Abstract
Peel colour is an important factor affecting the marketability of pomegranate fruits. Therefore, elucidating the genetic mechanism of fruit peel colour development may be useful for breeding pomegranate cultivars with enhanced fruit peel colours. In this study, we combined an iTRAQ-based proteome-level analysis with an RNA sequencing-based transcriptome-level analysis to detect the proteins and genes related to fruit peel colour development in pomegranate. We analysed the ‘Tunisia’ (red fruit) and ‘White’ (white fruit) pomegranate cultivars at two stages of fruit development. A total of 27 differentially abundant proteins (increased abundance) and 54 differentially expressed genes (16 up-regulated and 38 down-regulated) were identified from our proteomics and transcriptomics data. The identified proteins and genes contribute to pomegranate fruit peel colour by participating in the biosynthesis of anthocyanins, stilbenoids, diarylheptanoids, gingerols, flavonoids, and phenylpropanoids. Several candidate proteins and genes corresponded to enzymes related to general reactions (PAL, 4CL, DFR, LDOX/ANS, CHS, and F3′5′H) and glycosylation (GT1 and UGAT) of compounds and pigments related to the colour of pomegranate fruit peel. Complementary proteome- and transcriptome-level analyses revealed a complex molecular network controlling fruit peel colour. The candidate genes identified in this study may be useful for the marker-based breeding of new pomegranate cultivars.
Introduction
Pomegranate originated in Central Asia, including Iran, Afghanistan, and Caucasia, and is one of the oldest cultivated fruits. Pomegranate trees were introduced into China approximately 2000 years ago1, and their fruits are valued for their beauty and their desirable flavour, colour, and health benefits2. The peel colour of pomegranate fruits is a major factor affecting consumer acceptance and marketability. Thus, one of the main objectives of pomegranate breeders is to enhance fruit peel colour.
The biological and genetic factors regulating fruit peel colour have been studied extensively. A previous study revealed that storage temperature is the main factor affecting longkong fruit peel colour and physiology3. Light can also affect fruit colour, for example, light stimulates the synthesis of carotenoids in citrus fruit peel, resulting in the accumulation of β-cryptoxanthin during a specific stage of fruit development4. Changes in the carotenoid content of citrus fruit peel during fruit-colouring stages are significantly correlated with variations in peel colour5. In grape, a lack of light accelerates chlorophyll breakdown and induces carotenoid accumulation, resulting in intensely coloured fruits6. The development of peach fruit colour could be enhanced by bagging fruits in white non-woven polypropylene bags7. Researchers have also shown that light-responsive transcriptional regulatory factors regulate the anthocyanin levels in bagged Chinese sand pears8. In apple, light induces the expression of a MYB transcription factor gene associated with anthocyanin biosynthesis, and its expression level is related to the final fruit skin colour9. However, the pear MYB transcription factor gene PcMYB10 (an ortholog of MdMYBa/MdMYB10, which controls the pigmentation of apple fruit skin) is not directly responsible for mediating fruit colour9. Thus, fruit peel colour is a complex trait that involves physiological, biochemical, and molecular processes. At present, little is known about the development of pomegranate fruit peel colour.
High-throughput technologies for measuring gene expression levels and protein abundance have enabled transcriptome- and proteome-level analyses of developmental processes, gene functions, adaptations, and physiological stress responses in plants10. The genes related to the biosynthesis of natural products in the pomegranate fruit peel have been investigated based on a de novo transcriptome set11, but a comprehensive examination of the encoded proteins has not been conducted. The isobaric tags for relative and absolute quantitation (iTRAQ) method represents a powerful option for analysing actively produced proteins. This method allows for the simultaneous identification and quantification of proteins from multiple samples based on isotope labelling combined with multidimensional liquid chromatography and tandem mass spectrometry (LC-MS/MS)12. Because of its high sensitivity and accuracy, the iTRAQ method has been used to elucidate post-transcriptional regulatory activities in mango13, oriental melon14, strawberry15, and peach16. We recently sequenced and assembled the pomegranate genome (unpublished data). In the present study, we combined iTRAQ-based proteomics and RNA sequencing-based transcriptomics analyses to identify the proteins and genes related to fruit peel colour during two fruit development stages in two pomegranate cultivars, ‘Tunisia’ and ‘White’. Then, the differentially abundant proteins (DAPs) and differentially expressed genes (DEGs) were subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. Candidate genes related to fruit peel colour were predicted based on genome annotation and metabolic pathway information. In this study, we comprehensively characterised the development of pomegranate fruit peel colour at the proteome and transcriptome levels. The identified candidate genes may be useful for molecular marker-assisted pomegranate breeding.
Results
Identification of DAPs using iTRAQ technology
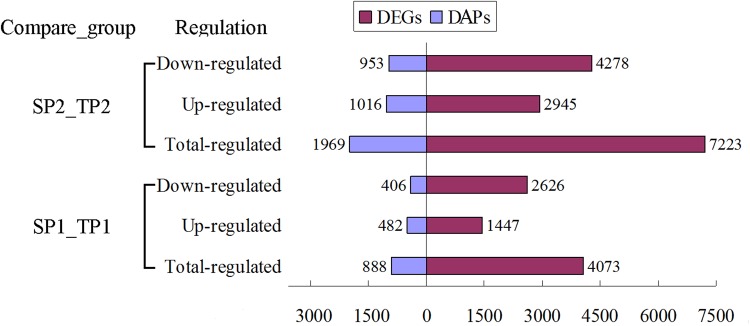
The DAPs in two groups during the fruit colouring (SP1_TP1) and ripening (SP2_TP2) stages were identified and quantified using iTRAQ and LC-MS/MS analysis. Accordingly, 237,421 spectra were generated, and 18,407 unique peptides and 5,357 proteins were identified with a false discovery rate (FDR) of ≤1% (Supplementary Table S1). Among these proteins, there were 888 and 1,969 DAPs in SP1_TP1 and SP2_TP2, respectively, with a fold-change >1.2 (mean value of all compared groups) or <0.85 and a P value (t-test of all comparison groups) of <0.05 (Fig. 1 and Table 1). In the comparison between SP1 and TP1 (SP1_TP1), 482 DAPs were more abundant and 406 DAPs were less abundant in TP1 than in SP1. Similarly, 1,016 DAPs were more abundant and 953 DAPs were less abundant in TP2 than in SP2. There were nearly twice as many DAPs in SP2_TP2 than in SP1_TP1. These results indicated that many proteins showed differential abundance between the two fruit development stages, with more DAPs at the second analysed stage.
Figure 1.
Differentially expressed genes and differentially abundant proteins in pomegranate fruit peels identified in different comparisons. SP1_TP1: comparison between ‘White’ and ‘Tunisia’ fruit peels during fruit colouring stage. SP2_TP2: comparison between ‘White’ and ‘Tunisia’ fruit peels during fruit ripening period.
Table 1.
Summary of proteins and transcripts detected from iTRAQ and RNA sequencing data.
| Proteins | mRNAs | |||
|---|---|---|---|---|
| SP1_TP1 | SP2_TP2 | SP1_TP1 | SP2_TP2 | |
| Unique proteins\genes detected | 5357 | 5357 | 18918 | 20545 |
| Significantly DAPs\DEGs | 888 | 1969 | 4073 | 7223 |
| Up-regulated | 482 | 1016 | 1447 | 2945 |
| Down-regulated | 406 | 953 | 2626 | 4278 |
| Shared proteins/genes | 552 | 1684 | ||
| Shared proteins/genes (up-regulated) | 311 | 304 | 655 | 636 |
| Shared proteins/genes (down-regulated) | 241 | 248 | 1029 | 1048 |
Transcriptional analysis of DEGs
To verify transcriptional-level changes, four cDNA libraries (i.e., SP1, SP2, TP1, and TP2) were constructed using total RNA extracted from the fruit peels of the two pomegranate cultivars. A total of 64.85, 61.99, 59.99, and 67.46 million raw sequence reads were generated from the SP1, SP2, TP1, and TP2 libraries, respectively (Supplementary Table S1). After removing low-quality reads and adaptor sequences, 37.43, 35.63, 34.68, and 38.76 million clean reads were obtained for SP1, SP2, TP1, and TP2, respectively, with 45.23%, 44.06%, 42.25%, and 41.89% of the reads mapped to the reference genome, respectively. The proportion of unique reads that could be aligned to the genome sequence ranged from 33.33% for TP2 to 37.34% for SP1. Finally, 18,918 and 20,545 unique genes were detected in SP1_TP1 and SP2_TP2, respectively. Compared with the SP group, the TP group had 4,073 (1,447 up-regulated and 2,626 down-regulated) significant DEGs (i.e., |log2(TP/SP)| > 1 and P ≤ 0.001) at the fruit colouring stage, and 7,223 (2,945 up-regulated and 4,278 down-regulated) significant DEGs at the ripening stage (Fig. 1 and Table 1). These results suggested that many genes were differentially expressed between the two fruit development stages in two pomegranate cultivars, with more DEGs at the fruit ripening stage.
