Abstract
Bacterial leaf blight is a major disease of eucalypt, especially under nursery conditions. Different bacterial species have been associated with the disease in several countries, and despite its importance worldwide, it is not clear to date whether similar disease symptoms are caused by the same or by different etiological agents. In this study, 43 bacterial strains were isolated from blighted eucalypt leaves collected in different geographic areas of Brazil and inoculated onto a susceptible eucalypt clone. Polyphasic taxonomy, including morphological, physiological, biochemical, molecular, and pathogenicity tests showed that only certain strains of Xanthomonas axonopodis caused symptoms of the disease. Strains varied in their aggressiveness, but no correlation with geographic origin was observed. MLSA-based phylogenetic analysis using concatenated dnaK, fyuA, gyrB and rpoD gene sequences allocated the strains in a well-defined clade, corresponding to Rade-marker’s group RG 9.6. Inoculation of nineteen plant species belonging to seven botanical families with representative strain LPF 602 showed it to be pathogenic only on Eucalyptus spp, and Corymbia spp. Based on distinct biochemical and pathogenic characteristics that differentiate the eucalypt strains from other pathovars of the X. axonopodis species, here we propose their allocation into the new pathovar X. axonopodis pv. eucalyptorum pv. nov.
Keywords: Eucalyptus spp., fatty acid profile, metabolism fingerprinting, multilocus sequence analysis, Xanthomonas axonopodis
Bacterial leaf blight is a major disease of eucalypt, especially under nursery conditions. In the period 2003–2008, the disease caused significant losses of clonal rooted cuttings and mini-stumps in several nurseries of Brazil, estimated at approximately 7.5 million dollars (Alfenas et al., 2009). Symptoms of the disease are characterized by the presence of water-soaked lesions, and interveinal necrosis, frequently accompanied by central perforations. The disease can also affect stems and branches (Neves et al., 2014). Intense defoliation is commonly observed in susceptible genotypes under conditions favorable for the disease.
In Brazil, the first records of bacterial leaf blight of eucalypt date back to the mid-1990s, when Pseudomonas cichorii (Pomella et al., 1995) and Xanthomonas sp. (Reis et al., 1996) were found to be associated with symptomatic nursery plants in the state of São Paulo. More recently, Gonçalves et al. (2008) reported that X. axonopodis, X. campestris, P. syringae, P. putida, P. cichorii, Erwinia sp. and strains belonging to the Rhizobiaceae family can also be recovered from diseased nursery and field plants in several regions of the country.
Different etiologic agents have been reported to be associated with eucalypt bacterial leaf blight in several other countries. Truman (1974) described similar disease symptoms affecting Corymbia citriodora (Hook) Hill and Johnson in Australia, and concluded that they were caused by X. deleyii subsp. eucalypti (Truman) Dye (synonym X. campestris pv. eucalypti). In South Africa, similar symptoms were observed in different eucalypt species, hybrids, and clones, and the disease was attributed to Pantoea ananatis (Serrano) Mergaert et al. (Coutinho et al., 2002) and, more recently, X. vasicola was also associated with similar disease symptoms in clones of Eucalyptus grandis W. Hill (Coutinho et al., 2015). In Thailand, a similar bacterial blight observed in seedling beds of Eucalyptus camaldulensis × E. deglupta and E. grandis under commercial production was attributed to X. axonopodis pv. eucalypti (Pothiluk et al., 2013).
Despite the high importance of leaf blight for eucalypt production worldwide, it remains to be conclusively determined whether similar symptoms are caused by the same or by different bacterial pathogens in several countries. In the study reported by Gonçalves et al. (2008), the authors found that isolates of X. axonopodis, P. cichorii and others belonging to the Rhizobiaceae family induced typical symptoms of the disease when spray inoculated onto a hybrid clone of E. urophylla × E. maidenii, which is susceptible to the disease upon natural infection. It is also not clear whether several bacterial pathogens have evolved to infect Eucalyptus spp. Recently, Coutinho et al. (2015) suggested that X. vasicola underwent a host jump from sugarcane to E. grandis in South Africa.
It is currently accepted that the genus Xanthomonas consists of at least 28 plant-associated species that infect more than 120 species of monocot and 260 species of dicot plants (Hayward, 1993; Samanta et al., 2013; Young et al., 2010). Successful classification of Xanthomonas at the infraspecific level has been accomplished using multilocus sequence analysis (MLSA) (Ah-You et al., 2009; Ferraz et al., 2017; Samanta et al., 2013; Young et al., 2008; 2010). MLSA is a simple and rapid technique that allows accurate allocation of strains in their respective taxonomic groups, in most cases reflecting their classification based on DNA-DNA reassociation techniques (Young et al., 2008).
We conducted an extensive sampling of eucalypt rooted cuttings with symptoms of bacterial leaf blight in different regions of Brazil and a polyphasic approach that included MLSA in order to identify the bacterium responsible for the disease. We found that Xanthomonas axonopodis was consistently associated with leaf blight and was the only species, among those recovered in this study, that caused such symptoms on eucalypt. Host range determination and cross-inoculation experiments revealed that the bacterium belongs to a pathovar not previously described.
Materials and Methods
Bacterial strains
Leaf samples were collected between September 2012 and June 2013 from Eucalyptus spp. plants showing leaf blight in five geographic regions of Brazil: Southeast, South, Midwest, North and Northeast. For confirmation of the bacterial etiology of the disease, symptomatic leaf tissue was subjected to the exudation test. To this end, fragments of approximately 0.5 × 0.5 cm were taken from the lesion margins and placed on a drop of sterile water on a glass slide and observed under a light microscope (magnification × 200). Leaf tissue showing bacterial exudation was washed with a neutral detergent, rinsed three times with sterile water, and macerated. Loopfuls of the macerate were streaked on solid 523 medium (Kado and Heskett, 1970) for bacterial isolation. Pure cultures were obtained from individual colonies after incubation at 28°C for 48 h. Bacterial strains were preserved in 30% glycerol stocks stored at −80°C for later use.
Pathogenicity tests
Clone CLR368, a E. urophylla × E. globulus hybrid previously characterized as highly susceptible to bacterial leaf blight (Clonar Resistência a Doenças Florestais, unpublished data), was used to determine the pathogenicity of the strains. For this, 60-day old cuttings were transplanted into 2-dm3 plastic bags containing the commercial substrate MecPlant (Mecprec Indústria e Comércio Ltda., Telêmaco Borba, PR, Brazil) supplemented with 6 kg m−3 of single superphosphate and fertilized with 1.5 kg m−3 Osmocote (19 : 6 : 10) (Scotts Australia Pty Ltd., Bella Vista, Australia). Thirty days after planting, cuttings of clone CLR368 were inoculated with suspensions of the bacterial strains prepared from a culture grown on solid 523 medium at 28°C for 24 h. The suspension was adjusted to an OD600 = 0.25 (equivalent to 108 cfu ml−1) and sprayed onto the eucalypt leaves using a De Vilbiss 1.5 HP air compressor (DeVilbiss Air Power Company, Jackson, TN, USA). Three individual cuttings were inoculated with each strain. The cuttings were maintained in a mist irrigation chamber at 26°C for 24 h before and for 24 h after inoculation. Subsequently, the plants were returned to a greenhouse until the appearance of bacterial blight symptoms. Percent of leaf area affected by the disease was determined using the diagrammatic scale proposed by Gonçalves (2003), choosing the ten leaves with the highest severity and then calculating the mean disease severity for each plant. Such a scale of disease severity assessment was developed by scanning symptomatic leaves at 300 dpi, saving the images in a bmp format and quantifying the affected leaf area with the program Quant v1.0 (Vale et al., 2003). The experiments were conducted in completely randomized designs, the data subjected to ANOVA, and the treatment means compared with the Scott-Knott grouping test (P ≤ 0.05). Statistical analyses were performed with the SAS software (SAS Institute Inc., Cary, NC, USA).
16S rDNA sequencing and analysis
For DNA extraction, bacterial strains were grown in 5 ml of liquid 523 medium at 28°C for 48 h in the dark. One ml of bacterial culture was transferred to a microfuge tube and centrifuged at 13,000 rpm for 3 min. The DNA was extracted from bacterial cells using the Wizard Genomic DNA Purification kit (Promega Corporation, Mannheim, Germany) according to the manufacturer’s instructions. The DNA of each isolate was diluted to a concentration of 10 ng μl−1 for conducting the PCR. Primers fd2 (5-AGAGTTTGATCCTGGCTCAG-3) and rP1 (5-ACGGTTACCTTGTTACGACTT-3) were used to PCR amplify the 16S rDNA region (Weisburg et al., 1991) in a reaction mixture containing 12.5 μl of Dream Taq PCR Master Mix 2X (MBI Fermentas, Hanover, MD), 1.5 μl of 10 μM of each primer, and 2 μl of genomic DNA in a total volume of 25 μl. PCR amplification was performed by an initial denaturation at 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 2 min, and a final extension at 72°C for 5 min. PCR products were confirmed for their expected sizes by running 5 μl on a 1.4% agarose gel at 100 V for 60 min, purified with the PCR kit GFX DNA and Gel Band Purification Kit (GE Healthcare Life Sci., Sao Paulo, SP, Brazil), and their sequences determined in an Applied Biosystems 3500XL Series Genetic Analyser (Applied BioSystems, Foster City, CA) using the same primers used for DNA amplification. The sequences were analyzed using Navigator version 1.0.1 (Applied BioSystems) and compared with sequences deposited in the GenBank (http://www.ncbi.nlm.nih.gov) using BLASTN (Altschul et al., 1997).
