Abstract
The advent of precision medicine for genetic diseases has been hampered by the large number of variants that cause familial and somatic disease, a complexity that is further confounded by the impact of genetic modifiers. To begin to understand differences in onset, progression and therapeutic response that exist among disease-causing variants, we present the proteomic variant approach (ProVarA), a proteomic method that integrates mass spectrometry with genomic tools to dissect the etiology of disease. To illustrate its value, we examined the impact of variation in cystic fibrosis (CF), where 2025 disease-associated mutations in the CF transmembrane conductance regulator (CFTR) gene have been annotated and where individual genotypes exhibit phenotypic heterogeneity and response to therapeutic intervention. A comparative analysis of variant-specific proteomics allows us to identify a number of protein interactions contributing to the basic defects associated with F508del- and G551D-CFTR, 2 of the most common disease-associated variants in the patient population. We demonstrate that a number of these causal interactions are significantly altered in response to treatment with Vx809 and Vx770, small molecule therapeutics that respectively target the F508del and G551D variants. ProVarA represents the first comparative proteomic analysis among multiple disease-causing mutations, thereby providing a methodological approach that provides a significant advancement to existing proteomic efforts in understanding the impact of variation in CF disease. We posit that the implementation of ProVarA for any familial or somatic mutation will provide a substantial increase in the knowledge base needed to implement a precision medicine-based approach for clinical management of disease.
Keywords: Cystic Fibrosis, Protein Interaction Profile, Lumacaftor, Ivacaftor, Disease-Associated Variants
Introduction
Disease-causing familial variation recorded in GnomAD [1] and ClinVar [2] now include more than 200,000 annotated variants in the human population that contribute to genetic diversity and healthspan [2–10]. Like rapidly evolving somatic variations observed in cancer, these mutations uncouple the affected protein from its normal community of interacting proteins, which are critical for its biogenesis and function in the cell. The inability of many disease-associated variants to properly interact with their respective components results in altered functional profiles leading to the disease phenotype. Therapeutic development in these genetic diseases is complicated by the fact that many are caused by different variants in a single protein and the fact that even patients with the same disease-causing allele are differentially impacted by modifier genes leading to a heterogeneity of responses to existing therapeutics [5]. These observations have led to the advent of high definition medicine [11] to better serve affected individuals from the perspective of the precision medicine initiative. However, the implementation of such a personalized medicine approach often suffers due to a lack of information pertaining to the etiology of the disease as well as a sparsity of data pertaining to the impact of the disease-causing mutation on the affected protein and its function.
Herein, we introduce the development of a proteomic methodology that captures critical information required to understand the onset and progression of variant-specific disease, referred to as the proteomic variant approach (ProVarA). Using affinity purification mass spectrometry (AP-MS) combined with genomic screening, we illustrate the utility of ProVarA in differentiating the impact of individual variants on disease etiology. To illustrate the utility of ProVarA to serve as a proteomic methodology for a complex familial disease, we focus on cystic fibrosis (CF). CF is caused by mutations in the CF transmembrane conductance regulator (CFTR) gene, which codes for a cAMP-regulated chloride channel expressed at the apical surface of epithelial cells [12–17] and is critical for the maintenance of proper chloride and bicarbonate balance in nearly all tissues. While more than 70% of CF patients carry at least one allele of a three-base pair deletion (delCTT) [18] resulting in the loss of phenylalanine at position 508 (F508del-CFTR), there are currently 2025 known CF-causing mutations in the patient population (http://www.genet.sickkids.on.ca; www.cftr2.org). These mutations are grouped into 1 of 6 classes including mutations that lead to a loss of CFTR production (Class I), misfolding and/or premature degradation (Class II), functional impairment (Class III), obstruction of the channel pore (Class IV), a reduction in the amount of CFTR produced (Class V) and destabilization of CFTR at the cell surface (Class VI) [19]. Recently a seventh class was proposed, which re-categorized those class I mutations that did not produce any mRNA into a class VII to reflect their lack of correctability by small molecule therapeutics [20]. Marson et al extended upon this 7 class suggestion by proposing that class VII be renamed as class IA and that class I proposed by De Boeck and Amaral be labelled as class IB to maintain the progressive decrease in disease severity historically associated with increasing class numbers in this CFTR classification system [21].
Herein we show how the application of ProVarA to CFTR variants can be used to interrogate the impact of these different CF-causing mutations on the functional interactions of the variant polypeptide chain with the binding proteins required for its normal function. These variants include the Class II variants, F508del, G85E, R560T and N1303K as well as the Class III variant G551D CFTR [19]. Profiling interactions through ProVarA allows us to identify both common and unique interactions that contribute to the development of CF disease in ER-restricted variants in comparison to those trafficking to the cell surface but exhibiting impaired function. We also generate protein interaction profiles (PIPs) in the presence of therapeutics to assess their impact on the profiles of the disease-causing variants. The high definition PIP of each variant and their causal impact on function provides a critical example of how ProVarA can be utilized for the advancement of a personalized medicine for any familial and/or somatic disease where alterations in the sequence impact human health.
Results
Development of the Proteome Variant Approach (ProVarA)
The absence of CFTR at the cell surface contributes to loss of ionic homeostasis and hydration of the epithelial lining of the lung and other affected tissues, triggering the progressive clinical pathology characteristic of CF. The F508del variant is the most common representative member of the Class II CF-causing mutations and produces a polypeptide chain that has been shown to exhibit aberrantly high affinities for chaperone proteins [22–24] leading to impaired recruitment of COPII components required for ER-to-Golgi trafficking [25] resulting in its rapid clearance by ER-associated degradation (ERAD) [13, 16, 26–36]. The largest resource of protein interaction data for CFTR has come from our recent characterization of the CFTR interactome of both the WT and F508del variants [23, 24]. This data has provided significant insight into the cohort of proteins that mediate CFTR biogenesis and function in the lung and has helped identify novel targets for correcting the defects associated with F508del-CFTR [23, 24, 37]. These studies reveal that many proteins exhibit a statistically significant difference in their interaction affinities for WT and F508del-CFTR, suggesting that a single point mutation can dramatically alter the community of interactions responsible for its biogenesis and function. While these data highlighted the complexities of the F508del-centric CF disease, they raised many questions as to the impact of other disease-causing mutations on their respective interactions, how these changes correlate with disease severity and progression and whether commonalities in PIP can be exploited to determine therapeutic effectiveness, to develop new therapeutics, or whether a completely personalized approach to the treatment of CF is required.
The CFTR protein contains 5 functional domains: 2 transmembrane domains (TMD1 and TMD2), 2 nucleotide binding domains (NBD1 and NBD2) and a regulatory domain (R). The 2025 CF-associated mutations found in the patient population map to all domains of the protein and are grouped into 7 classes based on the expression level, folding, function and stability of the resulting variant CFTR protein as well as their response to therapeutic intervention [19, 20]. To utilize ProVarA, we generated PIPs for 6 CFTR variants including 4 Class II CF-causing mutations, namely G85E (TMD1), F508del (NBD1), R560T (NBD1) and N1303K (NBD2), a Class III CF-causing mutation, G551D, and WT-CFTR. The mutations were selected based on their frequency in the patient population, with F508del, G551D, N1303K and G85E representing the 1st, 3rd, 4th and 15th most common alleles respectively [18], as well as their distribution along the polypeptide chain. The R560T variant was selected based on its extremely low abundance in the patient population [18] as well as a representation of a second class II NBD1 variant for our analyses.
To standardize our approach as well as avoid the many complexities of expression levels and evolvability of a cell that is found in laboratory-generated stable cell lines expressing different variants, we employed an adeno-viral vector (AVV) delivery system for rapid, transient expression of the CFTR variants in the CFBE41o-parental cell line, a bronchial epithelial cell line which expresses no detectable CFTR mRNA or protein. We observed that the transduction of null CFBE41o- cells resulted in a similar protein expression profile (Figure 1A upper panel) to that previously reported for these mutants [18]. These data indicate that the transient expression of these variants has little to no impact on their stability or trafficking, suggesting that the CFTR variants are properly engaging with the protein folding or proteostasis network (PN) components [13, 26, 38], supporting the use of this approach to generate representative PIPs for CF-causing variants as well as a versatile approach for any protein and its variant of interest as described below.
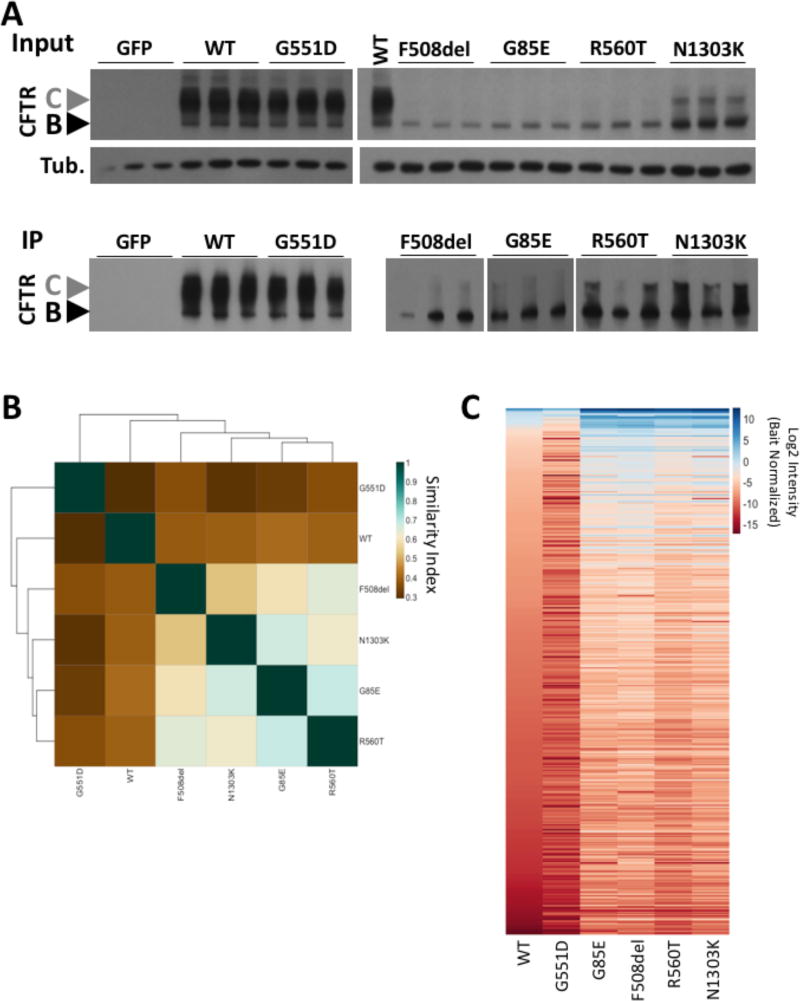
Figure 1. Characterization of the interaction PIPs of CFTR variants.
A. (Upper) Immunoblot analysis of CFTR and tubulin from a lysate prepared from CFBE41o- null cells transduced with the indicated CFTR variant. (Lower) Immunoblot analysis CFTR immunoprecipitation of samples shown in A (upper). B. Heat map of the Jaccard similarity indices for the indicated pairwise comparisons. C. Heat map of bait normalized protein recovery in the immunoprecipitation of the indicated CFTR variant. The data is presented as a log2 of the additive peptide intensity normalized to the additive CFTR peptide intensity and sorted from most to least abundant based on recovery with WT-CFTR.
The ProVarA Methodology
To identify the cohort of proteins that are differentially interacting with a broad spectrum of CFTR variants we employed the co-purifying protein identification technology (CoPIT) methodology [23], an immunoprecipitation based proteomic approach we have developed to compare the cohort of recovered protein with each of the CFTR variants of interest.
In order to validate our transient transduction system, we first compared our WT- and F508del-CFTR PIPs (Table S1 & S2) with the recently published WT- and F508del interactomes generated using CFBE41o- cell lines stably expressing WT or F508del transgenes, respectively [23]. A pair-wise comparison of the protein ID revealed a 37.2% and 30.7% overlap between WT and F508del PIPs respectively. In our previous study, we selected 52 proteins exhibiting a higher affinity for F508del- relative to WT-CFTR for siRNA-mediated knockdown experiments to assess their impact on F508del-CFTR trafficking and function, with 31 siRNAs correcting the trafficking defect associated with this disease-associated variant [23]. Our current F508del PIP recovered 19 (36.5%) of these proteins, 16 of which exhibit a higher affinity for F508del- relative to WT-CFTR, including 10 of the corrective siRNA targets. Furthermore, we identified 6 of 8 targets shown to restore F508del-CFTR chloride channel conductance in patient-derived primary human bronchial epithelial cells (hBE). Taken together, our current F508del PIP recapitulates many of the key features seen in CFBE41o- cell lines stably expressing WT- or F508del-CFTR transgenes [20]. The differences seen likely reflect the aberrantly elevated expression of the F508del transgene relative to the expression of the WT-CFTR transgene in the stable cell lines [20] a condition inconsistent with the low levels of F508del expression relative to that of WT-CFTR seen in patient derived primary human bronchial epithelial (hBE) cells but which is captured in our AVV transient expression system (Figure 1A) and reflected in our new F508del PIP.
