Abstract
Understanding protein-protein interactions (PPIs) in a cell is essential for learning protein functions, pathways, and mechanism of diseases. PPIs are also important targets for developing drugs. Experimental methods, both small-scale and large-scale, have identified PPIs in several model organisms. However, results cover only a part of PPIs of organisms; moreover, there are many organisms whose PPIs have not yet been investigated. To complement experimental methods, many computational methods have been developed that predict PPIs from various characteristics of proteins. Here we provide an overview of literature reports to classify computational PPI prediction methods that consider different features of proteins, including protein sequence, genomes, protein structure, function, PPI network topology, and those which integrate multiple methods.
Keywords: protein-protein interactions, PPI, computational methods, protein docking, bioinformatics, protein interaction network
INTRODUCTION
Identification of protein-protein interactions (PPIs) is important for understanding how proteins work together in a coordinated fashion in a cell to perform cellular functions. PPIs are essential for protein function, various cellular pathways, and development of diseases. PPIs are also important targets for drug design. Understanding how proteins interact can also lead to artificial design of protein interactions.
Individual PPIs are determined by experiments, such as co-immunoprecipitation (A. Guo et al., 2005), fluorescence resonance energy transfer (Kenworthy, 2001), and surface plasmon resonance (Nikolovska-Coleska, 2015). Ultimately, biophysical methods, such as nuclear magnetic resonance spectroscopy (NMR) (Vinogradova & Qin, 2011; Zuiderweg, 2002), X-ray crystallography (Kobe et al., 2008), and electron microscopy (Dudkina, Kouřil, Bultema, & Boekema, 2010), solve the tertiary structure of protein complexes, which can provide detailed atomic information about how the proteins interact. Moreover, from the mid 1990’s, PPIs were determined on a large-scale using the yeast-two-hybrid system (Fields & Sternglanz, 1994; Rajagopala et al., 2014; Rual et al., 2005; Walhout, Boulton, & Vidal, 2000), and affinity chromatography combined with mass spectrometry (Boeri Erba & Petosa, 2015; Dunham, Mullin, & Gingras, 2012; Guruharsha et al., 2011; Morris et al., 2014). However, experimental methods have several shortcomings for detecting PPIs. First, these experimental methods are time consuming and labor intensive. Second, the applicability of experimental methods depends on how effectively assay protocols are established in target organisms. Also, a method may not work on some classes of proteins (Piehler, 2005; Rao, Srinivas, Sujini, & Kumar, 2014). Third, it is known that experimental methods often have difficulty with identifying weak interactions, and leave out many transient interactions (Wetie et al., 2013). Fourth, it has been mentioned that results of large-scale methods often have substantial disagreement with each other, which may be partly due to false positives and false negatives (Gingras, Gstaiger, Raught, & Aebersold, 2007; H. Huang & Bader, 2009; Serebriiskii & Golemis, 2001).
In Table 1, databases of PPIs are listed. Most of the identified PPIs are from model organisms such as Escherichia coli, Homo sapiens, Mus musculus (mouse), Saccharomyces cerevisiae (baker’s yeast; yeast), Schizosaccharomyces pombe (fission yeast), Drosophila melanogaster (fruit fly), and Arabidopsis thaliana. Although large efforts have been made for detecting PPIs, there still exists a huge gap between the experimentally identified PPIs and actual PPIs. For example, it was estimated that humans have over 650,000 PPIs based on a statistical method that evaluates the number of undiscovered PPIs from the known human PPI network (Stumpf et al., 2008) whereas a little over 40,000 interactions have been identified based on the HPRD database (Prasad et al., 2009). Even for yeast, which is one of the most well studied organisms in terms of PPIs, 91,551 were identified based on the BioGrid database (Chatr-Aryamontri et al., 2017) whereas 240,000 PPIs were estimated. For Caenorhabditis elegans (roundworm), which is an important model organism, only 5,797 PPIs were identified among 220,000 estimated. Thus, currently identified PPIs derived from experiments only cover a small fraction in the entire all PPI networks. Hence, there is a strong need for computational methods for predicting PPIs and indeed many computational approaches have been developed to facilitate investigation of PPI networks in organisms.
Table 1.
List of available protein-protein interaction databases.
| Database | # of interactions | Description | Organisms | Website | Last update |
|---|---|---|---|---|---|
| BioGrid | 1,110,310 | Manually curated PPIs | 62 | https://thebiogrid.org/ | Mar 2017 |
| STRING | 932,553,897 | Protein associations including PPIs | 2,031 | http://string-db.org/ | Jan 2017 |
| DIP | 81,731 | Experimentally identified PPIs | 834 | http://dip.doe-mbi.ucla.edu/dip/Main.cgi | Mar 2017 |
| CORUM | 6,375 | Manually curated protein complexes in mammals | 10 | http://mips.helmholtz-muenchen.de/corum/ | Dec 2016 |
| IntAct | 718,180 | PPIs taken from literature and from user submissions | Model organisms including human, mouse, yeast, fruitfly, C. elegans, E. coli, A. thaliana | http://www.ebi.ac.uk/intact/ | Mar 2017 |
| MINT | 125,464 | Experimentally verified PPIs from literature | 611 | http://mint.bio.uniroma2.it/ | Mar 2017 |
| InnateDB | 367,478 | Manually curated PPIs for mammalian innate immune response | Human, mouse,B. taurus | http://www.innatedb.com/ | Nov 2016 |
| HPRD | 41,327 | PPI network of H. sapiens | human | http://www.hprd.org/ | April 2010 |
| EcoCyc | 6,399 | Manually curated PPIs in E. coli K-12 MG1655 | E. coli | https://ecocyc.org/ | Dec 2016 |
| TAIR | 8,826 | Experimentally identified PPIs in A. thaliana | A. thaliana | https://www.arabidopsis.org/ | Sep 2011 |
References of databases: BioGrid: (Chatr-Aryamontri et al., 2017); STRING: (Damian Szklarczyk et al., 2014); DIP: (Xenarios et al., 2002); CORUM: (P. Wong et al., 2008); IntAct: (Hermjakob et al., 2004); MINT: (Licata et al., 2012); InnateDB: (Breuer et al., 2013); HPRD: (Prasad et al., 2009); EcoCyc: (Keseler et al., 2016); TAIR: (Garcia-Hernandez et al., 2002).
Computational PPI prediction methods were reviewed in several earlier articles. Comparative genomics-based methods were reviewed in 2002; shortly after a couple of large-scale PPI networks emerged (Valencia & Pazos, 2002). Skrabanek et al. reviewed methods that use comparative genomics and gene expression data, as well as tools for visualizing PPIs (Skrabanek, Saini, Bader, & Enright, 2008). A review by Browne et al. focused on experimental methods for PPI detection and classified existing methods based on underlined machine learning algorithms (Browne, Zheng, Wang, & Azuaje, 2010). A review by Liu et al. discussed computational methods by classifying them into two groups, those which directly map information of known PPIs onto unknown protein pairs, and approaches that employs machine learning methods to classify protein pairs from a dataset of known PPIs and non-PPIs (Z.-P. Liu & Chen, 2012). Very recently, Chang et al. focused on methods that combine different types of evidence for predicting PPIs (Chang, Zhou, Ul Qamar, Chen, & Ding, 2016).
The current article classifies and reviews computational PPI prediction methods by features of proteins considered for prediction, which includes protein sequence-based, comparative genomics-based, gene expression-based, function-based, structure-based, and network-based prediction methods. This article has some overlaps in its scope with the previous review articles, but it is distinct from others by providing extensive discussion on protein sequence-based prediction methods and network-based prediction methods, and of course, by providing up-to-date information in this field. We also discuss applicability of each type of methods in genome-scale PPI predictions.
PPI PREDICTION METHODS
We classified PPI prediction methods into six large categories based on features of proteins considered as input information of the prediction. Below we discuss ideas behind methods that fall into each category. Most of the categories are further classified into sub-categories.
To develop a computational prediction method, one needs a dataset of known interacting protein pairs (a positive set) and a dataset of non-interacting protein pairs (a negative set), because the method needs to maximize its ability to distinguish between positive and negative datasets. A positive dataset is constructed from known PPIs stored in existing PPI databases (Table 1). On the other hand, constructing a negative dataset is not straightforward, because there are only few collections of protein pairs that are experimentally directly verified not to interact. To facilitate construction of a negative dataset, there is a database named Negatome, which collects protein pairs that are unlikely to interact by manual curation of literature and known protein complex structures (Blohm et al., 2014). Another commonly used strategy used to construct a negative dataset is to pair proteins from different cellular locations and a random pairing of proteins that appeared in the positive dataset excluding interacting pairs.
SEQUENCE-BASED METHODS
Many methods have been developed that use the amino acid sequence information of target proteins. The obvious advantage of using sequence information is that it is available for all proteins in an organism as long as its genome sequence is available.
Motif/Domain-based approach
The most straightforward approach in this category is to predict that two proteins interact with each other if they possess known sequence patterns of interacting proteins in their amino acid sequences. For example, Becerra et al. predicted PPIs between human immunodeficiency virus 1 (HIV-1) and human cells by detecting sequence motifs of protein interacting regions that have disordered structures (Becerra, Bucheli, & Moreno, 2017). Sequence patterns of known functional regions including PPI sites, which are called motifs or domains depending on the sequence length, are stored in public databases, such as ELM (Dinkel et al., 2012), InterPro (Finn et al., 2017), PROSITE (Sigrist et al., 2010), PRINTS (Attwood et al., 2012), Pfam (Finn et al., 2016), and ProDom (Bru et al., 2005; Corpet, Gouzy, & Kahn, 1998).
Instead of detecting specific motifs that are known as protein interaction sites, Sprinzak and Margalit computed the log-odds score of observing two motifs from the InterPro database in known interacting yeast protein pairs (Sprinzak & Margalit, 2001). The log-odds value was computed as log2(Pij/PiPj), where Pij is the observed frequency of motif pair (i, j) observed in interacting proteins, and Pi and Pj are the frequencies of motif i and j in the data, respectively. If a query protein pair contains at least one motif pair that has a log-odds value above a threshold, they are predicted as interacting. Later, essentially the same approach was taken to count motif pairs in interacting proteins in the DIP database (Kim, Park, & Suh, 2002). Above methods consider only a single motif pair from each protein pair. Chen and Liu extended the methods by considering contributions of all the possible pairs of 4293 Pfam domain combinations (X.-W. Chen & Liu, 2005). Each protein pair was represented with a 4293-dimensional vector with 0 indicating absence of a domain in either of the proteins, 1 indicating one of the proteins contains the domain, and 2 indicating presence of the domain in both proteins. Then protein pairs are predicted to interact or not to interact by classifying its feature vector using a machine learning method, random forest, which makes a prediction by voting from many decision trees.
Pitre et al. considered sequence similarity rather than detecting exact sequence patterns of interacting proteins (Pitre et al., 2006). The algorithm called Protein-Protein Interaction Prediction Engine (PIPE) they developed, considers the co-occurrence of all short subsequences. In this method, the query protein sequences A and B are fragmented into ai and bj using 20 amino acid-long sliding window. Then the fragment ai is compared with fragments of proteins in a known PPI network using the PAM120 amino acid similarity matrix. Once matched fragment of known proteins similar to ai are found, the known interacting partners to the matched proteins are compared with fragment bj using the PAM120 matrix. Finally, two proteins A and B are predicted to interact if frequency of matched fragment pairs from known PPIs is above a threshold (set to 10). Another similar method called D-MIST adopted position-specific scoring matrix (PSSM) to evaluate the similarity of motifs in a query protein pair to binding motifs in known PPIs with solved tertiary structures (Betel et al., 2007).
