SUMMARY
Viral envelope proteins are required for productive viral entry and initiation of infection. Although the humoral immune system provides ample evidence for targeting envelope proteins as an antiviral strategy, there are few pharmacological interventions that have this mode of action. In contrast to classical antiviral targets such as viral proteases and polymerases, viral envelope proteins as a class do not have a well-conserved active site that can be rationally targeted with small molecules. We previously identified compounds that inhibit dengue virus by binding to its envelope protein, E. Here, we show that these small molecules inhibit dengue virus fusion and map the binding site of these compounds to a specific pocket on E. We further demonstrate inhibition of Zika, West Nile, and Japanese encephalitis viruses by these compounds, providing pharmacological evidence for the pocket as a target for developing broad-spectrum antivirals against multiple, mosquito-borne flavivirus pathogens.
Keywords: antiviral, inhibitor of viral entry, E inhibitor, flavivirus inhibitor
eTOC blurb
Countermeasures against dengue, Zika, and other flaviviruses are a large, unmet medical need. de Wispelaere et al validate a conserved pocket of the flavivirus envelope protein as a target for small molecule antivirals with broad-spectrum activity against flaviviruses.
INTRODUCTION
Dengue virus (DENV) is a mosquito-borne virus from the genus Flavivirus, which includes several other human pathogens including yellow fever, West Nile, Japanese encephalitis, and Zika viruses (YFV, WNV, JEV, ZIKV, respectively). Over 300 million dengue infections occur annually (Bhatt, et al., 2013), resulting in disease that can escalate to dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS). We currently lack a broadly-protective vaccine or specific antiviral treatment to combat DENV, ZIKV, and many other flavivirus pathogens. Geographic spread of the Aedes mosquito species that transmits DENV and ZIKV and ZIKV’s recent explosive emergence in the Western Hemisphere have heightened the need for countermeasures that can reduce transmission and therapeutic interventions that can prevent severe disease resulting from infection. Due to the lack of success to date of efforts aimed at producing antivirals against the viral polymerase and protease, alternative antiviral strategies to inhibit infection are of growing relevance.
Small molecules that target the envelope protein on the surface of virions have the potential to engage their target extracellularly and to block the viral replication cycle at its earliest step. Although this antiviral strategy is well-validated by the humoral immune response to many viruses, there are few antivirals that act by this mode of action. A major barrier in developing antivirals targeting envelope proteins is the absence of a conserved active site analogous to those of viral proteases and polymerases. For DENV, the viral envelope protein, E, exists as 90 prefusion dimers on the surface of mature virions. Viral entry is initiated by engagement of E with attachment factors on the plasma membrane of the host cell, followed by uptake of the virion by a clathrin-dependent process (Acosta, et al., 2008; Krishnan, et al., 2007; van der Schaar, et al., 2008). Acidification of the endosomal compartment triggers significant structural changes leading to reorganization and refolding of E as a postfusion trimer (Allison, et al., 1995; Bressanelli, et al., 2004; Modis, et al., 2004). This structural transformation induces fusion of the viral and endosomal membranes and creates a pore that allows escape of the viral nucleocapsid into the host cytosol where the viral RNA genome can be expressed.
Over a decade ago, crystallization of the DENV serotype 2 (DENV2) E protein in the presence of millimolar concentrations of the detergent beta-octylglucoside (βOG) enabled Modis and colleagues to detect binding of a detergent molecule in a pocket between domains I and II (Modis, et al., 2003). The location of the detergent-binding site (the “βOG pocket”) at a hinge region for the major conformational changes that occur during E-mediated membrane fusion led to the proposal that small molecules that bind at this site might inhibit dengue virus entry. While multiple small molecules that inhibit dengue virus entry by binding E and/or by preventing fusion have been reported (Clark, et al., 2016; Kampmann, et al., 2009; Poh, et al., 2009; Schmidt, et al., 2012; Wang, et al., 2009; Zhou, et al., 2008), the structural basis for their inhibitory activities has not been determined. Furthermore, although residues lining this pocket appear to conserved across other flaviviruses, neither inhibitors of other flavivirus E proteins nor co-crystal structures of other flavivirus E proteins with βOG or other ligands have been reported. Validation of the “βOG pocket” as a bona fide pharmacological target mediating the inhibition of dengue virus and other flaviviruses has thus remained tantalizing but elusive.
We report here on 2,4-diamino pyrimidines and 4,6-disubstituted pyrimidines that target the prefusion form of the DENV E protein (DENV E) and block viral entry by inhibiting membrane fusion. A resistance mutation, E-M196V, located adjacent to the βOG pocket reduces small molecule-binding affinity and restores virus entry and infectivity in the presence of both types of pyrimidines, suggesting that this pocket may be the molecular target of these compounds. We confirmed this hypothesis by demonstrating crosslinking of a photoactivatable inhibitor to a region of E located in the βOG pocket and showing that other substitutions in the βOG-pocket introduced by site-directed mutagenesis affect inhibitor binding to recombinant, soluble E. Comparison of the effects of site-directed mutations on compounds representative of the two different pyrimidine inhibitors series as well as a previously reported cyanohydrazone inhibitor of DENV E suggests differences in how these inhibitors engage the βOG pocket. Consistent with conservation of residues in the βOG pocket, we demonstrate inhibition of ZIKV, WNV, and JEV by both pyrimidine and cyanohydrazone inhibitors with evidence for both DENV-specific antiviral activity as well as broad-spectrum activity against multiple flaviviruses. These findings establish the βOG pocket as a bona fide target for pharmacological inhibition of flavivirus entry and provide a foundation for defining structure-activity relationships for developing antivirals that target E.
RESULTS
Disubstituted pyrimidines inhibitors of DENV inhibit E-catalyzed membrane fusion
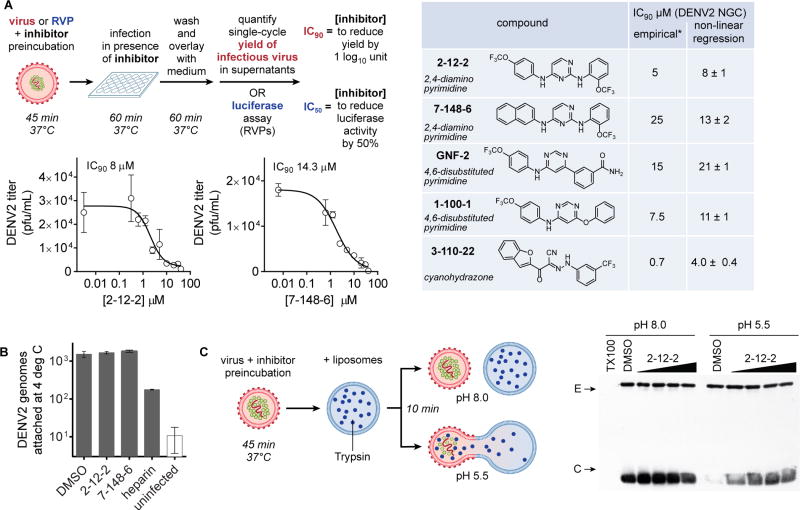
We recently reported the discovery and characterization of 2,4-diamino and 4,6-disubstituted pyrimidines that inhibit DENV entry by binding directly to the prefusion form of E (Clark, et al., 2016). For our current studies, we focused on representative compounds from each of these classes (Figure 1A). We additionally included the cyanohydrazone 3-110-22 as a control since we had previously shown it to inhibit dengue virus fusion although its binding site on E remains unknown (Schmidt, et al., 2012). To investigate how these compounds affect E’s function during viral entry, we first quantified antiviral activity against the New Guinea C strain of dengue virus serotype 2 (DENV2 NGC) for representative compounds using a previously established viral infectivity assay (Clark, et al., 2016) (Figures 1A and S1). We then determined how E’s functions in virion attachment and membrane fusion are affected at concentrations of the compounds sufficient to reduce infectivity by ten-fold (IC90). Since DENV virions attach to the plasma membrane but their clathrin-mediated uptake is inhibited at 4 °C (van der Schaar, et al., 2007), we quantified the number of viral genomes attached to cells at this temperature and found it to be unaffected by the inhibitors (Figure 1B). To determine how the inhibitors might affect E’s catalysis of membrane fusion, we monitored the low pH-triggered fusion of DENV virions with trypsin-loaded liposomes using digestion of the viral core protein, located on the interior of the viral lipid bilayer, as a marker for successful creation of a fusion pore (Figure 1C). Following incubation of DENV2 virions with liposomes, we detected pH-dependent digestion of core that was blocked in the presence of compounds 2-12-2, 7-148-6, and other pyrimidines (Figures 1C and S2). Protection of the viral capsid is concentration-dependent and correlates well with the inhibitor concentrations at which antiviral activity is observed, thus providing strong evidence that these compounds inhibit DENV by interfering with E’s function in the fusion step of viral entry.
Figure 1. Effects of inhibitors on the attachment and fusion steps of dengue virus (DENV) entry.
(A) Infectivity assays using live virus or single-cycle reporter virus were designed to assess inhibition of viral entry when compound treatment is limited to a preincubation with the inoculum and the initial one hour infection, after which non-adsorbed virus and compound were washed away. Productive viral entry was assessed by quantifying the single-cycle yield of infectious virus produced. IC90 values -- defined as the concentration of compound required to reduce single-cycle viral yield by 10-fold -- were previously estimated empirically (marked by “*”) (Schmidt et al., 2012; Clark, et al., 2016) and were determined here by titration and non-linear regression analysis. Values in the table are the average and standard deviation from n ≥ 2 independent experiments. Graphs are representative data from one of n ≥ 2 independent experiments. Graphs for 2,4-diamino pyrimidines 2-12-2 and 7-148-6 are shown here; graphs showing representative data for GNF2 and 1-100-1 are provided in Figure S1. (B) Effects of compounds on the attachment step of DENV2 entry were assessed by incubation of cells with virus at 4° C in the presence of representative inhibitors followed by washes to remove non-specifically bound material and RT-qPCR analysis of isolated RNA to quantify the number of attached DENV2 genomes. Heparin was used as a positive control. (C) Fusion of DENV2 virions with synthetic liposomes encapsulating trypsin is triggered by low pH. Formation of a fusion pore allows trypsin (blue dots) to access and digest core protein (green) from the interior of the virion while envelope protein (E, red ovals) on the exterior of the virion remains intact. Western blot analysis of the reactions for C and E shows that compound 2-12-2 protects core from digestion upon exposure of the virion-liposome mixture to acidic pH, indicating concentration-dependent inhibition of viral fusion. Representative data are shown for n ≥ 2 independent experiments. Final concentrations of 2-12-2 were 5, 15, 25, and 40 μM. Analogous data for other compounds are provided in Figure S2.
A mutation adjacent to the βOG-binding pocket confers antiviral resistance
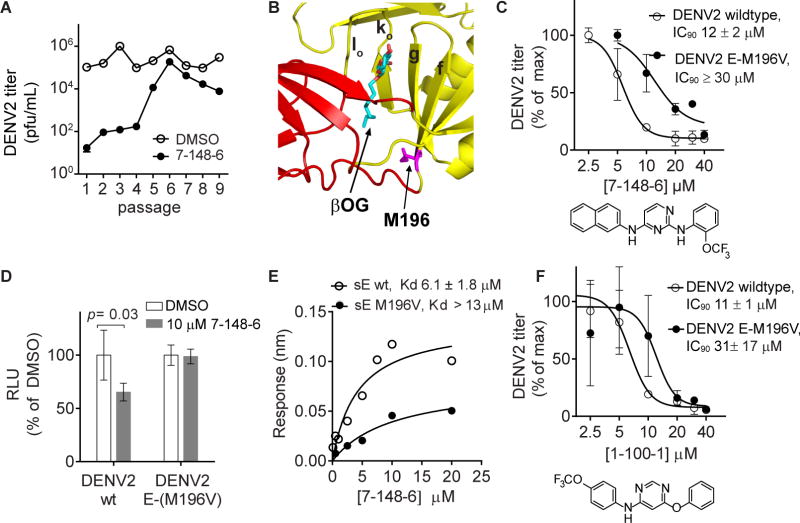
To understand how these compounds interact with DENV E to inhibit its fusogenic activity, we serially passaged DENV2 NGC in the presence of different inhibitors to select for resistance. While in most cases no resistance was observed after > 10 passages (data not shown), we did observe emergence of a resistant quasispecies after six passages in the presence of the 2,4-diamino pyrimidine 7-148-6 (Figure 2A). Sequencing of the quasispecies at passage 7 revealed an A to G mutation resulting in substitution of valine for methionine at residue 196 of E. This residue lies adjacent to the potential ligand-binding site occupied by the β-octyl-glucoside (βOG) detergent molecule (PDB 1OKE) in the DENV2 soluble, prefusion E dimer crystal structure (Modis, et al., 2003) (Figure 2B).
