Abstract
Microbial cells do not live in isolation in their environment, but rather they communicate with each other using chemical signals. This sophisticated mode of cell-to-cell signalling, known as quorum sensing, was first discovered in bacteria, and coordinates the behaviour of microbial population behaviour in a cell-density-dependent manner. More recently, these mechanisms have been described in eukaryotes, particularly in fungi, where they regulate processes such as pathogenesis, morphological differentiation, secondary metabolite production and biofilm formation. In this manuscript, we review the information available to date on these processes in yeast, dimorphic fungi and filamentous fungi. We analyse the diverse chemical ‘languages’ used by different groups of fungi, their possible cross-talk and interkingdom interactions with other organisms. We discuss the existence of these mechanisms in multicellular organisms, the ecophysiological role of QS in fungal colonisation and the potential applications of these mechanisms in biotechnology and pathogenesis.
Keywords: cell-to-cell signalling, yeast, dimorphic, filamentous fungi, farnesol, consortia
This review focuses on the complexity of quorum sensing mechanisms in fungi and its potential applications.
INTRODUCTION
What is quorum sensing?
Since the 1960s, it has been known that unicellular microorganisms do not live in isolation in their environment, but rather there is communication among cells that allows the population to coordinate its behaviour (Nealson, Platt and Hastings 1970). This sophisticated cell-to-cell signalling system, known as quorum sensing (QS), is mediated by small molecules that diffuse into the environment and accumulate during the growth of the microbial population. Each individual cell is able to sense the amount of quorum sensing molecules (QSMs) present in the medium which, upon reaching a threshold concentration, coordinate specific gene expression (Bandara et al.2012). Microbes monitor their population density through QS-mediated mechanisms that regulate a number of population density-related processes (Miller and Bassler 2001). Moreover, the QSMs are able to stimulate their own production, acting as ‘autoinducers’ (Bandara et al.2012). QS confers a great evolutionary advantage, allowing microbial populations to adapt to changing environments. Some authors suggest that these processes are neo-Darwinian mechanisms of evolution that may have played an important role in the appearance of the first multicellular organisms (Miller and Bassler 2001).
QS in prokaryotes
The QS phenomenon was first studied in bacteria, where it is known to be involved in the regulation of processes such as pathogenesis, symbiosis, competence, conjugation, nutrient uptake, morphological differentiation, secondary metabolite production and biofilms formation (Bandara et al.2012). The first organised response dependent on population density was reported in the luminous marine bacterium Aliivibrio fischeri (Nealson, Platt and Hastings 1970). This pioneer study on this Gram-negative bacterium showed that bioluminescence was regulated by a QS mechanism controlled by a two-component system integrated by a synthase (LuxI), responsible for the production of the QSMs N-acyl-homoserine-lactones (AHLs), and a sensor receptor (LuxR) the activity of which is dependent on the presence of the QSM. Subsequently, other forms of homoserine lactones and other molecules including quinolones, the Diffusible Signal Factor (DSF) and the Cholera Autoinducer 1, were identified as QSMs in Gram-negative bacteria (Papenfort and Bassler 2016).
QS-mediated signalling in Gram-positive bacteria is often regulated by small peptides after intracellular processing of certain polypeptides and secretion of the final molecule, which is detected by a sensor histidine-kinase (Hawver, Jung and Ng 2016). However, Gram-positive Streptomyces use γ-butyrolactones as QSMs (Biarnes-Carrera, Breitling and Takano 2015). Finally, the autoinducer 2 (AI-2) has been described as a QS system common to both Gram-positive and Gram-negative bacteria, although its role as a true QSM has been questioned for some microorganisms (Chen et al.2002; Winzer, Hardie and Williams 2002; Bandara et al.2012; Papenfort and Bassler 2016). This system gives rise to a family of molecules that are thought to serve as a common ‘language’ for most bacteria. Many bacteria possess more than one QS system. They can communicate with their own species using their particular QS system (e.g. DSF-type molecules), and also communicate with bacteria from other genera using AHLs or the AI-2 system, establishing a cross-talk among different bacterial species (Miller et al.2004; Ryan et al.2009; Dow et al.2016).
The study of QS in natural populations in environmentally complex settings is often complicated. This is because there are a variety of cell-intrinsic and cell-extrinsic factors that modulate the kinetics of QS signal production, detection and response (Fig. 1). Intrinsic factors include the roles of other regulators outside of the direct QS circuit that respond to environmental stimuli, such as phosphate availability (Blus-Kadosh et al.2013), metabolic pools that may influence signal synthesis (Bjarnsholt and Givskov 2007) and genetic differences between strains that alter the dynamics of QS activation. Extrinsic factors include QS interference or ‘quorum quenching’, in which QSMs are inactivated or degraded (Zhu and Kaufmann 2013), environmental factors that influence the stability and diffusion of QSM (Connell et al.2010), diffusion kinetics and population size. Furthermore, in some species, there are members of the population that lack certain components of the QS system. These cells may have lost the ability to produce exoproducts beneficial to the population and are referred to as ‘cheaters’ (Diggle et al.2007). Others may have lost their ability to synthesise QSMs (e.g. LuxI defective strains) but are still able to respond to QSMs; this is the case of bacterial strains harbouring LuxR ‘solos’ (Venturi and Ahmer 2015) that may be referred to as ‘listeners’. The ‘cheaters’ may have some fitness benefits in the presence of cells that are making QS-regulated public goods (Diggle et al.2007), while the ‘listeners’ may respond to external QSMs and express favourable traits induced by the population (Venturi and Ahmer 2015). On the other hand, QS-defective endemic strains are also present in the environment, suggesting that there may be specific niches in which QS is not required for fitness (Hammond et al.2016).
Figure 1.
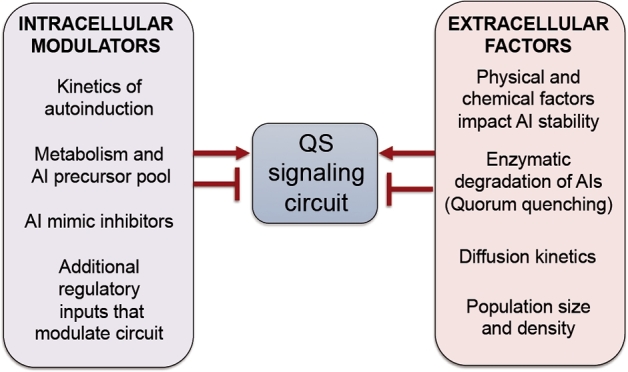
Summary of the intrinsic and extrinsic factors that modulate QS signalling in microbial cells. The intracellular modulators include the dynamics of signal detection and the activity of the positive feedback loop that characterises signal production, and this can vary between species or strains within a species. Metabolic state, and the effects of metabolism of autoinducer (AI) production, can also impact AI production. AI mimics that inhibit AI detection have been described. In bacteria, other environment-responsive regulatory elements, such as transcription factors and small RNAs, can positively or negatively tune the QS circuit. The extracellular factors that modulate QS signalling include physical and chemical elements that impact the stability of secreted AI molecules and the action of enzymes that can degrade AIs. In addition, the population size and factors that influence the diffusion and availability of the AI will influence the QS circuit.
QS in eukaryotes
It has long been known that eukaryotic cells are able to secrete and sense signalling molecules (Youk and Lim 2014). However, now we know that certain types of eukaryotic microorganisms monitor their population density through QS mechanisms (Hogan 2006a). This would be expected as many bacteria and eukaryotic microbes occupy similar ecological niches and face common challenges. QS in microscopic eukaryotes was first described in the pathogenic dimorphic fungus Candida albicans (Hornby et al.2001). Most of the published works on QS in eukaryotes relate to fungi, but it is unclear whether this is because QS regulation is more common in fungi or merely easier to detect and study. Nevertheless, the existence of QS mechanisms in the Protista kingdom has recently been claimed, particularly in the parasite Trypanosoma brucei (Mony et al.2014) and in the social amoeba Dictyostelium discoideum (Tarnita et al.2015).
