Abstract
In addition to regulating gene expression, RNA silencing is an essential antiviral defense system in plants. Triggered by double-stranded RNA, silencing results in degradation or translational repression of target transcripts. Viruses are inducers and targets of RNA silencing. To condition susceptibility, most plant viruses encode silencing suppressors that interfere with this process, such as the Tomato spotted wilt virus (TSWV) NSs protein. The mechanism by which NSs suppresses RNA silencing and its role in viral infection and movement remain to be determined. We cloned NSs from the Hawaii isolate of TSWV and using two independent assays show for the first time that this protein restored pathogenicity and supported the formation of local infection foci by suppressor-deficient Turnip mosaic virus and Turnip crinkle virus. Demonstrating the suppression of RNA silencing directed against heterologous viruses establishes the foundation to determine the means used by NSs to block this antiviral process.
Keywords: TSWV, NSs protein, Antiviral RNA silencing
INTRODUCTION
Tomato spotted wilt virus (TSWV) has a worldwide distribution and a host range that includes more than 1300 plant species (Parrella et al., 2003). In 2011, TSWV was ranked the second most important virus in molecular plant pathology (Scholthof et al., 2011). TSWV infection in plants causes a reduction in both yield and quality of fruit and vegetables, and induces developmental defects in ornamental species (Pappu et al., 2009). Among the most economically important plants susceptible to this virus are tomato (Solanum lycopersicum), lettuce (Lactuca sativa), pepper (Capsicum annuum), potato (S. tuberosum), onion (Allium cepa), peanut (Arachis hypogaea), tomatillo (Physalis ixocarpa Brot.), and chrysanthemum (Chrysanthemum morifolium; de la Torre-Almaráz et al., 2002; Parrella et al., 2003; Scholthof et al., 2011).
TSWV belongs to the genus Tospovirus in the family Bunyaviridae (Parrella et al., 2003). TSWV virions are 80–120 nm in diameter, contain several copies of RNA-dependent RNA polymerase, and are surrounded by an envelope formed of host-derived lipids and viral glycoproteins (Parrella et al., 2003). The TSWV genome consists of three negative-sense, single-strand RNAs designated large (L), medium (M), and small (S; Figure 1A; Parrella et al., 2003; Margaria and Rosa, 2015). RNA L (8.8 kb) encodes the RNA-dependent RNA polymerase (331.5 kDa) responsible for TSWV RNA replication and transcription (Margaria and Rosa, 2015). RNA M (4.8 kb) harbors the open reading frame of membrane glycoprotein Gc/Gn (127.4 kDa), which is cleaved by a host protease to form the Gc and Gn proteins that make up part of the virion envelope (Parrella et ak, 2003). In addition, RNA M is the template for a sub-genomic RNA encoding protein NSm (33.6 kDa), involved in virus cell-to-cell movement (Parrella et al., 2003). RNA S (2.9 kb) encodes the nucleocapsid protein N (29 kDa), which associates with genomic RNAs to form ribonucleoprotein complexes that are packaged into virions (Parrella et al., 2003). RNA S also serves as a template for a further sub-genomic RNA responsible for the production of the multifunctional protein NSs (52.4 kDa; Parrella et al., 2003).
Figure 1.
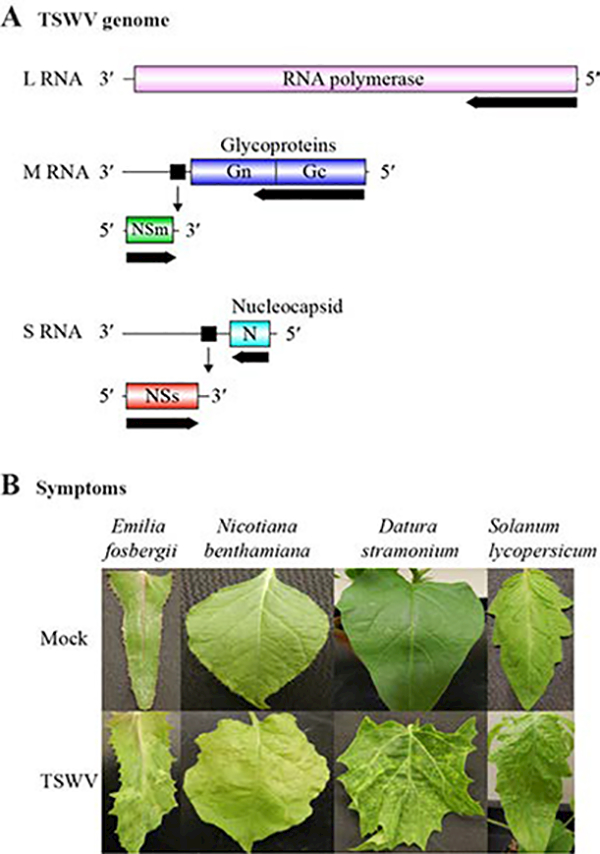
Tomato spotted wilt virus (TSWV) genome organization and systemic symptoms. A. Schematic representation of the TSWV genome. Colored cylinders indicate open reading frames. Lines indicate 3’ and 5’ untranslated regions and intergenic regions. Black squares indicate transcription termination hairpins. Black arrows indicate directions of translation. Sub-genomic transcripts are transcribed from M and S RNAs. B. Representative TSWV-induced systemic symptoms in selected hosts at 21 days post inoculation.
RNA silencing regulates gene expression in eukaryotes, including plants and insects (Ding and Voinnet, 2007), in which also functions as an inducible, potent, highly specific, and adaptable antiviral defense system (Ding and Voinnet, 2007; Garcia and Pallas, 2015). Most plant and insect viruses encode silencing suppressor proteins that inactivate RNA silencing to condition susceptibility and favor virus replication and spread (Powers et al., 2008; Qu et al., 2008; Garcia-Ruiz et al., 2010; Wang et al., 2010). In tospoviruses, protein NSs is an RNA silencing suppressor (Takeda et al., 2002; Bucher et al., 2003; Hedil et al., 2015) necessary for the establishment and maintenance of systemic infection in plants (Margaria et al., 2014) and for persistent infection and transmission by flower thrips (Frankliniella occidentalis; Whitfield et al., 2005; Margaria et al., 2014). TSWV NSs is also an avirulence determinant that triggers a hypersensitive response in resistant plants (de Ronde et al., 2013; de Ronde et al., 2014).
Variation in pathogenicity of this virus has been observed in pepper, tomatillo, and chrysanthemum (de la Torre-Almaráz et al., 2002), and in the former, such differences have been found to be due to NSs mutations (de Ronde et al., 2013). The mechanism by which tospoviral NSs inhibits RNA silencing, and its role in virus infection remain to be determined. In this paper, we show for the first time that TSWV NSs suppresses antiviral RNA silencing and restores the pathogenicity of two heterologous viruses: suppressor-deficient Turnip mosaic virus (TuMV) and Turnip crinkle virus (TCV).