Figure 2. Selection of a mutation conferring resistance to the 2,4-diamino pyrimidine 7-148-6.
(A) DENV2 NGC was serially passaged in the presence of 7-148-6 or vehicle control (“DMSO”) and titered by viral plaque formation assay after each passage. A resistant quasispeices was observed by passage 6. Sequencing of the resistant and vehicle control quasispecies identified a mutation leading to substitution of valine for methionine at position 196 of the viral E protein. (B) Structure of the DENV2 soluble E prefusion dimer structure (PDB 1OKE) with domains I and II highlighted in red and yellow. The E-M196V substitution at the base of beta strand f, highlighted in magenta, is located adjacent to the molecule of beta-octoglucoside (βOG, cyan). (C) IC90 values against DENV2 NGC and the DENV2 NGC E-M196V mutant virus were measured as described in Fig. 1A. Single-cycle viral yields were normalized to the titers of the DMSO-treated controls. The IC90 of 7-148-6 against DENV2 NGC is 13 ± 2 μM (average of 4 independent experiments; representative data shown). Although 7-148-6’s solubility under the conditions of the antiviral assays limited our ability to saturate inhibition of the E-M196V mutant in two of three independent experiments, we estimated the lower bound of the IC90 value to be 30 μM; representative data for the experiment in which we were able to saturate inhibition are shown. Analogous data for 2-12-2 are presented in Figure S3. (D) Recombinant, single-cycle reporter viruses for DENV2 wildtype and the E-M196V mutant were generated and used in the infectivity assay as outlined in Fig. 1A to confirm that the E-M196V mutation confers resistance to 7-148-6. Luminescence readings were normalized to the DMSO-treated controls. Representative data are shown for n ≥ 2 independent experiments. (E) Equilibrium dissociation constants for the interaction of wildtype recombinant, soluble DENV2 E protein (DENV2 sE) or the E-M196V mutant with 7-148-6 were measured by biolayer interferometry. KD values in the figure legend represent the average and standard deviation of 2 or more independent experiments, with the graphed data for one such experiment shown. Analogous data for 2-12-2 is presented in Figure S3. (F) IC90 values for 4,6-disubstituted pyrimidine 1-100-1 against DENV2 NGC and the DENV2 NGC E-M196V viruses were measured as described in Fig. 1A. Single-cycle viral yields were normalized to the titers of the DMSO-treated controls. Graph is of data from a representative experiment. IC90 values presented in the figure legend are the average and standard deviation of n ≥ 2 independent experiments. Analogous data for 4,6-disubstituted pyrimidine GNF2 are presented in Figure S4.
Following three rounds of virus plaque purification, we isolated and sequence-verified a DENV2 NGC E-M196V clone and confirmed that this virus has reduced sensitivity to 7-148-6. The IC90 of 7-148-6 against DENV2 NGC is 13 ± 2 μM (average of 4 independent experiments; representative experiment shown in Figure 2C). Although 7-148-6’s solubility under the conditions of the antiviral assays limited our ability to saturate inhibition of the E-M196V mutant in two of three independent experiments, we can estimate the lower bound of the IC90 value to be 30 μM (Figure 2C). The DENV2 E-M196V virus exhibits similar resistance to related 2,4-diamino pyrimidine 2-12-2 (Figure S3). We independently confirmed this by generating DENV2 single-cycle reporter virus particles (RVPs) (Ansarah-Sobrinho et al., 2008) to show that the E-M196V substitution significantly alleviates the inhibitory effects of 7-148-6 on RVP entry (Figure 2D).
We reasoned that the M196V mutation might confer resistance by interfering with binding of 7-148-6 and 2-12-2, by enabling E to mediate fusion despite binding of the inhibitor, and/or by increasing the proportion of non-infectious particles that can act as a “molecular sponge” to decrease inhibitor-binding by infectious virions. To examine the extent to which the mutation affects inhibitor-binding, we expressed soluble, prefusion DENV2 wildtype E and E-M196V proteins (sE and sE-M196V) and used biolayer interferometry to measure equilibrium dissociation constants (KD) for their interaction with 7-148-6 (Figure 2E) and 2-12-2 (Figure S3). The wildtype sE binds to both compounds with KD of 6.1 ± 1.8 μM for 7-148-6 and 4.2 ± 1.9 μM for 2-12-2 whereas sE-M196V has reduced affinity with KD of greater than 13 and 12 μM for 7-148-6 and 2-12-2, respectively. This demonstrates that the mutation affects the interactions of 7-148-6 and 2-12-2 with DENV2 sE through either a direct or an allosteric mechanism. Reduced sensitivity of the DENV2 E-M196V virus to the 4,6-disubstituted pyrimidines 1-100-1 and GNF2 (Figure 2F and S4) further suggests that this mutation may similarly affect the interaction of these inhibitors with E.
Disubstituted pyrimidine inhibitors of DENV E bind in the βOG pocket
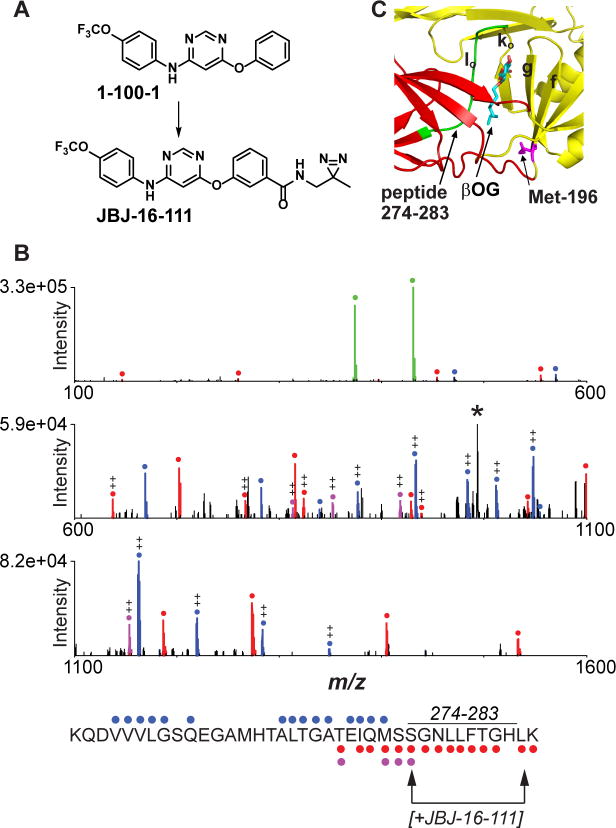
To probe the site of inhibitor-binding more directly, we synthesized JBJ-16-111, an analog of the 4,6-disubstituted pyrimidine 1-100-1 conjugated via linker to a diazirine group suitable for photocrosslinking experiments (Fig. 3A). A recombinant protein comprising DENV2 E domains I and II (DI-DII) was preincubated with JBJ-16-111 and then irradiated with long wave UV light to activate the photocrosslinking group. Following trypsin digestion of the crosslinking reaction, capillary electrophoresis - mass spectrometry (CE-MS) analysis detected crosslinking to a peptide corresponding to residues 247-284. Fragment ion data further suggest that the peptide is labeled between residues S274 and L283, which line one side of the βOG pocket (Fig. 3B-C).
Figure 3. Photocrosslinking of a 4,6-disubstituted inhibitor in the βOG pocket.
(A) Structures of inhibitor JBJ-16-111 and parental compound 1-100-1. (B) HCD MS/MS spectrum of the peptide KQDVVVLGSQEGAMHTALTGATEIQMSSGNLLFTGHLK (residues 247-284) acquired during CE-MS analysis of tryptic peptides from the UV crosslinking reaction between DI,II protein and JBJ-16-111. Unmodified b- and y- type ions are marked with blue and red circles, respectively, while y-ions modified by JBJ-16-111 are highlighted in purple. Abundant probe-related ions at m/z 431.12 and 374.07 (green), along with neutral-loss ion at m/z 993.24 (denoted ‘*’), suggest that JBJ-16-111 is readily lost during MS/MS. Fragment ion evidence suggests that the peptide is labeled between S274 and L283 (see arrows; assumes C-terminal lysine is not modified due to successful trypsin cleavage). ++, doubly charged ion. (C) DENV2 E structure (1OKE) with residues 274-283 highlighted in green.
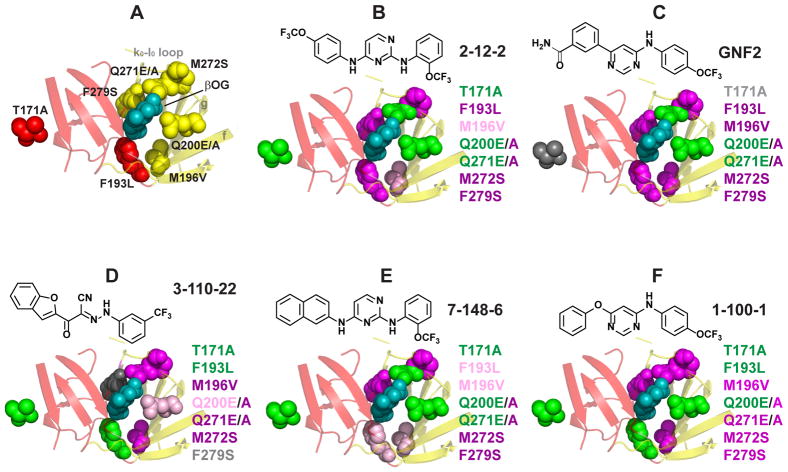
As a complementary approach to probe the interaction of the inhibitors with the βOG pocket, we used site-directed mutagenesis to introduce substitutions in this region of the protein. We successfully generated additional recombinant sE proteins bearing substitutions F193L, Q200A, Q200E, Q271A, Q271E, M272S, and F279S, all of which line the βOG pocket (Fig. 4A). We additionally produced sE-T171A, a substitution that lies distant from the βOG pocket but that may confer DENV2 resistance to 3-110-22, a cyanohydrazone that inhibits DENV fusion by targeting E (Schmidt, 2011; Schmidt, et al., 2012). To assess the impact of these point mutations on small molecule-binding, we measured equilibrium dissociation constants (KD) for compounds representative of the 2,4-diamino pyrimidine (2-12-2 and 7-148-6), the 4,6-disubstituted pyrimidine (GNF-2 and 1-100-1), and the cyanohydrazone (3-110-22) inhibitor series (Table S1).
Figure 4. Loss-of-affinity footprints of representative DENV E inhibitors.
The ligand-binding pocket of the DENV2 E structure (1OKE) is shown with βOG (cyan) bound. (A) Substitutions were introduced at seven different residues lining the pocket as shown. Residues 200 and 271 were mutated from glutamine to glutamate (E) and alanine (A). (B–F) Loss-of-affinity for each mutation is mapped onto the 1OKE structure for cyanohydrazone 3-110-22, 2,4-diaminopyrimidines 2-12-2 and 7-148-6, and 4,6-disubstituted pyrimidines GNF2 and 1-100-1. Magenta, > 5-fold loss of affinity; light pink, 3- to 5-fold loss of affinity; green, < 2-fold loss of affinity; gray, data indeterminate due to poor signal-to-background. Color-coding at residue 271 reflect the effects of the Q271E mutation because the Q271A mutant exhibited significant loss of binding with all inhibitors.KD values are presented in Table S1.
