Abstract
Permeabilization of the Gram negative bacterial outer membrane (OM) by antimicrobial peptides (AMPs) is the initial step enabling access of the AMP to the cytoplasmic membrane. We present a new single-cell, time-resolved fluorescence microscopy assay that reports on the permeabilization of the E. coli OM to small molecules, with time resolution of 3 sec or better. When the profluorophore JF646 (702 Da) crosses the outer membrane (OM) and gains access to the periplasm, it binds to localized HaloTag protein (34 kDa) and fluoresces in a characteristic hollow spatial pattern. Previous work used the much larger periplasmic GFP (27 kDa) probe, which reports on OM permeabilization to globular proteins. We test the assay on three cationic agents: the Gellman random β-peptide copolymer MM63:CHx37, the human AMP LL-37, and the synthetic hybrid AMP CM15. These results combined with previous work suggest a unifying sequence of OM and cytoplasmic membrane (CM) events that may prove commonplace in the attack of cationic peptides on Gram negative bacteria. The peptide initially induces gradual OM permeabilization to small molecules, likely including the peptide itself. After a lag time, abrupt permeabilization of the OM, abrupt re-sealing of the OM, and abrupt permeabilization of the CM (all to globular proteins) occur in rapid sequence. We propose a mechanism based on membrane curvature stress induced by time-dependent differential binding of peptide to the outer leaflet of the OM and CM. The results provide fresh insight into the critical OM permeabilization step leading to a variety of damaging downstream events.
TOC image
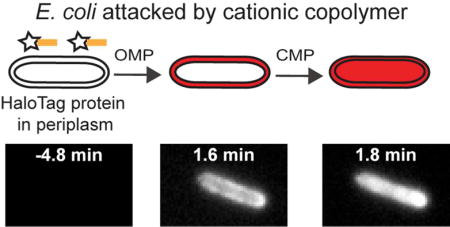
Antimicrobial peptides (AMPs), also called host-defense peptides, play a dual role in the innate immune system of many species, including humans, acting as both antimicrobial agents and signaling molecules.1,2 In this era of multi-drug resistant bacteria, AMPs may serve as useful lead compounds in the search for novel antibacterial agents. One widespread mechanism of action of natural AMPs is the permeabilization of bacterial membranes of both Gram positive and Gram negative species. Perhaps as a result, bacterial resistance to AMPs develops only slowly.
By now it is clear that the damage mechanisms induced by AMPs go well beyond membrane permeabilization and destruction of the trans-membrane potential that drives ATP production.3, 4 A wide variety of bulk culture and single-cell diagnostic assays have revealed numerous physical and biochemical effects of AMPs on cultured bacteria.5, 6 Our lab has been developing single-cell, time resolved, fluorescence-based assays that monitor a variety of AMP-induced “symptoms”.7–15 We have focused primarily on the Gram negative species E. coli under attack by the polycationic AMPs LL-37,7, 14 Cecropin A,8 CM15,11 and Melittin,15 as well as by the synthetic cationic random β-peptide copolymer MM63:CHx37 (Table 1).13 Thus far the methods enable direct determination of the timing of events such as outer membrane permeabilization (OMP) to GFP, cytoplasmic membrane permeabilization (CMP) to GFP and a Sytox dye, cell shrinkage, the halting of growth, and the onset of oxidative stress. Each antimicrobial agent exhibits a unique set of events.
Table 1.
Antimicrobial agents.
| Antimicrobial Agent | Sequence | Mass (Da) | Net Charge | MIC PM1 |
|---|---|---|---|---|
| LL-37 | LLGDFFRKSKEKIGKEFKRI-VQRIKDFLRNLVPRTES | 4493 | +6 | 4 |
| Cecropin A | KWKLFKKIEKVGQNIRDGII-KAGPAVAVVGQATQIAK-NH2 | 4404 | +7 | 0.9 |
| Melittin | GIGAVLKVLTTGLPALISWI-KRKRQQ-NH2 | 2846 | +6 | 5 |
| β-peptide copolymer1 MM63:CHx37 |
|
~7000 | 63% cationic sidechains | 25 μg/mL |
| CM15 | KWKLFKKIGAVLKVL-NH2 | 1770 | +6 | 5 |
Minimum inhibitory concentration (in μM) against WT MG1655 E. coli over 6-hr period in aerated EZRDM medium at 30°C, determined by OD for successive two-fold dilutions in 96 well plates. Copolymer MM63:CHx37 lacks a defined molar mass, so MIC is in μg/mL. The molecular weight of an “average” copolymer is ~7 kDa. Average is 35 subunits long, with 37% CHx (cyclohexyl) and 63% MM (mono-methyl) sidechains; see Ref. 13 for details. Thus an MIC of 25 μg/mL corresponds to a molar concentration of about 4 μM.
Our simplest assay uses a strain of E. coli MG1655 that expresses GFP bound to a TorA signal sequence, causing export of GFP from the cytoplasm to the periplasm via the Tat system.16 Periplasmic GFP creates a hollow, shell-like image in the 2D fluorescence microscope. This assay alone yields a surprising variety of behaviors for different cationic peptides. After initiation of the flow of LL-377,14 or Cecropin A8 across plated cells at t = 0, we observe a time lag followed by loss of the periplasmic GFP signal to the cell surround. The behavior is illustrated for a representative septating cell during attack by Cecropin A in Figure 1A. Evidently the AMP has induced OM permeabilization to small, globular proteins in localized fashion at the septal region. In sharp contrast, for the short, synthetic AMP CM15 and for the longer synthetic copolymer MM63:CHx37, the initial periplasmic GFP image abruptly evolves to a filled, cytoplasmic image. This alternative behavior is illustrated for a non-septating cell attacked by the copolymer in Figure 1B. Evidently the copolymer has first induced CM permeabilization to periplasmic GFP, which moves inward to fill the much larger cytoplasmic volume. Meanwhile, the total GFP fluorescence intensity remains essentially constant. GFP is lost to the surround only much later. Most recently, we found that Melittin induces behavior intermediate between that of Figure 1A and 1B.15 After a time lag, roughly half of the periplasmic GFP intensity is lost to the surround, and the remainder of the intensity soon exhibits a cytoplasmic spatial distribution. Complete loss of GFP occurs only much later. Evidently the OM has become transiently permeable to GFP and then re-sealed. Meanwhile the CM has become permeable to GFP. Such re-sealing behavior is reminiscent of the effects of AMPs on purified lipid vesicles and GUVs.17–24
Figure 1.