Comparative analysis between protein abundance and gene expression levels
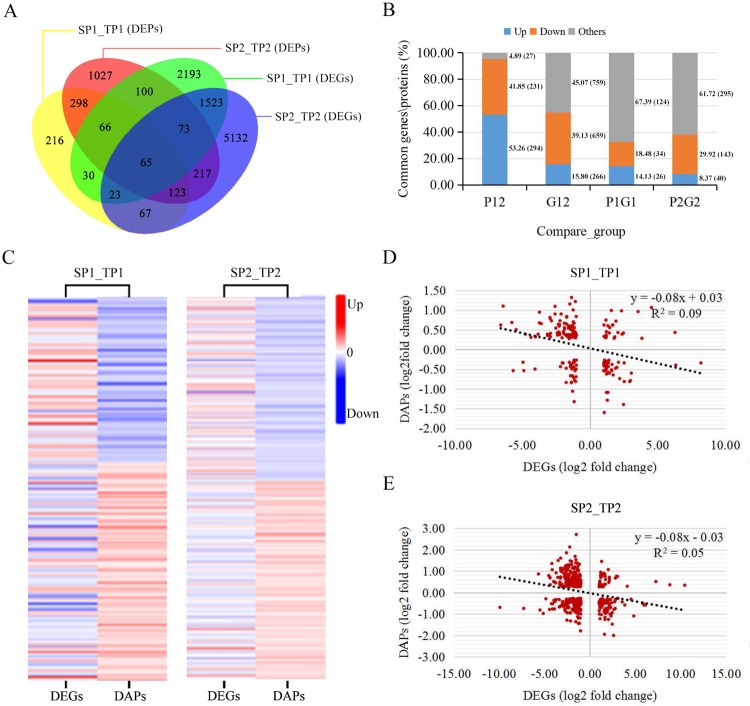
To evaluate the relationship between transcript levels and protein abundance, we compared the DAPs and DEGs between the two analysed fruit development stages. A total of 552 shared DAPs and 1,684 shared DEGs were identified during the comparison between SP1_TP1 and SP2_TP2 (Fig. 2A and Table 1). Among the shared DAPs, 311 were more abundant and 241 were less abundant in SP1_TP1 than in SP2_TP2, while 304 were more abundant and 248 were less abundant in SP2_TP2 than in SP1_TP1 (Fig. 2B). Of the common DEGs, 655 were up-regulated and 1,029 were down-regulated in SP1_TP1, whereas 636 were up-regulated and 1,048 were down-regulated in SP2_TP2 (Fig. 2B). Furthermore, 184 and 478 DAPs and their corresponding DEGs were identified in SP1_TP1 and SP2_TP2, respectively (Fig. 2C). Of these, only 60 DAPs (26 with increased abundance and 34 with decreased abundance) and 183 DAPs (40 with increased abundance and 143 with decreased abundance) were regulated in the same direction as their corresponding DEGs in SP1_TP1 and SP2_TP2, respectively (Fig. 2B,C). There were more DEGs than DAPs in both SP1_TP1 and SP2_TP2, with considerable differences between the trends in transcript levels and the trends in protein abundance. To analyse the consistency between transcriptomic and proteomic changes during the development of fruit peel colour, we conducted a correlation analysis using the quantitative data for DAPs and DEGs (Fig. 2D,E; Supplementary Table S2). Pearson’s correlation tests indicated that the fold-changes in the DAPs were significantly (P < 0.001) negatively correlated with the fold-changes in the corresponding DEGs (rpearson = −0.29 and −0.23 in SP1_TP1 and SP2_TP2, respectively). Thus, there was a poor correlation between transcript levels and protein abundance.
Figure 2.
Protein abundance and gene expression levels in different comparisons. (A) Venn diagram of proteins and genes. (B) Changes in the common DAPs and DEGs between groups. P12: common DAPs between SP1_TP1 and SP2_TP2; G12: common DEGs between SP1_TP1 and SP2_TP2; P1G1: SP1_TP1 genes in which associated transcript level and protein abundance exhibited opposite trends; P2G2: SP2_TP2 genes in which associated transcript level and protein abundance exhibited opposite trends. (C) Comparison of changes in transcript and protein levels of DEGs and DAPs. (D) Correlations between transcript levels and protein abundance of DEGs and DAPs in SP1_TP1. (E) Correlations between transcript levels and protein abundance of DEGs and DAPs in SP2_TP2.
KEGG pathway annotations for DAPs and DEGs
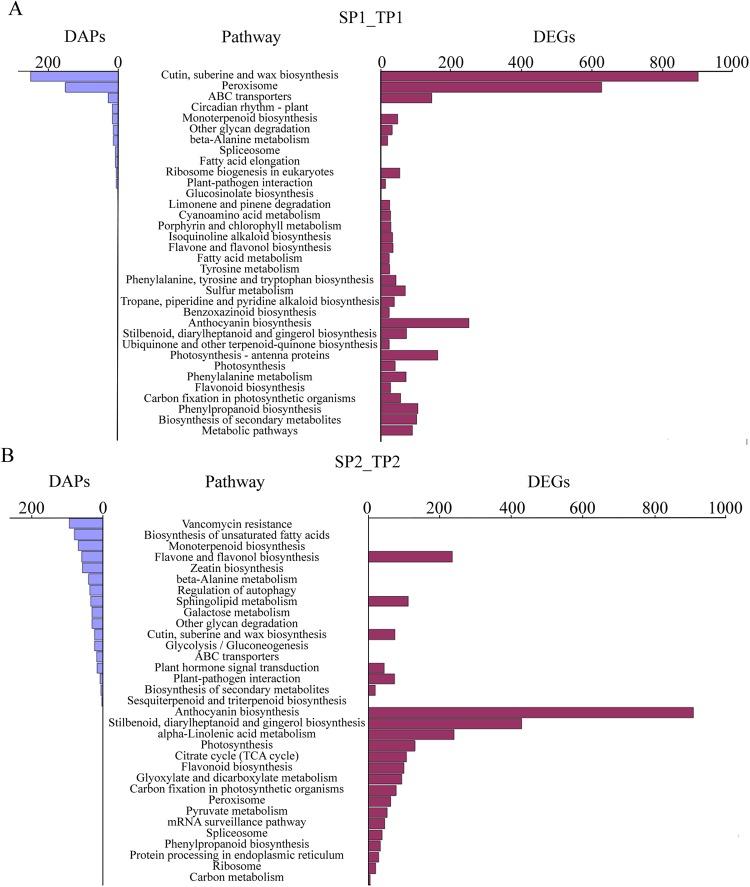
To annotate the functions of genes and proteins, we conducted a pathway enrichment analysis of the DAPs and DEGs in SP1_TP1 and SP2_TP2 based on the KEGG database (Fig. 3). In SP1_TP1, 521 DAPs were significantly enriched in 11 pathways (corrected P value < 0.05; Fig. 3A), while 3,181 DEGs were significantly enriched in 30 pathways (corrected P value < 0.05; Fig. 3B). The following pathways were enriched with both DAPs and DEGs in SP1_TP1: anthocyanin biosynthesis; stilbenoid, diarylheptanoid, and gingerol biosynthesis; photosynthesis; phenylalanine metabolism; flavonoid biosynthesis; phenylpropanoid biosynthesis; biosynthesis of secondary metabolites; and metabolic pathways. In SP2_TP2, 630 DAPs were significantly enriched in 14 pathways, while 2,936 DEGs were significantly enriched in 22 pathways (corrected P value < 0.05; Fig. 3). The following pathways were common to both the DAPs and DEGs in SP2_TP2: anthocyanin biosynthesis; stilbenoid, diarylheptanoid, and gingerol biosynthesis; photosynthesis; flavonoid biosynthesis; α-linolenic acid metabolism; and peroxisome and phenylpropanoid biosynthesis. An additional comparative analysis indicated that four pathways (i.e., anthocyanin biosynthesis; stilbenoid, diarylheptanoid, and gingerol biosynthesis; flavonoid biosynthesis; and phenylpropanoid biosynthesis) were common between SP1_TP1 and SP2_TP2. Thus, these four pathways were further investigated as candidate pathways related to the development of pomegranate fruit peel colour.
