resulting CPD bond is indicated as a red ellipsoid. (B) Laser-scanned fluorescent image of 2.0% agarose gel stained with ethidium bromide. Switch samples were irradiated at 310 nm for different periods of time and loaded on the gel. o and c, species of particles populating open and closed state, respectively. (C) Plot of the fraction of cross-linked switch particles as a function of time obtained from the gel in (B). The experiment was performed in triplicate; data points represent the mean, and error bars represent the SD. (D) Exemplary TEM micrographs. Top: Nonirradiated sample with particles populating the open state. Bottom: Irradiated (20 min at 310 nm) sample with particles locked in the closed conformational state. Scale bars, 100 nm. Inset: Average 2D particle micrograph of cross-linked particles. Scale bar, 20 nm. (E) Top left: Model of the multilayer DNA origami brick that polymerizes into linear filaments. Fields of view of TEM micrographs recorded at the indicated conditions. Scale bars, 100 nm.
In addition, we demonstrate the possibility of stabilizing higher-order assemblies with a previously described multilayer DNA origami brick (fig. S24) that oligomerizes at high ionic strength via shape-complementary base pair–stacking contacts into linear filaments (11). The filaments, by default, dissolve when the ionic strength of the solution is lowered again (Fig. 5E). By placing TT motifs at the base pair–stacking contacts, the higher-order filaments may also be covalently stabilized by simple UV irradiation. As a result, the filaments no longer dissociate when exposed again to low ionic strength conditions, as seen by TEM imaging (Fig. 5E). The possibility of stabilizing particular conformational states or higher-order assemblies may be especially useful to prepare containers or mechanisms built from many subunits for applications in low ionic strength conditions. Interior design, as shown in Fig. 1, and interfacial bonding schemes, as illustrated in Fig. 5, could be combined to yield subunits and higher-order assemblies that withstand a wide range of conditions.
DISCUSSION
Users of our method can simply define sites of covalent bonding in DNA assemblies by creating TT sequence motifs, where the two Ts need not be positioned within double-helical domains. The objects studied here featured, by default, several sites for CPD bond formation because the bacteriophage-derived scaffold strand itself already contained multiple TT and AA motifs. To suppress the formation of undesired CPD dimers upon irradiation and to avoid the extra T insertions if so desired, new custom scaffold sequences may be developed in the future. By design, these sequences could lack TT motifs and feature AA motifs in the regular intervals that correspond to the internal junction spacing rules in honeycomb- or square lattice–packing geometries. Scaffold-free DNA objects, such as tile-brick structures (15), may also be specifically designed with sequences that selectively place TT motifs at crossovers and at strand termini to enable covalent bonding by UV irradiation. Our results show that the mere proximity of thymidines is sufficient to template the formation of covalent linkages through UV irradiation. Moreover, the thymidines do not necessarily need to be placed within a double-helical context to form these linkages.
The cryo-EM maps presented here show that the DNA objects preserve their global shape after UV treatment. Our maps also add to the body of structural data in DNA nanotechnology and help understand the connection between design details and resulting shape. For example, we presented a multilayer DNA origami cryo-EM map at physiological ionic strength. Formerly, it was not possible to analyze these structures because the objects would “explode” under these conditions. Our cryo-EM map at physiological conditions reveals a substantial swelling behavior, which helps appreciate the contribution of electrostatics to global shape. Future designs for physiological conditions will need to consider the swelling behavior to produce shapes according to specifications.
Our method supports a broader applicability of DNA-based nanotechnology, in particular for the more structurally complex multilayer 3D DNA objects, which arguably offer attractive degrees of freedom to designers but tend to be more sensitive to environmental conditions. Because of the simplicity, sequence programmability, and scalability, covalent bonding by UV irradiation will help pave the way for applications of DNA nanostructures in a wide variety of conditions for a range of fields.
MATERIALS AND METHODS
Folding of DNA origami objects
The reaction mixtures contained scaffold DNA at a concentration of 20 nM and oligonucleotide strands at 200 nM each. The folding buffer included 5 mM tris, 1 mM EDTA, 5 mM NaCl (pH 8), and 20 mM MgCl2. The reaction mixtures were subjected to a thermal annealing ramp using Tetrad (MJ Research, now Bio-Rad) thermal cycling devices. Oligonucleotides were purchased from Eurofins MWG. See Table 1 for folding ramps used to assemble the objects described in this study.
