Abstract
We have previously shown myofibroblasts subjacent to the squamous epithelium in the normal human esophagus and an increase in esophagitis. Myofibroblast contribution to bone morphogenetic protein (BMP) signaling and to paracrine mediated epithelial-mesenchymal interactions in the human esophagus remains incompletely defined. We investigated BMP4 and BMP inhibitor GREM1 gene expression and protein levels in previously characterized human esophageal myofibroblast primary cultures and in a human esophageal myofibroblast cell line. We adapted human esophageal myofibroblast conditioned media into a 3D organotypic model to investigate the effect of myofibroblast secreted factors on squamous epithelial morphology, proliferation, differentiation and BMP signaling. Human esophageal myofibroblasts constitutively secrete GREM1 and increase BMP4 expression and BMP4 secretion in response to epithelial Hedgehog ligand SHH. Detection of secreted BMP4 is decreased in the presence of GREM1. Myofibroblast conditioned media increases epithelial proliferation and expression of basal markers p63 and CK14 leading to an overall increase in epithelial thickness. Epithelial BMP signaling increases with myofibroblast conditioned media. These findings were partially reversed with GREM1 inhibition. Our results demonstrate that myofibroblasts are potential sources of GREM1 and of BMP4 in the human esophagus and that human esophageal myofibroblast-epithelial paracrine interactions contribute in part to the regulation of epithelial growth.
Introduction
The bone morphogenetic protein (BMP) pathway is required for embryonic esophageal epithelial morphogenesis1 and activation of BMP signaling activity has been found in esophagitis2, Barrett’s esophagus (intestinal metaplasia of the squamous epithelium)3 and most recently eosinophilic esophagitis4. Recent studies using genetic mouse models have shown that hyper-activation of BMP signaling promotes squamous differentiation while inhibition promotes expansion of basal progenitor cells4. On the other hand, others have shown that an increase in BMP activity is associated with intestinal differentiation2. Overall, it appears that the regulation of esophageal epithelial basal layer proliferation and differentiation by BMP signaling may be context specific5. Secreted BMPs induce heterodimeric BMP receptor type 1 and 2 complexes on target cells, leading to phosphorylation of intracellular receptor SMAD 1/5/8 via BMPR1 activation6. P-SMAD 1/5/8 then translocates with SMAD4 to the nucleus to transcribe target genes including inhibitor of differentiation (ID) 1–27. BMP4 binds with greatest affinity to the type 1 BMP receptors BMPR1A and BMPR1B6 and its activity is regulated in part by secreted extracellular antagonists including Gremlin (GREM) 1, Follistatin (FST), Noggin (NOG), and Chordin (CHRD)8. BMP4 is also an end-target of the Hedgehog (Hh) signaling pathway9. Although BMP signaling has been described in the mouse esophageal epithelium1–4, the mesenchymal cell contribution to this pathway has been incompletely investigated in the human esophagus.
We have previously identified human esophageal myofibroblasts (HEMFs) subjacent to the squamous epithelium of the human esophagus and an increase in this population of cells in esophageal biopsies with histological characteristics of reflux esophagitis10. We have also previously established and characterized primary cultures of human esophageal myofibroblasts (HEMFs) and an immortalized HEMF cell line generated from normal human esophagus11. HEMFs secrete a number of inflammatory cytokines in response to stimulation10 and support the growth of squamous epithelial cells in 3D organotypic culture, in part by adding structural support to the collagen matrix11. The location of these cells coupled with their secretory capacity suggested a potential role for paracrine interactions between HEMFs and the squamous epithelium. We were curious whether HEMFs were a mesenchymal source of BMP4 and GREM1, a BMP inhibitor that has not been extensively described in the human esophagus. We were also interested whether paracrine mechanisms between HEMFs and the epithelium were involved in supporting the growth of the stratified squamous epithelium.
Results
BMP4 and GREMLIN1 gene expression and protein levels in unstimulated HEMFs
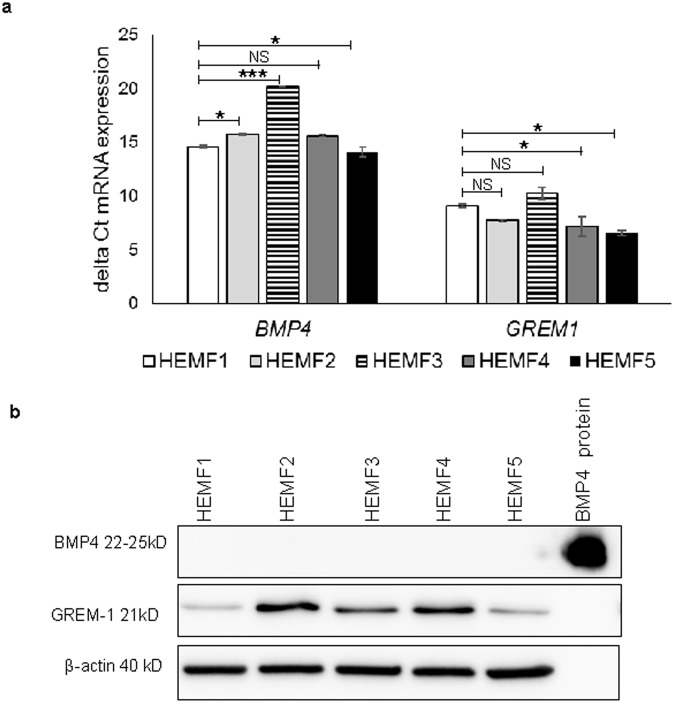
qRT-PCR showed baseline expression of BMP4 and GREM1 mRNA in primary HEMF cultures established from multiple normal human esophagi (HEMFs 1–4). As expected, absolute differences in mRNA expression of BMP4 and GREM1 were observed amongst HEMFs established from different individuals. However, BMP4 and GREM1 were readily detected in all samples. BMP4 and GREM1 mRNA expression were also confirmed in a previously described immortalized HEMF cell line (HEMF 5)11 (Fig. 1a). BMP4 and GREM1 protein expression were then evaluated in HEMF cell lysates. BMP4 protein could not be detected in untreated HEMFs. GREM1 protein expression was readily detected in all samples (Fig. 1b).
Figure 1.