In fungi, QS mechanisms have been described to regulate processes such as sporulation, secondary metabolite production, morphological transition and enzyme secretion. In this developing field, there are a range of fungal genera displaying these mechanisms and using a variety of signalling molecules with different structures. For instance, the inoculum size effect has been well studied in many fungi such as Ophiostoma (Ceratocystis) ulmi, Ophiostoma novo-ulmi, Ophiostoma piceae, Ophiostoma floccosum, Histoplasma capsulatum, Mucor rouxii, Aureobasidium pullulans, C.albicans, Cladosporium werneckii, Aspergillus flavus, Penicillium isariaeforme, Neurospora crassa and Schizophyllum commune (Nickerson, Atkin and Hornby 2006; Horowitz Brown et al.2008; Berrocal et al.2014; de Salas, Martínez and Barriuso 2015; Wedge et al.2016). Furthermore, fungi can communicate with bacteria, and even their plant or mammalian hosts.
QS mechanisms have emerged among the most important aspects in the study of bacterial behaviour, and are often well-characterised at the genetic and molecular levels due to their implication in biofilm formation and their potential as future drug targets to be used alone or in conjunction with antibiotics (Camara, Williams and Hardman 2002). In contrast, knowledge on QS mechanisms in fungi at the molecular, physiological and ecological levels is scarce. Here we aim to review the role of QS mechanisms in fungi due to their relevance in pathogenesis, ecology and biotechnology. In this context, we will discuss the intrinsic and extrinsic factors that modulate QS (Fig. 1). We will establish a correlation between the QSMs utilised in different fungal taxa or habitats to decipher the different ‘languages’ used by fungi, their evolution and their potential for cross-talk. We will discuss the prominence of these mechanisms in filamentous (multicellular) fungi, speculate on interkingdom communication and the formation of bacterial–fungal consortia, and propose future directions for the application of these mechanisms.
QS mechanisms in yeast and dimorphic fungi
Some evidence suggests that the yeast Saccharomyces cerevisiae exhibits a QS type of regulation mediated by aromatic alcohols derived from amino acids (Hogan 2006b). This budding yeast produces tryptophol and phenylethyl alcohol to control the formation of pseudohyphae, biofilm formation (Chen and Fink 2006) and perhaps virulence towards the grapevine Vitis vinifera (Gognies et al.2006). Some QSMs that were discovered first in other fungal species, such as C.albicans, have been subsequently described to have roles in S. cerevisiae. For example, S. cerevisiae can produce tyrosol (Avbelj et al.2015) and respond to farnesol (Egbe et al.2017), two QSMs first characterised in C. albicans as discussed in detail below.
On the other hand, the only fungus from the group of the basidiomycetes with described QS mechanisms is Cryptococcus neoformans, a human pathogen. This fungus, the aetiologic agent of cryptococcosis, undergoes cellular changes that result in the appearance of different cellular types during infection. After capsule enlargement and growth of the whole cell, ‘titan cells’ are formed (Albuquerque et al.2014). The main virulence factors in this fungus are the formation of the polysaccharide capsule and melanin accumulation, which are regulated by QS mechanisms. Madhani (2011) described QS regulation via peptides (QSP1–4) that are secreted outside the cell prior to uptake via the transporter Opt1. QSP1 is a central signalling molecule that regulates pathogenesis in C. neoformans, being a direct target of three transcription factors required for virulence. It is known that QSP1 regulates secreted protease activity and promotes cell wall function at high cell densities. Here, similar to Gram-positive bacteria, the signal production requires extracellular processing of the pro-peptide proQsp1 by a cell-associated protease (Homer et al.2016).
QS in C.albicans
QS mechanisms are best described in the dimorphic yeast C. albicans. This species is the causal agent of most common mycosis and is also a member of the resident flora in humans and animals (Nucci et al.2010). Under certain conditions, this organism undergoes a controlled, reversible interconversion of cell morphology, switching from yeast to hyphal form. This transformation is crucial to its pathogenicity and adaptation to the environment (Madhani 2011; Sudbery 2011). The morphological switch in dimorphic fungi is governed by many environmental signals, including temperature, pH, nutrients composition and concentration, CO2 levels, transition metals, chelating agents and cell population density (Romano 1966; Gauthier 2015). These stimuli act through several well-established signalling cascades (Lo et al.1997). In C. albicans two signalling compounds with opposite effects have been described as QSMs: farnesol and tyrosol. Farnesol is a 12-carbon backbone sesquiterpene alcohol ((2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol) made up of three isoprene units (Nickerson, Atkin and Hornby 2006). It is an intermediate in sterol biosynthesis that prevents differentiation from yeast to hyphae (Hornby et al.2001) and induces the switch from hypha back to yeast growth (Lindsay et al.2012). Tyrosol (2-(4-hydroxyphenyl) ethanol) stimulates the exit from lag phase such that germ-tube formation and hyphal development is accelerated in its presence (Chen et al.2004). Candidaalbicans QSMs also influence the formation and structure of biofilms (Ramage et al.2002; Cordeiro et al.2015) as well as the dispersal of cells from a biofilm; hence, these molecules play important roles in pathogenesis.
The regulation of morphological changes mediated by E,E-farnesol in C. albicans is complicated, but well studied. This molecule regulates hyphal development through the repression of the Ras1-cAMP/protein kinase A (PKA) signalling pathway (Davis-Hanna et al.2008). Evidence indicates that farnesol can affect filamentation by suppressing the cAMP pathway through inhibition of adenylyl cyclase (Cyr1) activity (Hall et al.2011). Activation of the cAMP-PKA pathway down-regulates the expression of Nrg1, the major repressor of hyphal development. Farnesol is able to repress the expression of the kinase required for Nrg1 degradation, Sok1 (Lu et al.2014). Moreover, the transcription repressor Cup9 has been found to be responsible for the regulation of SOK1 expression in response to farnesol inhibition. Interestingly, farnesol inhibits Cup9 degradation mediated by the N-end rule E3 ubiquitin ligase, Ubr1 (Lu et al.2014). The histidine kinase (Chk1) and the zinc finger 1 (Czf1) from C. albicans (Kebaara et al.2008) have also been identified as important response factors to farnesol (Langford et al.2013). Farnesol also modulates the transcription levels of TUP1, a major repressor of the morphological transition (Kebaara et al.2008). Nevertheless, the E,E-farnesol export pathways in the cell are unknown, but its secretion has been shown to vary across different strains (Weber, Schulz and Ruhnke 2010).
QS in other dimorphic fungi
Over the years, QS mechanisms have been identified in a few other dimorphic fungi. Many of these are pathogens of plants or animals, and many have applications in biotechnology. This is the case for fungi from the genus Ophiostoma, which includes plant pathogenic species such as O. ulmi and its aggressive sister species O. novo-ulmi, causal agents of Dutch elm disease. Both species belong phylogenetically to the so-called ‘O. piceae–O. ulmi complex’ (De Beer and Wingfield 2013; Forgetta et al.2013; Haridas et al.2013; Khoshraftar et al.2013). The effect of inoculum size on the morphology of O. ulmi as a manifestation of QS regulation has been studied. In 2004 it was shown that in medium inoculated with >106 spores per mL, cells initially developed as budding yeasts, while when inoculated at <106 spores per mL, cells developed as mycelia (Hornby et al. 2004). This inoculum size-dependent effect was related to the presence of QSMs that accumulated throughout the growth of the fungus. When the QSMs were extracted from spent medium with organic solvent and subsequently added back to fresh cultures, the QS activity demonstrated a dose-dependent effect specific for O. ulmi, without an appreciable effect on C. albicans. Similarly, O. ulmi did not respond to farnesol addition. Further studies indicated that the QSM mediating these processes could be 2-methyl-1-butanol (Berrocal et al.2012). Later, it was found that the O. ulmi strain used in this study actually belongs to O. novo-ulmi species (Naruzawa and Bernier 2014). Furthermore, inoculum size effects on yeast–mycelium dimorphism have been reported in several strains of O. novo-ulmi (Wedge et al.2016). This report revealed that different strains of this species respond differently to inoculum size, confirming a certain level of intra-specific variation. In addition, transcriptome analysis of O. novo-ulmi has confirmed that several genes in the cAMP and PKA pathways are differentially expressed during the morphological transition from yeast to hyphae, suggesting that some of these genes might be targets for QSMs, as is the case for C. albicans (Nigg and Bernier 2016).
