Abstract
Purpose
To develop a three dimensional adiabatic T1ρ prepared ultrashort echo time cones sequence (3D AdiabT1ρ UTE-Cones) for whole knee imaging on a clinical 3T scanner.
Methods
A train of adiabatic full passage pulses were used for spin locking, followed by time-efficient multispoke UTE acquisition to detect signals from both short and long T2 tissues in the whole knee joint. A modified signal model was proposed for multispoke UTE data fitting. The feasibility of this 3D AdiabT1ρ UTE-Cones technique was demonstrated through numerical simulation, phantom and ex vivo knee sample studies. The 3D AdiabT1ρ UTE-Cones technique was then applied to six in vivo knee joints of healthy volunteers to measure T1ρ values of quadriceps tendon, patellar tendon, anterior cruciate ligament (ACL), posterior cruciate ligament (PCL), meniscus, patellar cartilage and muscle.
Results
Numerical simulation, phantom and ex vivo knee sample studies demonstrated the feasibility of whole knee imaging using the proposed multispoke 3D AdiabT1ρ UTE-Cones sequence. The healthy volunteer knee study demonstrated an averaged T1ρ of 13.9±0.7 ms for the quadriceps tendon, 9.7±0.8 ms for the patellar tendon, 34.9±2.8 ms for the ACL, 21.6±1.4 ms for the PCL, 22.5±1.9 ms for the meniscus, 44.5±2.4 ms for the patellar cartilage and 43.2±1.1 ms for the muscle.
Conclusion
The 3D AdiabT1ρ UTE-Cones sequence allows volumetric T1ρ assessment of both short and long T2 tissues in the knee joint on a clinical 3T scanner.
Keywords: AdiabT1ρ, ultrashort echo time, multispoke, whole knee imaging
Introduction
Quantitative magnetic resonance imaging (MRI) of spin lattice relaxation in the rotating frame (T1ρ) has been proposed as a biomarker of cartilage degeneration (1–4). There is clinical interest in developing noninvasive biomarkers that are sensitive to the early degenerative changes in cartilage, including loss of proteoglycans (PGs) and changes in collagen, for the early diagnosis of osteoarthritis (OA) (1). It has been shown that T1ρ increases with cartilage degeneration (5–7), and spin lock at different frequencies has been used to detect changes in proteoglycans (PGs) or collagen (8–9).
A strong magic angle effect is an important limitation of quantitative continuous-wave (CW) T1ρ imaging of collagen-rich tissues such as cartilage, menisci and ligaments (10–13). The highly ordered collagen fibers in these tissues are subject to strong dipole-dipole interactions which are modulated by the term 3cos2(θ) − 1, where θ is the angle between the fiber orientation and the main magnetic field (14). Previous studies show that T1ρ values can increase more than 200% in the middle and deep zones of articular cartilage, and 300% in ligaments, when θ is oriented from 0° to 55° (12, 13). The significant T1ρ changes due to the magic angle effect make the evaluation of tissue degeneration extremely complicated.
Recently, a novel imaging technique was developed in which trains of adiabatic full passage (AFP) pulses are used to generate T1ρ relaxation (AdiabT1ρ) (15–21). AdiabT1ρ has been reported to be less sensitive to the magic angle effect compared with both CW-T1ρ and T2 relaxations in bovine cartilage studies (19, 20). Thus, AdiabT1ρ may be a more reliable biomarker of PG loss in collagen-rich tissues than conventional CW-T1ρ. In addition, AdiabT1ρ has other advantages over CW spin-lock sequences. Most notably, adiabatic pulses are less sensitive to the spatial inhomogeneity of the transmit radio-frequency (RF) magnetic field compared with CW spin-lock pulses, and the flexibility of AFP pulse design allows moderation of RF power deposition (15, 18, 20, 22). Moreover, an extended range of frequencies or correlation times are effectively involved in the spin lattice relaxation when using AFP pulses, which may provide more information on the physicochemical mechanisms underlying pathological changes in tissues.
Human knee joints are composed of many different tissues including articular cartilage, calcified cartilage, menisci, ligaments, tendons and bone, all of which are important for the health of the joint (23–25). However, both CW-T1ρ and AdiabT1ρ measurements based on conventional MRI pulse sequences (such as GRE and FSE) are of limited value for detecting early PG depletion in short T2 tissues or tissue components such as the deep radial and calcified cartilage, menisci, ligaments and tendons. These tissues or tissue components typically have T2s ranging from sub-milliseconds to several milliseconds and thus provide little or no detectable signal using conventional sequences (26–29).
To overcome this challenge, we propose a combination of a three dimensional ultrashort echo time sequence employing cones trajectories with an AdiabT1ρ preparation (3D AdiabT1ρ UTE-Cones) for volumetric T1ρ assessment of both short and long T2 tissues in the knee joint on a clinical 3T scanner. The details of 3D UTE-Cones sequence was described in the recent publications (30, 31). Multispoke acquisition after each AdiabT1ρ preparation was incorporated for time-efficiency. A modified signal model for multispoke acquisition was proposed for accurate T1ρ fitting. Both simulation and phantom studies were carried out to investigate the accuracy of the modified signal model. Next, the magic angle effect was investigated by the repeated imaging of a sliced human patellar cartilage sample at five angular orientations from 0° to 90° relative to the field. Finally, the new sequence was applied to four ex vivo human knee joint specimens and six in vivo knee joints of healthy volunteers for T1ρ measurements of quadriceps tendon, patellar tendon, anterior cruciate ligament (ACL), posterior cruciate ligament (PCL), meniscus, patellar cartilage and muscle.
Theory
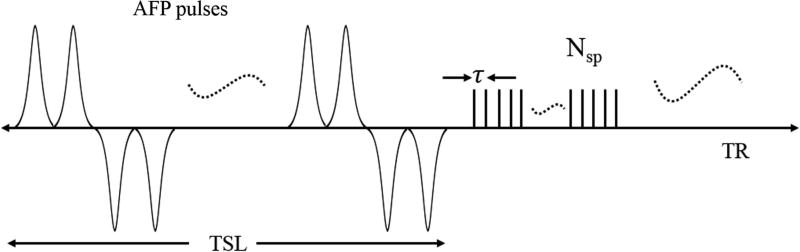
Features of the 3D AdiabT1ρ UTE-Cones pulse sequence used in this study are shown in Figure 1. An even number (NAFP) of AFP pulses are used for AdiabT1ρ preparation. When NAFP is a multiple of four, then every four consecutive AFP pulses follow a MLEV4 phase cycling scheme (32). When NAFP is equal to 4n+2 (n = 0, 1, 2 …), the first 4n AFP pulses follow a MLEV4 phase cycling scheme, and the amplitude of the remaining two AFP pulses can be arbitrarily positive or negative because the AFP pulse can invert the spins robustly when the adiabatic condition is satisfied (15). Here we use two positive AFP pulses. Following the AdiabT1ρ preparation are Nsp separate k-space spokes or acquisitions with an equal time interval τ for fast data acquisition.
Figure 1.
The 3D AdiabT1ρ UTE-Cones sequence employed a train of AFP pulses to generate T1ρ contrast, followed by 3D UTE-Cones data acquisition. To speed up data acquisition, multiple spokes were sampled after each AFP pulse train.
