Summary
In most eukaryotes, meiotic crossovers are essential for error-free chromosome segregation but are specifically repressed near centromeres to prevent missegregation. Recognized for >85 years, the molecular mechanism of this repression has remained unknown. Meiotic chromosomes contain two distinct cohesin complexes: pericentric complex (for segregation) and chromosomal arm complex (for crossing-over). We show that the pericentric-specific complex also actively represses pericentric meiotic double-strand break (DSB) formation and, consequently, crossovers. We uncover the mechanism by which fission yeast heterochromatin protein Swi6 (mammalian HP1-homolog) prevents recruitment of activators of meiotic DSB formation. Localizing missing activators to wild-type pericentromeres bypasses repression and generates abundant crossovers but reduces gamete viability. The molecular mechanism elucidated here likely extends to other species including humans, where pericentric crossovers can result in disorders such as Down syndrome. These mechanistic insights provide new clues to understand the roles played by multiple cohesin complexes, especially in human infertility and birth defects.
ETOC BLURB
Nambiar and Smith identify the molecular mechanisms repressing harmful meiotic crossovers around centromeres, which can produce disorders including Down syndrome. They show that presence of the mitotic cohesin subunit Psc3, instead of its meiotic paralog Rec11, in pericentric heterochromatin excludes Spo11-activating proteins and thereby blocks double-strand break formation and recombination.
Introduction
Meiosis is a special type of cell division essential for gamete formation in all sexually reproducing organisms. It involves one round of DNA replication followed by two rounds of nuclear division and chromosome segregation in order to reduce the chromosome number by half. Proper segregation of chromosomes during both nuclear divisions is essential to ensure healthy progeny. In most species, genetic recombination (crossover formation) between homologs along with sister chromatid cohesion is required to allow proper segregation of homologous chromosomes. The distribution of crossovers is not uniform across the genome, however, and certain regions such as the regions around centromeres (pericentric regions) have up to 100 times less crossing over per unit physical distance along the DNA than chromosomal arms (Beadle, 1932; Nambiar and Smith, 2016). Failure to prevent pericentric recombination events can be deleterious, causing missegregation of chromosomes that leads to aneuploidy, a hallmark of birth defects and several genetic disorders such as Down syndrome (Hassold and Hunt, 2001; Rockmill et al., 2006; Lamb et al., 2005). However, the molecular mechanism of this crucial regulation of repressing pericentric recombination during meiosis is unknown despite having been discovered over 85 years ago (Beadle, 1932; Sax, 1932; Mather, 1939).
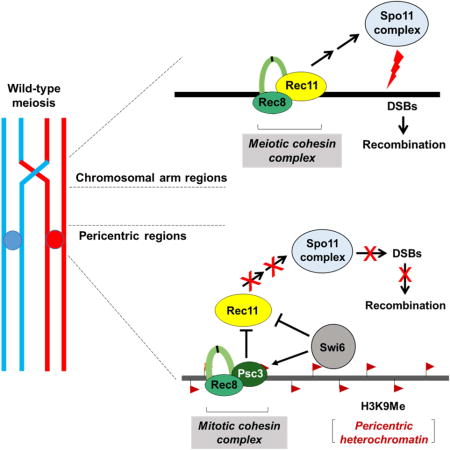
The genetically tractable fission yeast Schizosaccharomyces pombe has large (35 – 110 kb), repetitive, epigenetically maintained pericentromeres similar to those of less amenable multicellular eukaryotes (Grewal and Jia, 2007; Nambiar and Smith, 2016). The pericentric region contains H3 Lys9-(mono-, di-, or tri-)methylated histones (designated hereafter as H3K9me), which attracts many proteins to maintain and spread heterochromatin as well as to deposit sister-chromatid cohesins to facilitate segregation. RNAi facilitates the establishment of heterochromatin in S. pombe and directs the sole H3K9 methyl transferase Clr4 (SUV39H1) to the pericentromeres (Grewal and Jia, 2007) (here and below mammalian homologs are in parentheses at their first mention). H3K9me differentiates the pericentric region from chromosomal arms; it also specifically binds chromodomain (CD)-containing proteins such as heterochromatin protein (HP1) homologs Swi6 and Chp2. Swi6 recruits the mitotic cohesin complex Rad21(RAD21)-Psc3 (STAG1-STAG2) to the pericentric region through interactions with Psc3 and the cohesin loader Mis4 (SCC2) (Nonaka et al., 2002; Fischer et al., 2009) (Figure 1A). During meiosis, the meiosis-specific cohesin complex Rec8 (REC8)-Rec11 (STAG3) replaces its mitotic counterpart in the chromosomal arms (Kitajima et al., 2003) (Figure 1B). However, the meiotic Rec11 subunit fails to replace the mitotic Psc3 subunit at the pericentric regions, resulting in Rec8-Psc3 complex formation near meiotic centromeres.
Figure 1. Meiosis-specific linear-element protein Rec10 tethered to pericentric regions overcomes repression of meiotic recombination.
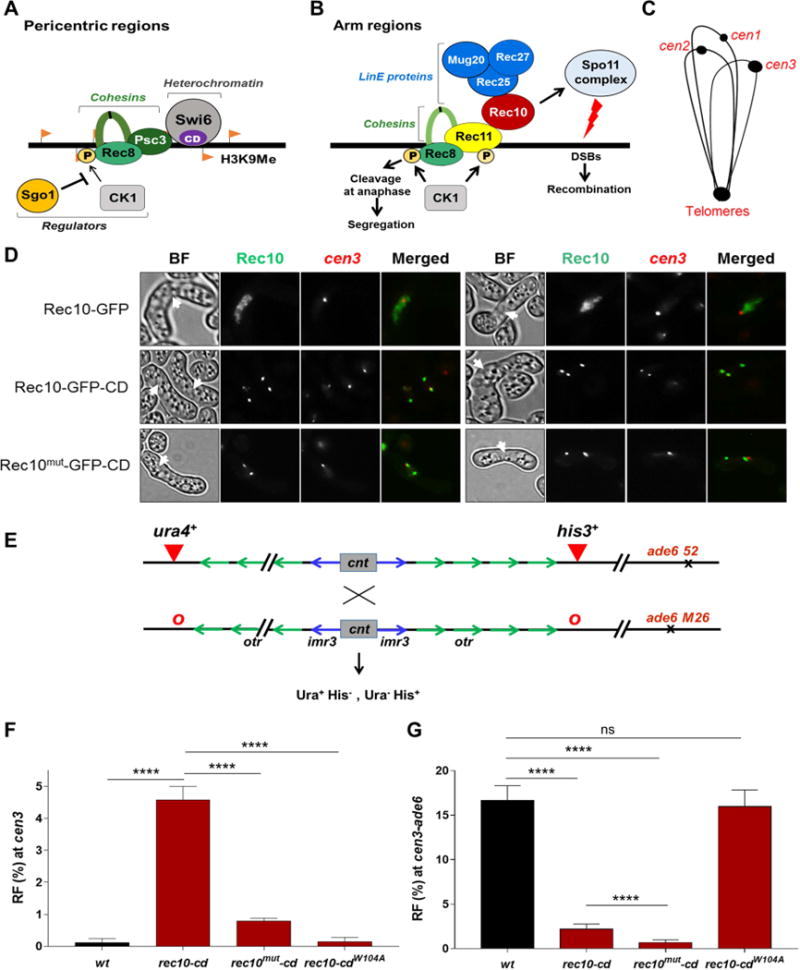
(A) Schematic representation of the composition of proteins present during meiosis at the pericentric regions in S. pombe. (B) Model for initiation of meiotic recombination by DSB formation across chromosomal arms in S. pombe. See text for explanation. (C) Cartoon of telomeric cluster during the horsetail stage of meiosis. (D) Fluorescent microscopic images of meiotic horsetail cells (marked with white arrowheads) expressing Rec10-GFP, Rec10-GFP-CD or Rec10MUT-GFP-CD (green). Rec10MUT is a recombination-deficient missense mutant (R184F D185T). cen3 (red) is at the nuclear pole opposite the telomeric cluster, which also binds CD due to heterochromatin. cen3 is labeled with tdTomato at the left innermost repeat (imr). BF, bright field. Yellow, colocalized signals when merged. Images are representative of at least 20-30 cells examined. (E) Assay for recombination across the pericentric region on chromosome 3 (cen3). cnt, central kinetochore-binding region; imr, innermost repeat; otr, outer repeat. Recombinants across cen3 were measured as Ura+ His− and Ura− His+, and chromosomal arm recombinants (cen3-ade6) as His+ dark red (ade6-M26) and His− light red (ade6-52). o, absence of the ura4+ or his3+ insertion. (F, G) Recombinant frequency (RF) across cen3 (F) and the cen3-ade6 arm interval (G). Data are mean ± SEM (n=4 to 8 experiments, assaying 654 to 3733 spore colonies). ****, p <0.0001 (two-tailed Fisher’s exact test). In this and all subsequent figures bar colors correspond to those of the recombination proteins in Figure 1B, with wild type (wt) as black. See also Figure S1, Tables S1, S2 and S3.
Meiotic recombination is frequent in chromosomal arms and begins with loading of meiosis-specific Rec8 and Rec11 during replication (Figure 1B). Casein kinase (CK1) phosphorylates Rec11 near its N-terminus to recruit linear element (LinE) protein Rec10 (distantly related to Red1 in budding yeast and SCP2 in humans) (Offenberg et al., 1998; Phadnis et al., 2015; Sakuno and Watanabe, 2015). Rec10 binds nearly uniformly across chromosomal arms and is essential for DSB-formation and recombination at all the genetic intervals measured in chromosomal arms (Ellermeier and Smith, 2005; Fowler et al., 2013). It associates with other LinE proteins Rec25, Rec27 and Mug20, which bind specifically to short chromosomal intervals at which DSBs form at high frequency (“hotspots”) and are required in a chromosomal region-specific manner for recombination (Davis et al., 2008; Fowler et al., 2013). DSBs are formed by Spo11 (SPO11; Rec12 in S. pombe), which acts in a putative complex with seven known partner proteins that are essential for its activity (Cromie and Smith, 2008). Spo11 can bind to DNA in regions without recombination, but Rec10 is essential for Spo11 to be activated to induce DSBs in cells (Ellermeier and Smith, 2005; Davis et al., 2008; Fowler et al., 2013). This is especially evident at the pericentric regions, where Spo11 is present in all three pericentric regions during meiosis (Ludin et al., 2008; Fowler et al., 2013), whereas the activator Rec10 is not (Sakuno and Watanabe, 2015). Rec10 likely activates Spo11 by directly binding to Rec15, a Spo11 partner protein essential for DSB formation and recombination (Ponticelli and Smith, 1989; DeVeaux and Smith, 1994; Lin and Smith, 1995; Ellermeier and Smith, 2005; Miyoshi et al., 2012).