Other species from the genus Ophiostoma are plant saprophytes, such as the dimorphic fungus O. piceae. QS mechanisms mediated by farnesol have been described in the strain O. piceae CECT 20 416. The accumulation of farnesol in the culture medium of this dimorphic fungus during the exponential growth phase coordinates the morphological transition from yeast to hyphae, and this is likely modulated by the transcription repressor Tup1 (de Salas, Martínez and Barriuso 2015). Furthermore, the addition of exogenous E,E-farnesol or spent medium to fresh cultures of O. piceae had a significant effect on fungal morphology; however, its effect was opposite to that produced by the same compounds in C. albicans, where it induced filamentation of the fungus (de Salas, Martínez and Barriuso 2015). Experiments with another saprophytic fungus, the albino strain O. floccosum FlF1A55 (Berrocal et al.2014) showed an inoculum size effect similar to that reported in C. albicans (Hornby et al.2001). In O. floccosum, three cyclic sesquiterpenes were described as QSMs in a defined culture medium with l-proline as a sole nitrogen source (Berrocal et al.2014). These studies have shown the diversity of QSMs in the phylogenetically related species O. piceae, O. floccosum and O. novo-ulmi. Further studies are needed to establish whether the ability of these fungi to colonise wood is influenced by the morphological switch and whether QSMs play a role in this process.
The presence of QS mechanisms was recently reported in another dimorphic fungus, the insect pathogen Metarhizium rileyi (Boucias et al.2016). The study revealed that the yeast-like hyphal bodies-to-mycelia transition, which occurs in the insect host at the late infection stage, is regulated by QS mechanisms. A QSM is produced when the density of hyphal bodies in the hemolymph reaches a threshold, triggering the synchronous switch in the tissue-invasive mycelia. In this case, the QSM is not identified yet, but lack of QS activity from of M. rileyi spent media suggests that the chemical signal mediating this response is produced by the host tissues in response to a high density of hyphal bodies (Boucias et al.2016).
QS in filamentous fungi
Filamentous fungi, developing mycelial growth, are not unicellular; hence the necessity of exporting a QSM to the environment in these organisms is controversial. However, QS mechanisms have been reported in this group, particularly in species from the genera Aspergillus and Penicillium (Raina, Odell and Keshavarz 2010; Sorrentino, Roy and Keshavarz 2010; Guo et al.2011; Raina et al.2012; Williams et al.2012). In these studies, fungi seem to induce secondary metabolism as a competition strategy. Penicillium sclerotiorum produces sclerotiorin, a secondary metabolite with antibiotic properties. In this fungus, γ-butyrolactone-containing molecules such as multicolic acid function as QSMs. The exogenous addition of extracts containing γ-butyrolactones produced by P. sclerotiorum enhances sclerotiorin production in P. sclerotiorum cultures (Raina, Odell and Keshavarz 2010). Similarly, the effect of farnesol on fungal morphology and cellulase production in Penicillium decumbens has been investigated. In this case, the addition of a high concentration of exogenous farnesol (1 mM) promoted hyphal growth facilitating higher cellulase secretion (Guo et al.2011). Further kinetic studies monitoring QSM production during the exponential growth phase are needed to determine the role of endogenous QS in the control of enzyme production and to corroborate the population density-dependent nature of this phenomenon.
In Aspergillus species, it has become apparent that QS mechanisms influence population-dependent behaviours, such as morphogenesis and secondary metabolite production. Oxylipins in A.flavus act as QSMs that regulate morphological differentiation and the production of either asexual spores or sclerotia. The specific structure of these oxylipins is still unknown (Horowitz Brown et al.2008). Similarly, oxylipins regulate a QS-dependent pathway in A. flavus controlling development and mycotoxin production (Affeldt, Brodhagen and Keller 2012). However, in this case, the production of oxylipins has not been demonstrated as a population-dependent phenomenon. Furthermore, the addition of linoleic acid, an oxylipin precursor, to cultures of O. novo-ulmi affected fungal morphology, but this effect was not regulated by QS mechanisms (Naruzawa, Malagnac and Bernier 2016). In Aspergillus terreus, the addition of linoleic acid improved production of the secondary metabolite lovastatin (a cholesterol-lowering drug) in stirred tank fermenters (Sorrentino, Roy and Keshavarz 2010). Later, a butyrolactone was proposed as a QSM in A. terreus for its ability to affect both lovastatin and its own production. Exogenous addition of 100 nM butyrolactone I resulted in a 2.5-fold increase in lovastatin production, regulating the transcription of genes involved in the lovastatin biosynthesis. Exogenous addition of butyrolactone I also increased the levels of endogenous butyrolactone I (2.5-fold), suggesting an auto-stimulatory function (Raina et al.2012). The strain AXTII9 of Aspergillusnidulans secretes a small diffusible molecule during its growth cycle, which accumulates at high population densities and alters growth profile and secondary metabolite production (Williams et al.2012). The addition of ethyl acetate extracts from stationary phase culture supernatants to low cell-density cultures of A. nidulans abolished the lag phase, decreased the final cell dry weight and increased penicillin production. The bioactive molecule present in the stationary phase extract was purified and identified as γ-heptalactone (Williams et al.2012).
The empirical evidence indicates the existence of QS in filamentous fungi, but the biological meaning is still unclear. Further studies are needed to elucidate whether these mechanisms are a form of communication among fungal microcolonies (de Bekker et al.2011) and/or are involved in the formation of biofilms in these fungi (Harding et al.2009).
FAMILIES OF QS MOLECULES
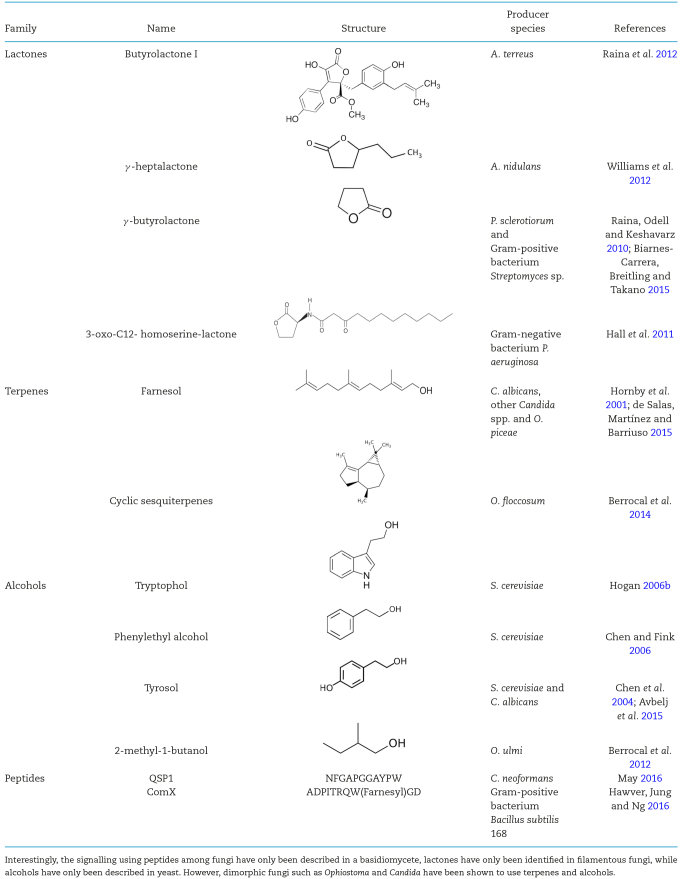
The signalling molecules described so far (Table 1) belong to four families of compounds. (i) Lactones, which are cyclic esters of hydroxycarboxylic acids (Safari et al.2014), including AHLs from Gram-negative bacteria; and butyrolactone I (Raina et al.2012), γ-heptalactone (Williams et al.2012) and γ-butyrolactones (Raina, Odell and Keshavarz 2010), exclusively from filamentous fungi. Interestingly, γ-butyrolactones exist in the filamentous bacterium Streptomyces sp. (Biarnes-Carrera, Breitling and Takano 2015), suggesting convergent evolution or horizontal gene transfer. (ii) Terpenes are derived biosynthetically from isoprene units, such as farnesol produced by the dimorphic fungi C. albicans (Hornby et al.2001) and O. piceae (de Salas, Martínez and Barriuso 2015), or the cyclic sesquiterpenes from the dimorphic fungus O. floccosum (Berrocal et al.2014). (iii) Alcohols are derived from amino acids, and are produced by yeast and dimorphic fungi. QS alcohols include tryptophol and phenylethyl alcohol in the baker's yeast S. cerevisiae (Hogan 2006b), tyrosol in S. cerevisiae and C. albicans (Chen et al.2004; Avbelj et al.2015), and 2-methyl-1-butanol in O. novo-ulmi (Berrocal et al.2012). (iv) Peptides, produced by one basidiomycete, C. neoformans (May 2016), as well as by several Gram-positive bacteria (Hawver, Jung and Ng 2016).
