Abstract
Background
Chimeric uPA/SCID mice reconstituted with humanized livers are useful for studying HBV infection in the absence of an adaptive immune response. However, the detailed characterization of HBV infection kinetics necessary to enable in-depth mechanistic studies in this novel in vivo HBV infection model is lacking.
Methods
To characterize HBV kinetics post-inoculation (p.i.) to steady state, 42 mice were inoculated with HBV. Serum HBV DNA was frequently measured from 1 minute to 63 days p.i. Total intrahepatic HBV DNA, HBV cccDNA, and HBV RNA was measured in a subset of mice at 2, 4, 6, 10, and 13 weeks p.i. HBV half-life (t1/2) was estimated using a linear mixed-effects model.
Results
During the first 6 h p.i. serum HBV declined in repopulated uPA/SCID mice with a t1/2=62 min [95%CI=59–67min]. Thereafter, viral decline slowed followed by a 2 day lower plateau. Subsequent viral amplification was multiphasic with an initial mean doubling time of t2=8±3 h followed by an interim plateau before prolonged amplification (t2=2±0.5 days) to a final HBV steady state of 9.3±0.3 log copies/ml. Serum HBV and intrahepatic HBV DNA were positively correlated (R2=0.98).
Conclusions
HBV infection in uPA/SCID chimeric mice is highly dynamic despite the absence of an adaptive immune response. The serum HBV t1/2 in humanized uPA/SCID mice was estimated to be ~1 h regardless of inoculum size. The HBV acute infection kinetics presented here is an important step in characterizing this experimental model system so that it can be effectively used to elucidate the dynamics of the HBV lifecycle and thus possibly reveal effective antiviral drug targets.
Keywords: HBV, Viral kinetics, uPA, SCID, HBV clearance rate, chimeric mice with humanized livers
Introduction
More than 2 billion people have been infected with hepatitis B virus (HBV) worldwide with ~240 million remaining chronically infected, making HBV a leading cause of morbidity and mortality in countries with high prevalence rates (1). During acute infection, HBV DNA reaches high levels in blood (up to 1010 copies/ml) within several weeks and almost all hepatocytes become infected (2). The age at which an individual becomes infected is highly associated with infection outcome (3) with chronic infection being established in more than 90% of infants and in up to 50% of children aged 1 to 5 years, but only in 5–15% of adults (4). Although the immune response has a pivotal role in controlling the infection, the precise mechanisms are still not known (5, 6).
The characterization of HBV kinetics during initial acute infection in humans is limited due to the uncertainty regarding the exact time of infection and lack of frequent data sampling (7, 8). Acute HBV infection has been studied in chimpanzees, but sampling was performed relatively infrequently, on a weekly basis (or longer time intervals) (9–12), and as of 2011 biomedical research involving chimpanzees has been restricted. Other animal models infected with HBV-related hepadnaviruses, such as woodchuck hepatitis virus, have also provided a limited kinetic picture but infection dynamics are known to vary among these related viruses (13). Major attempts have been made to develop small animal models of HBV infection, the most successful being based on liver-repopulation with primary human hepatocytes as these are the natural target of HBV [reviewed in (14)]. One example is the uPA/SCID mouse model, which lack functional B and T-cells and thus allow transplanted HBV-permissive human hepatocytes to be maintained in the liver (15). Such mouse models offer opportunities to study HBV kinetics during the early steps of infection and antiviral treatment in the absence of any confounding effects exerted by an adaptive immune response (14).
In this study, we analysed the detailed virus kinetics during acute HBV infection in chimeric uPA/SCID mice from infection initiation to steady state. We were able to identify distinct phases of infection, indicating a rapid initial virus decline from the serum and multiphasic resurgence until reaching steady state. Due to very frequent blood sampling, it was also possible to estimate the rate of HBV clearance from blood.
Materials and Methods
Mice
Mice with humanized liver were produced as described previously (15). Three different batches of frozen human hepatocytes (2 year old Hispanic female, 2YF [IL28B, rs12979860 genotype CC]; 5 year old African American male, 5YM (IL28B genotype TT); 1 year old Caucasian female, 1YF) were purchased from BD Biosciences Discovery Labware, San Jose, CA, USA (2YF and 5YM) and In Vitro Technologies, Baltimore, MD, USA (1YF). They were thawed and 1–2.5×105 viable hepatocytes were transplanted into 2–4 week-old uPA/SCID mice via splenic injection.
The study included 42 uPA/SCID mice with humanized livers divided into four experimental groups (Fig. 1A). Group A consisted of four humanized mice (donor 5YM) inoculated with 105/106/107/108 HBV DNA copies (cps) that were observed from baseline until 8 days post inoculation (p.i.). Group B comprised three humanized mice (donor 5YM) inoculated with 104/106/107 HBV DNA cps that were monitored from baseline until day 28 p.i. Group C consisted of four humanized mice (donor 2YF) inoculated with 108 HBV DNA cps and observed from baseline until day 51 p.i. Group D included 31 humanized mice transplanted with human hepatocytes from different donors (2YF: n=2; 5YM: n=25; 1YF: n=4). All animals in this group were inoculated with 104 HBV DNA cps. Longitudinal serum viral DNA was measured in these animals from day 7 to 63 p.i. or until they were sacrificed at 2, 4, 6, 8, 10 or 13 weeks p.i. supporting Table S1 only includes the 9 animals for which the complete time course of serum viral DNA measurements were obtained; the other 22 animals were sacrificed as indicated above in order to measure intrahepatic total HBV DNA, HBV cccDNA, and HBV RNA. Within the different groups, serum HBV DNA sampling frequency ranged between 30 min and 9 days with sampling in the different groups beginning as soon as 1 min p.i. and continuing through 63 days after inoculation. All animal protocols described in this study were performed in accordance with the Guide for the Care and Use of Laboratory Animals and approved by the Animal Welfare Committee of Phoenix Bio Co., Ltd.
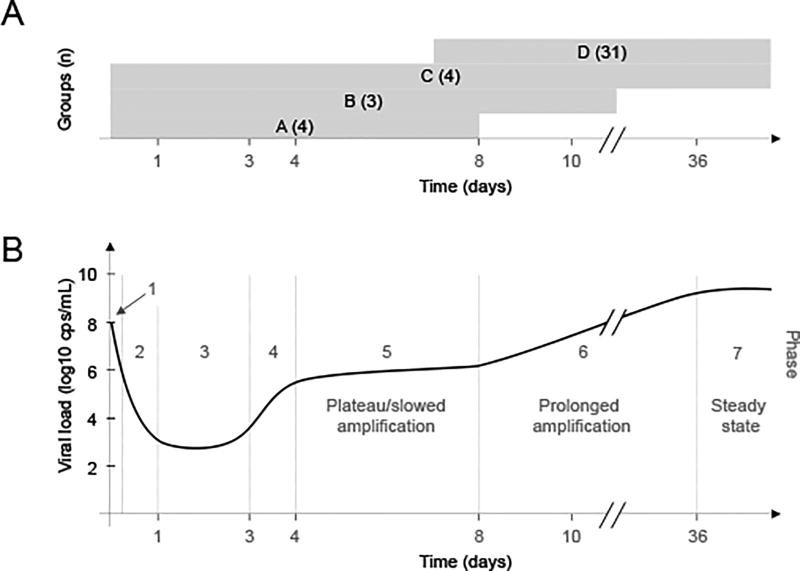
Figure 1. Overview of complex serum HBV kinetic pattern from inoculation to steady state.
