Highlights
-
•
We evaluated glyphosate, isoxaflutole, quizalofop-p-ethyl and mesotrione.
-
•
Gene expression alterations were measured in the HepaRG liver cell line.
-
•
Quizalofop-p-ethyl was the most toxic, causing alterations in lipid metabolism.
-
•
Isoxaflutole was less toxic, but was revealed as a possible PXR activator.
-
•
Glyphosate and mesotrione only caused subtle changes in transcriptome profiles.
Keywords: Glyphosate, Quizalofop, Mesotrione, Isoxaflutole, RNA-seq, HepaRG, Transcriptome, Metabolome, NAFLD
Abstract
Use and thus exposure to quizalofop-p-ethyl, isoxaflutole, mesotrione and glyphosate, which are declared as active principles in commercial formulations of herbicides, is predicted to rapidly increase in coming years in an effort to overcome the wide-spread appearance of glyphosate-resistant weeds, especially in fields where glyphosate-tolerant genetically modified crops are cultivated in the USA. Thus, there is an urgent need for an evaluation of metabolic effects of new pesticide ingredients used to replace glyphosate. As the liver is a primary target of chemical pollutant toxicity, we have used the HepaRG human liver cell line as a model system to assess the toxicological insult from quizalofop-p-ethyl, isoxaflutole, mesotrione and glyphosate by determining alterations in the transcriptome caused by exposure to three concentrations of each of these compounds, including a low environmentally relevant dose. RNA-seq data were analysed with HISAT2, StringTie and Ballgown. Quizalofop-p-ethyl was found to be the most toxic of the pesticide ingredients tested, causing alterations in gene expression that are associated with pathways involved in fatty acid degradation and response to alcoholism. Isoxaflutole was less toxic, but caused detectable changes in retinol metabolism and in the PPAR signalling pathway at a concentration of 1 mM. ToxCast data analysis revealed that isoxaflutole activated PPAR gamma receptor and pregnane X responsive elements in reporter gene assays. Glyphosate and mesotrione caused subtle changes in transcriptome profiles, with too few genes altered in their function to allow a reliable pathway analysis. In order to explore the effects of glyphosate in greater depth and detail, we undertook a global metabolome profiling. This revealed a decrease in free long chain fatty acids and polyunsaturated fatty acid levels at the lowest concentration (0.06 μM) of glyphosate, although no effects were detected at the two higher concentrations tested, perhaps suggesting a non-linear dose response. This surprising result will need to be confirmed by additional studies. Overall, our findings contribute to filling the knowledge gap regarding metabolic toxicity that can potentially arise from exposure to these four herbicide active principles.
1. Introduction
The application of genetic engineering in agriculture has resulted in the creation of agricultural crops modified to tolerate and accumulate a herbicide or/and to produce their own insecticides [1]. This has considerably modified pesticide use patterns. The use of the most toxic herbicides, such as alachlor or metolachlor, has decreased with the adoption of genetically modified (GM) crops [2]. In turn, glyphosate-based herbicides (GBH) uses have dramatically increased, by approximately 100-fold since the late 1970s [3]. GBHs are now associated with the cultivation of 80% of agricultural GM crops [3]. For example, as of 2017, 94% of soybeans and 89% of maize grown in the US is GM and mostly engineered to tolerate GBH application [4]. However, the growing reliance on a single herbicide in GM cropping systems has led to a widespread growth of glyphosate resistant weeds [5]. Since the introduction of glyphosate-tolerant GM crops in 1996 and the massive increase in GBH use, 14 plant species have evolved resistance to glyphosate in 32 states in the USA [1]. In an effort to control the very considerable economic damage caused by the rapid spread of glyphosate-resistant weeds, major biotechnology companies have developed new GM crops combining tolerances to several herbicide active principles. These new GM crops have been designed to tolerate the combined application of commercial formulations of pesticides including glyphosate, 2,4-D, dicamba, glufosinate, quizalofop-p-ethyl, mesotrione, and isoxaflutole [6]. This new generation of agricultural GM crops will in all likelihood lead to an increase in human exposure to these compounds, making the potential non-target health effects of their residues a burning question.
The liver is a very important target of the toxicity of chemical pollutants. Although it has long been known that acute exposure to chemicals in the workplace can cause damage to the liver, recent surveys show that the exposure to physiologically active environmental pollutants can be a risk factor in the development of chronic liver ailments such as non-alcoholic fatty liver disease (NAFLD) [7]. Exposure to chemical pollutants can also act as a second hit in the development of liver diseases in human populations having a liver function compromised by over-nutrition and a lack of exercise [8]. A recent review of rodent toxicology databases found that pesticides were the chemicals that were most frequently associated with fatty liver disease in rodents [9]. In addition, NAFLD is associated with great economic consequences as it currently affects 25% of the US population [10]. This makes the evaluation of liver toxicity a high priority for newly marketed pesticides.
We report here the use of terminally differentiated HepaRG cells to characterize the effects of quizalofop-p-ethyl, isoxaflutole, mesotrione and glyphosate on human liver metabolism. HepaRG cells are derived from the human hepatic HepaRG progenitor cell line [11]. HepaRG are more stable than other hepatic cell lines because the pool of cells used to produce the differentiated hepatocytes is consistently maintained at low passage. They can thus be used as an alternative to primary human hepatocytes in order to produce reproducible results by avoiding donor variation [11]. HepaRG cells express the most important liver nuclear receptors known to be targets of endocrine disrupting chemicals [12], such as the aryl hydrocarbon receptor (AhR) and the peroxisome proliferator-activated receptor γ (PPARγ). They are also metabolically competent and express most phase 1 (i.e., CYP1A2, CYP3 A4, or CYP2C9) and phase 2 (i.e., GSTA1/2/4, GSTM1, UGT1A1) enzymes at levels comparable to those found in freshly isolated human hepatocytes [13].
We performed transcriptomics experiments to identify gene networks involved in liver function disturbances induced by three concentrations of quizalofop-p-ethyl, isoxaflutole, mesotrione and glyphosate. Our results show that quizalofop-p-ethyl is the most potent toxicant in this assay affecting fatty acid degradation and bringing about a response akin to alcoholism. Isoxaflutole was less toxic, but caused detectable changes in retinol metabolism and in the PPAR signalling pathway. Glyphosate and mesotrione caused only subtle changes in transcriptome profiles that did not allow a reliable pathway analysis. A follow-up metabolomics analysis of cells treated with glyphosate revealed a decrease in the level of free long chain fatty acids (LCFAs) and polyunsaturated fatty acids (PUFAs) at the lowest concentration (0.06 μM) of glyphosate, perhaps suggesting a non-linear dose response.
