Abstract
Background
The clinical significance of LINC00996 in colorectal cancer (CRC) has not been verified. In the current study, the authors aimed to explore the expression of LINC00996 and its clinical significance in CRC based on the data mining of Gene Expression Omnibus (GEO) and the Cancer Genome Atlas (TCGA) datasets, as well as to elucidate the functions of its potential target genes.
Materials and methods
GEO and TCGA microarray datasets were used to evaluate the LINC00996 expression and its clinical significance in CRC. LINC00996 related genes were identified by Multi Experiment Matrix, RNA-Binding Protein DataBase, and The Atlas of Noncoding RNAs in Cancer. Subsequently, they were sent to gene ontology enrichment and Kyoto Encyclopedia of Genes and Genomes pathway analysis.
Results
LINC00996 is significantly decreased in CRC tissues compared with non-tumor tissues. Low level of LINC00996 is associated with remote metastasis and poor overall survival. However, LINC00996 has a minimal effect on gender, lymphatic invasion, tumor size, lymph node metastasis, and pathological stage. One hundred and forty-two LINC00996 related genes were identified; the results of functional analysis indicated that LINC00996 might repress tumorigenesis and metastasis via modulating the JAK-STAT, NF-κB, HIF-1, TLR, and PI3K-AKT signaling pathways.
Conclusion
Our study demonstrates that decreased LINC00996 expression may be involved in colorectal carcinogenesis and metastasis, and the depletion of LINC00996 is associated with a poor outcome in CRC patients. Moreover, the JAK-STAT, NF-κB, HIF-1, TLR, and PI3K-AKT pathways may be the key pathways regulated by LINC00996 in CRC.
Keywords: LINC00996, lncRNA, colorectal cancer, bioinformatics
Background
Colorectal cancer (CRC) is the third most common cancer worldwide, and it remains a major public health problem with an estimated one million new cases and half a million mortalities every year.1 The incidence of CRC is similar in both sexes, slightly higher in males for rectal cancer, and is higher in Western countries, especially in USA, Canada, Europe, New Zealand, and Australia.2 Despite the development of surgical techniques and clinical treatments, the overall cure rate of CRC has not been significantly improved in recent years. Numerous CRC patients are diagnosed with advanced stage or distant metastasis when they initially seek medical treatment, which accounts for a low (~10%) 5-year survival rate and 90% of cancer-related death.3,4 Thus, it is urgent to expand our knowledge of the molecular pathogenesis of CRC and identify new molecular targets for the development of clinical treatments.
Long non-coding RNAs (lncRNAs) are a class of non-coding RNAs that have no potential to code proteins and are more than 200 nucleotides in length. lncRNAs play an important role in regulating gene expression at various levels, including alternative splicing, regulation of protein activity, and alteration of protein localization, as well as chromatin modification, transcription, and posttranscriptional processing.5,6 Portions of lncRNAs can be specifically expressed in different tissues and various cancers.7,8 Recent studies have reported that the disorder of lncRNAs has a close relationship with the occurrence and progression of various cancers, such as lung cancer, leukemia, gastric cancer, colon cancer, liver cancer, and cholangiocarcinoma.9–14 However, the mechanism whereby lncRNAs regulate the progression of CRC remains elusive. In this study, we analyzed the Gene Expression Omnibus (GEO) and the Cancer Genome Atlas (TCGA) to explore the function of LINC00996 in the tumorigenesis and prognosis of CRC.
Materials and methods
Data acquisition
We extracted microarray data of LINC00996 in GEO profiles (http://www.ncbi.nlm.nih.gov/geoprofiles/) and GEO data-sets (http://www.ncbi.nlm.nih.gov/gds/) up to April 2018. The following search words were used: “colorectal neoplasm”, “colorectal cancer”, “lncRNA”, and “LINC00996”. Microarrays were included if they met the following inclusion criteria: 1) dataset included CRC tissues and non-tumor tissues and more than two samples were examined; 2) dataset provided overall survival (OS) information; 3) the expression profiling data of LINC00996 from the cases and the controls were provided or could be calculated; and 4) data were obtained from Homo sapiens.