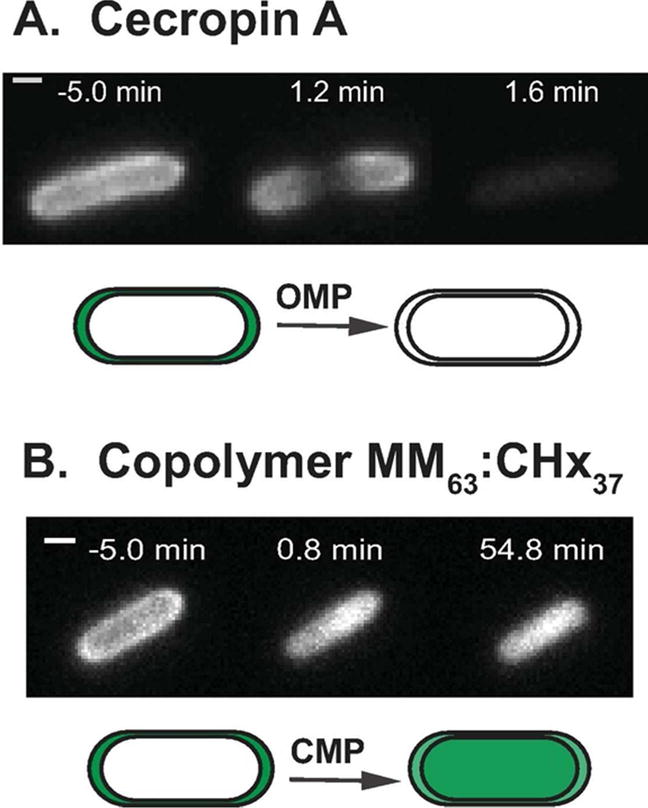
Single-cell, time-lapse assay imaging periplasmic GFP to detect membrane permeabilization events induced in live E. coli by antimicrobial agents. (A) Cecropin A at 0.9 μM (1X MIC) first induces outer membrane permeabilization to GFP, beginning at the septal region. Initial periplasmic halo image gradually disappears. (B) Gellman random β-peptide copolymer MM63:CHx37 at 30 μg/mL (1.2X MIC) first induces cytoplasmic membrane permeabilization to GFP. Initial periplasmic halo image evolves to a cytoplasmic filled image of which persists for at least 55 min.
GFP is a small, globular protein of mass 27 kDa, much larger than the typical antimicrobial agent of mass ~1-5 kDa. Before periplasmic GFP has moved inward to fill the cytoplasm, the antimicrobial agent must surely have translocated across the OM to gain access to the periplasmic space and to the CM itself. This passage is the key initial step in AMP activity against Gram negative bacteria. An important mechanistic question is: how does the cationic agent cross the OM? It is possible that linea r cationic peptides might thread their way through open porin channels in the OM, as seemingly occurs for colicin.25 The peptide might also transiently insert in the OM bilayer and subsequently de-insert into the periplasmic space, without having permeabilized the OM to other small molecules, much less to GFP. Finally, the peptide might first induce OM permeabilization to smaller species, including itself; these permeabilization sites may subsequently evolve to enable passage of small globular proteins such as GFP.
In order to dissect these possibilities, it is highly desirable to develop a single-cell assay that detects the onset of OM permeabilization to small molecules with good time resolution. Previously developed bulk assays used small profluorophores to detect permeabilization of the OM, but without spatiotemporal resolution.26–28 Here we describe a single-cell, time-resolved, fluorescence microscopy assay based on binding of a red-emitting profluorophore called JF646 ligand29 (mass 702 Da, Figure 2B) to HaloTag protein30 (34 kDa) that has been exported to the periplasm. When an antimicrobial agent induces OM permeabilization to the JF646 ligand, the ligand gains access to the periplasm, binds covalently to HaloTag protein, and fluoresces red with a periplasmic spatial distribution. The assay is sensitive enough to detect the onset of OM permeabilization to JF646 ligand with time resolution of 3 sec or better. The new results for copolymer MM63:CHx37 and for LL-37 strongly suggest that a common mechanism may apply to all five peptides in Table 1. Initial, gradual OM permeabilization to small species is followed by a time lag before abrupt OM permeabilization to globular proteins. Abrupt re-sealing of the OM and permeabilization of the CM to globular proteins follow shortly afterward. This may prove to be a fairly general sequence of events across a variety of cationic peptides, with timing details dependent on the specific peptide and its concentration. We present a mechanistic picture of membrane permeabilization and re-sealing in terms of time-dependent curvature stress caused by asymmetric binding of the peptide to the two leaflets of the membrane. This mechanism is borrowed from a large body of work on vesicles composed of pure lipids.17–22
Figure 2.
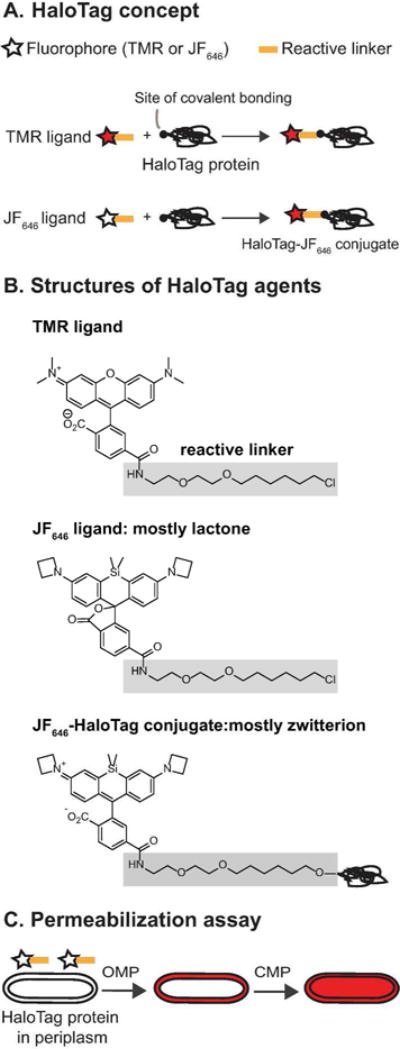
Schematic of HaloTag technique for detection of outer membrane permeabilization (OMP) to small molecules. (A) HaloTag ligand consists of a fluorophore (such as TMR) or a profluorophore (such as JF646) with a reactive chloroalkane linker (yellow line). HaloTag protein covalently binds to HaloTag ligand through the reactive linker. Free TMR ligand is already fluorescent, whereas JF646 ligand fluoresces much more strongly after binding to HaloTag protein. (B) Structures of free HaloTag TMR ligand, free HaloTag JF646 ligand and HaloTag-JF646 conjugate. The structure in the grey area is the reactive linker. (C) New OM permeabilization assay uses an E. coli strain carrying a plasmid that expresses HaloTag protein with a signal sequence causing efficient export to the periplasm. JF646 ligand is not permeable to intact E. coli. When an antimicrobial agent induces OMP to small molecules, JF646 ligand enters the periplasm, binds to HaloTag protein, and fluoresces in a periplasmic pattern. Subsequent CM permeabilization to globular proteins would cause the pattern to change to that of a filled cytoplasm.