Figure 3.
KEGG pathways associated with differentially expressed genes and differentially abundant proteins in (A) SP1_TP1 and (B) SP2_TP2. Number of proteins (left) and number of genes (right) associated with a pathway are indicated.
Identification of DAPs and DEGs associated with candidate pathways
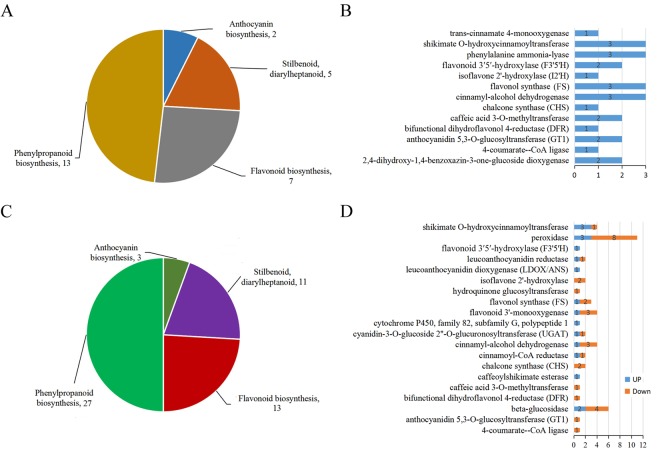
A total of 51 and 89 DAPs detected in SP1_TP1 and SP2_TP2, respectively, were associated with the four candidate pathways, with 34 DAPs common to both sample groups (Supplementary Table S3). Additionally, 241 and 397 DEGs identified in SP1_TP1 and SP2_TP2, respectively, were related to the four candidate pathways, including 100 DEGs common to both sample groups (Supplementary Table S4). After filtering out the DAPs and DEGs that were differentially regulated between SP1_TP1 and SP2_TP2, 27 DAPs with increasing abundance and 54 DEGs (16 up-regulated DEGs and 38 down-regulated DEGs) were annotated as being involved in anthocyanin biosynthesis; stilbenoid, diarylheptanoid, and gingerol biosynthesis; flavonoid biosynthesis; and phenylpropanoid biosynthesis (Fig. 4A,C; Tables 2 and 3). These DAPs and DEGs likely influence pomegranate fruit peel colour at the proteome and transcriptome levels. Except for Gglean022299.1, the DAPs represented 13 crucial enzymes (Fig. 4B). With the exception of Gglean025205.1, Gglean018889.1, and Gglean015246.1, the DEGs encoded 20 pivotal enzymes (Fig. 4D). Nine enzymes were affected by corresponding genes with differentially regulated trends at the transcriptome level. More DEGs than DAPs were involved in the formation of pomegranate fruit peel colour, and the DEGs showed complex differential regulation at the transcriptome level. Most of the genes encoding the DAPs were not included among the DEGs, consistent with the observed differences between the proteome and transcriptome data (Fig. 2B). Only four of the DEGs [i.e., two flavonoid biosynthesis genes (Gglean009951.1 and Gglean020650.1) and two phenylpropanoid biosynthesis genes (Gglean015001.1 and Gglean015003.1)] encoded DAPs. Further analyses revealed that the abundance of most of the flavonoid and phenylpropanoid biosynthesis enzymes increased but the expression levels of their corresponding genes decreased during fruit colour development.
Figure 4.
Summary of number of (A) differentially abundant proteins (DAPs) and (C) differentially expressed genes (DEGs) in each pathway. (B) Annotation and abundance changes of DAPs. (D) Annotation and expression level changes of DEGs. Numbers represent number of genes and proteins.
Table 2.
Pathways associated with candidate differentially abundant proteins (DAPs) between ‘Tunisia’ and ‘White’ pomegranate cultivars in TP1_SP1 and TP2_SP2 comparisons.
| Pathway | Common DAP | Regulation | KO id | Annotation [EC no.] | Fold (TP1/SP1) | P value | Fold (TP2/SP2) | P value |
|---|---|---|---|---|---|---|---|---|
| Anthocyanin biosynthesis | Gglean022014.1 | Up | K12938 | anthocyanidin 5,3-O-glucosyltransferase (GT1) [EC:2.4.1.-] | 1.46 | 0.03 | 1.38 | 0.00 |
| Gglean030887.1 | Up | K08237 | 1.59 | 0.05 | 1.36 | 0.00 | ||
| Stilbenoid, diarylheptanoid biosynthesis | Gglean022299.1 | Up | K00517 | — | 1.51 | 0.01 | 1.77 | 0.00 |
| Gglean017162.1 | Up | K13260 | isoflavone 2′-hydroxylase (I2,H) [EC:1.14.13.89] | 1.54 | 0.00 | 1.90 | 0.02 | |
| Gglean020115.1 | Up | K13065 | shikimate O-hydroxycinnamoyltransferase [EC:2.3.1.133] | 1.58 | 0.01 | 1.95 | 0.02 | |
| Gglean017092.1 | Up | 1.74 | 0.01 | 2.46 | 0.00 | |||
| Gglean030413.1 | Up | K00487 | trans-cinnamate 4-monooxygenase [EC:1.14.13.11] | 1.63 | 0.01 | 1.31 | 0.00 | |
| Flavonoid biosynthesis | Gglean024451.1 | Up | K13082 | bifunctional dihydroflavonol 4-reductase (DFR) [EC:1.1.1.219] | 1.86 | 0.04 | 1.96 | 0.00 |
| Gglean009951.1 | Up | K00660 | chalcone synthase (CHS) [EC:2.3.1.74] | 2.23 | 0.01 | 2.32 | 0.00 | |
| Gglean021885.1 | Up | K05278 | flavonol synthase (FS) [EC:1.14.11.23] | 1.57 | 0.00 | 1.76 | 0.03 | |
| Gglean002595.1 | Up | 1.21 | 0.04 | 1.40 | 0.02 | |||
| Gglean002270.1 | Up | 1.61 | 0.03 | 2.12 | 0.00 | |||
| Gglean024933.1 | Up | K00475 | flavonoid 3′5′-hydroxylase (F35H) [EC:1.14.11.9] | 2.08 | 0.01 | 2.41 | 0.00 | |
| Gglean020650.1 | Up | 2.33 | 0.00 | 2.16 | 0.00 | |||
| Phenylpropanoid biosynthesis | Gglean004667.1 | Up | K13229 | 2,4-dihydroxy-1,4-benzoxazin-3-one-glucoside dioxygenase [EC:1.14.20.2] | 1.64 | 0.01 | 1.42 | 0.03 |
| Gglean005438.1 | Up | 1.31 | 0.01 | 1.34 | 0.00 | |||
| Gglean008923.1 | Up | K01904 | 4-coumarate–CoA ligase [EC:6.2.1.12] | 1.24 | 0.01 | 2.22 | 0.00 | |
| Gglean004749.1 | Up | K01188 | beta-glucosidase [EC:3.2.1.21] | 1.25 | 0.01 | 1.30 | 0.01 | |
| Gglean030344.1 | Up | K13066 | caffeic acid 3-O-methyltransferase [EC:2.1.1.68] | 1.51 | 0.00 | 3.00 | 0.00 | |
| Gglean015152.1 | Up | 1.44 | 0.01 | 2.20 | 0.00 | |||
| Gglean015001.1 | Up | K00083 | cinnamyl-alcohol dehydrogenase [EC:1.1.1.195] | 1.46 | 0.01 | 1.78 | 0.00 | |
| Gglean015223.1 | Up | 1.26 | 0.01 | 1.29 | 0.03 | |||
| Gglean015003.1 | Up | 1.52 | 0.03 | 1.42 | 0.01 | |||
| Gglean023090.1 | Up | K10775 | phenylalanine ammonia-lyase [EC:4.3.1.24] | 1.60 | 0.02 | 1.37 | 0.00 | |
| Gglean026319.1 | Up | 1.98 | 0.01 | 1.41 | 0.00 | |||
| Gglean025624.1 | Up | 2.48 | 0.01 | 1.23 | 0.01 | |||
| Gglean017092.1 | Up | K13065 | shikimate O-hydroxycinnamoyltransferase [EC:2.3.1.133] | 1.74 | 0.01 | 1.73 | 0.00 |
Table 3.