Multilocus sequence analysis (MLSA)
PCR amplifications were carried out with primers specific for the four housekeeping genes dnaK, fyuA, gyrB and rpoD (Young et al., 2008) using the same reaction mixture as for amplification of the 16S rDNA region. PCR amplification was performed with initial denaturation at 94°C for 3 min, 30 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min, and final extension at 72°C for 10 min. The sequences of the PCR products were obtained and analyzed as for 16S rDNA and subsequently aligned using Clustal W v.1.5 (Thompson et al., 1994) followed by manual adjustments in MEGA 5.00 (Tamura et al., 2011), when necessary. Because dnaK, fyuA, gyrB and rpoD genes encode proteins, it was verified that the insertion of gaps in the alignment did not alter their protein sequences.
Phylogenetic trees were constructed with sequences of the four housekeeping genes obtained in this work, those reported by Young et al. (2008) for Xanthomonas spp., and others extracted from the Genbank (http://www.ncbi.nlm.nih.gov) (Table 1). In the first phylogenetic analysis, sequences from validly published Xanthomonas species were used (Bull et al., 2010; 2012). The second phylogenetic analysis was conducted using housekeeping gene sequences of several different X. axonopodis pathovars (Table 1). In this study, we adopted the Xanthomonas classification and nomenclature proposed by Vauterin et al (1995). When appropriate, the most recently published bacterial names are placed in parentheses.
Table 1.
Xanthomonas strains used in multilocus sequence analysis
| Bacterial strain | Species name | Synonym | GenBank accession number | |||
|---|---|---|---|---|---|---|
|
| ||||||
| dnaK | fyuA | gyrB | rpoD | |||
| ICMP 196a | Xanthomonas albilineans | - | - | EU498963 | EU499082 | |
| ICMP 35a | X. arboricola pv. juglandis | EU498750 | EU498852 | EU498951 | EU499070 | |
| ICMP 50a | X. axonopodis pv. axonopodis | EU498751 | EU498853 | EU498952 | EU499071 | |
| CFBP 6369 | X. axonopodis pv. allii | CM002866 | CM002866 | CM002866 | CM002866 | |
| ICMP 5718b | X. axonopodis pv. alfalfae | X. alfalfae subsp. alfalfae | EU498792 | EU498894 | EU499001 | EU499120 |
| ICMP 8432 | X. axonopodis pv. aurantifolii | X. fuscans subsp. aurantifolii | EU498811 | EU498913 | EU499027 | EU499146 |
| ICMP 194b | X. axonopodis pv. begoniae | EU498757 | EU498859 | EU498962 | EU499081 | |
| ICMP 444b | X. axonopodis pv. cajani | EU498767 | EU498869 | EU498973 | EU499092 | |
| ICMP 24a | X. axonopodis pv. citri | X. citri subsp. citri | EU498749 | EU498851 | EU498950 | EU499069 |
| ICMP 10009 | X. axonopodis pv. citrumelo | X. alfalfae subsp. citrumelonis | EU498826 | EU498926 | EU499042 | EU499162 |
| DXD 01 | X. axonopodis pv. commiphorae | JN898928 | JN621250 | JN621253 | JN621257 | |
| LMG 695 | X. axonopodis pv. dieffenbachiae | CP014347 | CP014347 | CP014347 | CP014347 | |
| ICMP 5732b | X. axonopodis pv. glycines | EU498794 | EU498896 | EU499003 | EU499122 | |
| ICMP 217 | X. axonopodis pv. malvacearum | X. citri subsp. malvacearum | EU498760 | EU498862 | EU498966 | EU499085 |
| LMG 941 | X. axonopodis pv. mangiferaeindicae | CAHO01000008 | HQ590941 | CAHO01000002 | CAHO01000012 | |
| ICMP 5741b | X. axonopodis pv. manihotis | EU498796 | EU498898 | EU499006 | EU499125 | |
| ICMP 5834b | X. axonopodis pv. phaseoli | X. phaseoli | EU498802 | EU498904 | EU499015 | EU499134 |
| ICMP 239a | X. axonopodis pv. phaseoli (biovar. fuscans) | X. fuscans subsp. fuscans | EU498761 | EU498863 | EU498967 | EU499086 |
| NCPPB 466 | X. axonopodis pv. punicae | JN790906 | JN790914 | JN790930 | JN790938 | |
| ICMP 3031 | X. axoopodisn pv. ricini | EU498782 | EU498884 | EU498991 | EU499110 | |
| ICMP 5757b | X. axonopodis pv. vasculorum | EU498798 | EU498900 | EU499011 | EU499130 | |
| ICMP 333b | X. axonopodis pv.vignicola | EU498764 | EU498866 | EU498970 | EU499089 | |
| ICMP 109a | X. axonopodis pv. vesicatoria | X. euvesicatoria | EU498754 | EU498856 | EU498955 | EU499074 |
| ICMP 16690a | X. axonopodis pv. vesicatoria | X. perforans | EU498844 | EU498944 | EU499059 | EU499179 |
| ICMP 12545a | X. bromi | EU498837 | EU498937 | EU499052 | EU499172 | |
| ICMP 6541 | X. campestris pv. campestris | EU498747 | EU498849 | EU498948 | EU499067 | |
| ICMP 204a | X. cassavae | EU498759 | EU498861 | EU498965 | EU499084 | |
| ICMP 9513a | X. codiaei | EU498822 | EU498922 | EU499038 | EU499158 | |
| ICMP 2299a | X. cucurbitae | EU498780 | EU498882 | EU498989 | EU499108 | |
| ICMP 16775a | X. cynarae | EU498846 | EU498946 | EU499061 | EU499181 | |
| ICMP 2415a | X. dyei pv. dysoxyli | EU498781 | EU498883 | EU498990 | EU499109 | |
| ICMP 5715a | X. frageriae | EU498791 | EU498893 | EU499000 | EU499119 | |
| ICMP 16689a | X. gardneri | EU498843 | EU498943 | EU499058 | EU499178 | |
| ICMP 453a | X. hortorum | EU498769 | EU498871 | EU498975 | EU499094 | |
| ICMP 189a | X. hyacinthi | - | - | EU498960 | EU499079 | |
| ICMP 8683 | X. melonis | EU498816 | EU498916 | EU499032 | EU499152 | |
| ICMP 3125a | X. oryzae pv. oryzae | EU498784 | EU498886 | EU498993 | EU499112 | |
| ICMP 570a | X. pisi | EU498770 | EU498872 | EU498976 | EU499095 | |
| ICMP 5816a | X. populi | EU498801 | EU498903 | EU499014 | EU499133 | |
| ICMP 16916a | X. sacchari | - | - | EU499063 | EU499183 | |
| ICMP 6774a | X. theicola | - | - | EU499020 | EU499139 | |
| ICMP 5752a | X. translucens pv. translucens | - | - | EU499009 | EU499128 | |
| ICMP 3103a | X. vasicola | EU498783 | EU498885 | EU498992 | EU499111 | |
| ICMP 63a | X. vesicatoria | EU498753 | EU498855 | EU498954 | EU499073 | |
| ICMP 17033a | Stenotrophomonas maltophilia | EU498848 | - | EU499066 | EU499186 | |
CFBP- Collection Française de Bactéries Phytopathogènes
ICMP- International Collection of Microorganisms from Plants
LMG- BCCM/LMG Bacteria Collection, Laboratory for Microbiology
NCPPB- National Collection of Plant Pathogenic Bacteria
Type strain of species
Pathotype strain
Bayesian Inference (BI) methods were used to construct phylogenetic trees of concatenated dnaK, fyuA, gyrB and rpoD gene sequences. BI was performed with MrBayes 3.1.2 (Bayesian Inference of Phylogeny) (Ronquist and Huelsenbeck, 2003). MrModelTest 3.2 was used to choose the substitution model based on the Akaike information criterion (AIC). The probability of an a posteriori tree distribution was calculated using an MCMC algorithm (Metropolis-coupled Markov Chain Monte Carlo) of two chains from a random tree with 10 million generations, discarding 25% of the first trees. The Tracer 1.4 program was used to examine the MCMC convergence and effective sample size. FigTree 1.3.1. (http://tree.bio.ac.uk/software) was used to view and edit the phylogenetic trees.
Biochemical and physiological tests
The following tests were performed on the eucalypt strains according to the Laboratory Guide for Identification of Plant Pathogenic Bacteria (Schaad et al., 2001): (i) Ryu’s non-staining KOH test for Gram determination; (ii) yellow pigmentation of colonies on YDC (yeast extract-dextrose-calcium carbonate agar) medium; (iii) xanthomonadin production; (iv) fluorescence production on King’s B medium; (v) anaerobic growth; (vi) oxidase test; (vii) catalase activity; (viii) utilization of asparagine as a sole carbon and nitrogen source; (ix) urease activity; (x) gelatin liquefaction; (xi) starch hydrolysis; (xii) H2S production from cysteine; (xiii) esculin hydrolysis; and (xiv) NO3 reduction.