Our earlier work showed that DSBs are undetectable and recombination is ~200-fold reduced across wild-type pericentromere 3 (cen3) during meiosis (Ellermeier et al., 2010), despite the presence of Spo11 protein at all pericentromeres (Ludin et al., 2008; Fowler et al., 2013). Furthermore, artificial tethering of Spo11 near centromeres in budding yeast did not increase DSB formation (Robine et al., 2007; Fukuda et al., 2008), perhaps because Spo11 cannot be activated there. Our earlier work also showed that heterochromatin mutants, such as clr4Δ, and RNAi mutants, such as dcr1Δ, are highly derepressed for cen3 recombination in a Rec8- and Rec10-dependent manner (Ellermeier et al., 2010). These results indicate that, in the absence of Clr4-mediated H3K9me formation, the pericentric regions behave like chromosomal arms with respect to meiotic recombination. In the present study, we unravel the molecular basis of pericentric repression of meiotic recombination in wild-type (heterochromatin-containing) strains and show that this repression results from lack of activation of the Spo11 protein complex to form DSBs. We uncover a complex, multi-layered interaction between the cohesin subunits Rec8, Psc3, and Rec11 and the heterochromatin protein Swi6 responsible for preventing recruitment of Spo11-activators and thus DSBs and deleterious crossovers at the pericentromeres. This repressive mechanism appears wide-spread among species, including mammals, and is crucial for formation of viable gametes.
Results
Overview
We first sought the molecular step(s) that limit DSB formation in pericentromeres by artificially localizing DSB- and recombination-promoting factors (Figure 1B) to the pericentromeres. The results showed that Rec10 is one limiting factor. Note that in a biochemical pathway more than one step can be limiting by the criterion that increasing the flux at that step increases the overall output. We then sought the molecular mechanism(s) that limit Rec10 action in pericentric regions. The results showed complex interactions among the cohesin subunits Rec8, Rec11, and Psc3 with the heterochromatin protein Swi6, which has both (a) negative and (b) positive effects on pericentric recombination. (a) By recruiting Psc3 to the pericentric region Swi6 excludes Rec11, the meiosis-specific paralog of Psc3, thereby reducing recombination. (b) By recruiting Psc3, which binds Rec8, Swi6 indirectly recruits Rec8, thereby potentially promoting recombination. Because recombination normally requires both Rec8 and Rec11 (Figures 1A and 1B), simple removal of Swi6 does not derepress pericentric recombination. These interactions, summarized in Figure 7, are important to keep in mind while considering the experiments below.
Figure 7. Model for repression of meiotic recombination in pericentric regions.
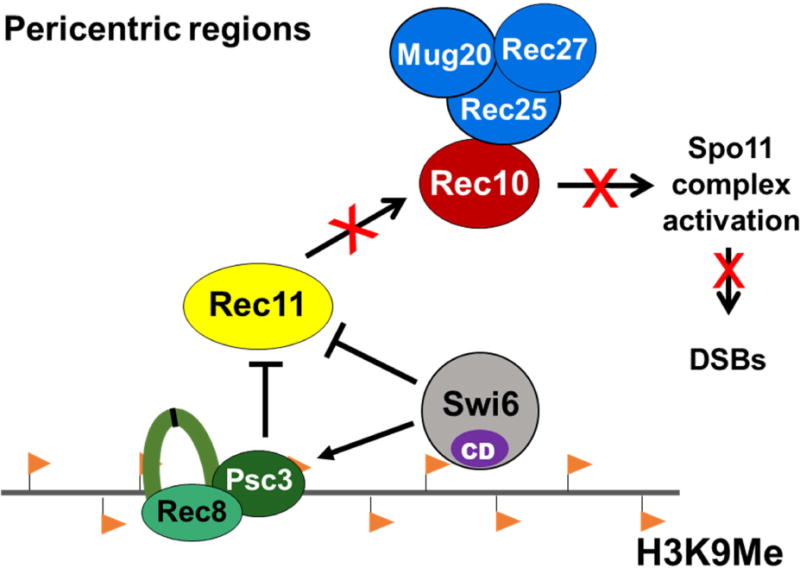
Histone H3K9me (orange flags) in pericentric regions binds the chromodomain (CD)-containing protein (Swi6) responsible for enriching cohesin subunits (Rec8-Psc3) specifically in pericentric regions. Both Psc3 and Swi6 exclude Rec11 from the pericentric regions and thereby prevent it from binding Rec8 and thus to recruit Rec10. Absence of Rec10 precludes activation of Spo11 complex to form DSBs that initiate recombination during meiosis.
LinE Protein Rec10 Is a Limiting Factor for Meiotic Recombination at a Pericentric Region
To determine the Spo11-activating factors limiting recombination in wild-type pericentromeres, we tethered proteins involved in the early steps of meiotic DSB formation to H3K9 methylated pericentric regions. For this, we fused an H3K9me-binding chromodomain (CD) to each protein expressed from its native locus on the chromosome. We first tested Rec10, because it activates the Spo11-complex and is essential for genome-wide recombination (Ellermeier and Smith, 2005; Fowler et al., 2013). We examined the localization of Rec10 (fused to GFP) during the horsetail stage of meiosis, during which telomeres cluster at one end of the elongated nucleus and centromeres at the other end (Figure 1C) (Chikashige et al., 1994). We labeled cen3 with a red fluorophore to visualize its location. Rec10 was uniformly distributed throughout the chromosomes and did not detectably localize to the pericentromeres in 23 out of 28 cells examined (Figure 1D). However, Rec10 fused with the CD (Rec10-CD) resulted in two sharp, discrete foci that appeared to correspond to the heterochromatic telomere cluster on one end of the horsetail and to the largest pericentromere (cen3) on the other end; Rec10-CD colocalized with the cen3 marker in 21 out of 26 cells examined (Figure 1D). In some cells, three foci were observed that could be due to separated homologs or centromere clusters (Figure 1D). This shows that, as expected, fusion with the CD localizes Rec10 to the pericentric regions and other H3K9me (heterochromatic) regions in the genome.
We next measured pericentric recombination between markers closely flanking cen3 (Ellermeier et al., 2010) (Figure 1E). In the presence of Rec10-CD there was a dramatic, >50-fold increase over wild type in cen3 recombination (to 4.8%) (Figures 1F and Table S1, Rows 1 and 2). The rec10-cd allele contained gfp, but GFP fusions alone to either Rec10 or other proteins had no significant effect on cen or arm recombination (Figure S1A and Table S2, Rows 1-6). There was also a concomitant 3- to 8-fold reduction, compared to wild type, in arm recombination at multiple intervals in cells with Rec10-CD (Figures 1G and S1C). This agrees with reduced Rec10 localization in chromosomal arms following CD fusion (Figure 1D). In a rec10+/rec10-cd heterozygous cross, recombination in chromosomal arms was restored due to the wild-type copy of rec10, while pericentric recombination also remained derepressed (Figure S1D and Table S3, Rows 1-3). Furthermore, elimination of H3K9me by clr4Δ abolished recruitment of Rec10-CD to pericentric regions and restored arm recombination as well (Figures S1D, S1E and Table S3, Rows 3-5), as expected from the lack of effect of CD in the absence of H3K9me.
These combined genetic and microscopic results show that localized recruitment of Rec10 is a limiting factor for recombination, most notably in the pericentric regions. This conclusion is consistent with previous observations that both Rec10 binding and DSB frequency are coordinately elevated at DSB hotspots in chromosomal arms (Davis et al., 2008; Fowler et al., 2013). Here, we observed that both recombination and Rec10-CD abundance were increased at the pericentric region but decreased in chromosomal arms, relative to Rec10 lacking the CD fusion. Collectively, these observations indicate that Rec10 binds where it acts and, conversely, acts where it binds.
To test directly the role of Rec10’s recombinogenic function in mediating pericentric recombination, we used a double missense mutant Rec10MUT (R184F, D185T). This mutant protein goes to the nucleus along with other linear element proteins such as Rec25 but strongly reduces arm recombination (Ma et al., 2017); it does not activate Spo11 (Rec12) to promote DSB formation or recombination in chromosomal arms or pericentric regions (Figures 1G and S1C). Like Rec10-CD, the Rec10MUT-CD localized to the pericentromeres (Figure 1D) but in contrast nearly abolished pericentric recombination (Figure 1F and Table S1, Rows 1-3). Additionally, fusion of Rec10 to CDW104A, a mutant CD that does not bind H3K9me (Jacobs and Khorasanizadeh, 2002), neither promoted recombination nor localized near centromeres and gave wild-type levels of arm recombination (Figures 1F, 1G, S1B and Table S1, Rows 1,2 and 4). These observations confirm that, as expected, Rec10-CD but not Rec10 itself binds to heterochromatic regions, including pericentric regions, and promotes recombination there. Thus, wild-type pericentric regions can undergo meiotic recombination but only if recombination-proficient Rec10 is localized there, suggesting an active mechanism for its exclusion from pericentric regions.
Presence of Rec10 in the Pericentric Region Generates DSBs
DSBs are a prerequisite for meiotic recombination. We tested the formation of DSBs across a 125 kb cen3 fragment upon induction of meiosis in wild-type, rec10-cd and rec10mut-cd strains. Analyses were done in rad50S mutants, in which DSBs are not processed and thus accumulate (Young et al., 2002). Pericentric DSBs were undetectable in wild-type even at late time points, although DSBs were readily observed at the ade6-3049 meiotic hotspot ~200 kb distant in the chromosomal arm (Figure 2). However, pericentric DSBs were strongly induced in the presence of Rec10-CD, further supporting the conclusion that repression of crossovers at the pericentric regions is due to the lack of Rec10 and failure to activate DSB formation. As expected, rec10mut-cd did not produce DSBs at either the pericentric region or the chromosomal arm site tested (Figure 2).
Figure 2. Linear element protein Rec10 tethered to pericentric regions induces DSB formation.
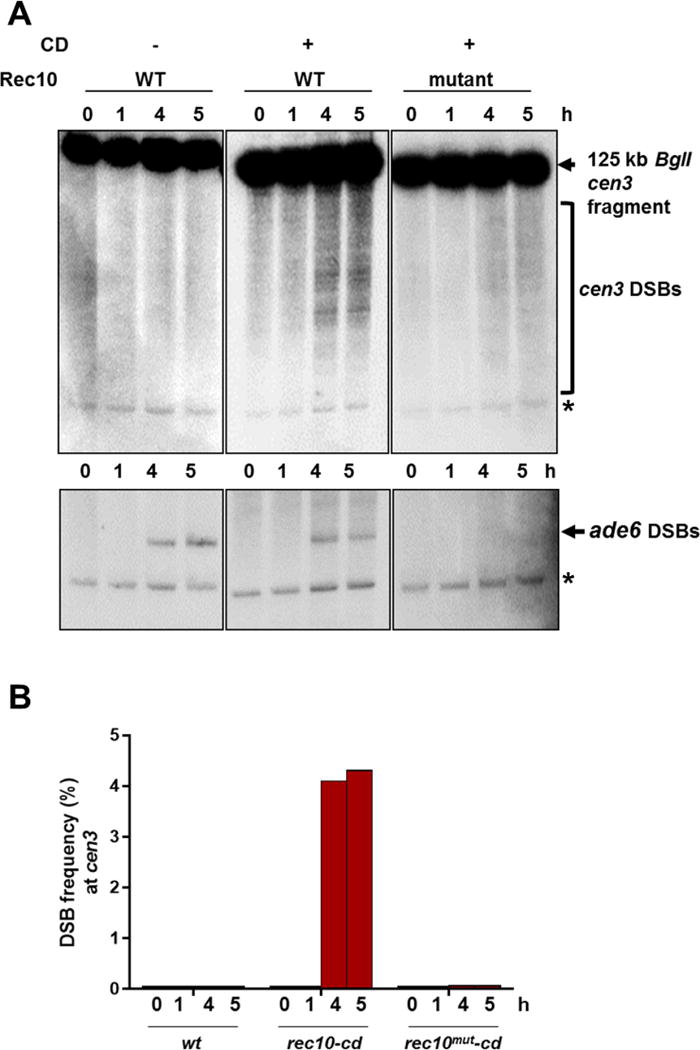
(A) Pulse-field gel electrophoresis showing cen3 DSBs at the indicated times after induction of meiosis in a rad50S mutant. The ~125 kb cen3 fragment from BglI digestion was detected on a Southern blot with a probe at the right end of the cen3 fragment (upper images). On the same blots DSBs were measured at the chromosomal arm hotspot ade6-3049 using a probe at the left end of the ~58 kb ade6 fragment. *, repetitive rDNA BglI fragment (10.9 kb) from cross-hybridization. Signals were analyzed by PhosphorImager. (B) Quantification of the blots in Figure 2A, using ImageQuant software. Bar colors correspond to those of the recombination proteins in Figure 1B.
