Abstract
Accumulating evidence suggests that genetic factors have a role in major depressive disorder (MDD). However, only limited MDD risk loci have been identified so far. Here we perform a meta-analysis (a total of 90,150 MDD cases and 246,603 controls) through combing three genome-wide association studies of MDD, including 23andMe (cases were self-reported with a clinical diagnosis or treatment of depression), CONVERGE (cases were diagnosed using the Composite International Diagnostic Interview) and PGC (cases were diagnosed using direct structured diagnostic interview (by trained interviewers) or clinician-administered DSM-IV checklists). Genetic variants from two previously unreported loci (rs10457592 on 6q16.2 and rs2004910 on 12q24.31) showed significant associations with MDD (P < 5 × 10−8) in a total of 336,753 subjects. SNPs (a total of 171) with a P < 1 × 10−7 in the meta-analysis were further replicated in an independent sample (GS:SFHS, 2,659 MDD cases (diagnosed with DSM-IV) and 17,237 controls) and one additional risk locus (rs3785234 on 16p13.3, P = 1.57 × 10−8) was identified in the combined samples (a total of 92,809 cases and 263,840 controls). Risk variants on the identified risk loci were associated with gene expression in human brain tissues and mRNA expression analysis showed that FBXL4 and RSRC1 were significantly upregulated in brains of MDD cases compared with controls, suggesting that genetic variants may confer risk of MDD through regulating the expression of these two genes. Our study identified three novel risk loci (6q16.2, 12q24.31, and 16p13.3) for MDD and suggested that FBXL4 and RSRC1 may play a role in MDD. Further functional characterization of the identified risk genes may provide new insights for MDD pathogenesis.
Introduction
Major depressive disorder (MDD) is a complex mental disorder with the highest prevalence (the lifetime prevalence of MDD is about 15% [1, 2]) among the psychiatric disorders [3]. In addition to high prevalence, MDD is also associated with substantial morbidity and mortality [4–6], which makes it the second leading cause of disability worldwide [7]. Despite the fact that MDD imposes great economic burden on society [7, 8], currently the pathogenesis of MDD remains largely unknown. The heritability of MDD is estimated to be around 30–40% [9, 10], indicating that genetic factors have a pivotal role in MDD. Though great effort has been made to investigate the genetic underpinnings of MDD, only limited risk variants and genes have been identified by genetic linkage and association studies [11–15]. The advent of GWAS provides an opportunity to explore the genetic basis of MDD. In 2015, the CONVERGE consortium successfully identified two genome-wide significant risk loci for MDD through using recurrent MDD cases [16]. In 2016, Hyde et al.[17] identified 15 genetic loci associated with risk of MDD through using a large cohort of MDD samples. Recently, Wray et al. conducted the largest GWAS meta-analysis of MDD so far and identified 44 risk loci [18].
To further identify novel risk variants for MDD, we performed a meta-analysis (a total of 336,753 subjects) through combining three independent GWAS of MDD (23andMe, Inc., a personal genetics company [17], the Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium (PGC) [20], and the CONVERGE consortium [16]). Novel genetic variants from ten independent loci showed significant association with MDD at genome-wide significance level (P < 5 × 10−8). SNPs with a P < 1 × 10−7 in the meta-analysis were further replicated in an independent sample, the Generation Scotland: Scottish Family Health Study (GS:SFHS), comprising 2659 MDD cases and 17,237 controls. We also performed eQTL analysis to explore the potential influence of the identified risk variants on gene expression. Our study identified three novel genetic loci (6q16.2, 12q24.31, and 16p13.3) associated with risk of MDD.
Materials and methods
GWAS datasets
We used three independent GWASs of MDD in this study. The first GWAS of MDD was obtained from a recent large-scale study conducted by Hyde et al.[17], which identified 15 genome-wide significant loci [17]. MDD cases and controls were ascertained from 23andMe and subjects who reported a history of clinical diagnosis (or treatment) of depression were included as MDD cases. Participants provided informed consent and participated in the research online, under a protocol approved by the external AAHRPP-accredited IRB, Ethical & Independent Review Services (E&I Review). SNPs were primarily genotyped with the Illumina HumanHap550 + BeadChip and the Illumina OmniExpress + BeadChip. In addition, custom arrays were also used. Logistic regression (additive allelic effects model) was used to test the association of SNPs with MDD. In brief, genome-wide association results from 75,607 MDD cases and 231,747 controls were used in this study. More detailed information about sample collection, SNP genotyping, quality control, and statistical analysis can be found in the original paper [17]. The second GWAS dataset is from the Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium [20]. This dataset contains genome-wide association results from 9240 MDD cases and 9519 controls. Cases were diagnosed with DSM-IV lifetime MDD using direct structured diagnostic interview (by trained interviewers) or clinician-administered DSM-IV checklists, and most of cases were from clinical sources [15]. Most of the controls were selected from the general population randomly and screened for lifetime history of MDD. All subjects were genotyped with Illumina or Affymetrix SNP arrays. Logistic regression was used to test the association between the SNPs and MDD (under an additive model). More detailed information about sample collection, diagnosis, genotyping, statistical analyses, and quality control can be found in the original paper [20]. The third GWAS dataset is from the CONVERGE consortium [16]. To reduce the phenotypic heterogeneity of MDD, CONVERGE consortium only used female MDD cases recruited from China. Briefly, 5303 female recurrent MDD cases and 5337 controls were included in this study and low-coverage whole-genome sequencing was used to genotype all of the subjects. The Composite International Diagnostic Interview (CIDI) (which used DSM-IV criteria) was used for MDD diagnosis. Linear mixed model was utilized to perform the genetic association analysis. More detailed information about sample recruitment, ascertainment, sequencing, genotype calling, quality control, and statistical analysis can be found in the original publication [16].