MATERIAL AND METHODS
TSWV mechanical inoculation
The Hawaii isolate of TSWV was transmitted by thrips (F. occidentalis) to Emilia fosbergii for maintenance and mechanically to other hosts for experiments. Using a mortar and pestle, inoculum was prepared by grinding 1 g systemically infected fresh leaves in 10 mL TSWV inoculation buffer (0.1 M potassium phosphate buffer, pH 7.0; Mandal et al., 2008), into which 10 mM sodium sulfite, 10 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride (PMSF) had been added before use. The extract was centrifuged at 4000 rpm for 10 min at 4°C, and the supernatant transferred to a new tube and kept on ice. Datura stramonium, Nicotiana benthamiana, and S. lycopersicum leaves were dusted with carborundum and rub-inoculated with 50 μL inoculum per leaf. Eighteen plants of each species were inoculated when they were 25–30 days old. In parallel, control plants were mock-inoculated with buffer. Plants were kept in a growth chamber at 22°C under a 16:8 h light:dark cycle. Systemically infected N. benthamiana, D. stramonium, and S. lycopersicum leaves (Figure 1B) were collected at 15, 20, and 25 days post inoculation (dpi), respectively. Leaf samples were then frozen in liquid nitrogen and stored at −80°C.
TSWV virion purification and RNA extraction
Two protocols (Lane, 1986; de Avila et al., 1990) were combined to purify TSWV virions. Frozen leaves (20 g) were ground with an electric blender in 100 mL cold TSWV inoculation buffer containing 10 mM sodium sulfite, 10 mM β-mercaptoethanol, and 1 mM PMSF. Extracts were filtered through four layers of cheese cloth and debris removed by spinning at 7700 g for 10 min. Aliquots of 50 and 800 μL were collected for total protein and total RNA extraction, respectively. The solution obtained was centrifuged at 50,000 g for 30 min and the supernatant discarded. The resulting pellet was re-suspended in 22.5 mL cold 0.1 M sodium citrate buffer, pH 5.5, before adding Triton X-100 to a final concentration of 2%, and mixing with a stir bar for 10 min at 4°C. After low-speed centrifugation (7700 g for 10 min), the supernatant was transferred to a new tube. Virions were then concentrated by centrifugation (35,000 rpm for 2 h in an L7–55 ultracentrifuge with a 50.2 Ti rotor, Beckman Coulter, Brea, CA, USA) through a 5-mL cushion of 20% sucrose in 0.1 M sodium citrate buffer, pH 5.5. The supernatant was discarded and virions were recovered by re-suspending the pellet in 800 μL suspension buffer (10 mM Tris-Cl, pH 8.0, 1 mM ethylenediaminetetraacetic acid, and 100 mM NaCl). Fifty microliters were taken for protein extraction, while the rest was used for virion RNA isolation. Total and virion protein samples were mixed with 50 μL 2X protein dissociation buffer containing bromophenol blue, and incubated at 100°C for 3 min, before being stored at −80°C until used for immunoblotting. Virion purification efficiency was determined by a dilution curve of total and virion proteins, and protein N abundance was estimated with respect to a two-fold dilution of the total protein sample.
Total and virion RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA), and concentration was measured with a NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA). Samples containing 1 mg RNA were run on a 1% agarose gel.
Western blotting
Protein samples were analyzed by western blotting. A Bradford assay revealed protein concentrations of 0.1–0.8 and 1.5–2 μg/μL for the total and virion extracts, respectively. Equal sample volumes (5 μL) were loaded on a 12% polyacrylamide gel and proteins were separated by electrophoresis at 150 V for 60 min. After being transferred to a membrane, proteins were stained with Ponceau S solution (Sigma, St. Louis, MO, USA). Protein N was detected using a primary antibody (1:64,000; Law and Moyer, 1990), and a goat anti-rabbit immunoglobulin G secondary antibody (1:10,000; NA934–1; GE Healthcare, Little Chalfont, UK). Chemiluminescence was detected with Clarity Western ECL Substrate and a ChemiDoc MP Imaging system (Bio-Rad, Hercules, CA, USA).
TSWV NSs cloning
Virion RNA from N. benthamiana was used to clone two versions of NSs, containing or lacking the 5’ untranslated region (UTR). Using Geneious 8.1.5 (Biomatters Ltd., Auckland, New Zealand), oligonucleotides were designed based on the consensus sequence of all S segments available on GenBank. First-strand complementary DNA (cDNA) was generated with Superscript III reverse transcriptase (Thermo Fisher Scientific). Virion RNA (4 μg) was treated with 2 U DNase (Ambion TURBO DNA-free; AM1907; Thermo Fisher Scientific), and reverse transcription (RT) was carried out at 50°C for 50 min using tospovirus universal primer TOS-R15 (5’-GGGAGAGCAATYGWGKYR-3’; Uga and Tsuda, 2005). First-strand cDNA was amplified by polymerase chain reaction (PCR) using TSWV-S14 (5’-CACCATGGAGAGCAATTGTGTC-3’) or TSWV_NSs (5’-CACCATGGAT GTCTTCAAGTGTTTATG-3’) as the forward primer and Stop_TSWV_ NSs (5’-ggccgctctAGATCATTTTGATCCTG-3’) as the reverse primer. TSWV-S14 and TSWV NSs, which add a topoisomerase recognition site, were used to generate an NSs clone that included or lacked the 5’ UTR, respectively. For cloning purposes, an NcoI site was formed by adding four base pairs (ATGG). Primer Stop TSWV NSs adds an XbaI site (lower case) after the stop codon in the NSs sequence.
The 50-μL PCR mix contained 2 μL cDNA, 1.5 mM MgCl2. 0.2 mM deoxynucleotides, 0.5 μΜ primers, 1X reaction buffer, and 1 μL PfuUltra II Fusion HS DNA polymerase (600672; Agilent Technologies, Santa Clara, CA, USA). Thermocycler conditions were as follows: initial denaturing at 95°C for 2 min, followed by 32 amplification cycles of denaturing at 95°C for 20 s, annealing at 53°C for 20 s, and extension at 72°C for 2 min, before a final extension at 72°C for 5 min.
Using a TOPO cloning kit (45–0218; Life Technologies, Carlsbad, CA, USA), RT-PCR products were inserted into pENTR plasmids, and clones were identified by EcoRI/ PvuI double digestion. One pENTR-NSs clone and one pENTR-5’ UTR-NSs clone were then transferred to pMDC32 vectors using Gateway LR Clonase II Enzyme Mix (11791–020; Life Technologies). pMDC32 clones were identified by EcoRI digestion. For each of the two NSs constructs, four independent clones were sequenced and their consensus sequence aligned to that of all 61 TSWV segment S sequences available on GenBank.
Agrobacterium transformation and agroinfiltration
By electroporation, Agrobacterium tumefaciens strain GV3101 was transformed with plasmids encoding NSs (pMDC32–5’ UTR-NSs or pMDC32-NSs), tombusviral P19 (pCB302-P19), beta-glucuronidase (pMDC32-GUS; Johansen and Carrington, 2001; Garcia-Ruiz et al., 2010), or a single-stranded green fluorescent protein (ssGFP) reporter (pPZP-35S-GFP; Tatineni et al., 2012). Cells were cultured on Luria broth (LB) plates containing rifampicin and kanamycin (for pCB and pMDC32 plasmids) or spectinomycin (for the pPZP plasmid) at 28°C for 48 h. Cells from a single colony were used to inoculate 1 mL LB medium containing the corresponding antibiotic(s), which was then incubated for 20 h at 28°C with shaking. Using 10 or 20 mL vir induction medium, consisting of 1X LB, 100 mg/mL rifampicin, 125 mg/mL kanamycin or spectinomycin, 10 mM 2-(N-morpholino) ethanesulfonic acid hydrate (MES), pH 5.2, and 100 μΜ acetosyringone, cultures were diluted to an optical density at 600 nm (OD600) of 0.0025 and incubated for 16 h at 28°C with shaking. Cells were pelleted by centrifugation at 6000 rpm for 10 min, re-suspended in an equal volume of infiltration solution, comprising 10 mM MgCl2,, 10 mM MES, pH 5.2, and 150 μΜ acetosyringone, and incubated in the dark at room temperature for 2 h before being used to infiltrate N. benthamiana leaves. Plants were at the five-to-six-leaf stage, and leaves three and four were infiltrated. Plants were kept in a growth chamber at 27°C under a 16:8-h light:dark cycle. Experiments were repeated two or three times.
