Abstract
Maintenance of a beneficial microbial community, especially in the rhizosphere, is indispensable for plant growth and agricultural sustainability. In this sense, plant growth-promoting rhizobacteria (PGPR) have been extensively studied for their role in plant growth promotion and disease resistance. However, the impact of introducing PGPR strains into rhizosphere microbial communities is still underexplored. We previously found that the Proteus vulgaris JBLS202 strain (JBLS202) promoted growth of Kimchi cabbage and altered the relative abundance of total bacteria and Pseudomonas spp. in the treated rhizosphere. To extend these findings, we used pyrosequencing to analyze the changes in bacterial communities in the rhizosphere of Kimchi cabbage after introduction of JBLS202. The alterations were also evaluated by taxon-specific real-time PCR (qPCR). The pyrosequencing data revealed an increase in total bacteria abundance, including specific groups such as Proteobacteria, Acidobacteria, and Actinobacteria, in the treated rhizosphere. Time-course qPCR analysis confirmed the increase in the abundance of Acidobacteria, Actinobacteria, Alphaproteobacteria, and Betaproteobacteria. Furthermore, genes involved in nitrogen cycling were upregulated by JBLS202 treatment indicating changes in ecological function of the rhizosphere soil. Overall, these results indicate that introduction of JBLS202 alters both the composition and function of the rhizosphere bacterial community, which can have direct and indirect effects on plant growth. Therefore, we propose that long-term changes in bacterial composition and community-level function need to be considered for practical use of PGPRs.
Keywords: microbiome, PGPR, pyrosequencing, rhizosphere
The composition and quantity of the microbial community in the plant rhizosphere depend on various factors including root exudates, the physical and chemical nature of the soil, climatic conditions, and the microbes themselves (Sugiyama et al., 2014). Therefore, the microbial community in the rhizosphere is altered by interactions between its constituent microbes, along with biotic and abiotic factors (Somers et al., 2004). In addition, human interventions such as fertilizer application and cultural practices tend to influence on the microbial community of the rhizosphere (Qiu et al., 2012; Shen et al., 2015). For instance, chemical fertilizers and pesticides that have been used to improve crop growth or reduce diseases perturbed the microbial community in the soil (Lo, 2010).
Over the last few decades, plant growth-promoting rhizobacteria (PGPR) have been studied and used as biofertilizers to stimulate growth and induce resistance in plants (Lugtenberg and Kamilova, 2009). Rhizosphere microorganisms, including PGPR, improve plant growth by enhancing phytohormone production, degrading phytotoxic compounds, solubilizing phosphates, and mobilizing nutrients (Lugtenberg and Kamilova, 2009). These microorganisms also play essential roles in biogeochemical processes such as nitrogen and phosphorous cycling (Nannipieri et al., 2003). Hence, the maintenance of a healthy rhizosphere microbiome is crucial for crop growth and development, as well as soil health (Bending et al., 2007; Lugtenberg and Kamilova, 2009).
Introduced/treated PGPRs face competition with the indigenous microbial population, which can alter the native microbial communities. For example, PGPR strains used as biocontrol agents eliminate root-dwelling pathogens through various mechanisms, including production of antibiotics and other bioactive compounds (Glick et al., 2007). This phenomenon might also remove other non-target microorganisms in addition to the harmful microbes of interest, thereby altering the microbiome (Trabelsi and Mhamdi, 2013). On the contrary, certain microorganisms might negatively affect PGPR function and survival (Dutta and Podile, 2010). PGPRs have also been reported to alter root exudates, which eventually affecting the surrounding microbes (Bais et al., 2008; Dutta and Podile, 2010; Dutta et al., 2013; Huang et al., 2014).
Because of complex roles of the microbial inoculants in the introduced rhizosphere, the effects of introduced PGPRs on the rhizosphere microbial communities are still controversial (Kang et al., 2013). For example, Chowdhury et al. (2013) reported that PGPRs cause minor changes in the microbial community and Kozdrój (2008) found that they cause significant shifts. In addition, Piromyou et al. (2011) demonstrated that the alteration of the microbial community was due to plant development rather than the inoculated PGPR. Considering these reports, detailed studies on the fate of introduced PGPRs in the context of the rhizosphere microbial community as well as their effects on the surrounding microbes, are essential for enhancing commercial application of PGPRs in the field.
Our earlier studies have shown that the PGPR strain P. vulgaris JBLS202 (JBLS202) promotes growth of Kimchi cabbage (Yu and Lee, 2013), and that its volatile compounds (VOCs) stimulate growth of Arabidopsis via auxin, cytokinin, and brassinosteroid pathways (Bhattacharyya et al., 2015). Further analysis revealed that the bacterization of Kimchi cabbage seeds with JBLS202 or exposure of soil to VOCs modulated the bacterial community (Bhattacharyya and Lee, 2016). Hence, the present study aimed to investigate the detailed changes of the overall bacterial community as well as changes in specific bacterial subgroups in Kimchi cabbage rhizosphere after treatment with JBLS202. We performed an in-depth phylum to species level analysis of the bacterial communities through high-throughput 454 pyrosequencing technology and assayed the time-course impact on the rhizosphere microbiome through a taxon-specific qPCR. Furthermore, the significance of the changes for plant growth was investigated by studying genes involved in nitrogen cycling.
Materials and Methods
Seed bacterization and rhizosphere soil sampling
The strain JBLS202, which was originally isolated from the rhizosphere of halophyte Glasswort (Salicornia hebacea L.), was maintained in Luria-Bertani (LB) broth (Difco Laboratories, MI, USA) containing 20% glycerol at −80°C and revived as necessary (Yu and Lee, 2013). JBLS202 was streaked on LB agar plates and incubated at 28°C overnight. Cells were harvested in sterile distilled water (DW) to make a suspension of 1 × 107 colony forming units (CFU) ml−1 (Bhattacharyya and Lee, 2016). Kimchi cabbage (Brassica rapa L. ssp. pekinensis) seeds were surface sterilized with 1% NaOCl for 15 min and then 70% ethanol for 2 min, after which they were rinsed three times with sterile DW. The disinfected seeds were bacterized with JBLS202 cell suspension for 2 h at 28°C with shaking. After carefully blotting on sterile tissue paper to remove excess inoculum, the seeds were sowed in pots (four seeds per pot) and cultivated at 25 ± 2°C. Control seeds were treated with sterile DW for 2 h. Soil samples from the treated and untreated rhizospheres were carefully collected by scraping the roots with forceps (1 mm of adherent soil) after 44 days of sowing following the method described by Trivedi et al (2012). At least eight replicate pots were maintained for each time-point. The isolated rhizosphere soil samples were stored at −80°C until DNA extraction.