Table 1. One-pot self-assembly of DNA origami objects.
| Object name | Denaturation temperature for 15 min (°C) | Folding ramp | Storage temperature (°C) | Scaffold |
| Brick-like, TT motifs 1–3 | 65 | [60–20°C]; 60 min/1°C | 20 | p7560 |
| Brick-like, TT motifs 1–4 | 65 | [60–20°C]; 60 min/1°C | 20 | p7560 |
| Pointer | 65 | [60–20°C]; 60 min/1°C | 20 | p7249 |
| Switch | 65 | [58–55°C]; 90 min/1°C | 25 | p8064 |
| Polymerization brick | 65 | [60–44°C]; 60 min/1°C | 25 | p8064 |
Purification and enrichment of DNA origami objects
After the folding reaction, all reaction products were purified using one round of PEG precipitation (63). The resulting pellet was dissolved in folding buffer (5 mM tris, 1 mM EDTA, and 5 mM NaCl) including 5 mM MgCl2. The final volume was chosen to get a monomer concentration of 100 nM. The samples were equilibrated at 30°C and 450 rpm overnight in a shaker incubator (Thermomix comfort from Eppendorf). All procedures were performed as previously described (64).
UV irradiation
For UV irradiation, we used a 300-W xenon light source (MAX-303 from Asahi Spectra) with a high transmission bandpass filter centered around 310 nm (XAQA310 from Asahi Spectra). We used a light guide (Asahi Spectra) to couple the light into the sample by placing it directly on top of a 0.65-ml reaction tube. Unless otherwise indicated, the brick-like samples were irradiated for 135 min, the pointer samples for 120 min, and the polymerizing brick samples for 30 min. Samples were irradiated in folding buffer (5 mM tris, 1 mM EDTA, and 5 mM NaCl) including 30 mM MgCl2, unless otherwise stated.
Ultrafiltration for enrichment and buffer exchange
All samples (cross-linked and uncross-linked) were subjected to three rounds of ultrafiltration (Amicon Ultra 500 μl with 100k cutoff). Ultrafiltration was carried out at 20°C and 7k relative centrifugal force (Eppendorf 5424R). The buffer was replaced by folding buffer (5 mM tris, 1 mM EDTA, and 5 mM NaCl; including 5 mM MgCl2), PBS, or double-distilled water supplemented with 300, 150, 100, 50, 25, and 0 mM NaCl. Samples used for cryo-EM were concentrated to 1000 nM.
Gel electrophoresis of DNA origami objects
Samples were electrophoresed on 2.0% agarose gels containing 0.5× tris-borate- EDTA and 5 mM MgCl2 for around 2 hours at 90-V bias voltage in a gel box immersed in a water or ice bath, unless otherwise stated. Samples were loaded on the gel at a monomer concentration of approximately 5 nM. The electrophoresed agarose gels were scanned using a Typhoon FLA 9500 laser scanner (GE Healthcare) at a resolution of 25 μm/pixel. The resulting 16-bit tif images were analyzed using ImageJ 1.440.
Negative-stain TEM: Preparation, acquisition, and data processing
Samples were adsorbed on glow-discharged, collodion-supported, carbon-coated (10 nm) Cu400 TEM grids (in-house production) and stained using a 2% aqueous uranyl formate solution containing 25 mM sodium hydroxide. Samples were incubated for 15 to 300 s depending on the buffer/solvent used. For samples dissolved in solvents including low concentrations of positively charged ions, we used higher monomer concentrations (50 nM) and longer incubation times. We used magnifications between ×10,000 and ×30,000 to acquire the data.
Imaging was performed on different microscopes (see Table 2). TEM micrographs used in the figures were high-pass–filtered to remove long-range staining gradients, and the contrast was autoleveled (Adobe Photoshop CS6).
Table 2. Negative-stain TEM settings.
| Microscope | Operating voltage (kV) | Camera | Objects |
| Philips CM 100 | 100 | AMT 4-megapixel charge-coupled device camera | Switch; polymerization brick |
| FEI Tecnai 120 | 120 | Tietz TemCam-F416 (4k × 4k) | Brick-like, TT motifs 1–3 Brick-like, TT motifs 1–4 Pointer |
For 2D image processing, libraries of individual particle micrographs were created by particle picking using the RELION-2 picking routine (65). Generation of average 2D particle micrographs was performed using RELION-2 (65). Typically, around 2000 individual particles were averaged.