2. Material and methods
2.1. Reagents
All reagents and chemicals, unless otherwise specified, were of analytical grade and were purchased from Sigma-Aldrich (Gillingham, Dorset, UK). The pesticides were N-(phosphonomethyl)glycine (CAS: 1071-83-6, purity≥96%), quizalofop-p-ethyl (CAS: 100646-51-3, purity≥98%, Santa Cruz Biotechnology), isoxaflutole (CAS: 141112-29-0, purity≥98%, Santa Cruz Biotechnology) and mesotrione (CAS: 104206-82-8, purity>99%, Santa Cruz Biotechnology). All these compounds were dissolved in dimethyl sulfoxide (DMSO) to prepare stock solutions. The Williams’ E medium + GlutaMAX™ were from Gibco (Thermo Fisher, Loughborough, UK). The supplements ADD670 and DMSO, were provided by Biopredic International (Rennes, France).
2.2. HepaRG cell culture
Differentiated HepaRG™ cells (HPR 116) were purchased from Biopredic International (Rennes, France). Cells were thawed, suspended and plated in the general purpose medium (Williams’ E medium + GlutaMAX™) containing the ADD670 supplement. A total of 72,000 and 2,000,000 cells per well were plated in collagen-coated 96 well-plates (Greiner Bio-One, Germany), and 6 well-plates (Biopredic), respectively. All cells were grown at 37 °C (5% CO2). The medium was refreshed at days-3, -5 and -7 following initial plating. The cells were kept in general purpose medium until day-8, when the culture becomes well organized and includes well-delineated trabeculae and many canaliculi-like structures. At this time, the culture is composed of primitive biliary epithelial cells and mature hepatocytes with basal metabolic activities similar to freshly isolated primary cells. From day-8 to day-14, cells were switched to the test medium composed of Williams’ E medium + GlutaMAX™ supplemented with 2% fetal bovine serum (FBS; GE Healthcare Life Sciences, Buckinghamshire, UK) and 1% DMSO, as well as different concentrations of pesticides or just solvent as a control. In order to ensure coverage of a wide range of potential biological effects, three concentrations of each active principle were tested; a concentration representative of low environmental exposure (0.06-0.1 μM), an intermediate concentration (6–10 μM) and a high concentration (600–1000 μM).
2.3. Sample quality control
The transcriptome of cells exposed to quizalofop-p-ethyl, isoxaflutole and mesotrione was analysed as follows. RNA extraction was performed using the Qiagen RNeasy kit (Qiagen, Manchester, UK) according to the manufacturer's instructions. The samples were checked for RNA quality using the Agilent 2100 Bioanalyzer (Agilent Technologies UK Limited, Craven Arms, UK) and quantified using a Nanodrop instrument (ND-1000 Spectrophotometer; Thermo Fisher Scientific Inc.). Out of 50 samples analysed, 48 showed good integrity with RIN numbers ranging from 7 to 10. The two remaining samples showed a small degree of degradation with RINs of 6.1 and 6.5. All samples were taken forward for RNA library preparation.
2.4. Library generation and RNA-sequencing
A 100 ng aliquot of total RNA from each sample was used to prepare total RNA libraries using the KAPA Stranded RNA-Seq Kit with RiboErase (KAPA Biosystems, Massachusetts, USA), and samples were randomised before preparation. Fragmentation prior to first strand cDNA synthesis was carried out using incubation conditions recommended by the manufacturer for intact RNA samples (94 °C for 6 min). Polymerase chain reaction (PCR) was performed for 14 cycles for final library amplification. Resulting libraries were quantified using the Qubit 2.0 spectrophotometer (Life Technologies, California, USA) and average fragment size assessed using the Agilent 2200 Tapestation (Agilent Technologies UK Limited). The transcriptome of HepaRG cells exposed to glyphosate was sequenced employing the same strategy, except that the libraries were prepared as previously described [14]. A total of three separate sequencing pools were created using equimolar quantities of each sample with compatible indexes; two with 17 samples each, and one with 16 samples. Paired-end reads of 75bp were generated for each library using the Illumina NextSeq®500 in conjunction with the NextSeq®500 v2 High-output 150-cycle kit (Illumina Inc., Cambridge, UK).
2.5. Mass spectrometry-based metabolomics
We harvested approximately 5,000,000 HepaRG cells per sample from the 6 well-plates in order to obtain a sufficient quantity of material to perform the metabolomics experiment. Cells were detached using 0.05% trypsin EDTA (Life Technologies), and then centrifuged in order to eliminate trypsin residues. Finally, cell pellets were frozen at −80 °C until use. Metabolomics analysis of the frozen cell pellets was conducted by Metabolon Inc. (Durham, NC, USA). Samples were prepared using the automated MicroLab STAR® system (Hamilton Company (Reno, NV, USA). Proteins were precipitated with methanol under vigorous shaking for 2 min using the Glen Mills GenoGrinder 2000 (Glen Mills, NJ, USA), followed by centrifugation. Samples were placed briefly on a TurboVap® (Zymark, Hopkinton, MA, USA)) to remove the organic solvent. The sample extracts were stored overnight under nitrogen before preparation for analysis. The resulting extract was analysed on four independent instrument platforms: two different separate reverse phase ultrahigh performance liquid chromatography-tandem mass spectroscopy analysis (RP/UPLC-MS/MS) with positive ion mode electrospray ionization (ESI), a RP/UPLC-MS/MS with negative ion mode ESI, as well as a by hydrophilic-interaction chromatography (HILIC)/UPLC-MS/MS with negative ion mode ESI as previously described [14].