Future extensions might combine the new HaloTag-based assay with addition of external Sytox Green or Sytox Orange, which bind to chromosomal DNA to signal CM permeabilization to small molecules. Such a combined single-cell assay could reveal the timing of OM and CM permeabilization events to both small species and globular proteins, all in one experiment. These methods may also find application in mechanistic studies of the effects of small-molecule antibiotics.
RESULTS AND DISCUSSION
The new assay detects the onset of OM permeabilization to small molecules using the HaloTag concept (Figure 2).30 This requires two components, an E. coli strain that exports the HaloTag protein to the periplasmic space and an impermeant profluorophore called JF646 ligand.29 The strains used in this work are listed in Table S1. Strain zy3720 carries a plasmid pMB2 that expresses HaloTag protein with the DsbA signal peptide appended to the N-terminus (ssDsbA-HaloTag). As a result, HaloTag protein is co-translationally exported to the periplasm, as previously shown.31 In aqueous solution, the JF646 ligand preferentially adopts the closed lactone form, which fluoresces only weakly (Figure 2B). When JF646 ligand binds covalently to the HaloTag protein to form the HaloTag-JF646 conjugate, the local environment around JF646 changes. The result is a large increase in red JF646 fluorescence.29,32 A hollow, shell-like spatial distribution signals the passage of JF646 ligand across the OM into the periplasm. At the same time, the OM presumably becomes permeable to other small molecules, likely including the antimicrobial agent itself. If the next step is CM permeabilization to globular proteins, the HaloTag-JF646 conjugate gains access to the cytoplasm, and the hollow spatial distribution fills in. If the next step is OM permeabilization to globular proteins, the conjugate leaves the cell envelope and the red fluorescence disappears. By measuring fluorescence intensity across the short axis of the cell, we can determine for each camera frame whether a fluorophore distributes predominantly in the periplasm or throughout the entire cell.
Minimum inhibitory concentration (MIC) results for the antimicrobial agents are shown in Table 1. Details of the microscopy experiments are described in SI. Briefly, exponentially growing cells are plated in a microfluidics chamber. At t = 0, the flow of aerated medium is switched to aerated medium plus a fixed concentration of antimicrobial agent and 100 nM JF646 ligand. For one-color imaging experiments using strain zy3720, we alternate fluorescence and phase contrast images, usually at 6 sec intervals (12 sec/cycle). The fluorescence image monitors the JF646 ligand binding to HaloTag protein and the phase contrast image monitors cell length. Two-color imaging experiments use strain zy3722 that contains a plasmid expressing both ssDsbA-HaloTag and ssTorA-GFP. Both HaloTag protein and GFP are exported to the periplasm. We sequentially acquire two fluorescence channels and the phase contrast channel, again at 12 sec/cycle. The green channel monitors GFP, the red channel monitors JF646 fluorescence, and the phase contrast channel monitors cell length. For each case, typically at least 60 cells in total were analyzed from three repeat experiments.
Preliminary tests
First we tested two strains for their ability to export HaloTag protein to the periplasm, zy3719 and zy3720 (Table S1). Strain zy3719 carries plasmid pMB1, expressing the HaloTag domain fused to the signal sequence of maltose-binding periplasmic protein (ssMalE-HaloTag). Strain zy3720 carries plasmid pMB2, expressing the HaloTag domain fused to the signal sequence of the periplasmic protein DsbA (ssDsbA-HaloTag). To determine where the HaloTag protein localizes inside E. coli cells, we added the permeable, red fluorescent tetramethylrhodamine ligand (TMR ligand).30 On excitation at 561 nm, TMR ligand emits at 580 nm. When supplied exogenously, TMR ligand permeates both membranes of living E. coli cells and covalently binds to HaloTag protein specifically. After rinsing with fresh medium, unbound TMR ligands are removed from the cells. The pattern of the residual fluorescence reports on the spatial distribution of the HaloTag protein.
Wild-type (WT) MG1655 cells do not express HaloTag protein and exhibited no significant TMR fluorescence after TMR ligand incubation and rinsing (Figure S1A). There is little or no permanent binding of TMR ligand to the cell envelope or to periplasmic or cytoplasmic components. Attempts at exporting HaloTag protein to the periplasm using ssMalE (strain zy3719) failed (Figure S1A). TMR fluorescence fills the entire cell, indicating that functional HaloTag protein remains primarily in the cytoplasm. In contrast, for strain zy3720 using the ssDsbA signal, TMR fluorescence exists predominately in the periplasm, indicating that HaloTag protein is exported efficiently to the periplasm (Figure S1A). Similarly, two-color imaging demonstrated that strain zy3722 efficiently exports both HaloTag protein and GFP to the periplasm (Figure S1A).
Next we tested the membrane permeability and toxicity of JF646 ligand towards E. coli. At t = 0, we flowed 100 nM JF646 ligand alone across E. coli zy3720 cells expressing periplasmic HaloTag protein. Fluorescence images and phase contrast images were interleaved, to monitor JF646 fluorescence and bacterial growth. During 1 hr of continuous flow, no obvious fluorescence was observed inside the cells (Figure S1B, Movie S1). Although some background fluorescence outside the cells appeared in the first few minutes, JF646 ligand at 100 nM does not permeate the OM of E. coli. A detailed description is included in SI. Meanwhile, JF646 ligand does not harm the growth of E. coli, as cells continue to grow and divide. The doubling time for E. coli zy3720 cells in the presence of 100 nM JF646 ligand is 52 ± 3 min, determined from cell length vs time. This is comparable to the 50 ± 3 min doubling time of WT E. coli cells plated in the microfluidic chamber when only EZ rich defined medium (EZRDM) is supplied.