Mechanism of Derepressed Pericentric Recombination Parallels That in Chromosomal Arms
Next, we addressed whether the observed pericentric recombination in the presence of Rec10 requires factors similar to those in chromosomal arms (Figure 1B). For recombination at the pericentric regions, Rec10-CD completely bypassed the requirement for the Rec11 cohesin subunit, the protein that recruits Rec10 (Phadnis et al., 2015; Sakuno and Watanabe, 2015) (Figure 3A and Table S1, Rows 2 and 5). This result indicates that Rec11 has little or no role other than the recruitment of Rec10 and is in accord with Rec10 acting after Rec11. In rec8Δ, however, there was an ~2-fold reduction in pericentric recombination, which might reflect Rec8 cohesin subunit aiding DSB repair with the homolog (L. Ma and G.R.S., unpublished observations). Rec10-CD-mediated recombination was slightly reduced in the absence of the DSB hotspot-specific LinE proteins Rec25 and Rec27: the data indicate a 1.6- and 1.2-fold reduction, respectively (Figure 3A and Table S1, Rows 2, 7 and 8). This slight reduction is not surprising because Rec25 and Rec27 bind specifically to hotspots and are not so strongly required for recombination in other non-hotspot DSB regions across the genome (Davis et al., 2008; Fowler et al., 2013). As expected, recombination was totally abolished in the absence of Spo11 (Figure 3A and Table S1, Rows 2 and 9), further demonstrating that the mechanism by which Rec10-CD promotes pericentric recombination is through DSB induction by Spo11.
Figure 3. Pericentric recombination mediated by pericentric-tethered Rec10 resembles chromosomal arm recombination.
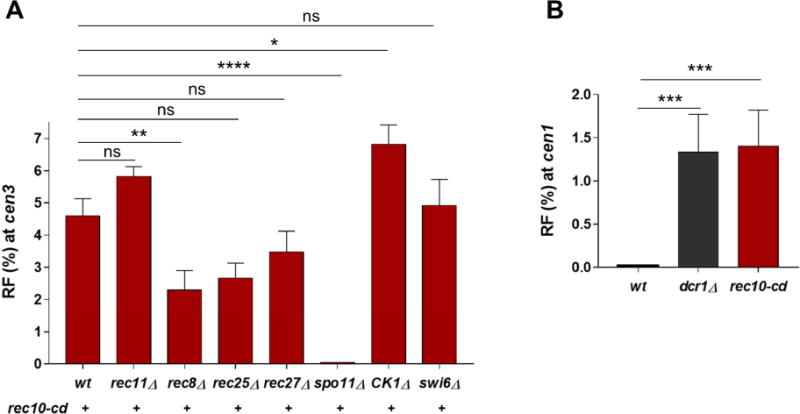
(A) Recombinant frequency (RF) across cen3 in strains expressing Rec10-CD in the absence of Rec8, Rec11, Rec25, Rec27, Spo11, CK1 or Swi6 as indicated. The RFs of rec10-CD rec11Δ and rec10-CD CK1Δ are not statistically different (p value >0.05; two-tailed Fisher’s exact test). (B) RF across cen1 (with flanking genetic markers as in Figure 1E) in wild-type, dcr1Δ and rec10-cd strains. Data are mean ± SEM (n=3 to 6 experiments, assaying 583 to 1284 spore colonies; for dcr1Δ and rec10-cd rec8Δ, n=2 experiments, assaying 749 and 707 spore colonies, respectively, and error bars indicate range). *, p = 0.0287 (two-tailed Fisher’s exact test); **, p = 0.002; ***, p <0.001; ****, p <0.0001; “ns”, p > 0.05 (not significant). Bar color (red) corresponds to that of Rec10 in Figure 1B, with wt and dcr1Δ in black. See also Tables S1 and S4.
However, in CK1Δ, cen3 recombination was slightly increased, perhaps because in the absence of CK1-phosphorylated Rec11 in the chromosomal arms, all the Rec10-CD is recruited to the pericentric region (Figure 3A and Table S1, Rows 2 and 10). As expected, arm recombination was abolished in CK1Δ (Table S1, Row 10).
Localizing Rec10 Derepresses Recombination around Another Centromere cen1
Pericentric repression in wild-type S. pombe was reported previously at cen2 and cen3 (Nakaseko et al., 1986; Ellermeier et al., 2010). We tested the frequency of recombination across the shortest pericentromere, cen1 (~35 kb), using flanking markers similar to those for cen3 (Figure 1E). The results showed that in wild type cen1 recombined at <0.3% frequency, which was >16-fold less than the genome-wide average for a similar-sized interval (~38 kb) (Young et al., 2002) (Figure 3B and Table S4, Row 1). Rec10-CD significantly increased the recombinant frequency over that of wild type, to 1.4%. A heterochromatin-deficient RNAi mutant, dcr1Δ, was also derepressed at cen1 (Figure 3B and Table S4), as at cen3 (Ellermeier et al., 2010). Thus, Rec10 appears to be a limiting factor for pericentric recombination in general.
Localizing Other LinE proteins, Rec25 and Mug20, Derepresses Pericentric Recombination in a Rec10-dependent Manner
Since other LinE proteins appear to form a complex with Rec10 in chromosomal arms (Davis et al., 2008; Fowler et al., 2013) (Figure 1B), we targeted them to the pericentromeres via a CD fusion and assayed for recombination. Rec25-CD and Mug20-CD increased the recombinant frequency across cen3 to 3.4% and 3.2%, respectively, and, like Rec10-CD, significantly reduced arm recombination concomitantly, likely because increasing their abundance in the pericentric regions decreased their abundance in arms (Figure 4A, 4B and Table S2, Rows 1, 7 and 9). Both Rec25-CD and Mug20-CD required Rec10 for this derepression at the pericentric region (Figure 4A and Table S2, Rows 7-10), reiterating the absolute requirement of Rec10 for genome-wide recombination. Both Rec25-CD and Mug20-CD formed discrete foci similar to those of Rec10-CD, as expected for binding to heterochromatic regions (Figure 4C). Surprisingly, in contrast to Rec25-CD and Mug20-CD, Rec27-CD neither localized specifically to the pericentromeres nor increased cen3 recombination, although it was as functional as wild-type Rec27 at chromosomal arms (Figure 4 and Table S2, Rows 6 and 11). This observation further supports the conclusion that the recombination activity of these LinE proteins is coincident with their binding to specific regions on the chromosome. It is plausible that Rec27’s position in the LinE complex prevents the CD moiety from efficiently binding to heterochromatin, leaving Rec27-CD indistinguishable from wild type.
Figure 4. Other linear element proteins tethered to pericentric regions also activate pericentric recombination in a Rec10-dependent manner.
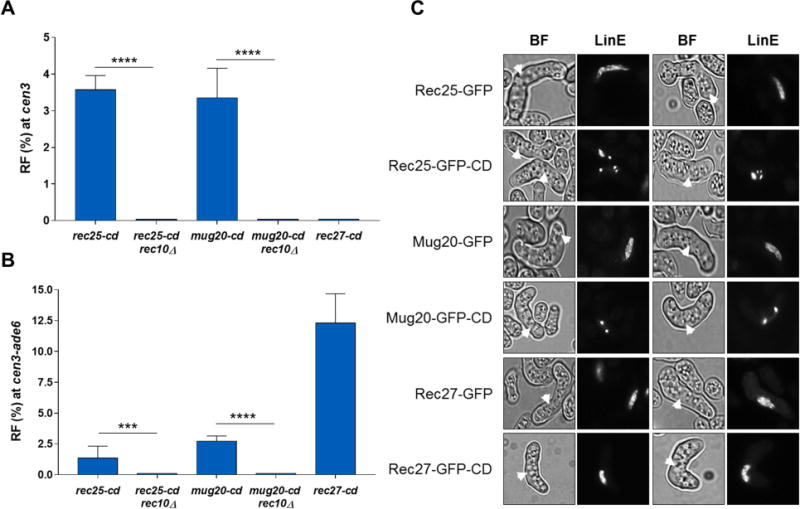
(A, B) RF across cen3 (A) and the cen3-ade6 interval (B). Data are mean ± SEM (n=3 to 6 experiments, assaying 546 to 878 spore colonies). ***, p <0.001 (two-tailed Fisher’s exact test); ****, p <0.0001. rec27-cd is not significantly different from wt (p=1.000) for RF at cen3 (Table S1). Bar color (blue) corresponds to that of Rec25 and Rec27 in Figure 1B. (C) Fluorescent microscopic images of meiotic horsetail cells (marked with white arrowheads) expressing Rec25-GFP, Mug20-GFP, or Rec27-GFP, each with or without CD. BF, bright field; LinE, linear element protein (Rec25, Rec27, or Mug20). Images are representative of at least 30-100 cells examined. See also Table S2.
Cohesin Subunit Rec11 Can Activate Pericentric Recombination but Only in the Absence of Heterochromatin Protein Swi6
The data above show that localization of Rec10 is a limiting factor for pericentric recombination, leading to the question of how Rec10 is excluded from these regions in wild type. Since Rec10 is recruited by phosphorylated Rec11 in the chromosomal arms (Phadnis et al., 2015; Sakuno and Watanabe, 2015) and Rec11 levels are low in the wild-type pericentric regions (Kitajima et al., 2003) (Figure S2A-C), we tethered Rec11 via CD to promote Rec10 recruitment in the pericentric regions. Rec11 is loaded by the cohesin loaders along chromosomal arms in wild-type cells (Bernard et al., 2006) (Figure 5A). As expected, in the presence of Rec11-CD, significant localization was also observed in the heterochromatic regions similar to that of the other CD fusions used above (Figure 5A). Surprisingly, however, despite availability of Rec11, recombination was absent at cen3 but was robust in chromosomal arms (Figure 5B, S2D and Table S5, Row 1). Swi6, which is found abundantly at the H3K9me-containing pericentric regions, recruits several proteins to repress gene expression (Grewal and Jia, 2007). Curiously, however, its removal alone does not derepress cen3 recombination, suggesting a separation of functions in repressing gene expression and in repressing recombination (Ellermeier et al., 2010) (Figure 5B). Low levels of Rec11 in swi6Δ cells at the pericentric regions provide an explanation for the absence of recombination in these regions, as compared to the chromosomal arms where Rec11 is present both in wild type and swi6Δ cells (Figure S2A-C, E).