All UPLC-MS/MS methods utilized a Waters ACQUITY ultra-performance liquid chromatography (UPLC) (Waters Corporation, Milford, MA, USA) and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer (Thermo Fisher Scientific Inc.) interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35,000 mass resolution. The sample extract was dried then reconstituted in solvents compatible to each of four methods used. Each reconstitution solvent contained a series of standards at fixed concentrations to ensure injection and chromatographic consistency. One aliquot was analysed using acidic positive ion conditions, chromatographically optimized for more hydrophilic compounds. In this method, the extract was gradient eluted from a C18 column (Waters UPLC BEH C18-2.1 x 100 mm, 1.7 μm) using water and methanol, containing 0.05% perfluoropentanoic acid (PFPA) and 0.1% formic acid (FA). Another aliquot was also analysed using acidic positive ion conditions, however it was chromatographically optimized for more hydrophobic compounds. In this method, the extract was gradient eluted from the same afore mentioned C18 column using methanol, acetonitrile, water, 0.05% PFPA and 0.01% FA and was operated at an overall higher organic content. Another aliquot was analysed using basic negative ion optimized conditions using a separate dedicated C18 column. The basic extracts were gradient eluted from the column using methanol and water, however with 6.5 mM Ammonium Bicarbonate at pH 8. The fourth aliquot was analysed via negative ionization following elution from a HILIC column (Waters UPLC BEH Amide 2.1 × 150 mm, 1.7 μm) using a gradient consisting of water and acetonitrile with 10 mM Ammonium Formate, pH 10.8. The MS analysis alternated between MS and data-dependent MSn scans using dynamic exclusion. The scan range varied slighted between methods but covered 70–1000 m/z.
Instrument variability was 5%. This was determined by calculating the median relative standard deviation (RSD) for the standards that were added to each sample prior to injection into the mass spectrometers. Overall process variability was 10%. This was determined by calculating the median RSD for all endogenous metabolites (i.e., non-instrument standards) present in 100% of the pooled matrix samples.
Raw data was extracted, peak-identified and QC processed using Metabolon Inc.’s hardware and software. Compounds were identified by comparison to library entries of purified standards or recurrent unknown entities. Biochemical identifications are based on three criteria: retention index within a narrow the retention time/index (RI) window of the proposed identification, accurate mass match to the library +/- 10 ppm, and the MS/MS forward and reverse scores between the experimental data and authentic standards. While there may be similarities between these molecules based on one of these factors, the use of all three data points can be utilized to distinguish and differentiate biochemicals. Peaks were quantified using the area-under-the-curve method.
2.6. ToxCast data mining
The data generated by the Toxicity Forecaster (ToxCast) programme was accessed by the iCSS ToxCast Dashboard (http:// actor.epa.gov/dashboard/; last accessed 18, May 2018) to search for isoxaflutole mode of action. Isoxaflutole scored positive in the pregnane X receptor (PXR) and peroxisome proliferator-activated receptor gamma (PPAR-γ) assays. These tests conducted by Attagene Inc were reporter gene assays in the human hepatocyte HepG2 cell line [15]. The concentration giving 50% activation in an assay (AC50 value) was considered as a quantitative measure to reflect the activity of isoxaflutole.
2.7. Statistics
The metabolome data analysis was performed using in-house services of Metabolon Inc. Biochemical data was normalized with respect to protein concentration as determined by Bradford assay to account for differences in metabolite levels due to differences in the amount of material present in each sample. The Welch’s two-sample t-test was used to test whether control and treatment group means are different from two independent populations. This version of the two-sample t-test allows for unequal variances. We used the FDR methods and estimated q-values in order to account for the highest number of false positive results caused by the high number of statistical tests [16].
The RNA-seq data analysis (Supplementary Material 1) was performed using the new version of the Tuxedo protocol with HISAT2, StringTie and Ballgown [17]. First, we analysed the quality scores and other metrics using FASTQC [18]. Contamination from rRNA was measured by aligning the sequences of human rRNA to the FASTQ files with the Burrows-Wheeler Alignment tool (BWA) by using the nucleotide sequences of the human rRNA as indexes. The transcriptome of HepaRG cells exposed to glyphosate was processed separately and differential gene expression quantified against their respective controls from the same sequencing batch. Sequences were aligned to the human genome using the hierarchical indexing for spliced alignment of transcripts programme HISAT2 [19]. For this purpose, we used prebuilt indexes (H. sapiens, GRCh38) downloaded from the HISAT2 website. Then, StringTie was used to assemble and quantify the transcripts in each sample using the Homo_sapiens.GRCh38.89 database [17]. Ultimately, the differential gene expression analysis was made with Ballgown [20] in R environment [21]. Low abundance transcripts with a variance across samples less than one were filtered. A standard linear model-based comparison of transcript abundance was performed without adjusting for other covariates to identify differentially expressed transcripts for each group. Although we tested 3 concentrations of each test compound, we have not used multigroup comparisons because we considered that the dose spacing was too large to allow reliable conclusions to be drawn from these methods. We thus used pair-wise comparisons. These RNA-seq data have been submitted to Gene Omnibus and are accessible through accession numbers GSE109565 and GSE114573.
3. Results
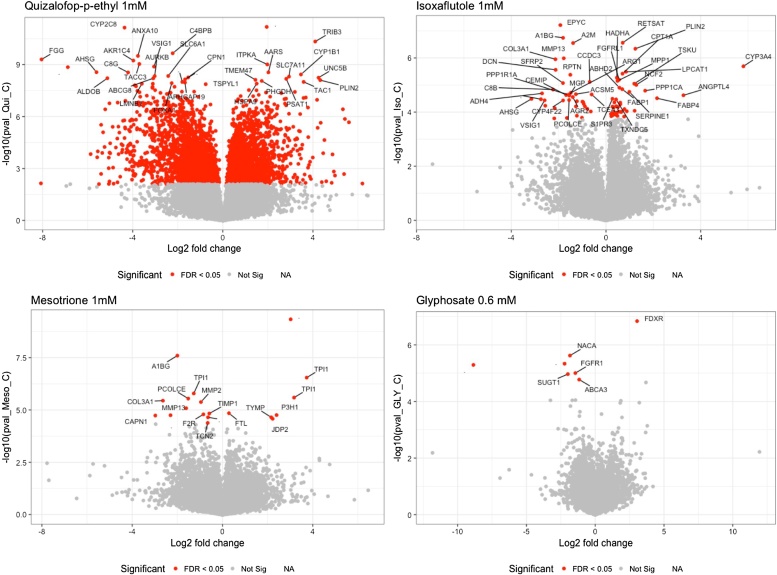
We determined transcriptome alterations in differentiated HepaRG human liver cells caused by exposure to the declared herbicide active principles glyphosate, quizalofop-p-ethyl, isoxaflutole and mesotrione (Fig. 1). Control, untreated cell cultures presented no visible signs of aging after a 6-day exposure (data not shown). Transcriptome profiles of HepaRG cells were then determined using the Illumina-based RNA sequencing platform. Only the highest concentration of each compound tested caused significant changes in transcriptome profiles (Supplementary Material 2). Alterations in gene expression caused by the two lowest concentrations (0.06-0.1 and 6–10 μM) failed to pass the statistical threshold that took into account the high number of tests performed. A total of 7, 19, 77 and 3833 transcripts had their levels altered (q<0.05) at the highest concentration tested namely 600 μM glyphosate, and 1000 μM of mesotrione, isoxaflutole and quizalofop-p-ethyl (Fig. 2).