Table 1.
QS molecules identified or active in fungi.
In this context, it could be hypothesised that fungi use a similar ‘language’ to communicate with related species, and a ‘common language’ to communicate to species from other phyla. However, this plausible scenario might prove to be an over-simplification; as new QSMs are discovered, a more detailed picture could emerge (Table 1).
In general, as QSMs are produced in small concentrations (nM–μM) researchers face an experimental problem when larger amounts are needed, e.g. for purification, identification and the discovery of new molecular structures. In this context, ways to increase their production (including molecular biology tools or scale up fermentations) will prove useful.
INTERKINGDOM COMMUNICATION
While bacterial species can sense different families of QSMs, there is no evidence of inter-species communication among fungi. However, this cross-talk has been described between bacteria and fungi, and even bacteria and fungi with their plants or mammalians hosts (Camara, Williams and Hardman 2002; Barriuso et al.2008; Pérez-Montaño et al.2013; Hargarten et al.2015). There are only a few reports dealing with interkingdom communication involving fungi, and most are focused on C. albicans. It is known that opaque cells of C. albicans are able to attract leukocytes to the site of infection by using the QSM farnesol (Langford et al.2013; Hargarten et al.2015). Moreover, farnesol can influence the behaviour of bacteria, inhibiting growth and virulence factors in some cases (Jabra-Rizk et al.2006). It has also been reported that co-cultivation of C. albicans and the nonpathogenic filamentous fungus A. nidulans results in farnesol-dependent inhibition of A. nidulans growth (Semighini et al.2006). Candidaalbicans may use farnesol to reduce competition from other microbes. However, it is not clear to what extent this is a population density-mediated process. The hydrophobic nature of farnesol and the high concentrations used in these examples may favour its accumulation in the membrane and its leakage.
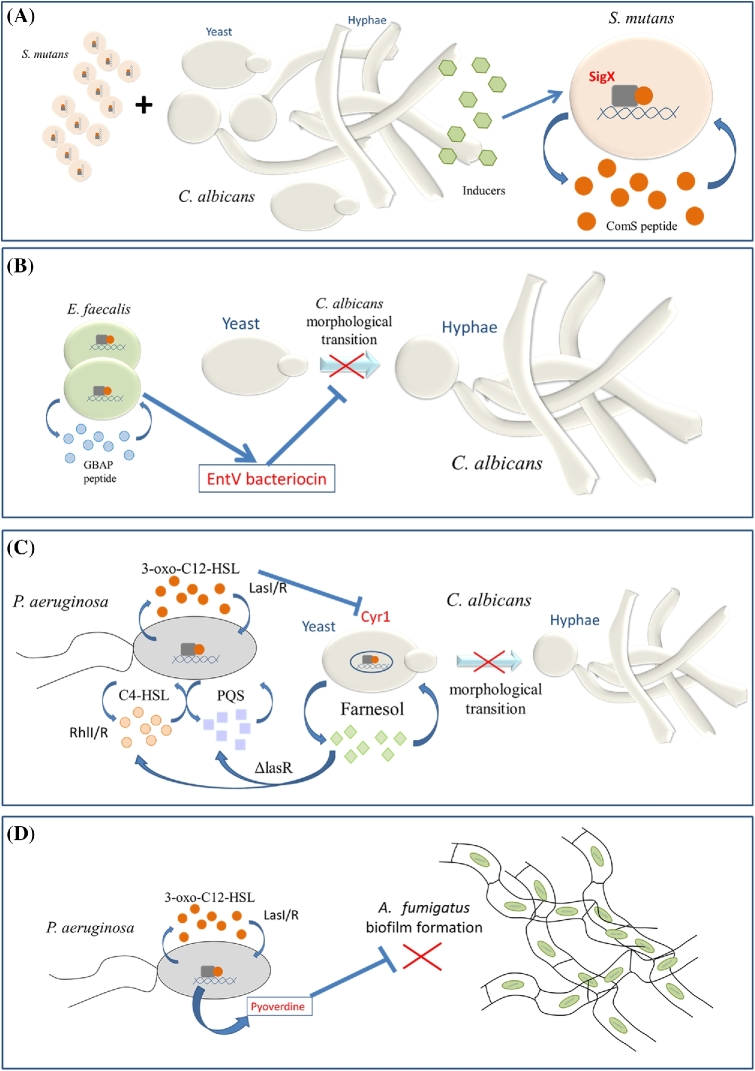
Interkingdom communication through QS systems has been reported in polymicrobial biofilms between the caries-promoting bacterium Streptococcus mutans and C. albicans, which are of medical importance (Sztajer et al.2014). In this case, dual-species biofilms reached higher biomass and cell numbers than mono-species biofilms. Spent media from mixed biofilms but not from C. albicans or S. mutans alone activated alternative sigma factor SigX of S. mutans, which is regulated by the bacterium's QSMs and influences its virulence properties (Sztajer et al.2014) (Fig. 2A). However, in this case, the signalling molecule involved has not been fully identified yet. In another example, the Gram-positive bacterium Enterococcus faecalis inhibits hyphal morphogenesis and virulence of C. albicans in the context of in vitro biofilm and in a Caenorhabditis elegans infection model (Cruz et al.2013). Both microbes are commensals in the same niche and frequently induce polymicrobial infections. The inhibition is partially dependent on the Fsr QS system of E. faecalis, specifically, two proteases regulated by Fsr, GelE and SerE are partially required. Preliminary characterisation of the inhibitory signal has revealed that it is heat resistant with a molecular weight between 3 and 10 kDa; later, this factor was identified as the EntV secreted bacteriocin (Graham et al.2017) (Fig. 2B).
Figure 2.
Communication between bacteria and fungi has co-evolved into polymicrobial biofilms. There are several examples of interkingdom cross-talk where bacteria and fungi exchange signal molecules depending on their population densities within a biofilm. (A) Spent medium from dual-species biofilms between S. mutans and C. albicans (not from C. albicans or S. mutans alone) activates alternative sigma factor SigX of S. mutans, which in turn is regulated by the bacterium's QSM ComS, resulting in a biofilm with higher cell number and biomass. (B) The bacterium E. faecalis inhibits hyphal morphogenesis of C. albicans by secreting the EntV bacteriocin in the context of in vitro biofilm. This inhibition is partly dependent on the Fsr QS system of E. faecalis. (C) The QSM from P. aeruginosa, 3-oxo-C12-HSL, inhibits the adenylyl cyclase (Cyr1) from C. albicans and prevents the morphological transition from yeast to hyphae in this fungus. On the other hand, the QSM from C. albicans, farnesol, is able to induce the production of the QSMs C4-HSL and quinolones (PQS) in biofilms of P. aeruginosa ΔlasR mutants. (D) Pseudomonas aeruginosa inhibits A. fumigatus biofilm formation by producing the siderophore pyoverdine, the production of which is regulated by the QS systems in P. aeruginosa.