Figure 5. Meiosis-specific cohesin subunit Rec11 tethered to pericentric regions promotes recombination during meiosis but only in the absence of Swi6 or when fused directly to Rec10.
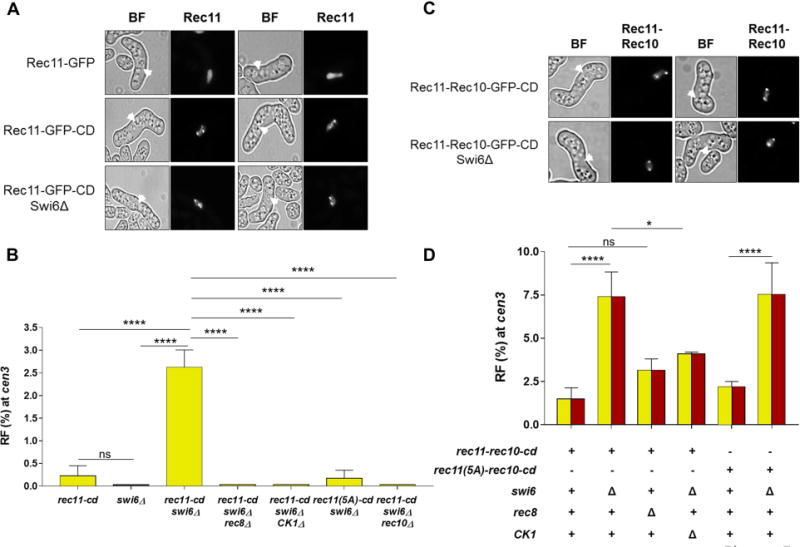
(A) Fluorescent microscopic images of meiotic horsetail cells (marked with white arrowheads) expressing Rec11-GFP or Rec11-GFP-CD in the presence and absence of Swi6. Images are representative of at least 25-100 cells examined. (B) RF across cen3 in the indicated mutants. Bar color (yellow) corresponds to that of Rec11 in Figure 1B. (C) Fluorescent microscopic images of meiotic horsetail cells (marked with white arrowheads) expressing Rec11-Rec10-GFP-CD in the presence and absence of Swi6. BF, bright field. Images are representative of at least 30-50 cells examined. (D) RF across cen3 in the indicated mutants. Yellow and red parts of the bars reflect the fusion of Rec11 (yellow) and Rec10 (red) in Rec11-Rec10-CD as in Figure 1B. Data are mean ± SEM (n = 3 to 9 experiments, assaying 577 to 1947 spore colonies). *, p = 0.0186 (two-tailed Fisher’s exact test); ****, p <0.0001; “ns”, p >0.05 (not significant). See also Figure S2 and Table S5.
Unexpectedly, the rec11-cd swi6Δ double mutant showed significantly more cen3 recombination (2.4%) than either single mutant (<0.3%) (Figure 5B and Table S5, Rows 1-3). However, this appeared not to be due to the increased abundance of Rec11-CD in swi6Δ cells at pericentric regions (Figure 5A). Pericentric recombination in rec11-cd swi6Δ was completely dependent on Rec8 (Figure 5B and Table S5, Rows 3 and 7), as is recombination in the arms (Figure S2D) (Ellermeier and Smith, 2005; Fowler et al., 2013) where Rec11 localization is abolished in rec8Δ (Ding et al., 2006). This suggests that Rec11 alone is not sufficient to promote recombination and may need to form a functional complex with Rec8 for recruiting Rec10. Phosphorylation of Rec11 by CK1 was still critical at pericentromeres, since either removal of CK1 or the mutational loss of five CK1-phosphorylation sites on Rec11 [Rec11 (5A)] (Phadnis et al., 2015; Sakuno and Watanabe, 2015) eliminated pericentric recombination in rec11-CD swi6Δ strains, similar to that in chromosomal arms (Figure 5B, S2D and Table S5, Rows 3-5). As expected, all recombination was eliminated in the absence of Rec10 (Figure 5B, S2D and Table S5, Rows 3 and 6).
Rec11 Recruitment of Rec10 Is the Rate-limiting Step for Meiotic Pericentric Recombination
Since localization of Rec11 via the CD to pericentric regions on its own was insufficient to activate pericentric recombination, we wondered if the rate-limiting step is recruitment of Rec10 by Rec11, which depends on Rec11’s phosphorylation by CK1. To test this hypothesis, we used a rec11-rec10 fusion allele (Sakuno and Watanabe, 2015) to bypass the phosphorylation step in order to recruit Rec10. Similar to Rec11-CD, Rec11-Rec10-CD showed significant localization to the heterochromatic regions, with or without Swi6 (Figure 5C). However, in contrast to Rec11-CD alone (i.e., in swi6+; 0.2% recombinant frequency), Rec11-Rec10-CD showed significantly higher recombinant frequency (1.6%) (Figure 5B, 5D and Table S5, Rows 1 and 8). Furthermore, unlike Rec11-CD, Rec11-Rec10-CD did not require Rec8 (Figure 5B, 5D and Table S5, Rows 3,7,8 and 12), further supporting our proposal above that a functional Rec8-Rec11 complex, not Rec11 alone, recruits Rec10 to initiate DSB formation. As expected, introducing the five CK1 phosphorylation-site mutations (5A) into Rec11-Rec10-CD left the cells recombination-proficient, reconfirming that fusion to Rec10 circumvents the need for Rec11 phosphorylation (Figure 5D and Table S5, Rows 1, 8 and 9).
Since there was a synergistic effect of removal of Swi6 in the presence of Rec11-CD on pericentric recombination (Figure 5B), we tested if Swi6 had any further role in blocking pericentric recombination when Rec11 was fused to Rec10. Remarkably, in combination with either wild-type or the 5A phosphorylation-deficient mutant Rec11-Rec10-CD, swi6Δ did further increase cen3 recombinant frequency to 7%, the highest frequency observed in this study (Figure 5D and Table S5, Rows 8-11). This indicates that Swi6 does not repress recombination simply by interfering with the Rec11-Rec10 interaction but in addition may reflect direct inhibition of Rec11 (and Rec11-Rec10 fusion) recruitment or activity. Rec11-Rec10-CD in the presence of CK1Δ still gave abundant cen3 recombinants, as expected (Figure 5D and Table S5, Rows 10 and 13). The partial dependence of recombination on CK1 here may reflect another function of CK1, such as phosphorylation of Rec8 (Ishiguro et al., 2010), in the pericentric region.
The Meiotic Cohesin Complex Rec8-Rec11, but not the Mitotic Complex Rec8-Psc3, Allows Pericentric Recombination
Since in wild-type meiotic cells the Rec8-Psc3 complex is present at pericentric regions and Psc3’s meiotic paralog Rec11 is at low level there (Kitajima et al., 2003) (Figure S2A-C), we tested if psc3Δ allows cen3 recombination. In the absence of Psc3, Rec11 might bind to the pericentric regions and thereby permit Rec8-Rec11 complex formation as in the chromosomal arms. During mitotic growth, constitutive Rec11 expression overcomes the lethality of psc3Δ, suggesting interchangeability of mitotic Psc3 and meiotic Rec11 for mitotic chromosome segregation (Kitajima et al., 2003). However, psc3Δ was not derepressed for pericentric recombination (Figure 6A, S2F and Table S6), probably due to low Rec8 deposition, which requires Psc3 deposition by Swi6 in the pericentric region (Nonaka et al., 2002); Rec8 is required for DSB formation and recombination in chromosomal arms (Ponticelli and Smith, 1989; Ellermeier and Smith, 2005; Fowler et al., 2013) (Figure 1B). To restore pericentric Rec8 levels, we employed Rec8-CD in either psc3Δ or swi6Δ and observed a low (~1%), but significantly higher, recombinant frequency compared to each single mutant (Figure 5B, 6A, Table S5, Row 2 and S7, Rows 1-4). This result suggests that exclusion of Rec11 by Psc3 from wild-type pericentromeres causes pericentric repression, as Psc3 has no function in promoting meiotic recombination in chromosomal arms (Kitajima et al., 2003) (Figure S2F and Table S6). Hence, competition by Psc3 for binding of Rec11 to pericentric regions may be the earliest step in repressing recombination during meiosis.
Figure 6. Both mitotic cohesin subunit Psc3 and Swi6 (HP1) repress meiotic recombination by blocking Rec11 at the pericentromeres.
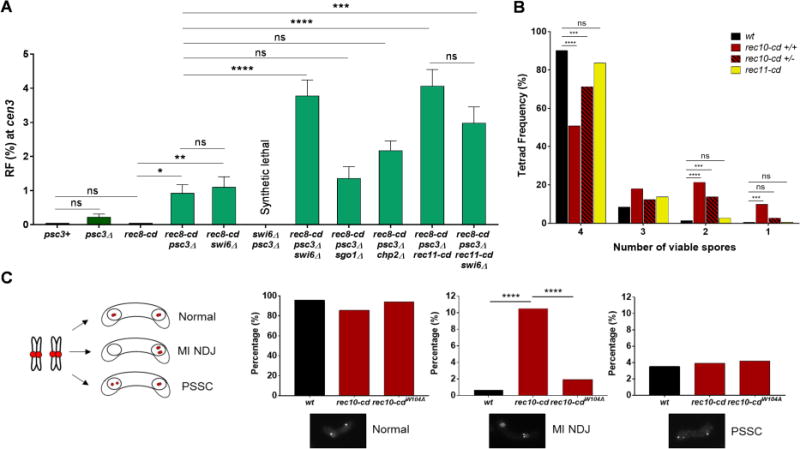
(A) RF across cen3 in the indicated mutants. psc3Δ swi6Δ is synthetically lethal and thus could not be tested. Data are mean ± SEM (n = 3 to 8 experiments, assaying 364 to 1529 spore colonies). (B) Frequency of dissected asci (tetrads) with 4, 3, 2 or 1 viable spore(s) in wild type, rec10-cd homozygous (+/+), rec10-cd heterozygous (+/−) and rec11-cd (homozygous) crosses. (C) Segregation errors in h90 mes1Δ cells at the end of MI in wt, rec10-cd and rec10-cdW104A strains (n >200 cells for each genotype). cen3 is labeled with tdTomato at the left innermost repeat (imr). Cells undergoing normal segregation show cen3 foci in both nuclei, while those with MI non-disjunction show two cen3 foci in the same nucleus. Premature separation of sister chromatids (PSSC) leads to more than one focus in the same nucleus. *, p = 0.0126 (two-tailed Fisher’s exact test); **, p = 0.0055; ***, p <0.001; ****, p <0.0001; “ns”, p >0.05 (not significant). Bar colors correspond to those of the recombination proteins in Figures 1A and 1B, with wt in black; striped bars indicate heterozygosity. See also Figure S2, Tables S3, S5, S6, S7 and S8.