Fig. 1.
Molecular structures of the herbicide ingredients tested in this study. These pesticides were chosen because their usage is increasing or is predicted to increase in order to deal with the rapid spread of weeds resistant to glyphosate-based commercial formulations.
Fig. 2.
Volcano plots showing the fold changes and statistical significance in the expression of genes affected by exposure to the major pesticide ingredients quizalofop-p-ethyl, isoxaflutole, mesotrione and glyphosate. This figure was created using R in-house functions and based on the statistics performed with the Ballgown R package.
We then studied the function of the genes having their expression altered by the pesticide ingredients tested. The number of genes disturbed by the exposure to glyphosate and mesotrione were insufficient to use a functional annotation tool for pathway enrichment analysis. It is not clear if these compounds lack hepatic toxic effects at these concentrations or if our experimental design lacks sensitivity to detect hepatic effects of weak toxicants. By contrast, isoxaflutole and quizalofop-p-ethyl at 1000 μM caused an enrichment in pathways associated with alterations in liver metabolism (Table 1).
Table 1.
Pathway analysis of HepaRG liver cells exposed to isoxaflutole and quizalofop-p-ethyl. This analysis was made using the DAVID gene functional classification tool to reveal the most affected KEGG pathways. Pvalue were adjusted using the Benjamini-Hochberg method.
| Terms for Isoxaflutole 1 mM group | Count | PValue | adj-PValue |
|---|---|---|---|
| hsa04610:Complement and coagulation cascades | 5 | 3E-04 | 3E-02 |
| hsa00830:Retinol metabolism | 4 | 4E-03 | 2E-01 |
| hsa03320:PPAR signalling pathway | 4 | 4E-03 | 1E-01 |
| hsa00071:Fatty acid degradation | 3 | 2E-02 | 4E-01 |
| hsa00140:Steroid hormone biosynthesis | 3 | 3E-02 | 5E-01 |
| hsa00982:Drug metabolism - cytochrome P450 | 3 | 4E-02 | 5E-01 |
| hsa00980:Metabolism of xenobiotics by cytochrome P450 | 3 | 5E-02 | 5E-01 |
| hsa05204:Chemical carcinogenesis | 3 | 6E-02 | 5E-01 |
| hsa01100:Metabolic pathways | 11 | 7E-02 | 5E-01 |
| hsa05146:Amoebiasis | 3 | 1E-01 | 6E-01 |
| Term for Quizalofop-p-ethyl 1 mM group | Count | PValue | Benjamini adj-Pvalue |
|---|---|---|---|
| hsa01100:Metabolic pathways | 275 | 4E-07 | 1E-04 |
| hsa05322:Systemic lupus erythematosus | 48 | 4E-07 | 6E-05 |
| hsa03010:Ribosome | 47 | 2E-06 | 2E-04 |
| hsa01200:Carbon metabolism | 40 | 7E-06 | 5E-04 |
| hsa00071:Fatty acid degradation | 21 | 1E-05 | 8E-04 |
| hsa05034:Alcoholism | 54 | 2E-05 | 9E-04 |
| hsa01130:Biosynthesis of antibiotics | 60 | 7E-05 | 3E-03 |
| hsa05204:Chemical carcinogenesis | 29 | 1E-04 | 4E-03 |
| hsa00280:Valine, leucine and isoleucine degradation | 20 | 1E-04 | 5E-03 |
| hsa00980:Metabolism of xenobiotics by cytochrome P450 | 27 | 2E-04 | 5E-03 |
Quizalofop-p-ethyl produced the strongest response. A total of 21 genes reflected an alteration of fatty acid degradation. In addition, alteration in expression of 54 genes were annotated to be involved in the response to alcoholism. Although the HepaRG cells were not exposed to ethanol, the phenotype of liver cells intoxicated by ethanol overlap with those identified in cases of NAFLD. The expression of the long-chain-fatty-acid-CoA ligases ACSL1, ACSL3 and ACSL5 were increased by 6.34 (q = 0.05), 2.1 (q = 0.01) and 4.7 (q = 0.06) fold respectively. Quizalofop-p-ethyl also caused alterations in the expression of several genes, which are members of the alcohol dehydrogenase family such as ADH1A (Fold Changes (FC) = -15.7, q = 0.003), ADH1B (FC = -12.9, q = 0.005), ADH1C (FC = -21, q = 0.001), ADH4 (FC = -7.1, q = 0.0007), ADH6 (FC = -5.95, q = 0.00004), and of the aldehyde dehydrogenases ALDH1A1 (FC = -3.2, q = 0.008), ALDH1B1 (FC = -2.6, q = 0.001), ALDH3 A1 (FC = -5.3, q = 0.02) and ALDH7A1 (FC = -2.2, q = 0.009). However, there were no indications that the effects of quizalofop-p-ethyl could have been mediated by a nuclear receptor.