In addition, the association between Candida and other pathogens, such as the bacterium Pseudomonas aeruginosa, increases the complexity of infections. In this case, the 12-carbon backbone QSM 3-oxo-C12-HSL secreted by P. aeruginosa inhibits the Cyr1 from the yeast by binding directly to the catalytic site, and prevents C. albicans filamentation (Hogan, Vik and Kolter 2004; Hall et al.2011). The fungal Cyr1 contains multiple domains acting as sensors for a diverse range of signals, including serum, peptidoglycan and elevated CO2 concentration (Klengel et al.2005). QSMs also bind to the catalytic domain of Cyr1p at a site distal to that of the CO2 binding site. Biochemical studies suggest that 3-oxo-C12-HSL is unique in its ability to mimic the actions of farnesol by binding to the farnesol inhibitory site in Cyr1p. Due to the importance of this signalling pathway in both fungal pathogenesis and interkingdom communication, the fungal soluble adenylyl cyclase has been proposed as a hub of signal sensing and integration (Hogan and Muhlschlegel 2011). On the other hand, farnesol is able to induce the production of the QSMs quinolone and C4-HSL in P. aeruginosa biofilms (Cugini, Morales and Hogan 2010) (Fig. 2C).
In addition to investigations on interkingdom cross-talk with C. albicans, there is an increasing number of publications describing the interactions of other fungi with bacteria. For example, P. aeruginosa, which causes biofilm lung infections in cystic fibrosis patients, is able to inhibit Aspergillus fumigatus biofilm formation by means of a secreted heat-stable soluble factor in a concentration-dependent manner (Ramage et al.2011). Co-culture of P. aeruginosa mutants in the master QS system LasI/R did not significantly inhibit A. fumigatus biofilms formation and filamentation to the same extent as did wild type. Subsequently, the secreted factor was determined to be the siderophore pyoverdine, the production of which is regulated by the QS systems in P. aeruginosa (Sass et al.2017) (Fig. 2D). The diffusible lipopeptide ralsolamycin, produced by the ubiquitous soil-borne plant pathogenic bacterium Ralstonia solanacearum, is a novel interkingdom interaction signal that induces chlamydospore development in a wide range of fungal taxa (Ascomycetes, Basidiomycetes and Zygomycetes) (Spraker et al.2016). Interestingly, the production of ralsolamycin is controlled in a bacterial population density-dependent manner by the PhcBSR QS system (Li et al.2017). It is thought that ralsolamycin contributes to the invasion of fungal hyphae, providing a specific niche for bacterial colonisation, but also diminishing survival of the fungus by preventing the production of protective agents against oxidative stress (Spraker et al.2016; Khalid et al.2018). Finally, it has recently been found that the ComX pheromone from Bacillus licheniformis (NCIMB-8874) inhibits growth of A.flavus strains (NRRL 3357 and ESP 15). This can lead to biotechnology-based strategies for the control of aflatoxin damage in agriculture, avoiding pre- and post-harvest aflatoxin contamination (Esmaeilishirazifard et al.2017).
These examples show how microorganisms use their QS systems to sense their population density as well as to directly interact with other microbes, or indirectly regulate the production of molecules that affect the survival of their neighbouring populations; and highlight the importance and timeliness of research to investigate how microorganisms communicate with each other and with their environment. Therefore, it is essential to examine other microbes in each fungal habitat and identify potential interactions among the microbial community (Johnston, Boddy and Weightman 2016).
ECOPHYSIOLOGICAL ROLE AND PATHOGENESIS
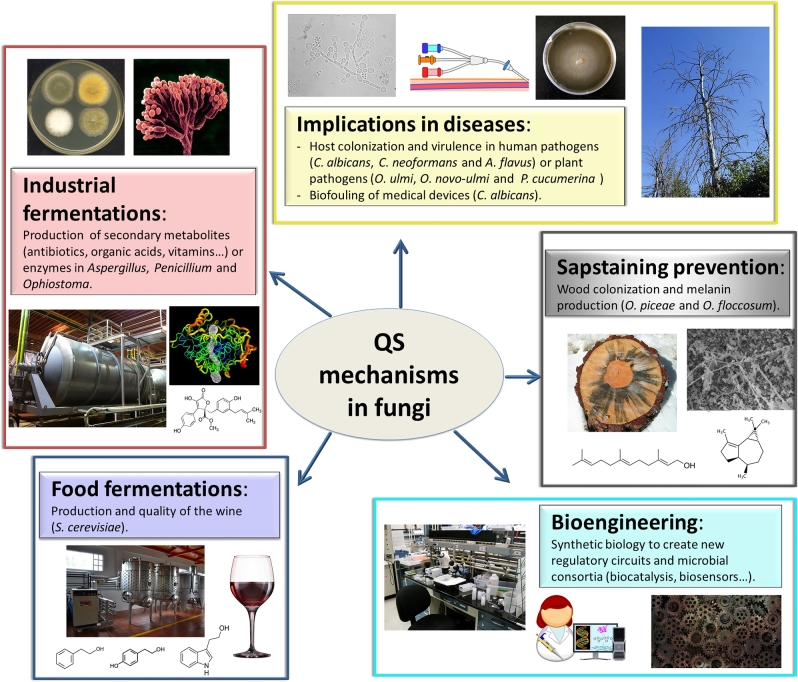
As has been shown, QS mechanisms play a central role in the ability of fungi to colonise their habitat, form biofilms and cause infections. In order to understand the clinical implications of QS and to exploit it as a therapeutic target, further studies of QS mechanisms in fungi are needed (Fig. 3).
Figure 3.
Potential biotechnological applications of the fungal QS mechanisms.
Candida albicans is a successful coloniser of the human host. Important virulence-associated traits of the fungus, such as the dimorphic switch and biofilm formation, are controlled by the QSM farnesol. Candidaalbicans also simultaneously produces and tolerates high farnesol concentrations that are lethal to other microbes (Polke et al.2017). The formation of biofilm structures in C. albicans has been documented as a form of resistance against antimycotic drugs (Nobile and Johnson 2015). Since traditional antimicrobial therapies are often ineffective against C. albicans biofilms, different approaches have been tested to enhance the susceptibility of biofilms. In this context, the clinical relevance of farnesol and tyrosol was demonstrated by the finding that the exogenous addition of these molecules inhibits biofilm formation in Candida species, enhancing their susceptibility to antifungals (Ramage et al.2002; Cordeiro et al.2015).
The study of QS processes in fungi may uncover strategies to prevent colonisation of hosts by not only human and animal pathogens, but also plant pathogens (Fig. 3). For example, O. ulmi and O. novo-ulmi, causal agents of two successive Dutch elm disease pandemics, are transported to elm trees by an intermediate vector, the elm bark beetle (Santini and Faccoli 2015). Fungal spores accumulate in the xylem vessels of the plant and disperse by translocation. The ability of these organisms to undergo morphological changes is thought to be a crucial factor for their virulence. Since QS regulation in O. novo-ulmi dimorphism has been identified (Wedge et al.2016), the pathogenesis of this fungus may be regulated by this mechanism, and thus may be interrupted by targeting this pathway. Other genera from the family Ophiostomataceae, such as Grosmannia and the anamorphic genus Sporothrix, contain saprophytic and pathogenic species of relevance that are dimorphic or polymorphic. QS mechanisms have not been described in these fungi but are worthy of investigation since the anamorphic genus Sporothrix, phylogenetically related to the teleomorphic genus Ophiostoma, includes several species that are pathogenic to humans or animals. In this sense, it has been described that spent medium from Sporothrix schenckii increased the growth of C.neoformans (Albuquerque et al.2014). On the other hand, Grosmannia clavigera is able to infect pine trees (DiGuistini et al.2011). The yeast is the infective form, while it usually grows as mycelia in the environment (López-Romero et al.2011). Population-level signalling, such as QS, that modulates morphology is yet to be described in these species, and warrants future studies.
Little is known with regards to the role of QS in filamentous fungi, although there is an example of population density-dependent phenomena in Plectosphaerella cucumerina regulating the infection strategy decision; however, the molecule regulating this process remains unknown (Pétriacq, Stassen and Ton 2016). In the case of A.flavus the role of oxylipins as QSMs has to do with the reproductive cycle that the fungus adopts to produce asexual spores or sclerotia (Horowitz Brown et al.2008). In other fungi, e.g. Aspergillus and Penicillium species, QS mechanisms are used to induce secondary metabolisms as a competition strategy (Raina, Odell and Keshavarz 2010; Sorrentino, Roy and Keshavarz 2010; Guo et al.2011; Raina et al.2012; Williams et al.2012).