To compare the composition of the PIPs of the CFTR variants (Tables S1–6), we used the list of proteins that exhibit a statistically significant difference in recovery relative to the control CFTR immunoprecipitation in GFP transduced CFBE41o- null cells to calculate the pairwise Jaccard similarity coefficients, a score which reflects the number of common proteins in the two datasets relative to the total proteins in the merged datasets where identical data sets would score as a 1 and completely unique datasets as a 0. Our analysis revealed that the Class II variants are more closely related to one another than to either WT- or G551D-CFTR (Figure 1B). An expected observation given the respective trafficking agendas of these variant that limit ER export, a feature evident by their accumulation in the ER-restricted band B N-linked glycoform and their lack of the post-Golgi band C glycoform relative to that seen with WT-CFTR (Figure 1A). To more accurately compare the PIPs of these CF-associated mutations we normalized the recovery of all proteins in each PIP of a given variant to the amount of the CFTR variant recovered (Tables S1–6). This bait normalization (BN) eliminates the differential recovery of common proteins due to differences seen in CFTR expression and recovery by immunoprecipitation among the variants (Figure 1A). A heatmap of the BN PIPs (Figure 1C) reveal clear differences between the ER-restricted Class II variants and both WT- and G551D-CFTR. While the G551D-CFTR variant exhibits a similar expression and trafficking profile to that seen for WT-CFTR (Figure 1A), we note that the composition of their interactomes are significantly different as determined by the Jaccard similarity index (Figure 1B & C). These results illustrate that PIPs can capture differences in the protein fold that reflect a WT-like spatial distribution of a non-functional variant.
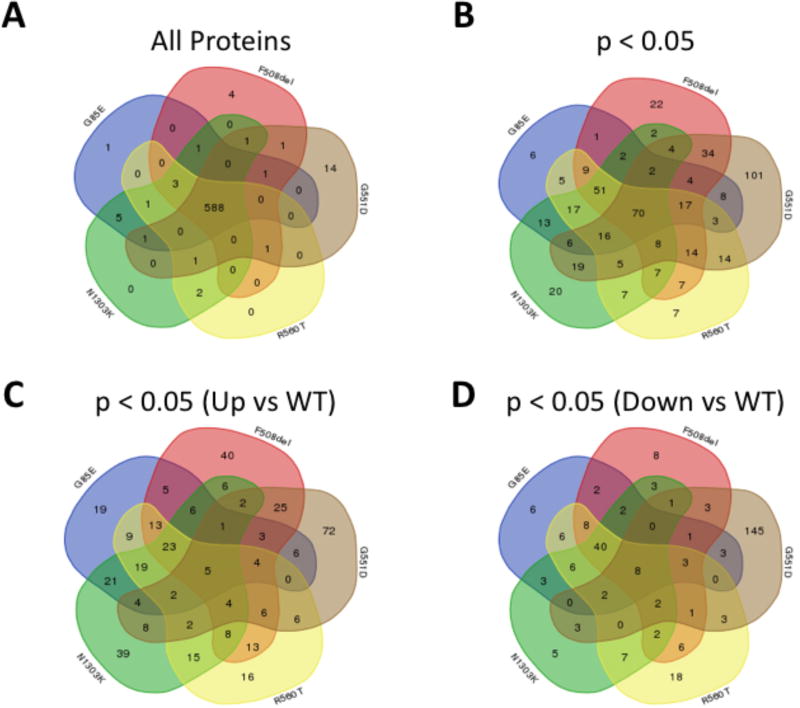
An analysis of the composition of the pairwise differential BN-PIPs (Tables S2–6), revealed 588 common proteins (Table 1, Figure 2A). These data indicate that many features of the CFTR fold are retained despite the presence of different disease-causing variations suggesting that a limited number of intermediate steps are impacting the ability of these mutants to achieve a functional fold. However, when we filtered the data for proteins that exhibit a statistically significant difference (p < 0.05) in the BN peptide intensity relative to that seen with WT-CFTR (Tables S2–6), we observed clear differences among the variants with 70 common proteins exhibiting a statistically significant difference in their affinity for the variants relative to that seen for WT-CFTR (Table 1, Figure 2B). A separation of these proteins into those that exhibit a significant increase (Figure 2C) or decrease (Figure 2D) in affinity for the variants, relative to WT-CFTR, highlight an additional layer of divergence between Class II and Class III variants. We observe that there are more proteins recovered with Class II variants that exhibit a higher binding affinity for the variant compared to WT (Figure 2C & D). This is consistent with the idea that these ER-restricted variants suffer from either a preponderance of aberrant interactions that WT-CFTR rarely or never sees, such as degradation machinery components [13, 16, 23, 24, 26–36] or that these variants suffer from an inability to navigate key folding intermediates and the PN components engaged in these key biogenesis steps are found in higher abundance with the variants [22, 39, 40]. Additionally, we also observe that the G551D variant suffers from an overall reduction in binding affinity relative to that seen with WT-CFTR (Figure 2C & D), suggesting that this variant is lacking key interactions despite its WT-like trafficking ability, which could, at least in part, explain its gating defect. These data also explain how both class II and class III variants can exhibit a low similarity to both WT-CFTR and to one another.
Table 1.
The PIPs of CFTR variants are significantly different
| Condition | All Proteins | p < 0.05 |
|---|---|---|
| G85E vs WT | 600 | 232 |
| F508del vs WT | 600 | 254 |
| G551D vs WT | 608 | 224 |
| R560T vs WT | 596 | 257 |
| N1303K vs WT | 603 | 249 |
| Total (All Combined) | 625 | 528 |
| Common | 588 | 70 |
Figure 2. The differential interaction PIPs of Class II CFTR variants identifies targets for correction of F508del-CFTR.
A. Venn diagram comparing the total protein content of the PIPs of the indicated CFTR variants. B. Venn diagram comparing the protein content of the PIPs of the indicated CFTR variants for proteins that exhibit a statistically significant difference in binding affinity relative to WT-CFTR. C. Venn diagram comparing the protein content of the PIPs of the indicated CFTR variants for proteins that exhibit a statistically significant increase in binding affinity relative to WT-CFTR. D. Venn diagram comparing the protein content of the PIPs of the indicated CFTR variants for proteins that exhibit a statistically significant decrease in binding affinity relative to WT-CFTR.
These data support the hypothesis that these PIPs can accurately characterize differences among diverse CF-causing variants and across multiple disease classes, an approach that will be useful to identify therapeutic targets for correction of either individual variants or for binning of variants with common PIP features, reflecting a common folding defect, an important step in determining the etiology of these respective variant-linked CF diseases. This represents a key step in advancing personalized medicine for CF as described below.
Using ProVarA to differentially profile multiple Class II variants
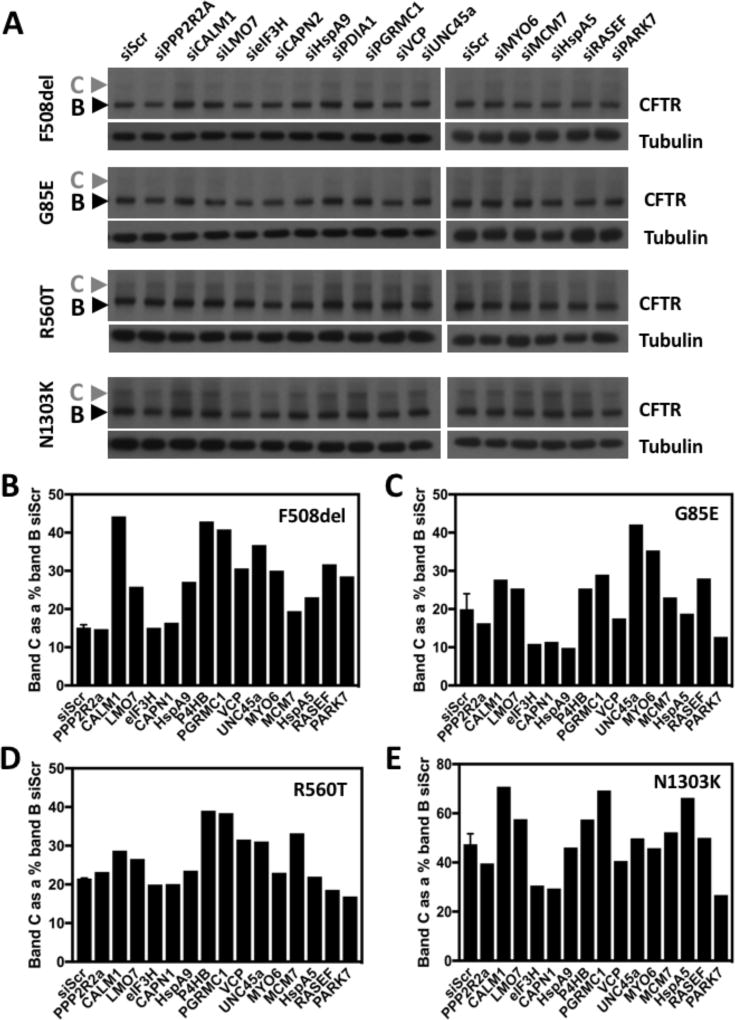
To assess the ability of ProVarA to identify potential therapeutic targets for the correction of individual or groups of CF-associated variants, we performed an alignment of proteins that exhibit a statistically significant difference in binding affinity for all 5 variants characterized above, relative to that seen with WT CFTR. We mined the dataset for proteins that exhibit increased binding to at least 3 of the 4 class II variants (F508del, G85E, R560T and N1303K) relative to that seen with WT-CFTR and no difference or decreased binding to G551D in the G551D versus WT differential PIP. The resulting network contained 60 proteins which satisfied these criteria, which map to diverse biological pathways. We selected a panel of 15 siRNAs targeting components involved in these diverse biological pathways, matching the diversity of the 60-protein network, to address if the identified proteins or their associated pathways might represent useful biological targets for the correction of these disease-causing CFTR variants. The selected siRNAs included components involved in transcriptional regulation (MCM7, LMO7), translational regulation (eIF3H), protein folding (HspA5, HspA9, P4HB, UNC45a, PARK7), protein degradation (VCP, CAPN1), post-translational modification (PPP2R2A), vesicle trafficking (RASEF, MYO6) and cellular signaling (PGRMC1, CALM1). Following siRNA-mediated silencing of these targets in CFBE41o- null cells transduced with each of the class II variants, we assessed if the silencing of any of these proteins impacted the trafficking ability of the variants as measured by changes in the amount of the post-Golgi CFTR fraction (band C). The analysis revealed that 12 out of the 15 tested siRNAs improved the trafficking of the F508del variant (Figure 3A & B) supporting the power of the ProVarA to identify proteins which can be targeted to correct the defects associated with CF-causing variants. The ability of these siRNAs to correct the trafficking of the 3 other variants were more modest, with 7/15 correcting the trafficking of each of the other class II variants (Figure 3A, C–E). All the siRNAs tested, with the exception of PPP2R2A, CAPN2 and eIF3H correct the trafficking of at least one of the class II variants tested and only siPARK7 and siHspA9 corrected a single variant, only targeting F508del-CFTR (Figure 3A–E). Of the targets tested, 5 siRNAs corrected at least 3 of the variants. These include the chaperone proteins unc45a, calmodulin 1 (CALM1) and PDIA1 (P4HB), LMO7, a protein with ubiquitin transferase activity and the progesterone receptor membrane component 1 (PGRMC1), an activator of Akt kinase [41], a negative regulator of CFTR expression and stability [42]. Interestingly, while the silencing of HspA5 resulted in an increase in band C for the F508del and N1303K variants (Figure 3A, B & E), we did note an improvement in the C/B ratio for all 4 variants (Data not shown), a trafficking index which measures the efficiency of trafficking. The differential response of these class II variants to the silencing of common, high affinity proteins likely stems from differences in their structural defects associated with the nature of the mutation. For example, while the deletion of an amino acid is intuitively thought to have severe implications for the resulting polypeptide, the F508del-NBD1 domain, while exhibiting decreased thermal stability [43], does not present extensive structural differences from that of the WT-NBD1 domain [44], suggesting a milder defect than initially predicted and increasing the likelihood of being a correctable mutation, a hypothesis supported by the extensive literature reporting correction of the trafficking and functional defects associated with the F508del variant and our data presented above showing a high rate of target identification for the correction of this CF-causing mutation (Figure 3A,B). Conversely, the G85E mutation has shown itself to be refractory to small molecule therapeutics [45] and represents a more severe CF-causing mutation. The G85E mutation is located in the N-terminal portion of the first membrane spanning helix of TMD1 and causes not only disruption of the helical structure of this transmembrane helix1 but also leads to a defect of the insertion of the helix1–helix2 hairpin structure [46, 47]. The ability of some of the targets discussed above to correct the G85E variant supports the use of ProVarA as a target identification methodology and speaks to the usefulness of comparative proteomics to identify therapeutic targets where high throughput small molecule screens have failed.
Figure 3. Correction of class II CFTR variants by siRNA knockdown of target proteins.
A. Immunoblot analysis of CFTR and tubulin from a lysate prepared from CFBE41o- null cells transfected with the indicated siRNA and transduced with the indicated CFTR variant. B–E. Bar graph of the amount of band C detected by quantitation of the immunoblot shown in panel A for F508del-CFTR (B), G85E-CFTR (C), R560T-CFTR (D) and N1303K-CFTR (E). The data shown in panels B–E represents the percentage of band C relative to band B in the control siRNA condition (siScr). The data for the siScr represents n = 2 and the data for the siRNA knockdown for the CFTR variants are a single replicate (n = 1).
The observation that several chaperone proteins exhibited increased affinity for these class II variants is consistent with our previous data demonstrating increased binding of components of the Hsp70/Hsp90 chaperone folding machinery to the F508del variant [22–24]. The results showing that siRNAs targeting these chaperone proteins can correct the defects associated with a number of class II CF-causing variants is in agreement with previous results, which have demonstrated that modulation of the expression level of chaperone proteins can provide functional correction of F508del [29, 31, 36, 48–54].