Cryo-EM: Preparation, acquisition, and processing of data
For the brick-like object with TT motifs 1 to 3, concentrations between 700 and 850 nM were used. The samples were applied to C-Flat 1.2/1.3, 1.2/1.3, 2/1, or 2/2 thick grids (Protochips). Plunge freezing was performed with an FEI Vitrobot Mark V instrument with a blot time of 3 s, a blot force of −1, and a drain time of 0 s under 95% humidity and 22°C.
For the brick-like object with TT motifs 1 to 4, concentrations between 560 and 800 nM were used. The samples were applied to C-Flat 1.2/1.3, 2/1, or 2/2 thick grids. Plunge freezing was performed with an FEI Vitrobot Mark V instrument with a blot time of 3 s, a blot force of −1, and a drain time of 0 s under 95% humidity and 22°C.
Automated data collection was performed on a Titan Krios G2 electron microscope (FEI) operated at 300 kV and equipped with a Falcon III direct detector (FEI). We used EPU for single particle and FEI tomography for tilt series acquisition. For all brick-like objects under different conditions, movies comprising 15 frames, 1.5- to 2-s exposure time, and a total dose of 60 e−/Å2 were recorded on a Falcon III (FEI) direct electron detection camera in fractioning mode at a calibrated magnification of ×29,000 with a magnified pixel size of 2.3 Å. Defocus values ranging from −1 to −3 μm were used.
The recorded movies were subjected to motion correction with MotionCor2 (66), and subsequently, contrast transfer function parameters were estimated with CTFFIND4.1 (67). All subsequent processing steps were performed in RELION-2.1 (65, 68). For each data set, references for automated picking were calculated from about 5000 manually selected particles. With the picked particles, multiple rounds of reference-free 2D classification were performed. The best 2D class averages, as judged by visual inspection, were selected. An initial model was produced from a bild file generated by CanDo. After multiple rounds of 3D classification, the classes showing the most features were selected for 3D autorefinement, and subsequently, post-processing for sharpening of the refined map was performed with different manually selected B factors.
A cryo-tomogram for validation of the twist direction was acquired with FEI tomography, with a defocus of −3 μm at a calibrated magnification of ×29,000, corresponding to a magnified pixel size of 2.3 Å. The session was set up as bidirectional tilting in increments of 2° up to 50°, and the dose per image was set to ~2 e−/Å2. The resulting tilt series was processed with the IMOD 4.9 routine (69).
Experiments
In Fig. 2, samples were folded and PEG-purified, and the MgCl2 concentration was adjusted to 30 mM. After UV irradiation, the buffer was exchanged to the target buffer/solvent by using ultracentrifugation. Before gel electrophoresis, the samples were incubated for around 2 to 3 hours at room temperature. Samples for the temperature screen were incubated for 30 min at the indicated temperatures. The samples for negative-stain TEM were prepared at a monomer concentration of 50 nM, with incubation on the grid for 3 to 5 min.
In Fig. 3B, the stability screen in folding buffer (5 mM MgCl2) supplemented with 10 % FBS (not heat-inactivated; Gibco; A3160801, Thermo Fisher Scientific) was performed at a monomer concentration of 20 nM at 37°C for the indicated time. The samples were frozen in liquid nitrogen and analyzed using agarose gel electrophoreses. In Fig. 3C, all nucleases were purchased from New England Biolabs and used at a concentration of 100 U/ml in the supplied manufacturer’s buffer. The samples (10 nM) were incubated at 37°C for 24 hours. In Fig. 3D, the time course of the stability against DNase I nuclease digestion was performed at a monomer concentration of 10 nM in the supplied DNase I buffer at 37°C.
In Fig. 5, the irradiation time screen for the switch was performed in triplicate. The irradiated volume was 25 μl at a monomer concentration of 5 nM. For the analysis of the gel shown in Fig. 5B, we calculated the ratio between the band including closed particles and the bands including open and closed particles. The grayscale values for each band were obtained by integration. The data points in Fig. 5C represent the average, and the error bars represent the SD of the three independent experiments. For the assembly of the filaments, monomers were folded and PEG-purified. The pellet was dissolved in folding buffer (5 mM MgCl2) to obtain a monomer concentration of 100 nM. After equilibration, the MgCl2 concentration was adjusted to 20 mM, and the sample was incubated at 40°C for 3 days in the Tetrad to obtain filaments. One part of the sample was irradiated at 310 nm for 30 min. The MgCl2 concentration was decreased to 5 mM by the addition of EDTA.