Using the same protocol, Agrobacterium tumefaciens cells were transformed with infectious clones of suppressor-deficient Turnip mosaic virus tagged with GFP (TuMV-AS9-GFP; Garcia-Ruiz et al., 2010) or GFP-labeled Turnip crinkle virus (TCV-GFP; Powers et al., 2008).
TuMV-AS9-GFP virion enrichment
TuMV-AS9-GFP was propagated by co-infiltrating N. benthamiana leaves with plasmids carrying TuMV-AS9-GFP (OD600 = 0.25) and P19 (OD600 = 0.5), as previously described (Garcia-Ruiz et al., 2010). Five days later, virions were enriched from 200 g infiltrated leaves by polyethylene glycol precipitation (Garcia-Ruiz et al., 2010). Enriched virions were stored at −20°C in 40% glycerol and titrated in Arabidopsis thaliana dcl2–1 dcl3–1 dcl4–2 triple-mutant plants (Garcia-Ruiz et al., 2010).
Transgene silencing
A standard assay based on the N. benthamiana 16c line, which expresses a GFP transgene (Johansen and Carrington, 2001), was used to measure transgene silencing suppression by NSs. Virus-induced gene silencing (Benedito et al., 2004) results when a fragment of viral RNA, in this case a GFP tag, exhibits sequence homology with a host gene, such as that encoding GFP in N. benthamiana 16c plants. Agrobacterium tumefaciens cells carrying an ssGFP construct (OD600 = 0.125) were infiltrated alone or in combination with NSs (OD600 = 0.5), while GUS and P19 were used as negative and positive controls, respectively. Eight plants were infiltrated per treatment, and the experiment was repeated twice. Induction of local and systemic transgene silencing was measured using photographs of infiltrated and non-infiltrated leaves taken under ultraviolet (UV) light.
GFP fluorescence and silencing index
GFP fluorescence intensity was determined from photographs taken under UV light using the green channel in ImageJ (Wayne Rasband, National Institutes of health, USA). Systemic silencing in transgenic N. benthamiana 16c plants was measured as the ratio of red to green light using the corresponding ImageJ channels. Green and red background levels were estimated using leaves from wild-type plants and those of untreated 16c plants, respectively. The former leaves were also used to estimate a wild-type red intensity value. The green signal from wild-type plants was divided by that from 16c plants, and the resulting value subtracted from 1. To estimate the change from GFP to red fluorescence (silenced) in 16c plants, a silencing index was estimated by subtracting the green from the red value dividing the result by the wild-type red intensity.
Antiviral RNA silencing assays
Suppressor-deficient TCV-GFP (Powers et al., 2008) and TuMV-AS9-GFP (Garcia-Ruiz et al., 2010) do not infect N. benthamiana and were used to measure antiviral RNA silencing by two complementary approaches: co-infiltration and infiltration followed by mechanical inoculation (Powers et al., 2008). TCV-GFP lacks the coat protein, which is a silencing suppressor (Powers et al., 2008; Qu et al., 2008), while TuMV-AS9-GFP harbors an inactivating mutation in the silencing suppressor HC-Pro (Garcia-Ruiz et al., 2010). Six plants were infiltrated per treatment, and experiments were repeated twice.
In co-infiltration assays, TCV-GFP or TuMV-AS9-GFP was infiltrated (OD600 = 0.0005) alone or in combination with NSs (OD600 = 0.5). GUS and P19 were used as negative and positive controls, respectively. Six days after infiltration, suppression of antiviral RNA silencing was measured by counting the number of local infection foci per leaf under UV light. TuMV-AS9-GFP coat protein levels in infiltrated leaves were measured by immunoblotting. To assess the accumulation of TuMV-derived small RNAs (sRNAs), the OD600 of the TuMV-AS9-GFP-carrying Agrobacterium suspension was increased to 0.125 and infiltrated leaves were collected for protein and RNA extraction.
In a complementary assay, NSs and controls were infiltrated at an OD600 of 1.0. Twenty-four hours later, infiltrated leaves were mechanically inoculated with TuMV-AS9-GFP virions. Six days after infiltration, suppression of antiviral RNA silencing was scored by counting the number of local infection foci per leaf.
To measure virus-induced RNA silencing, transgenic N. benthamiana 16c leaves were mechanically inoculated with TuMV-AS9-GFP virions 1 day after infiltration of NSs or controls. Silencing of the GFP transgene in non-inoculated leaves was measured at 10, 15, and 20 days after infiltration.
High molecular weight RNA and sRNA northern blotting
Small RNA accumulation was measured as previously described (Garcia-Ruiz et al., 2010). Using duplicate gels, TuMV-AS9-GFP-derived sRNAs were detected using probes to target the GFP tag or the cylindrical inclusion helicase (CI). Probes against GFP or CI consisted of PRC products labeled with digoxigenin (DIG)-labeled (11 175 033 910; Roche). Blots were hybridized for 16 h at 38°C and washed at 42°C. Chemiluminescence was detected using CDP-Star (12 041 677 001; Roche, Basel, Switzerland).
NSs transcript was detected by high molecular weight RNA northern blotting using 4 μg total RNA, a 1% agarose gel containing 1.1% formaldehyde, and 1X 4-(2-hydroxyethyl)-l-piperazineethanesulfonic acid running buffer. After capillary transfer to a nylon membrane (11 417 240 001; Roche), NSs RNA and ribosomal 18S RNA were detected with DIG-labeled PCR products amplified from pENTR-NSs and Arabidopsis thaliana genomic DNA, respectively. For the latter, the oligonucleotides 18S_rRNA_F(5’-CACCTCCCTTAACGAGGATCCATTGG-3’) and 18S_rRNA_R (5’-AGAGCGTAGGCTTGCTTTGAGCACTC-3’) were used and the PCR product was cloned into pENTR (pENTR-18SAtrDNA). Blots were hybridized for 16 h at 55°C and washed at 60°C.
RESULTS
TSWV virion purification
TSWV virions were purified from three plant species with similar results. Compared to the initial extracts, staining and immunoblots showed protein N enrichment and depletion of the large subunit of rubisco in samples purified for virions (Figure 2A). A dilution curve revealed a 16-fold enrichment of protein N after virion purification from N. benthamiana (Figure 2B). Using D. stramonium and S. lycopersicum, this protein was enriched by 8- and 2-fold, respectively (S1 Figure), suggesting that enrichment efficiency relates to TSWV abundance.
Figure 2.