Soil DNA isolation and pyrosequencing to assess the bacterial community
Soil samples collected from the replicate pots of each treatment were pooled separately, and DNA was extracted using an UltraCleanTM soil DNA isolation kit (MoBio, CA, USA) following the manufacturer’s instructions. The purity of the isolated DNA was analyzed using a Nanodrop spectrophotometer, and the DNA was pooled and pyrosequenced at Chunlab Inc. (Seoul, South Korea) (Kim et al., 2014) on a 454 GS FLX Titanium Sequencing System (Roche Applied Science, CT, USA). In brief, the V1–V3 region of the 16S rRNA gene was amplified from the extracted DNA using barcoded fusion universal primers 27F and 518R. These primers contained 454 adapters, a key sequence, and a linker (Chun and Goodfellow, 1995; Kwon et al., 2010). The amplified PCR products were purified using a PCR cleanup kit, and DNA library samples were subjected to pyrosequencing. All pyrosequencing reads were submitted to the NCBI sequence read archive (SRA) database under accession number SRP100670.
Taxonomic assignment and analysis of pyrosequencing data
The sequenced data were analyzed using software from ChunLab Inc. Raw pyrosequencing reads were sorted by their barcode sequences, barcode fusion primers were trimmed, and low-quality sequences (≥ 2 ambiguous base calls, read length < 300 bp, or average quality value < 25) were filtered out. Nontargeting 16S rRNA sequences and chimeric sequences were eliminated from the sequencing reads (Eddy, 2011). The resulting reads were taxonomically classified based on similarity scores in both the BLASTN searches and pair-wise alignments using the EzTaxon-e database (Myers and Miller, 1988; Chun et al., 2007). The cut-off values used for taxonomic assignments were as follows (x = similarity): species (x ≥ 97%), genus (97% > × ≥ 94%), family (94% > × ≥ 90%), order (90% > × ≥ 85%), class (85% > × ≥ 80%), and phylum (80 > × ≥ 75%). If the similarity was lower than the specific cut-off value, the sequence was assigned as ‘unclassified’ (Chun et al., 2007; Unno et al., 2010). For the taxonomic affiliation, if the scientific name for a taxon was unknown, the known name in the nomenclature was written first, after which a suffix was added at the end of the name after an underscore (e.g., if the class name was unknown, a ‘c’ was written after the phylum name: ‘Acidobacteria_c’; c = class, o = order, f = family, g = genus, and s = species) (Kim et al., 2012). A comparative study of the class Gammaproteobacteria from treated and untreated soil entailed a combined approach using species taxonomic composition and taxon exclusive/XOR analysis. In this approach, multiple communities can be compared to detect taxa (ranging from phylum to species) that vary in the selected sample sets.
All bacterial community analyses were carried out with CLcommunity software (Chunlab, Inc.). Operational taxonomic units (OTUs) were defined with the Cluster Database at High Identity with Tolerance (CD-HIT) program at 3% sequence dissimilarity (Li and Godzik, 2006). Rarefaction curves (Heck et al., 1975), richness indices, and diversity indices (abundance-based coverage-ACE, Chao1, Shannon, Simpson) were calculated using the Mothur platform (Schloss et al., 2009).
Taxon/genus-specific qPCR
To estimate the abundances of specific bacterial groups such as Acidobacteria, Actinobacteria, Firmicutes, Alphaproteobacteria, Bacteroidetes, and Betaproteobacteria, we performed qPCR at weekly intervals from 7 to 44 days following the procedures described by Trivedi et al (2012). The qPCR reactions were carried out according to the method of Bhattacharyya and Lee (2016) using TOP real qPCR 2 × PreMix (Enzynomics, Seoul, South Korea) on a Rotor-Gene 6000 qPCR machine (Corbett Research, Australia). DNA was extracted from soil samples at each time-point using an UltraCleanTM soil DNA isolation kit (MoBio, CA, USA) following the manufacturer’s instructions. Soil DNA was further separated on 0.8% agarose gels, purified using a PCR cleanup kit, and cloned into the pGEM-T Easy vector (Promega, WI, USA). Standard curves were prepared using plasmids with correct construct inserts. Target copy numbers were calculated from the standard curves, and the number of copies per gram of dry soil was determined. The primers and thermocycling conditions for qPCR are described in the supplementary data (Supplementary Table 1).
Quantification of genes involved in nitrogen cycling
To estimate the density of functional communities involved in nitrogen cycling, qPCR was performed to quantify genes encoding key enzymes of nitrate reduction (narG, which encodes the membrane-bound nitrate reductase) and denitrification (nirK and nirS, which encode the cd1 and copper nitrite reductase, respectively, and nosZ, which encodes the nitrous oxide reductase). Gene levels were assayed at weekly intervals for 44 days as described by Trivedi et al (2012). All experiments were run in triplicate. The primers used for qPCR and the thermocycling conditions are described in the supplementary data (Supplementary Table 1).
Statistical analysis
Data were subjected to analysis of variance using SAS JMP software (SAS Institute, NC, USA). Significant differences in the treatment means of each sample were determined using the least significant difference (LSD) test at P = 0.05. Student’s t-tests were used to determine the significance of paired values for a given time-point between treated and control conditions. All experiments were performed at least twice. Data from each experiment were analyzed separately.