Supplementary Material
Acknowledgments
We thank F. Praetorius for discussions and F. Engelhardt for auxiliary experiments. Funding: This work was supported by a European Research Council Consolidator Grant to H.D. (GA no. 724261) and the Deutsche Forschungsgemeinschaft through grants provided within the Gottfried-Wilhelm-Leibniz Program, the Excellence Clusters CIPSM (Center for Integrated Protein Science Munich), and the Technical University of Munich Institute for Advanced Study. Author contributions: T.G., M.K., and B.K. performed the research. H.D. designed the research. T.G. and H.D. wrote the manuscript and prepared the figures. All authors commented on the manuscript. Competing interests: T.G. and H.D. are inventors on a provisional patent related to this work that has been filed by the Technical University of Munich (“Novel method for stabilizing nucleic acid nanostructures,” submitted 5 June 2018). The authors declare no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Cryo-EM data that support the findings of this study have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-4354, EMD-0027, EMD-0028, EMD-4357, and EMD-0029. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/8/eaau1157/DC1
Fig. S1. Design diagram of the brick-like (TT motifs 1 to 3) object prepared using caDNAno.
Fig. S2. Exemplary negative-stained TEM micrographs of the brick-like object (TT motifs 1 to 4) in different buffers/solvents.
Fig. S3. Laser-scanned fluorescent image of a 2.0% agarose gel that was run in an ice-cooled water bath.
Fig. S4. Laser-scanned fluorescent image of a 2.0% agarose gel that was run in an ambient temperature water bath.
Fig. S5. Design diagram of the brick-like (TT motifs 1 to 4) object prepared using caDNAno.
Fig. S6. Laser-scanned fluorescent image of a 2.0% agarose gel placed in an ice-cooled water bath.
Fig. S7. Design diagram of the pointer object.
Fig. S8. Exemplary negative-stained TEM micrographs of the pointer object in different buffers/solvents.
Fig. S9. Laser-scanned fluorescent image of a 2.0% agarose gel placed in a water bath.
Fig. S10. Laser-scanned fluorescent image of a 2.0% agarose gel placed in a water bath.
Fig. S11. Laser-scanned fluorescent image of a 2.0% agarose gel placed in a water bath.
Fig. S12. Laser-scanned fluorescent image of a 2.0% agarose gel placed in an ice-cooled water bath.
Fig. S13. Laser-scanned fluorescent image of a 2.0% agarose gel placed in a water bath.
Fig. S14. Laser-scanned fluorescent image of a 2.0% agarose gel placed in a water bath.
Fig. S15. Laser-scanned fluorescent image of a 2.0% agarose gel placed in a water bath.
Fig. S16. Laser-scanned fluorescent image of a 2.0% agarose gel placed in a water bath.
Fig. S17. Cryo-EM data of the brick-like object with TT-motifs (1) – (3) before crosslinking in folding buffer.
Fig. S18. Cryo-EM data of the brick-like object with TT-motifs (1) – (3) after crosslinking in folding buffer.
Fig. S19. Cryo-EM data of the brick-like object with TT-motifs (1) – (3) after crosslinking in phosphate-buffered saline (PBS).
Fig. S20. Slice-by-slice visualization of cryo-EM maps determined from brick samples.
Fig. S21. Cryo-EM data of the brick-like object with TT-motifs (1) – (4) before crosslinking in folding buffer.
Fig. S22. Cryo-EM data of the brick-like object with TT-motifs (1) – (4) after crosslinking in folding buffer.
Fig. S23. Design diagram of the switch object.
Fig. S24. Design diagram of the polymerization brick object.
Table S1. Sequences of staple strands for all DNA objects used in this work.
Movie S1. Tomogram obtained from cryo-EM of the brick-like variant with thymines at all staple termini and with TT motifs at all crossover sites.
Movie S2. Corresponding movie to fig. S20 for the nonirradiated (uncross-linked) sample of the brick-like variant with thymines at all staple termini and with TT motifs at all crossover sites in folding buffer and in the presence of 5 mM MgCl2.
Movie S3. Corresponding movie to fig. S20 for the irradiated (cross-linked) sample of the brick-like variant with thymines at all staple termini and with TT motifs at all crossover sites in folding buffer and in the presence of 5 mM MgCl2.
Movie S4. Corresponding movie to fig. S20 for the irradiated (cross-linked) sample of the brick-like variant with thymines at all staple termini and with TT motifs at all crossover sites in PBS buffer and in the absence of MgCl2.