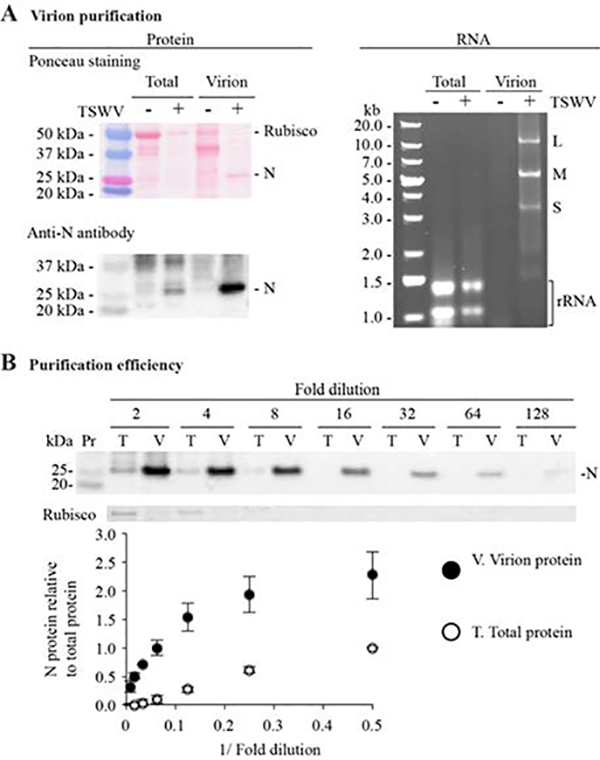
Tomato spotted wilt virus (TSWV) virion purification and genomic RNA extraction. Total and virion protein and RNA extracted from 20 g mock-inoculated (−) or TSWV-infected (+) Nicotiana benthamiana plants at 21 days post inoculation. A. Virion purification. Left panel: representative blot stained with Ponceau S solution and an immunoblot showing nucleocapsid protein (N). Precision Plus Dual Color (161–0374; Bio-Rad) was used as a size marker. Right panel: 1% agarose gel electrophoresis analysis of total and virion RNA. GeneRuler 1-kb Plus DNA ladder (SM1333; Thermo Fisher Scientific) was used as a marker. Positions of the large (L), medium (M), and small (S) TSWV genomic RNAs and ribosomal RNA (rRNA) are indicated. B. Virion purification efficiency evaluated by a dilution curve. The chart shows signal intensities of the N protein bands measured with ImageJ. Values are reported as averages ± standard errors of four biological replicates expressed relative to the two-fold dilution of the total protein sample.
In N. benthamiana, total RNA samples contained ribosomal RNA but TSWV genomic RNAs were not detected (Figure 2A). In contrast, a marked reduction in ribosomal RNA and an increase in TSWV genomic RNA was observed after virion purification (Figure 2A). Similar results were obtained for D. stramonium and S. lycopersicum (S1 Figure). In infected plants, RNA L is generally less abundant than the M and S transcripts (Hedil et al., 2014), as btained in the virion RNA isolated from D. stramonium and S. lycopersicum (S1 Figure). However, in N. benthamiana samples, RNA L was as abundant as RNA S (Figure 2A). RNA-dependent RNA polymerase (RDR1) is a critical component of antiviral RNA silencing in plants (Garcia-Ruiz et al., 2010; Wang et al., 2010), and N. benthamiana harbors a mutation in the gene by which it is encoded (NbRdRP1m; Yang et al., 2004). Thus, the higher levels of RNA L observed in N. benthamiana suggest that this RNA is preferentially targeted by RDR1-dependent silencing, as shown by sRNA sequencing in the absence of functional NSs (Margaria et al., 2015b).
NSs clones
Two NSs clones including or lacking the 5’ UTR were generated (Figure 3A). NSs cDNA was cloned into pENTR and inserted into pMDC32 for functional analysis (S2 Figure; Curtis and Grossniklaus, 2003). Nucleotide sequences of the NSs open reading frame were used to predict the corresponding amino acid sequence. The consensus sequence for all 61 TSWV segment S records available in GenBank was used as a reference. Compared to this reference sequence, the NSs clones described here exhibited 10 amino acid changes (Figure 3B) outside of conserved motifs, and hence not within the predicted essential RNA binding domains (de Ronde et al., 2014; Zhai et al., 2014; Hedil et al., 2015).
Figure 3.
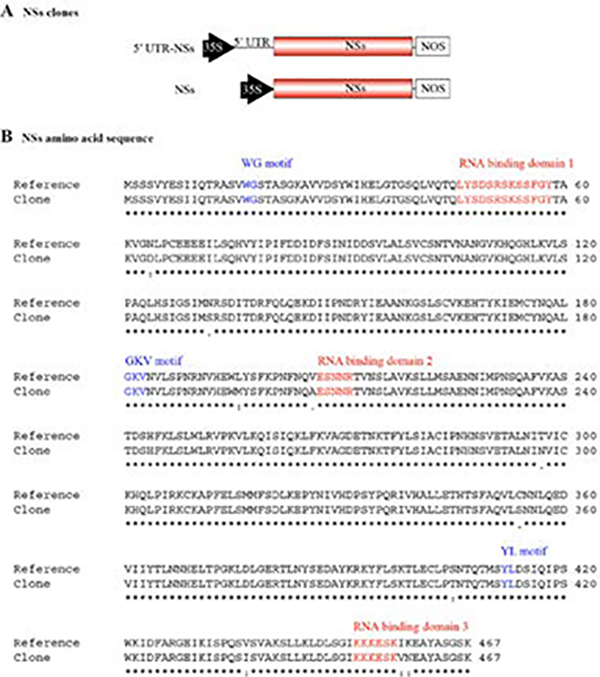
NSs clones and amino acid sequence. A. Representation of NSs clones in pMDC32, one containing and one lacking the 5’ untranslated region (UTR), placed between the 35S promoter and the nopaline synthase (NOS) terminator. B. Amino acid sequence of the NSs clone aligned to the consensus NSs sequence derived from all 61 Tomato spotted wilt virus segment S records in GenBank (Reference). Conserved WG, GKV, and YL motifs are indicated in blue. Three predicted RNA binding domains are indicated in red.
Local transgene silencing suppression
ssGFP was expressed alone or in the presence of NSs in N. benthamiana 16c plants. NSs prevented GFP silencing in the infiltrated zones (Figure 4). GFP fluorescence was 44 and 34% of that observed for P19 using the 5’ UTR-NSs and NSs clones, respectively. This level of local transgene silencing suppression agrees with previous results (Takeda et al., 2002).
Figure 4.