The T171A substitution had no effect on binding of all three structural classes of inhibitors, consistent with the idea that if this mutation confers resistance, it does so via a mechanism that does not affect inhibitor-binding. The other mutations had variable effects on binding of inhibitors to E. To visualize a mutational “footprint” for each inhibitor, we compared equilibrium dissociation constants for binding of each compound to wildtype and mutant proteins and mapped decreases in affinity onto the DENV2 sE – βOG co-crystal structure (PDB 1OKE) (Fig. 4B–F). Within this limited analysis, compounds in the same inhibitor class appeared to have footprints more similar to each other than to those of the other two structural classes. For example, 2,4-diaminopyrimidines 2-12-2 and 7-148-6, which differ only in the substituents at the 4-position are only modestly affected by the M196V mutation whereas binding of cyanohydrazone 3-110-22 and 4,6-disubstituted pyrimidines GNF-2 and 1-100-1 are more significantly affected. Similarly, binding of 3-110-22 appears more sensitive to changes made in beta strands f and g and the k0 loop, with M196V, Q200E/A, Q271E/A, and M272S all causing significant loss-of-binding whereas each of the pyrimidines appears to tolerate at least one substitution on this side of the pocket.
As a third example, the Q200A and Q271A substitutions cause significant loss of binding to all of the inhibitors. This could suggest participation of these residues in hydrogen bonds that are important for inhibitor-binding. This idea is consistent with the observation that the Q200E mutant binds to all of the compounds with affinities comparable to wildtype. Likewise, binding of the Q271E mutant to GNF-2, 7-148-6, or 2-12-2 occurs with affinity comparable to wildtype and may reflect a role of hydrogen bonding at this residue that impacts binding of these inhibitors but not 1-100-1 and 3-110-22. Collectively, these data support a model in which all three classes of inhibitors bind in the βOG pocket and suggest that each class makes specific – and distinct – contacts with the pocket. Since the M196V and M272S substitutions appeared to affect all of the inhibitors, we questioned whether they might have more global effects on E’s structure (e.g., dimerization, structural dynamics). This appears unlikely to be the case, since hydrogen-deuterium exchange mass spectrometry of the wildtype sE and sE-M196V and sE-M272S proteins detected no evidence for global structural changes (Data S1).
Targeting the ligand-binding pocket between domains I and II as a general strategy for inhibiting flaviviruses
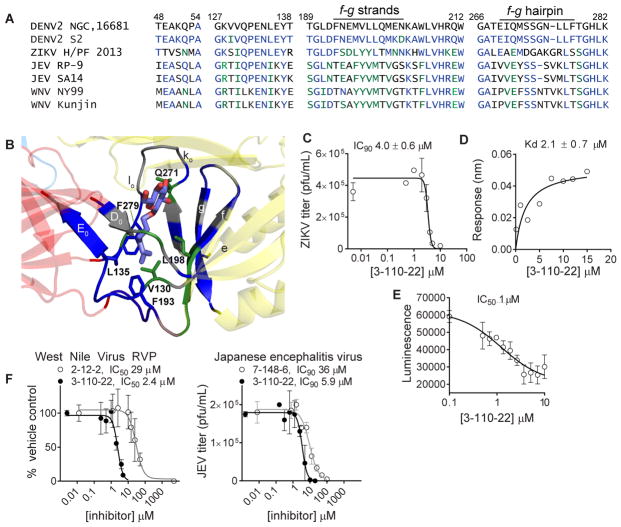
Although all characterized flavivirus E proteins share common domain structures and organization, small molecule inhibitors of other flavivirus E proteins have not been reported. Alignment of residues proximal to βOG in the DENV2 sE structure (1OKE) with the analogous residues of Zika, West Nile, and Japanese encephalitis viruses (Figure 5A) showed significant conservation. For example, the majority of residues that pack around the hydrocarbon tail of βOG deep in the pocket are identical (L135, F193, F279) or have conservative substitutions (V130, L198) in DENV2 and ZIKV E proteins while residues that interact with the glucoside at the top of the pocket are divergent, especially those in the fg beta-strands and the kl hairpin (Figure 5A and 5B). JEV and WNV E proteins exhibit similar patterns of conservation with DENV and ZIKV at the bottom of the pocket adjacent to the viral membrane and divergence in solvent-exposed residues at the top of the pocket (Figure 5A). Based on this analysis, we questioned whether the DENV E inhibitors might also inhibit other flaviviruses.
Figure 5. Pharmacological validation of the βOG pocket as a target for inhibition of Zika virus and other flaviviruses.
(A) Sequence alignment of the residues proximal to βOG in the high resolution structure of DENV2 S2 sE co-crystallized with βOG (PDB 1OKE and 1OAN). DENV2 NGC is used as a reference sequence. Blue indicates residues that are identical; green residues are conservative substitutions; gray residues are non-conserved substitutions. (B) The βOG-binding pocket of DENV2 sE showing βOG-proximal residues that are identical to ZIKV E (blue), conserved with ZIKV E (green), or non-conserved (gray). (C) 3-110-22 inhibits ZIKV PF13-251013-18 in the viral infectivity assay (Figure 1A) with IC90 value of 3.8 ± 0.8 μM. (D) 3-110-22 binds to recombinant ZIKV sE with KD 2.1 ± 0.7 μM. (E) 3-110-22 disrupts the interaction of ZIKV sE with GNF2-biotin in a competitive luminescence proximity assay with IC50 of approximately 1 μM. (F) Cyanohydrazone and pyrimidine inhibitors of DENV2 E also inhibit West Nile reporter virus and Japanese encephalitis virus. Figure S5 presents data for additional compounds that inhibit Zika and Japanese encephalitis viruses. IC90 and KD values presented in the text are the average and standard deviation for n ≥ 2 independent experiments. Graphs show representative data for an independent experiment with error bars reflecting the standard deviation for 2 or more experimental replicates.
To examine this possibility, we tested representative compounds from the 2,4-diamino pyrimidine series (2-12-2), the 4,6-disubstituted pyrimidine series (GNF-2), and the cyanohydrazone series (3-110-22) for activity against ZIKV PF13-251013-18 (Aubry, et al., 2016) using an infectivity assay analogous to that used to measure anti-DENV2 IC90 values as depicted in Figure 1A. Compound 3-110-22 inhibits ZIKV PF13-251013-18 with an IC90 of 4.0 ± 0.6 μM (Figure 5C), which is comparable to its activity against DENV2 NGC. Both 2-12-2 (IC90 46 μM) and GNF-2 (IC90 107 μM) have more modest effects on ZIKV PF13-251013 (Figure S5). Consistent with its anti-ZIKV activity, 3-110-22 interacts with recombinant, soluble ZIKV E (ZIKV sE) in biolayer interferometry experiments with a KD of 2.1 ± 0.7 μM (Figure 5D). Since 3-110-22’s anti-DENV2 activity is correlated with activity in a competitive luminescence proximity assay using recombinant DENV2 sE (Clark, et al., 2016), we additionally confirmed that 3-110-22’s activity against Zika sE in an analogous competitive luminescence proximity assay has an IC50 value of approximately 1 μM (Figure 5E). We also observed significant inhibition of both JEV and single-cycle reporter WNV (Pierson, et al., 2006) by one or more compounds (Figure 5F and Figure S5), albeit at concentrations generally higher than those required to inhibit authentic DENV2 infection. Together, these observations demonstrate that 3-110-22 and other inhibitors of DENV2 E can inhibit other flavivirus pathogens and suggest that the ligand-binding pocket between domains I and II is an antiviral target conserved across this viral genus.
DISCUSSION
The βOG-binding pocket of the DENV E protein as a pharmacological target mediating antiviral activity
Direct-acting antiviral drugs that target virally encoded enzymes such as viral polymerases or proteases have demonstrated enormous benefit to patients infected with HIV and HCV. While there is precedent for small molecule inhibitors of viral entry, these have more typically targeted host co-receptors (e.g., CXCR5) or the capsid proteins of non-enveloped viruses (e.g., pleconaril). In contrast, there is a paucity of examples of antiviral drugs that target the fusogenic activities of viral glycoproteins. Although biomimetic peptides have been shown to inhibit the entry of diverse enveloped viruses in cell culture by interfering with the conformational changes of the viral glycoprotein required for fusion (Kirschner, et al., 2007; Porotto, et al., 2006; Rapaport, et al., 1995; Schmidt, et al., 2010), inhibition of HIV gp41 by the peptide T20 (Kilby, et al., 1998) remains the only example of an approved drug that acts by this mechanism. Pursuit of small molecules that inhibit the membrane-fusing activity of viral glycoproteins has been limited by the absence of obvious drug-binding pockets on these proteins, and a paucity of target-based assays that can be used to discover and optimize small molecules that inhibit membrane fusion.
The discovery of the detergent βOG bound in a region of the DENV2 prefusion E protein (Modis, et al., 2003) that had previously been implicated in regulating the pH at which viral fusion occurs (Butrapet, et al., 2011; Lee, et al., 1997), suggested that this pocket might present a bona fide pharmacological target on E. Over the past decade, numerous virtual screening efforts (Kampmann, et al., 2009; Poh, et al., 2009; Wang, et al., 2009; Yennamalli, et al., 2009; Zhou, et al., 2008) have produced compounds that inhibit DENV entry, but clear evidence for the target site(s) of these compounds on E in the form of high resolution structural data, biochemical crosslinking experiments, or analysis of antiviral resistance has been elusive. To date, the best evidence for binding in the βOG pocket comes from work by Zhou and colleagues, who monitored the 1H spectrum of βOG in the presence of DENV E by saturation transfer difference NMR spectroscopy (Zhou, et al., 2008). They observed loss of specific resonances upon the addition of inhibitor P02, suggesting displacement of βOG by the inhibitor, but whether P02 directly or allosterically affects binding of βOG to E has remained unclear. We used photocrosslinking experiments to show that JBJ-16-111 interacts with residues of E contained within tryptic peptide 247-284 and that the likely site of crosslinking is within residues 274-283. These residues include the lo region of the protein immediately adjacent to the βOG molecule in the 1OKE structure. Loss of affinity caused by mutation of residues in the pocket was also observed for compounds from all three inhibitor series. Although it remains a formal possibility that the inhibitors bind elsewhere on E and are allosterically affected by the mutations, this seems unlikely since an allosteric mechanism would be expected to affect all compounds equivalently. The distinct differences in how mutations in the pocket affect each of the inhibitors are instead more consistent with a model in which the 2,4-diamino pyrimidine, 4,6-disubstituted pyrimidine, and cyanohydrazone inhibitors form specific and distinct interactions with the βOG pocket.
Mechanistic implications of the resistance profile for DENV2 E-M196V
While the absence of reported mutations conferring antiviral resistance can be a hindrance in identifying inhibitor target site(s), it also suggests that the natural barrier to resistance may be high for these compounds. Although we attempted to select for resistance against multiple pyrimidines (8 individual compounds, ≥ 10 passages), we obtained a resistant quasispecies only in the experiment that resulted in isolation of DENV2 E-M196V. This suggests that conservation of residues in the pocket is required to maintain E’s function in viral entry. We verified that the E-M196V mutation confers resistance to 7-148-6 and the related 2,4-diamino pyrimidine 2-12-2 and reduces the affinity of the interaction between both compounds and recombinant, soluble DENV2 E (sE). The DENV2 E-M196V mutation was also previously identified as one conferring resistance to multiple neutralizing antibodies targeting DENV2 (Sukupolvi-Petty, et al., 2010), suggesting that the E-M196V substitution perturbs E’s structure and/or conformational dynamics rather than by altering specific epitopes. This could suggest that the reductions in antiviral sensitivity and affinity caused by the E-M196V mutation are due to direct or allosteric effects of the mutation on accessibility of the βOG pocket through gross effects on E structure, dimerization, and conformational dynamics. Arguing against this, however, are the absence of changes in hydrogen-deuterium exchange kinetics we observe for sE-M196V versus wildtype protein (Data S1) and the greater effect of this substitution on binding of the 4,6-disubstituted pyrimidines and cyanohydrazone 3-110-22 (≥ 10-fold loss of affinity) versus the 2,4-diamino pyrimidines (≤ 5-fold loss of affinity). Since methionine and valine have different van der Waals volumes, an alternate hypothesis is that the methionine to valine substitution alters hydrophobic packing around the small molecule inhibitor. Although efforts to acquire high resolution structural data for all three inhibitor series bound to sE have been unsuccessful to date, we note that there have been no co-crystal structures of DENV E with other reported inhibitors (Clark, et al., 2016; Kampmann, et al., 2009; Poh, et al., 2009; Schmidt, et al., 2012; Wang, et al., 2009; Zhou, et al., 2008). A likely confounding factor has been the incompatibility of the conditions under which the protein crystallizes and the hydrophobicity of the small molecules. Our findings are currently driving renewed efforts in this area using inhibitors modified with solubilizing groups along with additional efforts to define resistance profiles for all three of our inhibitor series.