In general, a survey of the interactions impacting the function of a subset of Class II variants can lead to the identification of validated targets that could be exploited for the functional correction of multiple variants, demonstrating how ProVarA could be employed to assist in the development of a personalized approach to CF management of ER export and other phenotypic impacts of variation on disease presentation as described below.
Using ProVarA to profile the G551D-CFTR
To extend the utility of ProVarA beyond the Class II variant population, we focused on the Class III variant, G551D-CFTR, the third most prominent disease associated variant [18]. The G551D mutation produces a CFTR protein that exhibits WT-like trafficking (Figure 1A upper panel), where it accumulates in the post-Golgi band C glycoform but exhibits a defect in channel gating resulting in a CF phenotype in individuals carrying this mutation.
The differential PIP (G551D versus WT) identified proteins mapping to 597 genes with 49 uniquely bound to WT-CFTR and 20 uniquely bound to the G551D variant, leaving 528 overlapping proteins (Table 2; Table S6). While 88% of proteins were recovered with both WT-and G551D-CFTR, 325 proteins exhibited a statistically significant difference in affinity (Figure 1C; Table S6), a value that is consistent with the low pairwise Jaccard similarity index with WT-CFTR, but surprisingly large given the ability of both variants to navigate the cellular compartments associated with protein biogenesis and trafficking. Two previous studies used 2D-electrophoresis to identify G551D-CFTR interacting proteins [55, 56]. These studies noted that calumenin (CALU) and actin are recovered with increased affinity for the G551D variant relative to WT-CFTR [55, 56]. While our PIP analysis failed to identify actin as a differentially bound protein, we did observe an increased affinity of CALU for G551D-CFTR (Table S6).
Table 2.
Proteins recovered in the indicated pairwise analyses of CFTR variants
| G551D | WT | G551D + Vx770 | |
|---|---|---|---|
| Total Protein | 598 | ||
| p-Value < 0.05 | 325 | ||
| Total Protein | 598 | ||
| p-Value < 0.05 | 274 | ||
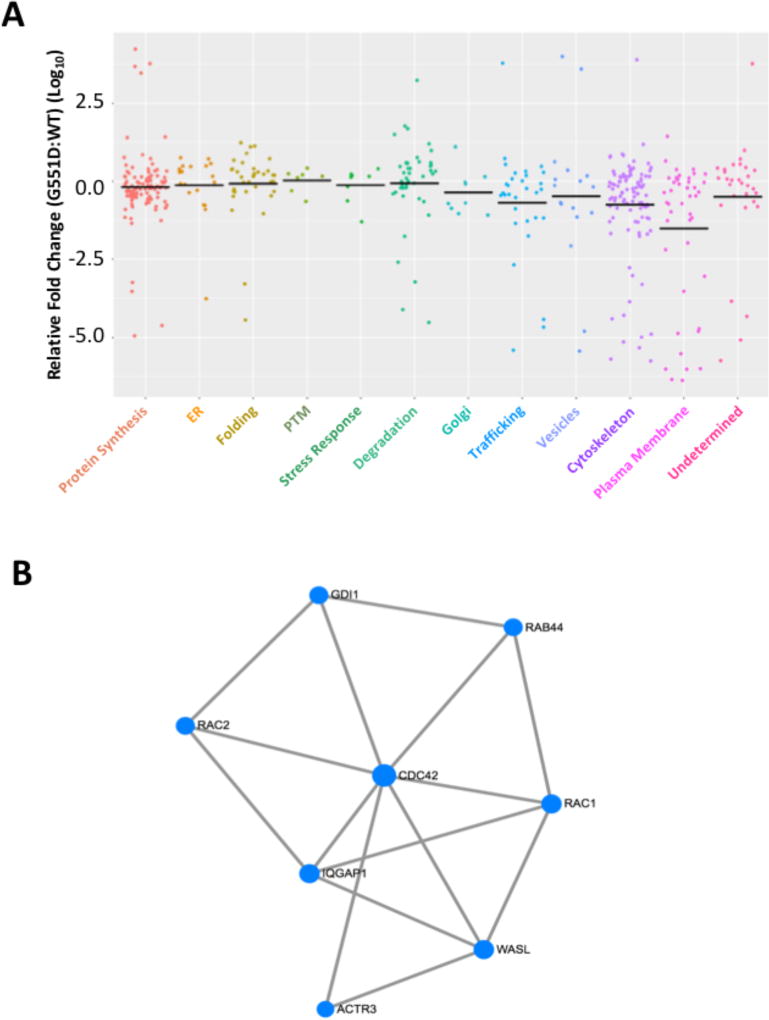
To provide insight into the specific differences that exist in the differential PIP comparing the WT and G551D variants, we binned the proteins into subgroups based on their associated cellular function and/or compartment of action and plotted the relative fold-change (FC) in the sum of BN peptide intensities for all protein recovered with G551D-CFTR relative to that seen with WT-CFTR. The subgroups were ordered along the biosynthetic pathway from protein synthesis to cell surface localization (Figure 4A). An analysis of the median log10-FC (G551D:WT) reveals a trend along the biosynthetic pathway revealing a median FC values approaching 1 until the cytoskeleton and plasma membrane compartments are reached where we see a preference of recovered proteins for WT over G551D (Figure 4A). This is consistent with the Venn diagram above showing that most proteins exhibiting a significant difference in affinity in the G551D versus WT differential PIP are proteins with reduced affinity for G551D (Figure 2D). These data are also in agreement with the ability of both proteins to escape the endoplasmic reticulum (ER) but exhibit a difference in their cell surface channel activity in response to stimuli, suggesting that the defect(s) associated with the loss of function of the G551D variant is associated with an inability to form key interactions critical for proper localization or insertion into the plasma membrane or for its functional response to stimuli (Figure 4A).
Figure 4. G551D-CFTR exhibits defective binding to trafficking and cytoskeletal components at the plasma membrane.
A. Scatter plot depicting the relative fold change in binding affinity between proteins recovered with G551D- and WT-CFTR. The recovered proteins were categorized based on their functional or subcellular site of action. The data is represented as the log10 of the fold change relative to WT (G551D:WT) and the black bars represent the median log10-FC value for each category. B. Minimal network depicting the connectivity between CDC42, RAC1, IQGAP1, GDI1 and ACTR3, involved in trafficking and cytoskeletal rearrangement.
A mining of this dataset revealed a small network of related proteins that exhibit a statistically significant difference in affinity between the G551D and WT variants. The network includes the GTPases, CDC42, and RAC1, as well as some of their interacting proteins including the Actin related protein (ARP) 2/3 complex, IQ-motif containing GTPase activating protein 1 (IQGAP1) and the guanine nucleotide dissociation inhibitor 1 (GDI1) (Figure 4B; Table S6). All of these proteins exhibit an increase affinity for WT-CFTR relative to the recovery seen with the G551D variant (Table S6). CDC42 has been shown to have a functional role in cytoskeletal remodeling [57, 58] and vesicle trafficking [39, 59–64]. Specifically, activated CDC42 recruits and activates neural Wiskott-Aldrich syndrome protein (N-WASP) to the sub-plasmalemmal region [39, 61, 63], which in turn activates the Arp2/3 complex which nucleates actin polymerization that helps bring secretory granules to the docking site at the plasma membrane [39, 61, 63]. The PIP data reveals that in addition to ACTR2 and ACTR3, we also co-purify the Arp2/3 proteins ARPC1A, ARPC1B, ARPC2, ARPC3, ARPC4 and ARPC5L with increased affinity for WT- relative to G551D-CFTR (Table S6). RAC1, has been reported to have a regulatory function in both exocytic [65–68] and endocytic trafficking [69–76]. Specifically, RAC1 participates in clathrin-mediated endocytosis where it associates with synaptojanin-2 to reduce clathrin-coated pit formation [66–68]. Additionally, RAC1 has been reported to promote exocytic trafficking in bovine chromaffin cells [71], as well as human pancreatic cells [69, 73–76]. IQGAP1 is a RHO/RAC GTPase interacting protein with a defective GTPase Activating Protein (GAP) domain that serves as a scaffolding protein for signaling complexes ([65, 77–79]). In addition to IQGAP1, we also recovered GDI1, a regulatory protein for numerous GTPase families whose role is to prevent the release of the bound nucleotide [80] and inhibit its GTPase activity [40, 80]. These proteins are all part of a small interaction network of inter-related proteins with a role in promoting trafficking to the plasma membrane or inhibiting endocytic removal of proteins from the plasma membrane, suggesting that part of the defect associated with the G551D variant includes incomplete delivery to or inappropriate retrieval of this CFTR mutant from the plasma membrane leading to the observed loss of cell surface chloride channel activity. Prince et al have previously shown that cAMP-mediated stimulation of CFTR-expressing cells inhibits the endocytic recycling of WT-CFTR but has no impact on the G551D variant [81], suggesting that activation of the chloride channel activity impedes the endocytic process or that the structural impact of the G551D mutation on the CFTR polypeptide alters the network of interacting proteins charged with the trafficking dynamics of this chloride channel. The data presented herein reveals that the PIP of the G551D-CFTR polypeptide is vastly different from that of its WT counterpart, which could account for the differential behavior of these variants at the PM. Furthermore, Trouve et al, showed that the G551D variant exhibits increased binding to actin relative to the affinity displayed by WT-CFTR and this actin binding is required to maintain the weak basal chloride channel activity exhibited by the G551D variant [56], suggesting that this actin binding is required to maintain the variant in the PM. The G551D PIP reveals reduced binding to actin related proteins suggesting that the G551D variant would not be maintained at the PM, but rather localized within endocytic recycling vesicles.
These data provide a functional example of how ProVarA can be mined for specific variants to provide insight into the causal events contributing to the development and severity of CF disease reflecting the unique PIP and functional defect in the protein fold. We posit that the data might also prove effective to address the mechanism of action of therapeutic interventions by determining the changes in the PIPs that contribute to the correction of the disease phenotype allowing us to then predict the response of other variants to this therapeutic based on the presence of fiduciary protein markers in their unique PIPs [82–84] as described below.
Characterizing the impact of Ivacaftor (Vx770) on G551D-CFTR
In 2012, the FDA granted approval for the use of Vx770/Ivacaftor (Kalydeco), a small molecule potentiator which increases the open probability (Po) of CFTR chloride channels [18, 85–88], in patients carrying at least one allele of the G551D mutation, which exhibits WT-like trafficking but defective cell surface channel activity. This approval was later expanded to include patients carrying at least one allele of 31 additional CF-causing Class III mutations [86]. While the broad efficacy of Vx770 suggests a common mechanism of action, it does not impact all Class III variants [86] and, moreover, has no corrective properties towards Class II variants [85, 86], suggesting that there are differences among CF variants within classes which remain to be elucidated. Investigation into the mechanism of action of Vx770 on the potentiation of G551D-CFTR channels revealed that the compound binds directly to CFTR [89] and activates ATP-independent channel opening as well as increases channel open time [89, 90]. This direct binding of Vx770 to G551D-CFTR, and likely other responsive CF-causing variants, suggests that this compound will alter the PIP of CFTR variants and that an understanding of these changes will impact our understanding of the mechanism of action for this small molecule.
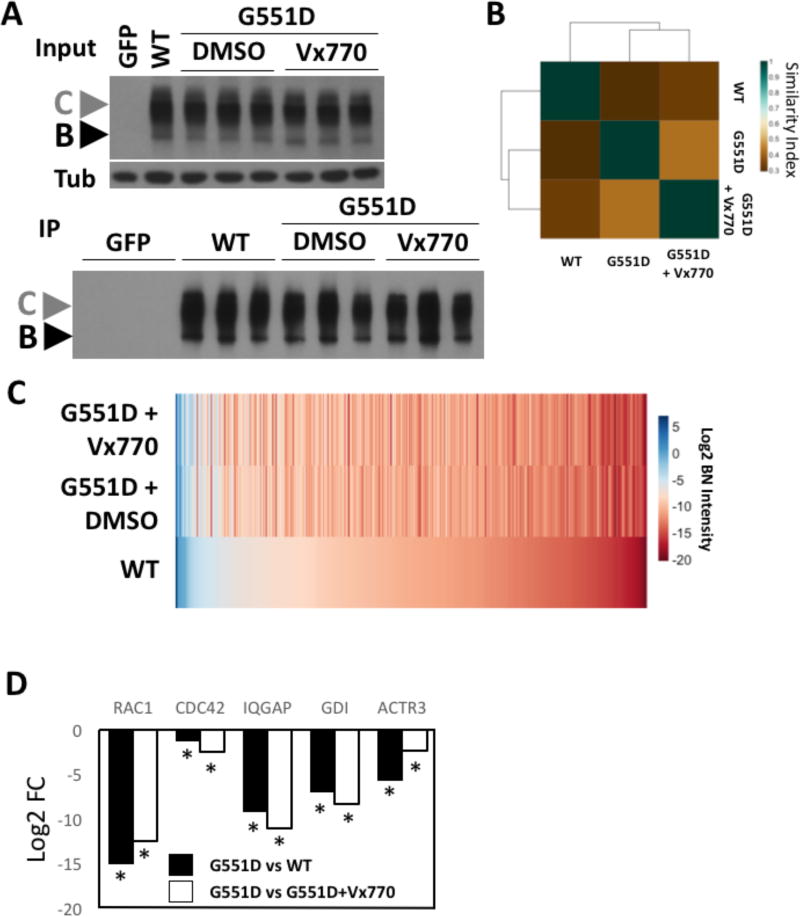
To assess the impact of Vx770 on the PIP of G551D-CFTR, we transduced parental CFBE41o- cells with AVV carrying the G551D cDNA (see Material and Methods) and treated cells for 24 h with 10 µM Vx770 (Figure 5A - upper panel), a dose previously shown to potentiate the cell surface chloride channel activity associated with the G551D variant [85]. G551D-CFTR was subsequently affinity purified from vehicle and Vx770 treated cell lysates (Figure 5A - lower panel) and subjected to ProVarA to determine co-purifying proteins. While a scan of the Jaccard plot shows only a modest improvement in the pairwise similarity index between Vx770-treated G551D and WT-CFTR relative to that seen between G551D and WT-CFTR (Figure 5B), an examination of the pairwise similarity score for Vx770-treated G551D vs non-treated G551D-CFTR (Figure 5B) reveals that these PIP are significantly different from one another suggesting that the compound is causing wide spread changes in the PIP for this CF-causing mutations. In fact, an analysis of the G551D vs WT differential PIP (Table S6) reveals 325 proteins exhibiting a statistically significant fold change in binding affinity between these 2 variants. The addition of Vx770 results in 126 of these 325 proteins no longer exhibiting statistically significant differences in their binding affinity relative to that seen for WT-CFTR (Table S8). These data reveal that Vx770 impacts the binding affinity of 38.8% of the differentially bound proteins in the G551D vs WT differential PIP, accounting for the low similarity index between Vx770-treated and non-treated G551D-CFTR (Figure 5B). This data is in agreement with our observation that the Vx770 treatment restores a number of protein interactions to WT-like levels (Figure 5C; Table 2; Table S8).