Movie S5. Corresponding movie to fig. S20 for the nonirradiated (uncross-linked) sample of the brick-like variant with thymines at all staple termini, with TT motifs at all crossover sites and additional 5-T loops in folding buffer and in the presence of 5 mM MgCl2.
Movie S6. Corresponding movie to fig. S20 for the irradiated (cross-linked) sample of the brick-like variant with thymines at all staple termini, with TT motifs at all crossover sites and additional 5-T loops in folding buffer and in the presence of 5 mM MgCl2.
Reference (70)
REFERENCES AND NOTES
- 1.Jones M. R., Seeman N. C., Mirkin C. A., Programmable materials and the nature of the DNA bond. Science 347, 1260901 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Seeman N. C., Nanomaterials based on DNA. Annu. Rev. Biochem. 79, 65–87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothemund P. W. K., Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Douglas S. M., Dietz H., Liedl T., Hogberg B., Graf F., Shih W. M., Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong F., Zhang F., Liu Y., Yan H., DNA origami: Scaffolds for creating higher order structures. Chem. Rev. 117, 12584–12640 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Dietz H., Douglas S. M., Shih W. M., Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–730 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han D., Pal S., Nangreave J., Deng Z., Liu Y., Yan H., DNA origami with complex curvatures in three-dimensional space. Science 332, 342–346 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Veneziano R., Ratanalert S., Zhang K., Zhang F., Yan H., Chiu W., Bathe M., Designer nanoscale DNA assemblies programmed from the top down. Science 352, 1534 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benson E., Mohammed A., Gardell J., Masich S., Czeizler E., Orponen P., Högberg B., DNA rendering of polyhedral meshes at the nanoscale. Nature 523, 441–444 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Han D., Pal S., Yang Y., Jiang S., Nangreave J., Liu Y., Yan H., DNA gridiron nanostructures based on four-arm junctions. Science 339, 1412–1415 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Gerling T., Wagenbauer K. F., Neuner A. M., Dietz H., Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 347, 1446–1452 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Funke J. J., Dietz H., Placing molecules with Bohr radius resolution using DNA origami. Nat. Nanotechnol. 11, 47–52 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Bai X.-C., Martin T. G., Scheres S. H. W., Dietz H., Cryo-EM structure of a 3D DNA-origami object. Proc. Natl. Acad. Sci. U.S.A. 109, 20012–20017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang F., Jiang S., Wu S., Li Y., Mao C., Liu Y., Yan H., Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat. Nanotechnol. 10, 779–784 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Wei B., Dai M., Yin P., Complex shapes self-assembled from single-stranded DNA tiles. Nature 485, 623–626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke Y., Ong L. L., Shih W. M., Yin P., Three-dimensional structures self-assembled from DNA bricks. Science 338, 1177–1183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagenbauer K. F., Sigl C., Dietz H., Gigadalton-scale shape-programmable DNA assemblies. Nature 552, 78–83 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Ong L. L., Hanikel N., Yaghi O. K., Grun C., Strauss M. T., Bron P., Lai-Kee-Him J., Schueder F., Wang B., Wang P., Kishi J. Y., Myhrvold C., Zhu A., Jungmann R., Bellot G., Ke Y., Yin P., Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components. Nature 552, 72–77 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knudsen J. B., Liu L., Bank Kodal A. L., Madsen M., Li Q., Song J., Woehrstein J. B., Wickham S. F. J., Strauss M. T., Schueder F., Vinther J., Krissanaprasit A., Gudnason D., Smith A. A. A., Ogaki R., Zelikin A. N., Besenbacher F., Birkedal V., Yin P., Shih W. M., Jungmann R., Dong M., Gothelf K. V., Routing of individual polymers in designed patterns. Nat. Nanotechnol. 10, 892–898 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Jahn K., Tørring T., Voigt N. V., Schøler Sørensen R., Bank Kodal A. L., Andersen E. S., Vesterager Gothelf K., Kjems J., Functional patterning of DNA origami by parallel enzymatic modification. Bioconjug. Chem. 22, 819–823 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Praetorius F., Dietz H., Self-assembly of genetically encoded DNA-protein hybrid nanoscale shapes. Science 355, eaam5488 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Ketterer P., Willner E. M., Dietz H., Nanoscale rotary apparatus formed from tight-fitting 3D DNA components. Sci. Adv. 2, e1501209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marras A. E., Zhou L., Su H.