Results and Discussion
Analysis of pyrosequencing data
Inoculation of PGPR(s) might change the structure of the indigenous microbial soil communities. To examine the influence of PGPRs on bacterial structure, we obtained pyrosequencing data from JBLS202-treated and untreated (control) rhizospheres of Kimchi cabbage. The rarefaction curve at 3% dissimilarity showed a maximum of 1,815 (out of 1,827) OTUs in the JBLS202-treated rhizosphere, compared with 1,446 (out of 1,460) OTUs in the non-bacterized control rhizosphere (Table 1). The total valid reads were 5,950 for control and 5,656 for JBLS202-treated rhizosphere. Although additional sequencing effort was necessary to capture the complete diversity of the bacterial communities (Supplementary Fig. 1), considering the reports of Qiu et al. (2012), we concluded that these data were sufficient to compare the changes in bacterial diversities between JBLS202-bacterized and non-bacterized rhizosphere soils.
Table 1.
Estimated OTU richness and diversity indices of the rhizospheres of JBLS202-treated and untreated Kimchi cabbage at 97% similarity
| Treatment | Community characteristics | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| OTU richness | OTU diversity | |||||
|
|
|
|||||
| Observed | Chao1 | ACE | Jackknife | Shannon | Simpson | |
| JBLS202-treated | 1,827 | 3,283.86 | 4,564.56 | 4,275.87 | 6.835 | 0.002 |
| Untreated control | 1,460 | 2,658.31 | 3,425.08 | 3,468.00 | 5.989 | 0.016 |
The Good’s coverage analysis revealed that 82.5% and 87.0% of the taxonomic richness was covered by sequencing efforts in the treated and control rhizospheres, respectively. Non-parametric analysis of other diversity indices for bacteria, e.g., ACE, Chao1, and Jack knife, indicated increased bacterial diversity in the treated soil compared to the untreated control soil (Table 1). The Shannon diversity index also indicated a more abundance and richness of bacterial community in the bacterized soil (6.84) compared to the control soil (5.99). In addition, the Simpson diversity index of dominant species was lower in the JBLS202-treated rhizosphere (Table 1), which indicating higher bacterial diversity in comparison to untreated rhizosphere. The increased diversity in the rhizosphere of treated soil compared to the non-treated rhizosphere might be due to the influence of nutrients such as carbon and nitrogen (Dutta and Podile, 2010; Liu et al., 2015), which might be increased by growth promotion of Kimchi cabbage.
Phylum-level taxonomic analyses of bacterial communities
Changes in bacterial abundance and community structure have a crucial impact on plant growth and development. We previously demonstrated that seed bacterization of Kimchi cabbage with JBLS202 increased the abundance of total bacteria in the bacterized rhizosphere compared to the control rhizosphere (Bhattacharyya and Lee, 2016). Shen et al. (2015) also suggested that introduction of biofertilizer in the rhizosphere increases the total bacterial population. The overall influence of PGPR inoculation on soil microbial communities has been suggested to be more effective and consistent if the introduced strain has a direct effect on plants. In addition, the plants indirectly contribute to the maintenance and colonization of inoculants (Trabelsi and Mhamdi, 2013). Therefore, the beneficial effects of PGPRs depend on intricate interactions between the introduced PGPRs, plants, and microbial communities in the rhizosphere. In this study, the introduction of JBLS202 and colonization in the rhizospheres of Kimchi cabbage (Bhattacharyya and Lee, 2016; Yu and Lee, 2013) might alter the abundance of total bacteria in two ways: that is to say, through a direct effect and through the plant, which might be mediated by changes in plant exudates (Dutta et al., 2013; Kamilova et al., 2006).
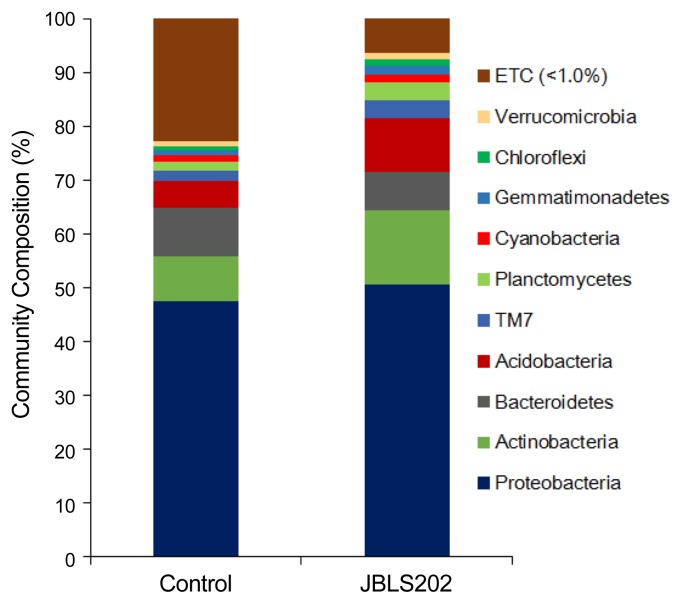
The functional diversity of microbial communities, along with the abundance and distribution of their resident species, plays important roles in the soil ecosystem. In this study, we performed an in-depth evaluation in abundance of the specific groups of bacteria after introduction of JBLS202 into the Kimchi cabbage rhizosphere. Out of the 27 identified phyla, 23 were present in both JBLS202-treated and control rhizospheres (Fig. 1). The most dominant phylum was Proteobacteria, followed by Actinobacteria, Acidobacteria, Bacteroidetes, TM7, and Planctomycetes (Fig. 1). The abundance in the treated soil was higher for Acidobacteria (85.67%), Planctomycetes (80.58%), TM7 (73.87%), Gemmatimonadetes (61.40%), and Actinobacteria (60.21%) compared to the control. In addition, the phyla Proteobacteria, Cyanobacteria, Gemmatimonadetes, Chloroflexi, Verrucomicrobia, Armatimonadetes, and Firmicutes were also more abundant in the JBLS202-treated rhizosphere. However, Bacteroidetes, Bacteria_uc, TM6, and Chlorobi were more abundant in control soil. The phyla BRC1, Spirochaetes, and JX172748 were exclusively present in the JBLS202-treated rhizosphere, whereas OP3 was detected only in control soil. The reason for difference in specific bacterial group needs more investigation.