Pathways associated with candidate differentially expressed genes (DEGs) between ‘Tunisia’ and ‘White’ pomegranate cultivars in TP1_SP1 and TP2_SP2 comparisons.
| pathway | Common DEGs | Regulation | KO id | Annotation [EC no.] | log2(TP1/SP1) | P value | log2(TP2/SP2) | P value |
|---|---|---|---|---|---|---|---|---|
| Anthocyanin biosynthesis | Gglean030299.1 | Down | K12938 | anthocyanidin 5,3-O-glucosyltransferase (GT1) [EC:2.4.1.-] | −1.39 | 0.00 | −1.54 | 0.00 |
| Gglean024784.1 | Down | K18823 | cyanidin-3-O-glucoside 2′′-O-glucuronosyltransferase (UGAT) [EC:2.4.1.254] | −3.84 | 0.00 | −1.92 | 0.00 | |
| Gglean024783.1 | Up | 1.20 | 0.00 | 1.66 | 0.00 | |||
| Stilbenoid, diarylheptanoid biosynthesis | Gglean025205.1 | Down | — | — | −6.44 | 0.00 | −2.28 | 0.00 |
| Gglean018889.1 | Up | — | — | 4.07 | 0.00 | 1.37 | 0.00 | |
| Gglean000381.1 | Up | K17961 | cytochrome P450, family 82, subfamily G, polypeptide 1 [EC:1.14.-.-] | 5.09 | 0.00 | 6.11 | 0.00 | |
| Gglean026629.1 | Up | K05280 | flavonoid 3′-monooxygenase [EC:1.14.13.21] | 1.32 | 0.00 | 1.64 | 0.00 | |
| Gglean019885.1 | Down | K13260 | isoflavone 2′-hydroxylase [EC:1.14.13.89] | −3.02 | 0.00 | −2.66 | 0.00 | |
| Gglean019884.1 | Down | −2.71 | 0.00 | −2.18 | 0.00 | |||
| Gglean015246.1 | Down | — | — | −2.53 | 0.00 | −1.81 | 0.00 | |
| Gglean015686.1 | Down | K13065 | shikimate O-hydroxycinnamoyltransferase [EC:2.3.1.133] | −3.22 | 0.00 | −1.77 | 0.00 | |
| Gglean020272.1 | Up | 1.96 | 0.00 | 2.92 | 0.00 | |||
| Gglean004433.1 | Up | 1.78 | 0.00 | 1.87 | 0.00 | |||
| Gglean013382.1 | Up | 1.19 | 0.00 | 1.07 | 0.00 | |||
| Flavonoid biosynthesis | Gglean024449.1 | Down | K13082 | bifunctional dihydroflavonol 4-reductase (DFR) [EC:1.1.1.219] | −1.10 | 0.00 | −3.98 | 0.00 |
| Gglean009951.1 | Down | K00660 | chalcone synthase (CHS) [EC:2.3.1.74] | −1.50 | 0.00 | −3.11 | 0.00 | |
| Gglean012297.1 | Down | −1.25 | 0.00 | −1.56 | 0.00 | |||
| Gglean025015.1 | Down | K05280 | flavonoid 3′-monooxygenase [EC:1.14.13.21] | −2.64 | 0.00 | −3.69 | 0.00 | |
| Gglean027001.1 | Down | −2.66 | 0.00 | −1.77 | 0.00 | |||
| Gglean000166.1 | Down | −2.59 | 0.00 | −1.32 | 0.00 | |||
| Gglean001689.1 | Up | K05278 | flavonol synthase (FS) [EC:1.14.11.23] | 1.49 | 0.00 | 2.12 | 0.00 | |
| Gglean002078.1 | Down | −2.69 | 0.00 | −2.19 | 0.00 | |||
| Gglean018019.1 | Down | −1.06 | 0.00 | −1.11 | 0.00 | |||
| Gglean026620.1 | Up | K05277 | leucoanthocyanidin dioxygenase (LDOX/ANS) [EC:1.14.11.19] | 2.57 | 0.00 | 4.83 | 0.00 | |
| Gglean024550.1 | Up | K13081 | leucoanthocyanidin reductase [EC:1.17.1.3] | 1.10 | 0.00 | 1.94 | 0.00 | |
| Gglean002570.1 | Down | −3.16 | 0.00 | −1.02 | 0.00 | |||
| Gglean020650.1 | Down | K00475 | flavonoid 3′5′-hydroxylase (F35H) [EC:1.14.11.9] | −1.09 | 0.00 | −3.18 | 0.00 | |
| Phenylpropanoid biosynthesis | Gglean004943.1 | Down | K01904 | 4-coumarate–CoA ligase [EC:6.2.1.12] | −1.37 | 0.00 | −2.12 | 0.00 |
| Gglean004745.1 | Up | K01188 | beta-glucosidase [EC:3.2.1.21] | 1.33 | 0.00 | 5.81 | 0.00 | |
| Gglean014679.1 | Down | −1.53 | 0.00 | −2.84 | 0.00 | |||
| Gglean025214.1 | Down | −7.85 | 0.00 | −2.79 | 0.00 | |||
| Gglean026132.1 | Down | −1.07 | 0.00 | −2.50 | 0.00 | |||
| Gglean028143.1 | Down | −2.66 | 0.00 | −1.88 | 0.00 | |||
| Gglean023909.1 | Up | 3.22 | 0.00 | 2.82 | 0.00 | |||
| Gglean015153.1 | Down | K13066 | caffeic acid 3-O-methyltransferase [EC:2.1.1.68] | −2.14 | 0.00 | −1.06 | 0.00 | |
| Gglean028708.1 | Up | K18368 | caffeoylshikimate esterase [EC:3.1.1.-] | 1.33 | 0.00 | 1.61 | 0.00 | |
| Gglean008959.1 | Down | K09753 | cinnamoyl-CoA reductase [EC:1.2.1.44] | −1.39 | 0.00 | −2.07 | 0.00 | |
| Gglean003647.1 | Up | 1.84 | 0.00 | 1.81 | 0.00 | |||
| Gglean015001.1 | Down | K00083 | cinnamyl-alcohol dehydrogenase [EC:1.1.1.195] | −1.60 | 0.00 | −3.79 | 0.00 | |
| Gglean015003.1 | Down | −1.59 | 0.00 | −4.39 | 0.00 | |||
| Gglean002744.1 | Down | −1.20 | 0.00 | −1.26 | 0.00 | |||
| Gglean017886.1 | Up | 3.65 | 0.00 | 1.87 | 0.00 | |||
| Gglean031042.1 | Down | K08237 | hydroquinone glucosyltransferase [EC:2.4.1.218] | −1.17 | 0.00 | −2.32 | 0.00 | |
| Gglean018745.1 | Down | K00430 | peroxidase [EC:1.11.1.7] | −1.15 | 0.00 | −2.60 | 0.00 | |
| Gglean004996.1 | Up | 1.61 | 0.00 | 1.14 | 0.00 | |||
| Gglean004579.1 | Down | −3.24 | 0.00 | −2.24 | 0.00 | |||
| Gglean027772.1 | Down | −1.58 | 0.00 | −3.38 | 0.00 | |||
| Gglean022575.1 | Down | −1.12 | 0.00 | −1.02 | 0.00 | |||
| Gglean018674.1 | Down | −1.11 | 0.00 | −3.38 | 0.00 | |||
| Gglean007149.1 | Down | −1.75 | 0.00 | −1.84 | 0.00 | |||
| Gglean016128.1 | Down | −2.31 | 0.00 | −2.94 | 0.00 | |||
| Gglean019365.1 | Up | 1.83 | 0.00 | 1.87 | 0.00 | |||
| Gglean022649.1 | Up | 5.02 | 0.00 | 4.12 | 0.00 | |||
| Gglean029864.1 | Down | −1.01 | 0.00 | −1.77 | 0.00 |
Pathway analysis of candidate DAPs and DEGs
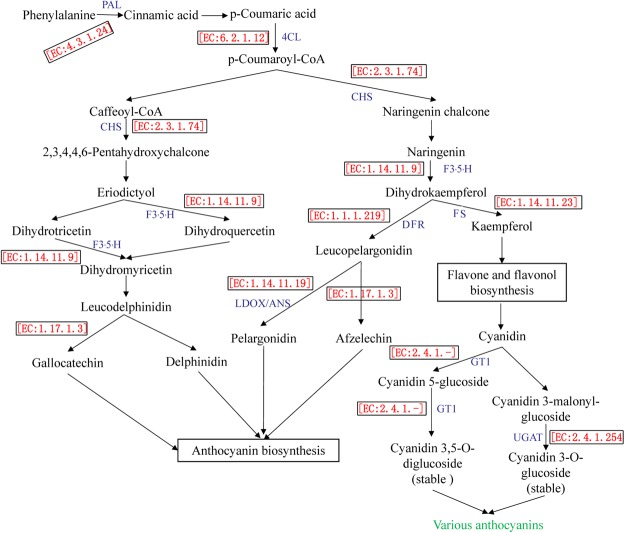