-J., Castro C. E., Programmable motion of DNA origami mechanisms. Proc. Natl. Acad. Sci. U.S.A. 112, 713–718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douglas S. M., Bachelet I., Church G. M., A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Douglas S. M., Chou J. J., Shih W. M., DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl. Acad. Sci. U.S.A. 104, 6644–6648 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berardi M. J., Shih W. M., Harrison S. C., Chou J. J., Mitochondrial uncoupling protein 2 structure determined by NMR molecular fragment searching. Nature 476, 109–113 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin T. G., Bharat T. A. M., Joerger A. C., Bai X.-c., Praetorius F., Fersht A. R., Dietz H., Scheres S. H. W., Design of a molecular support for cryo-EM structure determination. Proc. Natl. Acad. Sci. U.S.A. 113, E7456–E7463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derr N. D., Goodman B. S., Jungmann R., Leschziner A. E., Shih W. M., Reck-Peterson S. L., Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science 338, 662–665 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei R., Martin T. G., Rant U., Dietz H., DNA origami gatekeepers for solid-state nanopores. Angew. Chem. Int. Ed. Engl. 51, 4864–4867 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Langecker M., Arnaut V., Martin T. G., List J., Renner S., Mayer M., Dietz H., Simmel F. C., Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 338, 932–936 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Funke J. J., Ketterer P., Lieleg C., Schunter S., Korber P., Dietz H., Uncovering the forces between nucleosomes using DNA origami. Sci. Adv. 2, e1600974 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kilchherr F., Wachauf C., Pelz B., Rief M., Zacharias M., Dietz H., Single-molecule dissection of stacking forces in DNA. Science 353, aaf5508 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Nickels P. C., Wünsch B., Holzmeister P., Bae W., Kneer L. M., Grohmann D., Tinnefeld P., Liedl T., Molecular force spectroscopy with a DNA origami-based nanoscopic force clamp. Science 354, 305–307 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gopinath A., Miyazono E., Faraon A., Rothemund P. W. K., Engineering and mapping nanocavity emission via precision placement of DNA origami. Nature 535, 401–405 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Steinhauer C., Jungmann R., Sobey T. L., Simmel F. C., Tinnefeld P., DNA origami as a nanoscopic ruler for super-resolution microscopy. Angew. Chem. Int. Ed. Engl. 48, 8870–8873 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Bui H., Onodera C., Kidwell C., Tan Y. P., Graugnard E., Kuang W., Lee J., Knowlton W. B., Yurke B., Hughes W. L., Programmable periodicity of quantum dot arrays with DNA origami nanotubes. Nano Lett. 10, 3367–3372 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Acuna G. P., Möller F. M., Holzmeister P., Beater S., Lalkens B., Tinnefeld P., Fluorescence enhancement at docking sites of DNA-directed self-assembled nanoantennas. Science 338, 506–510 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Kuzyk A., Schreiber R., Fan Z., Pardatscher G., Roller E.-M., Högele A., Simmel F. C., Govorov A. O., Liedl T., DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 483, 311–314 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Roller E.-M., Argyropoulos C., Höegele A., Liedl T., Pilo-Pais M., Plasmon–exciton coupling using DNA templates. Nano Lett. 16, 5962–5966 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Roller E. M., Khorashad L. K., Fedoruk M., Schreiber R., Govorov A. O., Liedl T., DNA-assembled nanoparticle rings exhibit electric and magnetic resonances at visible frequencies. Nano Lett. 15, 1368–1373 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhler P., Roller E.-M., Schreiber R., Liedl T., Lohmüller T., Feldmann J., Plasmonic DNA-origami nanoantennas for surface-enhanced Raman spectroscopy. Nano Lett. 14, 2914–2919 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Schreiber R., Luong N., Fan Z., Kuzyk A., Nickels P. C., Zhang T., Smith D. M., Yurke B., Kuang W., Govorov A. O., Liedl T., Chiral plasmonic DNA nanostructures with switchable circular dichroism. Nat. Commun. 4, 2948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kershner R. J., Bozano L. D., Micheel C. M., Hung A. M., Fornof A. R., Cha J. N., Rettner C. T., Bersani M., Frommer J., Rothemund P. W. K., Wallraff G. M., Placement and orientation of individual DNA shapes on lithographically patterned surfaces. Nat. Nanotechnol. 4, 557–561 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Maune H. T., Han S.-., Barish R. D., Bockrath M., Goddard W. A. III, Rothemund P. W. K., Winfree E., Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates. Nat. Nanotechnol. 