The spin lock time TSL is defined as the total duration of the train of AFP pulses, i.e. TSL = NAFP×Tp (Tp is the duration of a single AFP pulse). TR defined in this study is the duration between the adjacent AdiabT1ρ preparations. A relatively short TR (e.g. several hundred milliseconds) is used in the proposed sequence to accelerate data acquisition. At steady state, the signal equation is expressed as follows when a single acquisition (Nsp = 1) is obtained after AdiabT1ρ preparation (27):
| [1] |
Where M0 is the equilibrium state magnetization and α is the excitation flip angle. A constant C is induced to account for non-T1ρ related factors such as background noise and artifacts associated with data acquisition and image reconstruction.
In our previous conventional CW-T1ρ study with five spokes per T1ρ preparation, acceptable T1ρ values were obtained by fitting the single spoke acquisition equation (Eq. [1]) (29). However, Eq. [1] will introduce increasing error as the number of excitation spokes per AdiabT1ρ preparation increases (i.e. Nsp > 5) because it does not model the saturation effect induced by the multiple excitations. Therefore, similar to our recent multispoke MT modeling study, we modified the single spoke equation Eq. [1] by simply changing cos(α) to cosNsp(α) to account for the saturation effect of Nsp acquisitions, which is expressed as follows (33):
| [2] |
The motivation for Eq. [2] can be understood when considering a tissue with very long T1 relative to the spoke time interval τ. In this case, the longitudinal magnetization will not substantially recover between consecutive excitations, resulting in progressive saturation between spokes. Thus, the net behavior of the multispoke excitations is analogous to a single excitation with a flip angle of acos(cosNsp(α)). In contrast, Eq. [1] assumes a short enough T1 for the longitudinal magnetization to fully recover between each spoke. In practice, the T1 values of most tissues are much longer than the spoke time interval τ (around 5 ms), so the signal model of Eq. [2] would be preferred. The two models were compared by simulation and phantom studies.
For both Eqs. [1] and [2], accurate T1 measurement is crucial for T1ρ calculation. Here a 3D UTE-Cones variable flip angle (VFA) method was used to measure T1 by fitting the following equation (34):
| [3] |
Where φ is the flip angle and TR is the repetition time. However, the VFA technique is very sensitive to B1 inhomogeneity. Thus, a 3D dual-TR UTE-Cones sequence was also developed for actual flip angle imaging (UTE-Cones AFI) (35) to obtain a B1 scaling factor by dividing the measured actual flip angle by the nominal flip angle. With a known B1 scaling factor, the flip angle φ in Eq. [3] can be corrected for accurate T1 fitting. The B1 scaling factor value can also be used to correct α in both Eqs. [1] and [2] for accurate T1ρ measurement. No B1 correction is needed for AdiabT1ρ preparation since the AFP pulses are insensitive to B1 inhomogeneity.
Methods
The 3D AdiabT1ρ UTE-Cones sequence (see Fig. 1) was implemented on a 3T whole body scanner (GE Healthcare Technologies, Milwaukee, WI). An 8-channel transmit/receive knee coil was used for both RF transmission and signal reception in the following experiments except as noted. The 3D UTE-Cones sequence used unique k-space trajectories that sampled data along evenly spaced twisting paths in the shape of multiple cones (31, 32). Data acquisition started as soon as possible after the RF excitation with a minimal nominal echo time of 32 µs. The nominal echo time is defined as the time between the end of the rectangular pulse and the k-space center. Both RF and gradient spoiling were used to crush the remaining transverse magnetizations after each data acquisition. The RF spoiling method used here is to increase the RF phase quadratically with a phase increment factor of 117°. The T2 weighted transverse magnetizations were crushed and will not contribute to steady state signals. Identical non-selective AFP pulses (hyperbolic secant type 1 pulse) with a duration of 6.048 ms, bandwidth of 1.643 kHz and maximum B1 amplitude of 17 µT were used to generate T1ρ contrast (36). Here, we used the shortest AFP pulse which can satisfy the adiabatic condition to increase the pulse bandwidth and get adequate TSLs for short T2 tissue imaging. A gradient following the train of AdiabT1ρ pulses was used to crush the remaining transverse magnetizations. The 3D AdiabT1ρ UTE-Cones sequence allows for anisotropic resolution (e.g., high in-plane resolution and thicker slices) for much improved SNR and reduced scan time relative to isotropic imaging (32, 36).
Simulation
Numerical simulation was carried out to investigate the accuracy of the fitting models of Eqs. [1] and [2] for multispoke acquisition. The simulated signal intensity of AdiabT1ρ preparation followed the mono-exponential function of e−TSL/T1ρ (18). The T1 value was set to 1000 ms and the AdiabT1ρ UTE-Cones sequence parameters were shown as follows: TR = 500 ms, excitation flip angle = 10°, acquisition interval between adjacent spokes τ = 5 ms, the gap between the end of the last AFP pulse and start of the excitation pulse was 8 ms, each AFP pulse duration Tp = 6 ms, and 8 different groups of AFP pulses in AdiabT1ρ preparation with NAFP = 0, 2, 4, 6, 8, 12 and 16. Eleven groups of data were generated by Bloch equation simulation with different numbers of acquisition spokes: Nsp = 1, 5, 10, 15, 20, 25, 30, 35, 40, 45 and 50. Both Eqs. [1] and [2] were used for data fitting.
Phantom Study
The phantom was prepared as 2% w/v agarose gel containing 0.1 mM MnCl2. In addition to the proposed 3D AdiabT1ρ UTE-Cones sequence, a 3D UTE-Cones AFI sequence and VFA UTE-Cones sequence were employed for B1 mapping and T1 measurement, respectively. B1 maps were used to correct both T1 and T1ρ calculation. The phantom was scanned using the same field of view (FOV) of 12 × 12 × 8 cm3 and receiver bandwidth of 166 kHz for all sequences. Other sequence parameters were: 1) 3D UTE-Cones AFI: TR1/TR2 = 20/100 ms, flip angle = 45°, acquisition matrix of 64 × 64 × 20 and a total scan time of 5 min 30 sec; 2) 3D VFA UTE-Cones: TR = 20 ms, flip angle = 5°, 10°, 20° and 30°, acquisition matrix of 128 × 128 × 20 and a total scan time of 5 min 48 sec; 3) 3D AdiabT1ρ UTE-Cones: TR = 500 ms, flip angle = 10°, acquisition matrix of 128 × 128 × 20 and NAFP = 0, 2, 4, 6, 8, 12 and 16. Six groups of AdiabT1ρ data were acquired with different Nsp of 1, 5, 15, 25, 35 and 45 per AdiabT1ρ preparation. The corresponding scan times were 253 min 59 sec, 50 min 52 sec, 17 min 23 sec, 10 min 37 sec, 7 min 49 sec and 6 min 11 sec, respectively. Reproducibility of the proposed AdiabT1ρ method was investigated using the agarose phantom. The protocol was repeated four times with the MR system reset before each 3D AdiabT1ρ UTE-Cones scan.