Meta-analysis
Genome-wide association results from 23andMe [17], PGC [20] and CONVERGE [16] (totaling 90,150 MDD cases and 246,603) were used to perform meta-analysis with the program PLINK (v1.9) [21]. Ancestry determination was performed and subjects who had >97% European ancestry were included in 23andMe study [17]. Genotype data of 23andMe were imputed (minimac2 software [22]) using the reference haplotypes from the 1000 Genomes project [23] (2013 September release). MDD cases and controls used in PGC study [20] were European ancestry and genotype data were imputed with Beagle (v3.0.4) [24] (the phased haplotypes of CEU + TSI from HapMap3 data were used as reference). Subjects used in CONVERGE study [16] were Han Chinese and whole-genome sequencing was used to genotype the samples. The number of SNPs used as input for the meta-analysis was as follows: 23andMe: 15,607,353 SNPs; CONVERGE: 5,992,772 SNPs; PGC 1,235,109 SNPs. We first performed a conversion so each SNP has the same effect allele in each GWAS study. Meta-analysis was then conducted (based on the same effect allele) using summary statistics (including odds ratio, P-value, standard error of odds ratio) from each GWAS. SNPs that were presented in at least two GWAS were included in the final meta-analysis. As described in most GWAS meta-analysis [20, 25], we used the fixed-effect model in this study. The fixed-effect model assumes that the effect of each SNP is the same across different studies. Compared with the random effect model, the fixed-effect model is more powerful for detecting association [25, 26]. I2 was used to quantify the heterogeneity of the meta-analysis [27]. We restricted our analysis on autosomal SNPs and we also validated our meta-analysis results using METAL software [28], which utilizes an inverse-weighted fixed-effects model.
Replication in GS:SFHS
Through combining samples from the 23andMe, CONVERGE and PGC, we identified 213 previously unreported SNPs that reached genome-wide significance level (P < 5 × 10−8). In addition, we also identified 171 SNPs that showed suggestive association (i.e., P < 1 × 10−7) in the meta-analysis (including 23andMe, CONVERGE, and PGC). To further explore if these 171 SNPs were associated with MDD in an independent sample, we tried to replicate these 171 SNPs in GS:SFHS, a family- and population-based Scottish cohort [29]. Due to that 43 SNPs were not available in GS:SFHS, a total of 128 SNPs (with a P < 1 × 10−7) were successfully interrogated in GS:SFHS finally. Briefly, 2659 MDD cases and 17,237 controls were included in GS:SFHS. All of the subjects were recruited from the United Kingdom and structured clinical interviews were applied for the diagnosis of MDD using DSM-IV criteria. The Illumina Human OmniExpressExome -8- v1.0 array was used for genotyping. More detailed information about sample collection, genotyping, quality control, and statistical analysis can be found in the original paper [29].
Linkage disequilibrium analysis
Linkage disequilibrium (LD) values (r2) among the studied SNPs were calculated using genotype data of 99 European subjects (Utah residents with northern and western European ancestry, CEU) from the 1000 Genomes project [23] (http://www.internationalgenome.org/). As the major MDD GWAS (including 23andMe [17], PGC [20], and GS:SFHS [29]) were from populations of European ancestry, we only calculated LD among the studied SNPs in Europeans. Haploview [30] was used to plot the LD pattern among the studied SNPs. LD block was defined with the confidence interval method as described by Gabriel et al. [31].
Frequency distribution of the risk variants in world populations
The frequency of the identified risk variants in different geographic populations was plotted using data from the 1000 Genomes project [32] and the Human Genome Diversity Project (HGDP) [33].
Prioritization of the potential functional variants
To pinpoint the potential functional SNPs at each identified risk loci, we conducted functional prioritization using LINSIGHT [34]. LINSIGHT predicts the functional consequence of the genetic variants using functional and population genomic data, including evolutional conservation (e.g., phyloP score and phastCons element), binding site (e.g., transcription binding site, miRNA binding site and splicing site), and regional annotation data (e.g., ChIP-seq peak of transcription factor, DNase-I hypersensitive site and histone modification). LINSIGHT combines these features using a linear model and scores each variant. The score of LINSIGHT ranges from 0 to 1 and a larger LINSIGHT score represents higher probability that this SNP is functional.
Functional fine-mapping using Probabilistic Annotation Integrator (PAINTOR)
In addition to LINSIGHT, we also used PAINTOR [35] to prioritize the possible causal variant (s) at each risk loci. PAINTOR prioritizes plausible causal variants through integrating genetic association signals (from GWAS) and functional annotation data (such as DNase hypersensitivity sites, enhancer, promoter, and etc.). For each input variant, PAINTOR calculates the probability that the variant is causal. The SNP with the smallest P-value at each identified risk loci was defined as index SNP, and SNPs that were in linkage disequilibrium with the index SNP (r2 > 0.7) were extracted using SNiPA (http://snipa.helmholtz-muenchen.de/snipa/index.php?task=about_snipa) [36]. European populations (CEU) from the 1000 Genomes project [23] were used to calculate the linkage disequilibrium values (r2). The index SNP and SNPs that were in linkage disequilibrium with the index SNP (r2 > 0.7) were used as input for functional fine-mapping. A higher PAINTOR score indicates a higher probability that the SNP is causal.
Pathway analysis
To explore if certain specific gene ontology (GO) categories or pathways were enriched in the identified MDD risk genes, we carried out pathway analysis. Briefly, we first performed LD analysis and SNPs linked with the identified risk SNPs (r2 > 0.3) were extracted. For each loci, the most significant SNP was defined as index SNP. We utilized PLINK (v1.09) [21] to calculate the LD values between the index SNP and nearby SNPs using genotype data of European populations (CEU, Phase I data) from the 1000 Genomes Project [23]. Genes covered by these extracted SNPs were then used for pathway analysis with DAVID [37].
Expression quantitative trait locus (eQTL) analysis
To explore if the identified SNPs are associated with the expression level of nearby genes, we performed eQTL analysis using the LIBD eQTL browser (http://eqtl.brainseq.org/phase1/eqtl/) [38, 39]. The LIBD eQTL browser included brain tissues (the dorsolateral prefrontal cortex, DLPFC) of 412 subjects (including 175 schizophrenia patients, and 237 controls). Gene expression was measured with RNA sequencing and an additive genetic effect model was used to test the association of genotyped SNPs with gene expression. We queried the most significant SNP (i.e., SNPs in Table 1 and Table 2) at each locus using LIBD eQTL browser and genes whose expression is associated with the query SNP were extracted. The P-values were extracted directly from the LIBD eQTL browser and were not corrected for multiple testing. Only significant associations with a P-value less than 1.0 × 10−4 and false discovery rate (FDR) <0.01 were retained. More detailed information about LIBD eQTL database can be found at http://eqtl.brainseq.org/phase1/eqtl/ [38, 39].
Table 1.