Figure 5. Vx770 restores a more WT-like PIPs to G551D-CFTR.
A. (Upper) Immunoblot analysis of CFTR and tubulin from a lysate prepare from CFBE41o- null cells transduced with the GFP, WT-CFTR or G551D-CFTR, the latter of which was treated with DMSO or 10 µM Vx770 for 24h. (Lower) Immunoblot analysis of CFTR immunoprecipitation from samples shown in panel A (upper). B. Heat map of the Jaccard similarity indices for the indicated pairwise comparisons. C. Heat map of bait normalized protein recovery in the immunoprecipitation of the indicated CFTR variant and treatment condition. The data is presented as a log2 of the additive peptide intensity normalized to the additive CFTR peptide intensity and sorted from most to least abundant based on recovery with WT-CFTR. D. Bar graph of the fold change in the binding affinity of the indicated proteins recovered in both the G551D versus WT and G551D+DMSO versus G551D+Vx770 differential PIPs. The data is shown as a log10 folding change of the total intensity for the indicated proteins with G551D relative to WT-CFTR (black) or Vx770 treated G551D (white).
Mining of this expanded G551D bioinformatic dataset revealed correction to WT-like interactions for the trafficking/cytoskeletal network discussed above (Figure 4B). Specifically, we observed that Vx770 restored binding to RAC1, IQGAP and GDI (Figure 5D, Table 3), where they were absent from the G551D PIP (Table S8). We also observed restoration of WT-like binding levels for CDC42 and ACTR3 where they exhibited lower binding affinities with the G551D variant than with WT-CFTR (Figure 5D, Table 3. These data support the interpretation that part of the functional defect associated with G551D is the improper engagement of the machinery that delivers and/or maintains CFTR in the plasma membrane [91, 92], a defect that is corrected, at least in part, by the CFTR potentiator Vx770.
Table 3.
Vx770 treatment restores the binding of Rac1, CDC42, IQGAP, GDI1 and ACTR to G551D-CFTR
| Protein | RAC1 | CDC42 | IQGAP | GDI | ACTR3 |
|---|---|---|---|---|---|
| Log2 FC (G551D vs WT) | −15.0 | −1.2 | −9.2 | −6.9 | −5.6 |
| p-Value | 0.004 | 0.009 | 0.004 | 0.01 | 0.01 |
| Log2 FC (DMSO vs 770) | −12.4 | −2.4 | −11.0 | −8.3 | −2.3 |
| p-Value | 5,2×10−5 | 0.005 | 7.1×10−5 | 0.0002 | 0.0005 |
These data highlight the potential impact of ProVarA as a methodological approach for identifying mechanisms to develop improved therapeutics or to expand therapeutic usage of Vx770 for other CF-causing variants that exhibit defective binding to this or other subnetworks of proteins mediating the action of this FDA-approved therapeutic at the cell surface. An expansion in the number of characterized PIPs for Vx770-responsive variants will provide a greater level of granularity into the full cohort of changes which correlate with potentiator activity leading to an improved understanding of the mechanism of action of this compound. A better understanding of the changes associated with the MoA of the compound will provide a better framework for personalizing the treatment of patients who might benefit from the clinical use of Vx770. The PIP of Vx770 on G551D provides a comparative framework to address its differential impact to the corrector Vx809 (Lumacaftor) that promotes ER export of Class II variants [19, 83, 93, 94] as described below.
Characterizing the impact of Lumacaftor (Vx809) on PIPs
While G551D-CFTR is the third most common CF-causing variant in the patient population, the F508del is the most common [18], with an allele frequency of 70% [18]. The F508del-CFTR variant has been the subject of countless efforts to identify small molecule therapeutics to correct the trafficking and functional defects associated with this mutation. Lumacaftor, also known as Vx809, is a small molecule therapeutic that has been shown to weakly correct the trafficking defect associated with F508del- and select other Class II variants at the bench and bedside [95]. Initial studies demonstrated that Vx809 stabilized N-terminal fragments of CFTR containing MSD1, suggesting that Vx809 binds directly to the first membrane spanning region (TMD1) of CFTR [96]. However, recent evidence has shown that Vx809 is minimally additive to revertant mutations such as R1070W and V510D, which have previously been shown to stabilize the interface between NBD1 and the fourth intracytoplasmic loop (ICL4) located in MSD2 (NBD1:ICL4) [97], but fully additive to revertant mutations that stabilize the NBD1:NBD2 interface [98], thereby suggesting that Vx809 acts upon the NBD1:ICL4 interface, a critical step in correcting the structural defects required for restoring trafficking to the F508del variant [97]. Despite these advancements in our understanding of the mechanism of action of Vx809, there remains a gap in our knowledge base to fully explain how the binding of this small molecule to F508del-CFTR might promote proper folding of the protein, escape from the ER associated degradation pathways and promote its targeting to ER export sites. Moreover, the effects of Lumacaftor were neither global nor of equal efficacy for all Class II variants [99], similar to what is seen with Vx770 in the potentiation of class III CF variants. Its use in combination with Vx770, referred to as Orkambi [93, 100–103], to potentiate the channel gating of the cell surface delivered F508del variant, was approved by the FDA for use in patients carrying at least one F508del variant allele in 2015. Taken together, these data highlight the need for a better understanding of the mechanism of action of therapeutics to predict the spectrum of variants for which Lumacaftor and Orkambi will be efficacious for personalized medicine-based approach for ER-restricted class II variants.
To begin to address this question from the perspective of ProVarA, we transduced parental CFBE41o- cells with AVV carrying the F508del cDNA (see Material and Methods) and treated cells with 3 µM Vx809 for 24 h, a dose previously shown to correct the trafficking and functional defects associated with the F508del variant (Figure 6A - upper panel) [95]. Vehicle-and Vx809-treated F508del-CFTR was subsequently affinity purified from the respective cell lysates (Figure 6A - lower panel) and subjected to MS/MS analysis to determine co-purifying proteins. As described above for the impact of Vx770 on the G551D variant, an analysis of the Jaccard plot does not show any obvious changes in the pairwise similarity indices between F508del and WT-CFTR and that of the Vx809-treated F508del and WT-CFTR. However, we do note that the pairwise similarity index between Vx809-treated and non-treated F508del-CFTR do show significant differences between their PIP. Given that this similarity index is based on the composition of the PIP protein lists, it suggests that while the composition of the overlapping list of common proteins may not be impacted by Vx809 (ie: a similar cohort of proteins are identified in both pairwise analyses), the binding affinity of the proteins within this PIP may change. Therefore, we undertook a more quantitative analysis of the impact of Vx809 treatment on the pairwise comparison between the differential PIPs of WT-CFTR and DMSO or Vx809 treated F508del-CFTR (Table S1, S2 and S7). A comparison of the differential PIP between F508del and WT CFTR identified 254 proteins exhibiting a statistically significant difference in protein binding (Table S2). The addition of Vx809 to the F508del-expressin CFBE41o- null cells resulted in 99 of these proteins no longer exhibiting a statistically significant difference in their binding affinity for the F508del variant relative to that seen with WT-CFTR. These data reveal that Vx809 alters the binding affinity of 39.0% of the differentially bound proteins in the F508del vs WT differential PIP (Table S2 and Table S7). These data and analyses are in agreement with the observations that a number of proteins are impacted by treatment with Vx809 (Figure 6B and Table S7).
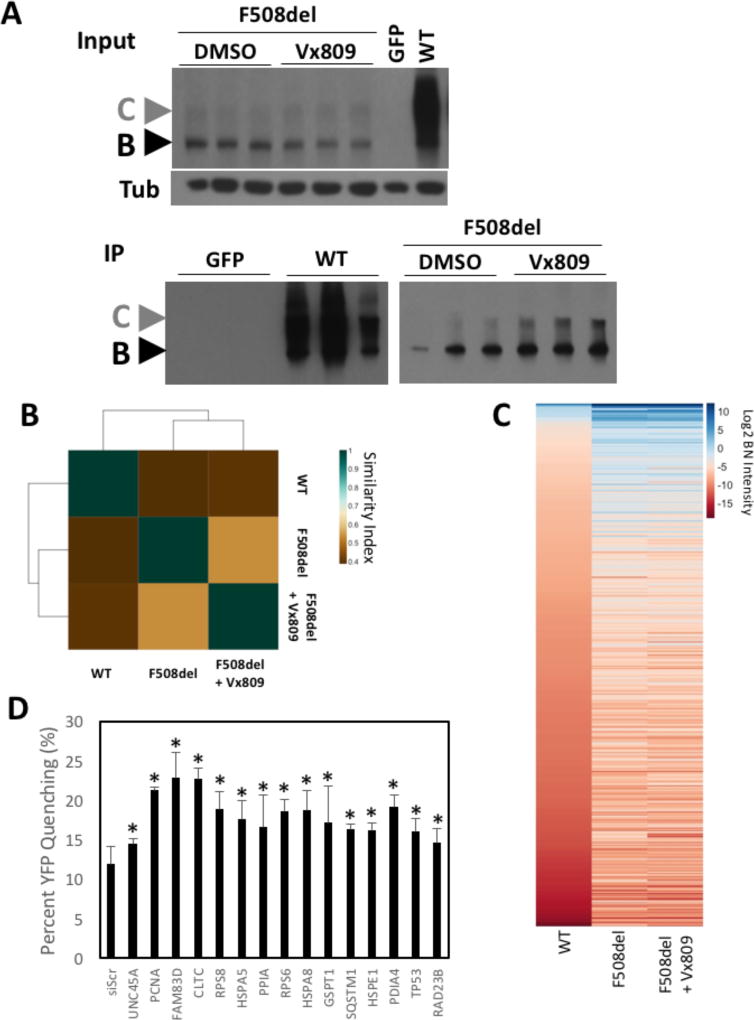
Figure 6. Vx809 restores a more WT-like PIPs to F508del-CFTR.
A. CFTR immunoblot analysis of CFTR immunoprecipitation samples from CFBE41o- cell lysates virally transduced with WT- and G551D-CFTR treated with DMSO or 3µM Vx809 for 24h. B. Heat map of the Jaccard similarity indices for the indicated pairwise comparisons. C. Heat map of bait normalized protein recovery in the immunoprecipitation of the indicated CFTR variant and treatment condition. The data is presented as a log2 of the additive peptide intensity normalized to the additive CFTR peptide intensity and sorted from most to least abundant based on recovery with WT-CFTR. D. Bar graph depicting the percentage of YFP-H148Q/I152L quenching seen in CFBE41o- cells expressing F508del-CFTR in response to the siRNA-mediated silencing of the indicated protein whose binding affinity were statistically significantly altered by the treatment with Vx809.
As described above, the F508del versus WT differential PIP identified 254 proteins exhibiting a statistically significant difference in binding (Table S2) with 99 proteins altered to a more WT-like binding affinity in response to Vx809 treatment. In order to assess the corrective potential of these 99 targets, we aligned this protein cohort with the dataset from a high throughput siRNA screen for functional correction of F508del-CFTR (Table S9) to assess if any of them provide corrective benefit that could account, at least in part, for the functional correction associated with Vx809. The assay measures the fluorescence of a halide sensitive Yellow Fluorescent Protein (YFP) variant (YFP-H148Q/I152L) which can be quenched in response to iodide influx which enters the cell through a functional, cell surface localized CFTR protein [104]. A comparison against our siRNA list revealed that 33 of the 99 proteins were tested, with 15 siRNA targets providing functional correction of F508del-CFTR (Figure 6D). These include the protein folding and degradation components Hsp A5, A8 & E1, unc45a PDIA4, cyclophilin (PPIA) and RAD23B, ribosomal proteins RPS6 and RPS8 as well as GSTP1, which is involved in the oxidative stress response pathway. Of this list, PDIA4 has previously been shown to have a higher affinity for F508del-CFTR relative to WT and siRNA targeting this protein provides functional correction of the F508del variant in both CFBE41o- and hBE cells [23]. The 40s ribosomal proteins, RPS6 & RPS8, are functionally related to the recently identified CFTR target, RPL12, whose silencing provides correction of F508del-CFTR in CFBE41o- and hBE cells. HspA8 also known as Hsc70 is a constitutively expressed Hsp70 family member which is critical for the biogenesis of CFTR and which has been shown to exhibit increased binding to F508del relative to WT [22–24]. The polyubiquitin binding, proteasome associated RAD23B is a component involved in regulating the delivery or ER localized proteins for ERAD. We also observed that 27 of the 99 Vx809 responsive proteins have been tested for their impact on Vx809-mediated correction of F508del-CFTR (Table S9). Here the silencing of PMSC4, a component of the 26S proteasome, provided functional correction only in combination with Vx809 suggesting that the impact of Vx809 on binding of PMSC4 to F508del-CFTR is not related to its mechanism of action (MoA) but rather a consequence of its improved folding by the drug. We found 8 siRNAs targets which showed no statistically significant differences in F508del-CFTR activity when combined with Vx809 compared to that seen with Vx809 alone, suggesting that they are involved in the MoA of the Vx809-mediated correction of F508del-CFTR. These include the chaperone proteins HspA8, HspE1 and cyclophilin, the ribosomal proteins RPS6 and RPS8, the oxidative stress response protein, GSTP1 and the ERAD associated protein RAD23B.