CRC-related LINC00996 data in tumor tissues and normal tissues was obtained from the TCGA database (http://cancergenome.nih.gov/) up to April 2018. All RNASeqV2 and clinical data were chosen, and then, Build Archive function was performed. The RNA expression data was classified as Level 3. All quantification files with “.txt” were selected, and all files in the metadata and clinical sections were examined.
LINC00996 related genes in CRC
To determine the genes related to LINC00996 in CRC, Multi Experiment Matrix (http://biit.cs.ut.ee/mem/), RNA-Binding Protein DataBase (RBPDB) (http://rbpdb.ccbr.utoronto.ca/), and The Atlas of Noncoding RNAs in Cancer (TANRIC) (http://ibl.mdanderson.org/tanric/_design/basic/index.html) were utilized. Herein, we recorded the overlapping genes more than two out of three programs as LINC00996 related genes in CRC.
The comprehensive analysis of LINC00996 related genes
We conducted gene ontology (GO) analysis using DAVID website (https://david.ncifcrf.gov/, version 6.8).15 The functions of differently expressed genes were then classified into three major groups: biological processes, cellular component, and molecular functions. Pathway analysis was applied in KOBAS website (http://kobas.cbi.pku.edu.cn/, version 3.0) for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment.16 Cytoscape 3.0 software (http://www.cytoscape.org/) with BINGO plugins were responsible for graphical display of the network of GO.17 In the networks, each node represents a biological process. The bigger node indicates more genes participating in the biological process, and the deeper color indicates the smaller P-value of the biological process. All colored nodes indicate statistical significance (P<0.05). White nodes were only used to connect the biological processes, and there is no statistical significance. The protein-to-protein network analysis was conducted by STRING website.18 In the networks, the nodes represent for proteins, and each color corresponds to a cluster. The edges indicate the predicted functional associations, which are colored by the types of predicted associations using different evidence. The red lines, green lines, blue lines, purple lines, yellow lines, light blue lines, and black lines indicate fusion evidence, neighborhood evidence, co-occurrence evidence, experimental evidence, text mining evidence, database evidence, and co-expression evidence, respectively. The line thickness is an indicator for the strength of the evidence.
Statistical analysis
Quantitative data of LINC00996 expression were presented as the mean ± SD, and SPSS 20.0 software was used for data analysis. Student’s t-test was performed for the analysis of LINC00996 expression between two groups and when it exceeded two groups, one-way ANOVA analysis of variance was employed. The Kaplan–Meier method and log-rank test were used to evaluate the relationship between LINC00996 expression and OS in CRC patients. The cutoff value of LINC00996 expression was determined by its median value.
Meta-analysis was performed using STATA 12.0 software. Continuous outcomes obtained from GEO profiles and GEO datasets were estimated as standard mean difference (SMD) with a 95% CI. HRs and 95% CIs were used to assess the prognostic value of LINC00996 in CRC. Effect sizes were pooled with a random or fixed effects model. HR>1 implied a poor prognosis for the group with low LINC00996 expression and would be considered to be statistically significant if the 95% CI did not overlap 1. P<0.05 was considered to a significant difference.