Association significance for representative SNPs reaching genome-wide significance level (P < 5 × 10−8) in the meta-analysis
| SNP | Loci | A1a | A2b | Minor allele: Freqc | ORd (23andMe) | P (23andMe) | OR (CONVERGE) | P (CONVERGE) | OR (PGC) | P (PGC) | DIRe | OR (Meta-analysis) | P (Meta-analysis) | I2 (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs12127789 | 1p31.1 | T | G | T:0.111 | 1.051 | 1.67 × 10−7 | NA | NA | 1.080 | 2.71 × 10−2 | +?+ | 1.053 | 1.67 × 10−8 | 0 |
| rs11682175 | 2p16.1 | T | C | C:0.446 | 0.968 | 9.10 × 10−8 | 0.935 | 1.57 × 10−2 | 0.986 | 5.02 × 10−1 | — | 0.968 | 1.21 × 10−8 | 9.53 |
| rs1193510 | 3q25.32 | G | A | G:0.445 | 1.033 | 1.50 × 10−7 | 1.071 | 1.76 × 10−2 | 1.024 | 2.73 × 10−1 | — | 1.033 | 1.09 × 10−8 | 0 |
| rs1501672 | 5q14.3 | G | A | G:0.144 | 1.047 | 1.30 × 10−7 | 1.059 | 4.66 × 10−2 | 1.047 | 1.37 × 10−1 | — | 1.048 | 4.84 × 10−9 | 0 |
| rs4543289 | 5q34 | T | G | T:0.476 | 0.970 | 5.68 × 10−7 | 0.981 | 5.75 × 10−1 | 0.947 | 1.04 × 10−2 | — | 0.969 | 3.36 × 10−8 | 0 |
| rs10457592 | 6q16.2 | G | A | A:0.224 | 0.967 | 2.18 × 10−6 | NA | NA | 0.921 | 9.22 × 10−4 | +?+ | 0.964 | 4.63 × 10−8 | 72.2 |
| rs2004910 | 12q24.31 | G | A | G:0.379 | 1.036 | 6.08 × 10−8 | 1.062 | 2.79 × 10−2 | NA | NA | —? | 1.037 | 8.16 × 10−9 | 0 |
| rs7140116 | 13q14.3 | T | C | C:0.474 | 0.968 | 7.92 × 10−8 | 0.970 | 3.67 × 10−1 | 0.954 | 3.07 × 10−2 | — | 0.967 | 4.97 × 10−9 | 0 |
| rs9540720 | 13q21.32 | G | A | G:0.494 | 1.030 | 1.07 × 10−6 | 1.077 | 6.00 × 10−3 | 1.041 | 5.50 × 10−2 | — | 1.033 | 1.62 × 10−8 | 22.4 |
| rs8037781 | 15q14 | C | A | A:0.401 | 0.967 | 5.76 × 10−8 | 1.012 | 6.98 × 10−1 | 0.960 | 6.25 × 10−2 | +−+ | 0.968 | 3.10 × 10−8 | 22.0 |
Only the most significant SNP on each risk loci was shown. The newly identified risk loci were shown in bold. I2 is an index of heterogeneity. Larger I2 indicates greater heterogeneity
NA not available
aReference allele
bAlternative allele
cMinor allele and frequency of the minor allele (based on Europeans (CEU) from the 1000 Genomes Project)
dOdds ratio is based on reference allele. A total of 75,607 MDD cases and 231,747 controls were included in 23andMe dataset, 5303 MDD cases and 5337 controls were included in CONVERGE dataset, 9240 MDD cases and 9519 controls were included in PGC dataset
eDirection of effect. ‘?’ indicates this SNP was not available in the corresponding dataset
Table 2.
Replication of SNPs with a P < 10−7 in GS:SFHS identifies one novel risk locus (16p13.3) for MDD
| SNP | Loci | A1a | A2b | Minor allele: freqc | OR (23andMe) | P (23andMe) | OR (CONVERGE) | P (CONVERGE) | OR (PGC) | P (PGC) | OR (GS:SFHS) | P (GS:SFHS) | DIRd | OR (meta) | I 2 | P (meta) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs6687024 | 1p31.1 | T | C | C: 0.365 | 1.035 | 5.00 × 10−8 | 1.011 | 7.60 × 10−1 | NA | NA | 1.024 | 4.45 × 10−1 | ++?+ | 1.034 | 0 | 4.45 × 10−8 |
| rs2717046 | 2p16.1 | T | C | T: 0.367 | 1.030 | 1.43 × 10−6 | 1.092 | 1.70 × 10−3 | NA | NA | 1.069 | 3.38 × 10−2 | ++?+ | 1.034 | 62.53 | 1.10 × 10−8 |
| rs1413573 | 13q21.32 | G | A | A: 0.492 | 0.972 | 3.23 × 10−6 | 0.927 | 5.42 × 10−3 | 0.961 | 5.78 × 10−2 | 0.954 | 1.26 × 10−1 | ++++ | 0.969 | 7.91 | 1.68 × 10−8 |
| rs1457257 | 15q14 | G | A | G: 0.366 | 1.036 | 3.22 × 10−8 | 1.006 | 7.55 × 10−1 | NA | NA | 1.060 | 6.24 × 10−2 | −−?− | 1.035 | 0 | 1.09 × 10−8 |
| rs3785234 | 16p13.3 | T | C | C: 0.376 | 1.035 | 3.06 × 10−8 | 0.992 | 9.99 × 10−1 | NA | NA | 1.070 | 3.74 × 10−2 | +−?+ | 1.035 | 10.65 | 1.57 × 10−8 |
| rs9623320 | 22q13.2 | G | A | A: 0.304 | 0.964 | 6.19 × 10−8 | 0.998 | 8.20 × 10−1 | NA | NA | 0.967 | 3.14 × 10−1 | ++?+ | 0.965 | 0 | 4.41 × 10−8 |
Odds ratio is based on reference allele. A total of 75,607 MDD cases and 231,747 controls were included in 23andMe dataset, 5303 MDD cases and 5337 controls were included in CONVERGE dataset, 9240 MDD cases and 9519 controls were included in PGC dataset, 2659 MDD cases and 17,237 controls were included in GS:SFHS. Only the most significant SNP at each risk loci were shown. I2 is an index that is used to quantify the heterogeneity
NA not available
aReference allele
bAlternative allele
cMinor allele and frequency of the minor allele (based on Europeans (CEU) from the 1000 Genomes Project)