Finally, we tested for JF646 fluorescence in WT cells (lacking HaloTag protein) after permeabilization by copolymer MM63:CHx37 at 1.2X MIC (Figure S1C). We observe only a weak burst of red intracellular fluorescence that begins after OM permeabilization (see below) and quickly dies away. This burst is at least 30-fold weaker than the JF646 fluorescence induced in zy3720 cells by the copolymer, as shown below.
Membrane permeabilization and re-sealing induced by copolymer MM63:CHx37
First we used the one-color assay to observe membrane permeabilization events induced by the copolymer MM63:CHx37 in E. coli cells. At t = 0, we flowed 30 μ,g/mL of MM63:CHx37 (1.2X MIC) plus 100 nM JF646 ligand across plated E. coli zy3720 cells that export HaloTag protein to the periplasm. In these one-color experiments, we monitored JF646 fluorescence and phase contrast every 12 s. Consistent with our previous study,13 nearly all cells began to shrink in length beginning within 24 sec of MM63:CHx37 addition. Later some cells partially recovered cell length, as before. For the representative cell in Figure 3A and Movie S2, the cell length decrease started at t1 = 0.2 min and continued until 1.4 min, when partial reversal occurred. Beginning at t2 = 0.4 min, red JF646 fluorescence increased abruptly and continued to rise until t = 1.8 min, when it reached a plateau (Figure 3C). The plateau may indicate that all the HaloTag protein has become bound by JF646 ligands. As shown from transverse intensity profiles (Figure 3B), the JF646 fluorescence at first accumulated in the periplasm, but began to fill the cytoplasm at t3 = 1.6—1.8 min.
Figure 3.
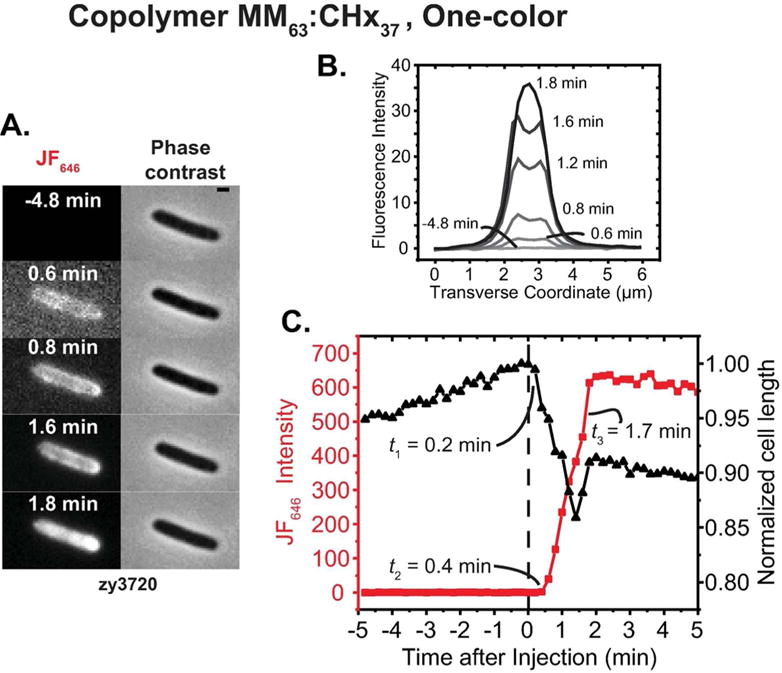
Application of one-color assay to membrane permeabilization by copolymer MM63:CHx37. (A) Red fluorescence and phase contrast snapshots of single zy3720 E. coli cell following addition of 30 μg/mL MM63:CHx37 (1.2X MIC) plus 100 nM JF646 ligand at t = 0. Time resolution is 12 sec/cycle = 0.2 min/cycle. Scale bar is 1 μm. (B) Quantitative transverse intensity profiles vs time. The profile is periplasmic from 0.6 min to 1.6 min and becomes cytoplasmic at 1.8 min. (C) Time dependence of cell length (from phase contrast images) and total HaloTag-JF646 fluorescence intensity for the same cell.
Copolymer MM63:CHx37 caused OM permeabilization to JF646 ligand, allowing it to enter the periplasm and bind to the HaloTag protein. Initially much or all of the HaloTag-JF646 conjugate remained in the periplasm. Later the CM was permeabilized to the HaloTag-JF646 conjugate, which began to fill the cytoplasm. Meanwhile, the OM remained essentially impermeable to the conjugate. We previously interpreted the shrinkage event as primarily an osmotic effect caused by entry of the highly cationic copolymer and its many counterions into the periplasm.13 The partial recovery of length may be due to import of K+ ions into the cytoplasm. Under the assumption that the OM was permeabilized to the copolymer (mean molar mass ~7 kDa) at the same time as the JF646 ligand (702 Da), the earlier explanation is corroborated. Notice that the onset of shrinkage matches the onset of HaloTag-JF646 conjugate fluorescence within 1 camera cycle = 0.2 min = 12 sec. The duration of the shrinkage event and the risetime of the fluorescence seem to match as well.
It is important to exclude the possibility that the JF646 fluorescence increase is produced by interaction of JF646 ligand with some intracellular species other than HaloTag protein. In Figure S1C (and Movie S3), we show a control experiment flowing 30 μ,g/mL MM63:CHx37 plus 100 nM JF646 ligand across wild type E. coli cells containing no HaloTag protein. A weak, transient, intracellular, cytoplasmic signal appears only after the shrinkage event. The peak fluorescence intensity in the control experiments (Figure S1C) is at least 30-fold smaller than the plateau value attained when HaloTag protein is present (Figure 3C). Evidently the bright periplasmic fluorescence observed in zy3720 cells is almost entirely due to the binding of JF646 ligand to the HaloTag protein following OM permeabilization.
We measure the mean values across cells of the timing events t1 (onset of cell shrinkage), (t2 - t1) (the additional lag time to the onset of HaloTag-JF646 conjugate fluorescence in the periplasm), and (t3 - t2) (the lag time between the onset of periplasmic fluorescence and entry of the conjugate into the cytoplasm). The corresponding detailed distributions are shown in Figure S2. Across 70 cells, <(t2 - t1)> = 0.2 ± 0.0 min and <(t3 - t2)> = 1.0 ± 0.7 min (± 1SD). Even at 1.2X MIC, the Gellman copolymer MM63:CHx37 permeabilizes the OM and CM in rapid succession. The new assay readily discerns both the onset of OM permeabilization to small molecules and the onset of CM permeabilization to globular proteins using a single fluorophore.