Methods that capture sequence features
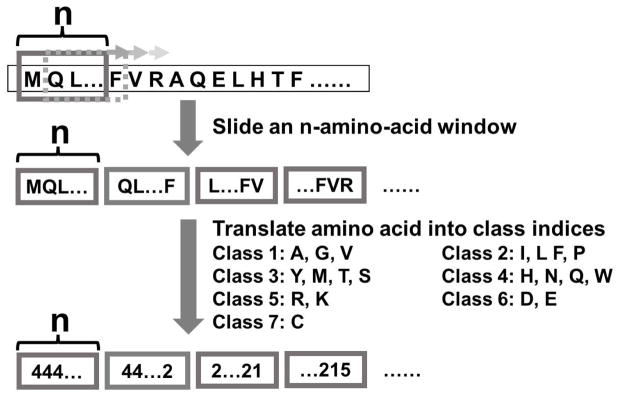
The motif/domain-based methods described in the previous section examine occurrence of known functional sequence motifs/domains in databases or in known interacting proteins. Sequence-based approaches can be extended to consider any sequence patterns including patterns that are not necessarily known to be involved in PPIs or in any function by simply extracting short sequences of a fixed length systematically from query protein sequences. A typical method in this category segments an amino acid sequence of a target protein into overlapping fragments (n-gram) by applying a small sliding window of a certain length (n), and to consider counts of sequence patterns of fragments as a feature vector of the protein (Fig. 1). Then, a machine learning method is trained on a dataset of feature vectors of known interacting proteins and non-interacting protein pairs so that the method distinguishes between the two datasets (Nanni, 2005; Shen et al., 2007). Instead of raw counts of sequence patterns, statistical significance of the counts relative to the background frequency of amino acids was also used (C.-Y. Yu, Chou, & Chang, 2010). Another variant of the n-gram approach was to consider sequence patterns that skip a certain number of sequence positions (L. Wei et al., 2017). Martin et al. used a so-called “signature molecular descriptor”, which considers the frequency of adjacent (i.e., preceding and following) amino acids for each amino acid, which essentially captures sequence patterns of 3-grams (Martin, Roe, & Faulon, 2005). Ding et al. considered both multivariate mutual information of 3-gram and mutual information of 2-gram, i.e.,
Figure 1. The n-gram features for a protein sequence.
The 20 amino acids are clustered into seven classes based on their physicochemical properties. A window of length n (e.g., n=3) slides along the sequence and captures amino acid class patterns in the window. Then the occurrences of every combination of amino acid class are counted to generate a feature vector for the sequence. For example, when n equals 3, the total number of combinations of amino acid class is 7*7*7=343.
| (1) |
where I (a,b,c) is the multivariate mutual information of 3-gram, I(a,b) is the mutual information of 2-gram, a, b, c are amino acid classes, and I(a,b|c) denotes the conditional mutual information of a and b given that c exists in the 3-gram (Ding, Tang, & Guo, 2016). Wong et al. considered amino acid pairs in a protein sequence (every pairs; including non-adjacent pairs) and represented it as an n*n matrix (n: the length of the protein), where each element is the sum of hydrophobicity value of every combination of two amino acids in the sequence (L. Wong, You, Li, Huang, & Liu, 2015). PSSM was used to represent a protein sequence, which considers similarity of 19 other amino acids at each position of a sequence (An et al., 2016). Using PSSM, 2-gram was represented as a 400-dimensional vector (=20*20), which was subject to the dimension reduction to 350 vectors.
The number of sequence combinations of n-grams is quite large, for example, there are 20*20*20 = 8000 combinations for 3-grams for protein sequences that consist of 20 different amino acids. A large number of combinations will generate unnecessarily long feature vectors for proteins and will causes a data sparseness problem when some sequence patterns are not well sampled. Therefore, for computing n-grams, it is common to reduce the number of letters in sequences by clustering amino acids into a smaller number of groups. Shen et al. classified amino acids to seven classes considering their polarity and volume (Shen et al., 2007), and several later papers used the classification.
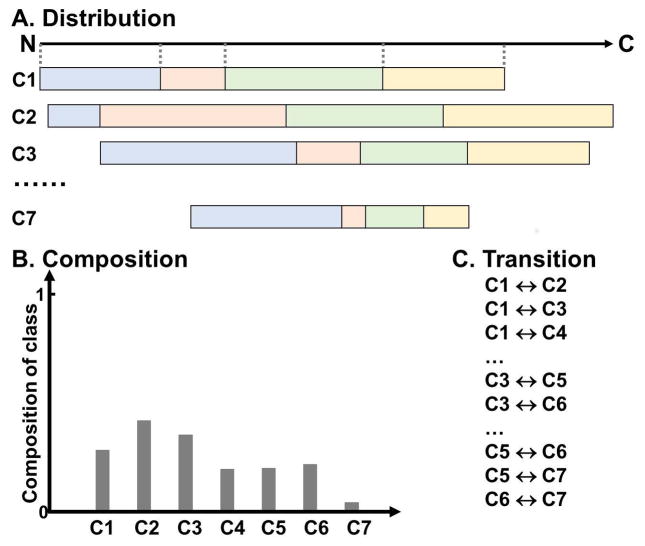
Besides using n-grams and its variants, there are several other ideas for capturing sequence patterns that were used for PPI prediction. To capture general characteristics of a protein sequence, a combination of three sequence features called the local descriptor was used (You, Chan, & Hu, 2015) (Yang, Xia, & Gui, 2010) (Y. Z. Zhou, Gao, & Zheng, 2011) (You, Lei, Zhu, Xia, & Wang, 2013) (Fig. 2). The features are the composition of amino acids, transition probabilities between two consecutive amino acids, and a feature called the distribution. The distribution describes the lengths of sequences from the N-terminus that contain the first, first 25%, 50%, 75%, and 100% of each amino acid (class) over the sequence (Dubchak, Muchnik, Holbrook, & Kim, 1995).
Figure 2. The local descriptors.
Amino acids are clustered into seven classes (C1-C7). A, The distribution of the lengths of sequences from the N-terminus that contain the first, first 25%, 50%, 75%, and 100% of each amino acid class in the protein sequence are represented in blue, pink, green, and yellow, respectively. The dotted line represents the position of the first, first 25%, 50%, 75%, and 100% of Class 1 in the local region. The number of distribution descriptor is 7 (classes) *5 (distribution values) =35 for a local region. B, The composition of each amino acid class in a local region is considered. C, The transition accounts for the frequency of the transition from one class to another. The number of transition descriptor is (7*6)/2=21. Therefore, each local region is represented by 35+7+21=63 descriptors.
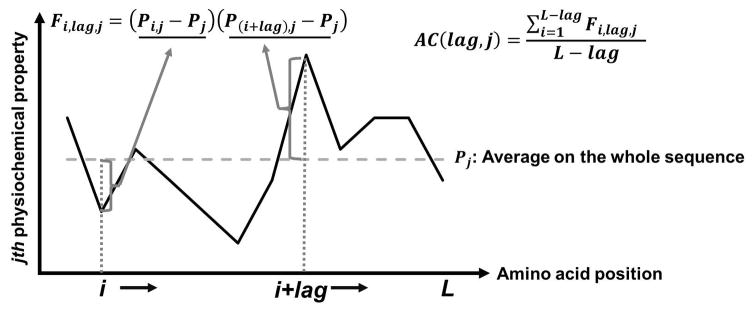
Guo et al. used a feature called auto covariance (AC) to represent protein sequences (Y. Guo, Yu, Wen, & Li, 2008). AC is intended to capture the periodicity of physicochemical properties along a protein sequence (Fig. 3). To compute AC of a protein sequence for a physicochemical property, amino acids are assigned with a property values, e.g., hydrophobicity, hydrophilicity, side-chain volume, polarity, solvent-accessible surface area, or the net charge index of side chain. Then, AC is defined as follows:
| (2) |
where lag is the distance between covariant residues to consider, which ranges from 1 to 30, j is the j-th physiochemical descriptor, i is the position in the sequence, and L is the length of sequence. Thus, AC of a property with a certain lag length will be large if amino acids with a large (or small) property value appear periodically with an interval of lag. There is a similar value called Moran auto correlation (MAC), which is defined as
Figure 3. Schematic view of calculating Auto-covariance (AC).
The black line in the plot represents the value of the j-th physiochemical property along the amino acid sequence. The dashed grey line represents the average value of the j-th physiochemical property. The grey bracket regions are the difference from the average value of i-th and (i+lag)-th amino acid, respectively. AC is the average of Fi,lag,j.
| (3) |
where d is the distance between covariant residues, which ranges from 1 to 30, Pj and Pj+d are the physiochemical property of j-th and (j+d)-th amino acid, respectively, N is the length of the protein sequence, is the average value of the physiochemical property (You et al., 2013). Thus, MAC is AC divided by variance of the physiochemical property, .
The intention behind computing the local descriptor, MC, and MAC is to capture global, long range sequence features of proteins, in contrast to n-gram and its variants, which captures local patterns of sequences. As these features are complementary to each other, often both types were combined (Ding et al., 2016). For example, in the method by You et al., there were four components in the protein sequence feature representation (You et al., 2013): 1) 3-grams. Amino acids were classified to seven classes and the frequency of 3-grams was considered as a feature of a protein. Thus, a protein pair is represented by a vector of 686 (= 2*7*7*7) features. 2) AC. Six physicochemical properties of amino acids were considered: hydrophobicity, side-chain volume, polarity, polarizability, solvent-accessible surface area, and the net charge of the side chains. For each of the properties, AC was computed using 1 to 30 lag values following Eq. 2. Thus, the length of the vector for a protein pair was 360 (= 2*6*30). 3) MAC. Similar to AC, a 360-dimension vector was constructed for a protein pair. 4) Local descriptors. Amino acids were classified to seven classes and the local descriptor, the composition, the transition, and the distribution, were computed for each of the seven amino acid classes for ten local regions in a protein. Thus, a pair of proteins were represented by a vector of 1260 = 2* 10* (7 compositions + 21 transitions + 35 distributions) values. Overall, considering all four features, a protein pair was represented by a vector of 2666 (= 686 + 360 + 360 + 1260) features.
With these sequence features, predictions of PPIs were made using various machine learning algorithms. Algorithms used include support vector machine (SVM) (Bock & Gough, 2001; Y. Guo et al., 2008; X. Liu et al., 2012; Martin et al., 2005; Shen et al., 2007; Y. Z. Zhou et al., 2011), relevance vector machine (An et al., 2016), random forest (X.-W. Chen & Liu, 2005; Ding et al., 2016; You et al., 2015), rotation forest (L. Wong et al., 2015), linear discriminant classifier and cloud points (Nanni, 2005), relaxed variable kernel density estimator (RVKDE) (C.-Y. Yu et al., 2010), an ensemble classifier (L. Wei et al., 2017), extreme learning machine (ELM) (You et al., 2013), and k-nearest neighbors (KNNs) (Yang et al., 2010).