The ligand-binding pocket targeted by DENV E inhibitors is conserved across flaviviruses
The high apparent barrier to antiviral resistance likely reflects the functional importance of the βOG-binding region of E in viral entry and/or in viral assembly. Consistent with this inference, we have found that many of the residues proximal to βOG in the crystal structure could not tolerate substitutions, as we were unable to recover infectious viral particles when we targeted the βOG-binding pocket by alanine scanning mutagenesis (unpublished data). Perhaps even more compelling is the observation of significant conservation of the DENV2 residues proximal to βOG when aligned with the analogous sequences from other members of the Flavivirus genus (Figure 5A). In particular, although residues that interact with the glucose ring near the top of the pocket are divergent, those deep within the pocket and proximal to the viral membrane exhibit conservation within DENV strains as well as within the ZIKV, WNV, and JEV.
Despite this apparent conservation of primary structure, co-crystallization of βOG with other flavivirus E proteins has not been reported despite deliberate efforts in some cases to obtain co-crystals with βOG (Kanai, et al., 2006; Modis, et al., 2005; Nybakken, et al., 2006; Zhang, et al., 2004). The absence of structural or pharmacological data indicating that the entry of other flaviviruses can be inhibited by this strategy has indirectly suggested that the pocket on other flavivirus E proteins is inaccessible and/or that the analogous sites on other flavivirus E proteins have insufficient molecular complementarity to bind to βOG or other candidate ligands. Our demonstration that cyanohydrazone 3-110-22 as well as compounds from both of the pyrimidine series inhibit ZIKV, WNV, and JEV and that 3-110-22 interacts with recombinant Zika sE in biochemical assays indicate that the ligand-binding pocket between domains I and II is a general feature of flavivirus E proteins and presents a defined molecular target for antiviral intervention.
Interestingly, the three inhibitor series we examined exhibit distinct viral specificity profiles, with the cyanohydrazone series in particular exhibiting broad-spectrum activity against multiple flaviviruses whereas both pyrimidine series exhibit greater selectivity for DENV. That the three inhibitor series also exhibit different sensitivities to mutations introduced to the βOG pocket reinforce the idea that specific interactions drive inhibitor-binding and motivate current efforts to understand the structure-activity relationships driving broad versus narrow spectrum activity against flaviviruses.
Antivirals that inhibit flavivirus fusion by targeting E
A major advantage of the viral envelope protein as a molecular target is that drugs can engage E extracellularly under conditions of physiological temperature and pH, and non-membrane permeant compounds can be devised to obviate the need for selectivity over the vast array of intracellular host targets. Conservation of residues in the βOG pocket open the possibility of discovery and development of antivirals efficacious against multiple flavivirus pathogens. Our demonstration that 3-110-22’s inhibition of multiple viruses is correlated with its binding to both DENV and ZIKV sE proteins ((Clark, et al., 2016) and Figure 5E) in a competitive luminescence proximity assay suggests that this may serve as a tool for target-based screening and medicinal chemistry optimization of inhibitors against these and other flavivirus E proteins. Collectively, these efforts provide pharmacological validation of the ligand-binding site between domains I and II as an anti-flaviviral target and provide a foundation for developing inhibitors to test whether inhibition of E’s fusogenic function is an efficacious antiviral strategy in preclinical murine models of flavivirus infection.
SIGNIFICANCE
Dengue virus (DENV), Zika virus (ZIKV) and other flaviviruses are human pathogens that cause significant disease. Due to their transmission by widespread mosquito species, flaviviruses can spread rapidly when introduced into new geographical locations where prior immunity does not exist. As illustrated by the explosive spread of Zika virus in South America and the Caribbean, rapid emergence of these viruses can have a devastating impact on public health. There are no approved drugs to counteract any of the flaviviruses. Dengvaxia, the only marketed vaccine against dengue, is effective in boosting natural immunity for those with prior dengue exposure, but actually sensitizes non-immune children to more severe disease and hospitalization once they are infected. Countermeasures to prevent or to treat dengue, Zika, and other flavivirus infections thus represent a large, unmet medical need, one even more critical by the lack of success of conventional vaccine and antivirals approaches to date. Viral envelope proteins have conserved structures and functions but lack “druggable” active sites that can be rationally targeted with small molecules. In addition, the surface of viral envelope proteins can be highly variable due to immune selection. In this study, we show that small molecules can bind extracellularly to the envelope protein, E, on the surface of virions and prevent infection by blocking E-mediated membrane fusion during viral entry. Our study demonstrates that these compounds bind in a pocket located between domains I and II that appears to be conserved across dengue, West Nile, Zika, and Japanese encephalitis viruses. The low frequency of resistance mutations in serial passaging and site-directed mutagenesis experiments supports the idea that the pocket is functionally important and thus conserved. These findings provide pharmacological evidence for the pocket as a target for developing broad-spectrum antivirals against multiple, mosquito-borne flavivirus pathogens.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Priscilla L. Yang (priscilla_yang@hms.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
African green monkey kidney-derived Vero cells (ATCC, RRID:CVCL_0059) and human kidney-derived HEK293T cells (ATCC, RRID:CVCL_0063) were cultured in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) at 37 °C with 5% CO2. Baby hamster kidney-derived BHK-21 cells (laboratory of Eva Harris, University of California, Berkeley) were maintained in Eagle minimum essential medium (MEM-α) supplemented with 5% FBS at 37 °C with 5% CO 2. C6/36 cells, a continuous mosquito cell line derived from Aedes albopictus (Diptera: Culicidae) embryonic tissue (ATCC, RRID:CVCL_Z230), were maintained in Leibovitz medium (L-15) containing 10% FBS at 28 °C. Spodoptera frugiperda cells (Sf9) (Cat # B825-01 Invitrogen/Thermo Fisher Scientific) were cultured in Sf-900™ II SFM media (Thermo Fisher Scientific) at 27 °C. Trichoplusia ni cells (High Five™, Thermo Fisher Scientific) were maintained at 27 °C in EX-CELL® 405 Serum-Free Medium (Sigma). The sex of the cell lines is not known.
METHOD DETAILS
Viruses
All work with infectious virus was performed in a biosafety level 2 (BSL2) laboratory using additional safety practices as approved by the Harvard Committee on Microbiological Safety.
Dengue virus serotype 2 strain New Guinea C (DENV2 NGC) was a kind gift from Lee Gehrke (Massachusetts Institute of Technology). Virus stocks were propagated in C6/36 cells.
The Zika virus (ZIKV) strain PF-251013-18 was obtained from Didier Musso (Institut Louis Malardé, Tahiti Island, French Polynesia), and was kindly provided by Nathalie Pardigon (Institut Pasteur, France). The ZIKV strain PF-251013-18 (Aubry, et al., 2016) was isolated from a viremic patient in French Polynesia in 2013, and was propagated three times on Vero cells. The viral stocks used in the current study were produced in C6/36 cells, and were not propagated on cells more than six times.
The Japanese encephalitis virus (JEV) vaccine strain SA14-14-2 was kindly provided by Gregory Gromowski (Walter Reed Army Institute of Research). The viral stocks used in the current study were produced in C6/36 cells, and were not propagated more than twice.
Antibodies
Monoclonal antibody 4G2 against the DENV E protein was produced from culture supernatants of hybridoma D1-4G2-4-15 (ATCC HB-112, RRID:CVCL_J890). Mouse hybridoma producing monoclonal antibody 6F3.1 against DENV2 core protein were generously provided by John Aaskov (Queensland University of Technology) (Bulich and Aaskov, 1992).
Compound synthesis and characterization
Unless otherwise noted, reagents and solvents were obtained from commercial suppliers and were used without further purification. 1H NMR spectra were recorded on 600 MHz (Varian AS600), and chemical shifts are reported in parts per million (ppm, δ) downfield from tetramethylsilane (TMS). Coupling constants (J) are reported in Hz. Spin multiplicities are described as s (singlet), brs (broad singlet), t (triplet), q (quartet), and m (multiplet). Mass spectra were obtained on a Waters Micromass ZQ instrument. Preparative HPLC was performed on a Waters Symmetry C18 column (19 x 50 mm, 5μM) using a gradient of 5–95% acetonitrile in water containing 0.05% trifluoacetic acid (TFA) over 8 min (10 min run time) at a flow rate of 30 mL/min. Purities of assayed compounds were in all cases greater than 95%, as determined by reverse-phase HPLC analysis.
Synthesis of JBJ-16-111
To a solution of 3-((6-((4-(trifluoromethoxy)phenyl)amino)pyrimidin-4-yl)oxy)benzoic acid (60 mg, 0.153 mmol), HATU (116 mg, 0.306 mmol) and DIEA (0.1 ml, 0.612 mmol) in N,N-dimethylformamide (1 ml) was added (3-methyl-3H-diazirin-3-yl)methanamine (20 mg, 0.230 mmol). After stirring for 30 min, the reaction mixture was diluted with EtOAc and washed with brine and water three times. The organic layer was dried over Na2SO4, filtered and concentrated under reduced pressure. The residue was purified by preparative HPLC to obtain N-((3-methyl-3H-diazirin-3-yl)methyl)-3-((6-((4-(trifluoromethoxy)phenyl)amino)pyrimidin-4-yl)oxy)benzamide (JBJ-16-111, 39 mg, 56 %). 1H NMR 500 MHz (DMSO-d6) δ 9.80 (s, 1H), 8.59 (t, J = 6.0 Hz, 1H), 8.39 (s, 1H), 7.77 – 7.70 (m, 3H), 7.62 (t, J = 1.8 Hz, 1H), 7.57 (t, J = 9.0 Hz, 1H), 7.40 (dd, J = 7.8, 2.0 Hz, 1H), 7.32 (d, J = 8.5 Hz, 2H), 6.17 (s, 1H), 3.29 (d, J = 6.1 Hz, 2H), 1.06 (s, 3H).
Other small molecule inhibitors were synthesized using previously reported methods (Clark, et al., 2016; Schmidt, et al., 2012; Adrian, et al., 2006).
Synthesis of 2-12-2 (Clark, et al., 2016)
1H NMR (600 MHz, CDCl3) δ 8.41 (dd, J = 9.0, 1.2 Hz, 1H), 8.04 (d, J = 5.4 Hz, 1H), 7.35 (m, 2H), 7.15–7.20 (m, 4H), 6.94 (dt, J = 9.0, 1.8 Hz, 1H), 6.54 (br s, 1H), 6.14 (d, J = 6.0 Hz, 1H).
Synthesis of 7-148-6 (Clark, et al., 2016)
1H NMR (600 MHz, DMSO) δ 8.17 (br s, 1H), 8.05 (d, J = 6.6 Hz, 1H), 7.85 (m, 4H), 7.55 (m, 4H),7.50–7.41 (m, 4H), 6.50 (d, J = 6.6 Hz, 1H).
Synthesis of 1-100-1 (Clark, et al., 2016)
1H NMR (600 MHz, CDCl3) δ 9.97 (br s, 1H), 8.34 (s, 1H), 7.43 (m, 2H), 7.36 (m, 2H), 7.28 (m, 3H), 7.11 (m, 2H), 6.24 (s, 1H).