Magic Angle Effect Study
Magic angle effect study for both T1ρ and T2 was carried out by imaging a sliced human patellar sample (around 4 mm thick) using a wrist coil (BC-10, Medspira, Minneapolis, MN) for both RF transmission and signal reception. The sample was imaged with five angular orientations (i.e. 0°, 30°, 55°, 70° and 90°) with respect to the normal direction of cartilage surface. For each angular orientation, 3D AdiabT1ρ UTE-Cones and 2D Carr-Purcell-Meiboom-Gill (CPMG) sequences were used for T1ρ and T2 measurements, respectively. The CPMG sequence is a multiple spin echo sequence to acquire a series of data with different echo times to quantitate T2 value. The phase encoding and phase rewinder gradients in this sequence are balanced to refocus the stimulated echoes from imperfect 180° pulses at the same time as the spin echoes. Since tissue T1 is not sensitive to the magic angle effect, T1 measurement with a 2D inversion recovery prepared fast spin echo (IR-FSE) sequence was performed only at angle 0°. A chemical shift saturation (FatSat) module (8 ms) located between AdiabT1ρ preparation and the data acquisition was employed for fat suppression. The sequence parameters were: 1) 3D AdiabT1ρ UTE-Cones with FatSat: FOV = 5 × 5 × 2 cm3, acquisition matrix of 128 × 128 × 10, receiver bandwidth = 83.3 kHz, TR = 1000 ms, flip angle = 10°, Nsp = 5 and NAFP = 0, 4, 8, 12, 16 and 20 each with a scan time of 6 min 45 sec; 2) 2D CPMG with FatSat: FOV = 5 × 5 cm2, acquisition matrix of 192 × 128, slice thickness = 4 mm, TR = 3000 ms, flip angle = 90°, TEs = 10.7, 21.3, 32.0, 43.7, 53.4, 64.0, 74.7 and 85.4 ms; 3) 2D IR-FSE: FOV = 5 × 5 cm2, acquisition matrix of 192 × 128, slice thickness = 4 mm, TR = 5000 ms, flip angle = 90°, TEs = 10.7 ms, TIs = 50, 150, 300, 500, 700, 1000, 1500, 2000 and 3000 ms.
Ex Vivo Knee Study
High resolution whole knee imaging was performed on four knee samples from four donors (aged 51–79 years, mean age 61.5 years; 1 male, 3 females). 3D UTE-Cones AFI, VFA and AdiabT1ρ UTE-Cones sequences were used to scan these knee samples using a common FOV of 15 × 15 × 12 cm3 and receiver bandwidth of 166 kHz. Other sequence parameters were: 1) 3D UTE-Cones AFI: TR1/TR2 = 20/100 ms, flip angle = 45°, acquisition matrix of 128 × 128 × 30 and a total scan time of 10 min 54 sec; 2) 3D VFA UTE-Cones: TR = 24 ms, flip angle = 4°, 8°, 16°, 24°, 32° and 40°, acquisition matrix of 256 × 256 × 60 and a total scan time of 8 min 19 sec; 3) 3D AdiabT1ρ UTE-Cones with FatSat: TR = 500 ms, flip angle = 10°, acquisition matrix of 256 × 256 × 60, Nsp = 21 and NAFP = 0, 2, 4, 6, 8, 12, 16 and 20 each with a scan time of 8 min 19 sec.
In Vivo Knee Study
In vivo whole knee imaging was carried out on six healthy volunteers (aged 23–42 years, mean age 30.3 years; 4 males, 2 females). Informed consent was obtained from all subjects in accordance with guidelines of the institutional review board. 3D UTE-Cones AFI, VFA and AdiabT1ρ UTE-Cones sequences were used to scan these knees, using a common FOV of 15 × 15 × 10.8 cm3 and receiver bandwidth of 166 kHz. Other sequence parameters were: 1) 3D UTE-Cones AFI: TR1/TR2 = 20/100 ms, flip angle = 45°, acquisition matrix of 128 × 128 × 18, acquisition stretch factor of 1.4 and a total scan time of 4 min 57 sec; 2) 3D VFA UTE-Cones: TR = 20 ms, flip angle = 5°, 10°, 20° and 30°, acquisition matrix of 256 × 256 × 36, undersampling factor of 0.9, acquisition stretch factor of 1.4 and a total scan time of 9 min 28 sec; 3) 3D AdiabT1ρ UTE-Cones with FatSat: TR = 500 ms, flip angle = 10°, acquisition matrix of 256 × 256 × 36, acquisition stretch factor of 1.6, Nsp = 25 and NAFP = 0, 2, 4, 6, 8, 12 and 16 each with a scan time of 2 min 34 sec.
Data Analysis
The Levenberg-Marquardt algorithm was used to fit Eqs. [1] to [3]. All analysis algorithms were written in Matlab (The MathWorks Inc., Natick, MA, USA) and were executed offline on the DICOM images obtained by the acquisition protocols described above. For each fitting of Eqs. [1] and [2], both the T1ρ value and its fitting error were calculated. ROIs were manually drawn for various tissues including quadriceps tendon, patellar tendon, ACL, PCL, meniscus, patellar cartilage and muscle in all ex vivo and in vivo knee joints.
Results
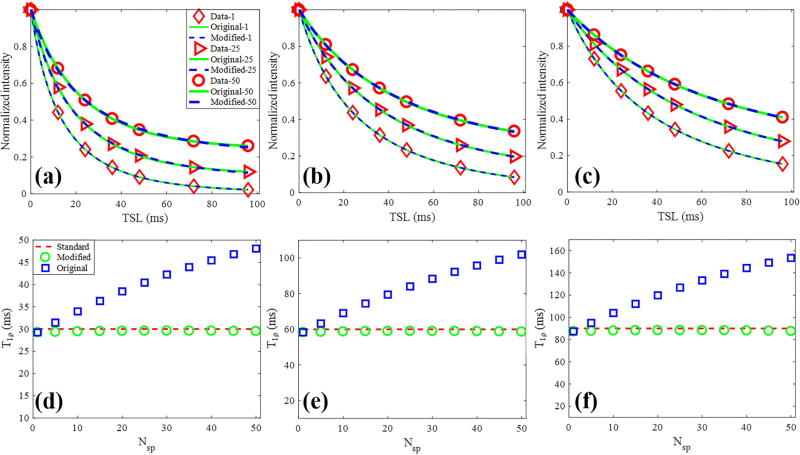
Numerical simulations for the fitting model study are shown in Figure 2. As shown in Figs. 2a to 2c, the Bloch equation simulated data with different Nsp per AdiabT1ρ preparation have different signal intensities. The data fitting with the original model of Eq. [1] and modified model of Eq. [2] were excellent in all simulations. However, as shown in Figs. 2d to 2f, the calculated T1ρ values using the original model were dependent on the Nsp. As expected, the calculated T1ρ values increased with higher Nsp using the original model. In contrast, T1ρ values calculated by the modified model were very close to the true simulated value independent of the Nsp, suggesting improved accuracy of the modified model.
Figure 2.
Comparison of multispoke fitting models by simulation. (a) – (c) Bloch equation simulated data with corresponding fitting curves of both original (Eq. [1]) and modified (Eq. [2]) signal models. Each series data with a specific Nsp was normalized. The red diamond, triangle, circle markers represented the simulated data with 1, 25 and 50 spokes per AdiabT1ρ preparation, respectively. (d) – (f) Calculated T1ρ values by the original and modified model as Nsp increases from 1 to 50. The red dashed line highlights the simulated T1ρ, and the blue squares and green circles represent the T1ρ values obtained from fitting the original and modified models, respectively. The columns represented three groups data with simulated T1ρ values of 30 (a and d), 60 (b and e) and 90 ms (c and f).