These sequence-based methods reported surprisingly high accuracies. For example, Shen et al., reported 83.90% accuracy on the HPRD dataset (Prasad et al., 2009; Shen et al., 2007). Yang et al. reported 86.15% accuracy on a yeast dataset (Yang et al., 2010). Yu et al. achieved 93.7% accuracy on a highly unbalanced HPRD dataset where positive-to-negative ratio was 1:15 (C.-Y. Yu et al., 2010). Zhu et al. reported over a 75% accuracy on five organisms including yeast, C. elegans, E. coli, human, and mouse (Y. Z. Zhou et al., 2011). Wong et al. achieved 93.92% on the S. cerevisiae dataset (L. Wong et al., 2015). You et al. achieved 93.46% to 97.01% accuracy on six different organisms including yeast, H. pylori, C. elegans, E. coli, human, and mouse (You et al., 2015). Ding et al. achieved 95.01% on the yeast dataset and 87.59% on the H. pylori dataset (Ding et al., 2016). An et al. achieved 94.57% and 90.57% on the S. cerevisiae and the H. pylori dataset, respectively, also 97.15% accuracy on an imbalanced yeast dataset (An et al., 2016). Wei showed over 81% accuracy using different features on the Negatome and the DIP dataset (Blohm et al., 2014; L. Wei et al., 2017; Xenarios et al., 2002). Although the reported accuracy values are high and encouraging, it needs to be noted that the datasets on which the methods are tested are limited to several organisms.
Using homology
So far we reviewed methods that use partial sequence patterns and statistical features in protein sequences. In this section, we introduce methods that use the similarity of entire protein sequences. Many functionally important proteins in an organism are conserved across species, which is the rationale of sequence similarity search for annotating function of genes (Chitale, Hawkins, Park, & Kihara, 2009; T. Hawkins, Chitale, Luban, & Kihara, 2009; Troy Hawkins & Kihara, 2007). Several databases, such as KEGG Orthology (Tanabe & Kanehisa, 2012), OrthoDB (Waterhouse, Tegenfeldt, Li, Zdobnov, & Kriventseva, 2013), OrthoMCL-DB (F. Chen, Mackey, Stoeckert, & Roos, 2006), HomoloGene (NCBI, 2016), and INPARANOID (Sonnhammer & Ostlund, 2015), contain lists of precomputed homologous genes in different species. As interactions with other proteins is a part of a protein’s function, it is known that PPIs are often conserved across species. These conserved interactions are noted as “interlogs” (Walhout, Sordella, et al., 2000). Matthew et al. mapped PPIs in the yeast interaction map to predict PPIs in C. elegans, and identified 257 potential interlogs (Matthews et al., 2001). Further experimental validation performed on 72 predicted interactions gave 19 positive results, which were roughly 25% among tested. The POINT web service provides human PPIs inferred from interlogs with mouse, fruitfly, yeast, and C. elegans (T.-W. Huang et al., 2004). Taking advantage of an increasing number of experimentally identified protein interactions, Lee et al. then expanded orthologous pairs to consider those from 18 eukaryotic species (Lee et al., 2008). The idea of interlogs was also applied to predict PPIs in the plant, A. thaliana, by considering homologs with yeast, fruitfly, human, and C. elegans (Geisler-Lee et al., 2007) (De Bodt, Proost, Vandepoele, Rouzé, & Van de Peer, 2009) and to a second plant, Oryza sativa (Asian rice), by considering interlogs with the six species including the same four species with E. coli and A. thaliana (Gu, Zhu, Jiao, Meng, & Chen, 2011). Dutkowski et al. developed a statistical model, which represents specification and duplication events of genes along an evolutionary tree, on which known interacting protein pairs in seven eukaryotic organisms were mapped and used for predicting PPIs (Dutkowski & Tiuryn, 2009). Interactome3D is a database that provides the tertiary structure models of protein complexes built based on known structure information of interlogs (Mosca, Pons, Céol, Valencia, & Aloy, 2013). Wang et al. merged prediction results from an interlog-based method and a motif-based method to cover a larger number of predicted PPIs in the pig proteome (Wang et al., 2012).
Codon usage
Interestingly, it was shown that the codon usage of genes can be used to predict PPIs. Using the difference of codon usage of protein pairs, Najafabadi et al. predicted PPIs in E. coli, yeast, and Plasmodium falciparum with reasonably good accuracy (Najafabadi & Salavati, 2008). For a pair of genes i and j the difference in usage of codon c among 64 codons is simply defined as
| (4) |
where fi(c) is the usage of codon c of gene i. Then, dij of each codon is binned into 50 intervals, and the likelihood ratio of the fraction in interacting and non-interacting proteins in a training dataset was computed. A PPI prediction for a protein pair is performed with a naïve Bayes approach using the likelihood ratio. Zhou et al. used SVM with the codon usage difference and applied to the yeast genome (Y. Zhou, Zhou, He, Song, & Zhang, 2012). One may wonder why codon usage is related to PPIs. But it is reasonable considering that codon usage is known to be correlated with gene expression levels (Jansen, Bussemaker, & Gerstein, 2003) and also that neighboring genes have similar codon usage. As we discuss later in this review, both gene expression level and conserved neighboring genes (gene order) have been successfully used to predict PPIs.
COMPARATIVE GENOMICS-BASED METHODS
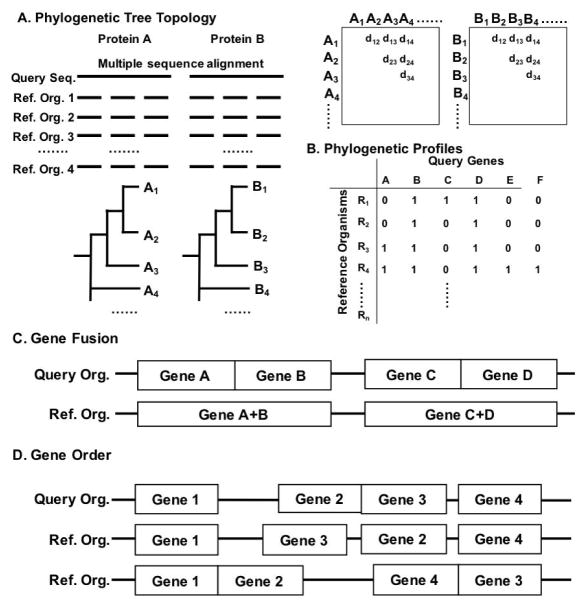
The last level of sequence information that can be used for PPI prediction is from genome sequences from various species. Since important features in a genome sequence are conserved during evolution, identifying such conserved features in genomes can be a clue for identifying proteins that are functionally related. Under this category, which we call the comparative genomics-based methods, we discuss four approaches, the phylogenetic tree topology analysis, the phylogenetic profile, considering gene fusion events, and conserved gene orders (Fig. 4). An important point to note is that these methods are not aimed toward predicting physical PPIs directly but for identifying functionally related proteins. Quite often, however, functionally related proteins do physically interact with each other. A strong advantage of the comparative genomics-based approaches is that, due to the increasing number of determined genome sequences, many proteins can now find related (and maybe interacting) proteins through these approaches (Huynen, Snel, von Mering, & Bork, 2003).
Figure 4. Comparative genomics-based methods.
A, Phylogenetic tree topology-based method. A pair of query sequences, A and B, are compared with n reference organisms and multiple sequence alignments are constructed. Based on the alignments, phylogenetic trees are computed, from which distances between all pairs of sequences are computed and stored in matrices. Then the similarity between two matrices is evaluated with a correlation coefficient. B, Phylogenetic profiles. Genes in a query organism is compared with n reference organisms by BLAST search. 1 represents the presence of the gene in the reference organism 0 for the absence. The table can also be filled with real-values, such as BLAST bit scores. C, Gene fusion. Two protein genes A and B are predicted as interacting if they fuse to form one protein gene AB in another organism. D, Gene order. If protein gene orders of proteins are conserved among different species, they are predicted as interacting proteins with each other.
Phylogenetic tree topology analysis
It has been observed that the phylogenetic trees between interacting proteins are more similar than a general divergence between the corresponding species (Goh, Bogan, Joachimiak, Walther, & Cohen, 2000; Goh & Cohen, 2002; Pazos & Valencia, 2001). The similarity between the phylogenetic trees of interacting proteins was explained as maintenance of the complex functionality and suffering similar evolutionary pressure.
The sequence signal of the coevolution is strong at the binding interface of proteins, but can also come from other regions of proteins (Kann, Shoemaker, Panchenko, & Przytycka, 2009).
The tree topology similarity can be measured as the correlation between the evolutionary distance matrices used to build the trees. The algorithm to calculate similarity of distance matrices is called the mirror tree method (Pazos & Valencia, 2001). It contains following steps (Fig. 4A): 1) To construct a multiple sequence alignment for each protein against a list of reference organisms; 2) To construct a phylogenetic tree for the proteins; 3) Then, for a pair of proteins in question, distances against orthologous proteins in different species are computed (distance matrices) and the correlation coefficient between two distance matrices is obtained. A protein pair is predicted to be interacting if the coefficient value is above a cut-off value, which is determined to distinguish known interacting and non-interacting proteins.
The mirror tree method was modified for improvement in several different ways. The method was extended to handle interacting protein families, such as a ligand family and a receptor family, to be able identify interacting specific protein pairs from the two families (Ramani & Marcotte, 2003). Sato et al. removed a background tree similarity that arises by the overall evolutionary distance of organisms from distance matrices of individual proteins, which yielded improvement of PPI prediction accuracy (Sato, Yamanishi, Kanehisa, & Toh, 2005). They further considered partial correlation of distance matrices that can more effectively remove background organism-level similarity from the tree similarity of a query protein pair, where the background organism-level similarity was represented by a linear combination of distance matrices of many proteins in the organisms (Sato, Yamanishi, Horimoto, Kanehisa, & Toh, 2006). Besides the background similarity of organisms, another source of noise in the mirror tree method is that a protein coevolves with multiple interacting proteins. Instead of evaluating tree similarity of a query protein pair, Juan et al. considered a network of similarities between all pairs of proteins simultaneously (Juan, Pazos, & Valencia, 2008). In the mirror tree method, selection of reference genomes is a key for successful prediction. Effective ways to select organisms for building trees were examined by Herman et al. (Herman et al., 2011). Instead of using correlation coefficient, SVM was also used to make predictions from distance matrices (Craig & Liao, 2007).
Phylogenetic profiles
Phylogenetically related and thus possibly interacting protein pairs can be identified in a simpler way of using comparative genomics. In the approach called the phylogenetic profiling co-presence and co-absence of orthologous proteins across organisms are examined rather than comparing phylogenetic trees of protein pairs as discussed in the previous section (Pellegrini, Marcotte, Thompson, Eisenberg, & Yeates, 1999). If two proteins are needed for realizing a certain biological function, an organism needs to possess both proteins if the function is required while both are not needed if it does not need the function. Coding one of the proteins only in its genome is meaningless.