Synthesis of GNF-2 (Adrian, et al., 2006)
GNF-2, [3-(6)-(4-Trifluoromethoxy-phenylamino)-pyrimidin-4-yl]-benzamide]: A mixture of (6-Chloro-pyrimidin-4-yl)-(4-trifluoromethoxy-phenyl)-amine (200 mg, 0.69 mmol), 3-carbamoylphenylboronic acid (115 mg, 0.69 mmol), palladium tetrakis (-triphenylphosphine) (40 mg, 0.034 mmol) and sodium carbonate (292 mg, 2.76 mmol) in acetonitrile/water (1:1, v/v, 10 mL) was heated to 90°C under an argon atmosphere. After 8h, the solution was cooled to room temperature and the solvent was removed in vacuo. The resulting crude product was purified by flash chromatography using ethyl acetate/hexanes (1:1, v/v) to yield 206 mg of GNF-2 (yield: 80 %). 1H NMR (400 MHz, DMSO-d6): δ8.79 (s, 1H), 8.53 (s, 1H), 8.23 (d, J = 8.5Hz, 1H), 7.96 (d, J = 5.1Hz, 1H), 7.85 (d, J = 6.9Hz, 2H), 7.75 (t, J = 7.9Hz, 1H), 7.48 (s, 2H), 7.36 (d, J =8.2Hz, 2H), 7.33 (s, 1H). MS: m/z 375.1 (M+H)+. GNF-2 is also available from commercial vendors (Sigma-Aldrich, catalog number G9420; Cayman Chemical catalog number 16253).
Synthesis of GNF-2-biotin (Clark, et al., 2016)
A reaction mixture of compound cGNF-2 3-(6-(4-(trifluoromethoxy)phenylamino)pyrimidin-4-yl)benzoic acid (61 mg, 0.18 mmol), 17-azido-3,6,9,12,15-pentaoxaheptadecan-1-amine (56 mg, 0.15 mmol), O-(7-Azabenzotriazol-1- yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HATU) (86 mg, 0.23 mmol), and DIEA (78 μL, 0.45 mmol) in DMF (3 mL) was stirred at room temperature overnight. Once the reaction was completed, the reaction mixture was diluted with ethyl acetate (20 mL) and washed with brine (20 mL). The organic phase was dried over Na2SO4 and evaporated. The residue was purified by flash chromatography to afford the desired product 13 N-(17- azido-3,6,9,12,15-pentaoxaheptadecyl)-3-(6-(4-(trifluoromethoxy)phenylamino)pyrimidin-4-yl)benzamide (57 mg, 58% yield). To a solution of N-(17-azido-3,6,9,12,15-pentaoxaheptadecyl)-3-(6-(4- (trifluoromethoxy)phenylamino)pyrimidin-4-yl)benzamide (17 mg, 0.026 mmol) in 5 mL of methanol was added 5 mg Pd/C. The resulting suspension was degassed three times and then was stirred with a balloon filled with H2 for 1 hour. The crude reaction mixture was filtered through a short pad of Celite, which was washed with 10 mL of methanol. The combined filtrates were concentrated and used for the next step without further purification. To a solution of above amine in DMF (2 mL) were added NHS-Biotin (Thermo Scientific), 2,5-dioxopyrrolidin-1-yl 5-(2-oxo-hexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanoate, 8.8 mg, 0.026 mmol) and DIEA (4.5 μL, 0.026 mmol). After stirring at room temperature for 2 h, the reaction mixture was concentrated and purified by prep-HPLC to afford GNF-2-biotin (6.8 mg, 31% yield). 1H NMR (600 MHz, DMSO-d6) δ 9.97 (s, 1H), 8.75 (s, 1H), 8.70 (t, J = 6.0 Hz, 1H), 8.51 (s, 1H), 8.17 (d, J = 8.4 Hz, 1H), 7.98 (d, J = 7.8 Hz, 1H), 7.83 – 7.85 (m, 2H), 7.80 (t, J = 7.8 Hz, 1H), 7.62 (t, J = 7.8 Hz, 1H), 7.33 – 7.35 (m, 2H), 6.39 (s, 1H), 6.33 (s, 1H), 5.74 (s, 1H), 4.26 – 4.30 (m, 1H), 4.09 – 4.13 (m, 1H), 3.49 – 3.57 (m, 20H), 3.36 (t, J = 6.0 Hz, 2H), 3.14 – 3.17 (m, 2H), 3.05 – 3.08 (m, 1H), 2.79 (dd, J = 12.0, 5.4 Hz, 1H), 2.53 – 2.57 (m, 1H), 2.04 (t, J = 7.2 Hz, 2H), 1.56 –1.60 (m, 1H), 1.41 – 1.49 (m, 3H), 1.24 – 1.30 (m, 2H).
Synthesis of 3-110-22 (Schmidt, et al., 2012)
To a solution of 3-(trifluoromethyl)aniline (0.45 mL, 3.6 mmol) in 6-N hydrochloric acid (10 mL) was added dropwise a solution of sodium nitrite (248 mg, 3.6 mmol) in water (5 mL) at 5 °C. After the addition, the mixture was stirred at ambient temperature for 15 min before the resulting solution was divided into 20 2-dram vials. To each vial was added a mixture of 2-aroylacetonitrile (0.15 mmol) and sodium acetate (74 mg, 0.90 mmol) in dioxane (1 mL). The vials were then vortexed at ambient temperature for 4 hrs, when the reaction mixtures were diluted by water (1 mL) and ethyl acetate (1 mL). The upper organic layer was taken by a pipette and transferred through a filter cartridge containing magnesium sulfate into a culture tube. The filtrates were concentrated in GeneVac and the residues were purified on silica SPE. 1H NMR (600 MHz, CDCl3) δ 8.30 (s, 1H), 7.80 (d, 1H, J=8.4), 7.75 (s, 1H), 7.67–7.63 (m, 2H), 7.60–7.53 (m, 3H), 7.39–7.36 (m, 1H).
Virus infections
All DENV2 and JEV infectivity assays were performed on BHK-21 cells, while ZIKV infectivity assays were performed on Vero cells. Confluent monolayers of cells were infected by incubation of viral inoculum diluted in Earle’s balanced salt solution (EBSS) at MOI of 1 (unless otherwise indicated) for 1 hour at 37 °C. The inoculum was then removed and the cells were washed twice with 1X PBS. The appropriate medium containing 2% FBS was then overlaid on cells. Unless stated otherwise, the viral supernatants were harvested at 24 hours post-infection and stored at −80 °C.
Viral plaque-formation assay
DENV2 and JEV titers were quantified by plaque-formation assay (PFA) on BHK-21 cells. ZIKV titers were quantified by PFA on Vero cells. Cells were seeded in 24-well plates and incubated overnight at 37 °C until formation of confluent monolayers. Aliquots from viral stocks or infections were thawed at room temperature. Ten-fold dilutions in EBSS were prepared in duplicate, and 100 μL of each dilution was added to the cells. The plates were incubated for 1 hour at 37 °C and rocked every 15 minutes. Non-adsorbed virus was removed, after which 1 mL of DMEM (Vero cells) or MEMα (BHK-21 cells) supplemented with 1.05% carboxymethyl cellulose (CMC), 44 mM sodium bicarbonate, and 2% FBS was added to each well, followed by incubation at 37 °C for 2.5 days (ZIKV), 3 days (JEV) or 4.5 days (D ENV2). The CMC overlay was aspirated, the cells were washed with 1X PBS and stained with crystal violet, and viral plaques were counted.
Viral infectivity assays for IC90 determination
Virus inocula were diluted in EBSS to achieve an MOI of 1, and were pre-incubated with the given small molecule at varying concentrations for 45 min at 37 °C. The mixture was then added to cells for 1 hour at 37 °C to allow infection, after which the inoculum was removed and the cells were washed with 1X PBS to remove unbound virus and compound. Cells were overlaid with medium lacking inhibitor and incubated at 37 °C for 24 hours, corresponding to a single cycle of infection. Culture supernatants were harvested at this time, and the yield of infectious particles produced was quantified by PFA.
Selection of DENV2 resistant to 7-148-6
DENV2 (MOI 0.1) was incubated with 20 μM 7-148-6 for 45 min at 37°C before addition to Vero cells in a T25 flask. After a 1 hour infection, 5 mL medium was added (2% FBS in DMEM) containing 20 μM 7-148-6 diluted from DMSO stock; the vehicle control was passaged in an equivalent percentage of DMSO. Infections proceeded for 4 days, at which point supernatant was harvested and spun briefly to remove cell debris. 500 μL of supernatant was used to infect a fresh T25 of Vero cells for 1 hour, at which point 4.5 mL of medium containing compound (or vehicle) was added. Viral titer of each passage was determined by PFA. Cell supernatants containing the resistant quasi-species or the corresponding DMSO-passaged control quasispecies were collected at passage 7, viral RNA was extracted (QIAamp Viral RNA kit, Qiagen), and cDNA was synthesized. Consensus sequencing of the cDNA sample (The Broad Institute Viral Genomics Sequencing Core) led to the identification of an A to T mutation resulting in substitution of methionine to valine (M196V) in the E protein. The mutant virus was triply plaque purified, and viral stocks of the clonal virus that contains the E-M196V mutation were propagated as described above. The presence of the mutation was monitored by extraction of viral RNA from the supernatants, followed by reverse transcription-PCR and sequencing.
Single-cycle reporter viral particles (RVP) production
To produce reporter viral particles (RVP), HEK293T cells were plated in a 10-cm dish and then co-transfected using Lipofectamine 2000 (Life Technologies), according to the manufacturer’s instructions, with two plasmids. The first plasmid encodes the structural proteins of either DENV2 (pCDNA6.2-D2.CprME (Ansarah-Sobrinho, et al., 2008)), or WNV (pWNI-CprME-IH2 (Pierson, et al., 2006)). The second plasmid encodes a WNV subgenomic replicon (pWIIrep-REN-IB (Pierson, et al., 2006)) that expresses the virus non-structural proteins and a Renilla luciferase reporter. To produce the mutant pCDNA6.2-D2.CprME that carries the E-M196V mutation, site-directed mutagenesis of the plasmid was performed using the Quikchange II kit according to manufacturer’s instructions (Agilent), and using the primers listed in Table S2.
To produce DENV2 RVP, the plasmids pCDNA6.2-D2.CprME and pWIIrep-REN-IB were transfected to cells at a 3:1 ratio. To produce WNV RVP, the plasmids pWNI-CprME-IH2 and pWIIrep-REN-IB were transfected to cells at a 1:2 ratio.
For all of the RVP preparations, fresh medium was added at 24 hours post-transfection. The supernatants were collected at 48 hours post-transfection, clarified by centrifugation at 1,000 x g for 5 minutes at 4 °C, and aliquots were stored at −80 °C. The genome copy number per mL was determined for each RVP stock by RT-qPCR on the supernatants using primers specific for the Renilla luciferase gene, listed in Table S2.
DENV2 RVP Titration Assay
BHK-21 cells were plated in 48-well plates the day before infection and incubated until ~70% confluency. On the day of infection, 80 μL of a dilution series of RVP was added to each well (in triplicate) and incubated for 1hour. Cells were then washed with 1X PBS and MEMα media supplemented with 2% FBS was added to the cells. At 24 hours post-infection, the cells were washed with 1X PBS and the samples were processed according to the manufacturer’s instructions of the Renilla luciferase assay system (Promega). The Renilla luciferase signal was read using a Perkin Elmer EnVision plate reader. Signal from non-infected sample was considered background. For DENV2 RVPs, the wells were scored as positive or negative for signal to determine the tissue culture infectious dose (TCID50).
Single-cycle reporter virus (RVP) infectivity assays
BHK-21 cells were seeded in 48-well tissue culture plates in MEMα supplemented with 2% FBS. The RVPs were diluted with EBSS, and treated with compounds for 45 minutes at 37 °C. For the DENV2 RVPs, the cells were infected at MOI of 0.007 based on TCID50 values. For the WNV RVPs, each well was infected with 4000 genome copies. The RVP-compound mixture was then added to the cells, and the plates were incubated for 1 hour at 37°C. Unadsorbed RVPs were washed with 1X PBS and removed, after which MEMα supplemented with 2% FBS was added to the cells, followed by incubation at 37°C. At 24 hours post-infection, the samples were processed according to the manufacturer’s instructions of the Renilla luciferase assay system (Promega). The Renilla luciferase signal was read using a Perkin Elmer EnVision plate reader.