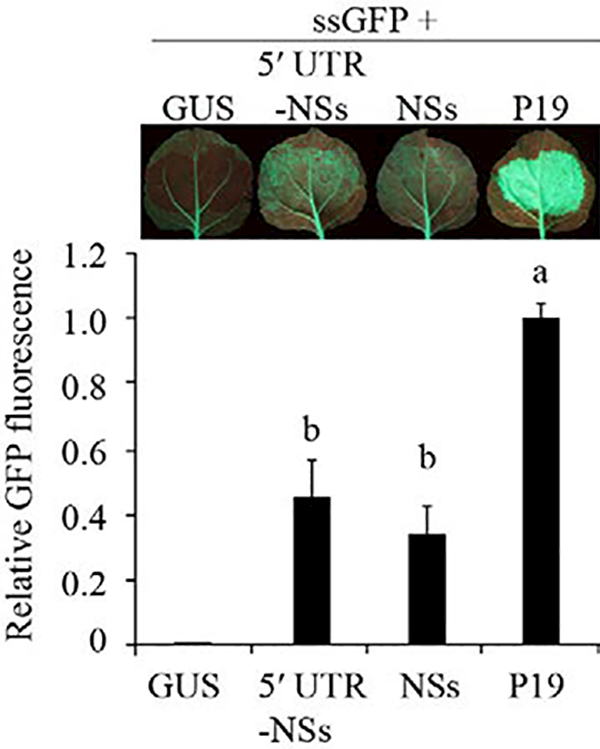
Suppression of transgene silencing by NSs. Single-stranded green fluorescent protein (ssGFP) was infiltrated alone or in combination with NSs in Nicotiana benthamiana 16c leaves. Beta-glucuronidase (GUS) and P19 were used as negative and positive controls, respectively. Five days after infiltration, GFP fluorescence was visualized under ultraviolet light. The histogram shows average GFP fluorescence ± standard error relative to leaves infiltrated with P19, based on 16 leaves from two independent experiments. Bars with the same letter do not significantly differ (Tukey’s test with α= 0.05). UTR = untranslated region.
Antiviral RNA silencing suppression
Tospoviral NSs clones have been characterized previously using transgene silencing and in vitro assays (Takeda et al., 2002; Bucher et al., 2003; Schnettler et al., 2010; de Ronde et al., 2013; de Ronde et al., 2014; Hedil et al., 2015). However, transgene silencing and antiviral RNA silencing differ in key genetic requirements (Ding and Voinnet, 2007). Moreover, the effect of NSs clones on the latter has not been demonstrated experimentally. We tested NSs antiviral RNA silencing activity using two complementary approaches and two silencing suppressor-deficient viruses unable to infect N. benthamiana. In co-infiltration assays, both 5’ UTR-NSs and NSs complemented suppressor-deficient TCV-GFP and TuMV-AS9-GFP, resulting in the formation of visible local infection foci (Figure 5). The number of infection foci per leaf for both viruses in the presence of NSs was between 40 and 60% of that observed using P19. Interestingly, the TCV-GFP infection foci formed after infiltration of the two NSs constructs were smaller (0.3 ± 0.1 nun in diameter) and dimmer than those seen following P19 treatment (1.0 ± 0.07 mm in diameter; Figure 5A). TuMV-AS9-GFP NSs and 5’ UTR-NSs infection foci were similar in size to those observed with P19, but of lower intensity (Figure 5B). Virus accumulation was measured by immunoblotting. With NSs, TuMV-AS9-GFP coat protein expression was 20% of the level observed using P19 (Figure 5B).
Figure 5.
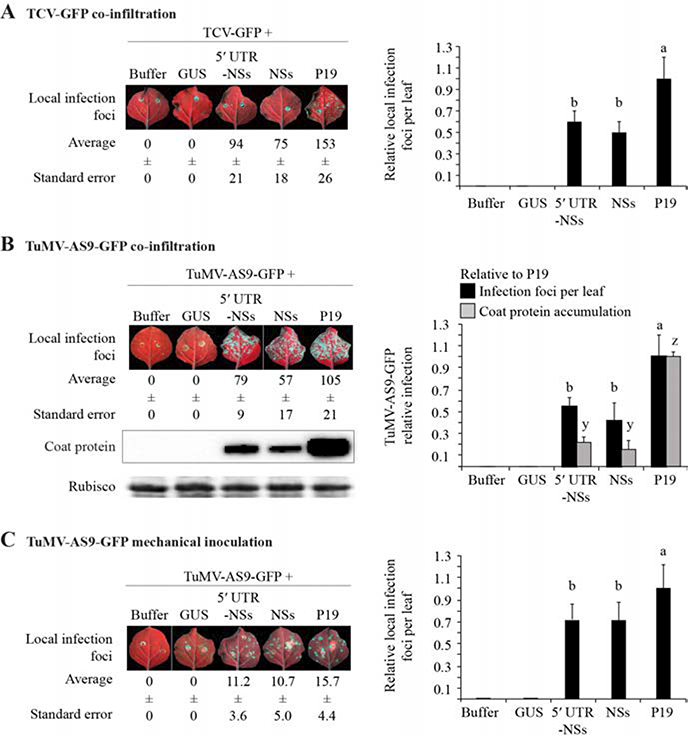
Complementation of suppressor-deficient Turnip crinkle virus-green fluorescent protein (TCV-GFP) and Turnip mosaic virus (TuMV)-AS9-GFP by NSs in Nicotiana benthamiana. Twelve leaves were infiltrated per treatment and visualized under ultraviolet light 6 days later. Green spots are local infection foci. Average numbers of visible local infection foci per leaf are given below pictures. The histogram shows average number of local infection foci ± standard error, relative to that observed on leaves infiltrated with P19. Bars with the same letter are not statistically different (Tukey’s test with α = 0.05). GUS = beta-glucuronidase; UTR = untranslated region. A. Co-infiltration of TCV-GFP with NSs or controls. B. Co-infiltration of TuMV-AS9-GFP with NSs or controls. The blot shows TuMV coat protein accumulation in the same leaves. The histogram shows average (± standard error) coat protein accumulation (four biological replicates) and local infection (12 leaves) foci number relative to that observed in leaves infiltrated with P19. C. Complementation of TuMV-AS9-GFP by NSs after mechanical inoculation. Leaves (12 per treatment) were infiltrated with NSs (at an optical density at 600 nm of 1.0) or controls, and mechanically inoculated with virions 1 day later. Data were collected and processed as in B.
Initiation of virus infection by agroinfiltration of binary vectors is sensitive to both transgene and antiviral RNA silencing (Powers et al., 2008). To exclude the possible effects of the former, we measured antiviral RNA silencing by first infiltrating NSs into N. benthamiana leaves and 1 day later, mechanically inoculating them with TuMV-AS9-GFP virions. Formation of local infection foci by TuMV-AS9-GFP confirmed NSs suppression of antiviral RNA silencing. The number of infection foci per leaf in the presence of NSs was 70% of that observed using P19 (Figure 5C).
Together, these results show that NSs is a suppressor of antiviral RNA silencing directed against two heterologous viruses: TCV-GFP and TuMV-AS9-GFP. However, NSs is less suppressive than P19. Inclusion of the 5’ UTR did not significantly affect NSs activity (Figures 4 and 5). In infiltrated leaves, RNA of both NSs clones accumulated to similar levels (Figure 6B), showing that the 5’ UTR did not significantly alter transcription or downstream events.
Figure 6.