(A) Graphical representation of time period for which serum viral load was measured in each humanized mouse group, A–D with size of group (n) indicated. Longitudinal serum viral DNA was measured in 9 out of 31 mice of group D. (B) HBV kinetic phases in one representative Group C mouse after inoculation with 108 copies HBV DNA: Phase 1-rapid decline; Phase 2-slower decline; Phase 3- lower viral plateau (or below detection); Phase 4-rapid increase; Phase 5-extremely slow increase or plateau; Phase 6-prolonged amplification; Phase 7-steady-state. Limit of HBV DNA detection (3.6 log cp/ml)
Inoculation with HBV DNA
The original virus (HBV genotype C), provided by Dr. Sugiyama (16) was used to create the viral stock for these experiments by amplifying the virus in uPA/SCID chimeric mice. Specifically, after observing stable high-level HBV viremia in inoculated mice, serum was collected and pooled for the present study. Serum containing 104–108 copies of HBV DNA was intravenously injected into humanized uPA/SCID mice. As in our previous study (17), mice with humanized livers had human albumin (hAlb) levels of 4.2–15.0 mg/mL (corresponding to a replacement index of 56–93%) at 7 to 11 weeks after human hepatocyte transplantation.
Extraction and Quantification of serum HBV DNA
DNA was extracted from 10 µL serum by Smitest Ex-R&D Nucleic Acid Extraction Kit (Medical & Biological Laboratories Co, Ltd, Nagoya, Japan) and was dissolved in 20 µL nuclease-free water (Life Technologies Japan Ltd., Tokyo, Japan). HBV DNA copy numbers were determined by quantitative real-time PCR (qRT-PCR) as reported previously (18).
Quantification of human serum albumin. Because human albumin concentration in the serum correlates with the degree of human hepatocyte repopulation observed by human-specific antibody staining of liver sections (15), hAlb concentration in mouse serum was measured by latex agglutination immunonephelometry test (LX Reagent “Eiken” Alb II; Eiken Chemical Co., Ltd., Tokyo, Japan). These hAlb levels were used to estimate the replacement index of human hepatocytes in mouse livers as previously described (19). Levels of human serum albumin were measured as soon as 1 min after inoculation and then at weekly intervals or less up to 63 days after HBV inoculation.
Extraction and Quantification of intrahepatic HBV DNA
Total DNA was extracted from approximately 20 mg of liver independently in two laboratories using DNeasy Blood and Tissue Kit (Qiagen K.K. Tokyo, Japan) according to the instruction provided by the supplier or by phenol chloroform extraction as previously described (20). DNeasy Blood and Tissue Kit isolated DNA (50 ng) was amplified by ABI Prism 7500 sequence detector system (Applied Biosystems Japan Ltd., Tokyo, Japan) using TaqMan PCR Core Reagents (Applied Biosystems Japan Ltd.). The amplification conditions for HBV DNA and cccDNA were described in previous reports respectively (18, 21). Using the following primers: total HBV (forward primer: 5’-CACATCAGGATTCCTAGGACC-3’, reverse primer: 5’-AGGTTGGTGAGTGATTGGAG-3’, TaqMan probe: 5’-CAGAGTCTAGACTCGTGGTGGACTTC-3’)(19), cccDNA (forward primer: 5’-CTCCCCGTCTGTGCCTTCT-3’, reverse primer: 5’-GCCCCAAAGCCACCCAAG-3’, TaqMan prove: 5’- CGTCGCATGGARACCACCGTGAACGCC -3’). Phenol chloroform isolated HBV DNA was quantified by qPCR amplification of 30ng liver DNA using an Applied Biosystems 7300 real-time thermocycler (Applied Biosystems) for total HBV (22), cccDNA (23) and human telomerase reverse transcriptase (hTERT) (forward primer: 5’- AAAATAGCTGGAACTGCAGACA-3’, reverse primer: 5’- AAGCAAAGCTACAGAAACACTCA -3’). HBV DNA levels were determined relative to standard curve comprised of serial dilutions of a plasmid containing the HBV genome. HBV DNA copies per cell were then determine relative to single genome copy human TERT gene in each sample.
Extraction and Quantification of intrahepatic HBV RNA. Total cellular RNA was isolated by the guanidine thiocyanate method using standard protocols (24). One µg of purified RNA was used for cDNA synthesis using the TaqMan reverse transcription reagents (Applied Biosystems), followed by SYBR green RTqPCR using the total HBV primers indicated above and an Applied Biosystems 7300 real-time thermocycler (Applied Biosystems). HBV RNA levels were determined relative to standard curve comprised of serial dilutions of a plasmid containing the HBV genome and normalized to human GAPDH RNA (forward primer: 5’-CAAGATCATCAGCAATGCCT-3’, reverse primer: 5’-AGGGATGATGTTCTGGAGAG-3’).
Immunostaining for hepatitis B core antigen (HBcAg)
Pieces of liver from HBV-infected mice were fixed with formalin, and embedded in paraffin blocks for immunohistochemical staining. Five micrometer thick liver sections were subjected to immunostaining using an antibody against HBcAg (DAKO Diagnostika, Hamburg, Germany). The antibodies were visualized with EnVision System™ HRP (DAKO) and DAB substrate (17).
Statistical analyses
Data is expressed as arithmetic mean ± standard deviation or median value with range. Logarithmic base 10 values of HBV DNA were used throughout the analysis. Statistical analyses and linear regression were computed with Excel 2010 and R (version 3). Linear regression and linear mixed-effects models (R version 3, package nlme) were used to estimate the slopes characterizing virus and albumin kinetics. P-values less than 0.05 were considered statistically significant. Kruskal-Wallis test were used to compare intergroup differences. Spearman rank correlation was used to evaluate the relationship between two quantitative variables.
Results
Monitoring HBV infection from inoculation to steady state revealed seven different viral kinetic phases independent of HBV inoculum dose or hepatocyte donor (Fig.1B). All of these distinct kinetic stages were observed in the Group C mice (phases 1–7) and confirmed by the early kinetics observed in Group A (phases 1–5) and Group B (phases 1–6) and the late kinetics observed in Group D (phases 6–7).