Virion attachment assay
DENV2 was incubated with either 5 μM of 2-12-2, 10 μM of 7-148-6, 20 μg/ml of heparin, or DMSO for 45 minutes at 37 °C, then added to BHK-21 cells (MOI 1) at 4°C for 1 hour with constant rocking. Cells were washed three times with cold 1X PBS to remove unbound virus and compound. For quantitation of viral genomes bound, total RNA was extracted using TRIzol (Thermo Fisher Scientific) according to the manufacturer’s instructions. Total RNA was used as a template for cDNA synthesis using iScript (Bio-Rad), according to the manufacturer’s instructions. The resulting cDNA was digested by RNaseH (NEB) for 1 hour at 37°C. Quantitative PCR (qPCR) assays utilized the iQSYBR green Supermix (Bio-Rad) and were performed on a MyiQ iCycler machine (Bio-Rad) as follow: 5 minutes at 95 °C and 40 cycles of 15 seconds at 95 °C and 30 seconds at 60 °C, followed by the melt curve that started at 50°C and increased 0.5 °C every 10 seconds for 90 cycles. The primers used targeted the DENV2 C gene (Table S2).
Capsid protection assay to monitor fusion of virions with liposomes
Liposomes were made with 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) (Avanti Polar Lipids), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) (Avanti Polar Lipids), and cholesterol (Avanti Polar Lipids) at 1:1:1 molar ratio in TAN buffer (20 mM triethanolamine, 100 mM NaCl, pH 8.0). Lipids were dried down and resuspended thoroughly by vortexing and sonication. Liposomes were prepared by extrusion through a 0.2 micron filter after five freeze/thaw cycles. For trypsin-containing liposomes, 10 mg of trypsin was added to 1 mL of lipids (3 mg each) after the third freeze/thaw cycle, prior to extrusion. Liposomes were separated from unincorporated trypsin by size-exclusion chromatography using a 10/300 Increase Superdex 200 column (GE Healthcare) on an Akta fast performance liquid chromatography (FPLC) system. Trypsin-containing liposomes were used within 24 hours.
Purified DENV2 virus (300 ng) was incubated with various concentrations of compounds (5–40 μM) for 45 min at 37 °C in TAN buffer (pH 8.0) prior to addition of liposomes (7.5 μL of peak fraction from column) for 10 minutes. After incubation with liposomes, 7.5 μL 1M sodium acetate (pH 5.0) was added to drop the pH to 5.5 for 10 minutes. Samples were back-neutralized with 15 μL 1M triethanolamine (unbuffered) and incubated for 45 minutes at 37°C to allow tryps in digestion. SDS sample buffer was added, and samples were boiled for 20 minutes before separation by SDS-PAGE on a 12% gel. Proteins were transferred to a nitrocellulose membrane using a semi-dry transfer apparatus. 4G2 and 6F3.1 antibodies were used for detection of DENV envelope and capsid protein, respectively.
Expression and purification of 6His-tagged DENV2 and ZIKV soluble envelope protein (sE) expression and purification
The coding sequence for the soluble portion of the DENV2 NGC envelope protein (sE), fused to a N-terminal signal peptide was codon optimized for expression in insect cells was synthesized and generously donated by Daryl Klein and Stephen C. Harrison. The coding sequences of full length or domain I and II of DENV2 envelope protein were cloned into a pFastBacTM/CT-TOPO® vector (Thermo Fisher Scientific) to enable expression of sE or DI-DII with a TEV-His6 tag at its C terminus. The sequence coding for the soluble portion of the mutant DENV2 NGC envelope protein carrying the E-M196V mutation (sE-M196V) and fused to an N-terminal signal peptide and to a C-terminal TEV-His6 tag was codon optimized for expression in insect cells and cloned into a pFastBac1 vector by Life Technologies (now Thermo Fisher Scientific). We then modified the pFastBac-DENV2-sE-His6Tag plasmid to introduce a C-terminal AviTagTM, as described previously (Clark, et al., 2016) to allow production of AviTagTM DENV2 sE protein. Site-directed mutagenesis was performed on this plasmid using primers listed in Table S2 to introduce the following mutations: Q52A, T171A, F193L, M196V, Q200A, Q200E, Q271A, M272S, and F279S.
The sequence coding for the soluble portion of the ZIKV H/PF/2013 envelope protein (E) (GenBank accession number KJ776791), fused to a N-terminal signal peptide and to a C-terminal TEV His6 tag, was codon-optimized for expression in insect cells and cloned into a pFastBac1 vector by GenScript.
The plasmids were used to transform DH10Bac cells (Thermo Fisher Scientific, 10361012). The transformed cells were grown in presence of 50 μg/mL kanamycin, 7 μg/mL gentamicin, 10 μg/mL tetracycline, 100 μg/mL Bluo-gal, and 40 μg/mL IPTG at 37°C for 2 to 3 days. The white colonies, containing the insert, were selected and grown in Luria broth supplemented with 50 μg/mL kanamycin, 7 μg/mL gentamicin, 10 μg/mL tetracycline overnight at 37°C in the dark. T he bacmids were extracted after lysis of cells with 50 mM Tris-Cl, pH 8.0, 10mM EDTA, 100μg/mL RNase A, followed by precipitation with 0.8 mL isopropanol. The pellets containing the bacmid DNA were washed with 70% ethanol, dried and dissolved in 40 μL TE buffer pH 8.
To produce baculoviruses expressing DENV2 or ZIKV proteins, 2–5 μg of the appropriate bacmid were used to transfect in 2x106 Sf9 cells in 5 mL Sf-900™ II SFM medium using Cellfectin II Reagent (Life Technologies). The cells were incubated at 27 °C for 3 days, and the supernatants that contained the first production of baculovirus (P1) were collected by centrifugation at 150 x g for 5 min at 4 °C. The baculoviruses were then amplified by inoculating 1 to 2 mL of P1 in 2x107 Sf9 cells in 30 mL of Sf-900™ II SFM medium. The supernatants containing the second passage of baculovirus (P2) were collected after 3 days and clarified by centrifugation at 150 x g for 10 min. The third passage of baculovirus (P3) was produced by inoculating 10–15 mL of P2 in 2x108 Sf9 cells in 400 mL of Sf-900™ II SFM medium and incubated under rotation at 130 rpm, at 27°C, for 3 days. The culture supernata nts were centrifuged at 1400 x g for 10 min, and stored at 4°C protected from light.
To express DENV2 sE or DI–DII proteins, High Five cells were cultured in 3–6 L of EX-CELL® 405 Serum-Free Medium until they reached a density of 1 to 1.5 million cells/mL, and infected with 20 mL of P3 baculoviruses per liter of medium. At 72 hours post-infection, the supernatants were collected by centrifugation at 3,000 x g for 20 min at 4 °C. Proteins were purified by passing over 25 mL TALON metal-affinity resin slurry (Clontech). The beads were washed once with 1X PBS pH 7.4, 10 mM imidazole and the proteins were eluted with 200 mM imidazole pH 8. The protein was further purified by size-exclusion chromatography using a Superdex 200 grade 10/300 GL column (GE Healthcare) with buffer containing 20 mM triethanolamine hydrochloride pH 8.0 and 100 mM NaCl. Fractions containing DENV2 sE or DI–DII were pooled and concentrated to approximately 1.2 to 2.5 mg/mL (20 to 54 μM), then flash-frozen in liquid nitrogen and stored at −80 °C.
Soluble ZIKV sE protein was expressed in the same manner as DENV2 sE protein, except only 10 mL of P3 baculovirus was used per liter of culture. The culture was harvested at 72 hours post-infection by centrifugation at 4000 x g for 30 minutes at 4 °C. The supernatants were then filtered through a 0.22 micron filter (Corning) before loading onto 15 mL Ni-NTA agarose (Qiagen) that had been pre-equilibrated in 1x PBS buffer pH 7.4. The unbound fraction was passed over the Ni-NTA resin a second time followed by 2 washes with 5 column volumes of 1X PBS pH 7.4, 10 mM imidazole. Bound protein was then eluted from the column with 2–3 column volumes of 200 mM imidazole pH 8.0. The buffer was exchanged to a buffer containing 20 mM triethanolamine hydrochloride pH 8.0 and 100 mM NaCl and concentrated down to 10 mL using a 30 kDa amicon filter (Millipore). Size-exclusion chromatography was used as a polishing step on a Superdex 200 grade 16/600 pg column (GE Healthcare) pre-equilibrated with 20 mM triethanolamine hydrochloride pH 8.0 and 100 mM NaCl. Fractions containing ZIKV sE protein were pooled and concentrated to 1.6–2.3 mg/mL (35–50 μM), flash frozen in liquid nitrogen, and stored at −80 °C.
Biotinylation of AviTagTM sE proteins
The purified AviTagTM DENV2 sE proteins were biotinylated using the BirA-500 kit (Avidity). Briefly, the protein was concentrated to around 400 μM using a 30 kDa Amicon filter (Millipore) and was then resuspended in 50 mM Biocine pH 8.5. The proteins were incubated for 60 minutes at 30 °C in the presence of 2.5 μg BirA per 10 nmol of purified protein, according to the manufacturer’s instructions. The biotinylated protein was then purified from the reaction using a Superdex 200 grade 10/300 GL column (GE Healthcare) pre-equilibrated with 20 mM triethanolamine hydrochloride (pH 8.0) and 100 mM NaCl.
Purified ZIKV sE protein was biotinylated as follows. 4 nmol of ZIKV sE protein was mixed with 20 nmol of biotin-PEG2-NHS (9-biotinlamino-4,7-dioxanonanoic acid N-hydroxysuccinimidyl ester, Broadpharm) in 1X PBS to which 20 mM NaHCO3, pH 8.3 was added to adjust the solution to slightly basic pH. The solution was incubated at 4 °C overnight. Excess biotin was removed by dialysis in 1X PBS buffer (Thermo Scientific Slide-A-Lyzer Dialysis Kit, 10000 MWCO, 0.1–0.5 mL).
Determination of dissociation constant (Kd) values by bio-layer interferometry (BLI)
BLI mixtures (80 μL) were prepared in wells of a 384-well black tilted-bottom plate (FortéBio) and the measures were monitored by Octet RED384 system (FortéBio).
Ni-NTA biosensor reactions for characterization of binding of small molecules 2-12-2 and 7-148-6 to DENV2 sE-wildtype and sE- M196V proteins carrying a 6His tag
1.6 μg of DENV2 sE protein was loaded on Ni-NTA biosensors (FortéBio) for 600 seconds and then quenched with 5 μM imidazole for 180 seconds. The Ni-NTA biosensors were then equilibrated in reaction buffer [1X Kinetic buffer (FortéBio), 20 mM triethanolamine hydrochloride pH 8, 100 mM NaCl, 2% DMSO] for 600 seconds prior to baseline collection. Small molecule concentrations ranged from 50 nM to 30 μM with association and dissociation times both set at 120 seconds.
Super streptavidin (SSA) biosensor reaction for determination of binding of small molecule inhibitors to biotinylated DENV2 and ZIKV sE proteins
1.6 μg of the biotinylated protein was loaded on an SSA biosensor tip (FortéBio) for 600 seconds and then quenched with 0.8 μg biocytin for 120 seconds. The SSA biosensors were then equilibrated in reaction buffer [1X Kinetic buffer (FortéBio), 1X PBS, 2% DMSO] for 180 seconds prior to baseline collection. For DENV2 sE proteins, association with small molecules was monitored for 120 seconds with inhibitor concentrations that ranged from 50 nM to 20 μM; dissociation was performed in reaction buffer and monitored for 120 seconds. For ZIKV sE protein, association with 3-110-22 was monitored for 150 seconds at inhibitor concentrations between 0.1 to 15 μM; dissociation was monitored in reaction buffer for 10 seconds.