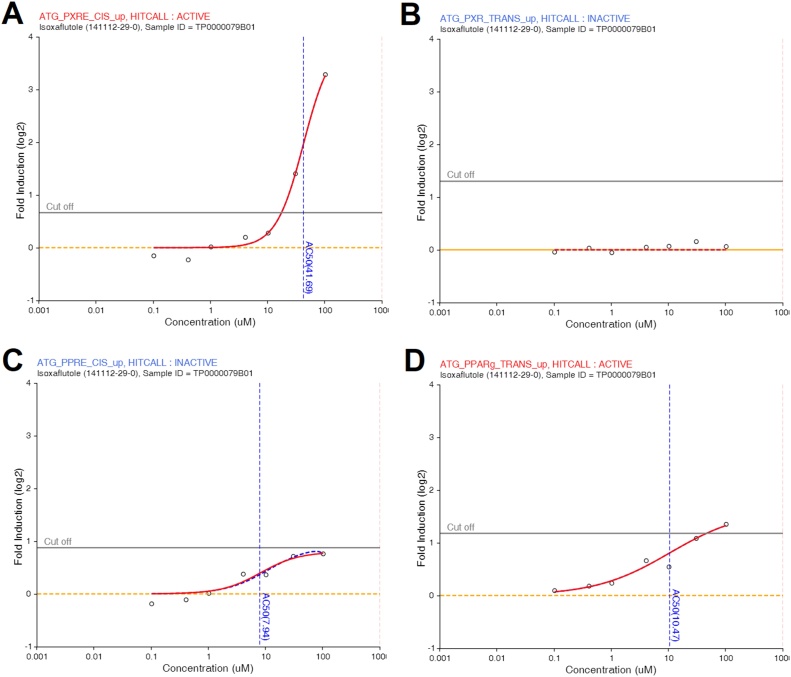
The gene expression profiles in HepaRG cells exposed to isoxaflutole suggested the involvement of a nuclear receptor inducing changes in lipid metabolism. This was reflected in the transcriptome profiles of HepaRG cells treated with 1000 μM of isoxaflutole being enriched in genes involved in retinol metabolism and in the PPAR signalling pathway, although these indications came from a low number of genes whose expression was disturbed. Among these, the genes coding for the fatty acid-binding proteins 1 and 4 (FABP1 and FABP2) had their expression increased by 1.6 (q = 0.01) and 4.4 (q = 0.02) fold respectively. Interestingly, the most affected gene was the cytochrome CYP3 A4 (FC = 55.8, q = 0.006), which is a known marker of PXR activation. Additionally, expression of the 17β-hydroxysteroid dehydrogenase 2 (HSD17B3) (FC = -1.95, q = 0.02) and 4 (HSD17B4) (FC = -4.1, q = 0.05) genes were decreased and could indicate alterations in hormone metabolism. We then investigated the activity of isoxaflutole towards nuclear receptors involved in the regulation of lipid metabolism by analysing data from the ToxCast programme (iCSS ToxCast Dashboard, accessed May 18, 2018). The ATG_PXRE_CIS_up assay suggested that isoxaflutole can activate PXR-mediated transcription (Fig. 3A), although the results of the transactivation assay ATG_PXR_TRANS_up were negative (Fig. 3B). Contrastingly, the results of the Attagene ATG_PPARg_Trans_up assay suggested that isoxaflutole can activate PPAR-γ (Fig. 3D), but the results of the peroxisome proliferator-activated receptor (PPAR) response element (PPRE) activation assay (ATG_PPRE_CIS_up) were negative (Fig. 3C).
Fig. 3.
Isoxaflutole activates PPAR gamma receptor and pregnane X responsive elements in reporter gene assays. Publically available Attagene FACTORIAL™ assay data from the ToxCast programme was scrutinised to identify nuclear receptors activated by isoxaflutole. The data for Attagene cis- and trans-FactorialTM assays ATG_PXRE_CIS_up (A) and ATG_PXR_TRANS_up (B), and ATG_PPRE_CIS_up (C) amd ATG_PPARg_Trans_up (D) were extracted using the iCSS ToxCast Dashboard (http:// actor.epa.gov/dashboard/; last accessed May, 16, 2018). Isoxaflutole dose response curve (0.1–100 μM) is expressed as fold induction.
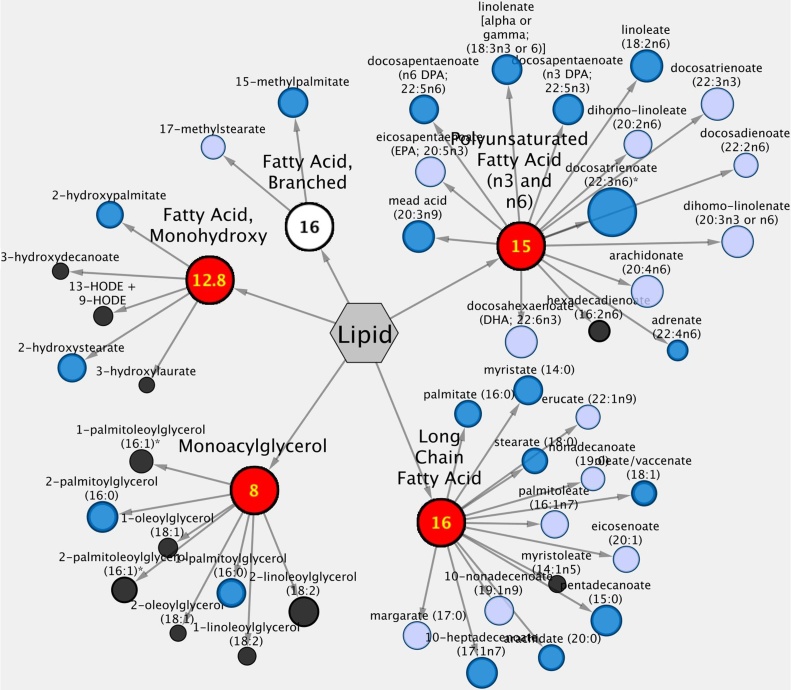
Previous results from our group indicated that chronic (2 year) exposure of rats to an extremely low dose of a commercial formulation of Roundup GBH, caused alterations in liver metabolism representative of NAFLD (Mesnage et al., 2017). In order to further explore changes in liver metabolism caused by glyphosate in greater detail, we performed a global metabolome profiling of HepaRG cells exposed to three concentrations of this herbicide (Supplementary Material 3). The Metabolon HD4 platform detected 802 named biochemicals in the HepaRG samples. Overall, glyphosate did not cause significant alterations in metabolome composition. However, exposure did cause a significant decrease in LCFAs and PUFAs. Notably, HepaRG cells exposed to the lowest concentration (0.06 μM) of glyphosate tested showed the most dramatic effects in the levels of these fatty acids as either significant or trends towards significant lower levels (Fig. 4). At the higher glyphosate concentrations of 6 μM and 600 μM, lower lipid levels were also observed but these did not reach statistical significance.