Fig. 1.
Relative abundance of major bacterial phyla in the rhizospheres of JBLS202-bacterized and untreated Kimchi cabbage. Each bar represents the relative mean abundance of triplicate samples. Kimchi cabbage seeds coated with Proteus vulgaris JBLS202 were sowed, after which DNA was isolated from the rhizosphere 44 days after sowing and pyrosequenced.
The results in phylum-level analysis indicate that Proteobacteria, Acidobacteria, and Actinobacteria were the dominant phyla in the rhizosphere. Proteobacteria can adapt to diverse plant rhizospheres, and Acidobacteria can degrade cellulose and lignin. These adaptabilities might also make both phyla to be most abundant in previous reports (Bulgarelli et al., 2013; Mendes et al., 2011). Moreover, bacteria belonging to Acidobacteria and Actinobacteria are known to be efficient in utilizing more oxidized forms of carbon (Zinger et al., 2009). Therefore, the increase in their abundance might alter the rhizodeposition and utilization of organic carbon, which can affect carbon storage and sequestration (Trivedi et al., 2012). Considering the role of these groups of bacteria in biogeochemical nutrient cycling, any change in their abundance will affect microbial structure and function, and consequently plant growth.
Comparison of bacterial communities in the class, order, and family levels
Among the 68 classes identified in this study, Alphaproteobacteria, Actinobacteria_c, and Acidobacteria_c were dominant and 52.08%, 44.77%, and 77.45% more abundant, respectively, in the JBLS202-treated rhizosphere compared to the control (Supplementary Fig. 2). In general, the abundance of bacteria in other classes was increased by treatment with JBLS202. Flavobacteria was detected only in the untreated rhizosphere.
Among the 132 orders identified, 91 were detected in both the bacterized and untreated rhizospheres. The abundance was higher in the treated rhizosphere in the orders Burkholderiales (9% increase over control), Rhizobiales (61.54%), Acidobacteriales (77.45%), Micrococcales (46.41%), Sphingomonadales (24.48%), and Planctomycetales (138.46%) (Supplementary Fig. 3). However, Pseudomonadales, Xanthomonadales, and Methylophilales showed higher abundance in the untreated rhizosphere compared with the JBLS202-treated rhizosphere and Flavobacteriales was detected only in the control rhizosphere.
Family-level analysis revealed that 156 families out of 266 were detected in both the treated and untreated rhizospheres. The families Acidobacteriaceae, Bradyrhizobiaceae, Burkholderiaceae, Sphingomonadaceae, Methylobacteriaceae, Planctomycetaceae, Solibacteraceae, and Gemmatimonadaceae were more abundant in the JBLS202-treated rhizosphere than the control rhizosphere (Supplementary Fig. 4), whereas Pseudomonadaceae, Xanthomonadaceae, and Sphingobacteriaceae were higher in the control rhizosphere.
Taken together, the composition of the bacterial community was reconstituted at all hierarchical levels in the rhizosphere of Kimchi cabbage by treatment of JBLS202. The groups of bacteria with lower abundance as well as higher abundance in the treated rhizosphere compared to the untreated rhizosphere also deserve attention. Xanthomonadaceae and Pseudomonadaceae were more abundant in the control rhizosphere than in JBLS202-treated soil. With the exception of Strenotrophomonas maltophilia, most of the members of Xanthomonadaceae are pathogenic to humans, plants, or both (LaSala et al., 2007). The reduction in the members of this family might suggest elimination of deleterious bacteria from the rhizosphere by treatment of JBLS202. Overall, the results of this study indicate that JBLS202 contributes to plant growth and health not only by restructuring of microbiome but also by removing harmful bacteria from the rhizosphere, which requires validation in further studies.
Comparison of bacterial communities in the genus and species levels
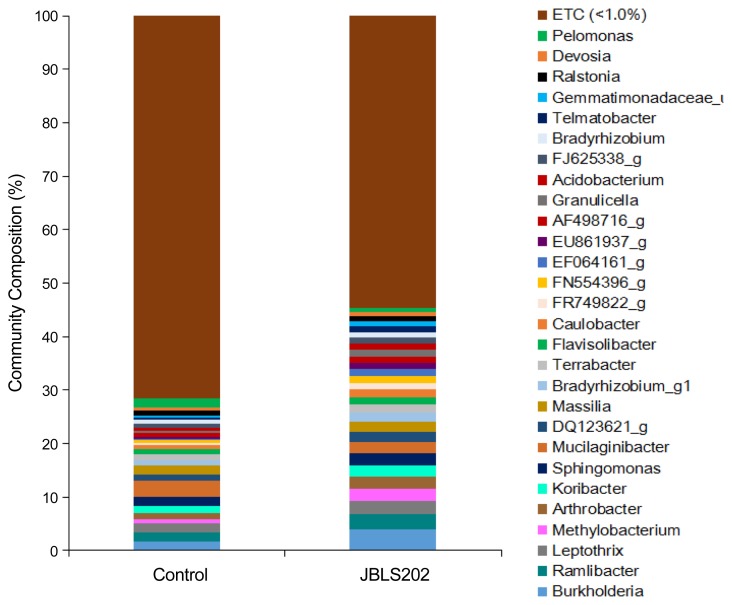
Colonization of JBLS202 in the rhizosphere also increased diversity and abundance in the genus-level bacteria (Fig. 2). Among 680 genera, the percentage increase in the treated rhizosphere was highest in Methylobacterium (165.31%), followed by Acidobacterium (153.85%), Burkholderia (128%), Arthrobacter (78.87%), Bradyrhizobium_g1 (72.23%), Koribacter (70.27%), Ramlibacter (64.95%), and Leptothrix (31.68%). In contrast, Ralstonia and Pelomonas were more abundant in the untreated rhizosphere.
Fig. 2.