BMP4 and GREM1 gene expression and protein levels in unstimulated HEMFs. (a) BMP4 and GREM1 mRNA expression were measured by qRT-PCR performed on RNA isolated from primary cultures of HEMFs (HEMFs 1–4) established from normal human esophagi and from a GFP-hTERT transfected immortalized HEMF cell line (HEMF 5). Absolute and relative gene abundance are shown with delta Cts (∆Ct) wherein a higher delta Ct indicates lower gene expression. *P ≤ 0.02; **P ≤ 0.001; NS, non-significant vs. HEMF1. Values represent the mean ± SEM of three individual experiments. (b) Representative immunoblots for BMP4 and GREM1 performed on HEMFs 1–5 with β-actin as the loading control are shown. Detection of BMP4 recombinant protein as a positive control is also shown.
BMP4 signaling in HEMFs
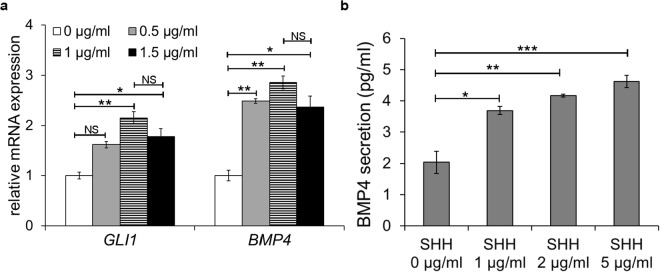
We were then interested in determining regulation of BMP4 gene expression and BMP4 protein secretion in HEMFs. Given that BMP4 has been shown to be an end-target of Hedgehog (Hh) signaling in multiple other cell types9, we wanted to determine whether HEMFs were similarly responsive to Hh signals with BMP4 an end-target. HEMFs were treated with increasing doses of the Hh ligand SHH and expression of GLI1, a direct Hh target5 was evaluated. HEMF expression of GLI1 mRNA increased to a peak of 2.1 fold in response to 1 µg/ml SHH and then plateaued. This finding suggested that the doses of SHH we were using were adequate to elicit a response. BMP4 mRNA expression was then evaluated. BMP4 mRNA increased in response to treatment with SHH and peaked and plateaued at 2.9 fold in response to a SHH dose of 1 µg/ml (Fig. 2a).
Figure 2.
The effect of SHH treatment on HEMF GLI1 expression and on BMP4 expression and BMP4 protein secretion. (a) GLI1 and BMP4 expression were evaluated by qRT-PCR in HEMFs treated with increasing doses of SHH. Relative change of the target gene is shown by the 2∆∆ (delta) CT method. HEMF expression of GLI1 and BMP4 increase in response to treatment with SHH. Results represent the means ± SEM of three individual experiments performed in HEMFs. *P ≤ 0.04; **P ≤ 0.02; NS, non-significant vs. no treatment for each gene per ANOVA for multiple comparisons (b) HEMF secretion of BMP4 in response to treatment with SHH was evaluated by ELISA. BMP4 secretion increases in response to treatment with SHH. Results represent the means ± SEM of three individual experiments performed in HEMFs. *P ≤ 0.02; **P ≤ 0.01; ***P ≤ 0.001 versus no treatment per ANOVA for multiple comparisons.
The increase in BMP4 mRNA expression in response to SHH, prompted us to evaluate BMP4 protein secretion from HEMFs treated with increasing doses of SHH (Fig. 2b). In the absence of treatment, secretion of BMP4 from HEMFs remained near the lower detection limits of the ELISA. BMP4 secretion increased in response to increasing doses of SHH and plateaued in response to doses of SHH greater than 1 µg/ml.
HEMFs secrete GREMLIN1
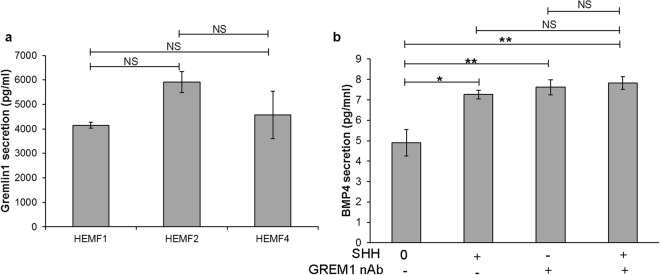
Given our observation that GREM1 protein was expressed in cell lysates of untreated HEMFs, we were interested in whether unstimulated HEMFs secreted this BMP inhibitor and evaluated secretion of GREM1 with ELISA. High concentrations of GREM1 protein were readily detected in conditioned media collected from untreated HEMFs. ELISA results are shown for primary HEMFs 1 and 2 and HEMF 5, an immortalized cell line (Fig. 3a). HEMF GREM1 expression and secretion were then evaluated in response to SHH. GREM1 mRNA expression in HEMFs was unchanged in response to SHH. GREM1 protein continued to be detected in conditioned media collected from HEMFs treated with SHH, however the response to SHH was variable and not reproducible. Given the consistently high levels of GREM1 detected in untreated HEMF conditioned media, we were curious whether HEMF GREM1 secretion interfered with detection of stimulated BMP4 secretion. Addition of GREM1 nAb to untreated or SHH treated HEMF conditioned media similarly slightly increased detection of BMP4 (Fig. 3b).
Figure 3.
HEMF Gremlin1 secretion and BMP4 detection. (a) HEMFs secretion of Gremlin (GREM)1 was evaluated by ELISA performed on conditioned media of untreated HEMFs. Results represent the means ± SEM of three individual experiments performed in previously described primary cultures (HEMF 1, 2) and a GFP-hTERT transfected immortalized HEMF cell line (HEMF 5). (b) Detection of BMP4 in HEMF conditioned media was with ELISA after the addition of GREM1 nAb. Detection of BMP4 increased with the addition of GREM1 nAb to HEMF conditioned media, both in the presence and absence of HEMF treatment with SHH. Results represent the means ± SEM of three individual experiments performed in HEMFs. *P ≤ 0.02; **P ≤ 0.01; NS, non-significant with ANOVA for multiple comparisons.