These results highlight the distinctive PIPs impacted by Vx809 and Vx770, emphasizing the high value of applying a rigorous ProVarA analysis for variants contributing to CF in order to provide an approach that provides a critical level of granularity to assess the role of diversity in disease presentation and its management in the clinic.
Discussion
Herein we describe the utility of ProVarA as a novel proteomic approach to provide a deeper understanding of how variant-specific PIP influence disease onset and progression and its response to FDA-approved therapeutics. ProVarA generates PIP based on binding affinities of proteins allowing us to classify disease causative mutations, determine causative events associated with these mutations, identify mutations that may be targeted by drugs as well as assess the mechanism of action of these small molecule therapeutics (Figure 7). In essence, ProVarA can be viewed as a protein-interaction extension of the Connectivity Map concept, which utilizes transcriptomic-based cellular signatures to catalog responses to genetic perturbations and pharmacological interventions [105, 106].
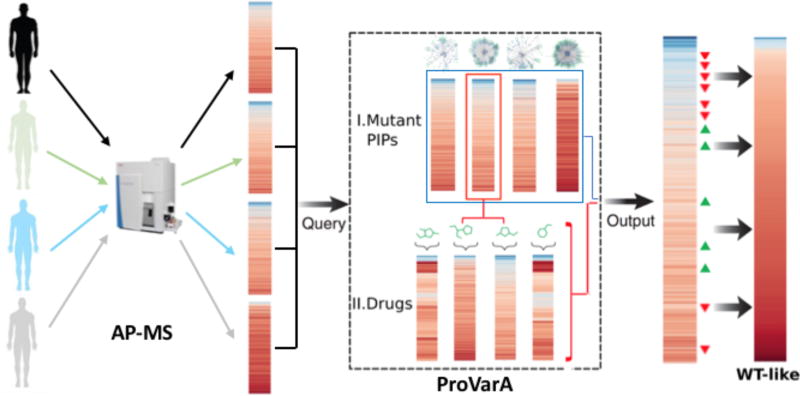
Figure 7. Workflow schematic for applying ProVarA to genetic diseases.
Schematic representation of how to apply ProVarA to human genetic diseases. In a first step a protein interaction profile (PIP) for the disease associated protein is generated from a patient. The resulting PIP is then queried against a database of established PIP for a panel of characterized disease-associated mutations in a 2-step process. The first step (I) aligns the patient or test PIP with the PIP of known mutations to identify target proteins that can be exploited for correction of the disease phenotype. In a second step (II) the response of the most similar reference PIP to a panel of small molecule therapeutics is mined to predict the most effective compounds for therapeutic intervention to restore a more WT-like PIP.
Our results highlight the analysis of 6 CF-causing variants of CFTR to demonstrate how ProVarA can be utilized to inform on the etiology of CF disease and to gain insight into key proteomic changes that are responsible, at least in part, for the therapeutic activity of the FDA-approved pharmaceuticals Ivacaftor (Vx770) and Lumacaftor (Vx809). Below we discuss the value of ProVarA for the CF community in evaluating the impact of variants on the disease state(s) and probable causes for response to therapeutics. We also briefly discuss the use of ProVarA for other familial/somatic diseases where mutation is an important feature of onset, progression, and therapeutic intervention reflecting the emergent genetic diversity in the population and its implications for precision medicine-based initiatives.
Application of ProVarA for the CF community
When the gene responsible for cystic fibrosis was identified in 1989 [107–109], it was believed that CF would represent the first genetic disease to be successfully treated, yet it took more than 20 years for the first therapeutic that addresses one of the basic defects of the disease to be approved by the FDA [85]. One of the reasons for this lag was the progressive realization of the vast array of CF-causing mutations and the heterogeneity of their etiologies reflecting genetic diversity. To date more than 2000 mutations have been documented in the clinic (http://www.genet.sickkids.on.ca; www.cftr2.org) which are grouped in 7 classes based on the molecular properties of the resulting protein [19, 20]. Early CF research took a mostly empirical medicine approach, believing that one drug could be developed to treat all patients. However, the expansion of our knowledge base pertaining to the array of disease causing variants leading to CF, paired with the unique properties of the resulting polypeptide chain, catapulted CF research into an era of stratified medicine, attempting to develop drugs for the different CF-specific classes. This led to the identification of non-sense read through compounds, such as PTC-124 [110–112], for the treatment of Class I mutations, correctors, such as Vx809 [95] and Vx661 [113], for the treatment of Class II ER-restricted variants and potentiators, such as Vx770 for the treatment of Class III and IV CFTR variants that exhibit defective channel gating [85, 86]. However, the heterogeneity of response to these compounds, even among patients carrying the same mutation [114, 115], has highlighted the need for the CF field to move into an era of personalized medicine. However, the implementation of such precision medicine for CF is currently impacted by a dearth of information as to causal events leading to variant specific properties, the impact of patient-specific modifier genes as well as the molecular events that mediate the efficacy of the various small molecule therapeutics under clinical use or investigation.
The complexity of the life cycle of the WT-CFTR protein is well established with the PIP exceeding 500 proteins (Table S1) [23]. An additional layer of complexity is introduced with a single point mutation, such as the deletion of F508 (F508del), leading to the most prevalent allele in the CF patient population. This F508del variant exhibits a significantly different PIP (Table S2) [23], leading to the identification of numerous targets which could be exploited to provide functional correction of F508del-CFTR [23, 24, 34, 54]. Now, with our characterization of PIPs for 3 additional Class II CF causing mutations, we increase our power to predict causal protein interactions that directly contribute to the ER retention of Class II variants not only allowing us to confirm the validity of previously identified corrective targets using stable expression system, but also to identify new ones. Our comparative analysis of the F508del- and WT-CFTR PIPs not only recapitulated the identification of PDIA4, LGALS3BP and PTBP1 as targets for the functional correction of the F508del variant [23], but also identified new targets such PDIA1 and unc45a, chaperone proteins with a known regulatory function in protein folding. While none of these specific targets have been directly linked to the correction of the defects associated with the F508del variant, we and others have previously shown that modulating the expression level of chaperone proteins can lead to the establishment of a corrective folding proteostasis environment that is permissive for improved folding and trafficking of F508del-CFTR [29, 31, 36, 48–54]. These previous observations provide supporting evidence for the validity of these newly identified targets supporting the usefulness of ProVarA for the CF community. A comparative analysis of the PIPs for the 4 ER-restricted variants revealed that many of the identified targets described above are also recovered with increased affinity for Class II mutants compared to the level seen with WT-CFTR, including HspA5, calmodulin, PDIA1, LMO7 and PGRMC1, suggesting that they may also be strong targets for the correction of other Class II variants. In contrast, others, such as HspA8, PDIA4, LGALS3BP, PTBP1, RPS2 and RPS6 were only recovered with increased affinity with the F508del, raising the possibility that they could represent high value variant-specific targets for functional correction of F508del. Additionally, each PIP includes its own unique protein set, but it remains to be determined if they represent variant-specific targets which could be exploited for therapeutic interventions for patients carrying these specific mutations.
In particular, we have demonstrated the utility of ProVarA in expanding our understanding of FDA-approved CF therapeutics. Often the determination of whether a drug is a viable therapeutic option for a given genotype comes from trial and error, a painstaking task in the best of conditions, but extremely difficult in CF, where 2025 mutations have been identified. Our new ProVarA-based assessment of the impact of Vx770 on the Class III variant, G551D-CFTR as well as for the impact of Vx809 on the Class II variant, F508del-CFTR yielded unique PIPs who’s binding to CFTR variants were affected by therapeutic intervention. We observed a reduction in the binding affinity of the chaperone proteins HspA5, PDIA1 and unc45a with F508del-CFTR in response to Vx809 treatment. Interestingly, combining the silencing of these targets with Vx809 treatment did not yield a statistically significant difference in the Vx809-mediated functional correction of F508del, supporting the hypothesis that they are, at least in part, contributing to the Vx809-mediated function of F508del-CFTR. With the recent FDA-approval of Symdeco, a combination-based therapeutic similar to Orkambi (Vx770/Vx809), which replaces Vx809 with the new corrector Vx661, there would be significant benefit to explore the impact of Vx661 on the PIP of F508del-CFTR as well as on other class II CF-causing variants to compare and contrast the changes in the PIP that are incurred in response to both of these small molecules. Such a ProVarA-based analysis would contribute to discerning the mechanism of action of this new corrector as well as assist in determining which variants might benefit from this compound in the clinic.
Moreover, our analysis of the impact of Vx770 on the G551D PIP identified protein cohorts involved in trafficking and cytoskeletal reorganization at the plasma membrane. Because these failed to properly engage G551D, we hypothesize that they contribute to the phenotype of this Class III variant. We observed that binding of the key components of this subnetwork are restored to a more WT-like affinity following treatment with the potentiator, Vx770, supporting our hypothesis that the absence of these proteins is causal in the etiology of G551D-CF disease. The ability of ProVarA to characterize the PIP of individual variants, compare variant specific PIP and to characterize the impact of small molecule therapeutics on the PIP of these variants will significantly impact the CF research community and our collective objective of continuing to develop clinically relevant therapeutics to benefit all patients affected by CF.
A challenge remains in terms of the best model to utilize to generate PIPs. The development of protocols to isolate and grow primary bronchial epithelial cells (hBE) from explanted lungs of CF patients [116, 117] opened the door for the development of precision medicine in CF. These cells allow for the testing of small molecule therapeutics using a relevant cell-based model that recapitulates the lung environment and have proven a valuable resource in the identification of existing approved therapeutics, including Ivacaftor and Lumacaftor. A limitation that arises is the fact that these cells are isolated from lungs removed during a lung transplant. While this patient will no longer suffer from the respiratory component of CF, they will still exhibit CF-associated intestinal and pancreatic disease, suggesting that the knowledge obtained from studying these patient derived cells will be applicable both on an individual patient basis as well as from a global understanding of CF disease. The recent development of protocols to allow for the expansion of primary bronchial cells (hBE) without the loss of electrophysiological properties, opens the door for the generation of the numbers of cells needed for the generation of PIP. A ProVarA-based analysis of PIP from patient derived hBE cells carrying the same CF-associated mutation would allow us to study the impact of genetic modifiers on the PIP of a given mutation, while a ProVarA-based analysis of the PIP from patients carrying different mutations would allow us to study the impact of variation on CFTR biology and identify both common and unique protein markers characteristic of these different disease-causing variants.
Additionally, the advent of protocols for isolating and growing of patient-derived intestinal and bronchial organoids make it possible to assess the therapeutic benefit of a panel of available compounds on a patient-to-patient basis. The in vitro data from these cells have been shown to correlate with clinical data [118–120] suggesting that this resource can be exploited for the benefit of precision medicine. However, their use purely as a screening tool fails to get to the underlying molecular cause(s) of CF disease for the different mutations nor do they provide insight into the mechanism of action of the very therapeutics they are screened against. Recently, the CF Canada-Sick Kids Program in Individual CF Therapy (CFIT) has proposed a more expansive program to determine gene expression profiles, whole genome sequencing (WGS) and response to therapeutics from patient derived nasal cells as well as generating and biobanking iPSC cells from these isolated cells [121]. While this effort aims to generate valuable data that extends beyond a purely electrophysiological read out of CFTR function in response to small molecules, the effort is currently limited to F508del patients and generates whole cell data that will be difficult to interpret without a direct readout on their impact on the CFTR polypeptide. For example, expression profiling data could identify countless changes in mRNA expression that result from dysfunction of CFTR, however the impact of these transcriptional differences on their respective protein expression are lacking and the impact of these differences on CFTR itself would make correlative interpretation difficult. Perhaps a further expansion of these efforts could combine the isolation of nasal and bronchial cells from a more diverse array of CF patients. Additionally, these cells can be expanded to levels that would allow not only for the screening efforts suggested by CFIT but would also allow for proteomic efforts aimed at understanding the impact of variation on the CFTR PIP of both a cohort of patients carrying the same mutations as well as those carrying a broader spectrum of CF-causing mutations and the impact of therapeutics on these PIPs. Combining whole cell data such as expression profiling and WGS with CFTR-centric studies such as protein interaction profiling combined with ProVarA-based analyses would allow for a more detailed understanding of the impact of these mutations on CFTR biogenesis as well as a better understanding of the mechanisms that dictate the successes and failures of therapeutics on a patient to patient basis.