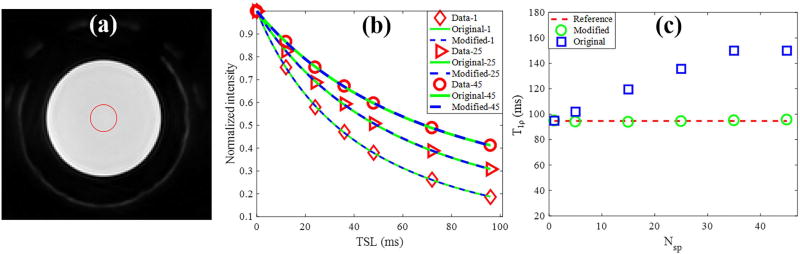
Figure 3 shows the agarose phantom results for the fitting model study. The data acquisition time was significantly reduced when a higher Nsp was used. As shown in Figs. 3b, the phantom data acquired with different Nsp per AdiabT1ρ preparation have different signal intensities, similar to the simulation study. The obtained T1ρ values calculated by the original model significantly increased with a higher Nsp. In contrast, the T1ρ values calculated by the modified model only increased very slightly with a higher Nsp, which further demonstrated the accuracy of the modified model. The average coefficient of variation for the AdiabT1ρ UTE-Cones scan of the agarose phantom on four repeated acquisitions was less than 3%, demonstrating good reproducibility of the technique.
Figure 3.
3D AdiabT1ρ UTE-Cones of a 2% agarose phantom with 0.1 mM MnCl2. (a) The region of interest for analysis is shown as the red circle on a selected AdiabT1ρ image (Nsp = 25, NAFP = 4). (b) Original and modified signal model fitting. The red diamond, triangle, circle markers represent the phantom data with 1, 25 and 45 spokes per AdiabT1ρ preparation, respectively. (c) Calculated T1ρ values by the original and modified models as Nsp increases from 1 to 50. The red dashed line is the reference T1ρ obtained with Nsp = 1, and the blue squares and green circles in represent the T1ρ values obtained from the fitting by original and modified model, respectively.
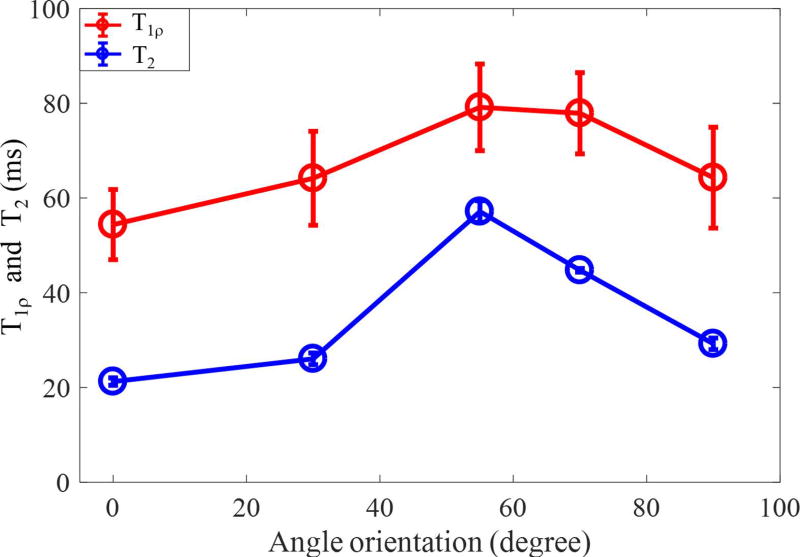
Supporting Figure S1 shows the magic angle effect in AdiabT1ρ UTE-Cones imaging of a sliced patellar sample. Significant signal intensity changes can be found in the localizer images with different angular orientations between the normal direction of cartilage surface and B⃗0. Excellent fitted curves using the modified signal model were obtained for each angle. Figure 4 shows how the calculated T1ρ values vary with orientation angle, with CPMG-derived T2 values included for comparison. While the T2 value increased by approximately 200 % as the angle increased from 0° to 55°, the calculated T1ρ values only increased by approximately 50%.
Figure 4.
Comparison of magic angle effect for cartilage T1ρ values from 3D AdiabT1ρ UTE-Cones and T2 values from a CPMG sequence.
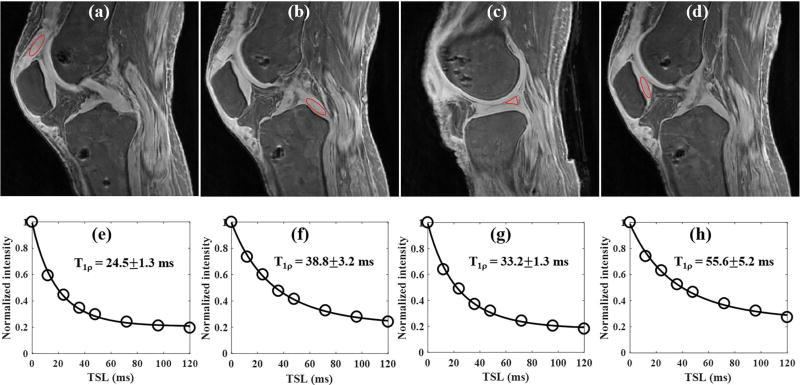
Figure 5 shows the results of the ex vivo whole knee study of a 63 year old female donor. The 3D AdiabT1ρ UTE-Cones sequence provides high signal and contrast imaging of long T2 tissues such as the articular cartilage and muscle, together with short T2 tissues of meniscus, quadriceps tendon, patellar tendon, ACL and PCL (Figs. 5a to 5c). Excellent T1ρ fitting of the 3D AdiabT1ρ UTE-Cones images with different TSLs (Figs. 5d to 5f) demonstrates a T1ρ of 24.5±1.3 ms for the quadriceps tendon, 38.8±3.2 ms for the PCL, 33.2±1.3 ms for the meniscus and 55.6±5.2 ms for the patellar cartilage.
Figure 5.
3D AdiabT1ρ UTE-Cones imaging of an ex vivo knee sample (63 year old female donor). Representative AdiabT1ρ images with regions of interest (red circles) and corresponding fitting curves of quadriceps tendon, PCL, meniscus and patellar cartilage are shown in the first and second rows, respectively. The T1ρ values of quadriceps tendon, PCL, meniscus and patellar cartilage were 24.5±1.3, 38.8±3.2, 33.2±1.3 and 55.6±5.2 ms, respectively.