HEMF secreted factors increase squamous epithelium thickness
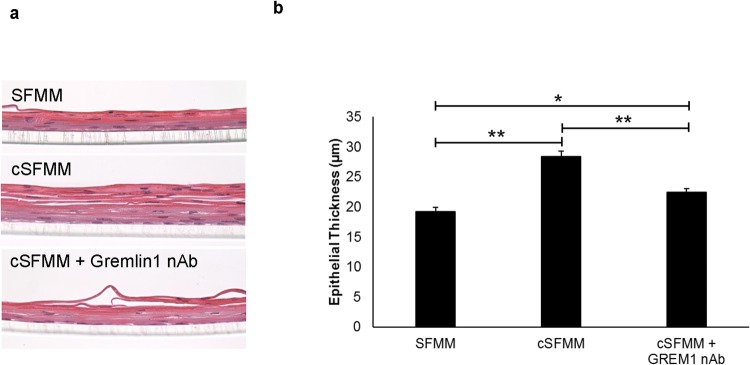
We were then interested in whether conditioned HEMF media had any effects on the growth, proliferation, or differentiation of the squamous epithelium. A simplified 3D organotypic air-liquid interface (ALI) culture system that recapitulates stratified squamous epithelium was used to study the effect of HEMF secretion on the epithelium. Epithelial thickness was visibly greater in the presence of conditioned HEMF compared to epithelial cells in ALI culture with control keratinocyte or serum-free myofibroblast media (SFMM) (Fig. 4a). Quantification of epithelial thickness demonstrated an increase in epithelial thickness in the presence of conditioned HEMF media (19.2 µm vs. 28.4 µm, p < 0.0001) (Fig. 4b).
Figure 4.
HEMF secreted factors increase epithelial cell thickness in 3D organotypic ALI culture. (a) H&E immunostaining of squamous epithelium harvested from 3D organotypic ALI culture shows the squamous epithelium in the presence of serum-free myofibroblast media (SFMM), conditioned serum-free myofibroblast media (cSFMM), or cSFMM plus the addition of GREM1 nAb. (b) Quantification demonstrates an increase in epithelial thickness 3D organotypic ALI culture with conditioned HEMF media. The increase in thickness is partially reversed with addition of GREM1 nAb. *P ≤ 0.02; **P ≤ 0.0001 with ANOVA for multiple comparisons.
Based on our results that showed that untreated HEMFs secreted GREM1, we were curious whether GREM1 was playing a role in this morphology. Addition of GREM1 nAb to HEMF conditioned media resulted in a decrease in epithelial thickness (28.4 µm vs. 22.4 µm, p < 0.0001) with a persistently prominent eosin stained suprabasal differentiated epithelial layer and thickening of the pseudo-keratinized layer with occasional visible sloughing at the top of the epithelium, suggestive of excessive squamous differentiation (Fig. 4a,b). Addition of GREM1 nAb to control SFMM had no effect on the squamous epithelium
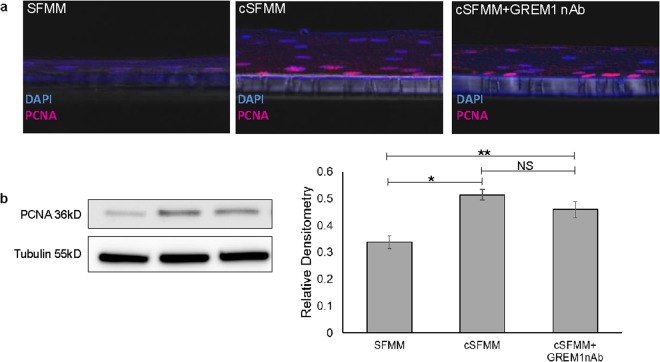
To investigate whether an increase in basal cell proliferation was responsible for the increase in epithelial thickness, we initially performed Ki67 immunostaining. Ki67 immunostaining was sparse under all conditions precluding further reliable quantification (Supplementary Fig. 1). We therefore, evaluated expression of another proliferation marker PCNA. Immunostaining and immunoblot for PCNA demonstrated an increase in epithelial proliferation with conditioned HEMF media. PCNA expression remained increased in the presence of GREM1 nAb (Fig. 5a,b).
Figure 5.
HEMF secreted factors regulate proliferation in squamous epithelial cells. (a) PCNA immunofluorescent staining (Cy5, purple) was performed on paraffin-embedded, formalin-fixed sections of 3D organotypic ALI culture established with control serum-free myofibroblast media (SFMM), conditioned SFMM (cSFMM), and cSFMM plus the addition of GREM1 nAb. Images were viewed with a confocal microscope. Magnification 40x. (b) Immunoblots for PCNA were performed on epithelium harvested from 3D organotypic ALI culture. Tubulin was used as the loading control. Representative images are shown of at least three individual 3D organotypic ALI cultures for each condition and their respective immunoblots. Samples from each experiment were derived at the same time and processed in parallel. Relative densitometry to loading control tubulin is shown. *P ≤ 0.01; **P ≤ 0.05; NS, non-significant with ANOVA for multiple comparisons.
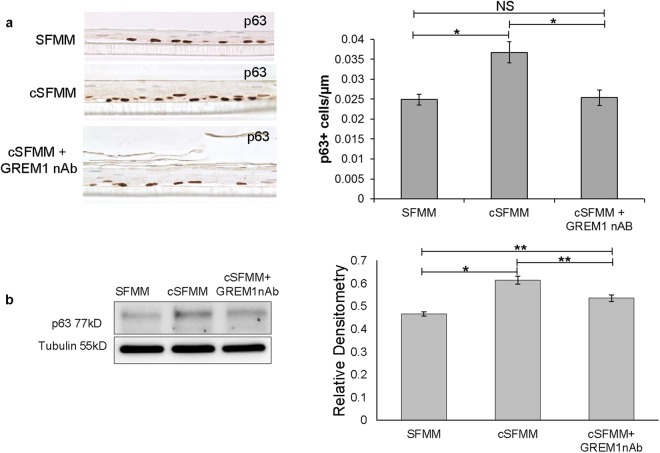
To further characterize the population of cells that accounted for the increase in thickness and proliferation of the squamous epithelium, we performed immunostaining for the basal cell marker p6312. An increase in p63 immunostaining was visible in the basal layer of the squamous epithelium in the presence of conditioned vs. control HEMF media (Fig. 6a). Immunoblot confirmed the increase in p63 expression in the presence of conditioned HEMF media (Fig. 6b). In the presence of GREM1 nAb, p63 expression decreased compared to conditioned HEMF media, although its expression remained greater than that in control conditions.
Figure 6.