There are three major steps to perform this method (Fig. 4B). The first step is to identify orthologous proteins for all the proteins in a query genome against other reference genomes by a sequence similarity search. Then, construct a phylogenetic profile for each protein in the query genome, which has binary values with 1 indicating the presence of an orthologous gene and 0 for the absence of the ortholog in a reference genome. Thus, the dimension of the profile is the number of reference genomes used. Finally, protein pairs that have similar profiles are predicted to be interacting (more precisely, functionally related). Similar to the phylogenetic tree topology methods, the choice of reference genome is crucial for this approach (J. Sun, Li, & Zhao, 2007). Also, a threshold value (E-value) in sequence similarity search for detecting orthologous proteins strongly affects the profiles, and thus the prediction performance of the method (Jingchun Sun et al., 2005). To accommodate the strong dependency of the performance to the threshold value in the similarity search, real value vectors of an alignment score was used for constructing profiles rather than binary values (T.-W. Lin, Wu, & Chang, 2013). In the method by de Vienne and Azé a combination of the phylogenetic tree topology and profile was used as features in a machine learning framework (de Vienne & Azé, 2012).
Gene fusion events
A gene fusion refers to an event in the comparative genomics where two individual genes in one organism fuse as a continuous sequence in another organism (Snel, Bork, & Huynen, 2000) (Fig. 4C). Fused genes are usually functionally related and further implies physical interactions between the proteins (Enright, Iliopoulos, Kyrpides, & Ouzounis, 1999; Marcotte et al., 1999; Yanai, Derti, & DeLisi, 2001). Computationally, fused genes can be found by gene sequence similarity search between genomes. It was reported that metabolic enzymes are frequently involved in gene fusions (Tsoka & Ouzounis, 2000).
Conserved gene orders
Through evolution, genomes undergo various rearrangements and transfers; therefore locations of genes in a genome tend to be shuffled unless an evolutionary pressure keeps the order of some genes together (Suyama & Bork, 2001) (Fig. 4D). Thus, conservation of gene orders, i.e., common local clusters of genes in genomes, indicates that there is a requirement or an advantage to keep the gene order for the organisms, and in fact in many cases, genes in a conserved cluster are involved in the same function (Tamames, Casari, Ouzounis, & Valencia, 1997). In bacterial and archaeal genomes, operon structures are conserved across many species, which code genes in the same pathways or complexes (Dandekar, Snel, Huynen, & Bork, 1998). After initial findings of the conserved gene orders, more systematic studies have been done (Fujibuchi, Ogata, Matsuda, & Kanehisa, 2000; Overbeek, Fonstein, D’souza, Pusch, & Maltsev, 1999). Similar to the other comparative genomics-based methods, a key for successful application of this analysis is to choose an appropriate set of reference genomes, which should not be too evolutionarily distant but not too close to each other, so that only clusters of functionally related genes are conserved. A related work was done by Kihara & Kanehisa where transmembrane protein complexes were predicted from genomes by identifying gene clusters that have predicted transmembrane domains (Kihara & Kanehisa, 2000).
FUNCTION-BASED METHODS
Since interacting proteins belong to the same pathway and share function, functional similarity of proteins can be a clue for predicting PPIs. Functional similarity of proteins are usually quantified by a similarity score of Gene Ontology (GO) terms (Consortium, 2017) that annotate the proteins. Similarity of GO terms are defined by the closeness of the terms on the GO hierarchy tree and/or the frequency of the GO terms in gene annotations observed in an protein annotation database, e.g., UniProt (D. Lin, 1998; Resnik, 1995; Schlicker, Domingues, Rahnenführer, & Lengauer, 2006) (Wu, Pang, Lin, & Pei, 2013). Interestingly, it was shown that considering common children terms of GO terms in addition to common parental GO terms, which are not used in the aforementioned functional similarity scores, improved PPI prediction accuracy (S.-B. Zhang & Tang, 2016). Jain and Bader defined a GO similarity score by considering the distance to the leaf nodes in order to reduce the influence of imbalanced branch depths in the GO hierarchy (Jain & Bader, 2010).
GO term similarity (or relevance) can be also defined by counting frequency of co-occurrence of GO term pairs in biological contexts, in gene annotation or PubMed abstracts (Chitale, Palakodety, & Kihara, 2011) or in known PPIs (Wei, Khan, Ding, Yerneni, & Kihara, 2017; Yerneni, Khan, Wei, & Kihara, 2015).
Since PPI prediction is a suitable and handy application of GO term similarity scores, all the GO term scores above have been tested and compared for their performance of PPI predictions (X. Guo, Liu, Shriver, Hu, & Liebman, 2006; Jain & Bader, 2010; Wu et al., 2013; Yerneni et al., 2015). Maetsche et al. showed that when using GO terms for PPI prediction in machine learning framework, induced GO term sets, e.g., common parental terms of annotated GO terms, performed better rather than using the original GO annotations of proteins (Maetschke, Simonsen, Davis, & Ragan, 2012).
GENE CO-EXPRESSION-BASED METHODS
Gene co-expression data such as microarray and RNA-sequencing data are valuable experimental data that can be used to infer PPIs. Intuitively, interacting protein pairs are expected to have similar gene expression levels across different conditions. Indeed significant correlation between the gene co-expression level and PPIs was shown in bacteriophage T7 (Grigoriev, 2001), yeast (Ge, Liu, Church, & Vidal, 2001; Jansen, Greenbaum, & Gerstein, 2002), human, mouse, and E. coli (Bhardwaj & Lu, 2005). Fraser et al. showed that gene expression level of interacting proteins co-evolve using four closely related yeast species, where the expression level was estimated by the codon usage (Fraser, Hirsh, Wall, & Eisen, 2004). Databases that provides large-scale gene co-expression information include GEO (Barrett et al., 2013), ATTED-II (Aoki, Okamura, Tadaka, Kinoshita, & Obayashi, 2016), and COXPRESdb (Okamura et al., 2014). ATTED-II and COXPRESdb are pre-calculated gene co-expression databases of plant organisms and animal species, respectively.
Although gene expression is shown to have significant correlation to PPIs, a major challenge is that co-expression data is noisy due to various types of systematic and stochastic fluctuations. Soong et al. adopted principle component analysis (PCA) and independent component analysis (ICA) to filter out noise in microarray data before feeding the data to SVM classifier (Soong, Wrzeszczynski, & Rost, 2008). As we see later in the section for integrated methods, gene expression is used frequently as one of the input features for proteins.
PROTEIN TERTIARY STRUCTURE-BASED METHODS
The tertiary (3D) structure of proteins can be important information to predict PPIs if available, or if the structures can be computationally reliably modelled. There are many computational methods developed that “docks” two protein structures to provide the tertiary structures of a protein complex from individual protein structures, which include LZerD (Esquivel-Rodriguez, Filos-Gonzalez, Li, & Kihara, 2014; Esquivel- Rodríguez, Yang, & Kihara, 2012; Peterson, Roy, Christoffer, Terashi, & Kihara, 2017; Venkatraman, Yang, Sael, & Kihara, 2009), GRAMM-X (Tovchigrechko & Vakser, 2006), ZDOCK (Pierce et al., 2014), RosettaDock (Lyskov & Gray, 2008), HADDOCK (Geng, Narasimhan, Rodrigues, & Bonvin, 2017), SwarmDock (Torchala & Bates, 2014), HEX (Ritchie & Kemp, 2000), and ClusPro (Kozakov et al., 2017). These docking methods build structure models of a protein complex given individual protein structures, which provide structural insights of the PPI. However, it should be noted that these docking methods do not predict whether a protein pair actually interacts or not.
Then how does one use structure information for predicting PPIs? There are two approaches explored. The first approach is to detect energetic characteristics of interacting protein pairs observed in protein docking prediction. A protein docking program generates typically over tens of thousands of different docking poses for a pair of input protein structures. Wass et al. reported the score distribution of docking poses of interacting protein pairs can be distinguished from those of non-interacting proteins, because the former distribution is skewed toward favorable scores (Wass, Fuentes, Pons, Pazos, & Valencia, 2011). This is an intriguing observation because a docking pose distribution include both near-native (i.e., almost correct) and incorrect poses, therefore, the report implies that even incorrect docking poses have relatively favorable scores (i.e., more favorable geometric complementary) in cases of interacting proteins. In MEGADOCK, a protein docking method aimed for fast, large-scale protein docking screening, a protein pair is predicted to be interacting if a pool of docking poses generated by the algorithm include clusters of similar poses that have significantly favorable docking scores in comparison with the rest of the poses (Ohue, Matsuzaki, Uchikoga, Ishida, & Akiyama, 2014).
The second approach to use protein structure information for PPI prediction is, for two query protein structures, to find similarity in known protein complexes. PRISM, developed by Keskin and his colleagues, is one of the first to take this approach (Aytuna, Gursoy, & Keskin, 2005; Tuncbag, Gursoy, Nussinov, & Keskin, 2011). PRISM takes two protein structures as input, and examines if surface shapes of the proteins have similarity to docking interfaces from known protein complexes structures. To perform this comparison, PRISM has a database of docking interface regions of known protein complexes extracted from the PDB database (Rose et al., 2017). Identified potential interface regions in the two query proteins that are identified by comparison to known interface regions are examined for structural similarity to the template, sequence conservation, and the binding energy. Although the prediction power of PRISM relies on the coverage of template dataset, the method will be able identify interactions between proteins that are globally dissimilar but have similar local interface regions to known protein complexes. PrePPI takes a similar approach PRISM (Q. C. Zhang et al., 2012). One difference is that PrePPI takes sequences of the query proteins and models their structures by homology modeling. Subsequently, the two structures are mapped to known protein complex structures, which are then evaluated by structure and sequence similarity scores to the known complex structures. Final prediction is made by a composite score that integrates five other features, gene co-expression, essentiality of the proteins, functional similarity, and the phylogenetic profile. Similarly, Coev2Net models a complex structure of two query proteins by mapping their sequences to a known complex structure with a threading method, and then evaluates the complex model by a logistic regression classifier that considers structural and sequence features taken from its interface (Hosur et al., 2012). In a recent method, InterPred, a similar approach is taken (Mirabello & Wallner, 2017): for a query protein sequence pair, structures are modelled, then known protein complexes are sought by structure comparison. Finally, the feasibility of the model is evaluated using a random forest classifier that considers interface structure and sequence features as well as overall structure similarity between individual models to the template complex structure.
Although protein structures can provide unique features for PPI prediction, a drawback is that not many proteins have known structures. In Table 2, the number of protein genes with GO terms, gene expression data, and experimentally determined/computationally-modelled protein structures for ten genomes are shown. Compared to GO terms and gene expressions, proteins with known structures are substantially fewer. This is more evident for genomes that are less studied. On the other hand, as shown in the right-most column, most of the protein structures can be computationally modelled (Kihara & Skolnick, 2004). Thus, there is room for new structure-based approaches that use modelled protein structures.
Table 2.
Lists of available information such as number of coding genes, number of genes with annotated GO terms, genes with co-expression information, number of solved structure, and number of redundant modeled structure, in each organism.
| Organism | # of genes with protein productsa | # of proteins with annotated GO termsb | # of genes with co-expression informationc | # of proteins with a solved structured | Fraction of modeled structure among all proteins (%)e(Pieper et al., 2006)(Pieper et al., 2006)(Pieper et al., 2006)(Pieper et al., 2006) |
|---|---|---|---|---|---|
| Human | 109,018 | 46,331 | 19,816 | 16620 | 82.21 |
| Yeast | 6,002 | 5,582 | 4,461 | 1340 | 90.25 |
| Mouse | 76,216 | 28,727 | 20,403 | 2891 | 84.07 |
| A. thaliana | 48,350 | 16,123 | 20,836 | 584 | 77.44 |
| Fruitfly | 30,482 | 6,886 | 13,099 | 875 | 88.92 |
| Asian rice | 28,555 | 100 | 20,625 | 1 | NAf |
| X. tropicalis | 39,662 | 1,998 | 11,095 | 10 | 91.06 |
| D. rerio | 46,451 | 2,723 | 10,112 | 26 | NA |
| C. annuum | 45,410 | 20 | 17,453 | 0 | NA |
| P. persica | 28,927 | 2 | 11 | 0 | NA |
Counted in the NCBI reference sequence (RefSeq) (Pruitt, Tatusova, & Maglott, 2005).