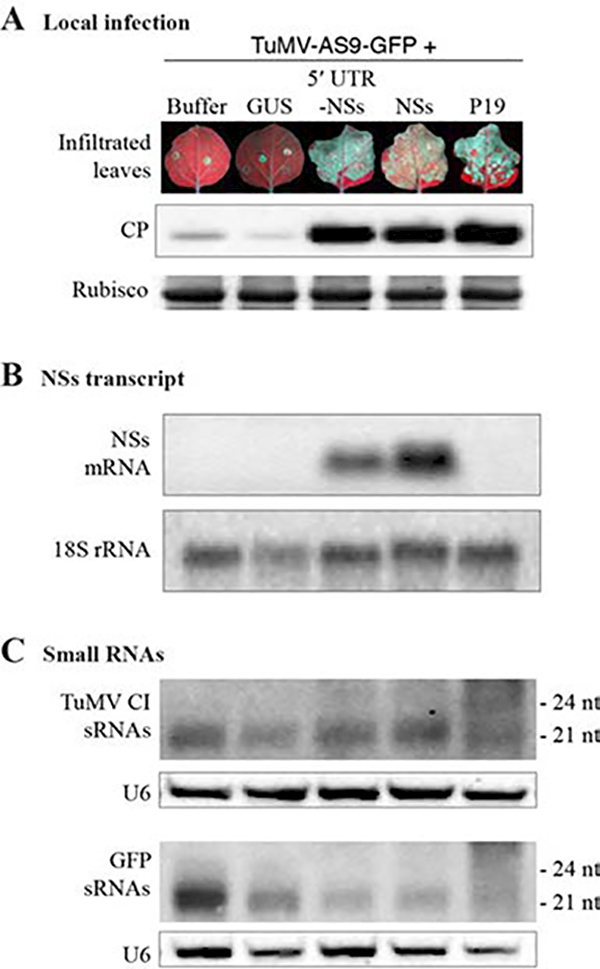
Virus and virus-derived small RNA (sRNA) accumulation. Turnip mosaic virus (TuMV)-AS9-green fluorescent protein (GFP) was co-infiltrated at an optical density at 600 nm (OD600) of 0.125 with NSs (OD600 = 0.5) or controls in Nicotiana benthamiana. Four days after infiltration, photographs were taken and samples collected for protein and RNA extraction. Accumulation of TuMV-AS9-GFP coat protein (CP), NSs transcript, and TuMV-AS9-GFP-derived small interfering RNA (siRNA) was determined in four biological replicates. GUS = beta-glucuronidase; UTR = untranslated region. A. Representative leaves showing local infection, and TuMV-AS9-GFP CP accumulation determined by immunoblotting. B. Accumulation of TuMV-AS9-GFP-derived sRNAs detected with probes against cylindrical inclusion helicase (CI) or GFP. U6 was used as a loading control. C. NSs transcript accumulation; 18S ribosomal RNA (rRNA) was used as a loading control.
Virus-derived sRNA accumulation
Viruses are inducers, targets, and suppressors of RNA silencing (Ding and Voinnet, 2007). Induction results in the formation of sRNAs of 21 to 24 nucleotides (nt) derived from viral RNA by Dicer-like proteins. Targeting is mediated by Argonaute proteins associated with virus-derived small interfering RNAs (sRNAs) and causes the degradation of viral RNA after slicing or translational repression (Ding and Voinnet, 2007; Carbonell et al., 2012; Carbonell and Carrington, 2015; Garcia-Ruiz et al., 2015). To gain insight into the mechanistic activity of NSs, we co-infiltrated it with TuMV-AS9-GFP and measured the accumulation of virus-derived sRNAs (Figure 6). sRNAs derived from the two areas of the TuMV-AS9-GFP genome examined (CI and GFP) were predominantly 21-nt long (Figure 6C). Despite greater viral presence (Figure 6A), and as observed for P19, NSs caused a reduction in the levels of virus-derived sRNA with respect to GUS treatment or leaves infiltrated with virus alone (Figure 6C).
Virus-induced GFP silencing
Accumulation of GFP-derived sRNAs in the presence of NSs (Figure 6C) offered an opportunity to measure the effect of this suppressor on the activity and movement of the silencing signal. Even in the presence of NSs, mechanical inoculation with TuMV-AS9-GFP induced systemic silencing of the N. benthamiana 16c GFP transgene (Figure 7), suggesting movement of the antiviral RNA silencing signal, TuMV-AS9-GFP, or both. Despite clear differences in local infection (Figure 7A), plants infiltrated with TuMV-AS9-GFP alone or with GUS displayed systemic GFP silencing at the same time point as those treated with NSs (Figure 7B). TuMV-AS9-GFP does not form local infection foci in the absence of a silencing suppressor (Figures 7A and 5C). Thus, induction of GFP silencing in non-inoculated leaves suggests systemic movement of the silencing signal.
Figure 7.
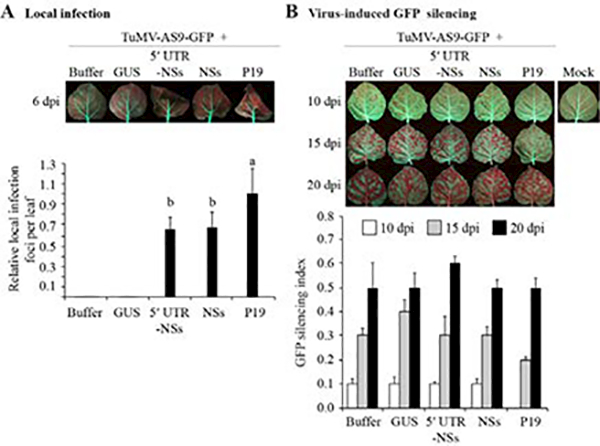
Virus-induced green fluorescent protein (GFP) silencing after mechanical inoculation of Turnip mosaic virus (TuMV)-AS9-GFP 1 day after infiltration of NSs or controls in Nicotiana benthamiana 16c plants. Photographs were taken and infection efficiency is expressed as in Figure 5. GUS = beta-glucuronidase; UTR = untranslated region. A. Local infection. Green spots are TuMV-AS9-GFP local infection foci. The histogram shows average number of local infection foci per leaf ± standard error, relative to P19 (14 ± 3.5). B. Virus-induced silencing of the GFP transgene in 16c plants. Photographs were taken under ultraviolet light at 10, 15, and 20 days post inoculation (dpi). Red areas indicate silencing of the GFP transgene. The histogram shows average GFP silencing index values ± standard error from eight plants.
Plants infiltrated with P19 showed a delay in the onset of systemic GFP silencing, which correlated with a reduction in the accumulation of GFP-derived sRNAs (Figure 6C). P19 is a strong silencing suppressor that binds sRNAs (Omarov et al., 2007). The delay observed suggests systemic movement of TuMV-AS9-GFP, followed by silencing of both viral RNA and the GFP transgene.
DISCUSSION
In plant-virus interactions, silencing suppressors are pathogenicity determinants involved in symptom development (Ding and Voinnet, 2007; Garcia-Ruiz et al., 2010; Garcia and Pallás, 2015), while avirulence factors trigger a hypersensitive response (de Ronde et al., 2014). TSWV NSs is both a silencing suppressor and an avirulence factor (de Ronde et al., 2014). Consistent with these models, variations in symptom development, differences in virus accumulation and clearance, and the emergence of TSWV isolates capable of overcoming resistance in pepper plants have been attributed to NSs mutations (de Ronde et al., 2013; de Ronde et al., 2014). Furthermore, NSs silencing suppression is necessary for persistent infection and transmission of TSWV by F. occidentalis (Whitfield et al., 2005; Margaria et al., 2014). Collectively, these observations imply that pathogenic differences in TSWV isolates, and possibly other tospoviruses, are likely caused by NSs genetic variation. The assays described here constitute valuable experimental tools for the characterization of NSs variants.