HEMF secreted factors increase epithelial basal cell p63 expression in 3D organotypic ALI culture. (a) Representative immunostaining for p63 performed in epithelium harvested from organotypic cultures is shown (n = 3 for each condition). Basal p63 expression in squamous epithelium is increased in the presence of conditioned HEMF media (cSFMM) compared to control (SFMM). Immunostaining for p63 is decreased in the presence of GREM1 nAb. Quantification is shown. (b) Immunoblot for p63 was performed on protein isolated from squamous epithelial cells harvested from 3D organotypic ALI culture. Tubulin was used as the loading control. Image shown is representative of at least three individual 3D organotypic ALI cultures for each condition and their respective immunoblots. Samples from each experiment were derived at the same time and processed in parallel. Epithelial p63 expression increases in the presence of cSFMM and partially decreases with the addition of GREM1 nAb. *P ≤ 0.01; **P ≤ 0.02; ***P < 0.05 with ANOVA for multiple comparisons.
To further evaluate the effect of HEMF conditioned media on expression of basal cell markers and on squamous differentiation we performed immunostaining for basal cell marker CK14 and differentiated suprabasal cells with CK 4, respectively13 (Fig. 7). We observed that the compared to control conditions, squamous epithelium in the presence of conditioned HEMF media had more prominent CK14 expression consistent with expansion of the basal layer. This finding was consistent with the observed increase in basal cell marker p63 expression with conditioned HEMF media. In the presence of GREM1 nAb, immunostaining for the basal layer was less intense compared to conditioned media with more prominent CK4 staining of the differentiated suprabasal layer.
Figure 7.
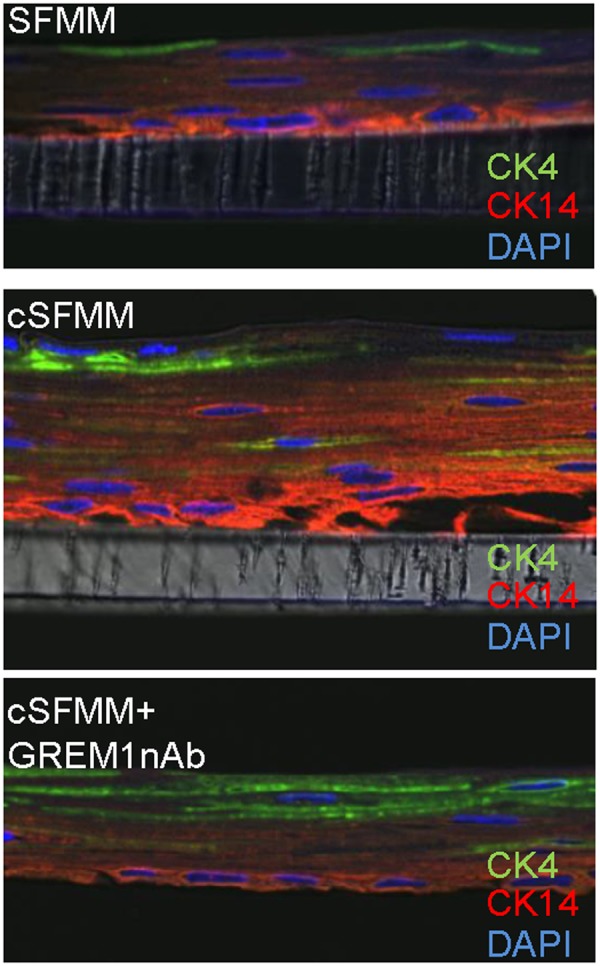
Differentiation of epithelial cells in 3D organotypic ALI culture in the presence of conditioned HEMF media and GREM1 nAb. Representative confocal images of immunostaining performed for basal cell marker CK14 (rhodamine; red) and suprabasal marker CK4 (Cy2; green) in squamous epithelium cultured with control serum-free myofibroblast media (SFMM), conditioned SFMM (cSFMM) and cSFMM plus GREM1 nAb (n = 3 each condition) are shown. Magnification 63X. Expression of basal marker CK14 is more prominent with cSFMM compared to control. Addition of GREM1 nAb decreases intensity of immunostaining for basal marker CK14 and increases intensity of suprabasal marker CK4.
HEMF secretion modulates esophageal epithelial BMP signals
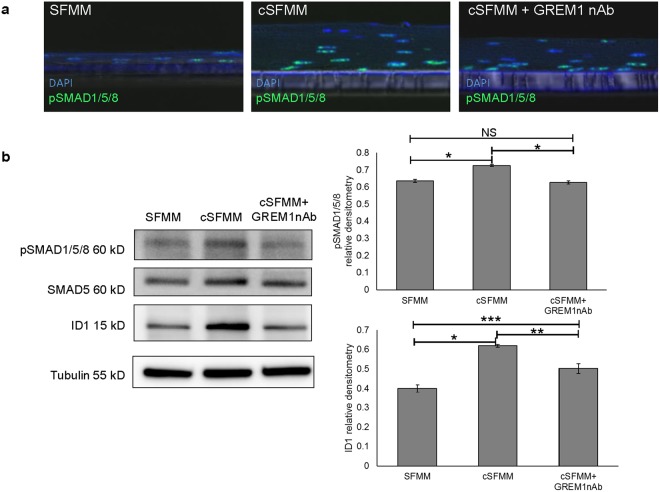
Given that GREM1 is a BMP inhibitor, we were curious whether phenotypic changes in the epithelium were a consequence of changes in BMP signaling. We therefore evaluated down-stream BMP targets in the squamous epithelium (Fig. 8a). Immunofluorescent staining for pSMAD1/5/8 showed weak expression of nuclear pSMAD1/5/8 in scattered cells of the squamous epithelium under control conditions. In the presence of conditioned HEMF media, pSMAD1/5/8 expression was intensely expressed in the nuclei of epithelial cells with occasional absent staining in the suprabasal layer. Immunofluorescent staining for pSMAD1/5/8 appeared to be weaker with the addition of GREM1 nAb to HEMF conditioned media. To better quantify pSMAD1/5/8 expression we performed immunoblot. Baseline expression of pSMAD1/5/8 increased in the presence of conditioned HEMF media. Inhibition of GREM1 decreased pSMAD1/5/8 expression similar to that in controls (Fig. 8b)
Figure 8.
HEMF secreted factors modulate BMP signaling in squamous epithelial cells. (a) pSMAD1/5/8 immunofluorescent staining (Cy2, green) was performed on paraffin-embedded, formalin-fixed sections of 3D organotypic ALI culture established with control serum-free myofibroblast media (SFMM), conditioned SFMM (cSFMM), and cSFMM plus the addition of GREM1 nAb. Representative images of three independent experiments is shown. Images were viewed with a confocal microscope. Magnification 40x. (b) Immunoblots for pSMAD1/5/8 and ID1 were performed on epithelium harvested from 3D organotypic ALI culture. Epithelial pSMAD1/5/8 and ID1 increase in the presence of cSFMM. Addition of GREM1 nAb inhibits the increase in pSMAD1/5/8 and ID1. Tubulin was used as the loading control. Representative immunoblots are shown of three individual experiments. Samples from each experiment were derived at the same time and processed in parallel. Relative densitometry to loading control tubulin is shown. *P ≤ 0.001; **P ≤ 0.02; **P ≤ 0.05; NS, non-significant with ANOVA for multiple comparisons.