Metabolic fingerprinting
The Biolog GN microPlate system (Microlog 2, Version 3.5, Biolog Inc.) was used to test the ability of the strains to metabolize different carbon sources. For that, trypticase soy agar (TSA) medium was inoculated with a single colony obtained from a nutrient agar (NA) culture and incubated at 28°C for 24 h. Then, a bacterial suspension was prepared in saline solution, the absorbance adjusted to match the turbidity standards of the Biolog GN MicroPlate system, and 150 μl pipetted into each well. The microplates were incubated at 28°C and the reactions rated as positive or negative at 48 h of incubation.
Fatty acid analysis
The Microbial Identification System (MIS; MIDI Inc., Newark, DE, USA) was used to determine the whole cell fatty acid methyl ester (FAME) profiles of the eucalypt strains. Fatty acids were extracted from cultures on trypticase soy agar (BD BBL, São Paulo, Brazil) grown at 28°C for one day according to the procedures described in the MIS Handbook. Extracts were analyzed with a Hewlett-Packard 7890 gas chromatograph (Hewlett Packard, Palo Alto, CA, USA), which provided automatic identification and quantification of fatty acids by comparison with internal FAME standards. Fatty acid profiles of eucalypt strains were compared to those of reference strains deposited in the MIS database. To this end, similarity indices were generated by the Sherlock MIS software comparing the fatty acid profiles of eucalypt strains with those of known Xanthomonas species and pathovars deposited in the MIS database.
Host range and cross-inoculation
Bacterial strains isolated from eucalypt leaves clustered together in Rademaker’s group RG 9.6 (Rademaker et al. 2005). Therefore, LPF 602 was selected to represent the strains isolated from Eucalyptus spp. Strain LPF 602 was used to inoculate plants belonging to different botanical families. Myrtaceae: Eucalyptus grandis W. Hill, E. urophylla S. T. Blake, E. robusta Sm., E. saligna Smith, E. globulus Labill., E. cloeziana F. Muell, E. camaldulensis Dehnh, Corymbia maculata (Hook.) K. D. Hill & L. A. S. Johnson, Psidium guajava L., Myrciaria jaboticaba (Vell.) O. Berg. and Eugenia jambolana Lam.; Rosaceae: Prunus persica (L.) Batsch; Caricaceae: Carica papaya L.; Fabaceae: Phaseolus vulgaris L. and Pisum sativum L.; Solanaceae: Solanum lycopersicum Mill.; Rutaceae: Citrus limon (L.) Burm. f.; Euphorbiaceae: Mabea fistulifera Mart. and Anacardiaceae: Schinus terebinthifolius Raddi. Twenty plants of each species were inoculated with bacterial suspensions of strain LPF 602.
Also, cross inoculation experiments were conducted with LPF 602 and strains of the X. axonopodis pathovars phylogenetically most closely related to the eucalypt strains and that clustered in Rademaker’s group RG 9.6. Based on the results of MLSA, it was decided to inoculate strain LPF 602 onto plants of common bean (Phaseolus vulgaris L.) and acid lime (Citrus × aurantiifolia (Christm.) Swingle). Representative strains of X. axonopodis pv. vignicola (IBSBF 1739 = NCPPB 1838 = ICMP 333 = ATCC 11648), X. axonopodis pv. aurantifolii (X. fuscans subsp. aurantifolii) (IBSBF 380 = NCPPB 3654 = CFBP 2905), and X. axonopodis pv. phaseoli var. fuscans (X. fuscans subsp. fuscans) (UNB 772) were inoculated onto rooted cuttings of clone CLR368 (E. urophylla × E. globulus) as well as on susceptible plants of their corresponding host species. Plants of common bean were inoculated separately with X. axonopodis pv. vignicola and X. axonopodis pv. phaseoli var. fuscans (X. fuscans subsp. fuscans), because it is susceptible to both pathovars. Plant maintenance, inoculum preparation and inoculation were conducted as described for pathogenicity tests. Bacterial strains inoculated onto their respective host species served as positive controls for disease.
Results
Bacterial isolation
Forty-three bacterial strains were obtained from eucalypt leaves with symptoms of bacterial blight. Yellow colonies were repeatedly isolated from symptomatic leaves after 24–48 h of incubation on solid 523 medium. In some cases, white cream colonies were also observed within the first 24 h of incubation. However, these bacterial colonies were not pathogenic to clone CLR368. Only pathogenic strains were selected for further characterization in this study.
Pathogenicity on eucalypt
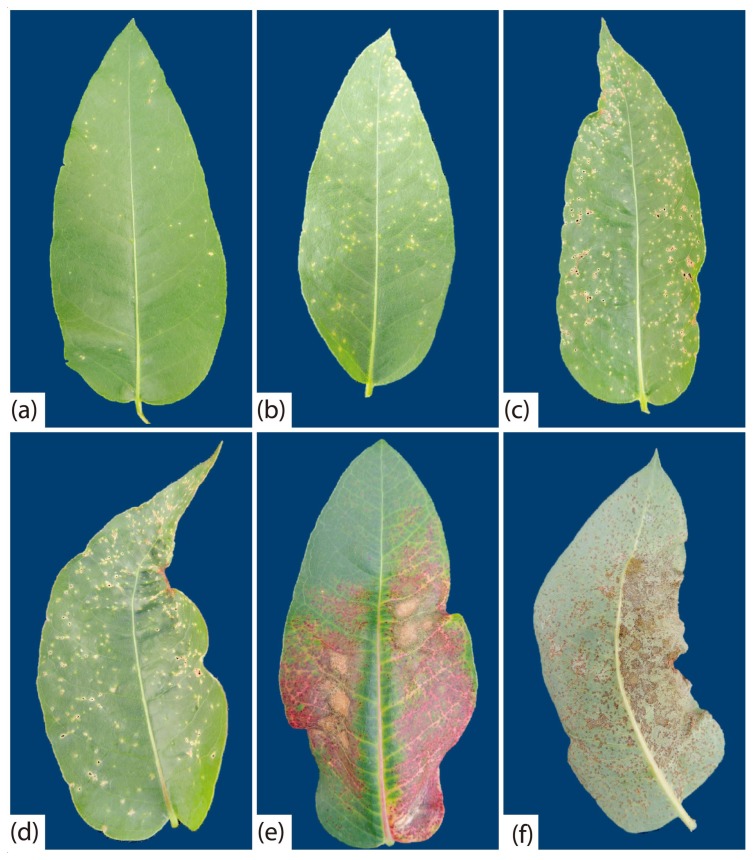
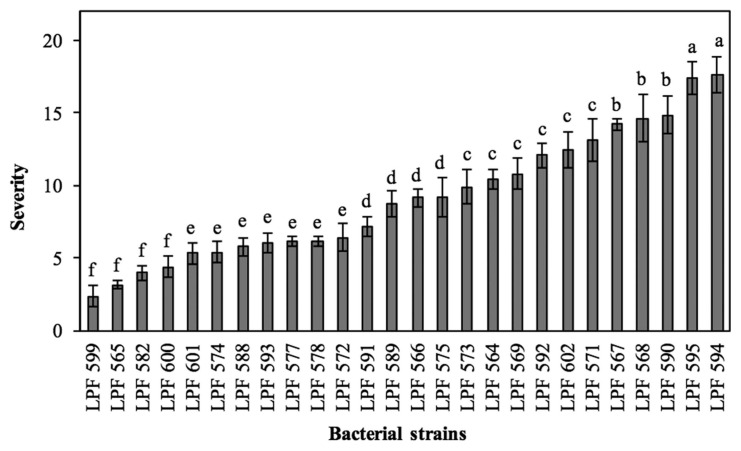
Twenty-six of the 43 bacterial strains obtained from symptomatic leaves were pathogenic when inoculated by spraying onto clone CLR368. The pathogenic strains were assigned accession numbers and deposited in the collection of phytopathogens of the Forest Pathology Laboratory at the Universidade Federal de Viçosa (Table 2). All strains caused the same symptoms, but they varied in aggressiveness (evolution rate of disease symptoms). The first symptoms of the disease were observed at seven days after inoculation (dai). Initially, lesions were small, chlorotic, and scattered on the leaf blade (Fig. 1A), and with the progress of the disease they became necrotic and coalescent (Fig. 1B). In some leaves, it was observed detachment of tissue from the center of the lesions, distortion of the leaf and tanning of the limb (Fig. 1C–E). On the abaxial side, necrotic and rough lesions were observed (Fig. 1F). No bacterial leaf blight symptoms were observed on control plants. Morphological characteristics and gyrB gene sequences of bacterial strains re-isolated in pure culture from inoculated plants were the same as those obtained from naturally infected tissue, in fulfilment of Koch’s postulate. The strains varied in aggressiveness, the most aggressive being LPF 594, and LPF 595, and the least aggressive being LPF 599, LPF 565, LPF 582 and LPF 600 (Fig. 2).
Table 2.