A pathway analysis was completed to characterise the functions of the candidate DAPs and DEGs more comprehensively. A total of 13 DAPs and 17 DEGs corresponded to enzymes involved in anthocyanin biosynthesis (Fig. 5), including phenylalanine ammonia-lyase (PAL, three proteins), 4-coumarate-CoA ligase (4CL, one protein and one transcript), dihydroflavonol 4-reductase (DFR, one protein), leucoanthocyanidin dioxygenase/anthocyanidin synthase (LDOX/ANS, one transcript), chalcone synthase (CHS, one protein and two transcripts), flavanone 3′,5′ hydroxylase (F3′5′H, two proteins and one transcript), [EC:1.1.1.234] (one transcript), [EC:1.14.11.23] (three proteins and three transcripts), [EC:1.14.13.21] (three transcripts), [EC:1.17.1.3] (two transcripts), GT1 (two proteins and one transcript), and UGAT (two transcripts). These genes and proteins represent the core enzymes in anthocyanin biosynthesis, which influence pomegranate fruit peel colour development. The DEGs Gglean009951.1 and Gglean020650.1 encoded the DAPs CHS and F3′5′H, respectively, while the DEGs Gglean015001.1 and Gglean015003.1 both encoded DAP cinnamyl-alcohol dehydrogenase (CAD) [EC:1.1.1.195] (Supplementary Fig. S1), which is involved in phenylpropanoid biosynthesis.
Figure 5.
Differentially abundant proteins (DAPs) and differentially expressed genes (DEGs) related to anthocyanin biosynthesis in pomegranate fruit peels. Numbers and abbreviations of key enzymes are shown in red and blue text, respectively.
Validation of gene expression levels
The expression levels of 12 genes, including Gglean009951.1, Gglean020650.1, Gglean015001.1, and Gglean015003.1, were determined by quantitative real-time polymerase chain reaction (qRT-PCR). The expression patterns were similar to those generated from high-throughput sequencing (Supplementary Fig. S2). The trends in the expression levels of Gglean009951.1, Gglean020650.1, Gglean015001.1, and Gglean015003.1 revealed by qRT-PCR were inconsistent with the changes in the abundance of their encoded proteins as detected in the iTRAQ analysis. These results suggested that deep sequencing represents an accurate and efficient method for analysing pomegranate gene expression levels and the abundance of their corresponding proteins.
Discussion
Peel colour is a key factor influencing the marketability of fruits. Thus, elucidating the genetic mechanism of the regulation of fruit peel colour may be useful for breeders interested in generating new pomegranate cultivars with enhanced fruit colours. Consequently, fruit peel development has been a topic of interest among fruit crop researchers and has been extensively studied. To date, however, no quantitative trait loci related to fruit colour have been identified, and none of the genes related to pomegranate fruit peel colour have been confirmed.
In this study, more DAPs and DEGs were detected between the ‘Tunisia’ and ‘White’ cultivars at the fruit ripening stage than at the fruit colouring stage, suggesting that greater changes in peel colour occurred at the ripening stage (Fig. 6). An integrated quantitative proteomics and transcriptomics analysis revealed that there were more DEGs than DAPs in both SP1_TP1 and SP2_TP2, with only a few of the DEGs encoding the DAPs. These results were similar to those reported for potato17 and orchid18. There were inconsistencies between the trends in transcript levels and the trends in protein abundance. These results may indicate that protein abundance is affected by post-translational modifications and splicing events in cells, rather than changes in gene transcript levels19. A correlation analysis revealed a negative correlation between protein abundance and the expression levels of the corresponding genes (Fig. 2D,E). An earlier study also reported a relatively poor correlation between transcriptome and proteome data (rpearson = 0.27–0.40)20. A possible explanation for the low correlation between transcript levels and protein abundance is that the transcription level can fluctuate more quickly than protein translation and modification processes. Thus, changes in the abundance of a protein occur after the level of its corresponding transcript has stabilised18. Our results suggest that iTRAQ and transcriptomic analyses are complementary methods for profiling candidate proteins mediating specific physiological processes, including pomegranate fruit peel colour development.
Figure 6.
Comparison of fruits from two pomegranate cultivars during fruit colouring and ripening stages. (A) ‘Tunisia’ and (B) ‘White’ fruits during fruit colouring stage. (C) ‘Tunisia’ and (D) ‘White’ fruits during fruit ripening stage.
A KEGG pathway enrichment analysis was completed to avoid false positive results, and the common pathways shared by DAPs and DEGs were selected for further study. Accordingly, we functionally annotated 27 proteins and 54 genes from among the thousands of detected DAPs and DEGs. These proteins and genes were associated with 13 and 20 critical enzymes, respectively, which contributed to pomegranate fruit peel colour mainly by catalysing reactions in pathways related to anthocyanin biosynthesis; stilbenoid, diarylheptanoid, and gingerol biosynthesis; flavonoid biosynthesis; and phenylpropanoid biosynthesis (Fig. 4; Tables 2 and 3). A total of 13 DAPs and 17 DEGs were detected as core regulators of anthocyanin biosynthesis in pomegranate fruit peel (Fig. 5), implying that the colour differences between white and red peels are due to variable anthocyanin production21.