To gain additional information, we also applied the two-color assay to the attack of copolymer MM63:CHx37 on E. coli cells. At t = 0, we flowed 30 μ,g/mL MM63:CHx37 (1.2X MIC) plus 100 nM JF646 ligand across E. coli zy3722 cells that export both HaloTag protein and GFP to the periplasm. In these experiments, we monitored green GFP fluorescence, red JF646 fluorescence, and phase contrast with a cycle time of 12 s. As in the one-color experiment, the JF646 signal begins to rise shortly after the onset of cell shrinkage, initially forming a periplasmic image. For the particular cell in Figure 4A and Movie S4, this occurs at t2 = 0.6 min, and the signal continues to rise gradually up until t3 = 5.2 min. The JF646 image remains periplasmic during this interval. Meanwhile, the periplasmic GFP image maintains its intensity (except for minor photobleaching) over the same period. This demonstrates clearly that the OM has initially become permeabilized to small molecules such as JF646 ligand, but not to small globular proteins such as periplasmic GFP.
Figure 4.
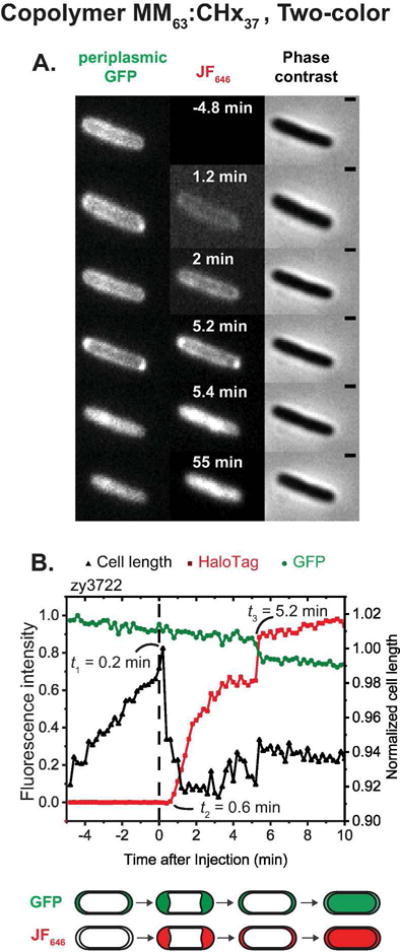
Application of two-color assay to membrane permeabilization by copolymer MM63:CHx37. (A) Phase contrast and fluorescence snapshots of single E. coli cell expressing periplasmic GFP and periplasmic HaloTag protein, following addition of 30 μg/mL MM63:CHx37 (1.2X MIC) plus 100 nM JF646 ligand at t = 0. (B) Time dependence of cell length (from phase contrast images) and of GFP and JF646 fluorescence intensity for the same cell. Scale bar is 1μm.
At t3 = 5.2 min, both GFP and JF646 images abruptly change from periplasmic to cytoplasmic. Interestingly, at the same moment the GFP intensity decreases by about 10% while the HaloTag-JF646 conjugate intensity increases by about 20%. Our interpretation is that the copolymer is inducing transient permeabilization of the OM to GFP, after which the OM re-seals to globular proteins of GFP size or larger. The transient opening lasts only about 3 frames = 36 sec. The same transient OM permeabilization event presumably also releases some of the (slightly larger) periplasmic HaloTag protein. However, during the transient event, the JF646 ligand can cross the OM more rapidly than before, resulting in a net abrupt increase in fluorescence from the HaloTag-JF646 conjugate. Essentially simultaneously with the OM re-sealing event, the CM becomes permeable to both GFP and HaloTag-JF646 conjugate and both images fill in. Meanwhile, the OM remains permeable to JF646 ligand, as evidenced by the slow continuing rise of conjugate fluorescence at t > 5.2 min.
Membrane permeabilization and re-sealing induced by LL-37
We also applied the one-color assay to the human AMP LL-37. In previous work imaging periplasmic GFP and Sytox Green in the same fluorescence channel, we had observed loss of roughly 90% of periplasmic GFP to the surround (signaling OM permeabilization to GFP) and the subsequent rise in Sytox Green fluorescence (CM permeabilization to Sytox Green).7,14 The very strong Sytox Green signal would have obscured any residual GFP that was trapped inside the cell by a membrane re-sealing event.
The one-color HaloTag assay reveals that an OM re-sealing event in fact occurs during the attack of LL-37. At t = 0 we initiated flow of 4 μM LL-37 (1X MIC) plus 100 nM JF646 across plated zy3720 cells. The typical behavior is shown in Figure 5 and Movie S5. Within 2 min, an intracellular, periplasmic signal from HaloTag-JF646 conjugate begins to rise gradually and cell growth begins to slow down. At t = 14.5 min, some 75% of the fluorescence is lost abruptly to the surround. The remaining 25% exhibits the cytoplasmic (filled cell) spatial pattern. Once again, our interpretation is that the OM is transiently permeabilized to the conjugate. At essentially the same time as the OM re-seals to globular proteins, the CM is permeabilized to the conjugate. A brief test of the two-color assay (green periplasmic GFP and red HaloTag-JF646 conjugate) on LL-37 confirmed that periplasmic GFP and HaloTag-JF646 conjugate move similarly in space and time (Figure S3). Most of the GFP and the conjugate intensity is lost at the same time, and both images change abruptly from the periplasmic to the cytoplasmic spatial distribution.
Figure 5.