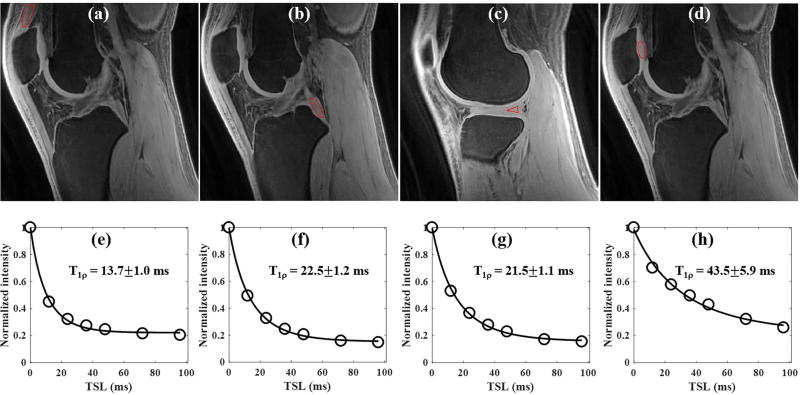
Figure 6 shows the results of an in vivo whole knee study of a 23 year old male volunteer. Similar to the ex vivo sample study, the 3D AdiabT1ρ UTE-Cones sequence also provides high signal and contrast imaging of both short and long T2 tissues in the whole knee joint (Figs. 6a to 6c). Excellent AdiabT1ρ fitting of the 3D AdiabT1ρ UTE-Cones images with different TSLs (Figs. 6d to 6f) demonstrates a T1ρ of 13.7±1.0 ms for the quadriceps tendon, 22.5±1.2 ms for the PCL, 21.5±1.1 ms for the meniscus and 43.5±5.9 ms for the patellar cartilage.
Figure 6.
3D AdiabT1ρ UTE-Cones imaging of an in vivo healthy knee (23 year old male volunteer). Representative AdiabT1ρ images with regions of interest (red circles) and corresponding fitting curves of quadriceps tendon, PCL, meniscus and patellar cartilage are shown in the first and second rows, respectively. The T1ρ values of quadriceps tendon, PCL, meniscus and patellar cartilage were 13.7±1.0, 22.5±1.2, 21.5±1.1 and 43.5±5.9 ms, respectively.
Table 1 summarizes the T1ρ values for quadriceps tendon, patellar tendon, ACL, PCL, meniscus, patellar cartilage and muscle for the four ex vivo knee images and the six in vivo knee images. Relatively consistent T1ρ values were derived for each knee tissue within each experiment, although the ex vivo T1ρ values are consistently higher than the corresponding in vivo T1ρ values.
Table 1.
T1ρ and its fitting standard errors (ms) of quadriceps tendon, patellar tendon, ACL, PCL, meniscus, patellar cartilage and muscles in four ex vivo human knee samples as well as their average and standard deviation values (ms).
| Ex vivo knee | # 1 | # 2 | # 3 | # 4 | Average |
|---|---|---|---|---|---|
| Quadriceps tendon | 24.5±1.3 | 21.9±1.8 | 23.8±2.0 | 19.5±1.5 | 22.5±2.2 |
| Patellar tendon | 22.5±1.5 | 26.4±3.1 | 23.2±2.5 | 17.5±1.7 | 22.4±3.7 |
| ACL | 45.8±2.0 | 53.2±8.9 | 49.3±5.3 | 45.2±5.4 | 48.4±3.7 |
| PCL | 38.8±3.2 | 38.9±4.5 | 44.3±3.9 | 38.5±4.0 | 40.1±2.8 |
| Meniscus | 33.2±1.3 | 36.8±3.7 | 33.7±2.9 | 34.8±3.6 | 34.6±1.6 |
| Patellar cartilage | 55.6±5.2 | 59.7±9.8 | 58.7±10.2 | 44.3±6.4 | 54.6±7.0 |
| Muscle | 60.9±4.1 | 55.3±8.1 | 58.6±6.4 | 51.8±6.3 | 56.7±4.0 |
Discussion
We have demonstrated in this study that the proposed 3D AdiabT1ρ UTE-Cones sequence can provide reliable volumetric T1ρ assessment of both short and long T2 tissues in whole knee imaging on a clinical 3T scanner. Our simulation and phantom studies suggest that the modified signal model is more preferred for the time-efficient multispoke acquisition than the original signal model. Furthermore, the magic angle study using a sliced human patellar cartilage sample demonstrated that the T1ρ values generated from the 3D AdiabT1ρ UTE-Cones technique were much less sensitive to the magic angle effect than CPMG-derived T2 values, and compared with CW-T1ρ values from previous studies (12, 13). Our ex vivo and in vivo whole knee studies demonstrate its feasibility in quantifying T1ρ for quadriceps tendon, patellar tendon, ACL, PCL, meniscus, patellar cartilage and muscle.
Knee OA is recognized as a whole organ disease. Previous studies have shown that failure of any component, such as meniscal positioning or collateral ligament damage, can lead to cartilage loss (24, 25, 37). In general, the deterioration or misalignment of any of the tissues comprising the knee joint can accelerate the progression of OA (23–25, 37). As such, it is essential to image all major components in the knee joint to allow for comprehensive assessment of OA. In this study, we demonstrate that the 3D AdiabT1ρ UTE-Cones sequence can image and calculate T1ρ values for all the major components in the knee joint including both short and long T2 tissues.
Recent studies have shown that AdiabT1ρ is sensitive to both ex vivo enzymatic cartilage degradation and in vivo articular cartilage degradation in OA patients (18, 38, 39). More importantly, the AdiabT1ρ is much less sensitive to the magic angle effect than both conventional CW-T1ρ and T2 as demonstrated in previous bovine cartilage studies (19, 20). Our human patellar cartilage study extends these findings to 3D UTE-Cones adapted AdiabT1ρ sequences. The combination of T1ρ preparation with UTE sequences can also be used to quantify other clinically meaningful short T2 tissues in the knee joint such as the meniscus (40–42). The meniscus plays an important role in normal knee function, and there is high interest in evaluating degenerative changes in the meniscus with T1ρ sequences (40–42). For example, Rauscher et al. reported a high correlation between meniscal T1ρ and clinical findings of OA, suggesting the importance of T1ρ imaging of the meniscus (40). However, only long T2 components in the meniscus could be quantified in that study because they utilized clinical gradient echo sequences with TEs around 4 ms, which are too long to detect the short T2 components that comprise a significant proportion of the meniscus. Therefore, the 3D AdiabT1ρ UTE-Cones sequence is likely to provide a more accurate assessment of cartilage degeneration in the meniscus compared to the magic angle sensitive CW-T1ρ sequence based on conventional gradient echo sequences (40–42).
In addition to lower RF power deposition compared with CW-T1ρ prepared sequences, the proposed AdiabT1ρ UTE-Cones sequence is more resistant to both B1 and B0 inhomogeneities because of the adiabatic pulse character and relatively broad pulse spectral coverage of 1.643 kHz. Thus, AdiabT1ρ prepared sequences will also be preferred over CW-T1ρ prepared sequences in high field MRI where these effects are more significant. In addition to the HS1 type of AFP pulse used in this study, other RF types such as HS4 and HS8 can also be designed for AdiabT1ρ preparation (15, 18, 20). Different T1ρ characters can be generated by different types of RF pulses. For example, T1ρ generated by HS4 type pulse trains is slightly more sensitive to cartilage degeneration but also more sensitive to the magic angle effect compared with the T1ρ generated by HS1 type pulse trains (20).