Consistent with previous descriptions of this protein from several tospoviruses (Takeda et al., 2002; Bucher et al., 2003; de Ronde et al., 2014; Margaria et al., 2014; Zhai et al., 2014; Hedil et al., 2015), NSs prevented local transgene silencing in the present study (Figure 4). However, the suppression of antiviral RNA silencing by NSs had not been demonstrated before. Using two independent assays, we showed that NSs rescued the pathogenicity of suppressor-deficient TCV-GFP and TuMV-AS9-GFP, resulting in the establishment of local infection (Figure 5). Accordingly, NSs suppresses antiviral RNA silencing directed against these viruses.
Transgene and antiviral silencing suppression assays showed NSs activity to be 30 to 70% of that of P19 (Figures 4 and 5). TCV-GFP and TuMV-AS9-GFP local infection foci were less abundant with NSs than P19 (Figure 5). In addition, TCV-GFP infection foci were smaller following co-infiltration with NSs than those formed in the presence of P19 (Figure 5A), and lower levels of TuMV-AS9-GFP accumulated in plants treated with NSs constructs than in those in which P19 was expressed (Figure 5B). Several models might explain this difference in the magnitude of silencing suppression. For instance, NSs may be less stable and accumulate to lower levels than P19, or could require viral or cellular co-factors not present in transient expression assays. A combination of such factors could also be responsible. An epitope-tagged NSs is currently being used to test these possibilities.
Antiviral RNA silencing is characterized by the production of virus-derived sRNAs that mediate targeting of viral transcripts (Ding and Voinnet, 2007; Carbonell et al., 2012; Carbonell and Carrington, 2015; Garcia-Ruiz et al., 2015). Leaves co-infiltrated with TuMV-AS9-GFP and NSs accumulated virus-derived sRNAs (Figure 6C), showing that this silencing suppressor does not prevent their formation. However, relative to the amount of virus present, NSs caused a reduction in the accumulation of TuMV-derived sRNAs. These observations suggest that NSs interferes with the amplification of the antiviral silencing response.
TuMV-AS9-GFP did not form local infection foci in the absence of a silencing suppressor (Figure 7A), but did induce systemic GFP transgene silencing in N. benthamiana 16c plants at the same rate in the presence or absence of NSs (Figure 7B). This suggests that NSs did not interfere with the systemic movement of the virus-induced GFP silencing signal. However, we cannot exclude the possibility that TuMV-AS9-GFP moved out of inoculated leaves in the presence of NSs or P19.
Several models have been proposed to explain RNA silencing suppression based on biochemical analyses and domain predictions of tospoviral NSs proteins. The presence of a WG motif (Figure 3B; de Ronde et al., 2014) suggests that NSs interacts with Argonaute proteins to trigger their degradation or inhibit their activity (Schnettler et al., 2010; Eifan et al., 2013; de Ronde et al., 2014). The Tomato ringspot virus coat protein contains a GW/ WG motif that mediates interaction with and triggers degradation of ARGONAUTE1 (AGO1; Karran and Sanfaçon, 2014). TCV capsid protein P38 also harbors two GW/WG motifs that facilitate association with AGO1, preventing loading of endogenous and virus-derived sRNAs (Azevedo et al., 2010). The existence of three RNA binding domains (Figure 3B; de Ronde et al., 2014) and the results of in vitro assays suggest that NSs binds long cellular and viral double-stranded RNAs (dsRNAs), such as microRNA (miRNA) precursors and viral replication intermediates (Schnettler et al., 2010; Eifan et al., 2013; de Ronde et al., 2014). These same domains may be involved in binding cellular miRNAs and virus-derived sRNAs (Schnettler et al., 2010; Eifan et al., 2013; de Ronde et al., 2014). Experimental evidence to support NSs-mediated degradation or inhibition of Argonaute proteins, binding of virus-derived sRNAs, association with miRNAs or their precursors, or interaction with viral dsRNA in vivo has not yet been produced. Similarly, the role of NSs silencing suppression in virus infection and movement has not been determined. An infectious TSWV clone is not available, limiting the experimental approaches by which these processes might be examined.
The NSs clones and assays described here establish a foundation to determine the mechanistic activity of NSs. TCV (Qu et al., 2008) and TuMV (Garcia-Ruiz et al., 2010) replicate in Arabidopsis thaliana, mutants of which lacking key RNA silencing components have been characterized (Qu et al., 2008; Garcia-Ruiz et al., 2010; Wang et al., 2010; Garcia-Ruiz et al., 2015). A genetically tractable system based on chimeric TuMV and Arabidopsis thaliana is being developed by our group to determine the mechanism by which NSs suppresses antiviral RNA silencing. The genetic features of this system will allow us to ascertain the contribution of NSs antiviral silencing suppression to the establishment of virus infection and movement and the development of symptoms. Similarly, it will be possible to identify plant genes that repress TSWV infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Thomas L. German for providing TSWV and the seed of several hosts, Feng Qu for donating TCV-GFP, and S. Tatineni for contributing pPZP-35s-GFP. This research was supported by the University of Nebraska-Lincoln Office of Research and Economic Development, the Institute for Agriculture and Natural Resources, and a First Award from the Nebraska-EPSCoR. Stella Uiterwaal and Aaron Knapp were supported by the Undergraduate Creative Activities Research Experience program. Tania Ocampo Ocampo was the recipient of a graduate student fellowship from CONACYT (#2014-MZ02015). We thank Stacie Skaff, Dalton Hegeholz, and Gwennan Plouzek for technical assistance.