Rapid serum HBV clearance (Phase 1)
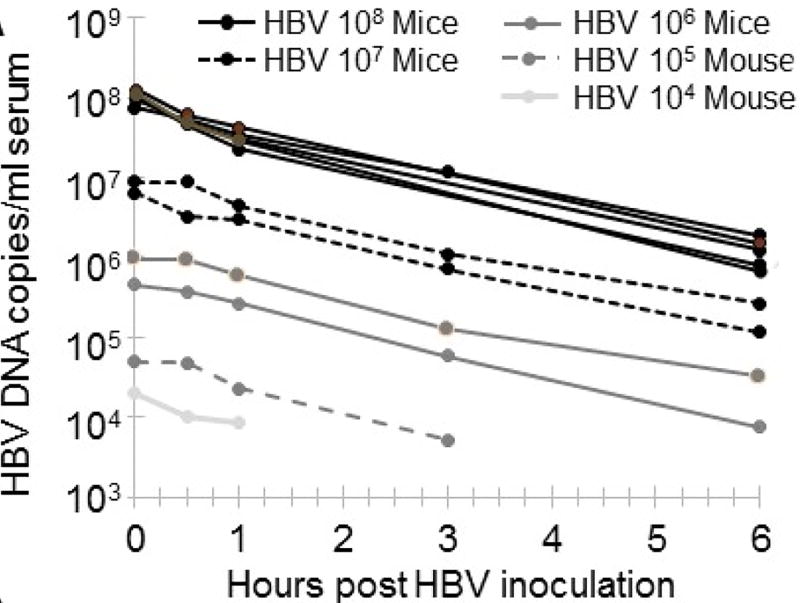
Serum HBV DNA in chimeric mice (Groups A, B, and C) was measured frequently (1 min, 30 min, and then 1, 2, 3, 5, 6, 12 and 18 h p.i.). While the mice across these 3 groups displayed dose-dependence in viral load, all exhibited a dose-independent rapid decline in HBV DNA levels within first 6 h p.i., with an average decrease rate of 7.3±0.9 log10/day corresponding to a mean half-life (t1/2) of 62±7 min (Fig. 2, Table S1). A mixed-effects model was used to calculate the initial 6 h exponential virus decay and estimate the HBV clearance rate and its confidence interval. These estimates correspond to a virus serum t1/2 of 63 min (95% confidence interval, CI, 59 to 67 min).
Figure 2. Serum HBV DNA clearance immediately following inoculation.
Clearance after inoculation with varying HBV doses. Chimeric uPA/SCID mice were inoculated with the indicated dose of HBV and serum levels of HBV were measured by RT-qPCR at the indicated times starting at 1 min post-inoculation and are graphed as HBV copies/ml serum.
Slowdown of serum HBV clearance followed by a lower viral plateau (Phases 2–3)
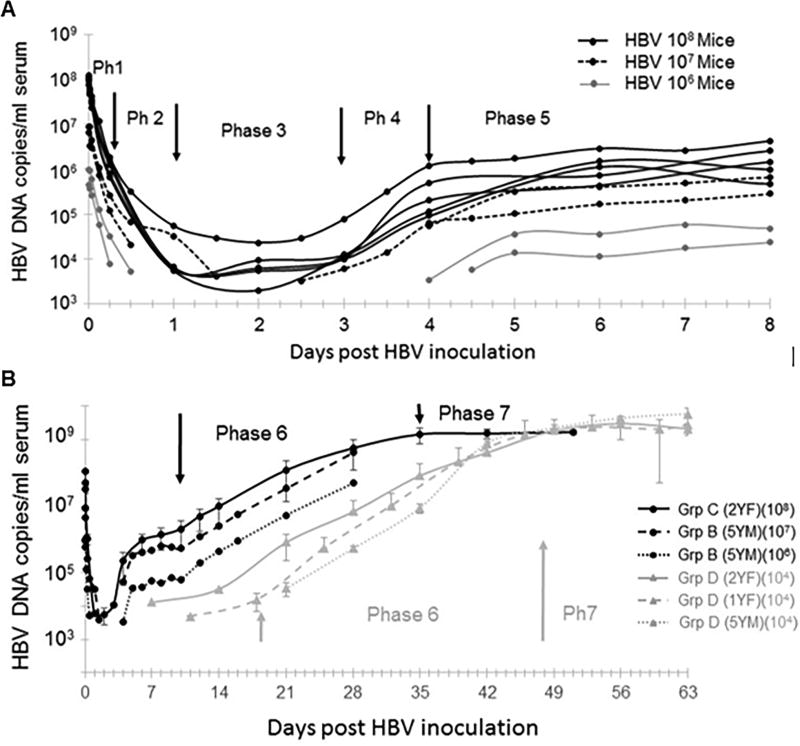
Between 6 – 24 h p.i. (18 hours) HBV could not be detected in 5 out of 11 Groups A–C mice monitored during this time period, but in the remaining mice the initial rapid decline of HBV slowed to 2.6±0.9 log10/day, which corresponds to a mean half-life of 3.6±1.8 h (Fig.3A, phase 2). Viral decline ended in a lower plateau that lasted from approximately day 1 to day 3 p.i. (Fig.3A, phase 3). Only the 5 mice that were inoculated with 108 HBV DNA copies remained HBV detectable during this 2 day viral plateau with an average 4±0.3 log10 HBV DNA cps/mL and slope constant between −0.01 and 0.17 log10/day (Table S1).
Figure. 3. Multiphasic HBV kinetic patterns in uPA/SCID chimeric mice from inoculation to steady state.
(A) Early HBV kinetic phases (1 to 5) in 9 individual mice (circles) during the first 8 days p.i.: 5 mice inoculated with 108 HBV genome equivalents (Black-solid lines), 2 mice were inoculated with 107 HBV genome equivalents (black-dashed lines), and 2 mice inoculated with 106 HBV genome equivalents (grey-solid lines). (B) Later HBV kinetic phases (6 and 7). Average serum HBV DNA +/− SD shown based on inoculum size and donor (as described in the Materials and Methods). Mice inoculated with high HBV doses (i.e. 108, 107, 106) and corresponding phases noted in black; mice inoculated with a low HBV dose (i.e. 104) and corresponding phases noted in grey. Ph, phase; Grp, Group)
Rapid resurgence of serum HBV and interim plateau (Phases 4–5)
After the above described lower viral plateau, serum HBV levels increased in the 9 mice that became productively infected (Figs.1B and 3A, phase 4). In mice inoculated with 107 or 108 HBV genome copies, this amplification phase could be clearly identified starting between days 2.5–3 p.i. and lasted 1–3 days (Fig.3A, black lines). While the two mice that received 106 HBV genome copies exhibited a delay in the initiation of this phase, the slope increase was similar in all animals with an average of 1.1±0.5 log10/day (doubling time, t2=8±3 h) regardless of HBV inoculum dose (Fig.3A, grey lines). Unexpectedly, after this brief exponential amplification, HBV DNA increased transiently, slowed and/or exhibited an interim plateau with a median duration of 3 days starting between day 4 and 6 p.i. This Phase 5 plateau was detected in all Group B and C mice observed throughout this time period (Fig.3A, phase 5). The median HBV doubling time during this period was 3 days with a median increase rate of 0.05 log10/day (ranging from −0.19 to 0.2 log10/day). Notably, serum HBV levels were recorded starting at day 7 p.i. in Group D mice and evidence of this plateau period was observed in some of these mice (Fig.3B, grey lines), but the data available was not complete enough to be included in the slope calculations for Phase 5 (Table S1).