Significant scan time reduction for the 3D AdiabT1ρ UTE-Cones sequence was achieved using multispoke data acquisition in this study. Both simulation and phantom studies demonstrated that the modified signal model of Eq. [2] was appropriate for T1ρ fitting by incorporating saturation effects during the multispoke acquisition. A low flip angle was used for signal excitation to avoid image artifacts induced by the signal intensity variations among the acquisition spokes. As can be seen from both phantom (Figs. 3) and knee data (Figs. 5 and 6), an interesting phenomenon was observed: the signal intensity of the data acquired with an AdiabT1ρ preparation of a non MLEV4 phase scheme (e.g. NAFP = 2 or 6) was still located properly along the fitting curve. This suggests that heteronuclear decoupling with a MLEV4 phase scheme is not necessary for AdiabT1ρ contrast generation.
There is an important difference between our sequence and the sequence recently reported by Zhang et al. for 3D adiabatic T1ρ mapping (21). In our sequence, in order to increase the SNR, there is a gap between the end of the acquisition spoke series and the next AdiabT1ρ preparation. However, no such gap exists in Zhang et al.’s sequence because they used a high field animal scanner which has much better SNR performance. In addition, while Zhang et al. used a semi-analytical approach, we used a simplified model since the accurate signal equation can be even more complicated than Zhang et al.’s signal equation. However, as demonstrated by both simulation and phantom studies, we can still get good fitting results with our simplified signal model. As shown in the fitting curves in both Figs. 5 and 6, the data of all the tissues in the knee fit the mono-exponential model well. Different proton pools in a tissue are likely to have different T1ρ values, and in particular, the extremely short T2 proton components may have shorter T1ρ values. However, AFP pulses cannot be made short enough to get the AdiabT1ρ signal decay curve for the extremely short T2 components due to the limitation of both RF peak power and specific absorption rate (SAR) levels in clinical scanners. A relatively high peak B1 value was used for the AFP pulses in order to shorten the RF duration so that more signals from the short T2 tissues (such as meniscus) can be acquired. SAR will be increased due to the relatively high RF power. However, the SAR level of the proposed protocol is still in the safe range for extremity imaging where the use of multispokes and a transmit/receive 8-channel knee coil help reduce SAR.
The FatSat pulse will saturate part of tissue magnetizations especially for short T2 tissues, leading to reduced SNR. However, in UTE imaging with radial or spiral sampling, chemical shift artifacts manifest as ringing artifacts, which will affect the quantitative measurement. Therefore, it is preferred to lose some image SNRs using a FatSat pulse rather than get inaccurate T1ρ values. Fat saturation time was counted in the non-spin-lock time (i.e. TR - TSL) in the equation. Moreover, with Bloch simulation, we found that placing the fat saturation time between the AdiabT1ρ preparation and the acquisition spokes has similar fitting results compared to when we placed the fat saturation time right before the AdiabT1ρ preparation. This is because the fat saturation time is much shorter than TR in this study. Additionally, coil ring-down time should be considered in the definition of the shortest echo time, which is very important for accurate T2* quantification of short T2 tissues. However, the majority of UTE papers published so far have used the definition of TE as the time between the end of the short rectangular pulse and the start of k-space center. We have chosen to follow that convention in this paper. Furthermore, since we used the same echo time for all the AdiabT1ρ preparations, the image contrasts are mainly generated by these AdiabT1ρ preparations. Therefore, even when contaminated with coil ring-down effects, we can still get accurate quantitative T1ρ values.
In general, as can be seen from Tables 1 and 2, the T1ρ values of the tissues measured in the in vivo knee study were consistently lower than the values in the ex vivo knee study. These differences likely reflect the differences in temperature during imaging. The faster T1ρ decay in vivo may be caused by the stronger spin or molecular fluctuations at the higher temperature of volunteer knee joints than cadaveric knee joints. Since this work focused on technical development, we only reported the feasibility of T1ρ quantification for most of the tissues about the knee joint, including tendons, ligaments, meniscus, cartilage and muscles. We are planning to perform a more systematic magic angle imaging study for cartilage, including the different layers (superficial, transitional, radial and calcified layers), with several quantitative MRI techniques including T1ρ, T2 and MT modeling (43).
Table 2.
T1ρ and its fitting standard errors (ms) of quadriceps tendon, patellar tendon, ACL, PCL, meniscus, patellar cartilage and muscles in six in vivo human knee samples as well as their average and standard deviation values (ms).
| In vivo knee | # 1 | # 2 | # 3 | # 4 | # 5 | # 6 | Average |
|---|---|---|---|---|---|---|---|
| Quadriceps tendon | 13.6±0.6 | 13.4±1.1 | 13.6±1.0 | 14.9±1.3 | 13.7±1.0 | 12.9±1.2 | 13.9±0.7 |
| Patellar tendon | 9.3±0.5 | 9.3±0.4 | 9.4±0.3 | 9.6±0.7 | 9.4±0.5 | 11.3±0.6 | 9.7±0.8 |
| ACL | 35.0±4.5 | 37.3±3.8 | 33.4±3.5 | 37.5±5.6 | 36.1±3.9 | 30.0±3.0 | 34.9±2.8 |
| PCL | 21.5±1.9 | 22.9±1.7 | 21.2±2.0 | 22.6±1.7 | 22.5±1.2 | 19.0±1.5 | 21.6±1.4 |
| Meniscus | 23.0±1.6 | 21.3±1.4 | 26.2±1.9 | 21.4±1.7 | 21.5±1.1 | 21.7±1.6 | 22.5±1.9 |
| Patellar cartilage | 42.3±6.5 | 47.7±6.9 | 47.3±7.5 | 42.5±6.6 | 43.5±5.9 | 44.1±6.2 | 44.5±2.4 |
| Muscle | 43.7±4.8 | 42.0±4.7 | 41.8±4.2 | 43.7±4.8 | 44.5±3.2 | 43.5±4.4 | 43.2±1.1 |
This study has several limitations. First, we have only demonstrated the technical feasibility of the 3D AdiabT1ρ UTE-Cones sequence in providing volumetric quantitative T1ρ imaging of both short and long T2 tissues both ex vivo and in vivo. No patients were studied in this work. Second, the sensitivity of 3D AdiabT1ρ UTE-Cones measurements to knee joint degeneration has not been investigated. It will be necessary to conduct a systematic study of knee joints with different degrees of degeneration followed by histological evaluation. Third, the 3D AdiabT1ρ UTE-Cones sequence used for in vivo imaging was approximately 18 min long, which is still relatively long for a patient study. The scan time can be further reduced with a smaller number of TSLs, such as 4. Moreover, fast 3D acquisition with acceleration techniques such as parallel imaging or compressed sensing can be used to further accelerate the data acquisition (44). Fourth, only patellar cartilage was used for this magic angle study. Other tissues, such as Achilles tendon, menisci, and ligaments, would also be very interesting for future magic angle studies (43, 45).
Conclusion
The 3D AdiabT1ρ UTE-Cones technique provides robust volumetric quantitative T1ρ measurement of both short and long T2 tissues including quadriceps tendon, patellar tendon, ACL, PCL, meniscus, patellar cartilage and muscle in the knee joint.
Supplementary Material
Supporting Figure S1. Patellar cartilage sample images used to study the magic angle effect on 3D AdiabT1ρ UTE-Cones. Localizers, AdiabT1ρ images, and corresponding fitting curves of five angular orientations (0°, 30°, 55°, 70° and 90° between the normal direction of cartilage surface and the main field B⃗0) are shown. The localizer was imaged by a conventional GRE sequence with TE/TR=3.2/8.6ms. The selected AdiabT1ρ images were acquired with NAFP = 0, 4, 8 and 16. The obtained T1ρ values are shown together with the fitting curves.