Relative abundances of major bacterial genera detected in the rhizospheres of JBLS202-bacterized and untreated Kimchi cabbage. Data are presented as mean; only classes whose relative abundance was above 1% are shown.
Among the 1,676 analyzed species, the abundances of Flavisolibater_uc, Bradyrhizobium jicamae, B. denitrificans, Terrabacter lapilli, Burkholderia grimmiae, B. jiangsuensis, Arthrobacter defluvii, and Methylobacterium persicinum were higher in the JBLS202-treated rhizosphere, whereas Pseudomonas nitroreducens, Pelomonas saccharophila, Streptomyces graminifolii, Mucilaginibacter_uc, and Methylotenera_uc were more abundant in the untreated rhizosphere. Furthermore, a detailed analysis of the species belonging to the class Gammaproteobacteria revealed that Pseudomonadales (17 species), Xanthomonadales (19), Enterobacteriales (10), Solimonas_o (3), Legionellales (2), AB096215_o (2), Rhizobiales (1), Chromatiales (1), and Alteromonadales (1) were present only in the control rhizosphere (Supplementary Table 2). The abundance of some species such as Pseudomonas monteilii, Pseudomonas beteli, Dyella_uc, and Coxiellaceae_uc_s was reduced by JBLS202 treatment. On the other hand, the abundance species such as Steroidobacter_o, Xanthomonadales, Solimonas_o, and Legionellales was increased in the treated rhizosphere (Supplementary Table 2).
The diversity indices indicated that the relative abundances of the bacterial groups found in both treated and untreated soil were higher in the JBLS202-treated rhizosphere. In our previous study (Bhattacharyya and Lee, 2016), the abundance of Pseudomonads was increased by JBLS202 treatment at 14 to 30 days after sowing (DAS), whereas it returned to the similar level to that in the control rhizosphere at 37 DAS. In the present study, the abundance of Pseudomonads at 44 DAS was reduced in the JBLS202-treated rhizosphere compared to the control rhizosphere. Trabelsi and Mhamdi (2013) suggested that a time-course study of microbiome alteration is necessary to evaluate the effect of introduced foreign microbes in the context of the buffering capacity of the ecosystem. Hirsch et al. (2010) reported that culture-independent studies might not provide an accurate picture of taxonomic groups because of biasness induced by DNA extraction and PCR amplification. Considering the previous reports, the dynamics of Pseudomonads abundance requires more intensive intervals with a couple of techniques using similar controlled environmental conditions (e.g., soil or plant growth stage, which can induce changes in nutrient content).
Kang et al. (2013) suggested that Bacillus pumilus might not maintain a high density population in the rhizosphere, but that its presence might have a crucial long-term impact on the surrounding microbiome, which consequently affects plant growth. Since the quality and quantity of root exudate nutrients can differ with plant age, plant growth stage also impacts the composition and abundance of the rhizosphere microbial community (Liu et al., 2015). Introduced bacterial strains have also been reported to change root exudates to suit their nutritional needs. In this study, we detected Proteus spp. at less than 1.0% at 44 DAS, which is in accordance with our previous results (Bhattacharyya and Lee, 2016). In our previous studies, the population levels of JBLS202 decreased to 1.9 × 105 cfu g−1 soil at initial 3 days after seed treatment. Although the survival rate was slightly decreased to 0.9 × 105 cfu g−1 at 7 DAS, the similar population maintained constantly till 21 DAS (Bhattacharyya and Lee, 2016), which corresponds to the pattern of present study. We hypothesize that the introduction of JBLS202 also induces a transient and short-term perturbation in bacterial community abundance, but the sequential plant and microbe interaction induces restructuring of the rhizosphere microbiome. In addition, we propose that the initial changes are important for long-term maintenance of beneficial microbes (Kang et al., 2013).
Time series estimation of bacterial groups through taxon/genus-specific qPCR
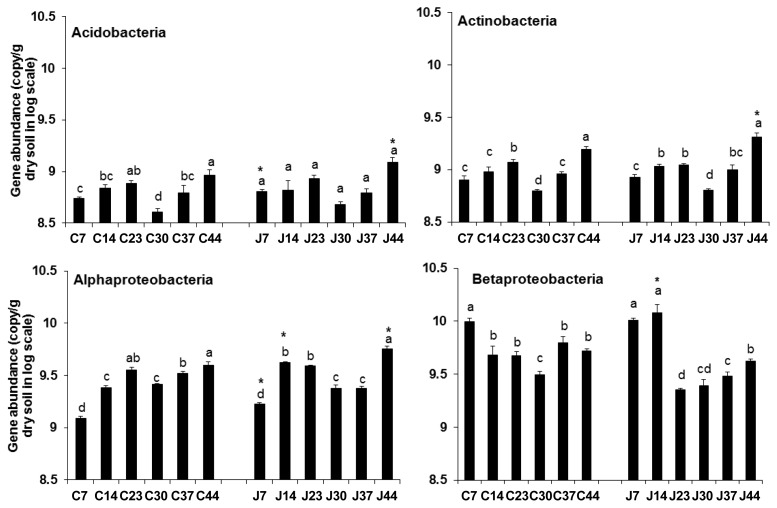
To analyze the alterations of major bacterial groups over time and to evaluate and validate the pyrosequencing data, we assayed the abundance of Acidobacteria, Actinobacteria, Alphaproteobacteria, and Betaproteobacteria using taxon-specific qPCR. The Acidobacteria data did not show any statistically significant changes between the JBLS202-treated and untreated rhizospheres, with the exception of increase at 7 and 44 days in the treated rhizosphere (Fig. 3A). In case of Actinobacteria, a gradual increase was observed with plant growth in both rhizospheres, followed by reductions at 30 and 37 DAS. Although the difference between the rhizospheres was not significant from 7 to 37 DAS, the gene copy number at 44 days was significantly higher in the rhizosphere of bacterized plants (Fig. 3B), which was consistent with the pyrosequencing results. Sanguin et al. (2009) previously found that both Acidobacteria and Actinobacteria are rich in disease-suppressive soils. Moreover, these taxa are responsible for elimination of disease-causing microbes. The changes could benefit the plant health by maintaining a disease-suppressive environment. Qiu et al. (2012) and Shen et al. (2015) also found simultaneous decreases in the rhizosphere fungal community accompanied by increases in Acidobacteria and Actinobacteria. In this sense, the long-term influence of fungal community changes on plant growth needs further study.