Protein Crosslinking and Capillary Electrophoresis/Mass Spectrometry Analysis
DENV2 DI-DII protein was treated with a five-fold molar excess of JBJ-16-111, incubated for 30 minutes at 37 ºC, and irradiated with long-wave UV light for 10 minutes using a Spectrolinker UV Crosslinker (model XL1000, Spectronics Corp., Westbury, NY). After irradiation, protein was denatured with Rapigest (0.1% final concentration; Waters, Milford, MA), reduced with 10 mM dithiothreitol for 30 minutes at 56 ºC, alkylated with 22.5 mM iodoacetamide for 30 minutes protected from light, and digested with trypsin overnight at 37 ºC. Rapigest was cleaved by adding 10% TFA and incubating at 37 ºC for 30 minutes. The digest was centrifuged for 10 minutes at 4 C and the supernatant subjected to desalting by C18. Eluted peptides were dried by vacuum centrifugation, reconstituted in 50% acetonitrile, 1% formic acid with 100 mM ammonium acetate and analyzed by CE-MS using a ZipChip CE system and autosampler (908 Devices, Boston, MA) interfaced to an Orbitrap Lumos mass spectrometer (Thermo Fisher Scientific). Peptide solutions were loaded for 30 seconds and separation performed at 500 V/cm for 10 minutes using an HR chip (22 cm separation channel) with a background electrolyte composed of 1% formic acid in 50% acetonitrile. Pressure assist was utilized and started at 1 minute. The mass spectrometer was operated in data-dependent mode, where the five most abundant ions in each MS scan (orbitrap detection, 120K resolution) were subjected to MS/MS (HCD, 30% collision energy, 2 Da isolation width, 2E5 target, 100 ms max fill time, FTMS detection at 15K resolution). Dynamic exclusion was enabled with a repeat count of 1 and an exclusion time of 6 seconds. To allow more accurate filtering of data to a 1% FDR, multiplierz (Alexander, et al., 2017) generated .mgf files were appended to an .mgf file derived from an analysis of K562 total protein tryptic peptides, and the merged .mgf was searched using Mascot against a forward-reverse human protein database (NCBI refseq) appended with the sequence of the DI-DII protein. Search parameters specified a precursor ion mass tolerance of 10 ppm, a product ion mass tolerance of 25 mmu, fixed carbamidomethylation of cysteine, variable oxidation of methionine, and variable JBJ-16-111 modification of lysine, serine, threonine, or tyrosine. Search results were filtered to 1% FDR using a multiplierz script. Spectral analysis was performed using mzStudio (Ficarro, et al., 2017).
AlphaScreen competition assay
7 pmol of ZIKV sE protein was mixed with AlphaScreen nickel chelate acceptor beads (15 ng/μL, PerkinElmer) in HEPES buffer (25 mM HEPES, 200 mM NaCl, pH 7.4), in the presence of 10.5 pmol of GNF-2-biotin (Choi, et al., 2009; Clark, et al., 2016) per well. The solution was dispensed at 30 μL/well into Perkin Elmer standard OptiPlates 384. Small molecule 3-110-22 was dispensed into the plate at concentrations ranging from 0.5 to 10 μM. The plate was centrifuged at 233 x g for 1 min and incubated at room temperature for 2.5 hours. AlphaScreen streptavidin donor beads (Perkin Elmer) were diluted at 15 ng/μL in HEPES buffer and 5 μL was distributed to the plate. The plate was centrifuged at 233 x g for 1 minute and incubated at room temperature for 2.5 hours. The fluorescence signal (excitation 680 nm, emission 520–620 nm) was measured using a Perkin Elmer EnVision plate reader.
Hydrogen-deuterium exchange (HDX) analysis
Stock solutions of DENV2 sE (wild-type 69.5 μM; M272S 68.6 μM; M196V 47.2 μM; Q52A 30 μM) were prepared in equilibration buffer (10 mM potassium phosphate, 100 mM NaCl, pH 7.0, H2O). To initiate labeling, 2 μL of each protein were independently diluted with 30 μL (15-fold) labeling buffer (10 mM potassium phosphate, 100 mM NaCl, pD 7.0, 99.8% D2O) and incubated for predetermined times (10 sec, 1 min, 10 min, 1 hour, and 4 hours). At the end of each labeling time, the entire deuterium labeling reaction was added to 30 μL of quench buffer (200 mM sodium phosphate, 4M guanidine hydrochloride, 0.72 M TCEP, pH 2.3, 0 °C) to quench the reaction. All steps from quenching forward were performed at 0 °C.
Quenched samples were immediately digested online with an immobilized pepsin column (prepared in house according to (Wang, et al., 2002) and directed into a Waters nanoAcquity UPLC with HDX technology (Wales, et al., 2008). Peptides were trapped on a Waters UPLC BEH C18 1.7 μm VanGuard BEH column and desalted with 0.1% formic acid in water for 3 minutes at 100 μL/min. Peptides were eluted over 10 minutes using a 5–35% gradient of water:acetonitrile with 0.1% formic acid flowing at a rate of 90 μL/min using a Waters HSS T3 1.8 μm C18, 1.0 mm x 50 mm analytical column. Deuterium incorporation was measured using a Waters Synapt G2Si equipped with a standard ESI source in HDMSE mode. Mass spectra were acquired over a m/z range of 50–2000, and mass accuracy was confirmed by calibration with 500 fmol/μL of human glu-fibrinopeptide. Peptic peptides were identified using ProteinLynx Global Server (PLGS) 3.0 (Waters) and deuterium incorporation measured using DynamX 3.0 (Waters).
QUANTIFICATION AND STATISTICAL ANALYSIS
Viral infectivity assays for IC90 determination
The yield of infectious particles produced was quantified by PFA and plotted relative to DMSO-treated samples. The concentrations that caused 90% JEV, ZIKV or DENV2 inhibition (IC90) were calculated using the nonlinear fit variable slope model (Prism, GraphPad Software). Differences in IC90 values for E-M196V mutant versus wildtype viruses were evaluated for significance using the Student’s T-test.
Determination of dissociation constant (KD) values by bio-layer interferometry (BLI)
Equilibrium dissociation constants (KD) values were determined by plotting the local fit maximum response (nm) as a function of small molecule concentrations (μM) using Octet Data Analaysis Software (FortéBio) and Prism (GraphPad Software). Titration curves were fit to the following steady-state analysis equation:
where Rmax is the local fit response maximum; Conc is the concentration of small molecule; and KD is the equilibrium dissociation constant.
Maximum compound concentrations employed in these experiments were set based on compound aggregation, as assessed by deterioration of signal-to-noise at higher concentrations in the BLI assay and empirical observation of compound aggregation by dynamic light scattering (data not shown). In several cases, we were unable to saturate inhibitor-binding with a given mutant. Where clear concentration-dependent interaction was observed, we have estimated a lower bound for the KD based on the fit of available data and the highest concentrations that yielded well-behaved data in the biolayer interferometry experiments. In other cases, we were unable to reliably detect signal over background, presumably due to low affinity of the E-inhibitor interaction and or due to aggregation of the compound at concentrations relevant for KD measurement. These are reported as “N.D.” in Table S1.
Hydrogen-deuterium exchange (HDX) analysis
As all experiments were comparisons between states and measurements were taken close in time using nearly identical experimental conditions, the deuterium levels were not corrected for back exchange and are reported as relative (Wales and Engen, 2006). All experiments were performed in duplicate. The average error for each data point in the instrumental system so described did not exceed ± 0.15 Da and is consistent with previously reported values (Houde, et al., 2011). Average differences exceeding 0.5 Da were considered significant.
Statistical Analysis
Statistical tests and the associated error bars are identified in the corresponding figure legends. “Independent” experiments describe the number of biological replicates, which were repeat experiments performed on different days. Statistical analysis was performed using Prism (GraphPad Software).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal antibody against DENV envelope protein | Supernatants of hybridoma D1- 4G2-4-15; ATCC | Cat#HB-112; RRID:CVCL_J890 |
| Mouse monoclonal antibody against DENV2 core protein | Supernatants of hybridoma 6F3.1; John Aaskov, Queensland University of Technology (Bulich and Aaskov, 1992) | N/A |
| Bacterial and Virus Strains | ||
| Dengue virus serotype 2 strain New Guinea C | Lee Gehrke, Massachusetts Institute of Technology | N/A |
| Zika virus strain PF-251013-18 | Didier Musso, Institut Louis Malardé (Aubry, et al., 2016) | N/A |
| Japanese encephalitis virus strain SA14-14-2 | Gregory Gromowski, Walter Reed Army Institute of Research (Gromowski et al., 2014) | N/A |
| DH10Bac | Thermo Fisher Scientific | Cat#10361012 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 2-12-2 | Clark, et al., 2016 | N/A |
| 7-148-6 | Clark, et al., 2016 | N/A |
| GNF-2 | Clark, et al., 2016 | N/A |
| GNF-2-biotin | Clark, et al., 2016 | N/A |
| 1-100-1 | Clark, et al., 2016 | N/A |
| 3-110-22 | Schmidt, et al., 2012 | N/A |
| JBJ-16-111 | This paper | N/A |
| 1-palmitoyl-2-oleoyl-sn-glycero-3- phosphocholine (POPC) | Avanti Polar Lipids | Cat#850457C |
| 1-palmitoyl-2-oleoyl-sn-glycero-3- phosphoethanolamine (POPE) | Avanti Polar Lipids | Cat#850757C |
| Cholesterol | Avanti Polar Lipids | Cat#700000P |
| Biotin-PEG2-NHS ester | Broadpharm | Cat#BP-22608 |
| Critical Commercial Assays | ||
| Renilla luciferase assay system | Promega | Cat#E2810 |
| BirA-500 kit | Avidity | Cat#BirA500 |
| Ni-NTA biosensors | PALL FortéBio | Cat#18-5101 |
| Super Streptavidin biosensors | PALL FortéBio | Cat#18-5057 |
| AlphaScreen histidine (nickel chelate) detection kit | PerkinElmer | Cat#6760619C |
| Experimental Models: Cell Lines | ||
| Vero | ATCC | Cat#CCL-81; RRID:CVCL_0059 |
| HEK293T | ATCC | Cat#CRL-3216; RRID:CVCL_0063 |
| BHK-21 | Eva Harris, University of California Berkeley | N/A |
| C6/36 | ATCC | Cat#CRL-1660; RRID:CVCL_Z230 |
| Sf9 | Invitrogen / Thermo Fisher Scientific | Cat#B825-01; RRID:CVCL_0549 |
| High Five™ | Thermo Fisher Scientific | Cat#B85502; RRID:CVCL_C190 |
| Oligonucleotides | ||
| Primers, see Table S2 | IDT DNA, this paper | N/A |
| Recombinant DNA | ||
| pCDNA6.2-D2.CprME | Theodore Pierson, N.I.H. (Ansarah-Sobrinho, et al., 2008) | N/A |
| pWNI-CprME-IH2 | Theodore Pierson, N.I.H. (Ansarah-Sobrinho, et al., 2008) | N/A |
| pWIIrep-REN-IB | Theodore Pierson, N.I.H. (Pierson, et al., 2006) | N/A |
| pFastBac-DENV2-sE-His6Tag | This paper | N/A |
| pFastBac-DENV2-DI-DII-His6Tag | This paper | N/A |
| pFastBac-DENV2-sE-AviTag | This paper | N/A |
| pFastBac-ZIKV-sE-AviTag | This paper | N/A |
| Software and Algorithms | ||
| Prism | GraphPad Software | RRID:SCR_002798 https://www.graphpad.com/scientific-software/prism/ |
| Octet Data Analysis Software | PALL FortéBio | N/A https://www.fortebio.com/octet-software.html |
| mzStudio | Ficarro, et al., 2017 | N/A |
| Mascot 2.6 | Matrix Science | RRID:SCR_014322 http://www.matrixscience.com |
| Multiplierz | Alexander, et al., 2017 | RRID:SCR_012058 |
| ProteinLynx Global Server (PLGS) 3.0 | Waters | N/A http://www.waters.com/waters/en_US/ProteinLynx-Global-SERVER-%28PLGS%29/nav.htm?cid=513821&locale=en_US |
| DynamX HDX Data Analysis Software 3.0 | Waters | N/A http://www.waters.com/waters/library.htm?cid=511436&lid=134832928&locale=en_US |
Supplementary Material
Deuterium uptake plots comparing relative deuterium levels measured for the wild-type DENV2 sE (black circles) and sE-M196V (red squares), DENV sE-M272S (blue squares), and DENV sE-Q52A (green squares), related to Figure 4.