Protein entries in RefSeq was mapped to UniProt, where the number of proteins with at least one annotated Gene Ontology term was counted (Boutet et al., 2016).
Gene expression data of human, yeast, mouse, fruitfly, and D. rerio were taken from COXPRESdb (Okamura et al., 2014). Data for A. thaliana and O. Sativa were taken from the ATTED-II database (Aoki et al., 2016). Expression data for the rest of the three organisms were taken from literature: X. tropicalis: (Yanai, Peshkin, Jorgensen, & Kirschner, 2011). C. annuum: (Sharma, 2016). P. persica: (Tong, Gao, Wang, Zhou, & Zhang, 2009).
After mapping RefSeq IDs of protein genes to UniProt, the number of proteins was counted with at least one crosslinked PDB entry (Boutet et al., 2016).
It is computed from the statistics provided in the ModBase base (Pieper et al., 2006).
NA indicates that no modeled structures provided on ModBase (but some proteins in a genome may be modelled by a standard modeling procedure).
PPI NETWORK TOPOLOGY-BASED METHODS
Methods in this category start from an existing PPI network of an organism, and predict new interactions between proteins by evaluating their network topology features. In the IRAP* method, a missing interaction is predicted if a protein pair has a high score that reflects the number of common neighbors between them in the current PPI network (J. Chen, Hsu, Lee, & Ng, 2006). Another idea by Yu et al. is to predict a PPI if two proteins are neighbors of a clique, a fully-connected graph, in the PPI network of the organism and connecting them would complete a larger clique, because most probably the two proteins are subunits of a protein complex (H. Yu, Paccanaro, Trifonov, & Gerstein, 2006). In the work by L. Wong and his colleagues, a prediction of a PPI is made using a combination of two scores, a score for capturing local network topology of proteins that is based on the number of common neighbors and a global topology-based score that accounts for the memberships of the proteins in protein groups where member proteins interact with each other (G. Liu, Li, & Wong, 2008). Kuchaiev et al. applied Multi-Dimensional Scaling (MDS), a dimension reduction method in statistics, to a PPI network, where distances are based on edge distances between proteins (Kuchaiev, Rašajski, Higham, & Pržulj, 2009). New PPIs are predicted if proteins are closer than a threshold in the projected space by MDS. Lei and Ruan applied a random walk-based approach, where the probability of reaching each node from each of the other nodes in the network is computed by assuming a random walk (Lei & Ruan, 2012). The resulting probability matrix contains information of the topology of the PPI network. Based on the probability matrix, protein pairs are connected if they are similar in their probability vectors to reach the other nodes.
INTEGRATION OF MULTIPLE FEATURES
PPIs can be predicted from different perspectives as discussed above. Naturally, there are methods that use multiple features to be able to combine strengths of different features and to increase the prediction confidence and coverage. Features can be combined using machine learning methods, such as random forest, Naïve Bayesian Network, artificial neural network, SVM, and logistic regression (Qi, Bar-Joseph, & Klein-Seetharaman, 2006). In Table 3 methods that use multiple features are summarized.
Table 3.
Features and algorithms used in multiple-feature integrative methods.
| Feature | Ben-Hur et al. a | Miller et al. b | Qi et al. c | Scott et al. d | Xu et al. e | PAIR f | PrePPI g | FpClass h | STRI-NG i | Taghi-pour et al. j | # of times used | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence | MOT/DOM | X | X | X | X | X | X | 6 | ||||
| NGM | X | 1 | ||||||||||
| PCH | X | 1 | ||||||||||
| HOM | X | X | X | X | X | X | X | 7 | ||||
| COD | X | 1 | ||||||||||
| PHP | X | X | X | X | X | 5 | ||||||
| FUS | X | X | X | 3 | ||||||||
| GNB | X | X | X | 3 | ||||||||
| PTM | X | X | 2 | |||||||||
| Function | GO | X | X | X | X | X | X | X | X | 8 | ||
| MIPS | X | 1 | ||||||||||
| Experiment | COX | X | X | X | X | X | X | X | X | X | 9 | |
| XPI | X | X | 2 | |||||||||
| CLA | X | 1 | ||||||||||
| ESN | X | X | X | 3 | ||||||||
| LOC | X | X | X | 3 | ||||||||
| COR | X | 1 | ||||||||||
| SNL | X | 1 | ||||||||||
| MUT | X | 1 | ||||||||||
| Structure | STR | X | 1 | |||||||||
| DIS | X | 1 | ||||||||||
| Network | NET | X | X | X | X | X | 5 | |||||
| Literature | TXT | X | 1 | |||||||||
| Integrating method | LPK | SVM | RF, KNN, NB, DT, LR, SVM | NB | NB | SVM | NB | NOR | LNR | NB, MCL |
References of each method:
From the table, we can see the most popular feature integrated was gene co-expression data (COX). The next most popular ones are GO functional similarity (GO), and homology (HOM). Several features in the table are not explained yet in this review. The physicochemical features (PCH) concerns features such as charge and aromaticity of amino acids in a protein sequence. The post-translational modification feature (PTM) indicates that PTM motifs are found in UniProt and HPRD. The disordered region (DIS) is a protein structure feature where non-structured regions in a protein can be predicted from its sequence. Thus, besides obvious sequence-based features, DIS, PCH, and PTM are features that are predicted from protein sequences. Direct experimental data of PPIs (EXP) used by Qi et al. were yeast-two-hybrid and mass spectrometry data (Qi et al., 2006), and those used by Miller et al. were data from yeast two-hybrid system (Miller et al., 2005). The protein functional class (CLA) in yeast are taken from the MIPS Protein Class Catalogue, which were determined by experiments (Mewes et al., 2004). Gene essentiality (ESN), synthetic lethality (SNL), and MIPS mutant phenotype (MUT) were determined by knockout mutants (Qi et al., 2006). Text mining (TXT) counts co-mentions of two proteins in PubMed abstracts.
Regarding combinations of features, methods by Ben-Hur et al., Xu et al. combine mostly sequence-based features (Ben-Hur & Noble, 2005; Xu et al., 2010). On the other hand, PrePPI (Q. C. Zhang et al., 2012), FpClass (Kotlyar et al., 2015), and Taghipour et al. (Taghipour, Zarrineh, Ganjtabesh, & Nowzari-Dalini, 2017) are intended to combine different types of features.
Turning our attention to algorithms used, naïve Bayes is the most frequently used among the multiple feature-based methods in Table 3. SVM was the next, used in the three methods. Qi et al. tested five integrating algorithms with different feature combinations (Qi et al., 2006).
DISCUSSION
The identification of PPIs is vital for a systems level understanding of molecular activity of living cells. To complement experimental approaches, we saw many computational tools, which use different types of protein features. Through writing this article, we felt that a wide variety of features were explored already, and development of novel computational approaches would need new types of experimental data. Also, we noticed that large scale PPI networks are experimentally revealed only for a limited number of organisms, and thus many computational methods were developed and benchmarked on those organisms. Therefore, for further advancement of PPI prediction, proteomics-scale PPIs of many more organisms would be needed.
Current PPI networks construct both experimental and computational methods, and only represent a static snapshot of interactions of proteins in a cell, which are dynamically changing over time, containing both transient and permanent interactions. Therefore, the next generation of PPI studies would aim to capture the time-dependent, dynamic aspects of PPIs. Computationally, this direction would eventually meet and be integrated with other computational approaches, such as pathway simulations and molecular dynamics simulation of molecules in a cell.
Acknowledgments
We acknowledge Natalie Tomoko Oda for proofreading the manuscript. This work was partly supported by grants from the National Institutes of Health (R01GM097528) and the National Science Foundation (DBI1262189, IIS1319551, IOS1127027, DMS1614777). ZD is supported by Purdue Research Foundation.