Fig. 3.
Time-course comparative analysis of different bacterial groups in the rhizospheres of JBLS202-treated and untreated Kimchi cabbage plants from 7 to 44 days after bacterization. Data are presented as the log of gene copies of each bacterial group per gram rhizosphere soil (C – Control, J – JBLS202-treated). The abundances of Acidobacteria, Actinobacteria, Alphaproteobacteria, and Betaproteobacteria are presented. Bars with the same letter(s) do not differ significantly at P = 0.05 for a given treatment. *denotes a statistically significant increase at a particular time-point. Vertical bars denote standard deviation.
The abundance of Alphaproteobacteria showed a similar pattern in both treated and untreated rhizospheres. But the abundance significantly increased at 7, 14, and 44 days in the JBLS202-treated rhizosphere (Fig. 3C) and the increase at 44 DAS was in accordance with the pyrosequencing data. In terms of gene copy number, Betaproteobacteria were gradually reduced from 7 to 44 days in both rhizospheres and the difference was not significant, except at 14 days (Fig. 3D).
Previous studies have indicated that the microbiome structure of the rhizosphere depends on plant growth stage (Sugiyama et al., 2014) and on the introduction of a foreign member into the root region. Since multiple factors are responsible for the microbiome structure in the rhizosphere, a change in any of the factors will alter the microbial community. JBLS 202 has been reported to emit VOCs, which have the potential to influence the rhizosphere microbiome (Bhattacharyya and Lee, 2016 ). Therefore, factors such as introduction of a foreign member, VOCs, soil, and growth conditions need to be considered in restructuring of rhizosphere microbiome. Taken together, the results of this study indicate that introduction of JBLS202 induced beneficial modulation of the surrounding microbiome. We believe that the results of this study would contribute to understand microbial changes by introduction of foreign PGPR and to tune microbiome in the rhizosphere. For that purpose, long-term time-course field studies are necessary.
Influence of JBLS202 treatment on functional communities involved in nitrogen cycling
Taxonomic perturbation of microbial community composition by the introduction of JBLS202 can influence on the function of soil microbial communities. To investigate whether functional shifts of the rhizosphere microbiome occur, we assayed the copy numbers of genes involved in nitrogen cycling. Despite fluctuations in copies of the denitrification gene nirK in the untreated control rhizosphere, the level of nirK was stably maintained in the treated rhizosphere, and a significant increase over the control rhizosphere was observed from 14 to 44 DAS, with the exception of 37 DAS. The copy number of the nitrate-reducing gene narG was significantly increased at 7, 14, and 44 DAS in the JBLS202-treated rhizosphere (Table 2). For nirS, no significant difference was observed in the quantities between the treated and control rhizosphere, except at the initial stage. The copy number of nosZ increased gradually from 7 to 44 days in the untreated control rhizosphere, and the copy number in the treated rhizosphere was significantly higher at each time-point, except 37 DAS.
Table 2.
Real-time PCR quantifications of genes encoding key enzymes of nitrate reduction (narG) and denitrification (nirK, nirS, nosZ)
| Gene | nirK | narG | ||
|---|---|---|---|---|
|
|
||||
| Days after treatment | Control | JBLS202-treated | Control | JBLS202-treated |
| 7 | 2.357 ± 3.40d | 5.198 ± 0.04cd | 6.026 ± 0.03e | 6.212 ± 0.02c* |
| 14 | 5.358 ± 0.06bc | 5.646 ± 0.12b* | 6.279 ± 0.02d | 6.662 ± 0.01a* |
| 23 | 5.250 ± 0.08bcd | 5.496 ± 0.09bc* | 6.385 ± 0.02cd | 6.424 ± 0.03b |
| 30 | 4.921 ± 0.23cd | 5.317 ± 0.04c* | 6.459 ± 0.05c | 6.462 ± 0.02b |
| 37 | 5.421 ± 0.03b | 5.431 ± 0.09c | 6.724 ± 0.05a | 6.631 ± 0.02a |
| 44 | 5.680 ± 0.07a | 5.830 ± 0.02a* | 6.589 ± 0.02b | 6.679 ± 0.05a* |
|
| ||||
| nirS | nosZ | |||
|
|
||||
| Control | JBLS202-treated | Control | JBLS202-treated | |
|
| ||||
| 7 | 5.024 ± 0.04e | 5.212 ± 0.02d* | 6.129 ± 0.06c | 6.459 ± 0.05b* |
| 14 | 5.544 ± 0.04d | 5.710 ± 0.03c* | 6.281 ± 0.03bc | 6.652 ± 0.02a* |
| 23 | 5.738 ± 0.05c | 5.805 ± 0.01b | 6.347 ± 0.01b | 6.675 ± 0.02a* |
| 30 | 5.956 ± 0.03a | 5.775 ± 0.01bc | 6.332 ± 0.01b | 6.448 ± 0.03b* |
| 37 | 5.957 ± 0.03a | 5.776 ± 0.02bc | 6.481 ± 0.06a | 6.338 ± 0.03c |
| 44 | 5.847 ± 0.02b | 5.898 ± 0.04a | 6.521 ± 0.05a | 6.624 ± 0.01a* |
Data represents mean of log copies of gene per g soil ± standard deviation. Values followed by the same letter in a column do not differ significantly at P = 0.05 according to the Tukey-Kramer HSD. In a given row for one gene at a given time-point, *denotes detection of statistically higher gene copies in the JBLS202-treated rhizosphere.