Xanthomonas axonopodis strains collected in Brazil
| Strain | Host species | Collection city/State* | Brazilian region | Growth conditions | GenBank accession number | ||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| 16S rDNA | dnaK | fyuA | gyrB | rpoD | |||||
| LPF564 | Eucalyptus urophylla | Curvelo/MG | Southeast | Nursery | KY288830 | KY287807 | KY287848 | KY287889 | KY460467 |
| LPF565 | Eucalyptus urophylla | Curvelo/MG | Southeast | Nursery | KY288831 | KY287808 | KY287879 | KY287890 | KY460503 |
| LPF566 | Eucalyptus urophylla | Curvelo/MG | Southeast | Nursery | KY288832 | KY287809 | KY287849 | KY287891 | KY460504 |
| LPF567 | Eucalyptus urophylla | Curvelo/MG | Southeast | Nursery | KY288833 | KY287810 | KY287880 | KY287892 | KY460468 |
| LPF568 | Eucalyptus urophylla | Curvelo/MG | Southeast | Field | KY288834 | KY287811 | KY287850 | KY287893 | KY460469 |
| LPF569 | Eucalyptus urophylla | Curvelo/MG | Southeast | Field | KY288835 | KY287812 | KY287851 | KY287894 | KY460470 |
| LPF571 | Eucalyptus urophylla | Niquelândia/GO | Midwest | Nursery | KY288837 | KY287814 | KY287852 | KY287896 | KY460472 |
| LPF572 | Eucalyptus urophylla | Niquelândia/GO | Midwest | Nursery | KY288838 | KY287815 | KY287853 | KY287897 | KY460473 |
| LPF573 | E. grandis x E. urophylla | Niquelândia/GO | Midwest | Field | KY288839 | KY287816 | KY287854 | KY287898 | KY460501 |
| LPF574 | E. grandis x E. urophylla | Niquelândia/GO | Midwest | Field | KY288840 | KY287817 | KY287855 | KY287899 | KY460474 |
| LPF575 | E. urophylla x E. maidenii | Guaíba/RS | South | Field | KY288841 | KY287818 | KY287856 | KY287900 | KY460475 |
| LPF577 | Eucalyptus grandis | Paracatu/MG | Southeast | Nursery | KY288842 | KY287819 | KY287857 | KY287901 | KY460476 |
| LPF578 | Eucalyptus grandis | Paracatu/MG | Southeast | Nursery | KY288843 | KY287820 | KY287858 | KY287902 | KY460477 |
| LPF582 | E. grandis x E. urophylla | Limeira/SP | Southeast | Nursery | KY288847 | KY287824 | KY287860 | KY287906 | KY460481 |
| LPF588 | E. grandis x E. urophylla | Aracruz/ES | Southeast | Field | KY288853 | KY287829 | KY287864 | KY287911 | KY460485 |
| LPF589 | E. grandis x E. urophylla | Aracruz/ES | Southeast | Nursery | KY288854 | KY287830 | KY287865 | KY287912 | KY460486 |
| LPF590 | E. grandis x E. urophylla | Santana/AP | North | Nursery | KY288855 | KY287831 | KY287866 | KY287913 | KY460487 |
| LPF591 | E. grandis x E. urophylla | Santana/AP | North | Nursery | KY288856 | KY287832 | KY287867 | KY287914 | KY460488 |
| LPF592 | Eucalyptus grandis | Santana/AP | North | Field | KY288857 | KY287833 | KY287868 | KY287915 | KY460489 |
| LPF593 | E. urophylla x E. globulus | São José do Triunfo/PR | South | Nursery | KY288858 | KY287844 | KY287869 | KY287927 | KY460490 |
| LPF594 | E. urophylla x E. globulus | São José do Triunfo/PR | South | Nursery | KY288859 | KY287845 | KY287870 | KY287916 | KY460491 |
| LPF595 | E. urophylla x E. globulus | São José do Triunfo/PR | South | Nursery | KY288860 | KY287834 | KY287871 | KY287917 | KY460492 |
| LPF599 | Eucalyptus saligna | São José do Triunfo/PR | South | Nursery | KY288864 | KY287838 | KY287874 | KY287921 | KY460495 |
| LPF600 | E. grandis x E. urophylla | São José do Triunfo/PR | South | Nursery | KY288865 | KY287839 | KY287875 | KY287922 | KY460496 |
| LPF601 | E. grandis x E. urophylla | Tocantins | North | Nursery | KY288866 | KY287840 | KY287876 | KY287923 | KY460497 |
| LPF602 | E. grandis x E. urophylla | Teixeira de Freitas/BA | Northeast | Field | KY288867 | KY287841 | KY287877 | KY287925 | KY460500 |
AP- Amapá, BA- Bahia, ES- Espírito Santo, GO- Goiás, MG- Minas Gerais, PR- Paraná, RS- Rio Grande do Sul and SP- São Paulo.
Fig. 1.
Symptoms caused by Xanthomonas axonopodis in clone CLR368 of Eucalyptus urophylla × E. globulus. (a) Small chlorotic lesions caused by LPF 588; (b) coalescing lesions caused by LPF601; (c) perforation in the centers of lesions caused by LPF 573; (d) tanned injuries caused by LPF 591; (e) and (f) necrotic and rough lesions on the adaxial and abaxial sides of the leaf caused by LPF 594. Pictures were taken at 23 days after inoculation.
Fig. 2.
Severity of bacterial leaf blight in rooted cuttings of clone CLR368 (Eucalyptus urophylla × E. globulus) inoculated with strains of Xanthomonas axonopodis. Bars represent the mean and vertical lines the standard error of the mean. Severity was determined as percent of leaf area affected by the disease as in Gonçalves (2003). Means followed by the same letter are not statistically different by the Scott-Knott test (P ≤ 0.05).
Analysis of partial 16S rDNA sequences
Partial 16S rDNA sequences of approximately 1200 bp were obtained for each strain. Blast searches in the NCBI data base (http://www.ncbi.nim.nih.gov) revealed that the 16S rDNA sequences of strains isolated from eucalypt show similarity with those of several Xanthomonas. For example, the LPF 565 sequence showed 100% identity with those of X. axonopodis pv. phaseoli var. fuscans (X. fuscans subsp. fuscans) (CP023294), X. axonopodis pv. vignicola (CP022270), and X. axonopodis (KY982974). The LPF 582 sequence showed 100% identity with those of X. axonopodis pv. phaseoli var. fuscans (X. fuscans subsp. fuscans) (CP023294), X. axonopodis pv. vesicatoria (X. euvesicatoria) (KX512836), and X. campestris (KU163445). And, the LPF 602 sequence showed higher than 99% identity with those of X. axonopodis pv. dieffenbachiae (KM576803), X. axonopodis pv. citrumelo (X. alfalfae subsp. citrumelonis) (CP002914), and X. campestris pv. viticola (JQ513818). Therefore, all strains were identified as belonging to the genus Xanthomonas.
Multilocus sequence analysis (MLSA)
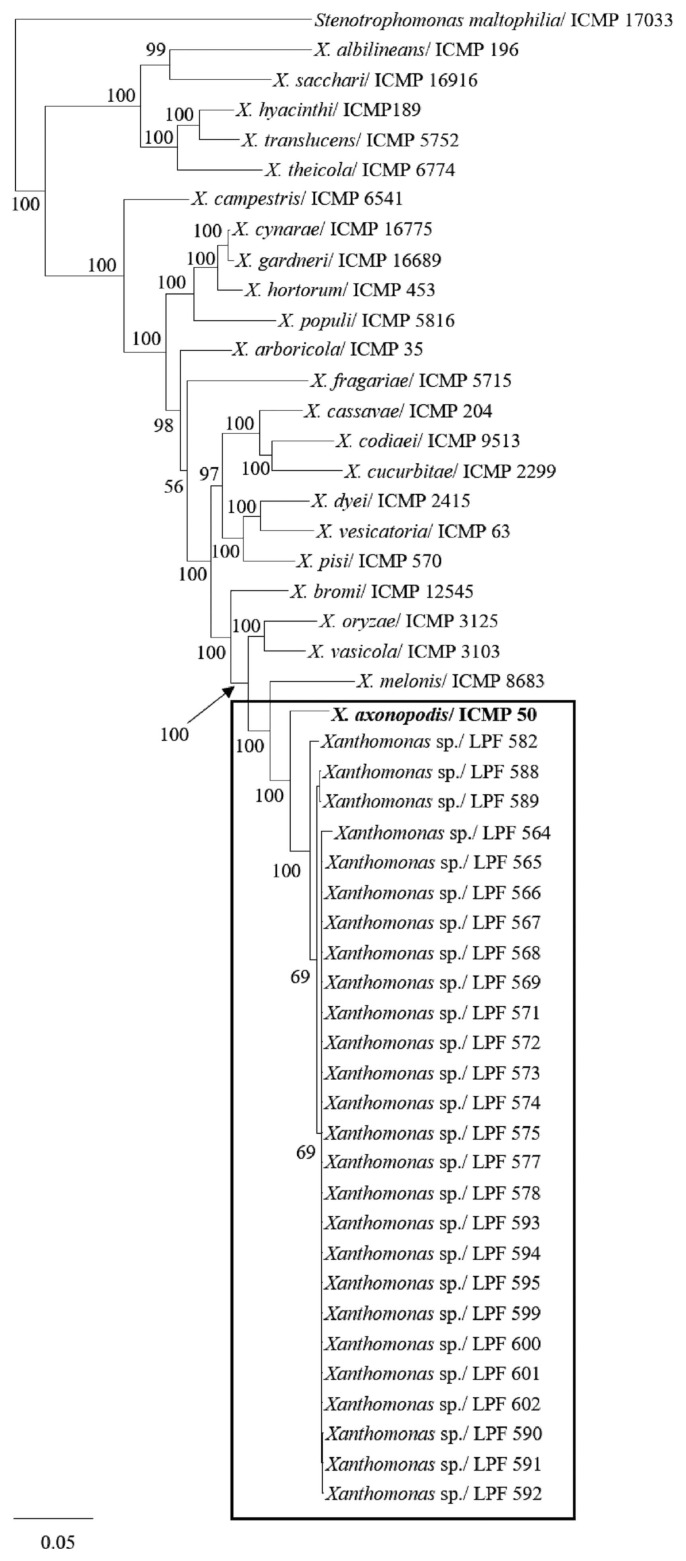
Partial sequences of 834, 657, 693, and 702 bp for dnaK, fyuA, gyrB and rpoD, respectively, were obtained for each strain and used in phylogenetic analyses. The total length of the alignment for the concatenated sequences of the four genes was 2886 bp. For the first phylogenetic analysis using sequences from validly published Xanthomonas species, the number of conserved sites was 648 for dnaK, 442 for fyuA, 321 for gyrB, and 402 for rpoD. The number of variable sites was 186 for dnaK, 215 for fyuA, 372 for gyrB, and 300 for rpoD, and the number of parsimony informative sites were 74, 144, 226, and 196 for dnaK, fyuA, gyrB, and rpoD, respectively. The best evolutionary model selected by MrModeltest 2.3 for Bayesian analysis by AIC was the General Time Reversible plus Invariant site plus Gamma (GTR + I + G) for all four genes. In this analysis, all pathogenic strains recovered from eucalypt grouped together with the type strain of the X. axonopodis species (Fig. 3).