BIOENGINEERING OF FUNGAL QS SYSTEMS
The exploitation of QS mechanisms as biotechnological tools is a developing field in bacteriology (Choudhary and Schmidt-Dannert 2010); however, there are few examples in mycology despite the fact that many fungi have biotechnological applications. For example, species such as O. piceae and O. floccosum produce the pigment melanin, which is responsible for sapstain conifers. Colonisation of wood by these fungi results in substantial economic losses in the forest industry products, making biological control of these fungi essential regarding both environmental and economic considerations (Krokene and Solheim 1998). Ophiostoma albino strains have been developed and are used as biocontrol agents (Held et al.2003), but the exploration of the QS systems of these fungi (Berrocal et al.2014; de Salas, Martínez and Barriuso 2015) may represent an alternative approach to avoid colonisation and hence the blue-staining (Fig. 3). Moreover, this review has addressed fungal species able to produce important secondary metabolites including antibiotics, organic acids and vitamins, and species capable of producing enzymes for decomposition of organic matter. For example, O. piceae CECT 20 416 is able to produce a versatile lipase/sterol esterase of biotechnological relevance that acts on triglycerides, esters of p-nitrophenol and cholesterol (Barriuso et al.2016). The modification of the QS parameters in fermentations with this fungus resulted in a 2-fold increased secretion of the enzyme (de Salas, Martínez and Barriuso 2015). This strategy has also been explored in P. sclerotiorum and A. terreus for the production of metabolites (Raina, Odell and Keshavarz 2010; Williams et al.2012) (Fig. 3).
Furthermore, some fungi take part in the production of foods; the most illustrative example is S. cerevisiae. It has been shown in this yeast that QS during fermentations may influence the production and quality of wine. In particular, ethanol stress during wine fermentation reduces the production of 2-phenylethanol, tryptophol and tyrosol, which opens new challenges in the control of food fermentations (Zupan et al.2013; Avbelj et al.2015) (Fig. 3).
QS systems from different microorganisms have been artificially modified by synthetic and systems biology techniques to engineer the communication among different species. Using the principles of engineering and standardisation, systems biology is taking advantage of microbial QS to create new regulatory circuits with new features and diverse biotechnological applications, allowing complex behaviours and the division of labour within a community (Purnick and Weiss 2009). In bacteria there are many examples of artificial QS circuits, biosensors based on QS mechanisms and even bacteria that are able to release a drug in tumour tissues in vivo using QS mechanisms (Choudhary and Schmidt-Dannert 2010). In fungi, there are some examples of artificial QS circuits depicting artificial intra- or interspecific communications. In this respect, Khakhar et al. (2016) developed a modified S. cerevisiae strain with an orthologous QS system, based on the transcription factor CRISPR and the plant hormone auxin. This engineered cell-to-cell communication system was composed of a sender strain able to produce auxin and a receiver strain with a broad range of auxin sensitivities based on a library of auxin-degradable CRISPR transcription factors (Fig. 3).
Oscillatory bacterial communities have been created already using QS mechanisms and systems with a bi-stable behaviour to increase the robustness of the system (Chen et al.2015), but the construction of new biological systems taking advantage of the QS systems of bacteria and fungi has never been explored.
One of the current challenges in biocatalytic processes is to improve their efficiency by using microbial consortia to carry out complex biotransformations. This strategy exploits the biotechnological potentials of different microorganisms that are put to work together. The interactions between fungi and bacteria would be of particular interest because both organisms have different, and in many cases complementary, biotechnological abilities such as the production of extracellular degrading enzymes or high added-value compounds (Fig. 3). In this context, it is worthwhile to study the pool of bacteria that surround and effectively interact with fungi in nature; in other words, the microbiome (Johnston, Boddy and Weightman 2016).
CONCLUSIONS
In this paper we have presented a broad overview of a novel field of research, QS mechanisms in fungi. This review tackles the fundamental issue of how microorganisms relate to each other and their environment. Since this is a maturing area of research, there is much to be discovered. It is now clear that there is no universal communicating ‘language’ among fungi, as QSMs are not generic across genera or species. Over the coming years, we will be able to elucidate whether fungal communication is common to taxonomic groups or is strain specific. In addition, an understanding of the molecular components of QS systems will be required to determine the extent of cross-talk among fungi, to decipher the evolutionary origin of the fungal ‘languages’ and to unravel the way fungi communicate with other organisms, such as bacteria, plants and animals. As our knowledge of the sophisticated microbial communication process increases and distils, we can determine whether fungal QS is modulated in response to environmental conditions through extrinsic and intrinsic factors, and whether microevolution of QS within fungal populations occurs. This knowledge will enhance the potential for targeting fungal QS processes for biotechnological and clinical purposes.
Acknowledgements
The authors would like to thank Dr E. Esmaeilishirazifard for her comments on cross-kingdom communication and B. Carro for her help editing the figures.
FUNDING
This work was supported by the Spanish Ministry of Economy, Industry and Competitiveness MEICOMP (grant numbers BIO2015–68 387-R and BIO2015–73 697-JIN co-financed with FEDER funds) and the US National Institutes of Health (grant numbers R01 AI127548).
Conflict of interest. None declared.
REFERENCES
- Affeldt KJ, Brodhagen M, Keller NP. Aspergillus oxylipin signaling and quorum sensing pathways depend on G protein-coupled receptors. Toxins 2012;4:695–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque P, Nicola AM, Nieves E et al. Quorum sensing-mediated, cell density-dependent regulation of growth and virulence in Cryptococcus neoformans. mBio 2014;31,5:e00986-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avbelj M, Zupan J, Kranjc L et al. Quorum-sensing kinetics in Saccharomyces cerevisiae: A symphony of ARO genes and aromatic alcohols. J Agric Food Chem 2015;63:8544–50. [DOI] [PubMed] [Google Scholar]
- Bandara HMHN, Lam OLT, Jin LJ et al. Microbial chemical signaling: a current perspective. Crit Rev Microbiol 2012;38:217–49. [DOI] [PubMed] [Google Scholar]
- Barriuso J, Solano BR, Fray RG et al. Transgenic tomato plants alter quorum sensing in plant growth-promoting rhizobacteria. Plant Biotech J 2008;6:442–52. [DOI] [PubMed] [Google Scholar]
- Barriuso J, Vaquero ME, Prieto A et al. Structural traits and catalytic versatility of the lipases from the Candida rugosa-like family: A review. Biotechnol Adv 2016;34:874–85. [DOI] [PubMed] [Google Scholar]
- Berrocal A, Navarrete J, Oviedo C et al. Quorum sensing activity in Ophiostoma ulmi: effects of fusel oils and branched chain amino acids on yeast-mycelial dimorphism. J App Microbiol 2012;113:126–34. [DOI] [PubMed] [Google Scholar]
- Berrocal A, Oviedo C, Nickerson KW et al. Quorum sensing activity and control of yeast-mycelium dimorphism in Ophiostoma floccosum. Biotechnol Lett 2014;36:1503–13. [DOI] [PubMed] [Google Scholar]
- Biarnes-Carrera M, Breitling R, Takano E. Butyrolactone signalling circuits for synthetic biology. Curr Opin Chem Biol 2015;28:91–98. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T, Givskov M. Quorum-sensing blockade as a strategy for enhancing host defences against bacterial pathogens. Philos Trans R Soc B Biol Sci 2007;362:1213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blus-Kadosh I, Zilka A, Yerushalmi G et al. The effect of pstS and phoB on quorum sensing and swarming motility in Pseudomonas aeruginosa. PLoS One 2013;8:e74444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucias D, Liu S, Meagher R et al. Fungal dimorphism in the entomopathogenic fungus Metarhizium rileyi: Detection of an in vivo quorum-sensing system. J Invertebr Pathol 2016;136:100–8. [DOI] [PubMed] [Google Scholar]