5, 61–66 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Hung A. M., Micheel C. M., Bozano L. D., Osterbur L. W., Wallraff G. M., Cha J. N., Large-area spatially ordered arrays of gold nanoparticles directed by lithographically confined DNA origami. Nat. Nanotechnol. 5, 121–126 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Li S., Jiang Q., Liu S., Zhang Y., Tian Y., Song C., Wang J., Zou Y., Anderson G. J., Han J.-Y., Chang Y., Liu Y., Zhang C., Chen L., Zhou G., Nie G., Yan H., Ding B., Zhao Y., A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 36, 258–264 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Praetorius F., Kick B., Behler K. L., Honemann M. N., Weuster-Botz D., Dietz H., Biotechnological mass-production of DNA origami. Nature 552, 84–87 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Cassinelli V., Oberleitner B., Sobotta J., Nickels P., Grossi G., Kempter S., Frischmuth T., Liedl T., Manetto A., One-step formation of “chain-armor”-stabilized DNA nanostructures. Angew. Chem. Int. Ed. Engl. 54, 7795–7798 (2015). [DOI] [PubMed] [Google Scholar]
- 49.De Stefano M., Gothelf K. V., Dynamic chemistry of disulfide terminated oligonucleotides in duplexes and double-crossover tiles. ChemBioChem 17, 1122–1126 (2016). [DOI] [PubMed] [Google Scholar]
- 50.O’Neill P., Rothemund P. W. K., Kumar A., Fygenson D. K., Sturdier DNA nanotubes via ligation. Nano Lett. 6, 1379–1383 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Rajendran A., Endo M., Katsuda Y., Hidaka K., Sugiyama H., Photo-cross-linking-assisted thermal stability of DNA origami structures and its application for higher-temperature self-assembly. J. Am. Chem. Soc. 133, 14488–14491 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Ponnuswamy N., Bastings M. M. C., Nathwani B., Hee Ryu J., Chou L. Y. T., Vinther M., Aileen Li W., Anastassacos F. M., Mooney D. J., Shih W. M., Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 8, 15654 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agarwal N. P., Matthies M., Gür F. N., Osada K., Schmidt T. L., Block copolymer micellization as a protection strategy for DNA origami. Angew. Chem. Int. Ed. Engl. 56, 5460–5464 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Taylor J. S., Unraveling the molecular pathway from sunlight to skin cancer. Acc. Chem. Res. 27, 76–82 (1994). [Google Scholar]
- 55.Lima-Bessa K. M., Menck C. F. M., Skin cancer: Lights on genome lesions. Curr. Biol. 15, R58–R61 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Lewis R. J., Hanawalt P. C., Ligation of oligonucleotides by pyrimidine dimers—A missing ‘link’ in the origin of life?. Nature 298, 393–396 (1982). [DOI] [PubMed] [Google Scholar]
- 57.Castro C. E., Kilchherr F., Kim D.-N., Lin Shiao E., Wauer T., Wortmann P., Bathe M., Dietz H., A primer to scaffolded DNA origami. Nat. Methods 8, 221–229 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Sobczak J.-P., Martin T. G., Gerling T., Dietz H., Rapid folding of DNA into nanoscale shapes at constant temperature. Science 338, 1458–1461 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Wagenbauer K. F., Wachauf C. H., Dietz H., Quantifying quality in DNA self-assembly. Nat. Commun. 5, 3691 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang Y., Rabbi M., Kim M., Ke C., Lee W., Clark R. L., Mieczkowski P. A., Marszalek P. E., UVA generates pyrimidine dimers in DNA directly. Biophys. J. 96, 1151–1158 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cherepanova A., Tamkovich S., Pyshnyi D., Kharkova M., Vlassov V., Laktionova P., Immunochemical assay for deoxyribonuclease activity in body fluids. J. Immunol. Methods 325, 96–103 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Chen H., Li R., Li S., Andréasson J., Choi J. H., Conformational effects of UV light on DNA origami. J. Am. Chem. Soc. 139, 1380–1383 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Stahl E., Martin T. G., Praetorius F., Dietz H., Facile and scalable preparation of pure and dense DNA origami solutions. Angew. Chem. Int. Ed. Engl. 53, 12735–12740 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wagenbauer K. F., Engelhardt F., Stahl E., Hechtl V. K., Stömmer P., Seebacher F., Meregalli L., Ketterer P., Gerling T., Dietz H., How we make DNA origami. ChemBioChem 18, 1873–1885 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Kimanius D., Forsberg B. O., Scheres S. H. W., Lindahl E., Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scheres S. H. W., Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kremer J. R., Mastronarde D. N., McIntosh J. R., Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 70.Douglas S. M., Marblestone A. H., Teerapittayanon S., Vazquez A., Church G. M., Shih W. M., Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/8/eaau1157/DC1
Fig. S1. Design diagram of the brick-like (TT motifs 1 to 3) object prepared using caDNAno.