Fig. 3.
Phylogenetic tree of concatenated dnaK, fyuA, gyrB and rpoD nucleotide sequences of diverse Xanthomonas species constructed by Bayesian inference. Strains isolated from eucalypt in this study are shown within the rectangle. Posterior probability values are indicated on the branches. Bars indicate the fraction of substitutions per site. Stenotrophomonas maltophilia (ICMP 17033) was used as an outgroup.
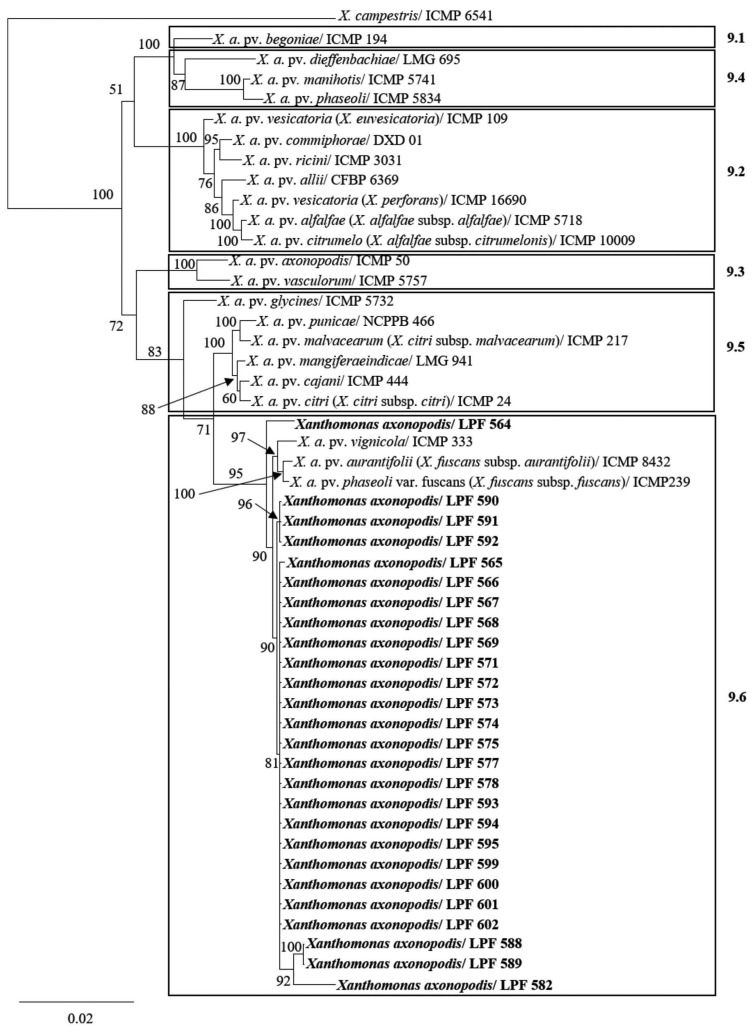
We then performed a phylogenetic analysis to position the eucalypt strains within the X. axonopodis species using gene sequences of pathovars previously described and available in the GenBank. In this case, the number of conserved sites was 749 for dnaK, 535 for fyuA, 534 for gyrB, and 599 for rpoD. The number of variable sites was 85 for dnaK, 122 for fyuA, 159 for gyrB, and 103 for rpoD, and the number of parsimony informative sites were 49, 57, 66, and 49 for dnaK, fyuA, gyrB, and rpoD, respectively. The best evolutionary model selected by MrModeltest 2.3 for Bayesian analysis by AIC were the GTR + I for dnaK and fyuA, and GTR + G for gyrB and rpoD. Our results indicate that all strains pathogenic to eucalypt cluster in RG 9.6 group (Fig. 4) corresponding to the allocation scheme of X. axonopodis pathovars proposed by Rademaker et al (2005).
Fig. 4.
Phylogenetic tree of concatenated dnaK, fyuA, gyrB and rpoD nucleotide sequences of Xanthomonas axonopodis pathovars constructed by Bayesian inference. Strains isolated from eucalypt in this study are shown in bold. Posterior probability values are indicated on the branches. X. axonopodis is divided in groups according to Rademaker et al. (2005) as RG 9.1 to RG 9.6. Bar indicates number of substitutions per site. X. campestris pv. campestris (ICMP 6541) was used as an outgroup.
Biochemical and physiological tests
Strains pathogenic to eucalypt were Gram-negative and on YDC agar medium colonies were yellow and mucilaginous with abundant slime formation. Bacterial strains were positive for catalase, starch utilization, H2S production, esculin hydrolysis, xanthomonadin production, and gelatin liquefaction. They were negative for growth under anaerobic conditions, production of fluorescent pigments on King’s B medium, oxidase, urease, nitrate reductase, and utilization of asparagine as a sole source of carbon and nitrogen.
Metabolic fingerprinting
We selected LPF 564, LPF 582, LPF 590, and LPF 602 pathogenic strains recovered from eucalypt to determine their profiles of carbon source utilization using the Biolog system. The profile of most strains was the same as the profile of X. axonopodis reported by Vauterin et al. (1995), the exceptions being LPF 582 that was not able to utilize methyl pyruvate, and LPF 582 and LPF 602 that were able to utilize glucuronamide (Table 3). Strains recovered from eucalypt were able to utilize 36 carbon sources and unable to utilize 47 carbon sources, whereas variable results were obtained for 12 carbon sources (Table 3). A reaction was considered positive when all strains metabolized the substrate and negative when no strain metabolized the substrate. A reaction was considered variable when at least one (but not all) strain utilized the substrate.
Table 3.