We next tested the effect of swi6Δ in the absence of Psc3. Removal of Swi6 in psc3Δ (even with mitotic Rec11 expression) is synthetically lethal (Nonaka et al., 2002), likely due to complete loss of pericentric cohesion, which in the absence of Psc3 is apparently maintained to some extent by the heterochromatin protein Swi6. We discovered, however, that constitutive expression of Rec8-CD rescued this mitotic growth defect. Rec8-CD alone expressed by either a constitutive (mitotic) or native (meiotic) promoter, however, did not allow pericentric recombination (Table S7, Rows 1 and 3). By contrast, in rec8-cd psc3Δ swi6Δ, pericentric recombination was much higher (4.1%) than in wild type or any single or double mutant (Figure 6A and Table S7, Rows 1-5). This result shows that Swi6 plays a second inhibitory role, in addition to its recruitment of Psc3, to cause pericentric repression. Eliminating the Swi6-localized protein Sgo1 (SHUGOSHIN) in rec8-cd psc3Δ had no significant effect on pericentric recombination (Figure 6A and Table S7, Rows 4 and 6), indicating that Swi6’s second inhibitory role is not recruitment of Sgo1, which blocks Rec8 cohesin phosphorylation by CK1 to maintain sister chromatid cohesion in meiosis I (Kitajima et al., 2004). Eliminating Chp2, the second HP1 homolog in S. pombe that binds H3K9me (Grewal and Jia, 2007), also had no significant effect, either in wild-type cells (Ellermeier et al., 2010) or in rec8-cd psc3Δ background (Figure 6A and Table S7, Rows 4 and 7). Chp2 is the less abundant HP1 homolog at S. pombe pericentric heterochromatin (Sadaie et al., 2008) and may have a partially overlapping effect along with the more ample Swi6. These results indicate that Swi6 has a second role, independent of Psc3, Sgo1, and Chp2, in blocking pericentric recombination.
What would happen to recombination levels if the pericentromere-specific cohesin complex (Rec8-Psc3) were experimentally replaced with that present in the arms (Rec8-Rec11)? We tested this by using Rec11-CD in rec8-cd psc3Δ in the presence and absence of Swi6. The presence of Rec8-CD is critical, because in psc3Δ pericentric Rec8 levels are low or nearly absent especially when Swi6 is absent, as noted above. Remarkably, when Psc3 was absent (rec8-cd psc3Δ), the addition of Rec11-CD increased the recombinant frequency to 4%, similar to that in rec8-cd psc3Δ swi6Δ (Figure 6A and Table S7, Rows 4,5 and 8). The recombinant frequency was not altered when Swi6 was further removed, strongly suggesting that the second inhibitory role of Swi6 (apart from Psc3 recruitment) is towards Rec11 recruitment or activity. Thus, when the pericentric-specific cohesin complex (containing Rec8-Psc3) is replaced with its counterpart from chromosomal arms (containing Rec8-Rec11), Swi6 does not detectably inhibit. rec8-cd rec11-cd together (in the presence of Swi6 and Psc3) showed much less pericentric recombination (Table S7, Row 10). This result reinforces our observations above that mere addition of Rec11 to the pericentromeres is not enough to derepress pericentric recombination. These results indicate that the second inhibitory role of Swi6 in pericentric repression is to block Rec11 recruitment or activity, perhaps directly, but independent of its inhibition via Psc3 (see Figure 7).
Pericentric Recombination Results in Gamete Inviability and Meiosis I Chromosomal Non-disjunction
Pericentric crossovers cause chromosomal missegregation and aneuploidy, which is not tolerated for most chromosomes and leads to cell or organismal inviability (Beadle, 1932; Nambiar and Smith, 2016). Dissection of meiotic tetrads from rec10-cd crosses showed that ~10% of the 4-spore-viable asci contained two reciprocal pericentric recombinants, confirming the generation of crossovers rather than gene convertants (non-reciprocal recombinants) in the derepressed condition (Table S8). This frequency is consistent with the frequency of recombinants among random viable spores – 4.2% (Figure 1F). More importantly, significantly more 2-spore-viable tetrads were observed in rec10-cd crosses (13/61 tetrads, or 21%) than in rec10+ crosses (1/71 tetrads, or 1%) (p=0.0003) (Figure 6B and Table S8), suggesting that pericentric recombination leads to increased gamete inviability.
Since Rec10-CD leads to reduced arm recombination (Figure 1G and S1C), which can also result in increased missegregation and inviability, we analyzed rec10-cd heterozygous crosses to restore arm recombination while maintaining pericentric recombination (Figure S1D). However, significant loss of 4-spore-viable tetrads was still observed compared to controls (Figure 6B). The frequency of 2-spore-viable tetrads was strikingly higher in rec10-cd/rec10-cd (21%) and rec10-cd/+ (14%) than in wild type (+/+) (~1%) (Figure 6B and Table S8). Thus, 2-spore-viable tetrads were observed at high frequency whenever there was high pericentric recombination (rec10-cd), in either the presence (rec10-cd/rec10+) or absence (rec10-cd/rec10-cd) of high arm recombination, fortifying our conclusion that dead spores arise due to pericentric recombination. The similar spore viability of tetrads from wild-type and rec11-cd, which localizes to pericentric regions but does not result in pericentric recombination (Figure 5A), ruled out any effect of pericentric CD itself in missegregation or spore death (Figure 6B and Table S8).
Curiously, we did not observe any cen3 recombinants among the 23 2-spore-viable tetrads obtained from rec10-cd crosses (Figure 6B). Chromatids with pericentric crossovers may remain entangled and unresolved in the cohesin-rich pericentric region and thereby produce one poorly-growing disomic spore, one dead nullisomic spore, and two viable haploid spores (event 1) or two poorly-growing disomic spores and two dead nullisomic spores (event 2) (Koehler et al., 1996). We did observe a few slow-growing chromosome 3 disomes in rec10-cd meioses; disomes for other chromosomes are inviable (Niwa and Yanagida, 1985). Alternatively, the entangled recombinant chromatids may be lost or become extra-nuclear and produce only two viable haploid spores (event 3). It is possible, however, that dead spores result from missegregation of chromatids that did not recombine. This possibility, however, does not account for 2-spore-viable tetrads being prevalent only in the genetic backgrounds with abundant pericentric recombination.
Events 2 and 3 could result from non-disjunction (NDJ) of homologs at meiosis I (MI), producing missegregation and aneuploidy. We tracked the segregation pattern of cen3-labeled homologs at the end of MI in mes1Δ arrested cells. Mes1 is essential to initiate meiosis II (MII) by blocking cyclin B proteolysis (Izawa et al., 2005). We observed normal segregation of the two homologs in wild type. However, in rec10-cd there was a dramatic (up to 15-fold) increase in the frequency of MI NDJ (10.5% in rec10-cd compared to 0.7% in wild type) (Figure 6C). As expected, NDJ was reduced to the wild-type level in the rec10-cdW104A mutant, which lacks localization of the CD to H3K9me (Jacobs and Khorasanizadeh, 2002). Thus, rec10-cd localization to pericentric regions and the resulting activation of recombination are responsible for increased MI NDJ during meiosis. Premature separation of sister chromatids (PSSC), which could result in event 1 described above, occurred with similar frequency in all the genotypes (Figure 6C). The increased MI NDJ is likely the cause of the increased frequency of inviable spores and 2-spore-viable tetrads in rec10-cd meioses. Hence, our observations are consistent with pericentric crossovers being deleterious during meiosis, as observed in other species (Koehler et al., 1996).
Discussion
Pericentric Meiotic Recombination Is Blocked by Dual Activities of Swi6 Dictating Cohesin Composition
Three major, structurally distinct cohesin complexes in cells can have differential effects on DNA recombination and chromosome segregation (Peters et al., 2008). During mitosis, the Rad21-Psc3 complex is present at both the pericentromeres and chromosomal arms and is responsible for proper chromosomal segregation. However, during meiosis, Rad21 is largely replaced by its meiosis-specific homolog Rec8 in both pericentromeres and arms. Rec8 forms two distinct complexes depending on its chromosomal location, and these complexes affect recombination and segregation in opposite ways. The Rec8-Psc3 complex at pericentromeres is essential for sister-chromatid cohesion and thereby proper segregation at the first meiotic division (Nonaka et al., 2002; Kitajima et al., 2003). In contrast, the Rec8-Rec11 complex in the chromosomal arms is needed for programmed meiotic recombination (Ellermeier and Smith, 2005), in addition to its role in maintaining sister chromatid cohesion until the end of anaphase I (Watanabe, 2004).
Our data establish that composition of the meiotic cohesin complex at the pericentric regions is the critical factor that prevents formation of meiotic DSBs and hence potentially harmful crossovers. The deposition of the meiosis-specific cohesin complex Rec8-Psc3 near centromeres is dependent on the H3K9me-binding Swi6 protein (Nonaka et al., 2002), which we find has dual roles, both positive and negative, in pericentric repression. Via its interaction with Psc3, Swi6 is essential to enrich Rec8 in the pericentric regions to allow proper segregation: in swi6Δ, the levels of Rec8 are reduced in the outer pericentric repeats (Nonaka et al., 2002; Kitajima et al., 2003). This is especially critical in the first meiotic division, when pericentric Rec8, unlike that in chromosomal arms, is protected from cleavage in order to maintain sister chromatid cohesion through the first meiotic division. Our results show that deposition of Rec8-Psc3 complex additionally helps in preventing deleterious crossovers at pericentromeres by confining Rec11 (and consequently recombination) to the chromosomal arms where crossovers are beneficial (Figure 7). This is the first level of control, wherein having a distinct class of cohesin composition excludes pro-recombinogenic proteins specifically in the pericentric region.
Additionally, as a second level of control, Swi6 appears to block directly any stray Rec11 at the pericentric region even in the absence of Psc3. This conclusion is consistent with the previous observation that Rec11 does not accumulate in the outer repeats in psc3Δ cells despite overexpression of Rec8 and Rec11 (Nonaka et al., 2002; Kitajima et al., 2003). This mechanism is also consistent with heterochromatin mutants such as clr4Δ being highly derepressed for pericentric recombination (Ellermeier et al., 2010), as lack of H3K9me makes the pericentric region equivalent to the chromosomal arms and allows Rec11 to promote recombination genome-wide, including that in pericentric regions. Supporting this argument is the requirement for Rec8, Rec11, and Rec10 to activate recombination to high levels in the pericentric regions in clr4Δ, just as they are required in chromosomal arms (Ellermeier et al., 2010) (Table S3, Rows 4 and 6). As expected, Rec10 is more abundant in the pericentric regions of clr4Δ than in clr4+ (Figure S3). In clr4Δ there is no H3K9me, and recombination occurs genome-wide at high level (Ellermeier et al., 2010).