We then looked at the transcriptional target of pSMAD1/5/8, ID114. Immunoblot demonstrates ID1 expression in the squamous epithelium established with control serum-free myofibroblast media. In the presence of conditioned HEMF media, baseline expression of ID1 also significantly increases. Expression of epithelial ID1 partially decreased with inhibition of GREM1 in HEMF conditioned media (Fig. 8b).
To begin to determine whether other HEMF secreted factors that could affect the BMP pathway were responsible for the observed changes in the squamous epithelium, we performed a screen for BMPs in HEMF conditioned media using a BMP array. Absence of BMP4 in conditioned media of untreated HEMFs was confirmed. BMPs 2, 5, 6, 7, 8, 9 and 11 were also not detected in conditioned media of untreated HEMF. We also performed 3D organotypic air-liquid interface (ALI) culture with the addition of BMP4 nAb to HEMF conditioned media. As expected, addition of BMP4 nAb to HEMF conditioned media did not result in morphologic changes to the squamous epithelium (Supplementary Fig. 2).
Conclusions
In this study, we investigated the contribution of previously characterized sub-epithelial mucosal HEMFs to the BMP pathway in the human esophagus, focusing on HEMF secretion of BMP4 and its inhibitor GREM1. In addition, we adapted HEMF secreted factors into a 3D organotypic model, which recapitulates stratified squamous epithelium, thereby providing a platform for understanding HEMF paracrine interactions with the esophageal epithelium.
In humans, esophageal biopsies show an increase in BMP4 expression in esophagitis2 and Barrett’s esophagus2,3 either by immunohistochemistry or mRNA, with no detection in normal human esophagus15. Our findings are consistent with these previous reports. Despite BMP4 mRNA expression in multiple cultures of HEMFs isolated from normal human esophagus we were unable to detect BMP4 protein expression or secretion from unstimulated HEMFs. Unstimulated HEMFs do not appear to be a source of BMP4. A BMP screen also did not show expression of other BMPs in unstimulated HEMF conditioned media.
We have shown, however, that BMP4 expression and secretion increase from HEMFs in response to Hh ligand SHH in a dose response manner. BMP4 mRNA expression plateaus above doses of 1 µg/ml, possibly due to a saturation effect. Upregulation of SHH expression is induced in epithelial cells in response to acid or bile exposure3. In disease states such as Barrett’s esophagus, normally absent epithelial Hh ligands are reactivated in metaplastic tissue along with BMP4 expression by mesenchymal cells16. We found that normal HEMFs respond to SHH with an increase in GLI1 and BMP4, known downstream targets of Hh signaling, demonstrating that HEMFs have the potential to be active participants in epithelial-stromal paracrine signaling in esophageal acid disorders.
Secreted active BMP4 can be inhibited at the extracellular level by interacting with secreted BMP4 antagonists such as GREM16. High levels of Gremlin expression have been reported in neurons, alveolar epithelial cells, and goblet cells17 and upregulation of GREM1 has been reported in fibrosis of the kidney, lung, heart, liver, and pancreas18. Immunohistochemistry performed on tissue microarrays has shown negative or weak GREM1 expression in the human esophagus19. However, expression of GREM1 in the esophageal stroma at the cellular level has not been previously reported in the normal human esophagus. We have demonstrated abundant GREM1 mRNA expression, GREM1 protein levels and secretion in HEMFs from the normal human esophagus.
Despite adequate stimulation by SHH, the quantity of BMP4 secretion as detected by ELISA in our hands remained low, suggesting possible interference from secreted GREM1. Our finding that detection of BMP4 was increased in the presence of GREM1 nAb is consistent with this possibility.
The molecular mechanisms by which GREM1 interferes with BMP activity were not fully addressed in the current study. However it is worthwhile to note that in addition to directly binding BMP4 to interfere with its interaction with the BMP receptor, Gremlin can interact with BMP4 precursor protein intracellularly and thereby inhibit the formation and secretion of mature and active BMP ligand in a tissue-specific manner17. Whether this mechanism is also present in HEMFs is not known. Interestingly, GREM1 may also participate in signaling pathways with endothelial and immune cells independent of the BMP pathway via interactions with VEGF receptors on endothelial cells or to Slit proteins in monocytes8. Furthermore, HEMFs may express additional secreted BMP antagonists that were not investigated in the current study.
We investigated the functional consequences of unstimulated HEMF secretion in an organotypic model in which upper chamber epithelial cells are co-cultured with conditioned myofibroblast media in the lower chamber to determine the effect of HEMF secreted factors on the epithelium. Epithelial cells are exposed to HEMF conditioned media at the time of initiation of the air-liquid interface (i.e. after epithelial cells have been seeded atop the transwell membrane and have already begun to proliferate). We observed that secreted factors in HEMF conditioned media increase epithelial thickness via an increase in epithelial proliferation and an increase in basal layer expansion, as reflected by the increase in basal cell markers p63 and CK14. We also observed an increase in epithelial pSMAD1/5/8 and ID1 in the presence of HEMF conditioned media, suggesting that secreted factors activate BMP signaling in epithelial cells. It is not clear from our results, however, that this increase is responsible for the observed increase in epithelial thickness and proliferation. For instance, BMP activation in the epithelium has recently been shown to promotes differentiation and not proliferation of basal progenitor cells in the esophagus4, suggesting the possibility that in our model, the increase in epithelial BMP signaling is a secondary response to activation of as yet uninvestigated pathways involved in the regulation of epithelial proliferation and differentiation (e.g. Wnt, Notch).