Acknowledgments
The authors acknowledge grant support from GE Healthcare, NIH (T32EB005970, 1R01 AR062581 and 1R01 AR068987 and) and the VA Clinical Science R&D Service (1I01CX001388).
References
- 1.Grushko G, Schneiderman R, Maroudas A. Some biochemical and biophysical parameters for the study of the pathogenesis of osteoarthritis: A comparison between the processes of aging and degeneration in human hip cartilage. Connect Tissue Res. 1989;19:149–176. doi: 10.3109/03008208909043895. [DOI] [PubMed] [Google Scholar]
- 2.Duvvuri U, Reddy R, Patel SD, Kaufman JH, Kneeland JB, Leigh JS. T1rho–relaxation in articular cartilage: Effects of enzymatic degradation. Magn Reson Med. 1997;38:863–867. doi: 10.1002/mrm.1910380602. [DOI] [PubMed] [Google Scholar]
- 3.Regatte RR, Akella SVS, Lonner JH, Kneeland JB, Reddy R. T1ρ relaxation mapping in human osteoarthritis (OA) cartilage: Comparison of T1ρ with T2. J Magn Reson Imaging. 2006;23:547–553. doi: 10.1002/jmri.20536. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Han ET, Ma B, Link TM, Newitt DC, Majumdar S. In vivo 3 T spiral imaging based multi-slice T1ρ mapping of knee cartilage in osteoarthritis. Magn Reson Med. 2005;54:929–936. doi: 10.1002/mrm.20609. [DOI] [PubMed] [Google Scholar]
- 5.Knispel RR, Thompson RT, Pintar MM. Dispersion of proton spin–lattice relaxation in tissues. J Magn Reson. 1974;14:44–51. [Google Scholar]
- 6.Regatte RR, Akella SVS, Borthakur A, Kneeland JB, Reddy R. Proteoglycan depletion–induced changes in transverse relaxation maps of cartilage. Acad Radiol. 2002;9:1388–1394. doi: 10.1016/s1076-6332(03)80666-9. [DOI] [PubMed] [Google Scholar]
- 7.Duvvuri U, Charagundla SR, Kudchodkar SB, et al. Human knee: In vivo T1ρ–weighted MR imaging at 1.5 T – Preliminary experience. Radiology. 2001;220:822–826. doi: 10.1148/radiol.2203001662. [DOI] [PubMed] [Google Scholar]
- 8.Mlynarik V, Szomolanyi P, Toffanin R, Vittur F, Trattnig S. Transverse relaxation mechanisms in articular cartilage. J Magn Reson. 2004;169(2):300–307. doi: 10.1016/j.jmr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Rautiainen J, Nissi MJ, Salo EN, Tiitu V, Finnila MA, Aho OM, Saarakkala S, Lehenkari P, Ellermann J, Nieminen MT. Multiparametric MRI assessment of human articular cartilage degeneration: Correlation with quantitative histology and mechanical properties. Magn Reson Med. 2015;74(1):249–259. doi: 10.1002/mrm.25401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akella SV, Regatte RR, Wheaton AJ, Borthakur A, Reddy R. Reduction of residual dipolar interaction in cartilage by spin-lock technique. Magn Reson Med. 2004;52:1103–1109. doi: 10.1002/mrm.20241. [DOI] [PubMed] [Google Scholar]
- 11.Wang N, Xia Y. Dependencies of multi-component T2 and T1ρ relaxation on the anisotropy of collagen fbrils in bovine nasal cartilage. J Magn Reson. 2011;212:124–132. doi: 10.1016/j.jmr.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao H, Pauli C, Li S, Ma Y, Tadros AS, Kavanaugh A, Chang EY, Tang G, Du J. Magic angle efect plays a major role in both T1rho and T2 relaxation in articular cartilage. Osteoarthritis Cartilage. 2017 doi: 10.1016/j.joca.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du J, Statum S, Znamirowski R, Bydder GM, Chung CB. Ultrashort TE T1ρ magic angle imaging. Magn Reson Med. 2013;1:69(3):682–7. doi: 10.1002/mrm.24296. [DOI] [PubMed] [Google Scholar]
- 14.Erickson SJ, Prost RW, Timins ME. The "magic angle" effect: background physics and clinical relevance. Radiology. 1993;188(1):23–5. doi: 10.1148/radiology.188.1.7685531. [DOI] [PubMed] [Google Scholar]
- 15.Garwood M, DelaBarre L. The return of the frequency sweep: designing adiabatic pulses for contemporary NMR. J Magn Reson. 2001;153(2):155–177. doi: 10.1006/jmre.2001.2340. [DOI] [PubMed] [Google Scholar]
- 16.Michaeli S, Grohn H, Grohn O, Sorce DJ, Kauppinen R, Springer CS, Jr, Ugurbil K, Garwood M. Exchange-influenced T2rho contrast in human brain images measured with adiabatic radio frequency pulses. Magn Reson Med. 2005;53(4):823–829. doi: 10.1002/mrm.20428. [DOI] [PubMed] [Google Scholar]
- 17.Michaeli S, Sorce DJ, Springer CS, Jr, Ugurbil K, Garwood M. T1rho MRI contrast in the human brain: modulation of the longitudinal rotating frame relaxation shutter-speed during an adiabatic RF pulse. J Magn Reson. 2006;181(1):135–147. doi: 10.1016/j.jmr.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Casula V, Autio J, Nissi MJ, Auerbach EJ, Ellermann J, Lammentausta E, Nieminen MT. Validation and optimization of adiabatic T1ρ and T2ρ for quantitative imaging of articular cartilage at 3 T. Magn Reson Med. 2017;77(3):1265–75. doi: 10.1002/mrm.26183. [DOI] [PubMed] [Google Scholar]
- 19.Nissi MJ, Mangia S, Michaeli S, Nieminen MT. Orientation anisotropy of rotating frame and T2 relaxation parameters in articular cartilage; Proceedings of the 21st Annual Meeting of ISMRM; Salt Lake City, Utah, USA. 2013. Abstract 3552. [Google Scholar]
- 20.Hänninen N, Rautiainen J, Rieppo L, Saarakkala S, Nissi MJ. Orientation anisotropy of quantitative MRI relaxation parameters in ordered tissue. Scientific Reports. 2017 doi: 10.1038/s41598-017-10053-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Nissi MJ, Idiyatullin D, Michaeli S, Garwood M, Ellermann J. Capturing fast relaxing spins with SWIFT adiabatic rotating frame spin–lattice relaxation (T1ρ) mapping. NMR in Biomedicine. 2016;1;29(4):420–30. doi: 10.1002/nbm.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen W. Errors in quantitative T1rho imaging and the correction methods. Quant Imaging Med Surg. 2015;5:583–591. doi: 10.3978/j.issn.2223-4292.2015.08.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandt KD, Radin EL, Dieppe PA, Putte L. Yet more evidence that osteoarthritis is not a cartilage disease (Editorial) Ann Rheum Dis. 2006;65:1261–1264. doi: 10.1136/ard.2006.058347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter DJ, Zhang YQ, Niu JB, Tu X, Amin S, Clancy M, Guermazi A, Grigorian M, Gale D, Felson DT. The association of meniscal pathologic changes with cartilage loss in symptomatic knee osteoarthritis. Arthritis Rheum. 2006;54:795–801. doi: 10.1002/art.21724. [DOI] [PubMed] [Google Scholar]