Anthocyanin biosynthesis is fairly complex, and is associated with diverse metabolites including phenylpropanoids and flavonoids22. To date, diverse genes (i.e., PAL, 4CL, DFR, LDOX/ANS, CHS, and F3′5′H) involved in the regulation of anthocyanin biosynthesis have been identified and their functions in the formation of pigments have been characterised in fruit crops including Rosaceae species23, apple24, peach25, and pear8. In the current study, the expression levels of genes encoding F3′5′H and LDOX/ANS were up-regulated and the abundance of their corresponding proteins also increased during fruit colour development (Fig. 4; Tables 2 and 3). In contrast, although the abundance of PAL, 4CL, CHS, and DFR proteins increased, their corresponding genes were down-regulated during fruit colour development (Fig. 4; Tables 2 and 3). Both PAL and 4CL are critical for the conversion of phenylalanine to anthocyanins13. A decrease in transcription but an increase in the abundance of the encoded proteins ensured a sufficient supply of anthocyanin precursors (e.g., p-coumaroyl-CoA) in ‘Tunisia’ fruit peels (Fig. 5). Similarly, CHS catalyses the conversion of p-coumaroyl-CoA to naringenin chalcone (Fig. 5). This represents a key regulatory step during the synthesis of flavonoids, which is a major pigment in many flowers, leaves, and fruits26,27. Naringenin is converted into several anthocyanin-related substances (e.g., dihydrokaempferol, leucopelargonidin, and pelargonidin) by the actions of F3′5′H, DFR, and LDOX/ANS (Fig. 5). These enzymatic reactions have been documented in sweet cherry28, apple24, mango13, and peach25. The results of a previous study indicated that a single nucleotide polymorphism in PgLDOX is responsible for the ‘white’ anthocyanin-less phenotype of pomegranate fruits21. Thus, we hypothesised that these genes may be conserved and play a crucial role in the anthocyanin biosynthesis related to pomegranate fruit peel colour development.
Caffeoyl-CoA, CHS, and F3′5′H catalyse a chain of reactions to generate dihydromyricetin, a coloured chemical compound related to leucoanthocyanidins29. Leucoanthocyanidin reductase [EC:1.17.1.3] converts leucoanthocyanidin to gallocatechin as well as eucopelargonidin to afzelechin (Fig. 5). These two reactions ultimately produce anthocyanins. It is possible that these pathways also contribute to pomegranate fruit peel colour development.
Plant scientists have long been interested in flavonoid pigments because they are responsible for the diverse colours (e.g., yellow, red, purple, and blue) of various organs, including fruits and flowers30,31. In this study, changes in FS transcript levels and the abundance of FS protein tended to induce the biosynthesis of flavones and flavonols (Fig. 5). Down- and up-regulated expression of Gglean024784.1 and Gglean024783.1, respectively, may help regulate the UGAT-catalysed conversion of cyanidin 3-malonylglucoside to the stable compound cyanidin 3,5-O-diglucoside. Additionally, GT1 can catalyse the glycosylation of the 5-OH and the 3-OH groups on the anthocyanidin molecule. Although the GT1 transcript levels decreased during fruit colour development, the abundance of GT1 protein increased, which likely promoted the conversion of cyanidin to cyanidin 5-glucoside and then to cyanidin 3,5-O-diglucoside. These reactions result in the production of various anthocyanins (Fig. 5). This is consistent with the findings of a previous study on anthocyanin biosynthesis in rose32. Our results also confirmed that the glucosyltransferase-catalysed glycosylation at two different positions on the precursor molecule is not limited to rose species32. Thus, we believe that genes regulating anthocyanin biosynthesis via glycosylation play important roles in the stabilization of pigments in pomegranate fruit peels.
In addition to anthocyanins and flavonoids, other secondary metabolites are produced in the phenylpropanoid biosynthesis pathway by the activities of enzymes such as CAD [EC:1.1.1.195]. We detected three CAD proteins with increased abundance and four CAD genes among the DEGs (one up-regulated and three down-regulated), including Gglean015001.1 and Gglean015003.1, during fruit colour development in pomegranate. An earlier study confirmed that CAD belongs to the polyphenol oxidase (PPO) family33, and catalyses the interconversion between aldehydes and alcohols (Supplementary Fig. S1). Disrupting the expression of CAD genes will result in the atypical incorporation of hydroxycinnamaldehyde into lignin, thereby modifying the cell wall structure34. A previous study revealed that PPO protects red pear fruit or leaf cells against pathogens by catalysing the synthesis of lignin and quinines35. Thus, CAD may induce the synthesis of lignin and quinines to alter the cell wall structure to enhance pathogen resistance, which may explain the relatively strong disease resistance of red pomegranate fruits36.
The regulatory functions of the other identified enzymes in pathways related to anthocyanin biosynthesis, especially those in the stilbenoid, diarylheptanoid, and gingerol biosynthesis pathways, have not been characterised. Combined analyses of metabolomics data may further clarify their functions in relation to the regulation of pomegranate fruit peel development. In conclusion, the transcriptome and proteome data generated in this study revealed a complex transcriptional and translational network regulating pomegranate fruit peel colour development. These candidate proteins and genes may be useful for marker-based breeding of new pomegranate cultivars.
Methods
Plant materials
The ‘Tunisia’ and ‘White’ pomegranate cultivars analysed in this study were grown and managed according to local production practices in Xingyang, China. Both cultivars were obtained from the Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou, China. The blooming stage was defined as the period during which 50% of the pomegranate flowers were open. According to our observations, the fruit development periods of the two cultivars were similar. The differences in the fruit peel colours between the two cultivars developed during the fruit colouring stage (60 days after blooming) and especially during the ripening stage (120 days after blooming). The peel of ‘Tunisia’ fruits reddened during the fruit colouring and ripening stages, in contrast to the white fruits of the ‘White’ cultivar (Fig. 6). During the fruit colouring and ripening periods, the peels of three normally growing fruits were removed and pooled. Three replicates were collected. The peels were immediately frozen in liquid nitrogen for 30 min and stored at −80 °C until use. For convenience, the ‘Tunisia’ and ‘White’ fruit peels were abbreviated as TP and SP, respectively.
Phenotypic variation, correlation, and linear regression analyses were completed using SPSS (version 19.0) (IBM Corp., Armonk, NY, USA).
Protein extraction
Proteins were extracted from two biological replicates of pomegranate fruit peels as previously described37. Briefly, Lysis Buffer 3 [8 M urea and 40 mM Tris-HCl containing 1 mM PMSF (phenylmethylsulfonyl fluoride), 2 mM EDTA (ethylenediaminetetraacetic acid), and 10 mM DTT (dithiothreitol), pH 8.5] was added to fruit peel samples, which were then ground. After centrifuging the ground material at 25,000 × g for 20 min at 4 °C, five volumes of 10% TCA/acetone with 10 mM DTT were added to the supernatant to precipitate proteins at −20 °C. The precipitation step was repeated with acetone alone until the supernatant became colourless. The precipitated proteins were air-dried and resuspended in Lysis Buffer 3. Samples were sonicated on ice for 5 min to resuspend proteins and then centrifuged as before. The resulting supernatants were incubated at 56 °C with 10 mM DTT for 1 h to reduce the proteins. The proteins were then alkylated with 55 mM iodoacetamide at room temperature for 45 min in darkness. Five volumes of acetone were added to samples to precipitate proteins at −20 °C. Proteins were resuspended in Lysis Buffer 3 and then sonicated on ice for 5 min. The protein concentration was determined by Bradford’s method using bovine serum albumin (BSA) as the standard.
Protein digestion and iTRAQ labelling
Protein solutions (100 µg) were diluted 4-times with 100 mM tetraethylammonium bromide, after which proteins were digested overnight with Trypsin Gold (Promega, Madison, WI, USA) at 37 °C (40:1, protein:trypsin). The peptides were desalted with a Strata X C18 column (Phenomenex, Torrance, CA, USA) and then vacuum-dried according to the protocol recommended by the manufacturer. Peptide samples were labelled using iTRAQ 8-plex kits (AB Sciex Inc., MA, USA). The two ‘Tunisia’ samples (TP1 and TP2) were labelled with iTRAQ tags 113 and 115, while the two ‘White’ samples (SP1 and SP2) were labelled with tags 117 and 119.