Given abundant GREM1 secretion from cultures of HEMFs we hypothesized that GREM1 was mediating some of these observed effects. GREM1 inhibition only partially decreased epithelial thickness and expression of p63 with a trend towards a decrease in epithelial proliferation. GREM1 inhibition also resulted in a squamous epithelium that had a more prominent differentiated suprabasal layer and less prominent basal layer. Furthermore, the increase in epithelial BMP signaling despite abundant HEMF GREM1 secretion and the incongruous decrease in BMP activation with GREM1 inhibition demonstrate the potential role of additional uninvestigated factors in HEMF conditioned media. Overall, our findings suggest that other as yet unidentified factors in conditioned HEMF media contribute to the observed phenotype. In addition, our work suggests that the effect of HEMF GREM1 secretion in our model may not be primarily via its role as a BMP inhibitor and suggests the possibility of a direct effect of GREM1 on the squamous epithelium. Others, for instance, have previously reported on the functions of GREM1 as a regulator of survival and proliferation independent of its effects on the BMP pathway20.
Our work has several limitations including the use of HEMF cell cultures and immortalized HEMF cell lines and limitations of the 3D organotypic model. Although HEMFs were derived from normal human esophagus, and in theory more reflective of human physiology than mouse cells, they are cells and may not fully recapitulate in vivo physiology. Furthermore, there may be underappreciated differences in secretion from primary cultures of HEMFs vs. from a cell line, although this cell line has been previously shown to behave similar to primary HEMFs11. While absolute expression values are expectedly different amongst primary cultures themselves and between primary cultures and cell line, the patterns are similar. We looked at the functional consequences of HEMF secretion in a modified 3D organotypic model using HEMF conditioned media. This model utilizes conditioned media from a well characterized HEMF cell line and does incorporate HEMFs into the collagen matrix of the traditional 3D OTC model. While stratified squamous epithelium is recapitulated in both models, the physical contribution of HEMFs working in concert with HEMF secreted factors is lacking. In addition, the squamous epithelium was only at the time of harvest on day 14 of culture. Off-target effects of the neutralizing antibody used may also be possible, although no difference in epithelial morphology was observed with addition of GREM1 to control serum-free myofibroblast media. These limitations along with investigation of the effect of stimulated HEMF secretion on the epithelium should be addressed in ongoing studies of paracrine interactions between HEMFs and the squamous epithelium using genetically modified HEMFs in both traditional and modified 3D OTC.
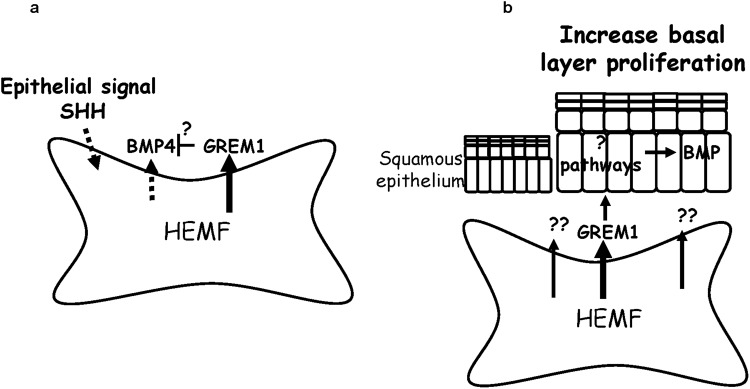
Investigation of human esophageal pathobiology has historically been limited to the squamous epithelium with little consideration of the microenvironment. Furthermore, profound differences between mouse and human esophagus may limit generalizability of signaling pathways detailed in the mouse esophagus and necessitate evaluation in human cells and tissue. Despite its limitations, our work is one of the first to look at BMP signaling in the HEMFs of the human esophagus and paracrine interactions between HEMFs and the squamous epithelium. We have demonstrated for the first time that HEMFs are a potential source of BMP4 in the setting of stimulation and that HEMF basal secretion of GREM1 secretion reduces detection of BMP4 (Fig. 9a). Our work shows that HEMF secreted factors regulate epithelial thickness in a 3D organotypic model at least in part via GREM1 secretion through as yet undetermined mechanisms, including the possibility of a direct effect on the epithelium (Fig. 9b). Our findings support additional investigation of the effects HEMF secretion on the squamous epithelium in normal and diseased esophagus.
Figure 9.
Schematic of proposed interactions between (a) SHH signaling and HEMF secretion of BMP4 and GREM1 and between (b) HEMF secreted factors, epithelial phenotypes and BMP signaling. (a) Untreated HEMFs constitutively secrete GREM1 and secrete BMP4 in response to SHH. GREM1 secretion may interfere with detection of BMP4 in stimulated HEMFs. (b) Unstimulated HEMF secreted factors increase epithelial cell thickness and basal cell proliferation in part via GREM1 secretion with contribution of additional unidentified factors. The effects of GREM1 in this model may be a direct effect on the squamous epithelium. Activated BMP signaling in the squamous epithelium may be a secondary consequence of unidentified pathway. HEMF, human esophageal myofibroblast.
Methods
Cell culture
Previously characterized primary HEMF cultures from normal human esophagi (HEMFs 1–3)10, a freshly generated HEMF culture (HEMF 4) from de-identified normal human esophageal specimen from donor without a known history of esophageal disorders, and an immortalized HEMF cell line (HEMF 5)11 with phenotypic, genotypic and functional similarity to primary cultures were cultured in myofibroblast media (DMEM with 10% FBS supplemented with 10 mg/ml insulin; and 10 µg/ml gentamicin, 2 ng/ml EGF, and 10 µg/ml transferrin) as previously described10 and incubated in 37 °C with 5% CO2. A well characterized human esophageal epithelial cell line EPC2 (telomerase reverse transcriptase-immortalized, gift from Dr. Anil Rustgi, University of Pennsylvania, Philadelphia, PA) cultured in keratinocyte serum-free media with 0.09 mM CaCl2 and supplemented with BPE and penicillin-streptomycin as previously described21. This protocol was approved by the Institutional Review Board of Keck School of Medicine of University of Southern California and deemed coded specimens/data. All methods were performed in accordance with relevant guidelines and regulations.
Treatment of HEMFs with SHH and ELISA
For treatment studies, HEMFs were grown in serum-free myofibroblast media (SFMM). Preliminary studies showed a similar response pattern to SHH treatment between HEMF primary cultures and immortalized cell line HEMF 5. Therefore, HEMF 5 was used for signaling studies and treated with SHH (0.5–5 µg/ml) and Gremlin (GREM) 1 neutralizing antibody (nAb) for 24 h. Pilot studies performed to determine the optimal concentration of Grem1 nAb showed that 3 µg/ml inhibited detection of Grem1 in HEMF conditioned media. This concentration was therefore used in subsequent studies. Conditioned media from untreated HEMFs was collected for evaluation of BMPs with the RayBio C-Series human BMP Related Array 1 according to the manufacturer’s instructions. Conditioned media from treated HEMFs was collected for evaluation of BMP4 (R&D Duoset, DY314) and Grem1 (Lifeome, E92128Hu-1) using enzyme-linked immunosorbent assay (ELISA) according to the manufacturer’s instructions. Protein levels in each sample were determined based on a standard curve generated by recombinant proteins provided with the ELISA kits. Results were expressed as the means ± SEM in pg/ml.