We suppose that Swi6 has dual roles as a fail-safe mechanism to ensure an exceptionally low level of pericentric recombination. Moreover, swi6Δ, like wild type, still has ample pericentric H3K9me (Sadaie et al., 2004; Stunnenberg et al., 2015) and lower levels of Rec11 at the pericentric regions than in the chromosomal arms (Figure S2A-C, E). This confirms that cohesin loading is prevented at the pericentric regions in the absence of Swi6, unlike the euchromatic chromosomal arms, which contain little H3K9me. Swi6 also assists in cohesin loading via its interaction with the cohesin loader Mis4 (Nonaka et al., 2002; Fischer et al., 2009). Therefore, in swi6Δ, due to limiting amounts of Rec8 (and consequently Rec11) at pericentric regions, recombination is not activated (Figure 5B and S2A, C, E) (Ellermeier et al., 2010). When we tethered either Rec11 or Rec8 to the H3K9me pericentric regions, we found a significant increase in recombination but only in swi6Δ (Figures 5B and 6A). Hence, our results show how structurally distinct protein complexes control two essential functions in meiosis – preventing crossovers near centromeres but promoting them in chromosomal arms for successful meiosis.
Meiosis-specific Cohesin Subunit Rec11 Is the Key Factor Governing Recombination
Rec11 is needed for the majority of recombination across the genome and is present at nearly uniform density except in the pericentric regions, where recombination is severely limited (Ponticelli and Smith, 1989; Ellermeier and Smith, 2005; Fowler et al., 2013). Our data presented here show that Rec11 is the pro-recombinogenic factor whose absence limits pericentric recombination. It is noteworthy that Rec11, with its partner Rec8, is the first factor in the biochemical pathway of meiotic recombination (Figure 1B) (Ellermeier and Smith, 2005; Fowler et al., 2013). This outcome is similar to that of other biochemical pathways in which regulation occurs at the first step, presumably a reflection of efficiency by not starting a metabolic pathway that should not be completed.
The second factor in the meiotic recombination pathway is Rec10 and its partner proteins Rec25, Rec27, and Mug20 (Figure 1B). As expected, pericentric recombination resulting from Rec11 or Rec10 experimentally bound to the pericentric regions requires the subsequent factors in the pathway. This outcome indicates that the mechanism of pericentric recombination is the same as that in chromosomal arms, once the limiting factor (Rec11 or Rec10) is provided. We note that Rec25 and Mug20 are only partially required for Rec10-CD-dependent pericentric recombination, but some regions of chromosomal arms are also only partially dependent on these factors (Davis et al., 2008; Estreicher et al., 2012). This result may reflect the specificity of these factors for DSB hotspots, which may be weak in the pericentric regions.
Mechanism of Pericentric Repression Is Conserved across Species
Discovered in the 1930’s (Beadle, 1932; Sax, 1932; Mather, 1939), repression of pericentric recombination was subsequently shown to be well-conserved among all species tested (Beadle, 1932; Nambiar and Smith, 2016). However, the levels of repression vary from 5- to >100-fold depending upon the complexity of the pericentromere structure. The molecular basis described here in fission yeast is likely similar to that in other species, including humans, with similarly complex, heterochromatic pericentromeres. Rec11 in S. pombe and its homolog in mammals, the meiosis-specific protein STAG3, share several characteristics. STAG3 in spermatocytes localizes similarly to Rec11, undergoes phosphorylation, and is essential for fertility in mice (Prieto et al., 2001; Fukuda et al., 2012). Drosophila has multiple cohesin complexes active during mitosis or meiosis or both, but their orthologous relations to cohesin subunits in other species are not clear, making it difficult to assign specific functions to these various complexes (Gyuricza et al., 2016). Nevertheless, there are meiosis-specific cohesin-like proteins, such as C(2)M, which appear to localize specifically to the chromosomal arms and are essential for crossing over in the arms (Manheim and McKim, 2003; Heidmann et al., 2004). It is also interesting that Drosophila Su(var) mutants lacking both Clr4 and HP1 homologs are derepressed for pericentric recombination and have increased pericentric γH2Av foci, presumably reflecting increased DSBs (Beadle, 1932; Westphal and Reuter, 2002; Peng and Karpen, 2009; Nambiar and Smith, 2016). These outcomes suggest a conserved mechanism of separately promoting meiotic segregation and recombination by having distinct cohesin complexes in the appropriate chromosomal regions distinguished by heterochromatin. Our study for the first time provides crucial insights on the necessity of multiple cohesin complexes in multicellular organisms.
In the budding yeast Saccharomyces cerevisiae, which contains simple (point) centromeres lacking heterochromatic pericentric regions, there is weaker (~5-fold) repression in a small (~10 kb) region around the centromere (Lambie and Roeder, 1986; Vincenten et al., 2015). Mutants lacking parts of the Ctf19 kinetochore subcomplex are derepressed for both centromeric recombination and DSB formation. It is proposed that the Ctf19 complex reduces DSB formation and, separately, reduces crossovers by deposition of cohesins and thereby favoring repair of DSBs with the sister chromatid rather than with the homolog (Vincenten et al., 2015). Although our results show that the meiotic pericentric cohesin complex (containing Rec8 and Psc3) acts at the stage of DSB formation (i.e., before DSB repair), the model proposed in budding yeast parallels our conclusion that cohesin composition regulates recombination near centromeres. However, due to the absence of heterochromatin in budding yeast, the molecular mechanism is markedly different. In addition, S. cerevisiae is unusual in not having a meiosis-specific homolog of Rec11 (STAG3) as found in fission yeast and mammals. Nevertheless, it is interesting that sister chromatid cohesins are the focal point of pericentric repression in these highly divergent yeasts. In all species, other chromosomal regions with repetitive DNA are also at risk from recombination, which can result in loss of repeats or produce other rearrangements. These changes can adversely affect the functions of repetitive DNA. Meiotic recombination in these regions may be limited by the same mechanism as we find for the repetitive pericentric regions, since the proteins known to bind to the H3K9me regions are similar.
Inaccessibility of Heterochromatin Does Not Adequately Explain Pericentric Repression
The role of heterochromatin in repressing gene expression and recombination is often explained by the “inaccessible,” compacted nature of DNA present in such special chromosomal regions (Beadle, 1932; Westphal and Reuter, 2002; Peng and Karpen, 2009; Nambiar and Smith, 2016). However, heterochromatin is accessible to the many proteins required for its maintenance and expansion, suggesting that such regions are not completely monolithic. There are special H3K9me-binding chromodomain proteins, such as Swi6, that recruit to heterochromatin effector molecules that either block or activate certain physiological processes.
Other proteins that do not require H3K9me for their activity, such as DNA and RNA polymerases, also act on heterochromatin (Volpe et al., 2002; Grewal and Jia, 2007). Most relevant to the current study is the presence of Rec12 (Spo11 homolog) in pericentric heterochromatic regions of S. pombe meiotic cells (Ludin et al., 2008; Fowler et al., 2013). The lack of pericentric DSBs and recombination is now explained not by the absence of Rec12 but by the absence of its activators, beginning with Rec11 (Figure 1A and 1B). Our data provide evidence that targeting just one protein (Rec10) to the pericentric region is sufficient to initiate a multi-protein pathway of DNA recombination.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Gerald R. Smith (gsmith@fredhutch.org).
Experimental Model and Subject Details
Strains and genetic methods
Genotypes of the S. pombe strains are described in Table S9. Growth media and methods used for meiotic crosses and random spore analysis were as previously described (Smith, 2009). Transformations used the lithium acetate method. Mutant alleles were generated by homology-directed integrations into the chromosome using DNA fragments generated by PCR containing 80 bp of homology with the targeted gene (Bähler et al., 1998). Oligonucleotides and plasmids are described in Tables S10 and S11, respectively.
Construction of plasmids and strains
Plasmid pMN1 contained the rec11 gene fused to GFP followed by a 442 bp fragment containing the swi6 chromodomain (CD; amino acids 89 to 134 of Swi6) from S. pombe. We subcloned the GFP-CD fragment from pMN1 into the BglII site of pMN2, which contains a hygromycin-resistance (HygR) determinant as a selectable marker to generate plasmid pMN6. The GFP-CD fusion and the HygR cassette were amplified using primers OL3370 and OL3391 and used to introduce the CD coding sequence into strains GP8538 and GP8541 (rec10-gfp-kanMX6); GP8438 and GP8536 (rec11-gfp-kanMX6); GP8543 and GP8544 (rec8-gfp-kanMX6); GP8637 (rec25-gfp-kanMX6) and GP8638 (rec27-gfp-kanMX6). Hygromycin-resistant (HygR) transformants were selected and scored for G418-sensitivity (G418S) to obtain the respective gfp-cd fusion strains. The mug20-gfp-cd strain was obtained by amplifying GFP-CD fusion DNA from pMN6 using primers OL3473 and OL3475 and transforming wild-type strains GP3303 and GP6898 to HygR. The rec10mut-gfp-cd strain was made by amplifying GFP-CD fusion DNA from pMN6 using primers OL3411 and OL3412 and transforming GP7714 containing rec10mut (R184F, D185T) to HygR. The CDW104A mutation was introduced into pMN6 with the Q5 site-directed mutagenesis kit (New England BioLabs) using primers OL3744 and OL3745 to get pMN7. The GFP-CDW104A fusion with HygR was amplified from pMN7 using primers OL3370 and OL3391; strains GP8539, GP8540 and GP8762 (rec10-gfp-kanMX6) were transformed to HygR with this DNA and scored for G418S to generate the rec10-gfp-cdW104A allele. Strains with rec11-gfp-rec10 and rec11(5A)-gfp-rec10 alleles were a gift from Y. Watanabe. We fused the CD to rec11-gfp-rec10 and rec11(5A)-gfp-rec10 by transforming GP8920 and GP9091, respectively with the PCR fragment obtained from pMN6 using primers OL3451 and OL3474 and selecting HygR transformants. The rec11(5A) mutations were generated in plasmid pYFL104 by Q5 site-directed mutagenesis using primers OL3421 and OL3422 to get pMN4. The mutation was introduced into the chromosome by replacing rec11∷ura4+ in GP8734 with rec11(5A) amplified from pMN4 using primers OL3342 and OL3435 and selecting 5-fluorouracil-resistant (FOAR) transformants. GFP-CD was fused to the rec11(5A) gene by transforming GP8738 with a PCR product from pMN6 using primers OL3465 and OL3466 and selecting HygR. In all the cases, the GFP-CD fusions and the point mutations generated were verified by PCR from the S. pombe transformant and sequencing. The strain containing psc3Δ with rec11 constitutively expressed from the Padh15 promoter was a gift from Y. Watanabe. This strain also contained rec8 constitutively expressed from the Padh1 promoter. We fused GFP-CD to this Padh1-rec8 allele by transforming GP9169 with a PCR fragment made from pMN6 using primers OL3516 and OL3517 and selecting FOAR. The swi6∷natR, sgo1∷natR and chp2∷natR alleles were generated by transforming GP9172 and GP9174 with PCR fragments from pSS16 to amplify the nourseothricin-resistance (NatR) determinant. The primers used were OL3647 and OL3648 for swi6; OL3667 and OL3668 for sgo1; and OL3725 and OL3726 for chp2. Integrations were verified by PCR.