Highlight points.
Small molecules targeting the dengue virus E protein inhibit membrane fusion
The target of these inhibitors is a pocket conserved in other flavivirus E proteins
Zika and other flaviviruses are also inhibited by compounds targeting this site
These compounds block infection without the risk of antibody-dependent enhancement
Acknowledgments
Stephen C. Harrison, Aaron Schmidt, Nicholas Kwiatkowski, and members of the Yang, Harrison, and Gray labs are thanked for scientific discussions. Stephen C. Harrison, Ian Walker, and Daryl Klein are also thanked for technical assistance in expression of recombinant sE proteins. We thank the following colleagues for sharing reagents: Gregory Gromowski for JEV SA14-14-2; Didier Musso and Nathalie Pardigon for ZIKV PF13-251013-18; John Aaskov for the hybridoma cell line producing Mab 6F3.1 against DENV core; Theodore Pierson for plasmids to produce DENV and WNV single-cycle reporter viruses; Lee Gehrke for DENV2 New Guinea C virus; Eva Harris for BHK-21 cells; Stephen C. Harrison for the synthetic DENV2 sE gene and cell lines. Kelly Arnett and the Center for Macromolecular Interactions and Jennifer Smith and the ICCB-Longwood Screening Facility provided access to instruments and technical support. We thank Matthew Henn, Sinead Chapman, and the Viral Genomics Sequencing group at The Broad Institute of Harvard and MIT for sequencing of dengue virus quasispecies. The Nancy Lurie Marks Medicinal Chemistry Laboratory at the Dana-Farber Cancer Institute provided access to compounds. We thank Congressman Michael Capuano and his staff for assistance importing ZIKV PF13-251013-18 virus and Wendy Jackelow for help with figure preparation. We thank Wendy Jackelow for assistance with graphic art and figure preparation. This work was supported by NIH R01AI095499 (Yang), NIH U19AI109740 (Whelan), NIH U54AI057159 (Kasper), NIH GM101135 (Engen), and a research collaboration with the Waters Corp (Engen). JDP was supported by NIH T32AI007245. JBK was supported by a Diversity Supplement to NIH R01AI095499 (Yang) and the Harvard Medical School Dean’s Postdoctoral Fellowship.
Footnotes
Declaration of Interests:
P.L.Y. and N.S.G. are co-inventors on patent application US20160031826A1.
Author Contributions
Conceptualization: PLY. Validation: MdW, WL, P-CL, JJ, SF, MJC, XZ, JBK, JDP, TEW. Formal analysis: MdW, WL, SP, P-CL, TEW. Investigation: MdW, WL, SP, P-CL, JJ, SF, MJC, JBK, JDP, TEW. Resources: JM, NSG, PLY. Data Curation: MdW, WL, JW. Writing – Original draft: PLY. Writing – Review and Editing: all authors. Visualization: SF, TEW, PLY. Supervision: JRE, JM, NSG, PLY. Funding acquisition: JRE, JM, NSG, PLY.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acosta EG, Castilla V, Damonte EB. Functional entry of dengue virus into Aedes albopictus mosquito cells is dependent on clathrin-mediated endocytosis. J Gen Virol. 2008;89:474–484. doi: 10.1099/vir.0.83357-0. [DOI] [PubMed] [Google Scholar]
- Adrián FJ, Ding Q, Sim T, Velentza A, Sloan C, Liu Y, Zhang G, Hur W, Ding S, Manley P, Mestan J, Fabbro D, Gray NS. Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nat Chem Biol. 2006;2:95–102. doi: 10.1038/nchembio760. [DOI] [PubMed] [Google Scholar]
- Alexander WM, Ficarro SB, Adelmant G, Marto JA. multiplierz v2.0: A Python-based ecosystem for shared access and analysis of native mass spectrometry data. Proteomics. 2017:17. doi: 10.1002/pmic.201700091. [DOI] [PubMed] [Google Scholar]
- Allison SL, Schalich J, Stiasny K, Mandl CW, Kunz C, Heinz FX. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J Virol. 1995;69:695–700. doi: 10.1128/jvi.69.2.695-700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansarah-Sobrinho C, Nelson S, Jost CA, Whitehead SS, Pierson TC. Temperature-dependent production of pseudoinfectious dengue reporter virus particles by complementation. Virology. 2008;381:67–74. doi: 10.1016/j.virol.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry M, Richard V, Green J, Broult J, Musso D. Inactivation of Zika virus in plasma with amotosalen and ultraviolet A illumination. Transfusion. 2016;56:33–40. doi: 10.1111/trf.13271. [DOI] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, Heinz FX, Rey FA. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 2004;23:728–738. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulich R, Aaskov JG. Nuclear localization of dengue 2 virus core protein detected with monoclonal antibodies. J Gen Virol. 1992;73:2999–3003. doi: 10.1099/0022-1317-73-11-2999. [DOI] [PubMed] [Google Scholar]
- Butrapet S, Childers T, Moss KJ, Erb SM, Luy BE, Calvert AE, Blair CD, Roehrig JT, Huang CY. Amino acid changes within the E protein hinge region that affect dengue virus type 2 infectivity and fusion. Virology. 2011;413:118–127. doi: 10.1016/j.virol.2011.01.030. [DOI] [PubMed] [Google Scholar]
- Choi Y, Seeliger MA, Panjarian SB, Kim H, Deng X, Sim T, Couch B, Koleske AJ, Smithgall TE, Gray NS. N-myristoylated c-Abl tyrosine kinase localizes to the endoplasmic reticulum upon binding to an allosteric inhibitor. J Biol Chem. 2009;284:29005–29014. doi: 10.1074/jbc.M109.026633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MJ, Miduturu C, Schmidt AG, Zhu X, Pitts JD, Wang J, Potisopon S, Zhang J, Wojciechowski A, Hann Chu JJ, et al. GNF-2 Inhibits Dengue Virus by Targeting Abl Kinases and the Viral E Protein. Cell Chem Biol. 2016;23:443–452. doi: 10.1016/j.chembiol.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro SB, Alexander WM, Marto JA. mzStudio: A Dynamic Digital Canvas for User-Driven Interrogation of Mass Spectrometry Data. Proteomes. 2017:5. doi: 10.3390/proteomes5030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromowski GD, Firestone CY, Bustos-Arriaga J, Whitehead SS. Genetic and phenotypic properties of Vero cell-adapted Japanese encephalitis virus SA14-14-2 vaccine strain variants and a recombinant clone, which demonstrates attenuation and immunogenicity in mice. The American Journal of Tropical Medicine and Hygiene. 2014;92(1):98–107. doi: 10.4269/ajtmh.14-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde D, Berkowitz SA, Engen JR. The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J Pharm Sci. 2011;100:2071–2086. doi: 10.1002/jps.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann T, Yennamalli R, Campbell P, Stoermer MJ, Fairlie DP, Kobe B, Young PR. In silico screening of small molecule libraries using the dengue virus envelope E protein has identified compounds with antiviral activity against multiple flaviviruses. Antiviral Res. 2009;84:234–241. doi: 10.1016/j.antiviral.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Kanai R, Kar K, Anthony K, Gould LH, Ledizet M, Fikrig E, Marasco WA, Koski RA, Modis Y. Crystal structure of west nile virus envelope glycoprotein reveals viral surface epitopes. J Virol. 2006;80:11000–11008. doi: 10.1128/JVI.01735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilby JM, Hopkins S, Venetta TM, DiMassimo B, Cloud GA, Lee JY, Alldredge L, Hunter E, Lambert D, Bolognesi D, et al. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- Kirschner AN, Lowrey AS, Longnecker R, Jardetzky TS. Binding-site interactions between Epstein-Barr virus fusion proteins gp42 and gH/gL reveal a peptide that inhibits both epithelial and B-cell membrane fusion. J Virol. 2007;81:9216–9229. doi: 10.1128/JVI.00575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan MN, Sukumaran B, Pal U, Agaisse H, Murray JL, Hodge TW, Fikrig E. Rab 5 is required for the cellular entry of dengue and West Nile viruses. J Virol. 2007;81:4881–4885. doi: 10.1128/JVI.02210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Weir RC, Dalgarno L. Changes in the dengue virus major envelope protein on passaging and their localization on the three-dimensional structure of the protein. Virology. 1997;232:281–290. doi: 10.1006/viro.1997.8570. [DOI] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J Virol. 2005;79:1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken GE, Nelson CA, Chen BR, Diamond MS, Fremont DH. Crystal structure of the West Nile virus envelope glycoprotein. J Virol. 2006;80:11467–11474. doi: 10.1128/JVI.01125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, Sanchez MD, Puffer BA, Ahmed AA, Geiss BJ, Valentine LE, Altamura LA, Diamond MS, Doms RW. A rapid and quantitative assay for measuring antibody-mediated neutralization of West Nile virus infection. Virology. 2006;346:53–65. doi: 10.1016/j.virol.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Poh MK, Yip A, Zhang S, Priestle JP, Ma NL, Smit JM, Wilschut J, Shi PY, Wenk MR, Schul W. A small molecule fusion inhibitor of dengue virus. Antiviral Res. 2009;84:260–266. doi: 10.1016/j.antiviral.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Porotto M, Doctor L, Carta P, Fornabaio M, Greengard O, Kellogg GE, Moscona A. Inhibition of hendra virus fusion. J Virol. 2006;80:9837–9849. doi: 10.1128/JVI.00736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Ovadia M, Shai Y. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 1995;14:5524–5531. doi: 10.1002/j.1460-2075.1995.tb00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG. Inhibition by Small Molecules and Peptides of Dengue-virus Fusion and Entry. Harvard University; Boston, MA: 2011. [Google Scholar]
- Schmidt AG, Lee K, Yang PL, Harrison SC. Small-molecule inhibitors of dengue-virus entry. PLoS Pathog. 2012;8:e1002627. doi: 10.1371/journal.ppat.1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Yang PL, Harrison SC. Peptide inhibitors of dengue-virus entry target a late-stage fusion intermediate. PLoS Pathog. 2010;6:e1000851. doi: 10.1371/journal.ppat.1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukupolvi-Petty S, Austin SK, Engle M, Brien JD, Dowd KA, Williams KL, Johnson S, Rico-Hesse R, Harris E, Pierson TC, et al. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J Virol. 2010;84:9227–9239. doi: 10.1128/JVI.01087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Schaar HM, Rust MJ, Chen C, van der Ende-Metselaar H, Wilschut J, Zhuang X, Smit JM. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 2008;4:e1000244. doi: 10.1371/journal.ppat.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Schaar HM, Rust MJ, Waarts BL, van der Ende-Metselaar H, Kuhn RJ, Wilschut J, Zhuang X, Smit JM. Characterization of the early events in dengue virus cell entry by biochemical assays and single-virus tracking. J Virol. 2007;81:12019–12028. doi: 10.1128/JVI.00300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol Cell Proteomics. 2002;1:132–138. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- Wang QY, Patel SJ, Vangrevelinghe E, Xu HY, Rao R, Jaber D, Schul W, Gu F, Heudi O, Ma NL, et al. A small-molecule dengue virus entry inhibitor. Antimicrob Agents Chemother. 2009;53:1823–1831. doi: 10.1128/AAC.01148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yennamalli R, Subbarao N, Kampmann T, McGeary RP, Young PR, Kobe B. Identification of novel target sites and an inhibitor of the dengue virus E protein. J Comput Aided Mol Des. 2009;23:333–341. doi: 10.1007/s10822-009-9263-6. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Khaliq M, Suk JE, Patkar C, Li L, Kuhn RJ, Post CB. Antiviral compounds discovered by virtual screening of small-molecule libraries against dengue virus E protein. ACS Chem Biol. 2008;3:765–775. doi: 10.1021/cb800176t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Deuterium uptake plots comparing relative deuterium levels measured for the wild-type DENV2 sE (black circles) and sE-M196V (red squares), DENV sE-M272S (blue squares), and DENV sE-Q52A (green squares), related to Figure 4.