Characterization of X. axonopodis strains based on the Biolog test
| Carbon source | LPF 564 | LPF 582 | LPF 590 | LPF 602 | Vauterin et al. (1995)* |
|---|---|---|---|---|---|
| α-cyclodextrin | − | − | − | − | − |
| Dextrin | + | + | + | + | + |
| Glycogen | + | + | + | + | V |
| Tween 40 | + | + | + | + | V |
| Tween 80 | + | + | + | + | V |
| N-Acetyl-D-galactosamine | − | − | − | − | V |
| N-acetyl-D-glucosamine | + | + | + | + | V |
| Adonitol | − | − | − | − | − |
| L-arabinose | − | − | − | − | V |
| D-arabitol | − | − | − | − | − |
| D-cellobiose | + | + | + | + | + |
| i-erythritol | − | − | − | − | − |
| D-fructose | + | + | + | + | + |
| L-fucose | + | + | + | + | V |
| D-galactose | + | + | + | + | V |
| Gentiobiose | + | + | + | + | + |
| α-D-glucose | + | + | + | + | + |
| m-inositol | − | − | − | − | − |
| α-D-lactose | − | + | − | − | V |
| Lactulose | + | + | + | + | V |
| Maltose | + | + | + | + | + |
| D-mannitol | − | − | − | − | V |
| D-mannose | + | + | + | + | + |
| D-melibiose | − | + | − | − | V |
| β-methyl-D-glucoside | − | − | − | − | − |
| D-psicose | + | + | + | + | + |
| D-raffinose | − | − | − | − | V |
| L-rhamnose | − | − | − | − | − |
| D-sorbitol | − | − | − | − | V |
| Sucrose | + | + | + | + | V |
| D-trehalose | + | + | + | + | + |
| Turanose | − | + | − | − | V |
| Xylitol | − | − | − | − | − |
| Methyl pyruvate | + | − | + | + | + |
| Mono-methyl-succinate | + | + | + | + | + |
| Acetic acid | − | − | − | − | V |
| Cis-aconitic acid | + | + | + | + | V |
| Citric acid | − | − | − | − | V |
| Formic acid | − | − | − | − | − |
| D-galactonic acid lactone | − | − | − | − | − |
| D-galacturonic acid | − | − | − | − | − |
| D-gluconic acid | − | − | − | − | V |
| D-glucosaminic acid | − | − | − | − | − |
| D-glucuronic acid | − | − | − | − | − |
| α-hydroxy butyric acid | − | − | − | − | V |
| β-hydroxy butyric acid | − | − | − | − | V |
| γ-hydroxy butyric acid | − | − | − | − | − |
| p-hydroxy phenylacetic acid | − | − | − | − | − |
| Itaconic acid | − | − | − | − | − |
| α-keto butyric acid | + | + | − | + | V |
| α-keto glutaric acid | + | + | + | + | + |
| α-keto valeric acid | − | − | − | − | − |
| D,L-lactic acid | − | − | − | − | V |
| Malonic acid | + | + | + | + | V |
| Propionic acid | − | − | − | − | V |
| Quinic acid | − | − | − | − | − |
| D-saccharic acid | + | + | + | + | V |
| Sebacic acid | − | − | − | − | − |
| Succinic acid | + | + | + | + | + |
| Bromo succinic acid | + | + | + | + | + |
| Succinamic acid | + | + | + | + | + |
| Glucuronamide | − | + | − | + | − |
| L-alaninamide | + | + | + | + | V |
| D-alanine | + | + | + | + | + |
| L-alanine | + | + | + | + | + |
| L-alanylglycine | + | + | + | + | + |
| L-asparagine | + | − | − | − | V |
| L-Aspartic acid | + | + | − | + | V |
| L-Glutamic acid | + | + | + | + | + |
| Glycyl-L-aspartic acid | + | + | + | + | V |
| Glycyl-L-glutamic acid | + | + | + | + | + |
| L-histidine | − | − | − | + | V |
| Hydroxy-L-proline | + | + | + | + | V |
| L-leucine | − | − | − | − | V |
| L-ornithine | − | − | − | − | V |
| L-phenylalanine | − | − | − | − | − |
| L-proline | + | + | + | + | V |
| L-pyroglutamic acid | − | − | − | − | − |
| D-serine | − | − | − | − | − |
| L-serine | + | + | + | + | V |
| L-threonine | + | + | − | − | V |
| D,L-carnitine | − | − | − | − | − |
| γ-amino butyric acid | − | − | − | − | − |
| Urocanic acid | − | − | − | − | V |
| Inosine | − | − | − | − | V |
| Uridine | − | − | − | − | V |
| Thymidine | − | − | − | − | − |
| Phenyethylamine | − | − | − | − | − |
| Putrescine | − | − | − | − | − |
| 2-Aminoethanol | − | − | − | − | − |
| 2,3-Butanediol | − | − | − | − | − |
| Glycerol | + | + | + | + | V |
| D,L-α-glycerol phosphate | + | + | − | − | V |
| Glucose-1-phosphate | + | + | − | − | V |
| Glucose-6-phosphate | − | − | − | − | V |
Strains of X. axonopodis used by Vauterin et al. (1995): (+) indicates that 90% or more of strains are positive, (−) indicates that 90% or more of strains are negative and (V) indicates that 11 to 89% of strains were able to metabolize the carbon source
Fatty acid analysis
Fatty acid profiles of the strains pathogenic to eucalypt were typical of X. axonopodis species. Average and standard deviation values of fatty acids identified for the twenty-six strains tested in this study are shown in Table 4. These values were compared with those obtained by Vauterin et al. (1996) for several Xanthomonas species. Fatty acid values were considered to be different from those of other Xanthomonas spp. when their averages differed by a magnitude greater than the sum of their standard deviations. The value of the fatty acid 17:0 iso 3OH differentiated strains pathogenic to eucalypt from all reported by Vauterin et al. (1996), except for X. codiaei, X. fragariae, and X. albilineans. The fatty acid 13:0 iso 2OH identified in all strains pathogenic to eucalypt is not in the list proposed by Vauterin et al (1996). The fatty acid profiles of the eucalypt strains were more similar to those of X. axonopodis pv. manihotis, X. axonopodis pv. phaseoli and X. axonopodis pv. glycines with average similarity indices of 0.660, 0.650 and 0.645, respectively.
Table 4.
Fatty acid profile of Xanthomonas axonopodis strains isolated from eucalypt
| Compound | Fatty acids (%) | Strains (%) |
|---|---|---|
| Saturated fatty acids | ||
| 10:0 | 0.897 ± 0.207 | 100 |
| 14:0 | 2.004 ± 0.333 | 100 |
| 16:0 | 4.899 ± 0.872 | 100 |
| 17:0 | 0.078 ± 0.005 | 15.4 |
| 18:0 | 0.681 ± 0.633 | 34.6 |
| Unsaturated fatty acids | ||
| 15:1 w6c | 0.52 ± 0.1559 | 100 |
| 16:1 w5c | 0.123 ± 0.023 | 61.5 |
| 16:1 w7c/16:1 w6c | 20.91 ± 1.315 | 100 |
| 16:0 10-methyl/17:1 iso w9c | 4.76 ± 0.697 | 100 |
| 17:1 w8c | 0.878 ± 0.101 | 100 |
| 17:1 w6c | 0.192 ± 0.074 | 61.5 |
| 18:1 w9c | 0.413 ± 0.089 | 96.2 |
| 18:1 w7c/18:1 w6c | 0.504 ± 0.096 | 100 |
| Hydroxy fatty acids | ||
| 10:0 3OH | 0.39 ± 0.064 | 84.6 |
| 10:0 2OH | 0.173 ± 0.023 | 23.1 |
| 11:0 3OH | 0.271 ± 0.049 | 76.9 |
| 12:0 3OH | 3.434 ± 0.414 | 100 |
| 16:0 3OH | 0.156 ± 0.040 | 53.8 |
| Branched-chain fatty acids | ||
| 11:0 iso | 4.765 ± 0.907 | 100 |
| 13:0 iso | 0.367 ± 0.058 | 84.6 |
| 14:0 iso | 0.395 ± 0.040 | 100 |
| 15:0 iso | 30.474 ± 1.533 | 100 |
| 15:0 anteiso | 8.504 ± 0.965 | 100 |
| 16:0 iso | 1.21 ± 0.244 | 100 |
| 17:0 iso | 4.922 ± 0.664 | 100 |
| 17:0 anteiso | 0.34 ± 0.058 | 88.5 |
| Branched-chain unsaturated fatty acids | ||
| 15:1 iso F | 0.462 ± 0.080 | 100 |
| Branched-chain hydroxy fatty acids | ||
| 11:0 iso 3OH | 2.062 ± 0.445 | 100 |
| 12:0 iso 3OH | 0.234 ± 0.034 | 53.8 |
| 13:0 iso 3OH | 4.998 ± 0.414 | 100 |
| 13:0 iso 2OH | 0.327 ± 0.054 | 77.0 |
| 17:0 iso 3OH | 0.326 ± 0.038 | 88.5 |
Host range and cross-inoculation
To gain insights into the host range of X. axonopodis strains isolated from eucalypt, we inoculated LPF 602 on different plant species. LPF 602 only caused disease on eucalypt plants belonging to the genera Eucalyptus and Corymbia. However, it was noted that there is intraspecific host variability in susceptibility to the disease. All plants of E. cloeziana and hybrid clones of E. grandis × E. urophylla and E. urophylla × E. maidenii were susceptible. At least 50% of the seedlings of C. maculata and E. globulus and 5% of E. grandis, E. urophylla, E. saligna, E. camaldulensis and E. robusta exhibited symptoms of bacterial blight. Plants of Psidium guajava, Myrciaria jaboticaba, Eugenia jambolana and other botanical families showed no disease symptoms. Bacteria were isolated from inoculated plants exhibiting disease symptoms and their colony morphologies were identical to those used for inoculation.
The results of cross-inoculation experiments show that X. axonopodis pv. vignicola (IBSBF 1739), X. axonopodis pv. phaseoli var. fuscans (X. fuscans subsp. fuscans) (UNB 772) and X. axonopodis pv. aurantifolii (X. fuscans subsp. aurantifolii) (IBSBF 380) do not cause disease symptoms when inoculated onto eucalypt rotted cuttings. Only strain LPF 602 caused disease in clone CLR368. Overall, the results of this study indicate that the strains pathogenic to Eucalyptus spp. belong to Xanthomonas axonopodis pv. eucalyptorum pv. nov. that have not been previously reported.
Discussion
From samples of eucalypt leaves with symptoms of bacterial blight collected in different geographic regions of Brazil, 26 strains were obtained, which had their pathogenicity confirmed by inoculation onto healthy plants. Symptoms developed on inoculated plants were similar to those observed under field and nursery conditions, and to those described in previous studies (Gonçalves et al., 2008; Neves et al., 2014), ranging from small necrotic lesions to angular leaf spots. However, the most frequently observed symptoms in the inoculated plants were small spots distributed in the leaf blade, whereas under natural field and nursery conditions, the lesions are generally larger and delimited by the leaf ribs. We observed that the strains isolated from eucalypt varied in aggressiveness when inoculated onto clone CLR368 (E. urophylla × E. globulus). This information is important for designing strategies to control the disease. For instance, selection of host genotypes resistant to bacterial leaf blight should preferably be conducted by challenging clones of different genetic backgrounds with the most aggressive strains.
Analysis of the 16S rDNA sequence indicated that all strains that caused disease on eucalypt belonged to the genus Xanthomonas, but sequence similarity was observed with strains of the species X. campestris and X. axonopodis. Thus, it was not sufficient to allocate strains in a particular bacterial species. The analysis of the 16S rDNA sequence is a simple method that allows rapid identification of strains at the genus level. According to Hauben et al. (1997) because Xanthomonas 16S rDNA sequences show limited variability, they do not provide sufficient resolution to differentiate among all species of the genus. It provides even less resolution to differentiate among pathovars of the same species.