Method Details
Genetic recombination assay
Recombination across cen3 was measured using selectable prototrophic markers (ura4+ and his3+) introduced into chk1 or near mid1, respectively, on chromosome 3 as described (Ellermeier et al., 2010). The two parental strains contained either ade6-52 (light pink colonies on YEA) or ade6-M26 (dark red colonies on YEA) to allow measurement of arm recombination between ade6 and his3. Since ura4+ and his3+ are tightly linked in wild type due to absence of recombination between them, the arm interval is referred to as cen3-ade6. Other intervals for arm recombination were ade6-arg1 on chromosome 3 and lys3-met5 on chromosome 1. For measuring recombination across cen1, his3+ was integrated between emc5 and IRC1-R on the right side of cen1, and ura4+ was integrated between spatrnaphe02 and IRC1-L on left side of cen1; the sites of insertion are ~38 kb apart. The primers used for ura4+ integration were OL3337 and OL3338; for his3+ they were OL3332 and OL3333. After mating of the two parental strains at 25 °C on supplemented SPA medium for 2-3 days, spores were harvested, plated, and incubated on YEA for 4-5 days. Random individual colonies were picked to YEA supplemented with adenine (100 μg/ml), allowed to grow for 1-2 days and replica-plated to different media to measure growth in the absence of required supplements. Recombinant frequencies between the markers defining different intervals were calculated, and the mean plotted as bar graphs. For each genotype, at least two or more independent crosses (biological replicates) were performed either using independent colonies or with independent strains as parents. Error bars are SEM for ≥3 or range for 2 crosses. Data for individual experiments are in Tables S1-S7.
Meiotic induction and DNA preparation
Meiotic induction and DNA preparation were performed as described (Hyppa and Smith, 2009). Briefly, synchronous meiosis was induced in pat1-114 strains after nitrogen starvation (G1-arrested cells) by raising the temperature to 34 °C and adding NH Cl as 4 a nitrogen source. Cells were collected at 0, 1, 4 and 5 h after induction, harvested by centrifugation, washed, embedded in agarose plugs and digested with lytic enzymes to break open the cells. The plugs were then treated with proteinase K, which was subsequently inactivated with PMSF, and washed with TE buffer multiple times. Meiotic induction was confirmed by measuring pre-meiotic replication by flow cytometry. Typically, replication began at 2 h post-induction and was completed by 3 h, as determined by analysis by flow cytometry of samples taken hourly.
DSB analysis by PFGE
DNA in the agarose plugs from each time point was digested with BglI overnight at 37 °C. The DNA fragments were separated by pulse-field gel electrophoresis using a BioRad CHEF-DR II or CHEF Mapper apparatus and the following conditions: run time of 20.8 h, 6V/cm, 120° angle, initial switch time of 0.5 s, and final switch time of 9.4 s. After alkaline denaturation, DNA was transferred from the gels to nylon membranes (Zeta-probe GT membrane, BioRad) and hybridized with 1 kb radiolabeled probes complementary to the right end of the 125 kb cen3 fragment or the left end of the 58 kb ade6 fragment. The probe for cen3 was generated by PCR amplification of wild-type genomic DNA using primers OL1558 and OL1559 (Ellermeier et al., 2010), and that for ade6 using primers OL3413 and OL3414; each was labeled with [α-32P] dCTP. Signals were detected using a Typhoon Trio PhosphorImager system (GE Healthcare).
Fluorescent microscopy Strains used were homothallic (h90) and expressed various proteins fused to GFP; some expressed tdTomato fused to TetR and had tandem repeats of tetO inserted into the innermost repeat (imrL) of cen3. Cells were grown to log phase in YEL medium at 30 °C, and the cells from 1 ml were washed twice in 1 ml water, spotted on MEA medium and incubated at room temperature (~22 °C) for 16 h to allow mating and meiosis to begin. Cells were observed under a Nikon Eclipse E800 microscope using either FITC or TRITC filters. Single focal-plane images were obtained at 60X magnification and analyzed using Image J software.
Tetrad analysis
Wild-type, rec10-cd (homozygous and heterozygous) and rec11-cd parental strains were mated on SPA at 25 °C for 2 days, and asci (tetrads) were isolated on YEA under a dissection microscope (Nikon). The plates were incubated at 37 °C for 3-4 h to allow autolysis of the ascal wall. Spores were isolated by micromanipulation and incubated at 32 °C for 3-4 days. The spore viability was scored as visible colonies, and the colonies further analyzed for recombination across cen3 (ura4+-his3+ interval) and in arms (cen3-ade6 and ade6-arg1 intervals), as described above.
Missegregation assay
We used h90 strains with rec10+, rec10-cd or rec10-cdW104A and mes1Δ to block meiosis after MI (Izawa et al., 2005). We used tdTomato fused to TetR and tetO inserted into the innermost repeat (imrL) of cen3 to follow the pericentric region. Cells were grown to log phase in YEL medium at 30 °C; the cells from 1 ml were washed twice in 1 ml of water, spotted on MEA medium and incubated at room temperature (~22 °C) for at least 36 h. This allowed mating, induction of meiosis, and completion of MI arrest. Asci (n>200) with two nuclei were observed under a Nikon Eclipse E800 microscope using TRITC filters. Single focal-plane images were obtained at 60X magnification and analyzed using Image J software. The cen3 labels in the arrested cells were counted and categorized as normal segregation (one focus in each of two nuclei), MI NDJ (both foci in the same nucleus) or premature separation of sister chromatids (PSSC; two foci in one or both nuclei).
Microarray analysis
Rec10-GFP ChIP microarray analysis was performed as described (Fowler et al., 2013). clr4+ and clr4Δ diploid meiotic cells were harvested, and the chromatin crosslinked with formaldehyde and immunoprecipitated for Rec10-GFP. Custom Agilent 4×44K S. pombe oligonucleotide microarrays were used that include 1125 additional probes for repetitive regions of the S. pombe genome as described (Cam et al., 2005). The microarray probe coordinates corresponded to RefSeq accessions NC_003424.2, NC_003423.2, and NC_003421.2 for chromosomes 1, 2, and 3, respectively. The data (log10 of the IP/WCE ratio for each probe) were genome-median normalized, and the LogRatios from the 0 hr (uninduced) samples were subtracted from the respective 3.5 hr (induced) samples. The probes were divided into those from the centromeres (cnt, imr and otr; total 330 probes) and the chromosomal arms (42,583 probes). Box-and-whisker plots were generated for each centromere and chromosome using GraphPad Prism software; data were analyzed in Excel.
Chromatin immunoprecipitation (ChIP) and qPCR for Rec11-GFP
Chromatin from pat1-114 strains GP8949 and GP9023 induced for 3.5 hr (two induced cultures of each) was extracted and immunoprecipitated as previously described (Fowler et al., 2013). Anti-GFP polyclonal antibodies (Living Colors Full-Length GFP Polyclonal Antibody, ClonTech) and Protein A Dynabeads (ThermoFisher) were used. DNA isolated from the whole-cell extracts and immunoprecipitates was analyzed via quantitative PCR (qPCR) with the OneStepPlus system (ABI) using Power SYBR Green master mix (ThermoFisher). The percent immunoprecipitate (IP) signal relative to the whole cell extract (WCE) signal was calculated using the Comparative CT method (ABI) with each locus done in triplicate for each qPCR run. Four qPCR runs were done for each strain at cnt and imr, and five at dg, dh, msp1, 3H7.03. These loci were assayed previously (Yokobayashi et al., 2003; Yokobayashi and Watanabe, 2005; Miyoshi et al., 2012; Sakuno and Watanabe, 2015), but we used different primer pairs (Table S10), except for msp1, to generate ~100 bp amplicons. Primer pairs for dg, dh, msp1, and 3H7.03 used in a previous study (Sakuno and Watanabe, 2015) gave similar results.
Quantification and Statistical Analysis
All recombination data were analyzed by using two-tailed Fisher’s exact test. The ChIP-chip data for Rec10 in wild type and clr4Δ cells was analyzed by two-tailed paired t-tests. The Rec11-GFP ChIP qPCR was analyzed by unpaired t-tests and two-tailed paired t-tests. All the analysis was done using GraphPad Prism software.
Data and Software Availability
Data Resources
The accession number for the raw data files for the microarray datasets reported in this paper is GSE115726.
Supplementary Material
Highlights.
Tethering Spo11-activators to pericentric heterochromatin promotes local crossovers
The meiotic cohesin subunit Rec11 is a limiting factor for pericentric recombination
Cohesin subunit Psc3 precludes activation of meiotic double-strand break formation
Pericentric crossovers increase gamete inviability and non-disjunction during meiosis
Acknowledgments
We thank Sue Amundsen, Sue Biggins, Randy Hyppa and Jeetu Thakur for helpful comments on the manuscript, Yoshinori Watanabe for strains and plasmids, Lijuan Ma for observations on rec8 mutants, Kyle Fowler for Rec10 ChIP-chip data, and Randy Hyppa and Mai-Chi Nguyen for technical assistance. This research was supported by NIH grants R01 GM032194 and R35 GM118120 to GRS, U54 DK106829 Pilot and Feasibility Study (Beverly Torok-Storb, PI) to MN, and P30 CA015704 to the Shared Resource Center of the Fred Hutchinson Cancer Research Center-University of Washington Cancer Consortium.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
M.N. conceived, designed, and conducted the experiments. M.N. and G.R.S. analyzed the data and wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Supplementary Data
Excel file, Table S9. [S. pombe strains used in the study.] Related to Star Methods.