dDirection of effect. ‘?’ indicates this SNP was not available in the corresponding dataset
Expression analysis of risk genes in MDD cases and controls
To explore whether nearby genes of the identified risk SNPs were dysregulated in MDD cases, we compared the expression of these genes in MDD cases with controls using expression data (GSE102556) from a recent study of Labonte et al. [40]. Briefly, six brain regions (including the dorsolateral PFC, ventromedial prefrontal cortex, orbitofrontal cortex, ventral subiculum, nucleus accumbens, and anterior insula) of 26 MDD cases (13 males and 13 females) and 22 controls (13 males and 9 females) were collected and genome-wide gene expression was measured with RNA sequencing method. In addition to human subjects, Labonte et al.[40] also established a stressed mice model (using chronic variable stress (CVS)) and measured the gene expression in brains of stressed mice (n = 10) and control mice (n = 10). As chronic stress is a well-characterized risk factor for depression, several rodent models (including chronic social defeat stress and chronic variable stress) have been introduced to uncover the role and mechanism of chronic stress in depression [41, 42]. Among these models, CVS has been proved to be a reliable paradigm and animals exposed to CVS exhibited symptoms parallel to human depression, including anxiety, depression-like behavior, and neurobiological alterations [43]. Labonte et al.[40] exposed the mice to CVS for 21 days and they showed that the stressed mice exhibited depression-and anxiety-like behaviors. Two representative brain regions (i.e., ventromedial prefrontal cortex (vmPFC) and nucleus accumbens (NAc)) implicated in stress responses in rodent models [44] were examined in stressed mice in the Labonte study. To assess if the expression of the identified risk genes was significantly different in MDD cases compared with controls, we extracted the P-values (uncorrected for multiple testing) of MDD risk genes directly from the study of Labonte et al. [40]. Labonte et al. [40] analyzed the males and females separately, and differentially expressed genes in female MDD cases (compared with healthy female controls) and male MDD cases (compared with healthy male controls) were identified separately. More detailed information about the human and mice subjects, RNA extraction, gene expression measurement, statistical analysis can be found in the original study of Labonte et al. [40].
Results
Meta-analysis identified two novel genetic loci associated with MDD
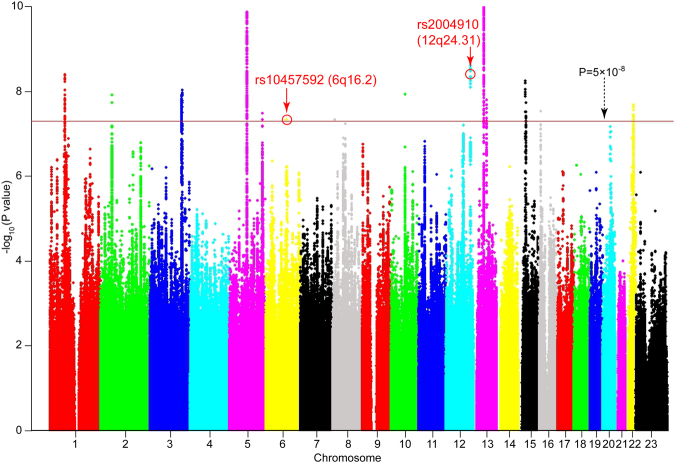
Genome-wide meta-analysis of 90,150 MDD cases and 246,603 controls (from 23andMe, PGC and CONVERGE) identified 213 previously unreported SNPs that showed significant association with MDD at genome-wide significance level (P < 5 × 10−8) (Fig. 1 and Supplementary Table S1). Quantile–quantile plot of the GWAS meta-analysis was shown in Supplementary Figure S1. Of note, these SNPs did not show significant associations (P < 5 × 10−8) with MDD in any of the three genome-wide association studies (Supplementary Table S1). These genome-wide significant SNPs are located in 10 independent genomic regions, including 1p31.1, 2p16.1, 3q25.32, 5q14.3, 5q34, 6q16.2, 12q24.31, 13q14.3, 13q21.32, and 15q14 (Fig. 2, Supplementary Figure S2 and S3). Genetic variants near 8 loci (1p31.1, 2p16.1, 3q25.32, 5q14.3, 5q34, 13q14.3, 13q21.32, and 15q14) have been reported to be associated with MDD previously [17, 45]. Nevertheless, no previous study has showed that genetic variants on 6q16.2 and 12q24.31 were associated with MDD. Thus, our results indicate that 6q16.2 and 12q24.31 are novel risk loci for MDD. The most significant SNP for each of the ten risk loci are listed in Table 1.
Fig. 1.
Meta-analysis results of three MDD GWAS. Novel genetic variants from ten independent loci (1p31.1, 2p16.1, 3q25.32, 5q14.3, 5q34, 6q16.2, 12q24.31, 13q14.3, 13q21.32, and 15q14) showed significant association with MDD (P < 5 × 10−8) in a total of 90,150 MDD cases and 246,603 controls. Two novel risk loci (6q16.2 and 12q24.31) showed significant association with MDD
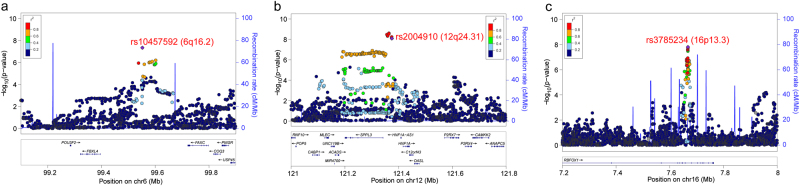
Fig. 2.