- 25.Tan AL, Toumi H, Benjamin M, Grainger AJ, Tanner SF, Emery P, McGonagle D. Combined high-resolution magnetic resonance imaging and histological examination to explore the role of ligaments and tendons in the phenotypic expression of early hand osteoarthritis. Ann Rheum Dis. 2006;65:1267–1272. doi: 10.1136/ard.2005.050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang EY, Du J, Bae WC, Chung CB. Qualitative and Quantitative Ultrashort Echo Time Imaging of Musculoskeletal Tissues. Semin Musculoskelet Radiol. 2015;19(4):375–386. doi: 10.1055/s-0035-1563733. [DOI] [PubMed] [Google Scholar]
- 27.Du J, Carl M, Diaz E, Takahashi A, Han E, Szeverenyi NM, Chung CB, Bydder GM. Ultrashort TE T1rho (UTE T1rho) imaging of the Achilles tendon and meniscus. Magn Reson Med. 2010;64(3):834–842. doi: 10.1002/mrm.22474. [DOI] [PubMed] [Google Scholar]
- 28.Du J, Carl M, Bae WC, Statum S, Chang EY, Bydder GM, Chung CB. Dual inversion recovery ultrashort echo time (DIR-UTE) imaging and quantification of the zone of calcified cartilage (ZCC) Osteoarthritis Cartilage. 2013;21(1):77–85. doi: 10.1016/j.joca.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma YJ, Carl M, Shao H, Tadros AS, Chang EY, Du J. Three-dimensional ultrashort echo time cones T1ρ (3D UTE-cones-T1ρ) imaging. NMR in Biomedicine. 2017 doi: 10.1002/nbm.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gurney PT, Hargreaves BA, Nishimura DG. Design and analysis of a practical 3D cones trajectory. Magn Reson Med. 2006;55:575–582. doi: 10.1002/mrm.20796. [DOI] [PubMed] [Google Scholar]
- 31.Carl M, Bydder GM, Du J. UTE imaging with simultaneous water and fat signal suppression using a time-efficient multispoke inversion recovery pulse sequence. Magn Reson Med. 2016;76:577–582. doi: 10.1002/mrm.25823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levitt MH, Freeman R, Frenkiel T. Supercycles for broad-band heteronuclear decoupling. J Magn Reson. 1982;50(1):157–160. [Google Scholar]
- 33.Ma YJ, Chang EY, Carl M, Du J. Quantitative magnetization transfer ultrashort echo time imaging using a time–efficient 3D multispoke Cones sequence. Magn Reson Med. 2017 doi: 10.1002/mrm.26716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fram EK, Herfkens RJ, Johnson GA, Glover GH, Karis JP, Shimakawa A, Perkins TG, Pelc NJ. Rapid calculation of T1 using variable flip angle gradient refocused imaging. Magn Reson Imaging. 1987;5:201–208. doi: 10.1016/0730-725x(87)90021-x. [DOI] [PubMed] [Google Scholar]
- 35.Ma YJ, Lv X, Carl M, Zhu Y, Szeverenyi NM, Bydder GM, Chang EY, Du J. Accurate T1 Mapping of Short T2 Tissues Using a Three-Dimensional Ultrashort Echo Time Cones Actual Flip Angle Imaging Variable Repetition Time (3D UTE-Cones AFI-VTR) Method. Magn Reson Med. 2017 doi: 10.1002/mrm.27066. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma YJ, Zhu Y, Lu X, Carl M, Chang EY, Du J. Short T2 imaging using a 3D double adiabatic inversion recovery prepared ultrashort echo time cones (3D DIR–UTE–Cones) sequence. Magn Reson Med. 2017 doi: 10.1002/mrm.26908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunter DJ, Zhang Y, Niu J, Tu X, Amin S, Goggins J, Lavalley M, Guermazi A, Gale D, Felson DT. Structural factors associated with malalignment in knee osteoarthritis: the Boston osteoarthritis knee study. J Rheumatol. 2005;32(11):2192–2199. [PubMed] [Google Scholar]
- 38.Nissi MJ, Salo EN, Tiitu V, Liimatainen T, Michaeli S, Mangia S, Ellermann J, Nieminen MT. Multi–parametric MRI characterization of enzymatically degraded articular cartilage. J Orthopaedic Research. 2016;34(7):1111–1120. doi: 10.1002/jor.23127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casula V, Nissi MJ, Podlipská J, Haapea M, Koski JM, Saarakkala S, Guermazi A, Lammentausta E, Nieminen MT. Elevated adiabatic T1ρ and T2ρ in articular cartilage are associated with cartilage and bone lesions in early osteoarthritis: A preliminary study. J Magn Reson Imaging. 2017;46(3):678–689. doi: 10.1002/jmri.25616. [DOI] [PubMed] [Google Scholar]
- 40.Rauscher I, Stahl R, Cheng J, Li X, Huber MB, Luke A, Majumdar S, Link TM. Meniscal measurements of T1rho and T2 at MR imaging in healthy subjects and patients with osteoarthritis. Radiology. 2008;249(2):591–600. doi: 10.1148/radiol.2492071870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolbos RI, Link TM, Ma CB, Majumdar S, Li X. T1rho relaxation time of the meniscus and its relationship with T1rho of adjacent cartilage in knees with acute ACL injuries at 3 T. Osteoarthritis Cartilage. 2009;17(1):12–18. doi: 10.1016/j.joca.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Chang G, Bencardino J, Babb JS, Krasnokutsky S, Abramson S, Regatte RR. T1rho MRI of menisci in patients with osteoarthritis at 3 Tesla: a preliminary study. J Magn Reson Imaging. 2014;40(3):588–595. doi: 10.1002/jmri.24437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma YJ, Shao H, Du J, Chang EY. Ultrashort echo time magnetization transfer (UTE-MT) imaging and modeling: magic angle independent biomarkers of tissue properties. NMR in Biomed. 2016;29:1546–1552. doi: 10.1002/nbm.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pandit P, Rivoire J, King K, Li X. Accelerated T1ρ acquisition for knee cartilage quantification using compressed sensing and data - driven parallel imaging: A feasibility study. Magn Reson Med. 2016;75(3):1256–61. doi: 10.1002/mrm.25702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henkelman RM, Stanisz GJ, Kim JK, Bronskill MJ. Anisotropy of NMR properties of tissues. Magn Reson Med. 1994;32(5):592–601. doi: 10.1002/mrm.1910320508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figure S1. Patellar cartilage sample images used to study the magic angle effect on 3D AdiabT1ρ UTE-Cones. Localizers, AdiabT1ρ images, and corresponding fitting curves of five angular orientations (0°, 30°, 55°, 70° and 90° between the normal direction of cartilage surface and the main field B⃗0) are shown. The localizer was imaged by a conventional GRE sequence with TE/TR=3.2/8.6ms. The selected AdiabT1ρ images were acquired with NAFP = 0, 4, 8 and 16. The obtained T1ρ values are shown together with the fitting curves.