Feature abbreviations
- MOT/DOM
protein motifs or domains
- NGM
n-gram
- PCH
physiochemical feature
- HOM
homologous interaction (interlogs)
- COD
codon usage
- PHP
phylogenetic profile and gene co-occurrence
- FUS
gene fusion
- GNB
gene neighbor
- PTM
post-translational modifications regions
- GO
gene ontology terms
- MIPS
Munich Information Centre for Protein Sequences (MIPS) functional similarity
- COX
gene/protein co-expression
- XPI
Experimental PPI detection, direct experimental data including two-hybrid screens and mass spectrometry
- CLA
protein functional class by MIPS Protein Class Catalogue
- ESN
essentiality
- LOC
protein localization
- COR
common co-regulators of genes
- SNL
synthetic lethality
- MUT
MIPS mutant phenotype
- STR
protein structure
- DIS
disordered region
- NET
protein-protein interaction network
- TXT
text mining
Integrating method abbreviations
- LPK
linear pairwise kernel
- SVM
support vector machine
- RF
random forest
- KNN
k-nearest neighbor
- NB
Naïve Bayes
- DT
decision tree
- LR
logistic regression
- NOR
noisy-OR model (a type of Bayesian network)
- LNR
linear regression
- MCL
Markov clustering algorithm
LITERATURE CITED
- An JY, Meng FR, You ZH, Chen X, Yan GY, Hu JP. Improving protein–protein interactions prediction accuracy using protein evolutionary information and relevance vector machine model. Protein Science. 2016;25(10):1825–1833. doi: 10.1002/pro.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Okamura Y, Tadaka S, Kinoshita K, Obayashi T. ATTED-II in 2016: A Plant Coexpression Database Towards Lineage-Specific Coexpression. Plant Cell Physiol. 2016;57(1):e5. doi: 10.1093/pcp/pcv165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwood TK, Coletta A, Muirhead G, Pavlopoulou A, Philippou PB, Popov I, … Mitchell AL. The PRINTS database: a fine-grained protein sequence annotation and analysis resource--its status in 2012. Database (Oxford), 2012. 2012:bas019. doi: 10.1093/database/bas019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aytuna AS, Gursoy A, Keskin O. Prediction of protein–protein interactions by combining structure and sequence conservation in protein interfaces. Bioinformatics. 2005;21(12):2850–2855. doi: 10.1093/bioinformatics/bti443. [DOI] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, … Holko M. NCBI GEO: archive for functional genomics data sets—update. Nucleic acids research. 2013;41(D1):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra A, Bucheli VA, Moreno PA. Prediction of virus-host protein-protein interactions mediated by short linear motifs. BMC bioinformatics. 2017;18(1):163. doi: 10.1186/s12859-017-1570-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Hur A, Noble WS. Kernel methods for predicting protein–protein interactions. Bioinformatics. 2005;21(suppl 1):i38–i46. doi: 10.1093/bioinformatics/bti1016. [DOI] [PubMed] [Google Scholar]
- Betel D, Breitkreuz KE, Isserlin R, Dewar-Darch D, Tyers M, Hogue CW. Structure-templated predictions of novel protein interactions from sequence information. PLoS Comput Biol. 2007;3(9):e182. doi: 10.1371/journal.pcbi.0030182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj N, Lu H. Correlation between gene expression profiles and protein–protein interactions within and across genomes. Bioinformatics. 2005;21(11):2730–2738. doi: 10.1093/bioinformatics/bti398. [DOI] [PubMed] [Google Scholar]
- Blohm P, Frishman G, Smialowski P, Goebels F, Wachinger B, Ruepp A, Frishman D. Negatome 2.0: a database of non-interacting proteins derived by literature mining, manual annotation and protein structure analysis. Nucleic Acids Res. 2014;42(Database issue):D396–400. doi: 10.1093/nar/gkt1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock JR, Gough DA. Predicting protein–protein interactions from primary structure. Bioinformatics. 2001;17(5):455–460. doi: 10.1093/bioinformatics/17.5.455. [DOI] [PubMed] [Google Scholar]
- Boeri Erba E, Petosa C. The emerging role of native mass spectrometry in characterizing the structure and dynamics of macromolecular complexes. Protein Science. 2015;24(8):1176–1192. doi: 10.1002/pro.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet E, Lieberherr D, Tognolli M, Schneider M, Bansal P, Bridge AJ, … Xenarios I. UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Methods Mol Biol. 2016;1374:23–54. doi: 10.1007/978-1-4939-3167-5_2. [DOI] [PubMed] [Google Scholar]
- Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, … Lynn DJ. InnateDB: systems biology of innate immunity and beyond--recent updates and continuing curation. Nucleic Acids Res. 2013;41(Database issue):D1228–1233. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne F, Zheng H, Wang H, Azuaje F. From experimental approaches to computational techniques: a review on the prediction of protein-protein interactions. Advances in Artificial Intelligence. 2010;2010:7. [Google Scholar]
- Bru C, Courcelle E, Carrere S, Beausse Y, Dalmar S, Kahn D. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 2005;33(Database issue):D212–215. doi: 10.1093/nar/gki034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JW, Zhou YQ, Ul Qamar MT, Chen LL, Ding YD. Prediction of Protein–Protein Interactions by Evidence Combining Methods. International Journal of Molecular Sciences. 2016;17(11):1946. doi: 10.3390/ijms17111946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, … Tyers M. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45(D1):D369–D379. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Stoeckert CJ, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic acids research. 2006;34(suppl 1):D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Hsu W, Lee ML, Ng SK. Increasing confidence of protein interactomes using network topological metrics. Bioinformatics. 2006;22(16):1998–2004. doi: 10.1093/bioinformatics/btl335. [DOI] [PubMed] [Google Scholar]
- Chen XW, Liu M. Prediction of protein–protein interactions using random decision forest framework. Bioinformatics. 2005;21(24):4394–4400. doi: 10.1093/bioinformatics/bti721. [DOI] [PubMed] [Google Scholar]
- Chitale M, Hawkins T, Park C, Kihara D. ESG: extended similarity group method for automated protein function prediction. Bioinformatics. 2009;25(14):1739–1745. doi: 10.1093/bioinformatics/btp309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitale M, Palakodety S, Kihara D. Quantification of protein group coherence and pathway assignment using functional association. BMC bioinformatics. 2011;12(1):373. doi: 10.1186/1471-2105-12-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GO. Expansion of the Gene Ontology knowledgebase and resources. Nucleic acids research. 2017;45(D1):D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F, Gouzy J, Kahn D. The ProDom database of protein domain families. Nucleic Acids Res. 1998;26(1):323–326. doi: 10.1093/nar/26.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig RA, Liao L. Improving Protein–Protein Interaction Prediction Based on Phylogenetic Information Using a Least-Squares Support Vector Machine. Annals of the New York Academy of Sciences. 2007;1115(1):154–167. doi: 10.1196/annals.1407.005. [DOI] [PubMed] [Google Scholar]
- Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends in biochemical sciences. 1998;23(9):324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- De Bodt S, Proost S, Vandepoele K, Rouzé P, Van de Peer Y. Predicting protein-protein interactions in Arabidopsis thaliana through integration of orthology, gene ontology and co-expression. BMC genomics. 2009;10(1):288. doi: 10.1186/1471-2164-10-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vienne DM, Azé J. Efficient prediction of co-complexed proteins based on coevolution. PloS one. 2012;7(11):e48728. doi: 10.1371/journal.pone.0048728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tang J, Guo F. Predicting protein-protein interactions via multivariate mutual information of protein sequences. BMC Bioinformatics. 2016;17(1):398. doi: 10.1186/s12859-016-1253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van Roey K, Altenberg B, … Gibson TJ. ELM--the database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40(Database issue):D242–251. doi: 10.1093/nar/gkr1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubchak I, Muchnik I, Holbrook SR, Kim SH. Prediction of protein folding class using global description of amino acid sequence. Proceedings of the National Academy of Sciences. 1995;92(19):8700–8704. doi: 10.1073/pnas.92.19.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkina NV, Kouřil R, Bultema JB, Boekema EJ. Imaging of organelles by electron microscopy reveals protein–protein interactions in mitochondria and chloroplasts. FEBS letters. 2010;584(12):2510–2515. doi: 10.1016/j.febslet.2010.03.027. [DOI] [PubMed] [Google Scholar]
- Dunham WH, Mullin M, Gingras AC. Affinity-purification coupled to mass spectrometry: Basic principles and strategies. Proteomics. 2012;12(10):1576–1590. doi: 10.1002/pmic.201100523. [DOI] [PubMed] [Google Scholar]
- Dutkowski J, Tiuryn J. Phylogeny-guided interaction mapping in seven eukaryotes. BMC bioinformatics. 2009;10(1):393. doi: 10.1186/1471-2105-10-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402(6757):86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- Esquivel-Rodriguez J, Filos-Gonzalez V, Li B, Kihara D. Pairwise and multimeric protein-protein docking using the LZerD program suite. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S.] Methods Mol Biol. 2014;1137:209–234. doi: 10.1007/978-1-4939-0366-5_15. [DOI] [PubMed] [Google Scholar]
- Esquivel-Rodríguez J, Yang YD, Kihara D. Multi-LZerD: Multiple protein docking for asymmetric complexes. Proteins: Structure, Function, and Bioinformatics. 2012;80(7):1818–1833. doi: 10.1002/prot.24079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends in Genetics. 1994;10(8):286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, … Mitchell AL. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45(D1):D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, … Bateman A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–285. doi: 10.1093/nar/gkv1344Fraser. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HB, Hirsh AE, Wall DP, Eisen MB. Coevolution of gene expression among interacting proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(24):9033–9038. doi: 10.1073/pnas.0402591101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujibuchi W, Ogata H, Matsuda H, Kanehisa M. Automatic detection of conserved gene clusters in multiple genomes by graph comparison and P-quasi grouping. Nucleic acids research. 2000;28(20):4029–4036. doi: 10.1093/nar/28.20.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Hernandez M, Berardini TZ, Chen G, Crist D, Doyle A, Huala E, … Zhang P. TAIR: a resource for integrated Arabidopsis data. Funct Integr Genomics. 2002;2(6):239–253. doi: 10.1007/s10142-002-0077-z. [DOI] [PubMed] [Google Scholar]
- Ge H, Liu Z, Church GM, Vidal M. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nature genetics. 2001;29(4):482–486. doi: 10.1038/ng776. [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, O’Toole N, Ammar R, Provart NJ, Millar AH, Geisler M. A predicted interactome for Arabidopsis. Plant Physiology. 2007;145(2):317–329. doi: 10.1104/pp.107.103465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng C, Narasimhan S, Rodrigues JP, Bonvin AM. Information-Driven, Ensemble Flexible Peptide Docking Using HADDOCK. Modeling Peptide-Protein Interactions: Methods and Protocols. 2017:109–138. doi: 10.1007/978-1-4939-6798-8_8. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nature reviews Molecular cell biology. 2007;8(8):645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- Goh CS, Bogan AA, Joachimiak M, Walther D, Cohen FE. Co-evolution of proteins with their interaction partners. Journal of molecular biology. 2000;299(2):283–293. doi: 10.1006/jmbi.2000.3732. [DOI] [PubMed] [Google Scholar]
- Goh CS, Cohen FE. Co-evolutionary analysis reveals insights into protein–protein interactions. Journal of molecular biology. 2002;324(1):177–192. doi: 10.1016/s0022-2836(02)01038-0. [DOI] [PubMed] [Google Scholar]
- Grigoriev A. A relationship between gene expression and protein interactions on the proteome scale: analysis of the bacteriophage T7 and the yeast Saccharomyces cerevisiae. Nucleic acids research. 2001;29(17):3513–3519. doi: 10.1093/nar/29.17.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Zhu P, Jiao Y, Meng Y, Chen M. PRIN: a predicted rice interactome network. BMC bioinformatics. 2011;12(1):161. doi: 10.1186/1471-2105-12-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, He K, Liu D, Bai S, Gu X, Wei L, Luo J. DATF: a database of Arabidopsis transcription factors. Bioinformatics. 2005;21(10):2568. doi: 10.1093/bioinformatics/bti334. [DOI] [PubMed] [Google Scholar]
- Guo X, Liu R, Shriver CD, Hu H, Liebman MN. Assessing semantic similarity measures for the characterization of human regulatory pathways. Bioinformatics. 2006;22(8):967–973. doi: 10.1093/bioinformatics/btl042. [DOI] [PubMed] [Google Scholar]