Long-term serum HBV amplification followed by viral steady state (Phases 6–7)
Following this interim viral plateau, all mice entered an extended phase of continuous viral expansion (Figs.1B and 3B, phase 6). In this phase, a clear correlation between virus level and inoculum dose was observed. For the mice in Groups B and C, this amplification period began 9 days p.i. (median; Fig.3B, black lines and arrows). In the Group D mice who received a lower 104 HBV DNA genome inoculum the beginning of Phase 6 was later (median day 18) and exhibited variability in the viral load and kinetics that appeared to be related in part to the different lots of human hepatocytes utilized (Fig.3B, grey lines and arrow). Strikingly however, the average slope of viral amplification in all 13 mice across groups was 0.15±0.04 log10/days (Table S1) corresponding to a mean viral doubling time of 2±0.5 days. The observation period for Group B mice ended during Phase 6, but the median duration of this period for the remaining 11 Group C and D mice who were observed through this period was 28 days.
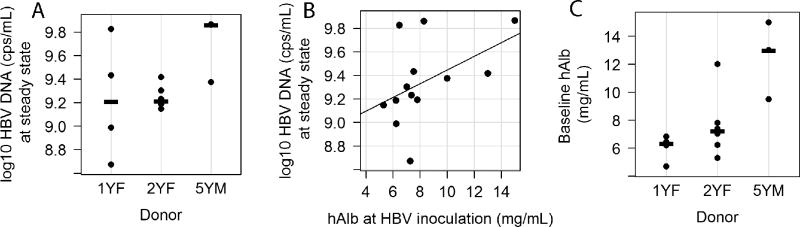
The final kinetic phase observed was viral steady state (Figs.1B and 3B, Phase 7). Steady state HBV serum levels were reached in all Group C mice between day 35 and 42 p.i. (median = day 35; Fig.3B, solid black line). Again, the kinetics were delayed and more variable in the low inoculum dose Group D mice where steady state infection was achieved between 42 and 56 days p.i. (median=day 49) (Fig.3B, grey lines). However, the 9.3±0.3 log10 HBV DNA copies/ml steady state levels obtained in all mice was independent of the initial HBV inoculation dose (Fig.3B, Table S1) or hepatocyte donor (Fig. 4A; P =0.13). A significant correlation (r=0.64, p=0.02) was found between HBV DNA level at steady state and hAlb at time of HBV inoculation (Fig.4B), which could partly explain the slightly higher HBV steady state levels (Fig.4A) in the mice that received the 5YM hepatocytes (p=0.03, Fig.4C).
Figure 4. Associations among hepatocyte donor, steady-state serum HBV DNA and baseline human albumin levels.
(A) Steady state HBV DNA levels in humanized chimeric mice by donor group (1YF: n=4, 2YF: n=6, 5YM: n=3)(p=0.06), (B) a significant correlation between HBV DNA level at steady state and hAlb at HBV inoculation (r=0.64, p=0.02, n=13), and (C) Baseline human serum albumin levels in humanized chimeric mice by donor group (1YF: n=4, 2YF: n=6, 5YM: n=3)(p=0.01). Medians in (A) and (C) are represented by horizontal lines.
As expected, serum hAlb levels remained relatively stable during the entire infection period with the change in hAlb during the long term observation period for Groups C and D fluctuating between −3.8 and 3.7 mg/mL with a median value of 0.0 mg/mL from baseline. Calculated individually, more than 80% of the mice observed throughout this extended period (11/13) exhibit a slope indistinguishable from 0 (i.e., p>0.05, mean=−0.01 mg/mL/day), two had decline rates between 0.05 – 0.06 mg/mL/day (not shown).
Correlation between serum and intrahepatic HBV levels
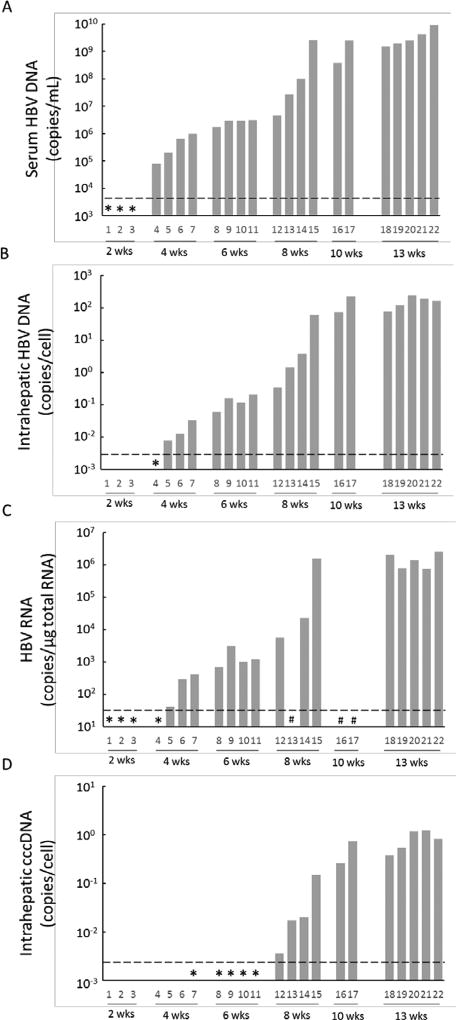
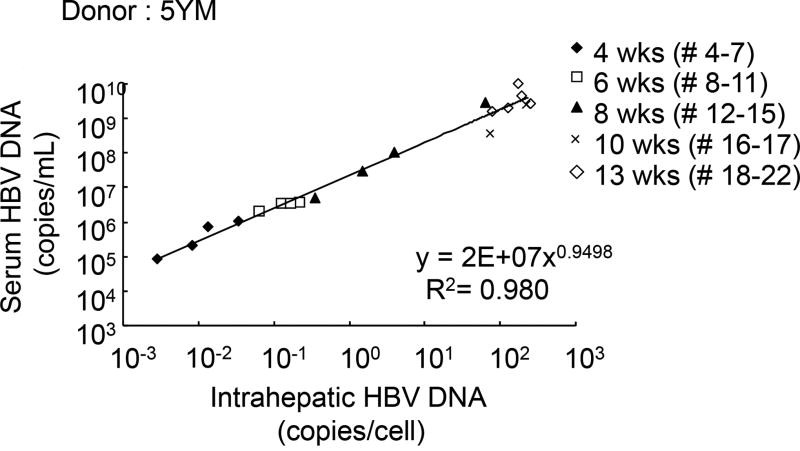
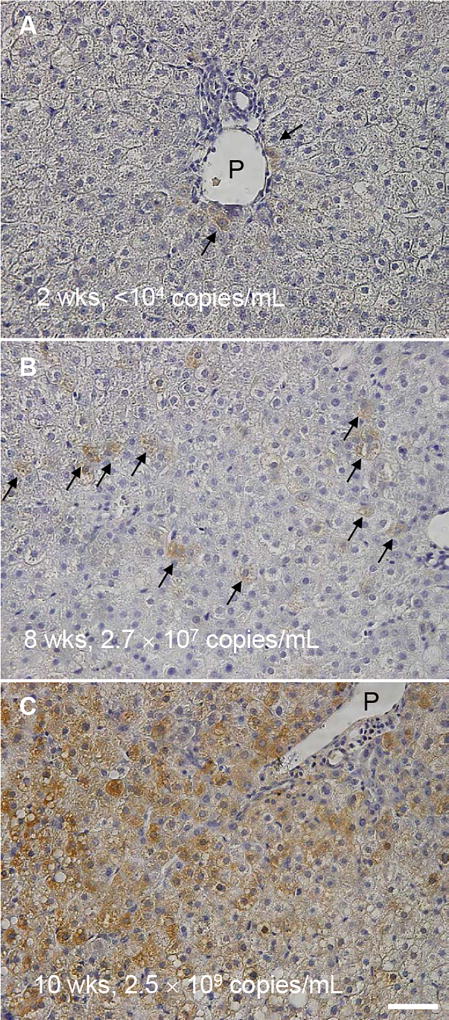
To examine the association among serum HBV DNA, intrahepatic total HBV DNA, HBV cccDNA, HBV RNA and intracellular HBcAg staining, Group D chimeric mice with humanized livers (donor 5YM) were sacrificed at 2 (n=3), 4 (n=4), 6 (n=4), 8 (n=4), 10 (n=2) and 13 (n=5) weeks after inoculation. Two weeks p.i. serum HBV DNA and intrahepatic HBV RNA were detectable; however were below the limit of quantification (Fig. 5A and C). After 4 weeks p.i. serum HBV DNA, total intrahepatic HBV DNA and intrahepatic HBV RNA could be quantified in all mice, whereas intrahepatic cccDNA was detected only in mouse #7 which exhibited the highest serum and intrahepatic HBV DNA (Fig. 5A–D). By week 6 p.i. all mice had cccDNA detected and from week 8 onwards cccDNA was quantifiable (Fig. 5D). Strong correlations were found among the levels of serum HBV DNA and intrahepatic HBV DNA (Fig. 6). To compare possible intracellular differences between group 4 mice that received hepatocytes from different donors, total intrahepatic HBV DNA, cccDNA and the ratio of cccDNA to HBV DNA in the liver of all mice sacrificed at week 13 groups [2YF (n=2), 5YM (n=5), and 1YF (n=4)] were compared, but no difference (P>0.05) among 3 different hepatocyte donor groups was observed (Fig. S1). Finally, immunostaining of liver samples for HBcAg performed in 3 mice at weeks 2, 8 and 10 p.i. (Fig. 7) qualitatively showed a similar pattern of slow HBV expansion. Specifically, on the cellular level, only a few HBV-positive cells could be found near the portal vein at week 2 p.i. (Fig. 7A, arrows) concomitant with detected but non-quantifiable serum HBV DNA. By week 8 when serum HBV DNA had increased to 2.7×107 copies/ml, an increased number of HBV-positive cells was observed which could be found throughout the liver (Fig. 7B, arrows). Then at week 10, when serum HBV DNA has reached steady state (2.5×109 copies/ml) the majority of cells were HBV-positive (Fig. 7C).
Figure 5. Kinetics of HBV in serum and liver of chimeric mice after HBV inoculation.