Peptide fractionation
Peptides were separated using a Shimadzu LC-20AB HPLC Pump system coupled with a high-pH RP column. The peptides were first reconstituted in Buffer A (5% ACN and 95% H2O, with the pH adjusted to 9.8 with ammonia) to a final volume of 2 ml, which was then loaded onto a column containing 5-μm particles (Phenomenex). Peptides were separated with a gradient of 5% Buffer B (5% H2O and 95% ACN, with the pH adjusted to 9.8 with ammonia) for 10 min, 5–35% Buffer B for 40 min, and 35–95% Buffer B for 1 min at a flow rate of 1 ml/min. The system was maintained in 95% Buffer B for 3 min, followed by a decrease to 5% Buffer B within 1 min and equilibration with 5% Buffer B for 10 min. The eluants were monitored by measuring the absorbance at 214 nm, and fractions were collected every 1 min. The eluted peptides were pooled as 20 fractions and vacuum-dried.
Analysis by LC-MS/MS
Each fraction was resuspended in Buffer A (2% ACN and 0.1% FA in water) and centrifuged at 20,000 × g for 10 min. The supernatant was loaded onto a C18 trap column (5 μl/min for 8 min) on an LC-20AD nano-HPLC instrument equipped with an autosampler (Shimadzu, Kyoto, Japan). The peptides were eluted from the column and separated using an analytical C18 column (75-μm inner diameter) packed in-house. The 8%–35% Buffer B (2% H2O and 0.1% FA in ACN) gradient was applied at 300 nl/min over 35 minutes, and then increased to 60% in 5 min, maintained at 80% for 5 min, and then decreased to 5% over 0.1 min. The column was equilibrated with 5% Buffer B for 10 min. The eluted peptides underwent nanoelectrospray ionisation before being analysed by MS/MS (Q-Exactive mass spectrometer; Thermo Fisher Scientific, San Jose, CA, USA) coupled with the HPLC system.
Protein identification and quantification based on iTRAQ data
Proteins were identified and quantified using the Mascot 2.3.02 search engine (Matrix Science, Boston, MA, USA). Our genome database was used to identify proteins with the IQuant program38. To assess the confidence of the peptide identifications, the PSMs were pre-filtered at a PSM-level FDR of 1%. The identified peptide sequences were then assembled into a set of accurately identified proteins based on the “simple principle”. To quantify proteins, peptides were automatically selected by calculating the reporter peak area using the default parameters of the Mascot software package. The resulting data set was auto-bias corrected, and the variations resulting from the unequal mixing of samples with different labels were eliminated. The DAPs between the TP and SP samples were defined as the proteins with a fold-change of >1.2 (mean value of all compared groups) or <0.85 and a P value (t-test of all comparison groups) of <0.05. To minimise the protein-level false-positive rates, a protein FDR of 1%, which was based on an established “picked” protein FDR strategy39, was estimated after proteins were tentatively identified (protein-level FDR ≤ 0.01).
Nuclear RNA extraction and sequencing
Total RNA was extracted from the fruit peels of the two pomegranate cultivars sampled at two fruit development stages for subsequent RNA sequencing analysis. The RNA was extracted from two biological replicates of frozen samples (100 mg) using the RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China). An ND 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) was used to evaluate the quality of the extracted RNA. Additionally, RNA with an RNA Integrity Number > 8 according to the 2100 Bioanalyzer (Agilent, USA) was used to prepare cDNA libraries with the RNA Library Prep Kit (Illumina, San Diego, CA, USA). The resulting libraries were sequenced on a HiSeq2000 platform (Illumina) to generate 100-bp paired-end reads.
Processing of sequence data and mapping reads to reference genome
The sequenced data were filtered by removing adaptor sequences, empty reads, reads with more than 5% unknown nucleotides, low-quality sequences (base quality ≤ 20), or sequences with >10% Ns using SOAPnuke (version 1.5.2)40. Clean reads were mapped to the reference genome sequence using HISAT (version 0.1.6-beta)41. The reads were assembled into transcripts and compared with reference gene models using Cufflinks42. Gene expression was quantified using RSEM (RNA-Seq by Expectation Maximization, version 1.2.12)43. The data were normalised as fragments per kilobase of transcript per million fragments mapped (FPKM)44. The differences in transcript abundance between two genotypes were calculated based on the ratio of FPKM values. The FDR control method was used to identify the threshold of the P value using Cuffdiff (included in the cufflinks package). Only transcripts with P ≤ 0.001 and |log2 (TP/SP)| > 1 were further analysed.
KEGG pathway enrichment analyses of DAPs and DEGs
The KEGG pathway enrichment analysis of the DAPs or DEGs was conducted using the KOBAS2.0 website (http://kobas.cbi.pku.edu.cn/)45. We identified significantly enriched metabolic pathways by using the hyper geometric test (http://en.wikipedia.org/wiki/Hypergeometric_distribution). The metabolic pathways that were significantly enriched compared with the whole genome background were considered with corrected values (P < 0.05).
Quantitative real-time PCR analysis
Total RNA was used as the template for cDNA synthesis with reverse transcriptase (Takara Bio, Kusatsu, Japan) following the manufacturer’s instructions. The qRT-PCR analyses were conducted using the SYBR premix Ex Taq™ kit (Takara, Dalian, China) and a LightCycler® 480 instrument (Roche, IN, USA), with gene-specific primers designed using Primer3 software46. The reactions were conducted in a 20-µl volume containing 10 µl 2 × SYBR Green Master Mix (Takara), 300 nM each primer (Supplementary Table S5), and 2 µl 10-fold diluted cDNA template. The qRT-PCR program was as follows: 95 °C for 10 s; and 40 cycles of 95 °C for 15 s and 60 °C for 30 s. The qRT-PCR analyses involved three biological replicates, each with two technical replicates. Relative gene expression levels were calculated according to the 2−ΔΔCt method47.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository under the dataset identifier PXD009129.
Electronic supplementary material
Acknowledgements
This work was funded by a grant from the Investigation and Collection of Indigenous Varieties of Deciduous Fruit Trees in Predominance Region (2012 FY110100). We thank Jennifer Smith, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac) for editing the English text of a draft of this manuscript.
Author Contributions
X.L. and S.C. designed the experiment. D.C., X.L. and J.N. collected the data. X.L. analysed the data, interpreted the results, and wrote the manuscript. All authors have read, edited, and approved the current version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiang Luo, Da Cao and Haoxian Li contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-30088-3.