- Camara M, Williams P, Hardman A. Controlling infection by tuning in and turning down the volume of bacterial small-talk. Lancet Infect Dis 2002;2:667–76. [DOI] [PubMed] [Google Scholar]
- Chen H, Fink GR. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev 2006;20:1150–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Fujita M, Feng Q et al. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc Natl Acad Sci USA 2004;101:5048–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Schauder S, Potier N et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 2002;415:545–9. [DOI] [PubMed] [Google Scholar]
- Chen Y, Kim JK, Hirning AJ et al. Emergent genetic oscillations in a synthetic microbial consortium. Science 2015;349:986–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary S, Schmidt-Dannert C. Applications of quorum sensing in biotechnology. Appl Microbiol Biotechnol 2010;86:1267–79. [DOI] [PubMed] [Google Scholar]
- Connell JL, Wessel AK, Parsek MR et al. Probing prokaryotic social behaviors with bacterial "Lobster Traps". mBio 2010;1:e00202–10-e00202–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro A, Teixeira CE, Brilhante RS et al. Exogenous tyrosol inhibits planktonic cells and biofilms of Candida species and enhances their susceptibility to antifungals. FEMS Yeast Res 2015;15:12. [DOI] [PubMed] [Google Scholar]
- Cruz MR, Graham CE, Gagliano BC et al. Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect Immun 2013;81:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugini C, Morales DK, Hogan DA. Candida albicans-produced farnesol stimulates Pseudomonas quinolone signal production in LasR-defective Pseudomonas aeruginosa strains. Microbiology 2010;156:3096–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Hanna A, Piispanen AE, Stateva LI et al. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol Microbiol 2008;67:47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beer ZW, Wingfield MJ. Emerging lineages in the Ophiostomatales. In: Ophiostomatoid fungi: expanding frontiers (Seifert KA, Wingfield MJ, eds). CBS Biodiversity Series2013;12:21–46. [Google Scholar]
- de Bekker C, van Veluw GJ, Vinck A et al. Heterogeneity of Aspergillus niger microcolonies in liquid shaken cultures. Appl Environ Microbiol 2011;77:1263–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Salas F, Martínez MJ, Barriuso J. Quorum-sensing mechanisms mediated by farnesol in Ophiostoma piceae: Effect on secretion of sterol esterase. Appl Environ Microbiol 2015;81:4351–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle SP, Griffin AS, Campbell GS et al. Cooperation and conflict in quorum-sensing bacterial populations. Nature 2007;450:411–4. [DOI] [PubMed] [Google Scholar]
- DiGuistini S, Wang Y, Liao NY et al. Genome and transcriptome analyses of the mountain pine beetle-fungal symbiont Grosmannia clavigera, a lodgepole pine pathogen. Proc Natl Acad Sci USA 2011;108:2504–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow JM, Naughton LM, Hollmann B et al. The diffusible signal factor family of bacterial cell-cell signals. Isr J Chem 2016; 56; 321–9. [Google Scholar]
- Egbe NE, Dornelles TO, Paget CM et al. Farnesol inhibits translation to limit growth and filamentation in C. albicans and S. cerevisiae. Microb Cell 2017;4:294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeilishirazifard E, De Vizio D Moschos SA et al. Genomic and molecular characterization of a novel quorum sensing molecule in Bacillus licheniformis. AMB Expr 2017;7:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgetta V, Leveque G, Dias J et al. Sequencing of the Dutch elm disease fungus genome using the Roche/454 GS-FLX Titanium System in a comparison of multiple genomics core facilities. J Biomol Tech 2013;24:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier GM. Dimorphism in fungal pathogens of mammals, plants, and insects. PLoS Pathog 2015;11:e1004608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gognies S, Barka EA, Gainvors-Claisse A et al. Interactions between yeasts and grapevines: filamentous growth, endopolygalacturonase and phytopathogenicity of colonizing yeasts. Microb Ecol 2006;51:109–16. [DOI] [PubMed] [Google Scholar]
- Graham CE, Cruz MR, Garsin DA et al. Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc Natl Acad Sci USA 2017;114:4507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ma A, Zhao G et al. Effect of farnesol on Penicillium decumbens’s morphology and cellulase production. Bioresources 2011;6:3252–9. [Google Scholar]
- Hall RA, Turner KJ, Chaloupka J et al. The quorum-sensing molecules farnesol/homoserine lactone and dodecanol operate via distinct modes of action in Candida albicans. Eukaryot Cell 2011;10:1034–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JH, Hebert WP, Naimie A et al. Environmentally endemic Pseudomonas aeruginosa strains with mutations in lasR are associated with increased disease severity in corneal ulcers. mSphere 2016;1:e00140–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding MW, Marques LL, Howard RJ et al. Can filamentous fungi form biofilms? Trends Microbiol 2009;17:475–80. [DOI] [PubMed] [Google Scholar]
- Hargarten JC, Moore TC, Petro TM et al. Candida albicans quorum sensing molecules stimulate mouse macrophage migration. Infect Immun 2015;83:3857–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haridas S, Wang Y, Lim L et al. The genome and transcriptome of the pine saprophyte Ophiostoma piceae, and a comparison with the bark beetle-associated pine pathogen Grosmannia clavigera. BMC Genomics 2013;14:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawver LA, Jung SA, Ng WL. Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol Rev 2016;40:738–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held BW, Thwaites JM, Farrell RL et al. Albino strains of Ophiostoma species for biological control of sapstaining fungi. Holzforschung 2003;57:237–42. [Google Scholar]
- Hogan DA. Talking to themselves: Autoregulation and quorum sensing in fungi. Eukaryot Cell 2006a;5:613–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DA. Quorum sensing: Alcohols in a social situation. Curr Biol 2006b;16:R457–8. [DOI] [PubMed] [Google Scholar]
- Hogan DA, Muhlschlegel FA. Candida albicans developmental regulation: adenylyl cyclase as a coincidence detector of parallel signals. Curr Opin Microbiol 2011;14:682–6. [DOI] [PubMed] [Google Scholar]
- Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol 2004;54:1212–23. [DOI] [PubMed] [Google Scholar]
- Homer CM, Summers DK, Goranov AI et al. Intracellular action of a secreted peptide required for fungal virulence. Cell Host Microbe 2016;19:849–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornby JM, Jensen EC, Lisec AD et al. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol 2001;67:2982–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornby JM, Jacobitz-Kizzier SM, Mcneel DJ et al. Inoculum size effect in dimorphic fungi: Extracellular control of yeast-mycelium dimorphism in Ceratocystis ulmi. Appl Environ Microbiol 2004;70:1356–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz Brown S, Zarnowski R, Sharpee WC et al. Morphological transitions governed by density dependence and lipoxygenase activity in Aspergillus flavus. Appl Environ Microbiol 200874:5674–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabra-Rizk MA, Meiller TF, James CE et al. Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob Agents Chemother 2006;50:1463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SR, Boddy L, Weightman AJ. Bacteria in decomposing wood and their interactions with wood-decay fungi. FEMS Microbiol Ecol 2016;92, doi: 10.1093/femsre/fiw179. [DOI] [PubMed] [Google Scholar]
- Kebaara BW, Langford ML, Navarathna DH et al. Candida albicans Tup1 is involved in farnesol-mediated inhibition of filamentous-growth induction. Eukaryot Cell 2008;7:980–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhar A, Bolten NJ, Nemhauser J et al. Cell-cell communication in yeast using auxin biosynthesis and auxin responsive CRISPR transcription factors. ACS Synth Biol 2016;5:279–86. [DOI] [PubMed] [Google Scholar]
- Khalid S, Baccile JA, Spraker JE et al. NRPS-Derived isoquinolines and lipopetides mediate antagonism between plant pathogenic fungi and bacteria. ACS Chem Biol 2018;19:171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshraftar S, Hung S, Khan S et al. Sequencing and annotation of the Ophiostoma ulmi genome. BMC Genomics 2013;14:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Liang WJ, Chaloupka J et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol 2005;15:2021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokene P, Solheim H. Pathogenicity of four blue-stain fungi associated with aggressive and nonaggressive bark beetles. Phytopathology 1998;88:39–44. [DOI] [PubMed] [Google Scholar]