Chimeric mice (5YM, n=22, #1–22) were infected with HBV and necropsied at 2 (n=3), 4 (n=4), 6 (n=4), 8 (n=4), 10 (n=2), and 13 (n=5) weeks after inoculation. Real-time qPCR was performed to measure the levels of (A) serum HBV DNA, (B) total intrahepatic HBV DNA, (C) total intrahepatic HBV RNA, and (D) intrahepatic cccDNA. Detected (*) but lower than quantification (----) and not determined (#).
Figure 6. Correlation between serum HBV DNA and total intrahepatic HBV DNA.
Twenty-two group D chimeric mice were sacrificed at weeks at 2 (n=3), 4 (n=4), 6 (n=4), 8 (n=4), 10 (n=2), and 13 (n=5) weeks after HBV inoculation. Real-time qPCR was performed to measure the level of serum HBV DNA and total intrahepatic HBV DNA.
Figure 7. Immunostaining of HBcAg in HBV infected chimeric mouse livers.
Representative images of group D chimeric mouse liver samples at (A) 2 weeks, (B) 8 weeks, and (C) 10 weeks post HBV inoculation. Serum HBV DNA levels of the corresponding mice are indicated. Arrows indicate HBV positive cells. P: portal vein. Bar, 50 µm.
Discussion
Currently, mice repopulated with human hepatocytes are the only small animal model available for the study of HBV infection; however, detailed analysis of HBV infection kinetics in this model has been lacking. To provide a more complete characterization of this valuable HBV infection model and enable future mechanistic studies and mathematical modelling necessary to elucidate the dynamics of the HBV infection in humanized chimeric mice we monitored HBV infection at frequent time intervals starting immediately post-inoculation. Quantification of serum HBV DNA allowed us to estimate the HBV clearance rate from circulation and characterize subsequent HBV amplification kinetics to steady state in uPA/SCID mice with humanized livers. While HBV acute infection was previously described in humans (7, 8, 25–28) and chimpanzees (10, 11, 29, 30) via infrequent sampling, the frequent sampling possible in this experimental system has allowed for the: (i) measurement of viral decline kinetics immediately following high dose inoculation, (ii) estimation of the eclipse phase length (i.e. the time between infection and detection of new virions released into circulation) and (iii) characterization of the kinetic pattern of subsequent HBV amplification to steady state, which we unexpectedly found to be multiphasic.
HBV clearance rates from blood of chronically infected subjects have been previously estimated during antiviral treatment in which intracellular HBV production was inhibited with antivirals such as interferon-alfa (IFNa), lamivudine or entecavir. The median t1/2 observed was ~21 hr and ~9 hr in HBeAg-positive and HBeAg-negative patients, respectively, with less than one order of magnitude variation among patients (summarized in (31)). In contrast, performing mathematical modeling of baseline serum and intrahepatic HBV DNA measured from 80 untreated chronically active HBV patients, Dandri et al. estimated a much shorter median t1/2 of ~46 min in HBeAg-positive patients and ~2.5 min in HBeAg-negative patients, respectively, with a strong inverse correlation between baseline viremia and HBV t1/2 (32). Using a different, more direct strategy, in the current study HBV clearance from blood was estimated based on the initial disappearance of virus after intravenous inoculation. During the first 6 h p.i., HBV was cleared from blood with a t1/2 of ~1 h independent of inoculum size which ranged from 104 to 108 copies/ml (Fig. 2A). These findings indicate a shorter HBV t1/2 than was estimated during the drug perturbation experiments in chronically infected patients. Notably, this difference could be related to the differences between acute and chronic infection and/or differences between the mice and humans. Our initial 1 minute post inoculation HBV measurements when extrapolated for total blood volume in the mice accurately reflect the input inoculation dose suggesting virtually instantaneous distribution of the viral inoculum throughout the blood. Theoretically clearance immediately after viral inoculation may include some initial re-distribution/clearance of virus from blood into tissues, a feature that does not occur during chronic infection in which viral equilibrium between blood and tissue has already been established. Interestingly, however, the short HBV t1/2 we observed herein is in line with the Dandri et al. modeling estimates in HBeAg-positive patients (32). Under these conditions in humanized mice, we did not observe any correlation between inoculation dose and HBV clearance rate. However, to confirm whether an inverse correlation between baseline viremia and HBV t1/2 uniquely exists after the establishment of HBV steady state (as suggested by Dandri et al. (32)) would require analogous drug perturbation experiments in the chimeric mouse model system. Notably, these should ideally be performed with antivirals that block HBV secretion as we recently demonstrated in the case of HCV (33). Specifically, an NS5A inhibitor, which rapidly/directly blocks HCV assembly/secretion was able to reveal a much shorter HCV t1/2 (~45 min) compared to previous HCV t1/2 estimates (~3 h) made using drugs such as IFN that mainly block intracellular HCV RNA replication (33).