Regional association plots for the three novel genome-wide significant loci. a The significant SNP (rs10457592) on 6q16.2 were located upstream of the FBXL4 gene. b The novel identified risk variant (rs2004910) on 12q24.32 were located in upstream of the SPPL3 gene. c The newly identified risk variant (rs3785234) on 16p13.3 were located in intron 7 of the RBFOX1 gene
Replication of SNPs with a P < 1 × 10−7 in GS:SFHS identified one additional novel risk locus for MDD
In addition to the 213 genome-wide significant SNPs (previously unreported) (Supplementary Table S1), we also identified a total of 171 SNPs that showed suggestive association (i.e., P < 1 × 10−7) with MDD in the meta-analysis (including 23andMe, CONVERGE and PGC). We interrogated these 171 SNPs in GS:SFHS and found that 128 SNPs were available in GS:SFHS. We thus performed a meta-analysis restricted to these 128 SNPs and 28 additional genome-wide significant SNPs (Pmeta < 5 × 10−8) were identified in the combined samples (including 23andMe, CONVERGE, PCG, and GS:SFHS, a total of 356,649 subjects (92,809 MDD cases and 263,840 controls)) (Supplementary Table S2). These newly identified significant SNPs were distributed in six genomic regions (Supplementary Table S2), including 1p31.1, 2p16.1, 13q21.32, 15q14, 16p13.3, and 22q13.2. Genetic variants near 1p31.1, 2p16.1, 13q21.32, 15q14, and 22q13.2 have been reported to be associated with MDD previously [17]. However, no previous study has shown the association between genetic variants on 16p13.3 and MDD. Thus, our study indicates that 16p13.3 is a novel risk locus for MDD. The genome-wide significant SNP on 16p13.3 is located in intron 7 of the RBFOX1 gene (Fig. 2c). The most significant SNP for each risk loci in the replication stage (including 23andMe, PGC, CONVERGE, and GS:SFHS) was listed in Table 2. Taken together, our study identified three novel MDD risk loci (i.e., 6q16.2, 12q24.31, and 16p13.3).
The identified risk SNPs did not show significant heterogeneity across studies
Considering that GWAS datasets from different populations (i.e., European and Chinese) were meta-analyzed with fixed-effect model, we also performed heterogeneity analysis. Among the 16 genome-wide significant SNPs, nine SNPs did not show heterogeneity (I2 = 0) and five SNPs showed low heterogeneity (I2 < 0.25) across studies (Tables 1 and 2). And two SNPs (rs10457592 and rs2717046) showed moderate to high heterogeneity (0.5 < I2 < 0.75). These results suggest that the identified SNPs may represent common risk variants for MDD in different populations. However, independent replication is needed to validate our findings.
Prioritization of potential functional SNP at each identified risk loci and pathway analysis
Our meta-analysis identified multiple independent risk loci for MDD (Tables 1 and 2). To further identify the possible functional (or causal) SNPs at each identified locus, we performed functional prediction using LINSIGHT [34]. We extracted the LINSIGHT scores of SNPs linked with the index SNP (r2 > 0.3). We found that 8 out of 10 risk loci have SNPs with a LINSIGHT score larger than 0.9, suggesting these SNPs may have functional consequences. The SNP with the largest LINSIGHT score at each risk locus was listed in Supplementary Table S3. We also performed functional fine-mapping using PAINTOR. The SNP with the highest PAINTOR score at each risk locus was listed in Supplementary Table S4. Of note, four SNPs have a PAINTOR score of 1, implying these SNPs may be functional. However, further experimental validation are needed. Finally, we conducted pathway analysis and found no pathways were significantly enriched in the identified risk genes.
Some of the identified risk SNPs showed significant association with gene expression in human brain (DLPFC)
To explore whether the identified risk variants are associated with gene expression in the DLPFC, we performed eQTL analysis. As the identified risk SNPs on each locus are in linkage disequilibrium (except for 1p31.1), we only selected the most significant SNP (i.e., SNPs in Tables 1 and 2) at each locus for eQTL analysis. SNP rs12127789 is associated with NEGR1 expression (P = 7.63 × 10−5), rs1193510 is associated with the expression of GFM1 (P = 5.49 × 10−6), RSRC1 (P = 5.63 × 10−5) and RARRES1 (P = 7.62 × 10−5), rs1501672 is associated with LINC00461 expression, rs2004910 is associated with SPPL3 expression (P = 9.16 × 10−13), rs9623320 is associated with the expression of L3MBTL2 (P = 1.64 × 10−7), XPNPEP3 (P = 2.84 × 10−7) and POLR3H (P = 3.41 × 10−5), and rs7140116 is associated with PCDH8P1 expression (P = 9.64 × 10−5) in the DLPFC (Supplementary Table S5). SNPs on five loci (rs4543289, rs10457592, rs9540720, rs8037781, and rs11682175) were not associated with gene expression in the LIBD eQTL database. These eQTL results suggest that the identified risk variants may modulate the expression level of nearby genes in the DLPFC.
Upregulation of FBXL4 and RSRC1 in brains of MDD cases compared with controls
Expression quantitative trait locus analysis showed that some of the identified risk variants were associated with gene expression in human brains (Supplementary Table S5), suggesting that the risk variants may confer risk of MDD through regulating gene expression. We thus examined the expression level of genes near the identified risk loci in MDD cases and controls using expression data (GSE102556) from Labonte et al. [40]. Only genes nearest to the identified risk SNP were examined. We found that NEGR1 (P = 0.038, uncorrected) was significantly downregulated in female MDD cases compared with controls. By contrast, FBXL4 (P = 0.0072, uncorrected) and RSRC1 (P = 0.042, uncorrected) were significantly upregulated in female MDD cases compared with controls. Consistent with the observation in female MDD cases, we found that Fbxl4 and Rsrc1 were also significantly upregulated in brains of stressed female mice (P = 0.019 and P = 8.50 × 10−4, respectively, uncorrected). The significant upregulation of FBXL4 and RSRC1 in both female MDD cases and stressed female mice suggest that dysregulation of these two genes may have a role in MDD.
Discussion
Accumulating evidence suggests that genetic factors play pivotal roles in MDD. However, currently the genetic basis of MDD remains largely unknown. Identification of MDD-associated genetic variants remains a major challenge as MDD is a moderately heritable, clinically heterogeneous condition with a complex genetic architecture [46]. Though previous GWAS have identified several genome-wide significant risk variants [16, 29, 47], most of the risk loci of MDD remain to be uncovered. To further identify new MDD-associated variants (which could not be detected in individual GWAS due to limited power), we tried to improve the power of this study through increasing sample size and utilizing a relatively powerful statistical method. First, considering that the effect size of most risk variants is relatively small, combining samples from different studies may help to identify new risk variants as the statistical power improves with the increase of sample size. Second, as reported in most previous GWAS [20, 25], we used the fixed-effect model in this study. The fixed-effect model assumes that the effects of the genetic variants are the same across studies, thus it is useful to identify novel risk variants through combining different studies. Compared with the random effect model, the fixed-effect model provides narrower confidence intervals and it is useful for detecting association [25, 26].