- Langford ML, Hargarten JC, Patefield KD et al. Candida albicans Czf1 and Efg1 coordinate the response to farnesol during quorum sensing, white-opaque thermal dimorphism, and cell death. Eukaryot Cell 2013;12:1281–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Yin W, Yan J et al. Modulation of Inter-kingdom communication by PhcBSR quorum sensing system in Ralstonia solanacearum phylotype I strain GMI1000. Front Microbiol 2017;8:1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay AK, Deveau A, Piispanen AE et al. Farnesol and cyclic AMP signaling effects on the hypha-to-yeast transition in Candida albicans. Eukaryot Cell 2012;11:1219–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HJ, Köhler JR, DiDomenico B et al. Nonfilamentous C. albicans mutants are avirulent. Cell 1997;90:939–49. [DOI] [PubMed] [Google Scholar]
- López-Romero E, Reyes-Montes Mdel R, Pérez-Torres A et al. Sporothrix schenckii complex and sporotrichosis, an emerging health problem. Future Microbiol 2011;6:85–102. [DOI] [PubMed] [Google Scholar]
- Lu Y, Su C, Unoje O et al. Quorum sensing controls hyphal initiation in Candida albicans through Ubr1-mediated protein degradation. Proc Natl Acad Sci USA 2014;111:1975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD. Quorum sensing in fungi: Q&A. PLoS Pathog 2011;7:e1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May RC. Custom-Made quorum sensing for a eukaryote. Dev Cell 2016;37:391–2. [DOI] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol 2001;55:165–99. [DOI] [PubMed] [Google Scholar]
- Miller ST, Xavier KB, Campagna SR et al. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell 2004;15:677–87. [DOI] [PubMed] [Google Scholar]
- Mony BM, MacGregor P, Ivens A et al. Genome-wide dissection of the quorum sensing signalling pathway in Trypanosoma brucei. Nature 2014;505:681–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruzawa ES, Bernier L. Control of yeast-mycelium dimorphism in vitro in Dutch elm disease fungi by manipulation of specific external stimuli. Fungal Biol 2014;118:872–84. [DOI] [PubMed] [Google Scholar]
- Naruzawa ES, Malagnac F, Bernier L. Effect of linoleic acid on reproduction and yeast–mycelium dimorphism in the Dutch elm disease pathogens. Botany 2016;96:31–39. [Google Scholar]
- Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol 1970;104:313–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson KW, Atkin AL, Hornby JM. Quorum sensing in dimorphic fungi: Farnesol and beyond. Appl Environ Microbiol 2006;72:3805–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg M, Bernier L. From yeast to hypha: defining transcriptomic signatures of the morphological switch in the dimorphic fungal pathogen Ophiostoma novo-ulmi. BMC Genomics 2016;15:920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Johnson AD. Candida albicans biofilms and human disease. Annu Rev Microbiol 2015;69:71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucci M, Queiroz-Telles F, Tobon AM et al. Epidemiology of opportunistic fungal infections in Latin America. Clin Infect Dis 2010;51:561–70. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Bassler BL. Quorum sensing signal–response systems in Gram-negative bacteria. Nat Rev Microbiol 2016;14:576–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Montaño F, Jiménez-Guerrero I, Contreras Sánchez-Matamoros R et al. Rice and bean AHL-mimic quorum-sensing signals specifically interfere with the capacity to form biofilms by plant-associated bacteria. Res Microbiol 2013;164:749–60. [DOI] [PubMed] [Google Scholar]
- Pétriacq P, Stassen JH, Ton J. Spore density determines infection strategy by the Plant Pathogenic Fungus Plectosphaerella cucumerina. Plant Physiol 2016;170:2325–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polke M, Leonhardt I, Kurzai O et al. Farnesol signalling in Candida albicans - more than just communication. Crit Rev Microbiol 2017;13:1–14. [DOI] [PubMed] [Google Scholar]
- Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol 2009;10:410–22. [DOI] [PubMed] [Google Scholar]
- Raina S, Odell M, Keshavarz T. Quorum sensing as a method for improving sclerotiorin production in Penicillium sclerotiorum. J Biotechnol 2010;148:91–98. [DOI] [PubMed] [Google Scholar]
- Raina S, De Vizio D, Palonen EK et al. Is quorum sensing involved in lovastatin production in the filamentous fungus Aspergillus terreus? Process Biochem 2012;47:843–52. [Google Scholar]
- Ramage G, Saville SP, Wickes BL et al. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl Environ Microbiol 2002;68:5459–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, Rajendran R, Gutierrez-Correa M et al. Aspergillus biofilms: clinical and industrial significance. FEMS Microbiol Lett 2011;324:89–97. [DOI] [PubMed] [Google Scholar]
- Romano A. Dimorphism. In: Ainsworth, GC, Sussman AS.. The Fungi. New York: Academic Press, 1966, 181–209. [Google Scholar]
- Ryan RP, McCarthy Y, Watt SA et al. Intraspecies signaling involving the diffusible signal factor BDSF (cis-2-dodecenoic acid) influences virulence in Burkholderia cenocepacia. J Bacteriol 2009;191:5013–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safari M, Amache R, Esmaeilishirazifard E et al. Microbial metabolism of quorum-sensing molecules acyl-homoserine lactones, γ-heptalactone and other lactones. Appl Microbiol Biotechnol 2014;98:3401–12. [DOI] [PubMed] [Google Scholar]
- Santini A, Faccoli M. Dutch elm disease and elm bark beetles: a century of association. iForest 2015;8:126–34. [Google Scholar]
- Sass G, Nazik H, Penner J et al. Studies of Pseudomonas aeruginosa mutants indicate pyoverdine as the central factor in inhibition of Aspergillus fumigatus biofilm. J Bacteriol 2017;200: e00345–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semighini CP, Hornby JM, Dumitru R et al. Farnesol-induced apoptosis in Aspergillus nidulans reveals a possible mechanism for antagonistic interactions between fungi. Mol Microbiol 2006;59:753–64. [DOI] [PubMed] [Google Scholar]
- Sorrentino F, Roy I, Keshavarz T. Impact of linoleic acid supplementation on lovastatin production in Aspergillus terreus cultures. Appl Microbiol Biotechnol 2010;88:65–73. [DOI] [PubMed] [Google Scholar]
- Spraker JE, Sanchez LM, Lowe TM et al. Ralstonia solanacearum lipopeptide induces chlamydospore development in fungi and facilitates bacterial entry into fungal tissues. ISME J 2016;10:2317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol 2011;9:737–48. [DOI] [PubMed] [Google Scholar]
- Sztajer H, Szafranski SP, Tomasch J et al. Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J 2014;8:2256–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnita CE, Washburne A, Martinez-Garcia R et al. Fitness tradeoffs between spores and nonaggregating cells can explain the coexistence of diverse genotypes in cellular slime molds. Proc Natl Acad Sci USA 2015;112:2776–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi V, Ahmer BMM. Editorial: LuxR solos are becoming major players in cell–cell communication in bacteria. Front Cell Infect Microbiol 2015;5:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K, Schulz B, Ruhnke M. The quorum-sensing molecule E,E-farnesol-its variable secretion and its impact on the growth and metabolism of Candida species. Yeast 2010;27:727–39. [DOI] [PubMed] [Google Scholar]
- Wedge MÈ, Naruzawa ES, Nigg M et al. Diversity in yeast–mycelium dimorphism response of the Dutch elm disease pathogens: the inoculum size effect. Can J Microbiol 2016;62:525–9. [DOI] [PubMed] [Google Scholar]
- Williams HE, Steele JC, Clements MO et al. γ-Heptalactone is an endogenously produced quorum-sensing molecule regulating growth and secondary metabolite production by Aspergillus nidulans. Appl Microbiol Biotechnol 2012;96:773–81. [DOI] [PubMed] [Google Scholar]
- Winzer K, Hardie KR, Williams P. Bacterial cell-to-cell communication: sorry, can’t talk now — gone to lunch! Curr Opin Microbiol 2002;5;216–22. [DOI] [PubMed] [Google Scholar]
- Youk H, Lim WA. Secreting and sensing the same molecule allows cells to achieve versatile social behaviors. Science 2014;343:1242782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Kaufmann GF. Quo vadis quorum quenching? Curr Opin Pharmacol 2013;13:688–98. [DOI] [PubMed] [Google Scholar]
- Zupan J, Avbelj M, Butinar B et al. Monitoring of quorum-sensing molecules during minifermentation studies in wine yeast. J Agric Food Chem 2013;61:2496–505. [DOI] [PubMed] [Google Scholar]