In the 5 mice inoculated with 108 HBV DNA copies viral decline p.i. ended in a low viral plateau that lasted approximately 1–3 days p.i. (mean 4±0.3 log copies/ml, Fig.3). Assuming that the observed viral plateau is maintained by equal viral production and clearance the duration of the cellular eclipse phase (defined as the time interval from when a cell gets infected until the time virus is secreted) would have to be approximately 1 day since the viral plateau initiated about 1 day p.i.. However, if the observed viral plateau is not maintained by viral production and clearance, but some other virion dynamics (e.g., HBV that initially bound to cells but then is released back into circulation as has been observed for HIV which reversibly binds to follicular dendritic cells (34, 35)) then an eclipse phase of at least 3 days is possible since 3 days p.i. rapid viral resurgence was observed which could only be explained by the production of new virions. If the low viral plateau were due to virus in the initial inoculum binding to cells and then being released, a higher inoculum would lead to more virus being bound and released and hence a higher plateau consistent with what was observed. Theoretically, now that we know the appropriate time window to study, the above bound-release hypothesis could be tested and rejected if the viral plateau is not observed when viral replication is blocked by antiviral treatment initiated before or at the time of infection.
The typical serum HBV increase observed in humans based on infrequent sampling from time of infection to peak viremia has been interpreted as exponential (i.e. monophasic on a log-linear scale) with average doubling time (t2) of ~3 days (range 2–4 days) (7, 8, 25–27). Monophasic HBV amplification has also been reported in acutely infected chimpanzees (10, 11). In contrast, the initial HBV amplification we observed in this immunocompromised humanized mouse model was fast with t2 =8±3 h for 1–3 days, followed by a 1–3 day interim plateau before a long-term HBV amplification with t2=2±0.5 days. Interestingly, the t2 during the long-term amplification in these mice is similar to the single phase t2 found in humans, suggesting that perhaps the fast HBV expansion (Phase 4) and interim plateau (Phase 5) detected in our study may have been missed in humans and chimpanzees due to a lack of sufficiently frequent sampling and/or lower dose infection which would potentially be too unsynchronized to detect the distinct phases observed here. In fact, looking carefully in some cases where chimpanzees have been inoculated with high dose HBV, there is evidence that a multiphasic viral expansion may have occurred [Fig.1a in (11) and Fig.1A in (10)]. The reason for the observed multiphasic HBV expansion in the circulation is not known, but can now be addressed by well-designed studies that include intrahepatic analysis at key time points. As a preliminary assay for possible NK cell mediate HBV-infected cell killing, we measured human alanine aminotransferase levels in the serum samples of the infected mice compared to uninfected controls, but no correlation between HBV levels and hALT levels were observed (Fig. S2). Although HBV is often referred to as a stealth virus because it does not induce significant innate immune signalling (36), one possible reason for the shift from rapid to slow multiphasic expansion could be an induction of a transient innate immune response reminiscent of the biphasic HCV increase with a transient viral decline (1 week p.i.) that was observed in acutely HCV-infected chimpanzees which was proposed to be caused by an observed induction of endogenous type I IFN at 1–2 weeks p.i. (37, 38). It would likewise be interesting to monitor intrahepatic IFN induction during acute HBV infection in future chimeric mouse studies as well as previous HBV chimpanzee experiments if samples are still available.
During the final HBV kinetic phase, virus reached high steady state levels in all uPA/SCID chimeric mice (9.3±0.3 log10 copies/ml) 35 to 56 days p.i. (Fig.3B). While, peak serum HBV DNA levels were observed 60 to 170 days p.i. in acutely HBV-infected humans (7, 8, 27, 39) and chimpanzees (10, 11), virus levels typically declined at some point in these immunocompetent host rather than remaining at high steady state levels as observed in the uPA/SCID mice consistent with the crucial role of the adaptive immune response in HBV clearance/control (2, 10–12). In addition, no evidence of a significant decline in hAlb was observed during the entire infection period in mice consistent with the notion that HBV itself is noncytopathic (40).
Interestingly, in chimpanzees the size of the viral inoculum was shown to contribute to the viral kinetics during acute infection and infection outcome, i.e., persistence or clearance (11). In the current study, we investigated the role of inoculum dose on viral kinetics in the absence of an adaptive immune response and found that inoculum size had no effect on HBV initial clearance rate from blood post inoculation, the viral doubling time during the HBV expansion phases (phases 4 and 6), the length of the interim plateau (phase 5), or viral steady state levels (phase 7). The most evident effect of low virus dose was simply a delay in detection of initial virus expansion (phase 4), which subsequently delayed all other kinetic phases. This seems to indicate that the adaptive immune response is not only important in HBV clearance, but also that the interplay between virus dose and the adaptive immune response is additionally a major determinant of HBV infection kinetics. While the development of an adaptive immune response to HBV is relatively slow and thus likely does not affect very early HBV infection kinetics, the chimeric uPA-SCID model does provides an advantage as it ensures that fundamental HBV infection dynamics can be analysed in the absence of this variable confounding factor. The study of HBV immunology certainly is of interest, but while such immunocompetent models are being developed the simplicity of the uPA-SCID model provides a particular advantage for many needed molecular virology studies.
While the distinct phases defined in this study highlight key time points post-inoculation that are of particular interest for more detailed intracellular analyses and provides a basis for determining what types of analyses might be most informative, our initial analysis found that intrahepatic total HBV DNA, cccDNA and RNA correlate with serum HBV DNA during infection in uPA/SCID mice (Fig.5 and 6), consistent with findings in 3 acutely HBV-infected chimpanzees (10). Whether intrahepatic HBV RNA and DNA levels exhibit the same multiphasic amplification pattern will require more frequent intrahepatic sampling, but meanwhile Northern Blot analysis suggests that early in infection expression levels of HBV 3.5kb RNA was more pronounced than the HBs mRNAs (Fig. S3), an unexpected finding that needs to be further explored. Also in agreement with chimpanzee data (2), at week 10 (at which time serum HBV levels had reached steady state) most cells were HBV positive. Notably, immunostaining for HBcAg in the liver at the interim weeks 2 and 8 reveal that during the rapid viral expansion (i.e., week 8) HBV-infected cells appear not to form clusters but rather were spatially distributed suggesting virus spread primarily occurs by diffusion of extracellular virus to non-adjacent cells as previously reported for in vitro duck hepatitis B virus (41). This is in contrast to the clusters/foci observed in HCV infected livers which has been attributed to direct cell-to-cell spread or virus released from one cell preferentially infecting its neighbours (42–44). Importantly, with the enhanced characterization of the uPA-SCID chimeric mouse HBV infection model presented here, we are now better poised to determine whether HBV has the capacity to spread cell-to-cell and address other clinically relevant questions.