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
REFERENCES
- Azevedo J, Garcia D, Pontier D, Ohnesorge S, et al. (2010). Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 24: 904–915. 10.1101/gad.1908710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Visser PB, Angenent GC and Krens FA (2004). The potential of virus-induced gene silencing for speeding up functional characterization of plant genes. Genet. Mol. Res 3: 323–341. [PubMed] [Google Scholar]
- Bucher E, Sijen T, De Haan P, Goldbach R, et al. (2003). Negative-strand tospoviruses and tenuiviruses carry a gene for a suppressor of gene silencing at analogous genomic positions. J. Virol 77: 1329–1336. 10.1128/JVI.77.2.1329-1336.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A and Carrington JC (2015). Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol 27: 111–117. 10.1016/j.pbi.2015.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A, Fahlgren N, Garcia-Ruiz H, Gilbert KB, et al. (2012). Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 24: 3613–3629. 10.1105/tpc.112.099945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD and Grossniklaus U (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. 10.1104/pp.103.027979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Avila AC, Huguenot C, Resende RdeO, Kitajima EW, et al. (1990). Serological differentiation of 20 isolates of tomato spotted wilt virus. J. Gen. Virol 71: 2801–2807. 10.1099/0022-1317-71-12-2801 [DOI] [PubMed] [Google Scholar]
- de la Torre-Almaráz R, Cervantes-Díaz L, Houston HA and Valverde R (2002). Variación fenotípica de algunos aislamientos mexicanos del virus marchitez manchada del tomate (TSWV). Agrociencia 36: 211–221. [Google Scholar]
- de Ronde D, Butterbach P, Lohuis D, Hedil M, et al. (2013). Tsw gene-based resistance is triggered by a functional RNA silencing suppressor protein of the Tomato spotted wilt virus. Mol. Plant Pathol 14: 405–415. 10.1111/mpp.12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ronde D, Pasquier A, Ying S, Butterbach P, et al. (2014). Analysis of Tomato spotted wilt virus NSs protein indicates the importance of the N-terminal domain for avirulence and RNA silencing suppression. Mol. Plant Pathol. 15: 185–195. 10.1111/mpp.12082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW and Voinnet O (2007). Antiviral immunity directed by small RNAs. Cell 130: 413–426. 10.1016/j.cell.2007.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eifan S, Schnettler E, Dietrich I, Kohl A, et al. (2013). Non-structural proteins of arthropod-borne bunyaviruses: roles and functions. Viruses 5: 2447–2468. 10.3390/v5102447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García JA and Pallás V (2015). Viral factors involved in plant pathogenesis. Curr. Opin. Virol 11: 21–30. 10.1016/j.coviro.2015.01.001 [DOI] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, et al. (2010). Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 22: 481–496. 10.1105/tpc.109.073056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Carbonell A, Hoyer JS, Fahlgren N, et al. (2015). Roles and programming of Arabidopsis ARGONAUTE proteins during Turnip mosaic virus infection. PLoS Pathog. 11: e1004755 10.1371/journal.ppat.1004755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedil M, Hassani-Mehraban A, Lohuis D and Kormelink R (2014). Analysis of the A-U rich hairpin from the intergenic region of tospovirus S RNA as target and inducer of RNA silencing. PLoS One 9: e106027 10.1371/journal.pone.0106027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedil M, Sterken MG, de Ronde D, Lohuis D, et al. (2015). Analysis of tospovirus NSs proteins in suppression of systemic silencing. PLoS One 10: e0134517 10.1371/journal.pone.0134517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen LK and Carrington JC (2001). Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 126: 930–938. 10.1104/pp.126.3.930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran RA and Sanfaçon H (2014). Tomato ringspot virus coat protein binds to ARGONAUTE 1 and suppresses the translation repression of a reporter gene. Mol. Plant Microbe Interact. 27: 933–943. 10.1094/MPMI-04-14-0099-R [DOI] [PubMed] [Google Scholar]
- Lane LC (1986). Propagation and purification of RNA plant viruses. Methods Enzymol. 118: 687–696. 10.1016/0076-6879(86)18109-2 [DOI] [Google Scholar]
- Law MD and Moyer JW (1990). A tomato spotted wilt-like virus with a serologically distinct N protein. J. Gen. Virol. 71: 933–938. 10.1099/0022-1317-71-4-933 [DOI] [Google Scholar]
- Mandal B, Csinos AS, Martinez-Ochoa N and Pappu HR (2008). A rapid and efficient inoculation method for Tomato spotted wilt tospovirus. J. Virol. Methods 149: 195–198. http://dx.doi.Org/10.1016/j.jviromet.2007.12.007 [DOI] [PubMed] [Google Scholar]
- Margaria P and Rosa C (2015). First complete genome sequence of a tomato spotted wilt virus isolate from the United States and its relationship to other TSWV isolates of different geographic origin. Arch. Virol 160: 2915–2920. 10.1007/s00705-015-2589-6 [DOI] [PubMed] [Google Scholar]
- Margaria P, Bosco L, Vallino M, Ciuffo M, et al. (2014). The NSs protein of tomato spotted wilt virus is required for persistent infection and transmission by Frankliniella Occidentalis.J. Virol. 88: 5788–5802. 10.1128/JVI.00079-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaria P, Ciuffo M, Rosa C and Turina M (2015a). Evidence of a tomato spotted wilt virus resistance-breaking strain originated through natural reassortment between two evolutionary-distinct isolates. Virus Res. 196: 157–161. 10.1016/j.virusres.2014.11.012 [DOI] [PubMed] [Google Scholar]
- Margaria P, Miozzi L, Rosa C, Axtell MJ, et al. (2015b). Small RNA profiles of wild-type and silencing suppressor-deficient tomato spotted wilt virus infected Nicotiana benthamiana. Virus Res. 208: 30–38. 10.1016/j.virusres.2015.05.021 [DOI] [PubMed] [Google Scholar]
- Omarov RT, Ciomperlik JJ and Scholthof HB (2007). RNAi-associated ssRNA-specific ribonucleases in Tombusvirus P19 mutant-infected plants and evidence for a discrete siRNA-containing effector complex. Proc. Natl. Acad. Sci. USA 104: 1714–1719. 10.1073/pnas.0608117104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu HR, Jones RA and Jain RK (2009). Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res. 141: 219–236. 10.1016/j.virusres.2009.01.009 [DOI] [PubMed] [Google Scholar]
- Parrella G, Gognalons P, Gebre-Selassie K, Vovlas C, et al. (2003). An update of the host range of Tomato spotted wilt virus. J. Plant Pathol. 85: 227–264. [Google Scholar]
- Powers JG, Sit TL, Qu F, Morris TJ, et al. (2008). A versatile assay for the identification of RNA silencing suppressors based on complementation of viral movement. Mol. Plant Microbe Interact. 21: 879–890. 10.1094/MPMI-21-7-0879 [DOI] [PubMed] [Google Scholar]
- Qu F, Ye X and Morris TJ (2008). Arabidopsis DRB4, AGO1, AG07, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. USA 105: 14732–14737. 10.1073/pnas.0805760105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnettler E, Hemmes H, Huismann R, Goldbach R, et al. (2010). Diverging affinity of tospovirus RNA silencing suppressor proteins, NSs, for various RNA duplex molecules. J. Virol. 84: 11542–11554. 10.1128/JVI.00595-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof KB, Adkins S, Czosnek H, Palukaitis P, et al. (2011). Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12: 938–954. 10.1111/j.1364-3703.2011.00752.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Sugiyama K, Nagano H, Mori M, et al. (2002). Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEES Lett. 532: 75–79. 10.1016/S0014-5793(02)3632-3 [DOI] [PubMed] [Google Scholar]
- Tatineni S, Qu F, Li R, Morris TJ, et al. (2012). Triticum mosaic poacevirus enlists P1 rather than HC-Pro to suppress RNA silencing-mediated host defense. Virology 433: 104–115. 10.1016/j.virol.2012.07.016 [DOI] [PubMed] [Google Scholar]
- Uga H and Tsuda S (2005). A one-step reverse transcription-polymerase chain reaction system for the simultaneous detection and identification of multiple tospovirus infections. Phytopathology 95: 166–171. 10.1094/PHYTO-95-0166 [DOI] [PubMed] [Google Scholar]
- Wang XB, Wu Q, Ito T, Cillo F, et al. (2010). RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 484–489. 10.1073/pnas.0904086107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield AE, Ullman DE and German TL (2005). Tospovirus-thrips interactions. Annu. Rev. Phytopathol. 43: 459–489. 10.1146/annurev.phyto.43.040204.140017 [DOI] [PubMed] [Google Scholar]
- Yang SJ, Carter SA, Cole AB, Cheng NH, et al. (2004). A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 101: 6297–6302. 10.1073/pnas.0304346101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Bag S, Mitter N, Turina M, et al. (2014). Mutational analysis of two highly conserved motifs in the silencing suppressor encoded by tomato spotted wilt virus (genus Tospovirus, family Bunyaviridae). Arch. Virol 159: 1499–1504. 10.1007/s00705-013-1928-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