We successfully identified three novel MDD-associated loci (6q16.2, 12q21.31, and 16p13.3). The newly identified SNP on 6q16.2 (rs10457592) is located upstream of the FBXL4 gene (Fig. 2a), which encodes a member of the F-box protein family. FBXL4 protein is found to be expressed in mitochondria and may play a pivotal role in the maintenance of mitochondrial DNA (mtDNA) [48]. Previous studies have showed that mutations in FBXL4 resulted in mitochondrial encephalopathy [48, 49], indicating the important role of FBXL4 in maintenance of mitochondrial function. In addition to the genetic evidence, expression analysis also suggests that FBXL4 may be involved in MDD. Compared with controls, FBXL4 was significantly upregulated in both female MDD cases and female stressed mice, implying dysregulation of FBXL4 in MDD.
In addition to 6q16.2, our study also suggests that 12q21.31 and 16p13.3 are novel risk loci for MDD. It should be noted that genetic variants near 12q21.31 and 16p13.3 showed significant associations with MDD in the discovery stage of Hyde et al.’s study [17]. However, they did not follow these SNPs as these SNPs were absent in PGC (Hyde et al. performed a meta-analysis through combining results from PGC and 23andMe, and only SNPs presented in both PGC and 23andMe were followed for downstream analysis). Accordingly, these two loci were not included in the final 15 loci reported by Hyde et al. [17].
We explored the genome-wide significant SNPs (rs12415800 and rs35936514, which located upstream of SIRT1 and intronic region of LHPP, respectively) reported by CONVERGE in the meta-analysis. Both rs12415800 and rs35936514 were not available in PGC dataset. We found that rs12415800 is also significantly associated with MDD in 23andMe (P = 0.041), with the same risk allele (i.e., A allele) in CONVERGE and 23andMe studies (Supplementary Table S6). In fact, SNP rs12415800 reached genome-wide significant level (P = 1.19 × 10−8) when samples from 23andMe and CONVERGE were combined. In addition, heterogeneity analysis showed that there was low heterogeneity (I2 = 0.11) in 23andMe and CONVERGE for SNP rs12415800, suggesting this SNP may represent a common risk variant in Chinese and European populations. SNP rs35936514 is not associated with MDD in 23andMe dataset. When samples from 23andMe and CONVERGE were combined, rs35936514 only showed marginal association with MDD (P = 0.0196). Heterogeneity analysis showed there was significant heterogeneity (I2 = 0.96) for rs35936514 in 23andMe and CONVERGE (Supplementary Table S6), implying that SNP may represent an Asian-specific susceptibility risk variant for MDD. In fact, we noted that the frequencies of the risk alleles of rs12415800 and rs35936514 are different in world populations (Supplementary Figure S4), further suggesting that population-specific risk variants may exist. However, more work is needed to verify this.
We also explored the potential functional consequences of the identified risk SNPs. Of note, the novel risk SNP (rs2004910) on 12q21.31 was associated with SPPL3 expression in human brain (Supplementary Table S5). SPPL3 encodes signal peptide peptidase like 3 (SPPL3), an intramembrane protease that cleaves several types of membrane signal peptides [50, 51]. Previous studies have showed the important functions of SPPL3 in eukaryotes [52]. Voss et al.[52] showed that SPPL3 regulates cellular N-glycosylation and downregulation of SPPL3 leads to a hyperglycosylation phenotype. In addition to regulation of glycosylation, recent studies also showed that SPPL3 is involved in immune response, including NFAT activation [53] and regulation of NK cell maturation and cytotoxicity [54]. Surprisingly, the activation of NFAT is not dependent on the proteolytic activity of SPPL3 [53]. A recent study also showed that genetic variant nearby SPPL3 is associated with the levels of markers of inflammation [55], consistent with SPPL3’s reported role in immunity and inflammation. Of note, immune dysfunction has been thought to be an important contributor to MDD [56, 57]. Our study suggests that SPPL3 may represent a novel risk gene for MDD.
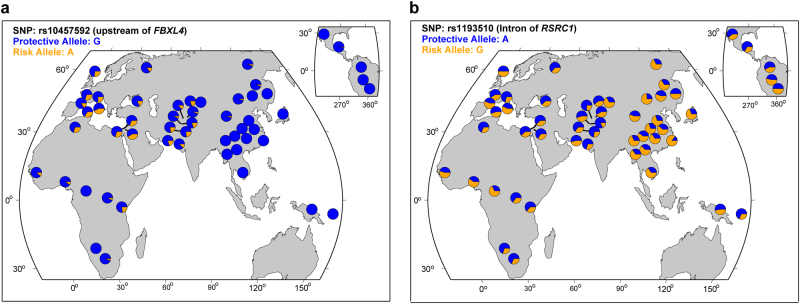
Another interesting gene is RSRC1 (also named SRrp53). Most of the newly identified risk SNPs on 3q25.32 are located in introns of RSRC1, and our eQTL analysis indicated that the most significant SNP (rs1193510) was associated with RSRC1 expression in DLPFC of human brain (Supplementary Table S5). We further showed that RSRC1 was significantly upregulated in brains of female MDD cases. Intriguingly, expression of Rsrc1 was also significantly upregulated in brains of stressed female mice. These results suggest that RSRC1 may have a role in MDD and genetic variants on 3q25.32 may confer risk of MDD through affecting the expression of RSRC1. RSRC1 encodes a member of the serine and arginine rich-related protein family that plays a pivotal role in mRNA splicing [58]. In addition to MDD, RSRC1 was also reported to be associated with schizophrenia [59] and height [60]. The frequency distribution of the risk alleles of FBXL4 and RSRC1 in global populations was shown in Figure 3.
Fig. 3.
The frequency distribution of the risk alleles of FBXL4 and RSRC1 in global populations. a Frequency distribution of the risk allele (A allele) of rs10457592 in global populations. b Frequency distribution of the risk allele (G allele) of rs1193510 in global populations
Taken together, our study identified three novel risk loci for MDD and our results suggest that these risk SNPs may contribute to MDD risk through modulating gene expression. Further verification of our findings in independent samples and functional characterization of the identified risk genes may provide potential targets for therapeutics and diagnostics.