In summary, we characterized HBV kinetics in blood and liver from the time of inoculation until steady state in uPA/SCID mice with humanized livers. We found that immediately after inoculation, the HBV half-life in blood is approximately 1 h. Our work also revealed an unexpectedly multiphasic viral amplification in the serum in the absence of adaptive immune response. This detailed viral kinetic analysis is an important starting point for developing mathematical models of HBV infection in the absence of an adaptive immune response, but in the presence of an innate response which perhaps contributes to the observed multiphasic pattern of HBV DNA amplification thus revealing further insights into the interaction between HBV and its host.
Supplementary Material
Acknowledgments
We thank Beverly Chrabot for technical assistance. Portion of this work were supported by PhoenixBio Co. Ltd., the Research Program on Hepatitis from the Japan Agency for Medical Research and Development, AMED (Grant no. 17fk0210104h0001), NIH grants R01-AI078881, R01-AI028433, R01-OD011095 and R01-AI116868 and done under the auspices of the U.S. Department of Energy under contract DE-AC52-06NA25396.
Abbreviations
- HBV
hepatitis B virus
- uPA
urokinase type plasminogen activator
- SCID
severely severe combined immunodeficiency
- cccDNA
covalently-closed-circular DNA
- HBcAg
hepatitis B core antigen
- p.i.
post inoculation
- CI
confidence interval
- cps
copies
- h
hours
- d−1
per day
- t1/2
half life
- t2
doubling time
Footnotes
Conflict of Interest Disclosures:
YI, HY and CT are employees of PhoenixBio Co. Ltd. None of the other authors has any financial interest or conflict of interest related to this research.
Reference list
- 1.Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 2.Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 3.McMahon BJ, Alward WL, Hall DB, Heyward WL, Bender TR, Francis DP, Maynard JE. Acute hepatitis B virus infection: relation of age to the clinical expression of disease and subsequent development of the carrier state. J Infect Dis. 1985;151:599–603. doi: 10.1093/infdis/151.4.599. [DOI] [PubMed] [Google Scholar]
- 4.Institute of Medicine (IOM) Hepatitis and liver cancer: a national strategy for prevenation and control of hepatitis B and C. Washington, DC: National Academies Press; 2010. [PubMed] [Google Scholar]
- 5.Protzer U, Maini MK, Knolle PA. Living in the liver: hepatic infections. Nat Rev Immunol. 2012;12:201–213. doi: 10.1038/nri3169. [DOI] [PubMed] [Google Scholar]
- 6.Guidotti LG, Chisari FV. Immunobiology and pathogenesis of viral hepatitis. Annu Rev Pathol. 2006;1:23–61. doi: 10.1146/annurev.pathol.1.110304.100230. [DOI] [PubMed] [Google Scholar]
- 7.Whalley SA, Murray JM, Brown D, Webster GJ, Emery VC, Dusheiko GM, Perelson AS. Kinetics of acute hepatitis B virus infection in humans. J Exp Med. 2001;193:847–854. doi: 10.1084/jem.193.7.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keating SM, Heitman JD, Wu S, Deng X, Stramer SL, Kuhns MC, Mullen C, et al. Cytokine and chemokine responses in the acute phase of hepatitis B virus replication in naive and previously vaccinated blood and plasma donors. J Infect Dis. 2014;209:845–854. doi: 10.1093/infdis/jit563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thimme R, Wieland S, Steiger C, Ghrayeb J, Reimann KA, Purcell RH, Chisari FV. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J Virol. 2003;77:68–76. doi: 10.1128/JVI.77.1.68-76.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray JM, Wieland SF, Purcell RH, Chisari FV. Dynamics of hepatitis B virus clearance in chimpanzees. Proc Natl Acad Sci U S A. 2005;102:17780–17785. doi: 10.1073/pnas.0508913102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asabe S, Wieland SF, Chattopadhyay PK, Roederer M, Engle RE, Purcell RH, Chisari FV. The size of the viral inoculum contributes to the outcome of hepatitis B virus infection. J Virol. 2009;83:9652–9662. doi: 10.1128/JVI.00867-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciupe SM, Ribeiro RM, Nelson PW, Perelson AS. Modeling the mechanisms of acute hepatitis B virus infection. Journal of theoretical biology. 2007;247:23–35. doi: 10.1016/j.jtbi.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korba BE, Cote PJ, Wells FV, Baldwin B, Popper H, Purcell RH, Tennant BC, et al. Natural history of woodchuck hepatitis virus infections during the course of experimental viral infection: molecular virologic features of the liver and lymphoid tissues. J Virol. 1989;63:1360–1370. doi: 10.1128/jvi.63.3.1360-1370.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dandri M, Lutgehetmann M. Mouse models of hepatitis B and delta virus infection. J Immunol Methods. 2014;410:39–49. doi: 10.1016/j.jim.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Tateno C, Yoshizane Y, Saito N, Kataoka M, Utoh R, Yamasaki C, Tachibana A, et al. Near completely humanized liver in mice shows human-type metabolic responses to drugs. Am J Pathol. 2004;165:901–912. doi: 10.1016/S0002-9440(10)63352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugiyama M, Tanaka Y, Kato T, Orito E, Ito K, Acharya SK, Gish RG, et al. Influence of hepatitis B virus genotypes on the intra- and extracellular expression of viral DNA and antigens. Hepatology. 2006;44:915–924. doi: 10.1002/hep.21345. [DOI] [PubMed] [Google Scholar]
- 17.Tsuge M, Hiraga N, Takaishi H, Noguchi C, Oga H, Imamura M, Takahashi S, et al. Infection of human hepatocyte chimeric mouse with genetically engineered hepatitis B virus. Hepatology. 2005;42:1046–1054. doi: 10.1002/hep.20892. [DOI] [PubMed] [Google Scholar]
- 18.Abe A, Inoue K, Tanaka T, Kato J, Kajiyama N, Kawaguchi R, Tanaka S, et al. Quantitation of hepatitis B virus genomic DNA by real-time detection PCR. J Clin Microbiol. 1999;37:2899–2903. doi: 10.1128/jcm.37.9.2899-2903.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masumoto N, Tateno C, Tachibana A, Utoh R, Morikawa Y, Shimada T, Momisako H, et al. GH enhances proliferation of human hepatocytes grafted into immunodeficient mice with damaged liver. J Endocrinol. 2007;194:529–537. doi: 10.1677/JOE-07-0126. [DOI] [PubMed] [Google Scholar]