Understanding the contribution of variation to human disease using ProVarA
ProVarA is generally applicable to any disease condition where a variant (familial or sporadic/somatic) contributes to pathology. Using comparative differential proteomics in the context of variation, revealed by ongoing WGS, whole exome sequencing (WES) and genotyping efforts, ProVarA provides a quantitative and robust approach to ascertain a plethora of unknown protein interactions driving disease etiology and would serve as a direct metric to ascertain the impact of expression profiling and WGS on the disease-associated protein. Once a database of PIP for a set of well characterized variants are established, a set of therapeutic biomarkers are extracted from the data and the impact of therapeutics on target biomarkers are established, the reference data set can be used to predict which small molecule(s) to utilize to make the necessary adjustments to the patient specific PIP to restore a more WT-like PIP and alleviate disease (Figure 7). We posit that ProVarA-based analyses can be used to better understand the underlying mechanistic details responsible for the basic defects associated with an individual variant and/or classes of variants based on their differential PIPs. By using a complete profile rather that individual hits, as is commonly practiced in familial/somatic disease analyses, ProVarA leverages the power of variation to help dissect both the common and specific interactions dictated by genetic diversity that drive the overall presentation of disease in the clinic. While additional work is needed to determine the validity and specificity of the multiple targets identified herein and to determine the number of PIPs needed to reach the necessary power to achieve maximal predictability, we believe that ProVarA, as implemented for 6 variants, already provides a significant advancement towards achieving precision medicine-based therapeutic in CF-disease.
Material and Methods
CFTR constructs
The cDNA constructs for the CFTR variants were a generous gift of Phil Thomas (University of Texas Southwestern) provided in pBL-CMV2. The variants were PCR amplified using CFTR specific primers (FRW: TCATGGTACCATGCAGAGGTCGCCT; REV: GCTGCTCGAGCTAAGCGTAATCTGGAACATCGTATGGGTAAAGCCTTGTATCTTG) and inserted into the KpnI/XhoI sites of pENTR1A shuttle vector. The DNA constructs were fully sequenced and recombined into the pAD-CMV-V5-DEST adenoviral vector (Invitrogen). Final constructs were sent to ViraQuest (North Liberty, IA) for viral particles generation and titration.
Cell Culture
The CFBE41o- cell line is an SV40 immortalized bronchial epithelial cell line derived from a F508del homozygous patient [122]. This original parental lineage stopped expressing the endogenous F508del variant providing a null CFTR background for expressing CF-causing tvaraint transgenes. The cells were cultured in alpha-MEM containing penicillin and streptomycin and supplemented with 10% FBS and 2mM L-glutamine.
Transduction
CBFE41o- cells were plated in 150mm culture dishes (1 × 150mm dish per replicate per condition) at a density of 6.4 × 106 cells per dish and cultured overnight as described above. Viral particles were diluted in complete culture media at a multiplicity of infection (MOI) of 200 and added to the cells. The cells were incubated with the virus for 18h. The cells were washed 2 × 20ml of PBS and replenished with complete culture media and incubated for 54 h before harvesting cells for immunoprecipitation (72h total culture time).
Immunoprecipitation
Transduced CFBE41o- cells were washed 2× with 10ml of PBS and lysed in ice cold lysis buffer (50mM Tris-HCl pH 7.4; 250mM NaCl; 1mM EDTA; 0.5% IGEPAL-CA630; 2 µg/ml of protease inhibitor cocktail (Roche)) [23] for 30 minutes on ice directly in the 150mm culture dish with rocking. The culture dishes were scraped, the lysate collected, and a protein assay was performed to determine the protein concentration. The lysate was pre-cleared using 50 µl of GammaBind Plus Sepharose beads for 1h at 4°C with mixing. The beads were pelleted at 500× g for 5 min at 4°C and the pre-cleared lysate transferred to a new tube. The CFTR immunoprecipitation was performed by adding the 3G11 CFTR antibody pre-crosslinked to GammaBind Plus Sepharose beads to the pre-cleared lysates (4mg total protein per IP). The lysate was incubated with the antibody overnight at 4°C with end-over-end mixing. The beads were pelleted at 500× g for 5 min at 4°C and washed twice with 10 bead volumes of lysis buffer and twice with 10 bead volumes of lysis buffer without IGEPAL-CA630.
Sample preparation
Following immunoprecipitation washes with lysis buffer and 50mM ammonium bicarbonate, proteins were digested directly on-beads. Briefly, proteins bound to the beads were resuspended with 8M urea, 50 mM ammonium bicarbonate, and cysteine disulfide bonds were reduced with 10 mM tris(2-carboxyethyl)phosphine (TCEP) at 30°C for 60 min followed by cysteine alkylation with 30 mM iodoacetamide (IAA) in the dark at room temperature for 30 min. Following alkylation, urea was diluted to 1 M urea using 50 mM ammonium bicarbonate, and proteins were finally subjected to overnight digestion with mass spec grade Trypsin/Lys-C mix (Promega, Madison, WI). Finally, beads were pulled down and the solution with peptides collected into a new tube. The beads were then washed once with 50mM ammonium bicarbonate to increase peptide recovery. The digested samples were desalted using a C18 TopTip (PolyLC, Columbia, MD), and the organic solvent was removed in a SpeedVac concentrator prior to LC-MS/MS analysis.
2DLC-MS/MS analysis
Dried samples were reconstituted in 100mM ammonium formate pH ~10 and analyzed by 2DLC-MS/MS using a 2D nanoACQUITY Ultra Performance Liquid Chromatography (UPLC) system (Waters corp., Milford, MA) coupled to a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific). Peptides were loaded onto the first-dimension column, XBridge BEH130 C18 NanoEase (300 µm × 50 mm, 5 µm) equilibrated with solvent A (20mM ammonium formate pH 10, first dimension pump) at 2 µL/min. The first fraction was eluted from the first dimension column at 17% of solvent B (100% acetonitrile) for 4 min and transferred to the second dimension Symmetry C18 trap column 0.180 × 20 mm (Waters corp., Milford, MA) using a 1:10 dilution with 99.9% second dimensional pump solvent A (0.1% formic acid in water) at 20 µL/min. Peptides were then eluted from the trap column and resolved on the analytical C18 BEH130 PicoChip column 0.075 × 100 mm, 1.7µm particles (NewObjective, MA) at low pH by increasing the composition of solvent B (100% acetonitrile) from 2 to 26% over 94 min at 400 nL/min. Subsequent fractions were carried with increasing concentrations of solvent B. The following 4 first dimension fractions were eluted at 19.5, 22, 26, and 65% solvent B. The mass spectrometer was operated in positive data-dependent acquisition mode. MS1 spectra were measured with a resolution of 70,000, an AGC target of 1e6 and a mass range from 350 to 1700 m/z. Up to 12 MS2 spectra per duty cycle were triggered, fragmented by HCD, and acquired with a resolution of 17,500 and an AGC target of 5e4, an isolation window of 2.0 m/z and a normalized collision energy of 25. Dynamic exclusion was enabled with duration of 20 sec.
Data analysis
All mass spectra from were analyzed with MaxQuant software version 1.5.5.1. MS/MS spectra were searched against the Homo sapiens Uniprot protein sequence database (version July 2016) and GPM cRAP sequences (commonly known protein contaminants). Precursor mass tolerance was set to 20ppm and 4.5ppm for the first search where initial mass recalibration was completed and for the main search, respectively. Product ions were searched with a mass tolerance 0.5 Da. The maximum precursor ion charge state used for searching was 7. Carbamidomethylation of cysteines was searched as a fixed modification, while oxidation of methionines and acetylation of protein N-terminal were searched as variable modifications. Enzyme was set to trypsin in a specific mode and a maximum of two missed cleavages was allowed for searching. The target-decoy-based false discovery rate (FDR) filter for spectrum and protein identification was set to 1%.
Peptide intensities were log2 transformed normalized to reduce systematic bias. An evaluation of different normalization options with the Normalyzer tool [123], it was determined that the Loess-R was the most optimal. Protein intensities were obtained by summing up all normalized peptide intensities, and bait-normalization was performed by dividing each protein intensity in each replicate by the corresponding CFTR protein intensity. Different replicates were aggregated by the median, requiring at least two values per condition. For proteins detected only in one condition, a pseudo-value was assigned to the missing condition in order to avoid indefinite fold changes. Statistical testing between conditions was conducted with a Wilcoxon signed-rank test at the peptide level (i.e. for each protein between two conditions, testing was performed between normalized log2 intensities of the corresponding peptides in the two conditions tested, requiring at least two peptides per condition). The test against the null condition (CFTR immunoprecipitation in CFBE41o- cells transduced with GFP) was used to assess statistical significance of recovered proteins. Proteostasis annotation was assembled using protein subcellular classification from the Protein Atlas [1] and LocDB [124]. All computations were done with R/Bioconductor [125], and network visualization with Cytoscape [126]. The networks generated in this study are availabile through the Network Data Exchange platform NDEx [127].
CFBE-YFP quenching assay
CFBE41o- cells stably expressing F508del and the halide sensitive YFP-H148Q/I152L (CFBE-YFP) [104], were reverse transfected with 50 nM final concentration of siRNA, and 0.09 µl of lipofectamine RNAiMax (Invitrogen) per well for a 384 plate. Cells were trypsinized, suspended in opti-MEM with 10% FBS and 6 × 103 cells added per well. Opti-MEM was replaced with growth media 24 h after transfection and the YFP-assay performed as previously described [128] 72 h after transfection.
Supplementary Material
Acknowledgments
This project was supported by grants from the NIH (HL095524, DK051870 & AG049665) as well as by a grant from the Tobacco Related Disease Research Program (TRDRP) (23RT-0012).