- Guo Y, Yu L, Wen Z, Li M. Using support vector machine combined with auto covariance to predict protein–protein interactions from protein sequences. Nucleic acids research. 2008;36(9):3025–3030. doi: 10.1093/nar/gkn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha K, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, … Cenaj O. A protein complex network of Drosophila melanogaster. Cell. 2011;147(3):690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins T, Chitale M, Luban S, Kihara D. PFP: automated prediction of gene ontology functional annotations with confidence scores using protein sequence data. Proteins: Struct, Funct, Bioinf. 2009;74 doi: 10.1002/prot.22172. [DOI] [PubMed] [Google Scholar]
- Hawkins T, Kihara D. Function prediction of uncharacterized proteins. Journal of bioinformatics and computational biology. 2007;5(01):1–30. doi: 10.1142/s0219720007002503. [DOI] [PubMed] [Google Scholar]
- Herman D, Ochoa D, Juan D, Lopez D, Valencia A, Pazos F. Selection of organisms for the co-evolution-based study of protein interactions. BMC bioinformatics. 2011;12(1):363. doi: 10.1186/1471-2105-12-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermjakob H, Montecchi-Palazzi L, Lewington C, Mudali S, Kerrien S, Orchard S, … Valencia A. IntAct: an open source molecular interaction database. Nucleic acids research. 2004;32(suppl 1):D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosur R, Peng J, Vinayagam A, Stelzl U, Xu J, Perrimon N, … Berger B. A computational framework for boosting confidence in high-throughput protein-protein interaction datasets. Genome biology. 2012;13(8):R76. doi: 10.1186/gb-2012-13-8-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Bader JS. Precision and recall estimates for two-hybrid screens. Bioinformatics. 2009;25(3):372–378. doi: 10.1093/bioinformatics/btn640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TW, Tien AC, Huang WS, Lee YCG, Peng CL, Tseng HH, … Huang CYF. POINT: a database for the prediction of protein–protein interactions based on the orthologous interactome. Bioinformatics. 2004;20(17):3273–3276. doi: 10.1093/bioinformatics/bth366. [DOI] [PubMed] [Google Scholar]
- Huynen MA, Snel B, von Mering C, Bork P. Function prediction and protein networks. Current opinion in cell biology. 2003;15(2):191–198. doi: 10.1016/s0955-0674(03)00009-7. [DOI] [PubMed] [Google Scholar]
- Jain S, Bader GD. An improved method for scoring protein-protein interactions using semantic similarity within the gene ontology. BMC bioinformatics. 2010;11(1):562. doi: 10.1186/1471-2105-11-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Bussemaker HJ, Gerstein M. Revisiting the codon adaptation index from a whole-genome perspective: analyzing the relationship between gene expression and codon occurrence in yeast using a variety of models. Nucleic acids research. 2003;31(8):2242–2251. doi: 10.1093/nar/gkg306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Greenbaum D, Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome research. 2002;12(1):37–46. doi: 10.1101/gr.205602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan D, Pazos F, Valencia A. High-confidence prediction of global interactomes based on genome-wide coevolutionary networks. Proceedings of the National Academy of Sciences. 2008;105(3):934–939. doi: 10.1073/pnas.0709671105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann MG, Shoemaker BA, Panchenko AR, Przytycka TM. Correlated evolution of interacting proteins: looking behind the mirrortree. Journal of molecular biology. 2009;385(1):91–98. doi: 10.1016/j.jmb.2008.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenworthy AK. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods. 2001;24(3):289–296. doi: 10.1006/meth.2001.1189. [DOI] [PubMed] [Google Scholar]
- Keseler IM, Mackie A, Santos-Zavaleta A, Billington R, Bonavides-Martínez C, Caspi R, … Krummenacker M. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Research. 2016:gkw1003. doi: 10.1093/nar/gkw1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara D, Kanehisa M. Tandem clusters of membrane proteins in complete genome sequences. Genome research. 2000;10(6):731–743. doi: 10.1101/gr.10.6.731. [DOI] [PubMed] [Google Scholar]
- Kihara D, Skolnick J. Microbial genomes have over 72% structure assignment by the threading algorithm PROSPECTOR_Q. Proteins: Structure, Function, and Bioinformatics. 2004;55(2):464–473. doi: 10.1002/prot.20044. [DOI] [PubMed] [Google Scholar]
- Kim WK, Park J, Suh JK. Large scale statistical prediction of protein-protein interaction by potentially interacting domain (PID) pair. Genome Informatics Series. 2002:42–50. [PubMed] [Google Scholar]
- Kobe B, Guncar G, Buchholz R, Huber T, Maco B, Cowieson N, … Forwood JK. Crystallography and protein–protein interactions: biological interfaces and crystal contacts. Portland Press Limited; 2008. [DOI] [PubMed] [Google Scholar]
- Kotlyar M, Pastrello C, Pivetta F, Lo Sardo A, Cumbaa C, Li H, … Jurisica I. In silico prediction of physical protein interactions and characterization of interactome orphans. Nat Methods. 2015;12(1):79–84. doi: 10.1038/nmeth.3178. [DOI] [PubMed] [Google Scholar]
- Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, … Vajda S. The ClusPro web server for protein-protein docking. Nature Protocols. 2017;12(2):255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchaiev O, Rašajski M, Higham DJ, Pržulj N. Geometric de-noising of protein-protein interaction networks. PLoS Comput Biol. 2009;5(8):e1000454. doi: 10.1371/journal.pcbi.1000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SA, Chan C-h, Tsai CH, Lai JM, Wang FS, Kao CY, Huang CYF. Ortholog-based protein-protein interaction prediction and its application to inter-species interactions. BMC bioinformatics. 2008;9(12):S11. doi: 10.1186/1471-2105-9-S12-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C, Ruan J. A random walk based approach for improving protein-protein interaction network and protein complex prediction. Paper presented at the Bioinformatics and Biomedicine (BIBM), 2012 IEEE International Conference on.2012. [Google Scholar]
- Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, … Cesareni G. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40(Database issue):D857–861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. An information-theoretic definition of similarity. Paper presented at the ICML.1998. [Google Scholar]
- Lin M, Shen X, Chen X. PAIR: the predicted Arabidopsis interactome resource. Nucleic acids research. 2011;39(suppl 1):D1134–D1140. doi: 10.1093/nar/gkq938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TW, Wu JW, Chang DTH. Combining phylogenetic profiling-based and machine learning-based techniques to predict functional related proteins. PloS one. 2013;8(9):e75940. doi: 10.1371/journal.pone.0075940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Li J, Wong L. Assessing and predicting protein interactions using both local and global network topological metrics. Genome Informatics. 2008;21:138–149. [PubMed] [Google Scholar]
- Liu X, Liu B, Huang Z, Shi T, Chen Y, Zhang J. SPPS: a sequence-based method for predicting probability of protein-protein interaction partners. PloS one. 2012;7(1):e30938. doi: 10.1371/journal.pone.0030938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z-P, Chen L. Proteome-wide prediction of protein-protein interactions from high-throughput data. Protein & cell. 2012:1–13. doi: 10.1007/s13238-012-2945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyskov S, Gray JJ. The RosettaDock server for local protein-protein docking. [Research Support, N.I.H., Extramural] Nucleic Acids Res. 2008;36(Web Server issue):W233–238. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetschke SR, Simonsen M, Davis MJ, Ragan MA. Gene Ontology-driven inference of protein–protein interactions using inducers. Bioinformatics. 2012;28(1):69–75. doi: 10.1093/bioinformatics/btr610. [DOI] [PubMed] [Google Scholar]
- Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285(5428):751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- Martin S, Roe D, Faulon JL. Predicting protein–protein interactions using signature products. Bioinformatics. 2005;21(2):218–226. doi: 10.1093/bioinformatics/bth483. [DOI] [PubMed] [Google Scholar]
- Matthews LR, Vaglio P, Reboul J, Ge H, Davis BP, Garrels J, … Vidal M. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs”. Genome research. 2001;11(12):2120–2126. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewes HW, Amid C, Arnold R, Frishman D, Güldener U, Mannhaupt G, … Stümpflen V. MIPS: analysis and annotation of proteins from whole genomes. Nucleic acids research. 2004;32(suppl 1):D41–D44. doi: 10.1093/nar/gkh092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JP, Lo RS, Ben-Hur A, Desmarais C, Stagljar I, Noble WS, Fields S. Large-scale identification of yeast integral membrane protein interactions. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(34):12123–12128. doi: 10.1073/pnas.0505482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello C, Wallner B. InterPred: a pipeline to identify and model protein-protein interactions. Proteins. 2017 doi: 10.1002/prot.25280. [DOI] [PubMed] [Google Scholar]
- Morris JH, Knudsen GM, Verschueren E, Johnson JR, Cimermancic P, Greninger AL, Pico AR. Affinity purification–mass spectrometry and network analysis to understand protein-protein interactions. Nature protocols. 2014;9(11):2539–2554. doi: 10.1038/nprot.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca R, Pons T, Céol A, Valencia A, Aloy P. Towards a detailed atlas of protein–protein interactions. Current opinion in structural biology. 2013;23(6):929–940. doi: 10.1016/j.sbi.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Najafabadi HS, Salavati R. Sequence-based prediction of protein-protein interactions by means of codon usage. Genome biology. 2008;9(5):R87. doi: 10.1186/gb-2008-9-5-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanni L. Fusion of classifiers for predicting protein–protein interactions. Neurocomputing. 2005;68:289–296. [Google Scholar]
- NCBI RC. Database resources of the National Center for Biotechnology Information. Nucleic acids research. 2016;44(D1):D7. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolovska-Coleska Z. Studying protein-protein interactions using surface plasmon resonance. Protein-Protein Interactions: Methods and Applications. 2015:109–138. doi: 10.1007/978-1-4939-2425-7_7. [DOI] [PubMed] [Google Scholar]
- Ohue M, Matsuzaki Y, Uchikoga N, Ishida T, Akiyama Y. MEGADOCK: an all-to-all protein-protein interaction prediction system using tertiary structure data. Protein and peptide letters. 2014;21(8):766–778. doi: 10.2174/09298665113209990050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura Y, Aoki Y, Obayashi T, Tadaka S, Ito S, Narise T, Kinoshita K. COXPRESdb in 2015: coexpression database for animal species by DNA-microarray and RNAseq-based expression data with multiple quality assessment systems. Nucleic acids research. 2014:gku1163. doi: 10.1093/nar/gku1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Fonstein M, D’souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proceedings of the National Academy of Sciences. 1999;96(6):2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos F, Valencia A. Similarity of phylogenetic trees as indicator of protein–protein interaction. Protein engineering. 2001;14(9):609–614. doi: 10.1093/protein/14.9.609. [DOI] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(8):4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson LX, Roy A, Christoffer C, Terashi G, Kihara D. Modeling disordered protein interactions from biophysical principles. PLOS Computational Biology. 2017;13(4):e1005485. doi: 10.1371/journal.pcbi.1005485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehler J. New methodologies for measuring protein interactions in vivo and in vitro. Current opinion in structural biology. 2005;15(1):4–14. doi: 10.1016/j.sbi.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Pieper U, Eswar N, Davis FP, Braberg H, Madhusudhan MS, Rossi A, … Eramian D. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic acids research. 2006;34(suppl 1):D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30(12):1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitre S, Dehne F, Chan A, Cheetham J, Duong A, Emili A, … Krogan N. PIPE: a protein-protein interaction prediction engine based on the re-occurring short polypeptide sequences between known interacting protein pairs. BMC bioinformatics. 2006;7(1):365. doi: 10.1186/1471-2105-7-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad TK, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, … Venugopal A. Human protein reference database—2009 update. Nucleic acids research. 2009;37(suppl 1):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic acids research. 2005;33(suppl 1):D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Bar-Joseph Z, Klein-Seetharaman J. Evaluation of different biological data and computational classification methods for use in protein interaction prediction. Proteins: Structure, Function, and Bioinformatics. 2006;63(3):490–500. doi: 10.1002/prot.20865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopala SV, Sikorski P, Kumar A, Mosca R, Vlasblom J, Arnold R, … Ceol A. The binary protein-protein interaction landscape of Escherichia coli. Nature biotechnology. 2014;32(3):285–290. doi: 10.1038/nbt.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani AK, Marcotte EM. Exploiting the co-evolution of interacting proteins to discover interaction specificity. Journal of molecular biology. 2003;327(1):273–284. doi: 10.1016/s0022-2836(03)00114-1. [DOI] [PubMed] [Google Scholar]