Fig. 4.
Metabolome analysis of HepaRG liver cells treated with 60 nM glyphosate shows a decrease in levels of free long chain fatty acids and polyunsaturated fatty acids. Visualization of lipid metabolites were created using Cytoscape. Blue nodes represent significantly decreased metabolites compared to control cultures (p < 0.05). There was no increased metabolite. The size of the node is proportional to the fold change relative to the negative control. Nodes with a red background and yellow characters display enriched pathways.
4. Discussion
We present here an in-depth investigation of the transcriptome profile alterations in the HepaRG human liver cell line caused by exposure to four chemical compounds declared as active principles in commercial formulations of herbicides (Fig. 1). Our results show that isoxaflutole and quizalofop-p-ethyl are potent toxicants in this tissue culture model system as they caused transcriptome profile alterations consistent with dysregulation of lipid metabolism (Fig. 2; Table 1). In contrast, we found mesotrione and glyphosate to be only weakly toxic inducing little change in transcriptome profiles (Fig. 2; Table 1). Interestingly, a follow-up metabolomics analysis of HepaRG cells treated with the lowest (0.06 μM) concentration of glyphosate revealed a significant decrease in levels of LCFAs and PUFAs (Fig. 3). In general, the tissue culture cell model system we have employed in this study has demonstrated that it can be used in the context of the establishment of high-throughput assays, which are advocated as promising tools to reduce the use of animals in toxicology [22].
Although these findings from an in vitro tissue culture model system cannot be readily translated to effects in vivo, they are nevertheless indicative of differences in toxicity potency between these four pesticide ingredients. Glyphosate was the least toxic of the compounds tested in this study. The most toxic was quizalofop-p-ethyl inducing large scale alterations in the transcriptome profile. However, it is not clear if these alterations are indicative of a potential to induce an NAFLD phenotype. One of the strongest effects of quizalofop-p-ethyl was a global decrease in the expression of genes encoding enzymes involved in alcohol metabolism, such as ADH. However, it has been reported in other studies that the ADH family of genes have their expression increased in cases of steatohepatitis [23] and NAFLD [24]. It could thus be hypothesised that quizalofop-p-ethyl could cause toxicant-associated fatty liver disease and different metabolic diseases related to lipid metabolism dysregulations such as obesity.
The toxicity of quizalofop-p-ethyl is poorly studied compared to glyphosate. Although the use of quizalofop-based herbicides has decreased following the introduction of “Roundup-Ready” glyphosate tolerant GM crops, an increase in application can be observed in recent years as revealed by US government pesticide national synthesis project (water.usgs.gov/nawqa/pnsp/). Quizalofop is increasingly being used in combination with glyphosate and this use will be further increased by the cultivation of crops such as DAS– 40,278–9 GM maize carrying an aad-1 transgene introduced to detoxify both 2,4-D and quizalofop herbicides [6]. Sprout Intelligence has estimated the global quizalofop-p-ethyl market to be more than $21.32 million [25] and this is now set to increase markedly in the coming years.
The liver is the main target for quizalofop-p-ethyl toxic health effects as revealed by an analysis of the studies used to establish regulatory guidance values for this compound [26]. It was observed that quizalofop-p-tefuryl resulted in hypertrophy and hyperplasia of hepatocytes, as well as bile stasis and hepatocellular adenomas/carcinomas in males and females administered with 39.5 and 48.7 mg/kg bw/day, respectively, in a chronic toxicity study in rats [26]. A human case report has confirmed that commercial formulations of pesticides containing quizalofop-p-ethyl is a probable inducer of occupational liver injury [27]. Interestingly, quizalofop-p-ethyl may also be an endocrine disruptor as suggested by an investigation showing estrogenic effects in zebrafish [28]. Health consequences of environmental levels of exposure in human populations are unclear and evidently require further research.
The mechanism by which isoxaflutole, brings about plant death is via inhibition of the enzyme 4-hydroxyphenylpyruvate dioxygenase (HPPD), a component of the biochemical pathway that converts tyrosine to plastoquinone and α-tocopherol that is necessary for carotenoid biosynthesis [29]. Again, it is expected that the use of isoxaflutole will increase with the introduction of isoxaflutole-tolerant GM crops, including the soybean variety FG72 developed by Bayer CropScience, which carries a modified HPPD gene from Pseudomonas fluorescens strain A32 [6]. Our study reveals that isoxaflutole can cause metabolic alterations in liver cells, and that these effects could be due to the activation of nuclear receptors PXR and/or PPAR-γ. Future studies would have to be undertaken to confirm these findings as the reliability of the ToxCast database to predict PPAR-γ activation is debated [30]. However, CYP3 A4 was the most affected gene and the measurement of CYP3 A4 mRNA has been found to be a reliable marker of PXR activation [31]. Since the role of PXR in activation of hepatocyte proliferation has been shown to influence tumorigenesis [32], it is interesting to note that possible carcinogenic effects of isoxaflutole are documented by the US EPA. In particular, a dose-related increased incidence of hepatocellular adenomas was observed in CD-1 mice following a 78-week exposure to isoxaflutole, starting at a concentration of 3.6 mg/kg/day [33]. A possible role of isoxaflutole in the activation of PXR remains to be ascertained as there was a discrepancy between the results of the cis- and the trans- Attagene transactivation assays (Fig. 3).
Our study reported here failed to provide an indication of the NAFLD phenotypes caused by a glyphosate-based herbicide (Roundup) after a two-year exposure in rats [34]. Surprisingly, the lowest concentration of glyphosate tested (0.06 μM) caused a decrease in a large number of lipid species (Fig. 4). The exact nature of this low dose response cannot be determined from this single experiment, but it is possible that at higher concentrations, more overtly toxic mechanisms are masking the effects on lipids. Another possibility is that a saturation effect is occurring once the low dose is exceeded bearing in mind that glyphosate levels found in the HepaRG cells during the metabolomics analysis increased by 3.7 and 336.35 fold at the intermediate and highest concentrations tested compared to the negative controls (Supplementary Material 3). Moving forward, it might be interesting to perform a time course study of the lower and middle range concentrations of glyphosate treatment to determine the effects of long-term, low dose exposure. The effects observed here may be exacerbated by longer exposure, and possible mechanisms of action may be elucidated by this type of investigation. The results obtained with treatment of HepaRG cells with glyphosate also raise the intriguing possibility that the NAFLD phenotype we observed in rats following chronic exposure to an ultra-low dose of Roundup GBH (Mesnage et al., 2017), occurs indirectly rather than as a consequence of a direct effect of this herbicide active principle on the liver. In addition, at present we cannot exclude the possibility that the adjuvants and other co-formulants present in the Roundup ingested by the rats contributed to the causation of NAFLD. Further studies are clearly needed to resolve this issue.