References
- 1.Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, et al. A subcellular map of the human proteome. Science. 2017;356 doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 2.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–8. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karczewski KJ, Weisburd B, Thomas B, Solomonson M, Ruderfer DM, Kavanagh D, et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 2017;45:D840–D5. doi: 10.1093/nar/gkw971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hindorff LA, Bonham VL, Brody LC, Ginoza MEC, Hutter CM, Manolio TA, et al. Prioritizing diversity in human genomics research. Nat Rev Genet. 2017 doi: 10.1038/nrg.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long T, Hicks M, Yu HC, Biggs WH, Kirkness EF, Menni C, et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet. 2017;49:568–78. doi: 10.1038/ng.3809. [DOI] [PubMed] [Google Scholar]
- 7.Price ND, Magis AT, Earls JC, Glusman G, Levy R, Lausted C, et al. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat Biotechnol. 2017;35:747–56. doi: 10.1038/nbt.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D. Methods of integrating data to uncover genotype-phenotype interations. Nature Rev Genetics. 2015;16:85–97. doi: 10.1038/nrg3868. [DOI] [PubMed] [Google Scholar]
- 9.Consortium GT, Laboratory DA, et al. Coordinating Center -Analysis Working G, Statistical Methods groups-Analysis Working G, Enhancing Gg, Fund NIHC. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–13. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sosnay PR, Cutting GR. Interpretation of genetic variants. Thorax. 2014;69:295–7. doi: 10.1136/thoraxjnl-2013-204903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torkamani A, Andersen KG, Steinhubl SR, Topol EJ. High-Definition Medicine. Cell. 2017;170:828–43. doi: 10.1016/j.cell.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amaral MD, Balch WE. Hallmarks of therapeutic management of the cystic fibrosis functional landscape. J Cyst Fibros. 2015;14:687–99. doi: 10.1016/j.jcf.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balch WE, Roth DM, Hutt DM. Emergent properties of proteostasis in managing cystic fibrosis. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callebaut I, Chong PA, Forman-Kay JD. CFTR structure. J Cyst Fibros. 2017 doi: 10.1016/j.jcf.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Hartl D, Amaral M. Cystic fibrosis -- From basic science to clinical benefit: A review series. J Cyst Fibros. 2015;14:415–6. doi: 10.1016/j.jcf.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 16.McClure ML, Barnes S, Brodsky JL, Sorscher EJ. Trafficking and function of the cystic fibrosis transmembrane conductance regulator: a complex network of posttranslational modifications. Am J Physiol Lung Cell Mol Physiol. 2016;311:L719–L33. doi: 10.1152/ajplung.00431.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spielberg DR, Clancy JP. Cystic Fibrosis and Its Management Through Established and Emerging Therapies. Annu Rev Genomics Hum Genet. 2016 doi: 10.1146/annurev-genom-090314-050024. [DOI] [PubMed] [Google Scholar]
- 18.Sosnay PR, Siklosi KR, Van Goor F, Kaniecki K, Yu H, Sharma N, et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat Genet. 2013;45:1160–7. doi: 10.1038/ng.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veit G, Avramescu RG, Chiang AN, Houck SA, Cai Z, Peters KW, et al. From CFTR biology toward combinatorial pharmacotherapy: expanded classification of cystic fibrosis mutations. Mol Biol Cell. 2016;27:424–33. doi: 10.1091/mbc.E14-04-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Boeck K, Amaral MD. Progress in therapies for cystic fibrosis. Lancet Respir Med. 2016;4:662–74. doi: 10.1016/S2213-2600(16)00023-0. [DOI] [PubMed] [Google Scholar]
- 21.Marson FAL, Bertuzzo CS, Ribeiro JD. Classification of CFTR mutation classes. Lancet Respir Med. 2016;4:e37–e8. doi: 10.1016/S2213-2600(16)30188-6. [DOI] [PubMed] [Google Scholar]
- 22.Coppinger JA, Hutt DM, Razvi A, Koulov AV, Pankow S, Yates JR, 3rd, et al. A chaperone trap contributes to the onset of cystic fibrosis. PLoS One. 2012;7:e37682. doi: 10.1371/journal.pone.0037682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pankow S, Bamberger C, Calzolari D, Martinez-Bartolome S, Lavallee-Adam M, Balch WE, et al. F508 CFTR interactome remodelling promotes rescue of cystic fibrosis. Nature. 2015;528:510–6. doi: 10.1038/nature15729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Venable J, LaPointe P, Hutt DM, Koulov AV, Coppinger J, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–15. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Matteson J, An Y, Moyer B, Yoo JS, Bannykh S, et al. COPII-dependent export of cystic fibrosis transmembrane conductance regulator from the ER uses a di-acidic exit code. J Cell Biol. 2004;167:65–74. doi: 10.1083/jcb.200401035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–9. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 27.Brodsky JL, Wojcikiewicz RJ. Substrate-specific mediators of ER associated degradation (ERAD) Curr Opin Cell Biol. 2009;21:516–21. doi: 10.1016/j.ceb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–57. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong X, Ahner A, Roldan A, Lukacs GL, Thibodeau PH, Frizzell RA. Non-native Conformers of Cystic Fibrosis Transmembrane Conductance Regulator NBD1 Are Recognized by Hsp27 and Conjugated to SUMO-2 for Degradation. J Biol Chem. 2016;291:2004–17. doi: 10.1074/jbc.M115.685628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahner A, Gong X, Frizzell RA. Divergent signaling via SUMO modification: potential for CFTR modulation. Am J Physiol Cell Physiol. 2016;310:C175–80. doi: 10.1152/ajpcell.00124.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahner A, Gong X, Schmidt BZ, Peters KW, Rabeh WM, Thibodeau PH, et al. Small heat shock proteins target mutant cystic fibrosis transmembrane conductance regulator for degradation via a small ubiquitin-like modifier-dependent pathway. Mol Biol Cell. 2013;24:74–84. doi: 10.1091/mbc.E12-09-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saxena A, Banasavadi-Siddegowda YK, Fan Y, Bhattacharya S, Roy G, Giovannucci DR, et al. Human heat shock protein 105/110 kDa (Hsp105/110) regulates biogenesis and quality control of misfolded cystic fibrosis transmembrane conductance regulator at multiple levels. J Biol Chem. 2012;287:19158–70. doi: 10.1074/jbc.M111.297580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt BZ, Watts RJ, Aridor M, Frizzell RA. Cysteine string protein promotes proteasomal degradation of the cystic fibrosis transmembrane conductance regulator (CFTR) by increasing its interaction with the C terminus of Hsp70-interacting protein and promoting CFTR ubiquitylation. J Biol Chem. 2009;284:4168–78. doi: 10.1074/jbc.M806485200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hutt DM, Roth DM, Chalfant MA, Youker RT, Matteson J, Brodsky JL, et al. FK506 binding protein 8 peptidylprolyl isomerase activity manages a late stage of cystic fibrosis transmembrane conductance regulator (CFTR) folding and stability. J Biol Chem. 2012;287:21914–25. doi: 10.1074/jbc.M112.339788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran JR, Tomsic LR, Brodsky JL. A Cdc48p-associated factor modulates endoplasmic reticulum-associated degradation, cell stress, and ubiquitinated protein homeostasis. J Biol Chem. 2011;286:5744–55. doi: 10.1074/jbc.M110.179259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahner A, Nakatsukasa K, Zhang H, Frizzell RA, Brodsky JL. Small heat-shock proteins select deltaF508-CFTR for endoplasmic reticulum-associated degradation. Mol Biol Cell. 2007;18:806–14. doi: 10.1091/mbc.E06-05-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koulov AV, LaPointe P, Lu B, Razvi A, Coppinger J, Dong MQ, et al. Biological and structural basis for Aha1 regulation of Hsp90 ATPase activity in maintaining proteostasis in the human disease cystic fibrosis. Mol Biol Cell. 2010;21:871–84. doi: 10.1091/mbc.E09-12-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 39.Gasman S, Chasserot-Golaz S, Malacombe M, Way M, Bader MF. Regulated exocytosis in neuroendocrine cells: a role for subplasmalemmal Cdc42/N-WASP-induced actin filaments. Mol Biol Cell. 2004;15:520–31. doi: 10.1091/mbc.E03-06-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hart MJ, Maru Y, Leonard D, Witte ON, Evans T, Cerione RA. A GDP dissociation inhibitor that serves as a GTPase inhibitor for the Ras-like protein CDC42Hs. Science. 1992;258:812–5. doi: 10.1126/science.1439791. [DOI] [PubMed] [Google Scholar]
- 41.Hand RA, Craven RJ. Hpr6.6 protein mediates cell death from oxidative damage in MCF-7 human breast cancer cells. J Cell Biochem. 2003;90:534–47. doi: 10.1002/jcb.10648. [DOI] [PubMed] [Google Scholar]
- 42.Reilly R, Mroz MS, Dempsey E, Wynne K, Keely SJ, McKone EF, et al. Targeting the PI3K/Akt/mTOR signalling pathway in Cystic Fibrosis. Sci Rep. 2017;7:7642. doi: 10.1038/s41598-017-06588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Estacio SG, Martiniano HF, Faisca PF. Thermal unfolding simulations of NBD1 domain variants reveal structural motifs associated with the impaired folding of F508del-CFTR. Mol Biosyst. 2016;12:2834–48. doi: 10.1039/c6mb00193a. [DOI] [PubMed] [Google Scholar]
- 44.Lewis HA, Wang C, Zhao X, Hamuro Y, Conners K, Kearins MC, et al. Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry. J Mol Biol. 2010;396:406–30. doi: 10.1016/j.jmb.2009.11.051. [DOI] [PubMed] [Google Scholar]
- 45.Lopes-Pacheco M, Boinot C, Sabirzhanova I, Morales MM, Guggino WB, Cebotaru L. Combination of Correctors Rescue DeltaF508-CFTR by Reducing Its Association with Hsp40 and Hsp27. J Biol Chem. 2015;290:25636–45. doi: 10.1074/jbc.M115.671925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Decaestecker K, Decaestecker E, Castellani C, Jaspers M, Cuppens H, De Boeck K. Genotype/phenotype correlation of the G85E mutation in a large cohort of cystic fibrosis patients. Eur Respir J. 2004;23:679–84. doi: 10.1183/09031936.04.00014804. [DOI] [PubMed] [Google Scholar]
- 47.Patrick AE, Karamyshev AL, Millen L, Thomas PJ. Alteration of CFTR transmembrane span integration by disease-causing mutations. Mol Biol Cell. 2011;22:4461–71. doi: 10.1091/mbc.E11-05-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bagdany M, Veit G, Fukuda R, Avramescu RG, Okiyoneda T, Baaklini I, et al. Chaperones rescue the energetic landscape of mutant CFTR at single molecule and in cell. Nat Commun. 2017;8:398. doi: 10.1038/s41467-017-00444-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grumbach Y, Bikard Y, Suaud L, Chanoux RA, Rubenstein RC. ERp29 regulates epithelial sodium channel functional expression by promoting channel cleavage. Am J Physiol Cell Physiol. 2014;307:C701–9. doi: 10.1152/ajpcell.00134.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ihrig V, Obermann WMJ. Identifying Inhibitors of the Hsp90-Aha1 Protein Complex, a Potential Target to Drug Cystic Fibrosis, by Alpha Technology. SLAS Discov. 2017;22:923–8. doi: 10.1177/2472555216688312. [DOI] [PubMed] [Google Scholar]
- 51.Kakoi S, Yorimitsu T, Sato K. COPII machinery cooperates with ER-localized Hsp40 to sequester misfolded membrane proteins into ER-associated compartments. Mol Biol Cell. 2013;24:633–42. doi: 10.1091/mbc.E12-08-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsumura Y, Sakai J, Skach WR. Endoplasmic reticulum protein quality control is determined by cooperative interactions between Hsp/c70 protein and the CHIP E3 ligase. J Biol Chem. 2013;288:31069–79. doi: 10.1074/jbc.M113.479345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young JC. The role of the cytosolic HSP70 chaperone system in diseases caused by misfolding and aberrant trafficking of ion channels. Dis Model Mech. 2014;7:319–29. doi: 10.1242/dmm.014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roth DM, Hutt DM, Tong J, Bouchecareilh M, Wang N, Seeley T, et al. Modulation of the maladaptive stress response to manage diseases of protein folding. PLoS Biol. 2014;12:e1001998. doi: 10.1371/journal.pbio.1001998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teng L, Kerbiriou M, Taiya M, Le Hir S, Mignen O, Benz N, et al. Proteomic identification of calumenin as a G551D-CFTR associated protein. PLoS One. 2012;7:e40173. doi: 10.1371/journal.pone.0040173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trouve P, Kerbiriou M, Teng L, Benz N, Taiya M, Le Hir S, et al. G551D-CFTR needs more bound actin than wild-type CFTR to maintain its presence in plasma membranes. Cell Biol Int. 2015;39:978–85. doi: 10.1002/cbin.10456. [DOI] [PubMed] [Google Scholar]
- 57.Ho HY, Rohatgi R, Lebensohn AM, Le M, Li J, Gygi SP, et al. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 2004;118:203–16. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 58.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 59.Egorov MV, Capestrano M, Vorontsova OA, Di Pentima A, Egorova AV, Mariggio S, et al. Faciogenital dysplasia protein (FGD1) regulates export of cargo proteins from the golgi complex via Cdc42 activation. Mol Biol Cell. 2009;20:2413–27. doi: 10.1091/mbc.E08-11-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ellis S, Mellor H. Regulation of endocytic traffic by rho family GTPases. Trends Cell Biol. 2000;10:85–8. doi: 10.1016/s0962-8924(99)01710-9. [DOI] [PubMed] [Google Scholar]
- 61.Garrett WS, Chen LM, Kroschewski R, Ebersold M, Turley S, Trombetta S, et al. Developmental control of endocytosis in dendritic cells by Cdc42. Cell. 2000;102:325–34. doi: 10.1016/s0092-8674(00)00038-6. [DOI] [PubMed] [Google Scholar]
- 62.Kroschewski R, Hall A, Mellman I. Cdc42 controls secretory and endocytic transport to the basolateral plasma membrane of MDCK cells. Nat Cell Biol. 1999;1:8–13. doi: 10.1038/8977. [DOI] [PubMed] [Google Scholar]
- 63.Malacombe M, Ceridono M, Calco V, Chasserot-Golaz S, McPherson PS, Bader MF, et al. Intersectin-1L nucleotide exchange factor regulates secretory granule exocytosis by activating Cdc42. EMBO J. 2006;25:3494–503. doi: 10.1038/sj.emboj.7601247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu WJ, Erickson JW, Lin R, Cerione RA. The gamma-subunit of the coatomer complex binds Cdc42 to mediate transformation. Nature. 2000;405:800–4. doi: 10.1038/35015585. [DOI] [PubMed] [Google Scholar]
- 65.Izumi G, Sakisaka T, Baba T, Tanaka S, Morimoto K, Takai Y. Endocytosis of E-cadherin regulated by Rac and Cdc42 small G proteins through IQGAP1 and actin filaments. J Cell Biol. 2004;166:237–48. doi: 10.1083/jcb.200401078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lamaze C, Chuang TH, Terlecky LJ, Bokoch GM, Schmid SL. Regulation of receptor-mediated endocytosis by Rho and Rac. Nature. 1996;382:177–9. doi: 10.1038/382177a0. [DOI] [PubMed] [Google Scholar]