- Rao VS, Srinivas K, Sujini G, Kumar G. Protein-protein interaction detection: methods and analysis. International journal of proteomics, 2014. 2014 doi: 10.1155/2014/147648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnik P. Using information content to evaluate semantic similarity in a taxonomy. arXiv preprint cmp-lg/9511007 1995 [Google Scholar]
- Ritchie DW, Kemp GJ. Protein docking using spherical polar Fourier correlations. Proteins: Structure, Function, and Bioinformatics. 2000;39(2):178–194. [PubMed] [Google Scholar]
- Rose PW, Prlic A, Altunkaya A, Bi C, Bradley AR, Christie CH, … Burley SK. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017;45(D1):D271–D281. doi: 10.1093/nar/gkw1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, … Ayivi-Guedehoussou N. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437(7062):1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Sato T, Yamanishi Y, Horimoto K, Kanehisa M, Toh H. Partial correlation coefficient between distance matrices as a new indicator of protein–protein interactions. Bioinformatics. 2006;22(20):2488–2492. doi: 10.1093/bioinformatics/btl419. [DOI] [PubMed] [Google Scholar]
- Sato T, Yamanishi Y, Kanehisa M, Toh H. The inference of protein–protein interactions by co-evolutionary analysis is improved by excluding the information about the phylogenetic relationships. Bioinformatics. 2005;21(17):3482–3489. doi: 10.1093/bioinformatics/bti564. [DOI] [PubMed] [Google Scholar]
- Schlicker A, Domingues F, Rahnenführer J, Lengauer T. A new measure for functional similarity of gene products based on gene ontology. BMC Bioinf. 2006;7 doi: 10.1186/1471-2105-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MS, Barton GJ. Probabilistic prediction and ranking of human protein-protein interactions. BMC bioinformatics. 2007;8(1):239. doi: 10.1186/1471-2105-8-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebriiskii IG, Golemis EA. Two-Hybrid System and False Positives: Approahes to Detection and Elimination. Two-Hybrid Systems: Methods and Protocols. 2001:123–134. doi: 10.1385/1-59259-210-4:123. [DOI] [PubMed] [Google Scholar]
- Sharma A. Computational gene expression profiling under salt stress reveals patterns of co-expression. Genomics data. 2016;7:214–221. doi: 10.1016/j.gdata.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Zhang J, Luo X, Zhu W, Yu K, Chen K, … Jiang H. Predicting protein–protein interactions based only on sequences information. Proceedings of the National Academy of Sciences. 2007;104(11):4337–4341. doi: 10.1073/pnas.0607879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38(Database issue):D161–166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrabanek L, Saini HK, Bader GD, Enright AJ. Computational prediction of protein–protein interactions. Molecular biotechnology. 2008;38(1):1–17. doi: 10.1007/s12033-007-0069-2. [DOI] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen M. Genome evolution. Trends in genetics. 2000;16(1):9–10. doi: 10.1016/s0168-9525(99)01924-1. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Ostlund G. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015;43(Database issue):D234–239. doi: 10.1093/nar/gku1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soong T-t, Wrzeszczynski KO, Rost B. Physical protein–protein interactions predicted from microarrays. Bioinformatics. 2008;24(22):2608–2614. doi: 10.1093/bioinformatics/btn498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzak E, Margalit H. Correlated sequence-signatures as markers of protein-protein interaction. Journal of molecular biology. 2001;311(4):681–692. doi: 10.1006/jmbi.2001.4920. [DOI] [PubMed] [Google Scholar]
- Stumpf MP, Thorne T, de Silva E, Stewart R, An HJ, Lappe M, Wiuf C. Estimating the size of the human interactome. Proceedings of the National Academy of Sciences. 2008;105(19):6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Li Y, Zhao Z. Phylogenetic profiles for the prediction of protein-protein interactions: how to select reference organisms? Biochem Biophys Res Commun. 2007;353(4):985–991. doi: 10.1016/j.bbrc.2006.12.146. [DOI] [PubMed] [Google Scholar]
- Sun J, Xu J, Liu Z, Liu Q, Zhao A, Shi T, Li Y. Refined phylogenetic profiles method for predicting protein–protein interactions. Bioinformatics. 2005;21(16):3409–3415. doi: 10.1093/bioinformatics/bti532. [DOI] [PubMed] [Google Scholar]
- Suyama M, Bork P. Evolution of prokaryotic gene order: genome rearrangements in closely related species. Trends in Genetics. 2001;17(1):10–13. doi: 10.1016/s0168-9525(00)02159-4. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, … Tsafou KP. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic acids research. 2014:gku1003. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, … von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghipour S, Zarrineh P, Ganjtabesh M, Nowzari-Dalini A. Improving protein complex prediction by reconstructing a high-confidence protein-protein interaction network of Escherichia coli from different physical interaction data sources. BMC Bioinformatics. 2017;18(1):10. doi: 10.1186/s12859-016-1422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamames J, Casari G, Ouzounis C, Valencia A. Conserved clusters of functionally related genes in two bacterial genomes. Journal of molecular evolution. 1997;44(1):66–73. doi: 10.1007/pl00006122. [DOI] [PubMed] [Google Scholar]
- Tanabe M, Kanehisa M. Using the KEGG database resource. Curr Protoc Bioinformatics. 2012;Chapter 1(Unit1 12) doi: 10.1002/0471250953.bi0112s38. [DOI] [PubMed] [Google Scholar]
- Tong Z, Gao Z, Wang F, Zhou J, Zhang Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC molecular biology. 2009;10(1):71. doi: 10.1186/1471-2199-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchala M, Bates PA. Predicting the structure of protein–protein complexes using the SwarmDock web server. Protein Structure Prediction. 2014:181–197. doi: 10.1007/978-1-4939-0366-5_13. [DOI] [PubMed] [Google Scholar]
- Tovchigrechko A, Vakser IA. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006;34(Web Server issue):W310–314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoka S, Ouzounis CA. Prediction of protein interactions: metabolic enzymes are frequently involved in gene fusion. Nature Genetics. 2000;26(2):141–142. doi: 10.1038/79847. [DOI] [PubMed] [Google Scholar]
- Tuncbag N, Gursoy A, Nussinov R, Keskin O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nature protocols. 2011;6(9):1341–1354. doi: 10.1038/nprot.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia A, Pazos F. Computational methods for the prediction of protein interactions. Curr Opin Struct Biol. 2002;12(3):368–373. doi: 10.1016/s0959-440x(02)00333-0. [DOI] [PubMed] [Google Scholar]
- Venkatraman V, Yang YD, Sael L, Kihara D. Protein-protein docking using region-based 3D Zernike descriptors. BMC bioinformatics. 2009;10(1):407. doi: 10.1186/1471-2105-10-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova O, Qin J. NMR of Proteins and Small Biomolecules. Springer; 2011. NMR as a unique tool in assessment and complex determination of weak protein–protein interactions; pp. 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJ, Boulton SJ, Vidal M. Yeast two-hybrid systems and protein interaction mapping projects for yeast and worm. Yeast. 2000;17(2):88–94. doi: 10.1002/1097-0061(20000630)17:2<88::AID-YEA20>3.0.CO;2-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, … Vidal M. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287(5450):116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- Wang F, Liu M, Song B, Li D, Pei H, Guo Y, … Zhang D. Prediction and characterization of protein-protein interaction networks in swine. Proteome science. 2012;10(1):2. doi: 10.1186/1477-5956-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wass MN, Fuentes G, Pons C, Pazos F, Valencia A. Towards the prediction of protein interaction partners using physical docking. Molecular systems biology. 2011;7(1):469. doi: 10.1038/msb.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Tegenfeldt F, Li J, Zdobnov EM, Kriventseva EV. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic acids research. 2013;41(D1):D358–D365. doi: 10.1093/nar/gks1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Xing P, Zeng J, Chen J, Su R, Guo F. Improved prediction of protein-protein interactions using novel negative samples, features, and an ensemble classifier. Artif Intell Med. 2017 doi: 10.1016/j.artmed.2017.03.001. [DOI] [PubMed] [Google Scholar]
- Wei Q, Khan IK, Ding Z, Yerneni S, Kihara D. NaviGO: interactive tool for visualization and functional similarity and coherence analysis with gene ontology. [journal article] BMC Bioinformatics. 2017;18(1):177. doi: 10.1186/s12859-017-1600-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetie N, Armand G, Sokolowska I, Woods AG, Roy U, Loo JA, Darie CC. Investigation of stable and transient protein–protein interactions: past, present, and future. Proteomics. 2013;13(3–4):538–557. doi: 10.1002/pmic.201200328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L, You Z-H, Li S, Huang Y-A, Liu G. Detection of protein-protein interactions from amino acid sequences using a rotation forest model with a novel PR-LPQ descriptor. Paper presented at the International Conference on Intelligent Computing.2015. [Google Scholar]
- Wong P, Althammer S, Hildebrand A, Kirschner A, Pagel P, Geissler B, … Schmidt T. An evolutionary and structural characterization of mammalian protein complex organization. Bmc Genomics. 2008;9(1):629. doi: 10.1186/1471-2164-9-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Pang E, Lin K, Pei ZM. Improving the measurement of semantic similarity between gene ontology terms and gene products: insights from an edge-and IC-based hybrid method. PloS one. 2013;8(5):e66745. doi: 10.1371/journal.pone.0066745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenarios I, Salwinski L, Duan XJ, Higney P, Kim SM, Eisenberg D. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic acids research. 2002;30(1):303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Li G, Zhao C, Li Y, Li P, Cui J, … Shi T. Global protein interactome exploration through mining genome-scale data in Arabidopsis thaliana. BMC genomics. 2010;11(2):S2. doi: 10.1186/1471-2164-11-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Derti A, DeLisi C. Genes linked by fusion events are generally of the same functional category: a systematic analysis of 30 microbial genomes. Proceedings of the National Academy of Sciences. 2001;98(14):7940–7945. doi: 10.1073/pnas.141236298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Developmental cell. 2011;20(4):483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xia JF, Gui J. Prediction of protein-protein interactions from protein sequence using local descriptors. Protein and Peptide Letters. 2010;17(9):1085–1090. doi: 10.2174/092986610791760306. [DOI] [PubMed] [Google Scholar]
- Yerneni S, Khan I, Wei Q, Kihara D. IAS: Interaction specific GO term associations for predicting Protein-Protein Interaction Networks. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2015 doi: 10.1109/TCBB.2015.2476809. [DOI] [PubMed] [Google Scholar]
- You ZH, Chan KC, Hu P. Predicting protein-protein interactions from primary protein sequences using a novel multi-scale local feature representation scheme and the random forest. PLoS One. 2015;10(5):e0125811. doi: 10.1371/journal.pone.0125811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You ZH, Lei YK, Zhu L, Xia J, Wang B. Prediction of protein-protein interactions from amino acid sequences with ensemble extreme learning machines and principal component analysis. BMC bioinformatics. 2013;14(8):S10. doi: 10.1186/1471-2105-14-S8-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Chou LC, Chang DTH. Predicting protein-protein interactions in unbalanced data using the primary structure of proteins. BMC bioinformatics. 2010;11(1):167. doi: 10.1186/1471-2105-11-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Paccanaro A, Trifonov V, Gerstein M. Predicting interactions in protein networks by completing defective cliques. Bioinformatics. 2006;22(7):823–829. doi: 10.1093/bioinformatics/btl014. [DOI] [PubMed] [Google Scholar]
- Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, … Hunter T. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490(7421):556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SB, Tang QR. Protein–protein interaction inference based on semantic similarity of Gene Ontology terms. Journal of theoretical biology. 2016;401:30–37. doi: 10.1016/j.jtbi.2016.04.020. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhou YS, He F, Song J, Zhang Z. Can simple codon pair usage predict protein–protein interaction? Molecular BioSystems. 2012;8(5):1396–1404. doi: 10.1039/c2mb05427b. [DOI] [PubMed] [Google Scholar]
- Zhou YZ, Gao Y, Zheng YY. Advances in Computer Science and Education Applications. Springer; 2011. Prediction of protein-protein interactions using local description of amino acid sequence; pp. 254–262. [Google Scholar]
- Zuiderweg ER. Mapping protein− protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41(1):1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]