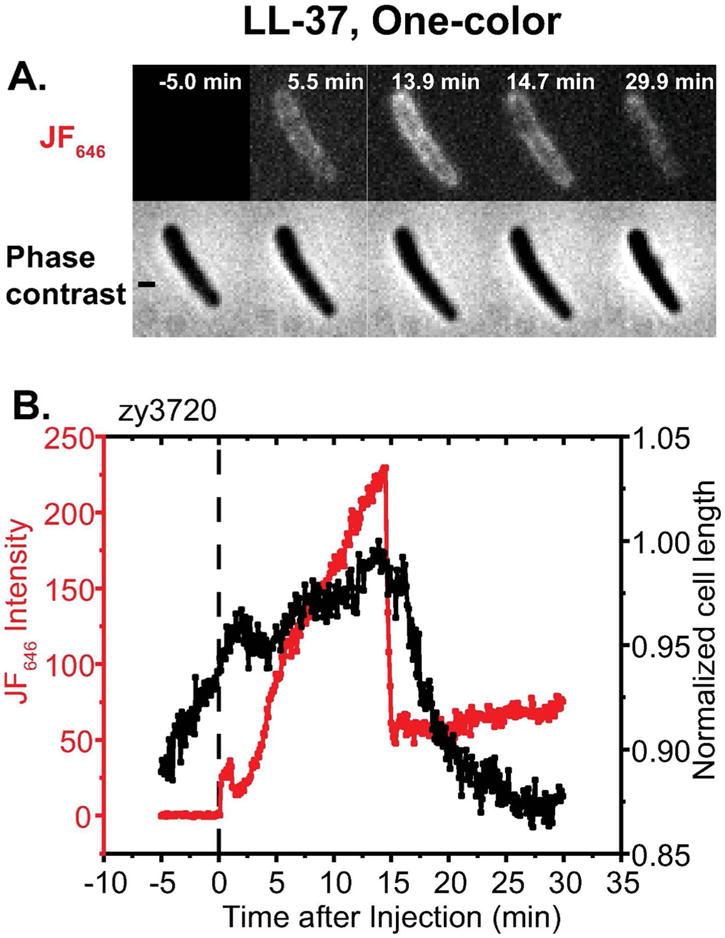
Application of one-color assay to membrane permeabilization by the antimicrobial peptide LL-37. Top: Red fluorescence and phase contrast snapshots of single zy3720 E. coli cell following addition of 4 μM LL-37 (1X MIC) plus 100 nM JF646 ligand at t = 0. Time resolution is 6 sec/cycle = 0.1 min/cycle. Bottom: Time dependence of cell length (from phase contrast images) and HaloTag-JF646 conjugate fluorescence intensity for the same cell. The fluorescence builds up in the periplasm for ~10 min, prior to the onset of significant cell shrinkage. Abrupt loss of some 75% of the intensity is attributed to OM permeabilization to globular proteins including GFP, HaloTag protein and HaloTag-JF646 conjugate. Simultaneous loss of GFP and HaloTag-JF646 conjugate was demonstrated in the two-color experiment of Figure S3. Scale bar is 1 μm.
Rapid CM permeabilization induced by CM15
Finally, we applied the one-color assay to the short, synthetic antimicrobial peptide CM15, which was previously shown to induce very rapid migration of periplasmic GFP into the cytoplasm.11 Flow of 10 μM CM15 (2X MIC) plus 100 nM JF646 ligand across zy3720 cells began at t = 0. In an attempt to estimate the kinetic response time of the new assay, the time resolution was increased to 3 sec/cycle. For the representative cell in Figure 6, shrinkage of cell length by some 20% began abruptly at t1 = 39 sec and was essentially complete in only 4 frames = 12 sec. The onset of JF646 fluorescence was essentially simultaneous with t1. We infer very rapid passage of CM15 across the OM and into the periplasm. Evidently the assay is sufficiently sensitive to detect the onset of OM permeabilization to small molecules to within 3 sec or better. The shift of JF646 fluorescence from a periplasmic distribution to a cytoplasmic distribution occurs within one camera cycle, at t2 = 0.70-0.75 min = 42-45 sec. This event is also detected with time resolution of 3 sec or better. The rising signal reached 2/3 of its plateau value in 10 frames = 30 sec. This is a convolution of the timescale of passage of JF646 ligand into the periplasm and the timescale for the bimolecular reaction between JF646 ligand and the periplasmic HaloTag protein. Thus we take 30 sec as an upper bound on the kinetic response time of the new assay under the conditions described. The comparable fluorescence risetime for the copolymer experiment in Figure 3C is much longer, ~60 sec. Accordingly, the cell shrinkage event is slower, itself requiring ~60 sec. We interpret this to mean that the copolymer and JF646 ligand gain access to the periplasm less rapidly than do CM15 and JF646 ligand.
Figure 6.
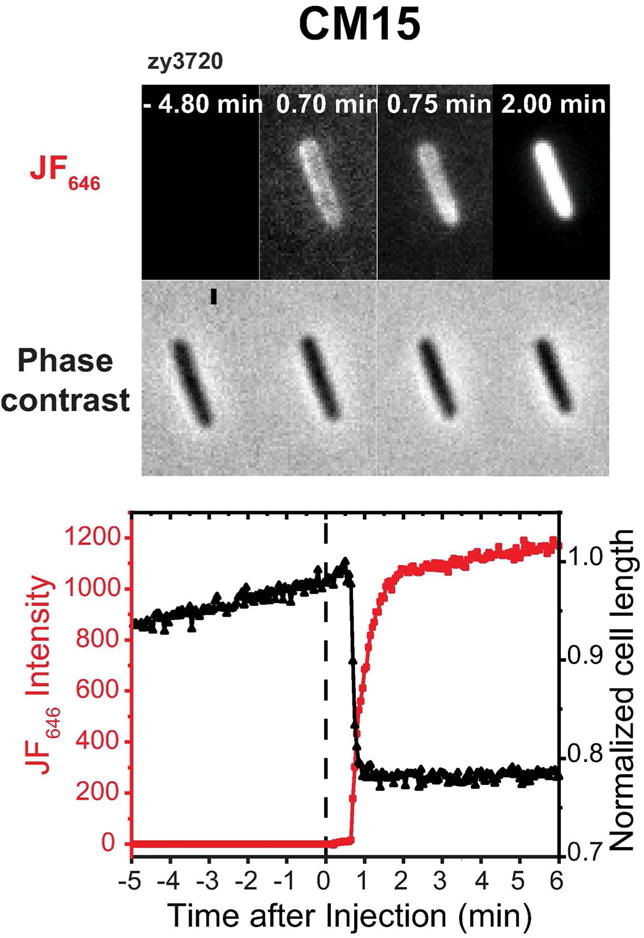
Application of one-color assay to membrane permeabilization by the antimicrobial peptide CM15. Top: Red fluorescence and phase contrast snapshots of single zy3720 E. coli cell following addition of 10 μM CM15 (2X MIC) plus 100 nM JF646 ligand at t = 0. Time resolution is 3 sec/cycle = 0.05 min/cycle. Bottom: Time dependence of cell length (from phase contrast images) and HaloTag-JF646 fluorescence intensity for the same cell. There is evidence of a transient periplasmic image at 0.70-0.75 min. The onset of red fluorescence and of cell shrinkage are simultaneous within one camera frame (3 sec).