Perhaps what is most important to undertake is to compare effects between these isolated pesticide ingredients and their respective whole commercial formulations, which contain additional complex mixtures of chemicals used as adjuvants. Chemical components declared as active ingredients in commercial pesticides are never used alone but always in combination with co-formulants and adjuvants. There have been numerous studies reported, which have investigated the toxic effects of GBH adjuvant co-formulants. It has been shown that commercial formulations including glyphosate can be up to 1000 times more toxic than glyphosate alone on human tissue culture cells, and that this toxicity can be attributed to ethoxylated surfactants [35]. This is the case for most commercial formulations of pesticides which were up to 1056 times more toxic than their active principles in a comparison of 3 major insecticides, fungicides and herbicide formulations [36]. The long-term consequences of the effects of co-formulants are unclear, although some epidemiological studies have suggested that they can be responsible of congenitial malformations [37] or adverse respiratory effects [38]. Generally, adjuvant toxicity is poorly researched and a largely missing component in regulatory health risk assessment of pesticide active ingredients, which remains to be addressed [39].
Our study revealed that isoxaflutole and quizalofop-p-ethyl are inducing toxic effects on liver, which are reflected by large scale transcriptome profile alterations in lipid metabolism. The use of these pesticide ingredients is being expected to increase and there is an urgent need for an evaluation of their effects on human metabolism.
Authors’ contributions
RM participated in the cell culture experiment, performed the transcriptome data analysis, and drafted the manuscript. MB performed the cell culture experiment and the RNA extraction. EW, TX and CAM performed the RNA-seq. MNA coordinated the investigation and assisted in the drafting of the manuscript.
Competing interests
The authors declare they have no competing interests.
Transparency document
Acknowledgements
This work was funded by the Sustainable Food Alliance (USA) whose support is gratefully acknowledged.
References
- 1.Bonny S. Genetically modified herbicide-tolerant crops, weeds, and herbicides: overview and impact. Environ. Manage. 2016;57:31–48. doi: 10.1007/s00267-015-0589-7. [DOI] [PubMed] [Google Scholar]
- 2.Coupe R.H., Capel P.D. Trends in pesticide use on soybean, corn and cotton since the introduction of major genetically modified crops in the United States. Pest Manage. Sci. 2016;72:1013–1022. doi: 10.1002/ps.4082. [DOI] [PubMed] [Google Scholar]
- 3.Benbrook C.M. Trends in glyphosate herbicide use in the United States and globally. Environ. Sci. Eur. 2016;28:3. doi: 10.1186/s12302-016-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.USDA, Acreage Report. Released June 30, 2017, by the National Agricultural Statistics Service (NASS), Agricultural Statistics Board, United States Department of Agriculture (USDA) Available at http://usda.mannlib.cornell.edu/usda/current/Acre/Acre-06-30-2017.pdf, 2017.
- 5.Heap I. Global perspective of herbicide-resistant weeds. Pest Manage. Sci. 2014;70:1306–1315. doi: 10.1002/ps.3696. [DOI] [PubMed] [Google Scholar]
- 6.ISAAA . 2017. GM Approval Database.http://www.isaaa.org/gmapprovaldatabase/default.asp [Google Scholar]
- 7.Foulds C.E., Trevino L.S., York B., Walker C.L. Endocrine-disrupting chemicals and fatty liver disease. Nat. Rev. Endocrinol. 2017;13:445–457. doi: 10.1038/nrendo.2017.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wahlang B., Song M., Beier J.I., Cameron Falkner K., Al-Eryani L., Clair H.B., Prough R.A., Osborne T.S., Malarkey D.E., Christopher States J., Cave M.C. Evaluation of Aroclor 1260 exposure in a mouse model of diet-induced obesity and non-alcoholic fatty liver disease. Toxicol. Appl. Pharmacol. 2014;279:380–390. doi: 10.1016/j.taap.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Eryani L., Wahlang B., Falkner K.C., Guardiola J.J., Clair H.B., Prough R.A., Cave M. Identification of environmental chemicals associated with the development of toxicant-associated fatty liver disease in rodents. Toxicol. Pathol. 2015;43:482–497. doi: 10.1177/0192623314549960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vernon G., Baranova A., Younossi Z.M. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults, Aliment. Pharmacol. Ther. 2011;34:274–285. doi: 10.1111/j.1365-2036.2011.04724.x. [DOI] [PubMed] [Google Scholar]
- 11.Guillouzo A., Corlu A., Aninat C., Glaise D., Morel F., Guguen-Guillouzo C. The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem. Biol. Interact. 2007;168:66–73. doi: 10.1016/j.cbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Heindel J.J., Blumberg B., Cave M., Machtinger R., Mantovani A., Mendez M.A., Nadal A., Palanza P., Panzica G., Sargis R., Vandenberg L.N., Vom Saal F. Metabolism disrupting chemicals and metabolic disorders. Reprod. Toxicol. 2017;68:3–33. doi: 10.1016/j.reprotox.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aninat C., Piton A., Glaise D., Le Charpentier T., Langouet S., Morel F., Guguen-Guillouzo C., Guillouzo A. Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab. Dispos. 2006;34:75–83. doi: 10.1124/dmd.105.006759. [DOI] [PubMed] [Google Scholar]
- 14.Mesnage R., Biserni M., Balu S., Frainay C., Poupin N., Jourdan F., Wozniak E., Xenakis T., Mein C.A., Antoniou M.A. Integrated transcriptomics and metabolomics reveal signatures of lipid metabolism dysregulation in HepaRG liver cells exposed to PCB 126. bioRxiv. 2018;92(Aug. (8)):2533–2547. doi: 10.1007/s00204-018-2235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richard A.M., Judson R.S., Houck K.A., Grulke C.M., Volarath P., Thillainadarajah I., Yang C., Rathman J., Martin M.T., Wambaugh J.F., Knudsen T.B., Kancherla J., Mansouri K., Patlewicz G., Williams A.J., Little S.B., Crofton K.M., Thomas R.S. ToxCast chemical landscape: paving the Road to 21st century toxicology. Chem. Res. Toxicol. 2016;29:1225–1251. doi: 10.1021/acs.chemrestox.6b00135. [DOI] [PubMed] [Google Scholar]