RNA isolation and rt-PCR
RNA was harvested from HEMFs using GeneElute Mammalian Total RNA miniprep Kit (Sigma-Aldrich, St. Louis, MO) per manufacturer’s recommendations and further cleaned with DNA-free (Ambion, Austin, TX). cDNA was synthesized and mRNA expression of the following genes was evaluated: BMP4 (5′ CTGGTCTTGAGTATCCTGAGCG 3′; 3′CTGGTCTTGAGTATCCTGAGCG 5′), GREM1 (5′ TCATCAACCGCTTCTGTTACGGC 3′; 3′ CAGAAGGAGCAGGACTGAAAGG 5′), GREM2 (5′ TTTCCCTGTCCTTGTTCCTG 3′; 3′ TGCACCAGTCACTCTTGAGG 5′)and Gli1 (5′ GCCGTGCTAAAGCTCCAGTGA 3′; 3′ CTGCCCTATGTGAAGCCCTATTTG 5′). Human 18 s (5′ CCA TGA AGA GGT GAG CGG GGA TTG; 3′ ATT AAG TCC CTG CCC TTT GTA CAC 5′) was used as an endogenous control to normalize the samples using the ∆Ct and ∆∆Ct methods of relative quantitation, where Ct is the critical threshold cycle. Target gene expression was normalized by Ct of the endogenous control (18 s rRNA) from that of the target gene (ΔCT). For comparisons, relative change of the target gene equals 2ΔΔCT, where ΔΔ CT = ΔCT of the tested sample - ΔCT of the control sample. Experiments included no template and no reverse transcriptase controls for each gene.
3D organotypic Air-liquid interface culture
A 3D organotypic air-liquid interface (ALI) culture protocol was performed as previously described22. Briefly, immortalized esophageal epithelial cells (EPC2-hTERT) were seeded onto semi-permeable membranes (0.4 μm, Corning, Corning NY) and grown to confluence in the presence of low-calcium media (keratinocyte serum free media, 0.09 mM calcium). Epithelial differentiation was induced by the addition of extracellular calcium (1.8 mM final concentration) for the next five days (day 3 to 8). Stratification was induced by removing the media from the upper chamber and exposing the cells to the ALI for a period of 6 days (day 8 to 14). At the start of the ALI exposure (day 8), different treatment regimens were added to the lower chamber and continued until the day of harvest (day 14). Epithelial cells had a similar morphology with keratinocyte serum free media and serum-free myofibroblast media (SFMM) in the lower chamber. SFMM therefore served as the control medium and comparator in subsequent studies. Treatment regimens included HEMF conditioned SFMM (cSFMM) and HEMF conditioned media containing Gremlin1 nAb (cSFMM + GREM1 nAb). Conditioned media was prepared by growing 8 × 105 immortalized HEMFs in serum-free myofibroblast medium in a 75 cm2 flask with and without the addition of GREM1 nAb and collection of media after 24 hours. Lower chamber media was replaced with fresh conditioned media every 3 days. Cultures were fixed with 10% neutralized buffered formaldehyde in the plate at 4 °C for 1 hour and submitted for histology.
Protein isolation and immunoblot
The squamous epithelium in 3D ALI culture was harvested by stripping the epithelial layer off the membrane with forceps. Cells were collected into RIPA lysis buffer and sonicated. Protein was quantified by the Bradford assay; 20 µg of whole cell lysates were subjected to 4–20% SDS-PAGE gel and transferred to PVDF membrane. The membrane was blocked with 5% milk or BSA and incubated overnight with rabbit anti-p63 (0.33 µg/ml, Genetex, #GTX102425), mouse anti-PCNA (1:400, BD Transduction, cat#610664), rabbit anti-pSMAD1/5/8 monoclonal (1:500, Cell Signaling, cat# 9516), rabbit anti-SMAD5 monoclonal (1:1000, Cell Signaling, cat# 12534 S), or rabbit anti-ID1 polyclonal antibody (0.5 µg/ml, Santa Cruz #sc-488). Membranes were washed with TBST and incubated with donkey anti-mouse-HRP (1:5000, Santa Cruz, cat#sc-2314) or donkey anti-rabbit IgG-HRP (1:5000, Santa Cruz, cat#sc-2317) for 1 hour at room temperature. Immunoreactive bands were detected by SuperSignal West Pico PLUS chemiluminescent substrate (Thermo Scientific, cat#34577). Membranes were stripped and reprobed with mouse anti-tubulin (1:5k,Santa Cruz, sc-5286) or rabbit β-actin polyclonal (1:10k, Abcam (ab8227) antibodies as loading controls. Protein expression was quantified using NIH Image J software and the relative quantity of protein with respect to tubulin was calculated.
Immunohistochemistry and immunofluorescence
Paraffin-embedded, formalin-fixed sections of 3D organotypic ALI cultures were used for immunohistochemistry and immunofluorescence as previously described10. Primary antibodies used for immunohistochemistry were mouse monoclonal anti-p63 (1:200, BioCare Medical, CM163A] and rabbit polyclonal anti-Ki-67 (1:500, Leica Biosystems Ki67P-CE). The slides were examined with a Nikon microscope. To quantify epithelial thickness, 5 µm sections stained with hematoxylin-eosin and evaluated under a microscope (Nikon) with a 40x objective. For each field of vision, epithelial thickness was measured from each end of the visible strip of tissue as well as the middle, for a total of 3 measurements. The entire length of each available strip was evaluated in this manner. p63 positive cells were counted in each field of vision (FOV) under the microscope at 40x. The length of the epithelial cell strip in each FOV was measured and the number of p63-positive cells counted. The number of p63+ cells per um of tissue was then calculated. The primary antibodies used for immunofluorescence were mouse anti-CK14 (1:600, Abcam, ab7800), rabbit anti-CK4 (1:100, Abcam, ab51599), mouse anti-PCNA (1:100, BD Transduction, cat#610664), rabbit anti-pSMAD1/5/8 (1:100, Cell Signaling Technology, cat# 9516) and followed by goat anti-rabbit CY2 (1:1000), goat anti-mouse CY5 (1:600), or rodamine goat anti-mouse (1:200) secondary antibodies (Jackson ImmunoResearch). Immunofluorescent slides were examined with a confocal microscope (Leice TCS SP8-Leica microsystems).