References
- 1.Yuan Z, Yin Y, Qu J, Zhu L, Li Y. Population Genetic Diversity in Chinese Pomegranate (Punica granatum L.) Cultivars Revealed by Fluorescent-AFLP Markers. Journal of Genetics and Genomics. 2007;34:1061–1071. doi: 10.1016/S1673-8527(07)60121-0. [DOI] [PubMed] [Google Scholar]
- 2.Levin GM. Pomegranate (Punica granatum L.) plant genetic resources in Turkmenistan. Plant Genetic Resources Newsletter. 1994;20:31–36. [Google Scholar]
- 3.Lichanporn I, Srilaong V, Wongsaree C, Kanlayanarat S. Effect of storage temperature on peel color and physiological changes of longkong fruit (Aglaia dookkoo Griff) Acta Horticulturae. 2008;804:599–604. doi: 10.17660/ActaHortic.2008.804.88. [DOI] [Google Scholar]
- 4.Tao J, Zhang S, An X, Zhao Z. Effects of light on carotenoid biosynthesis and color formation of citrus fruit peel. Chinese Journal of Applied Ecology. 2003;14:1833. [PubMed] [Google Scholar]
- 5.Tao J, Zhang S, Zhang L, An X, Liu C. Relation ship Between Color Formation and Change in composition of Carotenoids in Peel of Citrus fruit. Acta Photophysiologica Sinica. 2003;29:121–126. [Google Scholar]
- 6.Lado J, et al. Fruit shading enhances peel color, carotenes accumulation and chromoplast differentiation in red grapefruit. Physiol Plant. 2015;154:469–484. doi: 10.1111/ppl.12332. [DOI] [PubMed] [Google Scholar]
- 7.Liu T, et al. Improved peach peel color development by fruit bagging. Enhanced expression of anthocyanin biosynthetic and regulatory genes using white non-woven polypropylene as replacement for yellow paper. Scientia Horticulturae. 2015;184:142–148. [Google Scholar]
- 8.Bai S, et al. Transcriptome analysis of bagging-treated red Chinese sand pear peels reveals light-responsive pathway functions in anthocyanin accumulation. Scientific reports. 2017;7:63. doi: 10.1038/s41598-017-00069-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda C, et al. Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiology & Biochemistry. 2002;40:955–962. doi: 10.1016/S0981-9428(02)01454-7. [DOI] [Google Scholar]
- 10.Voelckel C, Gruenheit N, Lockhart P. Evolutionary Transcriptomics and Proteomics: Insight into Plant Adaptation. Trends in plant science. 2017;22:462. doi: 10.1016/j.tplants.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Ono NN, et al. Exploring the Transcriptome Landscape of Pomegranate Fruit Peel for Natural Product Biosynthetic Gene and SSR Marker Discovery(F) Journal of integrative plant biology. 2011;53:800–813. doi: 10.1111/j.1744-7909.2011.01073.x. [DOI] [PubMed] [Google Scholar]
- 12.Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Molecular & Cellular Proteomics. 2004;3:1154. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Wu HX, et al. Transcriptome and proteomic analysis of mango (Mangifera indica Linn) fruits. Journal of proteomics. 2014;105:19–30. doi: 10.1016/j.jprot.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 14.Guo X, Xu J, Cui X, Chen H, Qi H. iTRAQ-based Protein Profiling and Fruit Quality Changes at Different Development Stages of Oriental Melon. BMC plant biology. 2017;17:28. doi: 10.1186/s12870-017-0977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, et al. Quantitative proteomic investigation employing stable isotope labeling by peptide dimethylation on proteins of strawberry fruit at different ripening stages. Journal of proteomics. 2013;94:219–239. doi: 10.1016/j.jprot.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, Wu X, Zhang Z, Gao Z. Comparative proteomic analysis of floral color variegation in peach. Biochemical and biophysical research communications. 2015;464:1101–1106. doi: 10.1016/j.bbrc.2015.07.084. [DOI] [PubMed] [Google Scholar]
- 17.Li LQ, et al. Comparative Morphology, Transcription, and Proteomics Study Revealing the Key Molecular Mechanism of Camphor on the Potato Tuber Sprouting Effect. International journal of molecular sciences. 2017;18:2280. doi: 10.3390/ijms18112280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, et al. iTRAQ and RNA-Seq Analyses Provide New Insights into Regulation Mechanism of Symbiotic Germination of Dendrobium officinale Seeds (Orchidaceae) Journal of proteome research. 2017;16:2174–2187. doi: 10.1021/acs.jproteome.6b00999. [DOI] [PubMed] [Google Scholar]
- 19.Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000;405:837. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 20.Muers M. Gene expression: Transcriptome to proteome and back to genome. Nature Reviews Genetics. 2011;12:518. doi: 10.1038/nrg3037. [DOI] [PubMed] [Google Scholar]
- 21.Ben-Simhon Z, et al. A “White” Anthocyanin-less Pomegranate (Punica granatum L.) Caused by an Insertion in the Coding Region of the Leucoanthocyanidin Dioxygenase (LDOX; ANS) Gene. Plos One. 2015;10:e0142777. doi: 10.1371/journal.pone.0142777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tunen AJ, Mur LA, Recourt K, Gerats AG, Mol JN. Regulation and manipulation of flavonoid gene expression in anthers of petunia: the molecular basis of the Po mutation. The Plant cell. 1991;3:39–48. doi: 10.1105/tpc.3.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin-Wang K, et al. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC plant biology. 2010;10:50. doi: 10.1186/1471-2229-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takos AM, et al. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant physiology. 2006;142:1216–1232. doi: 10.1104/pp.106.088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuda T, Yamaguchi M, Honda C, Moriguchi T. Expression of anthocyanin biosynthesis genes in the skin of peach and nectarine fruit. Journal of the American Society for Horticultural Science American Society for Horticultural Science. 2004;129:857–862. [Google Scholar]
- 26.Li SJ, Deng XM, Mao HZ, Hong Y. Enhanced anthocyanin synthesis in foliage plant Caladium bicolor. Plant Cell Reports. 2005;23:716–720. doi: 10.1007/s00299-004-0871-2. [DOI] [PubMed] [Google Scholar]
- 27.Liu XJ, et al. Methylation effect on chalcone synthase gene expression determines anthocyanin pigmentation in floral tissues of two Oncidium orchid cultivars. Planta. 2012;236:401–409. doi: 10.1007/s00425-012-1616-z. [DOI] [PubMed] [Google Scholar]
- 28.Wei H, et al. Comparative Transcriptome Analysis of Genes Involved in Anthocyanin Biosynthesis in the Red and Yellow Fruits of Sweet Cherry (Prunus avium L.) Plos One. 2015;10:e0121164. doi: 10.1371/journal.pone.0121164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakshminarayana S, Mathew AG. Leucoanthocyanidins of Sapola Fruit. Journal of Food Science. 2010;32:451–452. doi: 10.1111/j.1365-2621.1967.tb09708.x. [DOI] [Google Scholar]
- 30.Quattrocchio F, Verweij W, Koes R. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science. 2005;10:236. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Grotewold E. The genetics and biochemistry of floral pigments. Annual Review of Plant Biology. 2006;57:761. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- 32.Ogata J, Kanno Y, Itoh Y, Tsugawa H, Suzuki M. Anthocyanin biosynthesis in roses. Nature. 2005;435:757–758. doi: 10.1038/nature435757a. [DOI] [PubMed] [Google Scholar]
- 33.Thygesen PW, Dry IB, Robinson SP. Polyphenol oxidase in potato. A multigene family that exhibits differential expression patterns. Plant Physiology. 1995;109:525–531. doi: 10.1104/pp.109.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson NA, et al. Manipulation of guaiacyl and syringyl monomer biosynthesis in an arabidopsis cinnamyl alcohol dehydrogenase mutant results in atypical lignin biosynthesis and modified cell wall structure. Plant Cell. 2015;27:2195–2209. doi: 10.1105/tpc.15.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu M, Qiu Z, Zhou P, Xu L, Zhang J. Proteomic analysis of ‘Zaosu’ pear (Pyrus bretschneideri Rehd.) and its red skin bud mutation. Proteome Science. 2012;10:1–15. doi: 10.1186/1477-5956-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ben-Simhon Z, et al. A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta. 2011;234:865–881. doi: 10.1007/s00425-011-1438-4. [DOI] [PubMed] [Google Scholar]
- 37.Unwin RD, Griffiths JR, Whetton AD. Simultaneous analysis of relative protein expression levels across multiple samples using iTRAQ isobaric tags with 2D nano LC-MS/MS. Nature Protocols. 2010;5:1574. doi: 10.1038/nprot.2010.123. [DOI] [PubMed] [Google Scholar]
- 38.Wen B, et al. IQuant: an automated pipeline for quantitative proteomics based upon isobaric tags. Proteomics. 2014;14:2280–2285. doi: 10.1002/pmic.201300361. [DOI] [PubMed] [Google Scholar]
- 39.Savitski MM, Wilhelm M, Hahne H, Kuster B, Bantscheff M. A Scalable Approach for Protein False Discovery Rate Estimation in Large Proteomic Data Sets. Molecular & Cellular Proteomics Mcp. 2015;14:2394. doi: 10.1074/mcp.M114.046995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 41.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nature Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5:621. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 45.Kanehisa FM, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics (Oxford, England) 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository under the dataset identifier PXD009129.