Electronic supplementary material
Acknowledgements
This study was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13000000 to X.-J.L), the National Key Research and Development Program of China (Stem Cell and Translational Research) (2016YFA0100900 to X.-J.L), the National Natural Science Foundation of China (31722029 to X.-J.L, 81471358 and 81671326 to C.Z.), and the Key Research Project of Yunnan Province (2017FA008 to X.-J.L). X.-J.L was also supported by the 1000 Young Talents Program. Generation Scotland received core support from the Chief Scientist Office of the Scottish Government Health Directorates [CZD/16/6] and the Scottish Funding Council [HR03006]. Genotyping of the GS:SFHS samples was carried out by the Genetics Core Laboratory at the Wellcome Trust Clinical Research Facility, Edinburgh, Scotland and was funded by the Medical Research Council UK and the Wellcome Trust (Wellcome Trust Strategic Award “STratifying Resilience and Depression Longitudinally” (STRADL) Reference 104036/Z/14/Z). We would like to thank the research participants and employees of 23andMe for making this work possible. We thank the following members of the 23andMe Research Team: Michelle Agee, Babak Alipanahi, Adam Auton, Robert K. Bell, Katarzyna Bryc, Sarah L. Elson, Pierre Fontanillas, Nicholas A. Furlotte, David A. Hinds, Karen E. Huber, Aaron Kleinman, Nadia K. Litterman, Jennifer C. McCreight, Matthew H. McIntyre, Joanna L. Mountain, Elizabeth S. Noblin, Carrie A.M. Northover, Steven J. Pitts, J. Fah Sathirapongsasuti, Olga V. Sazonova, Janie F. Shelton, Suyash Shringarpure, Chao Tian, Joyce Y. Tung, Vladimir Vacic, and Catherine H. Wilson, who generated and made the summary statistics available for us, which made this work possible.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiaoyan Li, Zhenwu Luo.
Contributor Information
Chen Zhang, Email: zhangchen645@163.com.
Xiong-Jian Luo, Phone: +86-871-68125413, Email: luoxiongjian@mail.kiz.ac.cn.
Electronic supplementary material
The online version of this article (10.1038/s41386-018-0078-9) contains supplementary material, which is available to authorized users.
References
- 1.Hasin DS, Goodwin RD, Stinson FS, Grant BF. Epidemiology of major depressive disorder: results from the National Epidemiologic Survey on Alcoholism and Related Conditions. Arch Gen Psychiatry. 2005;62:1097–106. doi: 10.1001/archpsyc.62.10.1097. [DOI] [PubMed] [Google Scholar]
- 2.Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R) JAMA. 2003;289:3095–105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- 3.Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–51. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angst F, Stassen HH, Clayton PJ, Angst J. Mortality of patients with mood disorders: follow-up over 34-38 years. J Affect Disord. 2002;68:167–81. doi: 10.1016/S0165-0327(01)00377-9. [DOI] [PubMed] [Google Scholar]
- 5.Judd LL. The clinical course of unipolar major depressive disorders. Arch Gen Psychiatry. 1997;54:989–91. doi: 10.1001/archpsyc.1997.01830230015002. [DOI] [PubMed] [Google Scholar]
- 6.Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006;367:1747–57. doi: 10.1016/S0140-6736(06)68770-9. [DOI] [PubMed] [Google Scholar]
- 7.Ferrari AJ, Charlson FJ, Norman RE, Patten SB, Freedman G, Murray CJ, et al. Burden of depressive disorders by country, sex, age, and year: findings from the global burden of disease study 2010. PLoS Med. 2013;10:e1001547. doi: 10.1371/journal.pmed.1001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenberg PE, Fournier AA, Sisitsky T, Pike CT, Kessler RC. The economic burden of adults with major depressive disorder in the United States (2005 and 2010) J Clin Psychiatry. 2015;76:155–62. doi: 10.4088/JCP.14m09298. [DOI] [PubMed] [Google Scholar]
- 9.Corfield EC, Yang Y, Martin NG, Nyholt DR. A continuum of genetic liability for minor and major depression. Transl Psychiatry. 2017;7:e1131. doi: 10.1038/tp.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson RE, Cai N, Bigdeli TB, Li Y, Reimers M, Nikulova A, et al. The genetic architecture of major depressive disorder in Han Chinese Women. JAMA Psychiatry. 2017;74:162–8. doi: 10.1001/jamapsychiatry.2016.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breen G, Webb BT, Butler AW, van den Oord EJ, Tozzi F, Craddock N, et al. A genome-wide significant linkage for severe depression on chromosome 3: the depression network study. Am J Psychiatry. 2011;168:840–7. doi: 10.1176/appi.ajp.2011.10091342. [DOI] [PubMed] [Google Scholar]
- 12.Flint J, Kendler KS. The genetics of major depression. Neuron. 2014;81:484–503. doi: 10.1016/j.neuron.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knowles EE, Kent JW, Jr., McKay DR, Sprooten E, Mathias SR, Curran JE, et al. Genome-wide linkage on chromosome 10q26 for a dimensional scale of major depression. J Affect Disord. 2016;191:123–31. doi: 10.1016/j.jad.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo X, Stavrakakis N, Penninx BW, Bosker FJ, Nolen WA, Boomsma DI, et al. Does refining the phenotype improve replication rates? A review and replication of candidate gene studies on Major Depressive Disorder and Chronic Major Depressive Disorder. Am J Med Genet B Neuropsychiatr Genet. 2016;171B:215–36. doi: 10.1002/ajmg.b.32396. [DOI] [PubMed] [Google Scholar]
- 15.Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2012;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CONVERGE consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–91. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR, et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016;48:1031–6. doi: 10.1038/ng.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. First published online April 26, 2018; 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed]
- 19.The Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium (2018). Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depressive disorder. BioRxiv preprint first posted online Jul 24, 2017 doi: 101101/167577.
- 20.Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–4. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–23. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeggini E, Ioannidis JP. Meta-analysis in genome-wide association studies. Pharmacogenomics. 2009;10:191–201. doi: 10.2217/14622416.10.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Begum F, Ghosh D, Tseng GC, Feingold E. Comprehensive literature review and statistical considerations for GWAS meta-analysis. Nucleic Acids Res. 2012;40:3777–84. doi: 10.1093/nar/gkr1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genome-wide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng Y, Navarro P, Shirali M, Howard DM, Adams MJ, Hall LS, et al. Genome-wide regional heritability mapping identifies a locus within the TOX2 gene associated with major depressive disorder. Biol Psychiatry. 2017;82:312–21. doi: 10.1016/j.biopsych.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 31.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 32.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–37. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang YF, Gulko B, Siepel A. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat Genet. 2017;49:618–24. doi: 10.1038/ng.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kichaev G, Yang WY, Lindstrom S, Hormozdiari F, Eskin E, Price AL, et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 2014;10:e1004722. doi: 10.1371/journal.pgen.1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnold M, Raffler J, Pfeufer A, Suhre K, Kastenmuller G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics. 2014;31:1334–6. doi: 10.1093/bioinformatics/btu779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 38.BrainSeq: A Human Brain Genomics Consortium. BrainSeq: neurogenomics to drive novel target discovery for neuropsychiatric disorders. Neuron. 2015;88:1078–83. doi: 10.1016/j.neuron.2015.10.047. [DOI] [PubMed] [Google Scholar]
- 39.Birnbaum R, Jaffe A, Q C, Shin J, Consortium B, Kleinman J, et al. Investigating the neuro-immunogenic architecture of schizophrenia. Mol Psychiatry. Advance online publication 9 May 2017; doi: 2010.1038/mp.2017.2089.