Statistics/Analysis
All experiments were performed in triplicate and data presented as means ± SE. Data were analyzed using Student’s two-tailed type 2 t-test or ANOVA with a Tukey’s or Dunnett’s post hoc test, as appropriate with GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA). A value of P < 0.05 indicated statistical significance.
Materials, data, and associated protocols are available upon request.
Electronic supplementary material
Acknowledgements
This study was supported by the DeGregorio Family foundation for Gastric and Esophageal Cancer Research, the Price Family Foundation, and the National Cancer Institute R03CA198692 (AS). We thank Dr. Anil Rustgi (University of Pennsylvania) for the EPC2-human telomerase reverse transcriptase esophageal epithelial cell line and Meng Li from USC Libraries Bioinformatics Service for comments and discussions. Microscopy and histology services were provided by the Cell and Tissue Imaging Core of USC Research Center for Liver Diseases (NIH P30 DK048522).
Author Contributions
A.S. conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, and wrote the main manuscript text. C.Z. and C.N. performed the experiments, analyzed data, and prepared figures. K.Y. analyzed data and prepared figures. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-30799-7.
References
- 1.Rodriguez P, et al. BMP signaling in the development of the mouse esophagus and forestomach. Development. 2010;137:4171–4176. doi: 10.1242/dev.056077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milano F, et al. Bone morphogenetic protein 4 expressed in esophagitis induces a columnar phenotype in esophageal squamous cells. Gastroenterology. 2007;132:2412–2421. doi: 10.1053/j.gastro.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 3.Wang DH, et al. Aberrant epithelial-mesenchymal Hedgehog signaling characterizes Barrett’s metaplasia. Gastroenterology. 2010;138:1810–1822. doi: 10.1053/j.gastro.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang M, et al. BMP-driven NRF2 activation in esophageal basal cell differentiation and eosinophilic esophagitis. The Journal of clinical investigation. 2015;125:1557–1568. doi: 10.1172/JCI78850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosekrans SL, Baan B, Muncan V, van den Brink GR. Esophageal development and epithelial homeostasis. American journal of physiology. Gastrointestinal and liver physiology. 2015;309:G216–228. doi: 10.1152/ajpgi.00088.2015. [DOI] [PubMed] [Google Scholar]
- 6.Ali IH, Brazil DP. Bone morphogenetic proteins and their antagonists: current and emerging clinical uses. British journal of pharmacology. 2014;171:3620–3632. doi: 10.1111/bph.12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyazono K, Miyazawa K. Id: a target of BMP signaling. Science’s STKE: signal transduction knowledge environment. 2002;2002:pe40. doi: 10.1126/stke.2002.151.pe40. [DOI] [PubMed] [Google Scholar]
- 8.Brazil DP, Church RH, Surae S, Godson C, Martin F. BMP signalling: agony and antagony in the family. Trends in cell biology. 2015;25:249–264. doi: 10.1016/j.tcb.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Roberts DJ, et al. Sonic hedgehog is an endodermal signal inducing Bmp-4 and Hox genes during induction and regionalization of the chick hindgut. Development. 1995;121:3163–3174. doi: 10.1242/dev.121.10.3163. [DOI] [PubMed] [Google Scholar]
- 10.Gargus M, et al. Human esophageal myofibroblasts secrete proinflammatory cytokines in response to acid and Toll-like receptor 4 ligands. American journal of physiology. Gastrointestinal and liver physiology. 2015;308:G904–923. doi: 10.1152/ajpgi.00333.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu C, Chauhan U, Gargus M, Shaker A. Generation and Characterization of an Immortalized Human Esophageal Myofibroblast Line. PloS one. 2016;11:e0153185. doi: 10.1371/journal.pone.0153185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeWard AD, Cramer J, Lagasse E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell reports. 2014;9:701–711. doi: 10.1016/j.celrep.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Dop WA, et al. Hedgehog signalling stimulates precursor cell accumulation and impairs epithelial maturation in the murine oesophagus. Gut. 2013;62:348–357. doi: 10.1136/gutjnl-2011-301141. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, et al. Id proteins are critical downstream effectors of BMP signaling in human pulmonary arterial smooth muscle cells. American journal of physiology. Lung cellular and molecular physiology. 2013;305:L312–321. doi: 10.1152/ajplung.00054.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Baal JW, et al. A comparative analysis by SAGE of gene expression profiles of Barrett’s esophagus, normal squamous esophagus, and gastric cardia. Gastroenterology. 2005;129:1274–1281. doi: 10.1053/j.gastro.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 16.Wang DH, et al. Hedgehog signaling regulates FOXA2 in esophageal embryogenesis and Barrett’s metaplasia. The Journal of clinical investigation. 2014;124:3767–3780. doi: 10.1172/JCI66603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun J, et al. BMP4 activation and secretion are negatively regulated by an intracellular gremlin-BMP4 interaction. The Journal of biological chemistry. 2006;281:29349–29356. doi: 10.1074/jbc.M603833200. [DOI] [PubMed] [Google Scholar]
- 18.Brazil DP. Gremlin1 and chronic pancreatitis: a new clinical target and biomarker? Journal of molecular medicine. 2015;93:1057–1060. doi: 10.1007/s00109-015-1335-6. [DOI] [PubMed] [Google Scholar]
- 19.Laurila R, Parkkila S, Isola J, Kallioniemi A, Alarmo EL. The expression patterns of gremlin 1 and noggin in normal adult and tumor tissues. International journal of clinical and experimental pathology. 2013;6:1400–1408. [PMC free article] [PubMed] [Google Scholar]
- 20.Han EJ, et al. GREM1 Is a Key Regulator of Synoviocyte Hyperplasia and Invasiveness. The Journal of rheumatology. 2016;43:474–485. doi: 10.3899/jrheum.150523. [DOI] [PubMed] [Google Scholar]
- 21.Kalabis J, et al. Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nature protocols. 2012;7:235–246. doi: 10.1038/nprot.2011.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kc K, Rothenberg ME, Sherrill JD. In vitro model for studying esophageal epithelial differentiation and allergic inflammatory responses identifies keratin involvement in eosinophilic esophagitis. PloS one. 2015;10:e0127755. doi: 10.1371/journal.pone.0127755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.