The identity of the strains causing eucalypt bacterial blight as X. axonopodis was confirmed by results of metabolic and FAME profiling. As expected, we observed metabolic variations among strains, as indicated by the fact that LPF 582 was unable to metabolize methyl pyruvate. We also found that LPF 582 and LPF 602 were able to metabolize glucuronamide. According to Vauterin et al. (1995) methyl pyruvate is used as a carbon source by 99% of X. axonopodis strains, whereas glucuronamide is utilized as a sole carbon and nitrogen source by only 8% of X. axonopodis strains. The same authors also reported that the fatty acid 13:0 isso 2OH is not commonly found in X. axonopodis strains. However, 76.9% of strains recovered in the present study had such a fatty acid. Here, we found that the percentage of 17:0 isso 3OH of the strains pathogenic to eucalypt was different from those of the X. axonopodis strains studied by Vauterin et al (1996). These results indicate that strains that cause disease on eucalypt plants have distinct phenotypic characteristics that distinguish them from other X. axonopodis pathovars.
Moreover, MLSA also indicated that the eucalypt strains belong to the species X. axonopodis. Other bacterial species that have been reported to be associated with similar disease symptoms in Brazil (Gonçalves et al., 2008; Pomella et al., 1995) and in other countries (Coutinho et al., 2002, 2015) were not found in this study. Despite finding other bacterial species associated with the disease symptoms, Gonçalves et al. (2008) pointed X. axonopodis as the predominant species causing eucalypt bacterial leaf blight in Brazil and our results confirm such an interpretation. MLSA is considered a more robust alternative to 16S rDNA sequencing and more flexible than DNA-DNA reassociation methods for classification of bacterial species (Ah-You et al., 2007; Ferraz et al., 2017; Young et al., 2008, 2010). Young et al. (2008) showed that MLSA based on the dnaK, fyuA, gyrB and rpoD genes sequences allow discrimination of Xanthomonas species with results similar to those obtained by Vauterin et al. (1995) using a DNA-DNA reassociation method. Furthermore, MLSA using these four genes allows separation of X. axonopodis pathovars into the six subgroups obtained by Rademaker et al. (2005) using rep-PCR.
When comparing the concatenated dnaK, fyuA, gyrB and rpoD gene sequences, we observed some genetic variability among X. axonopodis strains associated with bacterial leaf blight of eucalypt (Supplementary Fig. 1). However, MLSA revealed that these strains clustered together in only one distinct clade in the phylogenetic tree, corresponding to Rademaker’s group RG 9.6 (Rademaker et al. 2005). There are several proposals to reclassify some pathovars of this X. axonopodis RG group in other species. For instance, X. axonopodis pv. aurantifolii and X. axonopodis pv. phaseoli var. fuscans were proposed to be reclassified as X. fuscans subsp. aurantifolii and X. fuscans subsp. fuscans, respectively (Schaad et al., 2005, 2006). Pathovars from other RG groups have also been proposed to be reclassified. For example, X. axonopodis pv. vesicatoria, X. axonopodis pv. alfalfae, and X. axonopodis pv. citrumelo, belonging to RG 9.2, were proposed to be reclassified as X. euvesicatoria, X. alfalfae subsp. alfalfae and X. alfalfae subsp. citrumelo, respectively (Jones et al., 2004; Schaad et al., 2005). In contrast, no reclassifications have been proposed for pathovars of RG 9.4, in which X. axonopodis pv. phaseoli and X. axonopodis pv. manihotis are included.
Some of the species recently proposed by Jones et al. (2004) and Schaad et al. (2005, 2006), such as X. alfalfae, X. citri, X. euvesicatoria, X. fuscans, and X. perforans, are not clearly differentiated from X. axonopodis (Young et al., 2008). The separation of these newly proposed species from X. axonopodis was mainly based on results of DNA-DNA reassociation studies. Young et al. (2008) hypothesized a possible disagreement between MLSA and the DNA-DNA reassociation method used by Jones et al. (2004) and Schaad et al. (2005) with regard to the circumscription of species. According to Young et al. (2008), the stringency of the DNA-DNA reassociation method, and perhaps its high experimental error, may be responsible for the discrepancy. There is also the possibility that MLSA and DNA-DNA reassociation methods do not give equivalent results when studying some groups of strains (Young et al., 2008), and this could be the case for X. axonopodis. Therefore, the taxonomic classification and nomenclature widely accepted and used by Vauterin et al. (1995) and Rademaker et al. (2005) were adopted to allocate the strains pathogenic to Eucalyptus spp. isolated in this study to the species Xanthomonas axonopodis.
MLSA analysis showed that strains pathogenic to Eucalyptus spp. are more closely related to the X. axonopodis pathovars vignicola, aurantifolii (X. fuscans subsp. aurantifolii), and phaseoli var. fuscans (X. fuscans subsp. fuscans). LPF 602, a representative of the pathogenic strains isolated in this study, caused disease only in plants belonging to the genera Eucalyptus and Corymbia, which suggests specialization of the pathogen to the Myrtaceae family. Cross-inoculation experiments revealed that the host range of strain LPF 602 is different from any other pathovar that has been previously reported, and most importantly, is distinctively pathogenic to the plant species from which it was isolated.
The first description of a bacterium causing leaf blight of eucalypt was made by Truman (1974). The bacterium was classified as Xanthomonas eucalypti Truman, then as X. campestris pv. eucalypti (Truman, 1974) Dye 1978 and finally transferred to X. dyei pv. eucalypti (Truman, 1974) Young et al., 2010. Our phylogenetic analyses indicate that the strains of X. axonopodis pathogenic to Eucalyptus and Corymbia reported in this study are distantly related to X. dyei. Hence, we propose to accommodate strains isolated from eucalypt and belonging to Rademaker’s group RG 9.6 (Rademaker et al., 2005) as X. axonopodis pv. eucalyptorum pv. nov.
Description of Xanthomonas axonopodis pv. eucalyptorum pv. nov
Xanthomonas axonopodis pv. eucalyptorum (eu’ca’lip’to’rum. N.L. gen. n. eucalyptorum of Eucalyptus, referring to the isolation source of the type strain).
The phenotypic description of the pathovar is based on pathotype strain LPF 602 (Table 2). Bacterial strains are Gram negative by KOH test. Colonies are yellow and mucilaginous with abundant slime formation on YDC agar. Strains are positive for H2S, and xanthomonadin production, catalase, gelatin liquefaction, starch utilization, and esculin hydrolysis. They are negative for growth under anaerobic conditions, production of fluorescent pigments on King’s B medium, utilization of asparagine as a sole source of carbon and nitrogen, oxidase, urease, and NO3 reductase.
Strain gives positive auxanographic reactions for dextrin, glycogen, Tween 40, Tween 80, N-acetyl-D-glucosamine, D-cellobiose, D-fructose, L-fucose, D-galactose, gentiobiose, α-D-glucose, lactulose, maltose, D-mannose, D-psicose, sucrose, D-trehalose, methyl pyruvate, mono-methylsuccinate, cis-aconitic acid, α-keto butyric acid, α-keto glutaric acid, malonic acid, D-saccharic acid, succinic acid, bromo succinic acid, succinamic acid, glucuronamide, L-alaninamide, D-alanine, L-alanine, L-alanylglycine, L-aspartic acid, L-glutamic acid, glycyl-L-aspartic acid, glycyl-L-glutamic acid, L-histidine, hydroxyl-L-proline, L-proline, L-serine and glycerol. Negative reactions are given by α-cyclodextrin, N-acetyl-D-galactosamine, adonitol, L-arabinose, D-arabitol, i-erythritol, m-inositol, α-D-lactose, D-mannitol, D-melibiose, β-methyl-D-glucoside, D-raffinose, L-rhamnose, D-sorbitol, turanose, xylitol, acetic acid, citric acid, formic acid, D-galactonic acid lactone, D-galacturonic acid, D-gluconic acid, D-glucosaminic acid, D-glucuronic acid, α-hydroxy butyric acid, β-hydroxy butyric acid, γ-hydroxy butyric acid, p-hydroxy phenylacetic acid, itaconic acid, α-keto valeric acid, D,L-lactic acid, propionic acid, quinic acid, sebacic acid, L-asparagine, L-leucine, L-ornitine, L-phenylalanine, L-pyroglutamic acid, D-serine, L-threonine, D,L-carnitine, γ-amino butyric acid, urocanic acid, inosine, uridine, thymidine, phenyethylamine, putrescine, 2-aminoethanol, 2,3-butanediol, D,L,-α glycerol phosphate, glucose-1-phosphate and glucose-6-phosphate.
Strains are distinguished from other Xanthomonas species by MLSA based on concatenated dnaK, fyuA, gyrB and rpoD gene sequences. They can be differentiated from other X. axonopodis strains because of their proportion of 17:0 iso 3OH and 13:0 iso 2OH fatty acids. Pathotype strain is LPF 602, which was isolated from infected eucalypt tissue in the municipality of Teixeira de Freitas in the state of Bahia, Brazil, and causes bacterial leaf blight on Eucalyptus spp. and Corymbia spp. GeneBank accession numbers of the 16S rDNA, rpoD, dnaK, gyrB, and fyuA sequences of the pathotype strain are KY288867, KY287841, KY287877, KY287925, and KY460500, respectively.