- 16.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrews S. 2017. FastQC A Quality Control Tool for High Throughput Sequence Data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ DOI: citeulike-article-id:11583827. [Google Scholar]
- 19.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frazee A.C., Pertea G., Jaffe A.E., Langmead B., Salzberg S.L., Leek J.T. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 2015;33:243–246. doi: 10.1038/nbt.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R. C. Team . R Foundation for Statistical Computing; Vienna, Austria: 2017. R: A Language and Environment for Statistical Computing.https://www.r-project.org/ [Google Scholar]
- 22.EPA U.S. 2015. Endocrine Disruptor Screening Program: Use of High Throughput Assays and Computational Tools. Notice. Federal Register (2015-15182, Jun 19, 2015) (FRL-9928-69) [Google Scholar]
- 23.Baker S.S., Baker R.D., Liu W., Nowak N.J., Zhu L. Role of alcohol metabolism in non-alcoholic steatohepatitis. PLoS One. 2010;5:e9570. doi: 10.1371/journal.pone.0009570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu R., Baker S.S., Moylan C.A., Abdelmalek M.F., Guy C.D., Zamboni F., Wu D., Lin W., Liu W., Baker R.D., Govindarajan S., Cao Z., Farci P., Diehl A.M., Zhu L. Systematic transcriptome analysis reveals elevated expression of alcohol-metabolizing genes in NAFLD livers. J. Pathol. 2016;238:531–542. doi: 10.1002/path.4650. [DOI] [PubMed] [Google Scholar]
- 25.Intelligence S. 2016. Market Research Report. Quizalofop-e: Global Product Intelligence; pp. 2015–2020.https://www.giiresearch.com/report/spro368112-quizalofop-e-global-product-intelligence.html Available at. [Google Scholar]
- 26.European Food A., Safety Conclusion regarding the peer review of the pesticide risk assessment of the active substance quizalofop-P. Efsa J. 2009;7 doi: 10.2903/j.efsa.2008.147r. 205r-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elefsiniotis I.S., Liatsos G.D., Stamelakis D., Moulakakis A. Case report: mixed cholestatic/hepatocellular liver injury induced by the herbicide quizalofop-p-ethyl. Environ. Health Perspect. 2007;115:1479–1481. doi: 10.1289/ehp.9968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu L.Z., Qi S.Z., Cao F.J., Mu X.Y., Yang Y., Wang C. Quizalofop-P-ethyl exposure increases estrogen axis activity in male and slightly decreases estrogen axis activity in female zebrafish (Danio rerio) Aquat. Toxicol. 2017;183:76–84. doi: 10.1016/j.aquatox.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell G., Bartlett D.W., Fraser T.E., Hawkes T.R., Holt D.C., Townson J.K., Wichert R.A. Mesotrione: a new selective herbicide for use in maize. Pest Manage. Sci. 2001;57:120–128. doi: 10.1002/1526-4998(200102)57:2<120::AID-PS254>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 30.Houck K.A., Judson R.S., Knudsen T.B., Martin M.T., Richard A.M., Crofton K.M., Simeonov A., Paules R.S., Bucher J.R., Thomas R.S. Comment on “On the utility of ToxCast and ToxPi as methods for identifying new obesogens”. Environ. Health Perspect. 2017;125:A8–A11. doi: 10.1289/EHP881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fahmi O.A., Kish M., Boldt S., Obach R.S. Cytochrome P450 3A4 mRNA is a more reliable marker than CYP3A4 activity for detecting pregnane X receptor-activated induction of drug-metabolizing enzymes. Drug Metab. Dispos. 2010;38:1605–1611. doi: 10.1124/dmd.110.033126. [DOI] [PubMed] [Google Scholar]
- 32.Pondugula S.R., Pavek P., Mani S. Pregnane X receptor and Cancer: context-Specificity is key. Nucl. Receptor Res. 2016;3 doi: 10.11131/2016/101198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.U. EPA, Memorandum . 1997. Carcinogenicity Peer Review of Isoxaflutole. Office of Prevention, Pesticides, and Toxic Susbtances.https://www3.epa.gov/pesticides/chem_search/cleared_reviews/csr_PC-123000_6-Aug-97_075.pdf Available at. [Google Scholar]
- 34.Mesnage R., Renney G., Séralini G.-E., Ward M., Antoniou M.N. Multiomics reveal non-alcoholic fatty liver disease in rats following chronic exposure to an ultra-low dose of Roundup herbicide. Sci. Rep. 2017;7:39328. doi: 10.1038/srep39328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mesnage R., Bernay B., Seralini G.E. Ethoxylated adjuvants of glyphosate-based herbicides are active principles of human cell toxicity. Toxicology. 2013;313:122–128. doi: 10.1016/j.tox.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Mesnage R., Defarge N., Spiroux de Vendômois J., Séralini G.E. Major pesticides are more toxic to human cells than their declared active principles. Biomed Res. Int. 2014;2014 doi: 10.1155/2014/179691. Article ID 179691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carmichael S.L., Yang W., Roberts E.M., Kegley S.E., Wolff C., Guo L., Lammer E.J., English P., Shaw G.M. Hypospadias and residential proximity to pesticide applications. Pediatrics. 2013 doi: 10.1542/peds.2013-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoppin J.A., Umbach D.M., Long S., London S.J., Henneberger P.K., Blair A., Alavanja M., Freeman L.E., Sandler D.P. Pesticides are associated with allergic and non-allergic wheeze among male farmers. Environ. Health Perspect. 2017;125:535–543. doi: 10.1289/EHP315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mesnage R., Antoniou M.N. Ignoring adjuvant toxicity falsifies the safety profile of commercial pesticides. Front. Public Health. 2017;5:361. doi: 10.3389/fpubh.2017.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.