References
- Bähler J, Wu J-Q, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Beadle GW. A Possible Influence of the Spindle Fibre on Crossing-Over in Drosophila. Proc Natl Acad Sci U S A. 1932;18:160–165. doi: 10.1073/pnas.18.2.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Drogat J, Maure JF, Dheur S, Vaur S, Genier S, Javerzat JP. A screen for cohesion mutants uncovers Ssl3, the fission yeast counterpart of the cohesin loading factor Scc4. Curr Biol. 2006;16:875–881. doi: 10.1016/j.cub.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Ding D-Q, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- Cottarel G, Beach D, Deuschle U. Two new multi-purpose multicopy Schizosaccharomyces pombe shuttle vectors, pSP1 and pSP2. Curr Genet. 1993;23:547–548. doi: 10.1007/BF00312650. [DOI] [PubMed] [Google Scholar]
- Cromie GA, Smith GR. Meiotic recombination in Schizosaccharomyces pombe: A paradigm for genetic and molecular analysis. In: Egel R, Lankenau D-H, editors. Recombination and meiosis: Models, means, and evolution. Berlin: Springer-Verlag; 2008. pp. 195–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Rozalén AE, Moreno S, Smith GR, Martin-Castellanos C. Rec25 and Rec27, novel components of meiotic linear elements, link cohesin to DNA breakage and recombination in fission yeast. Curr Biol. 2008;18:849–854. doi: 10.1016/j.cub.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Smith GR. Non-random homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics. 2003;163:857–874. doi: 10.1093/genetics/163.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeaux LC, Smith GR. Region-specific activators of meiotic recombination inSchizosaccharomyces pombe. Genes Dev. 1994;8:203–210. doi: 10.1101/gad.8.2.203. [DOI] [PubMed] [Google Scholar]
- Ding DQ, Sakurai N, Katou Y, Itoh T, Shirahige K, Haraguchi T, Hiraoka Y. Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J Cell Biol. 2006;174:499–508. doi: 10.1083/jcb.200605074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C, Higuchi EC, Phadnis N, Holm L, Geelhood JL, Thon G, Smith GR. RNAi and heterochromatin repress centromeric meiotic recombination. Proc Natl Acad Sci USA. 2010;107:8701–8705. doi: 10.1073/pnas.0914160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C, Smith GR. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 2005;102:10952–10957. doi: 10.1073/pnas.0504805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estreicher A, Lorenz A, Loidl J. Mug20, a novel protein associated with linear elements in fission yeast meiosis. Curr Genet. 2012;58:119–127. doi: 10.1007/s00294-012-0369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah JA, Hartsuiker E, Mizuno K-I, Ohta K, Smith GR. A 160-bp palindrome is a Rad50•Rad32-dependent mitotic recombination hotspot in Schizosacchromyces pombe. Genetics. 2002;161:461–468. doi: 10.1093/genetics/161.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, Veenstra TD, Grewal SI. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci USA. 2009;106:8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler KR, Gutiérrez-Velasco S, Martín-Castellanos C, Smith GR. Protein determinants of meiotic DNA break hotspots. Mol Cell. 2013;49:983–996. doi: 10.1016/j.molcel.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Kugou K, Sasanuma H, Shibata T, Ohta K. Targeted induction of meiotic double-strand breaks reveals chromosomal domain-dependent regulation of Spo11 and interactions among potential sites of meiotic recombination. Nucleic Acids Res. 2008;36:984–997. doi: 10.1093/nar/gkm1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Pratto F, Schimenti JC, Turner JM, Camerini-Otero RD, Hoog C. Phosphorylation of chromosome core components may serve as axis marks for the status of chromosomal events during mammalian meiosis. PLoS Genet. 2012;8:e1002485. doi: 10.1371/journal.pgen.1002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Gyuricza MR, Manheimer KB, Apte V, Krishnan B, Joyce EF, McKee BD, McKim KS. Dynamic and Stable Cohesins Regulate Synaptonemal Complex Assembly and Chromosome Segregation. Curr Biol. 2016;26:1688–1698. doi: 10.1016/j.cub.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Heidmann D, Horn S, Heidmann S, Schleiffer A, Nasmyth K, Lehner CF. The Drosophila meiotic kleisin C(2)M functions before the meiotic divisions. Chromosoma. 2004;113:177–187. doi: 10.1007/s00412-004-0305-5. [DOI] [PubMed] [Google Scholar]
- Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast. 2005;22:1013–1019. doi: 10.1002/yea.1291. [DOI] [PubMed] [Google Scholar]
- Hyppa RW, Smith GR. In: Using Schizosaccharomyces pombe meiosis to analyze DNA recombination intermediates. Meiosis S Keeney., editor. Totowa, NJ: Humana Press; 2009. pp. 235–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro T, Tanaka K, Sakuno T, Watanabe Y. Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat Cell Biol. 2010;12:500–506. doi: 10.1038/ncb2052. [DOI] [PubMed] [Google Scholar]
- Izawa D, Goto M, Yamashita A, Yamano H, Yamamoto M. Fission yeast Mes1p ensures the onset of meiosis II by blocking degradation of cyclin Cdc13p. Nature. 2005;434:529–533. doi: 10.1038/nature03406. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Yokobayashi S, Yamamoto M, Watanabe Y. Distinct cohesin complexes organize meiotic chromosome domains. Science. 2003;300:1152–1155. doi: 10.1126/science.1083634. [DOI] [PubMed] [Google Scholar]
- Koehler KE, Hawley RS, Sherman S, Hassold T. Recombination and nondisjunction in humans and flies. Human Mol Genet. 1996;5:1495–1504. doi: 10.1093/hmg/5.supplement_1.1495. [DOI] [PubMed] [Google Scholar]
- Lamb NE, Yu K, Shaffer J, Feingold E, Sherman SL. Association between maternal age and meiotic recombination for trisomy 21. Am J Hum Genet. 2005;76:91–99. doi: 10.1086/427266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambie EJ, Roeder GS. Repression of meiotic crossing over by a centromere (CEN3) in Saccharomyces cerevisiae. Genetics. 1986;114:769–789. doi: 10.1093/genetics/114.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Numata M, Wahls WP, Smith GR. Region-specific meiotic recombination in S. pombe: the rec11 gene. Mol Microbiol. 1997;23:869–878. doi: 10.1046/j.1365-2958.1997.2691632.x. [DOI] [PubMed] [Google Scholar]
- Limbo O, Porter-Goff ME, Rhind N, Russell P. Mre11 nuclease activity and Ctp1 regulate Chk1 activation by Rad3ATR and Tel1ATM checkpoint kinases at double-strand breaks. Mol Cell Biol. 2011;31:573–583. doi: 10.1128/MCB.00994-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Smith GR. Molecular cloning of the meiosis-induced rec10 gene of Schizosaccharomyces pombe. Curr Genet. 1995;27:440–446. doi: 10.1007/BF00311213. [DOI] [PubMed] [Google Scholar]
- Iino Y, Yamamoto M. Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol Gen Genet. 1985;198:416–421. doi: 10.1007/BF00332932. [DOI] [PubMed] [Google Scholar]
- Ludin K, Mata J, Watt S, Lehmann E, Bahler J, Kohli J. Sites of strong Rec12/Spo11 binding in the fission yeast genome are associated with meiotic recombination and with centromeres. Chromosoma. 2008;117:431–444. doi: 10.1007/s00412-008-0159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Fowler KR, Martin-Castellanos C, Smith GR. Functional organization of protein determinants of meiotic DNA break hotspots. Sci Rep. 2017;7:1393. doi: 10.1038/s41598-017-00742-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manheim EA, McKim KS. The Synaptonemal complex component C(2)M regulates meiotic crossing over in Drosophila. Curr Biol. 2003;13:276–285. doi: 10.1016/s0960-9822(03)00050-2. [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C, Blanco M, Rozalen AE, Perez-Hidalgo L, Garcia AI, Conde F, Mata J, Ellermeier C, Davis L, San-Segundo P, et al. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr Biol. 2005;22:2056–2062. doi: 10.1016/j.cub.2005.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather K. Crossing over and Heterochromatin in the X Chromosome of Drosophila Melanogaster. Genetics. 1939;24:413–435. doi: 10.1093/genetics/24.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Ito M, Kugou K, Yamada S, Furuichi M, Oda A, Yamada T, Hirota K, Masai H, Ohta K. A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol Cell. 2012;47:722–733. doi: 10.1016/j.molcel.2012.06.023. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y, Adachi Y, Funahashi S, Niwa O, Yanagida M. Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO J. 1986;5:1011–1021. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nambiar M, Smith GR. Repression of harmful meiotic recombination in centromeric regions. Sem Cell Dev Biol. 2016;54:188–197. doi: 10.1016/j.semcdb.2016.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Yanagida M. Triploid meiosis and aneuploidy in Schizosaccharomyces pombe: an unstable aneuploid disomic for chromosome III. Curr Genet. 1985;9:463–470. [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Offenberg HH, Schalk JA, Meuwissen RL, van Aalderen M, Kester HA, Dietrich AJ, Heyting C. SCP2: a major protein component of the axial elements of synaptonemal complexes of the rat. Nucleic Acids Res. 1998;26:2572–2579. doi: 10.1093/nar/26.11.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 2009;5:e1000435. doi: 10.1371/journal.pgen.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- Phadnis N, Cipak L, Polakova S, Hyppa RW, Cipakova I, Anrather D, Karvaiova L, Mechtler K, Smith GR, Gregan J. Casein kinase 1 and phosphorylation of cohesin subunit Rec11 (SA3) promote meiotic recombination through linear element formation. Plos Genet. 2015;11:e1005225. doi: 10.1371/journal.pgen.1005225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli AS, Smith GR. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics. 1989;123:45–54. doi: 10.1093/genetics/123.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto I, Suja JA, Pezzi N, Kremer L, Martinez AC, Rufas JS, Barbero JL. Mammalian STAG3 is a cohesin specific to sister chromatid arms in meiosis I. Nat Cell Biol. 2001;3:761–766. doi: 10.1038/35087082. [DOI] [PubMed] [Google Scholar]
- Robine N, Uematsu N, Amiot F, Gidrol X, Barillot E, Nicolas A, Borde V. Genome-wide redistribution of meiotic double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:1868–1880. doi: 10.1128/MCB.02063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Voelkel-Meiman K, Roeder GS. Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics. 2006;174:1745–1754. doi: 10.1534/genetics.106.058933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Iida T, Urano T, Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Kawaguchi R, Ohtani Y, Arisaka F, Tanaka K, Shirahige K, Nakayama J. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol Cell Biol. 2008;28:6973–6988. doi: 10.1128/MCB.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuno T, Watanabe Y. Phosphorylation of cohesin Rec11/SA3 by casein kinase 1 promotes homologous recombination by assembling the meiotic chromosome axis. Dev Cell. 2015;32:220–230. doi: 10.1016/j.devcel.2014.11.033. [DOI] [PubMed] [Google Scholar]
- Sax K. Chromosome structure and the mechanism of crossing over. J Arnold Arb. 1932;11:193–220. [Google Scholar]
- Smith GR. Genetic analysis of meiotic recombination. In: Meiosis S Keeney., editor. Schizosaccharomyces pombe. Totowa, NJ: Humana Press; 2009. pp. 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner WW, Smith GR. Optimizing the nucleotide sequence of a meiotic recombination hotspot in Schizosaccharomyces pombe. Genetics. 2005;169:1973–1983. doi: 10.1534/genetics.104.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunnenberg R, Kulasegaran-Shylini R, Keller C, Kirschmann MA, Gelman L, Buhler M. H3K9 methylation extends across natural boundaries of heterochromatin in the absence of an HP1 protein. EMBO J. 2015;34:2789–2803. doi: 10.15252/embj.201591320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunder S, Greeson-Lott NT, Runge KW, Sanders SL. A new method to efficiently induce a site-specific double-strand break in the fission yeast Schizosaccharomyces pombe. Yeast. 2012;29:275–291. doi: 10.1002/yea.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenten N, Kuhl LM, Lam I, Oke A, Kerr AR, Hochwagen A, Fung J, Keeney S, Vader G, Marston AL. The kinetochore prevents centromere-proximal crossover recombination during meiosis. eLife. 2015;4 doi: 10.7554/eLife.10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- Watanabe Y. Modifying sister chromatid cohesion for meiosis. J Cell Sci. 2004;117:4017–4023. doi: 10.1242/jcs.01352. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Westphal T, Reuter G. Recombinogenic effects of suppressors of position-effect variegation in Drosophila. Genetics. 2002;160:609–621. doi: 10.1093/genetics/160.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S, Watanabe Y. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell. 2005;123:803–817. doi: 10.1016/j.cell.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Yokobayashi S, Yamamoto M, Watanabe Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol Cell Biol. 2003;23:3965–3973. doi: 10.1128/MCB.23.11.3965-3973.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Schreckhise RW, Steiner WW, Smith GR. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol Cell. 2002;9:253–263. doi: 10.1016/s1097-2765(02)00452-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