Supplementary data
Acknowledgments
The authors are grateful to the biotechnological company Clonar Resistência a Doenças Florestais for providing the plant material and the inoculation facilities, to Minas Gerais State Foundation for Research Development (FAPEMIG) and National Council for Scientific and Technological Development (CNPq) for financial support. The authors would like to thank Dra. Marisa Alvares Velloso Ferreira for kindly providing strain UNB 772 for cross inoculation experiments.
References
- Ah-You N, Gagnevin L, Chiroleu F, Jouen E, Neto JR, Pruvost O. Pathological variations within Xanthomonas campestris pv. mangiferaeindicae support its separation into three distinct pathovars that can be distinguished by amplified fragment length polymorphism. Phytopathology. 2007;97:1568–1577. doi: 10.1094/PHYTO-97-12-1568. [DOI] [PubMed] [Google Scholar]
- Ah-You N, Gagnevin L, Grimont PAD, Brisse S, Nesme X, Chiroleu F, Bui Thi Ngoc L, Jouen E, Lefeuvre P, Vernière C, Pruvost O. Polyphasic characterization of xanthomonads pathogenic to members of the Anacardiaceae and their relatedness to species of Xanthomonas. Int J Syst Evol Microbiol. 2009;59:306–318. doi: 10.1099/ijs.0.65453-0. [DOI] [PubMed] [Google Scholar]
- Alfenas AC, Zauza EAV, Mafia RG, Assis TF. Clonagem e doenças do eucalipto. Editora Universidade Federal de Viçosa; Viçosa, Brazil: 2009. [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull CT, De Boer SH, Denny TP, Firrao G, Fischer-Le Saux M, Saddler GS, Scortichini M, Stead DE, Takikawa Y. List of new names of plant pathogenic bacteria (2008–2010) J Plant Pathol. 2012;94:21–27. [Google Scholar]
- Bull CT, De Boer SH, Denny TP, Firrao G, Fischer-Le Saux M, Saddler GS, Scortichini M, Stead DE, Takikawa Y. Comprehensive list of names of plant pathogenic bacteria, 1980–2007. J Plant Pathol. 2010;92:551–592. [Google Scholar]
- Coutinho TA, Preisig O, Mergaert J, Cnockaert MC, Riedel K-H, Swings J, Wingfield MJ. Bacterial blight and dieback of Eucalyptus species, hybrids, and clones in South Africa. Plant Dis. 2002;86:20–25. doi: 10.1094/PDIS.2002.86.1.20. [DOI] [PubMed] [Google Scholar]
- Coutinho TA, Westhuizen L, Roux J, McFarlane SA, Venter SN. Significant host jump of Xanthomonas vasicola from sugarcane to a Eucalyptus grandis clone in South Africa. Plant Pathol. 2015;64:576–581. doi: 10.1111/ppa.12298. [DOI] [Google Scholar]
- Ferraz HGM, Guimarães LMS, Badel JL, Tótola MR, Alfenas AC. Leaf blight and defoliation caused by two new pathovars of Xanthomonas axonopodis on Schinus terebinthifolius and Mabea fistulifera. Plant Pathol. 2017;66:1527–1538. doi: 10.1111/ppa.12705. [DOI] [Google Scholar]
- Gonçalves RC. Doctor thesis. Federal University of Viçosa; Viçosa, Brazil: 2003. Etiologia da mancha bacteriana do eucalipto no Brasil. (in Portuguese, English summary). [Google Scholar]
- Gonçalves RC, Douglas L, Oliveira JR, Maffia LA, Cascardo J, Alfenas AC. Etiology of bacterial leaf blight of eucalyptus in Brazil. Trop Plant Pathol. 2008;33:180–188. doi: 10.1590/S1982-56762008000300002. [DOI] [Google Scholar]
- Hauben L, Vauterin L, Swings J, Moore ERB. Comparison of 16S ribosomal DNA sequences of all Xanthomonas species. Int J Syst Evol Microbiol. 1997;47:328–335. doi: 10.1099/00207713-47-2-328. [DOI] [PubMed] [Google Scholar]
- Hayward AC. The hosts of Xanthomonas. In: Swings JG, Civerolo EL, editors. Xanthomonas. Springer; Dordrecht, Netherlands: 1993. pp. 1–119. [Google Scholar]
- Jones JB, Lacy GH, Bouzar H, Stall RE, Schaad NW. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst Appl Microbiol. 2004;27:755–762. doi: 10.1078/0723202042369884. [DOI] [PubMed] [Google Scholar]
- Kado CI, Heskett MG. Selective media for isolation of Agrobacterium, Corynebacterium, Erwinia, Pseudomonas and Xanthomonas. Phytopathology. 1970;60:969–976. doi: 10.1094/Phyto-60-969. [DOI] [PubMed] [Google Scholar]
- Neves DA, Guimarães L, Ferraz HGM, Alfenas AC. Favorable conditions for Xanthomonas axonopodis infection in Eucalyptus spp. Trop Plant Pathol. 2014;39:428–433. doi: 10.1590/S1982-56762014000600002. [DOI] [Google Scholar]
- Pomella AWV, Romeiro RS, Ferreira FA, Oliveira JR. Lesões foliares em viveiro de eucalipto incitadas por uma espécie fluorescente de Pseudomonas. Fitopatol Bras. 1995;20(Suppl):374. [Google Scholar]
- Pothiluk T, Thowthampitak J, Kasem S, Prathuangwong S. Identification and aggressiveness of phytopathogenic bacterium caused angular leaf spot of eucalyptus seedling. Proceedings of the 51st Kasetsart University Annual Conference; February 5–7, 2013; Annual Conference; Bangkok, Thailand. 2013. p. 0181. ref. 10. [Google Scholar]
- Rademaker JLW, Louws FJ, Schultz MH, Rossbach U, Vauterin L, Swings J, De Bruijn FJ. A comprehensive species to strain taxonomic framework for Xanthomonas. Phytopathology. 2005;95:1098–1111. doi: 10.1094/PHYTO-95-1098. [DOI] [PubMed] [Google Scholar]
- Reis AV, Souza RM, Castro HA, Cardoso M, Kobayasti L. Uma nova bacteriose em mudas de eucalipto incitada por Xanthomonas campestris. Fitopatol Bras. 1996;21(Suppl):342. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Samanta JN, Mandal K, Maiti S. A novel pathovar of Xanthomonas axonopodis causes gumming of Guggal (Commiphora wightii) Eur J Plant Pathol. 2013;135:115–125. doi: 10.1007/s10658-012-0070-x. [DOI] [Google Scholar]
- Schaad NW, Jones JB, Chun W. Laboratory guide for the identification of plant pathogenic bacteria. American Phytopathological Society (APS Press); MN, USA: 2001. [Google Scholar]
- Schaad NW, Postnikova E, Lacy G, Sechler A, Agarkova IV, Stromberg PE, Stromberg VK, Vidaver AM. Emended classification of xanthomonad pathogens on citrus. Syst Appl Microbiol. 2006;29:690–695. doi: 10.1016/j.syapm.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Schaad NW, Postnikova E, Lacy GH, Sechler A, Agarkova I, Stromberg PE, Stromberg VK, Vidaver AK. Reclassification of Xanthomonas campestris pv. citri (ex Hasse 1915) Dye 1978 forms A, B/C/D, and E as X. smithii subsp. citri (ex Hasse) sp. nov. nom. rev. comb. nov., X. fuscans subsp. aurantifolii (ex Gabriel 1989) sp. nov. nom. rev. comb. nov., and X. alfalfae subsp. citrumelo (ex Riker and Jones) Gabriel et al., 1989 sp. nov. nom. rev. comb. nov.; X. campestris pv malvacearum (ex Smith 1901) Dye 1978 as X. smithii subsp. smithii nov. comb. nov. nom. nov.; X. campestris pv. alfalfae (ex Riker and Jones, 1935) Dye 1978 as X. alfalfae subsp. alfalfae (ex Riker et al., 1935) sp. nov. nom. rev.; and “var. fuscans” of X. campestris pv. phaseoli (ex Smith, 1987) Dye 1978 as X. fuscans subsp. fuscans sp. nov. Syst Appl Microbiol. 2005;28:494–518. doi: 10.1016/j.syapm.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman R. Die-back of Eucalyptus citriodora caused by Xanthomonas eucalypti sp. n. Phytopathology. 1974;64:43–44. doi: 10.1094/Phyto-64-143. [DOI] [Google Scholar]
- Vale FR, Fernandes Filho EL, Liberato JR. QUANT: a software for plant disease severity assessment. Abstracts of the 8th international congress of plant pathology; Christchurch, New Zealand. 2003. p. 105. [Google Scholar]
- Vauterin L, Hoste B, Kersters K, Swings J. Reclassification of Xanthomonas. Int J Syst Evol Microbiol. 1995;45:472–489. doi: 10.1099/00207713-50-2-665. [DOI] [PubMed] [Google Scholar]
- Vauterin L, Yang P, Swings J. Utilization of fatty acid methyl esters for the differentiation of new Xanthomonas species. Int J Syst Evol Microbiol. 1996;46:298–304. [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Park D-C, Shearman HM, Fargier E. A multilocus sequence analysis of the genus Xanthomonas. Syst Appl Microbiol. 2008;31:366–377. doi: 10.1016/j.syapm.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Young JM, Wilkie JP, Park D, Watson DRW. New Zealand strains of plant pathogenic bacteria classified by multi-locus squence analysis; proposal of Xanthomonas dyei sp. nov. Plant Pathol. 2010;59:270–281. doi: 10.1111/j.1365-3059.2009.02210.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.