Fig. S2. Exemplary negative-stained TEM micrographs of the brick-like object (TT motifs 1 to 4) in different buffers/solvents.
Fig. S3. Laser-scanned fluorescent image of a 2.0% agarose gel that was run in an ice-cooled water bath.
Fig. S4. Laser-scanned fluorescent image of a 2.0% agarose gel that was run in an ambient temperature water bath.
Fig. S5. Design diagram of the brick-like (TT motifs 1 to 4) object prepared using caDNAno.
Fig. S6. Laser-scanned fluorescent image of a 2.0% agarose gel placed in an ice-cooled water bath.
Fig. S7. Design diagram of the pointer object.
Fig. S8. Exemplary negative-stained TEM micrographs of the pointer object in different buffers/solvents.
Fig. S9. Laser-scanned fluorescent image of a 2.0% agarose gel placed in a water bath.
Fig. S10. Laser-scanned fluorescent image of a 2.0% agarose gel placed in a water bath.
Fig. S11. Laser-scanned fluorescent image of a 2.0% agarose gel placed in a water bath.
Fig. S12. Laser-scanned fluorescent image of a 2.0% agarose gel placed in an ice-cooled water bath.
Fig. S13. Laser-scanned fluorescent image of a 2.0% agarose gel placed in a water bath.
Fig. S14. Laser-scanned fluorescent image of a 2.0% agarose gel placed in a water bath.
Fig. S15. Laser-scanned fluorescent image of a 2.0% agarose gel placed in a water bath.
Fig. S16. Laser-scanned fluorescent image of a 2.0% agarose gel placed in a water bath.
Fig. S17. Cryo-EM data of the brick-like object with TT-motifs (1) – (3) before crosslinking in folding buffer.
Fig. S18. Cryo-EM data of the brick-like object with TT-motifs (1) – (3) after crosslinking in folding buffer.
Fig. S19. Cryo-EM data of the brick-like object with TT-motifs (1) – (3) after crosslinking in phosphate-buffered saline (PBS).
Fig. S20. Slice-by-slice visualization of cryo-EM maps determined from brick samples.
Fig. S21. Cryo-EM data of the brick-like object with TT-motifs (1) – (4) before crosslinking in folding buffer.
Fig. S22. Cryo-EM data of the brick-like object with TT-motifs (1) – (4) after crosslinking in folding buffer.
Fig. S23. Design diagram of the switch object.
Fig. S24. Design diagram of the polymerization brick object.
Table S1. Sequences of staple strands for all DNA objects used in this work.
Movie S1. Tomogram obtained from cryo-EM of the brick-like variant with thymines at all staple termini and with TT motifs at all crossover sites.
Movie S2. Corresponding movie to fig. S20 for the nonirradiated (uncross-linked) sample of the brick-like variant with thymines at all staple termini and with TT motifs at all crossover sites in folding buffer and in the presence of 5 mM MgCl2.
Movie S3. Corresponding movie to fig. S20 for the irradiated (cross-linked) sample of the brick-like variant with thymines at all staple termini and with TT motifs at all crossover sites in folding buffer and in the presence of 5 mM MgCl2.
Movie S4. Corresponding movie to fig. S20 for the irradiated (cross-linked) sample of the brick-like variant with thymines at all staple termini and with TT motifs at all crossover sites in PBS buffer and in the absence of MgCl2.
Movie S5. Corresponding movie to fig. S20 for the nonirradiated (uncross-linked) sample of the brick-like variant with thymines at all staple termini, with TT motifs at all crossover sites and additional 5-T loops in folding buffer and in the presence of 5 mM MgCl2.
Movie S6. Corresponding movie to fig. S20 for the irradiated (cross-linked) sample of the brick-like variant with thymines at all staple termini, with TT motifs at all crossover sites and additional 5-T loops in folding buffer and in the presence of 5 mM MgCl2.
Reference (70)