- 20.Uprichard SL, Boyd B, Althage A, Chisari FV. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc Natl Acad Sci U S A. 2005;102:773–778. doi: 10.1073/pnas.0409028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takkenberg RB, Zaaijer HL, Molenkamp R, Menting S, Terpstra V, Weegink CJ, Dijkgraaf MG, et al. Validation of a sensitive and specific real-time PCR for detection and quantitation of hepatitis B virus covalently closed circular DNA in plasma of chronic hepatitis B patients. J Med Virol. 2009;81:988–995. doi: 10.1002/jmv.21477. [DOI] [PubMed] [Google Scholar]
- 22.Pasquetto V, Wieland SF, Uprichard SL, Tripodi M, Chisari FV. Cytokine-sensitive replication of hepatitis B virus in immortalized mouse hepatocyte cultures. J Virol. 2002;76:5646–5653. doi: 10.1128/JVI.76.11.5646-5653.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malmstrom S, Larsson SB, Hannoun C, Lindh M. Hepatitis B viral DNA decline at loss of HBeAg is mainly explained by reduced cccDNA load--down-regulated transcription of PgRNA has limited impact. PLoS One. 2012;7:e36349. doi: 10.1371/journal.pone.0036349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 25.Biswas R, Tabor E, Hsia CC, Wright DJ, Laycock ME, Fiebig EW, Peddada L, et al. Comparative sensitivity of HBV NATs and HBsAg assays for detection of acute HBV infection. Transfusion. 2003;43:788–798. doi: 10.1046/j.1537-2995.2003.00424.x. [DOI] [PubMed] [Google Scholar]
- 26.Yoshikawa A, Gotanda Y, Itabashi M, Minegishi K, Kanemitsu K, Nishioka K Japanese Red Cross NATSRG. HBV NAT positive [corrected] blood donors in the early and late stages of HBV infection: analyses of the window period and kinetics of HBV DNA. Vox Sang. 2005;88:77–86. doi: 10.1111/j.1423-0410.2005.00602.x. [DOI] [PubMed] [Google Scholar]
- 27.Yoshikawa A, Gotanda Y, Minegishi K, Taira R, Hino S, Tadokoro K, Ohnuma H, et al. Lengths of hepatitis B viremia and antigenemia in blood donors: preliminary evidence of occult (hepatitis B surface antigen-negative) infection in the acute stage. Transfusion. 2007;47:1162–1171. doi: 10.1111/j.1537-2995.2007.01234.x. [DOI] [PubMed] [Google Scholar]
- 28.Stramer SL, Wend U, Candotti D, Foster GA, Hollinger FB, Dodd RY, Allain JP, et al. Nucleic acid testing to detect HBV infection in blood donors. N Engl J Med. 2011;364:236–247. doi: 10.1056/NEJMoa1007644. [DOI] [PubMed] [Google Scholar]
- 29.Wieland SF, Spangenberg HC, Thimme R, Purcell RH, Chisari FV. Expansion and contraction of the hepatitis B virus transcriptional template in infected chimpanzees. Proc Natl Acad Sci U S A. 2004;101:2129–2134. doi: 10.1073/pnas.0308478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabuchi A, Tanaka J, Katayama K, Mizui M, Matsukura H, Yugi H, Shimada T, et al. Titration of hepatitis B virus infectivity in the sera of pre-acute and late acute phases of HBV infection: transmission experiments to chimeric mice with human liver repopulated hepatocytes. J Med Virol. 2008;80:2064–2068. doi: 10.1002/jmv.21320. [DOI] [PubMed] [Google Scholar]
- 31.Dahari H, Cotler SJ, Layden TJ, Perelson AS. Hepatitis B virus clearance rate estimates. Hepatology. 2009;49:1779–1780. doi: 10.1002/hep.22874. author reply 1780–1771. [DOI] [PubMed] [Google Scholar]
- 32.Dandri M, Murray JM, Lutgehetmann M, Volz T, Lohse AW, Petersen J. Virion half-life in chronic hepatitis B infection is strongly correlated with levels of viremia. Hepatology. 2008;48:1079–1086. doi: 10.1002/hep.22469. [DOI] [PubMed] [Google Scholar]
- 33.Guedj J, Dahari H, Rong L, Sansone ND, Nettles RE, Cotler SJ, Layden TJ, et al. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc Natl Acad Sci U S A. 2013;110:3991–3996. doi: 10.1073/pnas.1203110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cavert W, Notermans DW, Staskus K, Wietgrefe SW, Zupancic M, Gebhard K, Henry K, et al. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV-1 infection. Science. 1997;276:960–964. doi: 10.1126/science.276.5314.960. [DOI] [PubMed] [Google Scholar]
- 35.Hlavacek WS, Wofsy C, Perelson AS. Dissociation of HIV-1 from follicular dendritic cells during HAART: mathematical analysis. Proc Natl Acad Sci U S A. 1999;96:14681–14686. doi: 10.1073/pnas.96.26.14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wieland SF, Chisari FV. Stealth and cunning: hepatitis B and hepatitis C viruses. J Virol. 2005;79:9369–9380. doi: 10.1128/JVI.79.15.9369-9380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dahari H, Major M, Zhang X, Mihalik K, Rice CM, Perelson AS, Feinstone SM, et al. Mathematical modeling of primary hepatitis C infection: noncytolytic clearance and early blockage of virion production. Gastroenterology. 2005;128:1056–1066. doi: 10.1053/j.gastro.2005.01.049. [DOI] [PubMed] [Google Scholar]
- 38.Major ME, Dahari H, Mihalik K, Puig M, Rice CM, Neumann AU, Feinstone SM. Hepatitis C virus kinetics and host responses associated with disease and outcome of infection in chimpanzees. Hepatology. 2004;39:1709–1720. doi: 10.1002/hep.20239. [DOI] [PubMed] [Google Scholar]
- 39.McIntyre N. Clinical presentation of acute viral hepatitis. Br Med Bull. 1990;46:533–547. doi: 10.1093/oxfordjournals.bmb.a072414. [DOI] [PubMed] [Google Scholar]
- 40.Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 41.Funk A, Hohenberg H, Mhamdi M, Will H, Sirma H. Spread of hepatitis B viruses in vitro requires extracellular progeny and may be codetermined by polarized egress. J Virol. 2004;78:3977–3983. doi: 10.1128/JVI.78.8.3977-3983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wieland S, Makowska Z, Campana B, Calabrese D, Dill MT, Chung J, Chisari FV, et al. Simultaneous detection of hepatitis C virus and interferon stimulated gene expression in infected human liver. Hepatology. 2014;59:2121–2130. doi: 10.1002/hep.26770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graw F, Balagopal A, Kandathil AJ, Ray SC, Thomas DL, Ribeiro RM, Perelson AS. Inferring viral dynamics in chronically HCV infected patients from the spatial distribution of infected hepatocytes. PLoS Comput Biol. 2014;10:e1003934. doi: 10.1371/journal.pcbi.1003934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kandathil AJ, Graw F, Quinn J, Hwang HS, Torbenson M, Perelson AS, Ray SC, et al. Use of laser capture microdissection to map hepatitis C virus-positive hepatocytes in human liver. Gastroenterology. 2013;145:1404–1413. e1401–1410. doi: 10.1053/j.gastro.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.