These results indicate that the abundance of genes involved in nitrogen cycling was higher in the JBLS202-treated rhizosphere than control rhizosphere, which corresponds with the results observed by taxon-specific qPCR. Root exudates and available nutrients in the rhizosphere are both essential for growth of nitrogen fixers and denitrifiers (Burgmann et al., 2005; Hai et al., 2009). The increased levels of genes involved in the nitrogen cycle can be beneficial for plant growth because the amount of available nitrogen might increase. The increase of gene copies might indicate changes in the root exudation due to the introduction of JBLS202, which eventually contribute to maintain a beneficial microbial community in the rhizosphere. An increase in the functional genes was responsible for increase in both soil respiration and nitrification efficiency in maize plants indicating that ecological functioning can be influenced by changes in microbial communities and network interactions (Wang et al., 2015). Taken together, the results of this study indicate that introduction of beneficial bacteria can play a crucial role in the composition and function of the rhizosphere microbial community. Further studies on the complicated interactions between the introduced inoculant, plant growth stage, and resident microbes are needed to understand more about the interactions in the systems.
Supplementary data
Acknowledgments
This study was supported by a grant from Next-Generation BioGreen 21 Program (Project No. PJ01316301), Rural Development Administration. We also gratefully acknowledge a grant from Nakdonggang National Institute of Biological Resources (2017-075) and the National Research Foundation of Korea (NRF) grant (Project No. 2017R1A2B2002221) funded by the Korean government (MSIT), Republic of Korea.
References
- Bais HP, Broeckling CD, Vivanco JM. Root exudates modulate plant-microbe interactions in the rhizosphere. In: Karlovsky P, editor. Soil Biol. Springer; Berlin, Germany: 2008. pp. 241–252. [DOI] [Google Scholar]
- Bending GD, Rodriguez-Cruz MS, Lincoln SD. Fungicide impacts on microbial communities in soils with contrasting management histories. Chemosphere. 2007;69:82–88. doi: 10.1016/j.chemosphere.2007.04.042. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya D, Lee YH. The bacterial community in the rhizosphere of Kimchi cabbage restructured by volatile compounds emitted from rhizobacterium Proteus vulgaris JBLS202. Appl Soil Ecol. 2016;105:48–56. doi: 10.1016/j.apsoil.2016.03.020. [DOI] [Google Scholar]
- Bhattacharyya D, Garladinne M, Lee YH. Volatile indole produced by rhizobacterium Proteus vulgaris JBLS202 stimulates growth of Arabidopsis thaliana through auxin, cytokinin, and brassinosteroid pathways. J Plant Growth Regul. 2015;34:158–168. doi: 10.1007/s00344-014-9453-x. [DOI] [Google Scholar]
- Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- Burgmann H, Meier S, Bunge M, Widmer F, Zeyer J. Effects of model root exudates on structure and activity of a soil diazotroph community. Environ Microbiol. 2005;7:1711–1724. doi: 10.1111/j.1462-2920.2005.00818.x. [DOI] [PubMed] [Google Scholar]
- Chowdhury SP, Dietel K, Rändler M, Schmid M, Junge H, Borriss R, Hartmann A, Grosch R. Effects of Bacillus amyloliquefaciens FZB42 on lettuce growth and health under pathogen pressure and its impact on the rhizosphere bacterial community. PloS One. 2013;8:e68818. doi: 10.1371/journal.pone.0068818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J, Goodfellow M. A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int J Syst Bacteriol. 1995;45:240–245. doi: 10.1099/00207713-45-2-240. [DOI] [PubMed] [Google Scholar]
- Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007;57:2259–2261. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- Dutta S, Podile AR. Plant growth promoting rhizobacteria (PGPR): Bugs to debug the root zone. Crit Rev Microbiol. 2010;36:232–244. doi: 10.3109/10408411003766806. [DOI] [PubMed] [Google Scholar]
- Dutta S, Rani TS, Podile AR. Root exudate-induced alterations in Bacillus cereus cell wall contribute to root colonization and plant growth promotion. Plos One. 2013;8:e78369. doi: 10.1371/journal.pone.0078369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Accelerated profile HMM Searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BR, Todorovic B, Czarny J, Cheng Z, Duan J, McConkey B. Promotion of plant growth by bacterial ACC deaminase. Crit Rev Plant Sci. 2007;26:227–242. doi: 10.1080/07352680701572966. [DOI] [Google Scholar]
- Hai B, Diallo NH, Sall S, Haesler F, Schauss K, Bonzi M, Assigbetse K, Chotte JL, Munch JC, Schloter M. Quantification of key genes steering the microbial nitrogen cycle in the rhizosphere of sorghum cultivars in tropical agroecosystems. Appl Environ Microbiol. 2009;75:4993–5000. doi: 10.1128/AEM.02917-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck KL, Jr, Van Belle G, Simberloff D. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology. 1975;56:1459–1461. doi: 10.2307/1934716. [DOI] [Google Scholar]
- Hirsch PR, Mauchline TH, Clark IM. Culture-independent molecular techniques for soil microbial ecology. Soil Biol Biochem. 2010;42:878–887. doi: 10.1016/j.soilbio.2010.02.019. [DOI] [Google Scholar]
- Hou J, Liu W, Wang B, Wang Q, Luo Y, Franks AE. PGPR enhanced phytoremediation of petroleum contaminated soil and rhizosphere microbial community response. Chemosphere. 2015;138:592–598. doi: 10.1016/j.chemosphere.2015.07.025. [DOI] [PubMed] [Google Scholar]
- Huang XF, Chaparro JM, Reardon KF, Zhang R, Shen Q, Vivanco JM. Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany. 2014;92:267–275. doi: 10.1139/cjb-2013-0225. [DOI] [Google Scholar]
- Kamilova F, Kravchenko LV, Shaposhnikov AI, Azarova T, Makarova N, Lugtenberg B. Organic acids, sugars, and L-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol Plant-Microbe Interact. 2006;19:250–256. doi: 10.1094/MPMI-19-0250. [DOI] [PubMed] [Google Scholar]