- 67.Malecz N, McCabe PC, Spaargaren C, Qiu R, Chuang Y, Symons M. Synaptojanin 2, a novel Rac1 effector that regulates clathrin-mediated endocytosis. Curr Biol. 2000;10:1383–6. doi: 10.1016/s0960-9822(00)00778-8. [DOI] [PubMed] [Google Scholar]
- 68.Ridley AJ. Rho proteins: linking signaling with membrane trafficking. Traffic. 2001;2:303–10. doi: 10.1034/j.1600-0854.2001.002005303.x. [DOI] [PubMed] [Google Scholar]
- 69.Bi Y, Williams JA. A role for Rho and Rac in secretagogue-induced amylase release by pancreatic acini. Am J Physiol Cell Physiol. 2005;289:C22–32. doi: 10.1152/ajpcell.00395.2004. [DOI] [PubMed] [Google Scholar]
- 70.Doussau F, Gasman S, Humeau Y, Vitiello F, Popoff M, Boquet P, et al. A Rho-related GTPase is involved in Ca(2+)-dependent neurotransmitter exocytosis. J Biol Chem. 2000;275:7764–70. doi: 10.1074/jbc.275.11.7764. [DOI] [PubMed] [Google Scholar]
- 71.Li Q, Ho CS, Marinescu V, Bhatti H, Bokoch GM, Ernst SA, et al. Facilitation of Ca(2+)-dependent exocytosis by Rac1-GTPase in bovine chromaffin cells. J Physiol. 2003;550:431–45. doi: 10.1113/jphysiol.2003.039073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Momboisse F, Ory S, Ceridono M, Calco V, Vitale N, Bader MF, et al. The Rho guanine nucleotide exchange factors Intersectin 1L and beta-Pix control calcium-regulated exocytosis in neuroendocrine PC12 cells. Cell Mol Neurobiol. 2010;30:1327–33. doi: 10.1007/s10571-010-9580-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sylow L, Jensen TE, Kleinert M, Hojlund K, Kiens B, Wojtaszewski J, et al. Rac1 signaling is required for insulin-stimulated glucose uptake and is dysregulated in insulin-resistant murine and human skeletal muscle. Diabetes. 2013;62:1865–75. doi: 10.2337/db12-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sylow L, Kleinert M, Pehmoller C, Prats C, Chiu TT, Klip A, et al. Akt and Rac1 signaling are jointly required for insulin-stimulated glucose uptake in skeletal muscle and downregulated in insulin resistance. Cell Signal. 2014;26:323–31. doi: 10.1016/j.cellsig.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 75.Sylow L, Nielsen IL, Kleinert M, Moller LL, Ploug T, Schjerling P, et al. Rac1 governs exercise-stimulated glucose uptake in skeletal muscle through regulation of GLUT4 translocation in mice. J Physiol. 2016;594:4997–5008. doi: 10.1113/JP272039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ueda S, Kitazawa S, Ishida K, Nishikawa Y, Matsui M, Matsumoto H, et al. Crucial role of the small GTPase Rac1 in insulin-stimulated translocation of glucose transporter 4 to the mouse skeletal muscle sarcolemma. FASEB J. 2010;24:2254–61. doi: 10.1096/fj.09-137380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hedman AC, Smith JM, Sacks DB. The biology of IQGAP proteins: beyond the cytoskeleton. EMBO Rep. 2015;16:427–46. doi: 10.15252/embr.201439834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nouri K, Fansa EK, Amin E, Dvorsky R, Gremer L, Willbold D, et al. IQGAP1 Interaction with RHO Family Proteins Revisited: KINETIC AND EQUILIBRIUM EVIDENCE FOR MULTIPLE DISTINCT BINDING SITES. J Biol Chem. 2016;291:26364–76. doi: 10.1074/jbc.M116.752121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rittmeyer EN, Daniel S, Hsu SC, Osman MA. A dual role for IQGAP1 in regulating exocytosis. J Cell Sci. 2008;121:391–403. doi: 10.1242/jcs.016881. [DOI] [PubMed] [Google Scholar]
- 80.Nomanbhoy TK, Cerione R. Characterization of the interaction between RhoGDI and Cdc42Hs using fluorescence spectroscopy. J Biol Chem. 1996;271:10004–9. doi: 10.1074/jbc.271.17.10004. [DOI] [PubMed] [Google Scholar]
- 81.Prince LS, Workman RB, Jr, Marchase RB. Rapid endocytosis of the cystic fibrosis transmembrane conductance regulator chloride channel. Proc Natl Acad Sci U S A. 1994;91:5192–6. doi: 10.1073/pnas.91.11.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cutting GR. Treating Specific Variants Causing Cystic Fibrosis. JAMA. 2017;318:2130–1. doi: 10.1001/jama.2017.16823. [DOI] [PubMed] [Google Scholar]
- 83.Oliver KE, Han ST, Sorscher EJ, Cutting GR. Transformative therapies for rare CFTR missense alleles. Curr Opin Pharmacol. 2017;34:76–82. doi: 10.1016/j.coph.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cutting GR. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet. 2015;16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van Goor F, Hadida S, Grootenhuis PD, Burton B, Cao D, Neuberger T, et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc Natl Acad Sci U S A. 2009;106:18825–30. doi: 10.1073/pnas.0904709106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Goor F, Yu H, Burton B, Hoffman BJ. Effect of ivacaftor on CFTR forms with missense mutations associated with defects in protein processing or function. J Cyst Fibros. 2014;13:29–36. doi: 10.1016/j.jcf.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 87.Fidler MC, Beusmans J, Panorchan P, Van Goor F. Correlation of sweat chloride and percent predicted FEV1 in cystic fibrosis patients treated with ivacaftor. J Cyst Fibros. 2017;16:41–4. doi: 10.1016/j.jcf.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 88.Yu H, Burton B, Huang CJ, Worley J, Cao D, Johnson JP, Jr, et al. Ivacaftor potentiation of multiple CFTR channels with gating mutations. J Cyst Fibros. 2012;11:237–45. doi: 10.1016/j.jcf.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 89.Eckford PD, Li C, Ramjeesingh M, Bear CE. Cystic fibrosis transmembrane conductance regulator (CFTR) potentiator VX-770 (ivacaftor) opens the defective channel gate of mutant CFTR in a phosphorylation-dependent but ATP-independent manner. J Biol Chem. 2012;287:36639–49. doi: 10.1074/jbc.M112.393637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jih KY, Hwang TC. Vx-770 potentiates CFTR function by promoting decoupling between the gating cycle and ATP hydrolysis cycle. Proc Natl Acad Sci U S A. 2013;110:4404–9. doi: 10.1073/pnas.1215982110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Okiyoneda T, Barriere H, Bagdany M, Rabeh WM, Du K, Hohfeld J, et al. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science. 2010;329:805–10. doi: 10.1126/science.1191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wolde M, Fellows A, Cheng J, Kivenson A, Coutermarsh B, Talebian L, et al. Targeting CAL as a negative regulator of DeltaF508-CFTR cell-surface expression: an RNA interference and structure-based mutagenetic approach. J Biol Chem. 2007;282:8099–109. doi: 10.1074/jbc.M611049200. [DOI] [PubMed] [Google Scholar]
- 93.Bulloch MN, Hanna C, Giovane R. Lumacaftor/ivacaftor, a novel agent for the treatment of cystic fibrosis patients who are homozygous for the F580del CFTR mutation. Expert Rev Clin Pharmacol. 2017;10:1055–72. doi: 10.1080/17512433.2017.1378094. [DOI] [PubMed] [Google Scholar]
- 94.Rowe SM, McColley SA, Rietschel E, Li X, Bell SC, Konstan MW, et al. Lumacaftor/Ivacaftor Treatment of Patients with Cystic Fibrosis Heterozygous for F508del-CFTR. Ann Am Thorac Soc. 2017;14:213–9. doi: 10.1513/AnnalsATS.201609-689OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Van Goor F, Hadida S, Grootenhuis PD, Burton B, Stack JH, Straley KS, et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc Natl Acad Sci U S A. 2011;108:18843–8. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ren HY, Grove DE, De La Rosa O, Houck SA, Sopha P, Van Goor F, et al. VX-809 corrects folding defects in cystic fibrosis transmembrane conductance regulator protein through action on membrane-spanning domain 1. Mol Biol Cell. 2013;24:3016–24. doi: 10.1091/mbc.E13-05-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Serohijos AW, Hegedus T, Aleksandrov AA, He L, Cui L, Dokholyan NV, et al. Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc Natl Acad Sci U S A. 2008;105:3256–61. doi: 10.1073/pnas.0800254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Farinha CM, King-Underwood J, Sousa M, Correia AR, Henriques BJ, Roxo-Rosa M, et al. Revertants, low temperature, and correctors reveal the mechanism of F508del-CFTR rescue by VX-809 and suggest multiple agents for full correction. Chem Biol. 2013;20:943–55. doi: 10.1016/j.chembiol.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 99.Dekkers JF, Gogorza Gondra RA, Kruisselbrink E, Vonk AM, Janssens HM, de Winter-de Groot KM, et al. Optimal correction of distinct CFTR folding mutants in rectal cystic fibrosis organoids. Eur Respir J. 2016;48:451–8. doi: 10.1183/13993003.01192-2015. [DOI] [PubMed] [Google Scholar]
- 100.Baatallah N, Bitam S, Martin N, Servel N, Costes B, Mekki C, et al. Cis variants identified in F508del complex alleles modulate CFTR channel rescue by small molecules. Hum Mutat. 2017 doi: 10.1002/humu.23389. [DOI] [PubMed] [Google Scholar]
- 101.Harutyunyan M, Huang Y, Mun KS, Yang F, Arora K, Naren AP. Personalized Medicine in CF: From Modulator Development to Therapy for Cystic Fibrosis Patients with Rare CFTR Mutations. Am J Physiol Lung Cell Mol Physiol. 2017 doi: 10.1152/ajplung.00465.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Molinski SV, Ahmadi S, Ip W, Ouyang H, Villella A, Miller JP, et al. Orkambi(R) and amplifier co-therapy improves function from a rare CFTR mutation in gene-edited cells and patient tissue. EMBO Mol Med. 2017;9:1224–43. doi: 10.15252/emmm.201607137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ruffin M, Roussel L, Maille E, Rousseau S, Brochiero E. Vx-809/Vx-770 treatment reduces inflammatory response to Pseudomonas aeruginosa in primary differentiated cystic fibrosis bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2017 doi: 10.1152/ajplung.00198.2017. [DOI] [PubMed] [Google Scholar]
- 104.Galietta LV, Jayaraman S, Verkman AS. Cell-based assay for high-throughput quantitative screening of CFTR chloride transport agonists. Am J Physiol Cell Physiol. 2001;281:C1734–42. doi: 10.1152/ajpcell.2001.281.5.C1734. [DOI] [PubMed] [Google Scholar]
- 105.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 106.Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell. 2017;171:1437–52. e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–80. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 108.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–73. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 109.Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–65. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 110.Lentini L, Melfi R, Di Leonardo A, Spinello A, Barone G, Pace A, et al. Toward a rationale for the PTC124 (Ataluren) promoted readthrough of premature stop codons: a computational approach and GFP-reporter cell-based assay. Mol Pharm. 2014;11:653–64. doi: 10.1021/mp400230s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pibiri I, Lentini L, Melfi R, Gallucci G, Pace A, Spinello A, et al. Enhancement of premature stop codon readthrough in the CFTR gene by Ataluren (PTC124) derivatives. Eur J Med Chem. 2015;101:236–44. doi: 10.1016/j.ejmech.2015.06.038. [DOI] [PubMed] [Google Scholar]
- 112.Pibiri I, Lentini L, Tutone M, Melfi R, Pace A, Di Leonardo A. Exploring the readthrough of nonsense mutations by non-acidic Ataluren analogues selected by ligand-based virtual screening. Eur J Med Chem. 2016;122:429–35. doi: 10.1016/j.ejmech.2016.06.048. [DOI] [PubMed] [Google Scholar]
- 113.Rowe SM, Daines C, Ringshausen FC, Kerem E, Wilson J, Tullis E, et al. Tezacaftor-Ivacaftor in Residual-Function Heterozygotes with Cystic Fibrosis. N Engl J Med. 2017;377:2024–35. doi: 10.1056/NEJMoa1709847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Boyle MP, Bell SC, Konstan MW, McColley SA, Rowe SM, Rietschel E, et al. A CFTR corrector (lumacaftor) and a CFTR potentiator (ivacaftor) for treatment of patients with cystic fibrosis who have a phe508del CFTR mutation: a phase 2 randomised controlled trial. Lancet Respir Med. 2014;2:527–38. doi: 10.1016/S2213-2600(14)70132-8. [DOI] [PubMed] [Google Scholar]
- 115.Clancy JP, Johnson SG, Yee SW, McDonagh EM, Caudle KE, Klein TE, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for ivacaftor therapy in the context of CFTR genotype. Clin Pharmacol Ther. 2014;95:592–7. doi: 10.1038/clpt.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. Methods Mol Med. 2005;107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- 117.Neuberger T, Burton B, Clark H, Van Goor F. Use of primary cultures of human bronchial epithelial cells isolated from cystic fibrosis patients for the pre-clinical testing of CFTR modulators. Methods Mol Biol. 2011;741:39–54. doi: 10.1007/978-1-61779-117-8_4. [DOI] [PubMed] [Google Scholar]
- 118.Cholon DM, Gentzsch M. Recent progress in translational cystic fibrosis research using precision medicine strategies. J Cyst Fibros. 2018;17:S52–S60. doi: 10.1016/j.jcf.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dekkers JF, Berkers G, Kruisselbrink E, Vonk A, de Jonge HR, Janssens HM, et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci Transl Med. 2016;8:344ra84. doi: 10.1126/scitranslmed.aad8278. [DOI] [PubMed] [Google Scholar]
- 120.Dekkers JF, Wiegerinck CL, de Jonge HR, Bronsveld I, Janssens HM, de Winter-de Groot KM, et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med. 2013;19:939–45. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 121.Eckford PDW, McCormack J, Munsie L, He G, Stanojevic S, Pereira SL, et al. The CF Canada-Sick Kids Program in individual CF therapy: A resource for the advancement of personalized medicine in CF. J Cyst Fibros. 2018 doi: 10.1016/j.jcf.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 122.Ehrhardt C, Collnot EM, Baldes C, Becker U, Laue M, Kim KJ, et al. Towards an in vitro model of cystic fibrosis small airway epithelium: characterisation of the human bronchial epithelial cell line CFBE41o. Cell Tissue Res. 2006;323:405–15. doi: 10.1007/s00441-005-0062-7. [DOI] [PubMed] [Google Scholar]
- 123.Chawade A, Alexandersson E, Levander F. Normalyzer: a tool for rapid evaluation of normalization methods for omics data sets. J Proteome Res. 2014;13:3114–20. doi: 10.1021/pr401264n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rastogi S, Rost B. LocDB: experimental annotations of localization for Homo sapiens and Arabidopsis thaliana. Nucleic Acids Res. 2011;39:D230–4. doi: 10.1093/nar/gkq927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–21. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pratt D, Chen J, Welker D, Rivas R, Pillich R, Rynkov V, et al. NDEx, the Network Data Exchange. Cell Syst. 2015;1:302–5. doi: 10.1016/j.cels.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Calamini B, Silva MC, Madoux F, Hutt DM, Khanna S, Chalfant MA, et al. Small-molecule proteostasis regulators for protein conformational diseases. Nat Chem Biol. 2012;8:185–96. doi: 10.1038/nchembio.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.