General mechanistic insights
The new HaloTag-JF646 assay reveals that after an initial time delay, both the copolymer MM63:CHx37 and the natural AMP LL-37 induce a brief period of OM permeability to globular proteins followed by re-sealing of the OM and abrupt permeabilization of the CM to globular proteins. A similar sequence of events was recently elucidated for the antimicrobial peptide Melittin using periplasmic GFP as the probe.15 For MM63:CHx37 and for LL-37, the new HaloTag assay also enables direct observation of permeabilization of the OM to the much smaller JF646 ligand during the initial lag time leading up to OM permeabilization to globular proteins. It is highly likely that the OM has become permeable to the antimicrobial agent itself during the same initial period. We have not tested the timing of JF646 permeability induced by Melittin.
Evidently all three of these cationic peptides attack the E. coli cell envelope in a similar sequence of mechanistic steps. During the initial period, the peptide binds to and penetrates the anionic lipopolysaccharide (LPS) layer, eventually permeabilizing the OM to small molecules such as JF646 ligand, and presumably to the peptide itself. Our methods do not directly speak to the molecular-level details of the initial permeabilization mechanism, which could involve “detergent-like” disruption of OM structure33,34 or formation of small toroidal pores.35,36 It would be interesting to learn whether the initial permeabilization occurs locally or globally; such observations would require faster imaging than we have employed here. As more and more cationic peptide binds to the outer leaflet of the OM, this generates increasing curvature stress. The stress eventually causes abrupt OM permeabilization to globular proteins such as GFP, HaloTag protein, and the HaloTag-JF646 conjugate, enabling partial loss to the cell surround. This larger-scale disruption of the OM begins and ends abruptly in comparison with the earlier, gradual, continual leakage of small molecules across the OM. Earlier work showed that the larger disruption is often localized at curved membrane surfaces; see the septating cell attacked by Cecropin A in Figure 1. These details suggest a slow nucleation event preceding the formation of pores sufficiently large to pass globular proteins. The larger-scale disruption event then enables much more rapid translocation of the peptide into the periplasm, where it binds to the inner leaflet of the OM. This relieves the curvature stress and enables effective re-sealing of the OM to globular proteins.
During the disruption of the OM to globular proteins, the peptide concentration can build rapidly in the periplasm. The same process of differential curvature stress repeats itself due to peptide binding to the outer leaflet of the CM, which then abruptly becomes permeable to globular proteins. The proteins that did not escape the periplasm during the transient OM permeabilization event thus gain entry to the cytoplasm. The rapid build-up of peptide concentration in the periplasm while the OM is open to passage of globular proteins explains why OM re-sealing and CM permeabilization to such proteins seem coupled in time. In the specific case of Melittin, detailed analysis showed that subsequently the CM also re-sealed to GFP, presumably by the same curvature relief mechanism. Seemingly analogous peptide-induced permeabilization and re-sealing events have been observed in studies of pure lipid vesicles and GUVs. Indeed, we have borrowed the curvature stress mechanism from that body of work17–24
This detailed mechanistic picture of gradual permeabilization of the OM to small molecules followed by abrupt, transient permeabilization of the OM to globular proteins followed by permeabilization of the CM may prove to be fairly general in describing the attack of cationic peptides on E. coli, and perhaps on other Gram negative species as well. Similarly detailed studies of additional AMPs would test the generality of these events. Previous work shows that the timing of such events depends on the bulk peptide concentration.7,8 It is easy to imagine that different peptides will carry out these events at different rates. The HaloTag assay described here provides a new way to probe size- and time-dependent membrane permeabilization events in living cells in real time.
METHODS
The strains and plasmids used in this study are summarized in Table S1. Details of strain construction and induction conditions are provided in SI. Bulk cultures were grown in EZRDM37 from glycerol frozen stock to stationary phase overnight at 30°C. Subcultures were grown to exponential phase (OD = 0.2-0.6 at 600 nm) before sampling for the microscopy experiments at 30°C. The antimicrobial agents are described in Table 1; additional details are found in SI. The aerobic MIC values for the various antimicrobial agents were determined using the broth microdilution method as previously described.7 Imaging of individual cells was carried out at 30°C in a simple microfluidics chamber as previously described.11 Fresh, aerated medium flows over the cells continuously. The cells were imaged for ~5 min before switching to fresh medium containing the compounds under study (antimicrobial agent, JF646 ligand). Details of the microscopy procedure and setup are in SI. The concentration of each peptide was chosen to be sufficiently high to cause significant antimicrobial action on a 30-min timescale, but low enough to enable the methods to resolve sequential events in time.
Supplementary Material
Acknowledgments
We thank B. Mehmet (New England Biolabs) for providing the plasmids pMB1 and pMB2, L. Lavis (Janelia Research Campus) for providing JF646 ligand, and H. Choi (Janelia Research Campus) for helpful discussion. The Gellman lab provided the MM63:CHx37 copolymer sample.
FUNDING SOURCES
This research was supported by the National Institute of General Medical Sciences of the National Institutes of Health under awards R01GM094510 (to JCW as PI) and R01GM093265 (to JCW and Samuel Gellman as co-PIs). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
SUPPORTING INFORMATION
Supporting Information Available: This material is available free of charge via the Internet. Details on methods, strains and plasmids used in this work (Table S1). Control experiments for the HaloTag-based membrane permeabilization assay (Figure S1). Histograms of lag times for one-color experiment after MM63:CHx37 addition (Figure S2). An example of two-color experiment after LL-37 addition (Figure S3). Five movies under various conditions.