- Kang Y, Shen M, Wang H, Zhao Q. A possible mechanism of action of plant growth promoting rhizobacteria (PGPR) strain Bacillus pumilus WP8 via regulation of soil bacterial community structure. J Gen Appl Microbiol. 2013;59:267–277. doi: 10.2323/jgam.59.267. [DOI] [PubMed] [Google Scholar]
- Kim M, Yoon H, Kim YE, Kim YJ, Kong WS, Kim JG. Comparative analysis of bacterial diversity and communities inhabiting the fairy ring of Tricholoma matsutake by barcoded pyrosequencing. J Appl Microbiol. 2014;117:699–710. doi: 10.1111/jam.12572. [DOI] [PubMed] [Google Scholar]
- Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultures species. Int J Syst Evol Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- Kozdrój J. Microbial community in the rhizosphere of young maize seedlings is susceptible to the impact of introduced pseudomonads as indicated by FAME analysis. J Gen Appl Microbiol. 2008;54:205–210. doi: 10.2323/jgam.54.205. [DOI] [PubMed] [Google Scholar]
- Kwon S, Kim TS, Yu GH, Jung JH, Park HD. Bacterial community composition and diversity of a full-scale integrated fixed-film activated sludge system as investigated by pyrosequencing. J Microbiol Biotechnol. 2010;20:1717–1723. [PubMed] [Google Scholar]
- LaSala PR, Segal J, Han Faye S, Tarrand Jeffrey J, Han Xiang Y. First reported infections caused by three newly described genera in the family Xanthomonadaceae. J Clin Microbiol. 2007;45:641–644. doi: 10.1128/JCM.01938-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WZ, Godzik A. Cd-hit: a fast program for clustering and comparing large setsof protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Liu D, Liu Y, Fang S, Tian Y. Tree species composition influenced microbial diversity and nitrogen availability in rhizosphere soil. Plant Soil Environ. 2015;61:438–443. doi: 10.17221/94/2015-PSE. [DOI] [Google Scholar]
- Lo CC. Effect of pesticides on soil microbial community. J Environ Sci Health B. 2010;45:348–359. doi: 10.1080/03601231003799804. [DOI] [PubMed] [Google Scholar]
- Lugtenberg B, Kamilova F. Plant-growth promoting rhizobacteria. Annu Rev Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JH, Piceno YM, DeSantis TZ, Anderson GL, Bakker PA, Raaijmakers JM. Deciphering the rhizosphere microbiome. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- Myers EW, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Nannipieri P, Ascher J, Ceccherini MT, Landi L, Pietramellara G, Renella G. Microbial diversity and soil functions. Eur J Soil Sci. 2003;54:655–670. doi: 10.1046/j.1351-0754.2003.0556.x. [DOI] [Google Scholar]
- Piromyou P, Buranabanyat B, Tantasawat P, Tittabutr P, Boonkerd N, Teaumroong N. Effect of plant growth promoting rhizobacteria (PGPR) inoculation on microbial community structure in rhizosphere of forage corn cultivated in Thailand. Eur J Soil Biol. 2011;47:44–54. doi: 10.1016/j.ejsobi.2010.11.004. [DOI] [Google Scholar]
- Qiu M, Zhang R, Xue C, Zhang S, Li S, Zhang N, Shen Q. Application of a novel bio-organic fertilizer can control Fusarium wilt by regulating the microbial community of cucumber rhizosphere soils. Biol Fert Soils. 2012;48:807–816. doi: 10.1007/s00374-012-0675-4. [DOI] [Google Scholar]
- Sanguin H, Sarniguet A, Gazengel K, Moenne-Loccoz Y, Grundmann G. Rhizosphere bacterial communities associated with disease suppressiveness stages of take-all decline in wheat monoculture. New Phytol. 2009;184:694–707. doi: 10.1111/j.1469-8137.2009.03010.x. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Ruan Y, Chao X, Zhang J, Li R, Shen Q. Rhizosphere microbial community manipulated by 2 years of consecutive biofertilizer application associated with banana Fusarium wilt disease suppression. Biol Fert Soils. 2015;51:553–562. doi: 10.1007/s00374-015-1002-7. [DOI] [Google Scholar]
- Somers E, Vanderleyden J, Srinivasan M. Rhizosphere bacterial signalling: a love parade beneath our feet. Crit Rev Microbiol. 2004;30:205–240. doi: 10.1080/10408410490468786. [DOI] [PubMed] [Google Scholar]
- Sugiyama A, Ueda Y, Zushi T, Takase H, Yazaki K. Changes in the bacterial community of soybean rhizospheres during growth in the field. PLoS One. 2014;9:e100709. doi: 10.1371/journal.pone.0100709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabelsi D, Mhamdi R. Microbial inoculants and their impact on soil microbial communities: A review. Biomed Res Int. 20132013:863240. doi: 10.1155/2013/863240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi P, He Z, Van Nostrand JD, Albrigo G, Zhou J, Wang N. Huanglongbing alters the structure and functional diversity of microbial communities associated with citrus rhizosphere. ISME J. 2012;6:363–383. doi: 10.1038/ismej.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno T, Jang J, Han D, Kim JH, Sadowsky MJ, Kim OS, Chun J, Hur HG. Use of barcoded pyrosequencing and shared OTUs to determine sources of fecal bacteria in watersheds. Environ Sci Technol. 2010;44:7777–7782. doi: 10.1021/es101500z. [DOI] [PubMed] [Google Scholar]
- Wang F, Liang Y, Jiang Y, Yang Y, Xue K, Xiong J, Zhou J, Sun B. Planting increases the abundance and structure complexity of soil core functional genes relevant to carbon and nitrogen cycling. Sci Rep. 2015;5:14345. doi: 10.1038/srep14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SM, Lee YH. Plant growth promoting rhizobacterium Proteus vulgaris JBLS202 stimulates the seedling growth of Chinese cabbage through indole emission. Plant Soil. 2013;370:485–495. doi: 10.1007/s11104-013-1652-x. [DOI] [Google Scholar]
- Zinger L, Shanhavaz B, Baptist F, Geremia RA, Choler P. Microbial diversity in alpine tundra soils correlates with snow cover dynamics. ISME J. 2009;3:850–859. doi: 10.1038/ismej.2009.20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.