- 40.Labonte B, Engmann O, Purushothaman I, Menard C, Wang J, Tan C, et al. Sex-specific transcriptional signatures in human depression. Nat Med. 2017;23:1102–11. doi: 10.1038/nm.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katz RJ, Hersh S. Amitriptyline and scopolamine in an animal model of depression. Neurosci Biobehav Rev. 1981;5:265–71. doi: 10.1016/0149-7634(81)90008-7. [DOI] [PubMed] [Google Scholar]
- 42.Katz RJ, Roth KA, Carroll BJ. Acute and chronic stress effects on open field activity in the rat: implications for a model of depression. Neurosci Biobehav Rev. 1981;5:247–51. doi: 10.1016/0149-7634(81)90005-1. [DOI] [PubMed] [Google Scholar]
- 43.Scheich B, Cseko K, Borbely E, Abraham I, Csernus V, Gaszner B, et al. Higher susceptibility of somatostatin 4 receptor gene-deleted mice to chronic stress-induced behavioral and neuroendocrine alterations. Neuroscience. 2017;346:320–36. doi: 10.1016/j.neuroscience.2017.01.039. [DOI] [PubMed] [Google Scholar]
- 44.Russo SJ, Nestler EJ. The brain reward circuitry in mood disorders. Nat Rev Neurosci. 2013;14:609–25. doi: 10.1038/nrn3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao X, Zheng F, Chang H, Ma Y, Yao YG, Luo XJ, et al. The Gene Encoding Protocadherin 9 (PCDH9), a Novel Risk Factor for Major Depressive Disorder. Neuropsychopharmacology. 2017; In Press: 10.1038/npp.2017.1241. [DOI] [PMC free article] [PubMed]
- 46.Bigdeli TB, Ripke S, Peterson RE, Trzaskowski M, Bacanu SA, Abdellaoui A, et al. Genetic effects influencing risk for major depressive disorder in China and Europe. Transl Psychiatry. 2017;7:e1074. doi: 10.1038/tp.2016.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng Y, Navarro P, Fernandez-Pujals AM, Hall LS, Clarke TK, Thomson PA, et al. A combined pathway and regional heritability analysis indicates NETRIN1 pathway is associated with major depressive disorder. Biol Psychiatry. 2017;81:336–46. doi: 10.1016/j.biopsych.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonnen PE, Yarham JW, Besse A, Wu P, Faqeih EA, Al-Asmari AM, et al. Mutations in FBXL4 cause mitochondrial encephalopathy and a disorder of mitochondrial DNA maintenance. Am J Hum Genet. 2013;93:471–81. doi: 10.1016/j.ajhg.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gai X, Ghezzi D, Johnson MA, Biagosch CA, Shamseldin HE, Haack TB, et al. Mutations in FBXL4, encoding a mitochondrial protein, cause early-onset mitochondrial encephalomyopathy. Am J Hum Genet. 2013;93:482–95. doi: 10.1016/j.ajhg.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nyborg AC, Ladd TB, Jansen K, Kukar T, Golde TE. Intramembrane proteolytic cleavage by human signal peptide peptidase like 3 and malaria signal peptide peptidase. FASEB J. 2006;20:1671–9. doi: 10.1096/fj.06-5762com. [DOI] [PubMed] [Google Scholar]
- 51.Voss M, Schroder B, Fluhrer R. Mechanism, specificity, and physiology of signal peptide peptidase (SPP) and SPP-like proteases. Biochim Biophys Acta. 2013;1828:2828–39. doi: 10.1016/j.bbamem.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 52.Voss M, Kunzel U, Higel F, Kuhn PH, Colombo A, Fukumori A, et al. Shedding of glycan-modifying enzymes by signal peptide peptidase-like 3 (SPPL3) regulates cellular N-glycosylation. EMBO J. 2014;33:2890–905. doi: 10.15252/embj.201488375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makowski SL, Wang Z, Pomerantz JL. A protease-independent function for SPPL3 in NFAT activation. Mol Cell Biol. 2015;35:451–67. doi: 10.1128/MCB.01124-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hamblet CE, Makowski SL, Tritapoe JM, Pomerantz JL. NK cell maturation and cytotoxicity are controlled by the intramembrane aspartyl protease SPPL3. J Immunol. 2016;196:2614–26. doi: 10.4049/jimmunol.1501970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naitza S, Porcu E, Steri M, Taub DD, Mulas A, Xiao X, et al. A genome-wide association scan on the levels of markers of inflammation in Sardinians reveals associations that underpin its complex regulation. PLoS Genet. 2012;8:e1002480. doi: 10.1371/journal.pgen.1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. 2008;9:46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Otte C, Gold SM, Penninx BW, Pariante CM, Etkin A, Fava M, et al. Major depressive disorder. Nat Rev Dis Prim. 2016;2:16065. doi: 10.1038/nrdp.2016.65. [DOI] [PubMed] [Google Scholar]
- 58.Cazalla D, Newton K, Caceres JF. A novel SR-related protein is required for the second step of Pre-mRNA splicing. Mol Cell Biol. 2005;25:2969–80. doi: 10.1128/MCB.25.8.2969-2980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Potkin SG, Turner JA, Fallon JA, Lakatos A, Keator DB, Guffanti G, et al. Gene discovery through imaging genetics: identification of two novel genes associated with schizophrenia. Mol Psychiatry. 2009;14:416–28. doi: 10.1038/mp.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berndt SI, Gustafsson S, Magi R, Ganna A, Wheeler E, Feitosa MF, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45:501–12. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.