References
- 1.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Wimley WC, Hristova K. Antimicrobial peptides: successes, challenges and unanswered questions. J Membr Biol. 2011;239:27–34. doi: 10.1007/s00232-011-9343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 4.Hancock RE, Sahl HG. New strategies and compounds for anti-infective treatment. Current opinion in microbiology. 2013;16:519–521. doi: 10.1016/j.mib.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Munoz A, Read ND. Live-cell imaging and analysis shed light on the complexity and dynamics of antimicrobial Peptide action. Front Immunol. 2012;3:248. doi: 10.3389/fimmu.2012.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi H, Rangarajan N, Weisshaar JC. Lights, camera, action! Antimicrobial peptide mechanisms imaged in space and time. Trends Microbiol. 2016;24:111–122. doi: 10.1016/j.tim.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sochacki KA, Barns KJ, Bucki R, Weisshaar JC. Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proc Natl Acad Sci US A. 2011;108:E77–81. doi: 10.1073/pnas.1101130108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rangarajan N, Bakshi S, Weisshaar JC. Localized permeabilization of E. coli membranes by the antimicrobial peptide Cecropin A. Biochemistry. 2013;52:6584–6594. doi: 10.1021/bi400785j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barns KJ, Weisshaar JC. Real-time attack of LL-37 on single Bacillus subtilis cells. Biochim Biophys Acta. 2013;1828:1511–1520. doi: 10.1016/j.bbamem.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakshi S, Choi H, Rangarajan N, Barns KJ, Bratton BP, Weisshaar JC. Nonperturbative imaging of nucleoid morphology in live bacterial cells during an antimicrobial peptide attack. Appl Environ Microbiol. 2014;80:4977–4986. doi: 10.1128/AEM.00989-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi H, Yang Z, Weisshaar JC. Single-cell, real-time detection of oxidative stress induced in Escherichia coli by the antimicrobial peptide CM15. Proc Natl Acad Sci USA. 2015;112:E303–310. doi: 10.1073/pnas.1417703112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barns KJ, Weisshaar JC. Single-cell, time-resolved study of the effects of the antimicrobial peptide alamethicin on Bacillus subtilis. Biochim Biophys Acta. 2016;1858:725–732. doi: 10.1016/j.bbamem.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi H, Chakraborty S, Liu R, Gellman SH, Weisshaar JC. Single-cell, time-resolved antimicrobial effects of a highly cationic, random nylon-3 copolymer on live Escherichia coli. ACS Chem Biol. 2016;11:113–120. doi: 10.1021/acschembio.5b00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi H, Yang Z, Weisshaar JC. Oxidative stress induced in E. coli by the human antimicrobial peptide LL-37. PLoS Pathog. 2017;13:e1006481. doi: 10.1371/journal.ppat.1006481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, Choi H, Weisshaar JC. Melittin-induced permeabilization, re-sealing, and re-permeabilization of E. coli Membranes. Biophys J. 2018;114:368–379. doi: 10.1016/j.bpj.2017.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sochacki KA, Shkel IA, Record MT, Weisshaar JC. Protein diffusion in the periplasm of E. coli under osmotic stress. Biophys J. 2011;100:22–31. doi: 10.1016/j.bpj.2010.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rex S, Schwarz G. Quantitative studies on the Melittin-induced leakage mechanism of lipid vesicles. Biochemistry. 1998;37:2336–2345. doi: 10.1021/bi971009p. [DOI] [PubMed] [Google Scholar]
- 18.Gregory SM, Pokorny A, Almeida PF. Magainin 2 revisited: a test of the quantitative model for the all-or-none permeabilization of phospholipid vesicles. Biophys J. 2009;96:116–131. doi: 10.1016/j.bpj.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krauson AJ, He J, Wimley WC. Determining the mechanism of membrane permeabilizing peptides: identification of potent, equilibrium pore-formers. Biochim Biophys Acta. 2012;1818:1625–1632. doi: 10.1016/j.bbamem.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiedman G, Herman K, Searson P, Wimley WC, Hristova K. The electrical response of bilayers to the bee venom toxin melittin: evidence for transient bilayer permeabilization. Biochim Biophys Acta. 2013;1828:1357–1364. doi: 10.1016/j.bbamem.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cruz J, Mihailescu M, Wiedman G, Herman K, Searson PC, Wimley WC, Hristova K. A membrane-translocating peptide penetrates into bilayers without significant bilayer perturbations. Biophys J. 2013;104:2419–2428. doi: 10.1016/j.bpj.2013.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savini F, Bobone S, Roversi D, Mangoni ML, Stella L. From liposomes to cells: Filling the gap between physicochemical and microbiological studies of the activity and selectivity of host-defense peptides. Peptide Science. 2018:e24041. [Google Scholar]
- 23.Wimley WC. How does Melittin permeabilize membranes? Biophys J. 2018;114:251–253. doi: 10.1016/j.bpj.2017.11.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wheaten SA, Ablan FD, Spaller BL, Trieu JM, Almeida PF. Translocation of cationic amphipathic peptides across the membranes of pure phospholipid giant vesicles. J Am Chem Soc. 2013;135:16517–16525. doi: 10.1021/ja407451c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zakharov SD, Sharma O, Zhalnina M, Yamashita E, Cramer WA. Pathways of colicin import: utilization of BtuB, OmpF porin and the TolC drug-export protein. Biochemical Society transactions. 2012;40:1463–1468. doi: 10.1042/BST20120211. [DOI] [PubMed] [Google Scholar]
- 26.Loh B, Grant C, Hancock RE. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy. 1984;26:546–551. doi: 10.1128/aac.26.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehrer RI, Barton A, Ganz T. Concurrent assessment of inner and outer membrane permeabilization and bacteriolysis in E. coli by multiple-wavelength spectrophotometry. Journal of Immunological Methods. 1988;108:153–158. doi: 10.1016/0022-1759(88)90414-0. [DOI] [PubMed] [Google Scholar]
- 28.Junkes C, Wessolowski A, Farnaud S, Evans RW, Good L, Bienert M, Dathe M. The interaction of arginine- and tryptophan-rich cyclic hexapeptides with Escherichia coli membranes. J Pept Sci. 2008;14:535–543. doi: 10.1002/psc.940. [DOI] [PubMed] [Google Scholar]
- 29.Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH, Lionnet T, Lavis LD. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods. 2015;1:2, 244–250. doi: 10.1038/nmeth.3256. 243 p following 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chemical Biology. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 31.Ke N, Landgraf D, Paulsson J, Berkmen M. Visualization of periplasmic and cytoplasmic proteins with a self-labeling protein Tag. J Bacteriol. 2016;198:1035–1043. doi: 10.1128/JB.00864-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimm JB, Muthusamy AK, Liang Y, Brown TA, Lemon WC, Patel R, Lu R, Macklin JJ, Keller PJ, Ji N, Lavis LD. A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat Methods. 2017;14:987–994. doi: 10.1038/nmeth.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladokhin AS, White SH. ‘Detergent-like’ permeabilization of anionic lipid vesicles by Melittin. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2001;1514:253–260. doi: 10.1016/s0005-2736(01)00382-0. [DOI] [PubMed] [Google Scholar]
- 34.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 35.Lee MT, Chen FY, Huang HW. Energetics of pore formation induced by membrane active peptides. Biochemistry. 2004;43:3590–3599. doi: 10.1021/bi036153r. [DOI] [PubMed] [Google Scholar]
- 36.Yamaguchi S, Hong T, Waring A, Lehrer RI, Hong M. Solid-state NMR investigations of peptide-lipid interaction and orientation of a β-Sheet antimicrobial peptide, Protegrin. Biochemistry. 2002;41:9852–9862. doi: 10.1021/bi0257991. [DOI] [PubMed] [Google Scholar]
- 37.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