Results
Expression of LINC00996 in CRC in GEO and TCGA databases
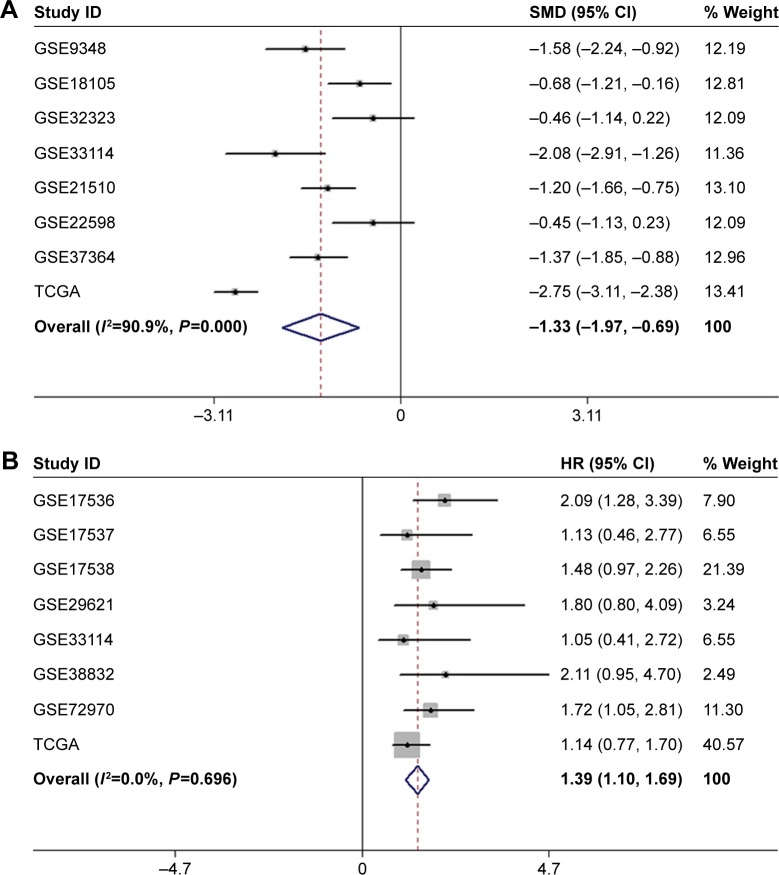
As shown in Figure 1, a total of 13 GEO series (GSE) in the GEO database were extracted for the present study according to the inclusion criteria. Among those, seven series detected both tumor samples and non-tumor control samples; seven series contained data about LINC00996 expression and OS information; one series contained LINC00996 expression data in both tumor and control samples and OS information. Combining GEO and TCGA data, a total of 1,813 CRC patients were enrolled. Sample sizes ranged from 34 to 497 patients. No pharmacological treatments were used when the tissue was obtained. Detailed information is shown in Tables 1 and 2. As shown in Figure 2A, meta-analysis revealed the expression level of LINC00996 was significantly downregulated in CRC tissues compared with non-tumor tissues in seven GEO datasets and TCGA datasets (SMD =−1.33, 95% CI: −1.97 to −0.69, P<0.001, random).
Figure 1.
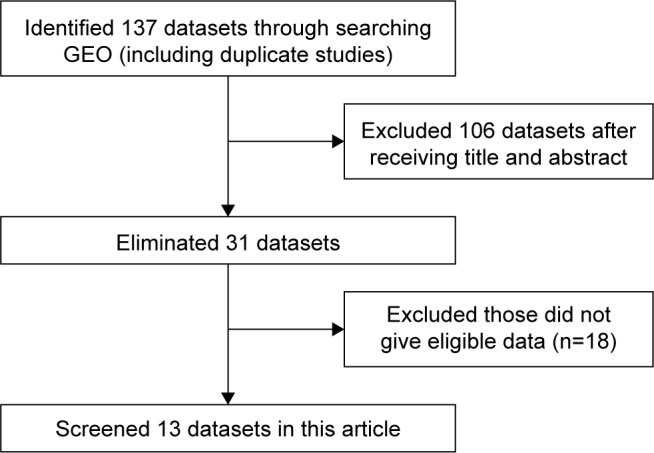
The process of data identification and acquisition.
Abbreviation: GEO, Gene Expression Omnibus.
Table 1.
Characteristics of LINC00996 expression profiling datasets obtained from GEO and TCGA
| Accession | No of tumor samples | No of normal samples | Tumor tissue
|
Normal tissue
|
||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| GSE9348 | 70 | 12 | 39.0650 | 34.9879 | 101.4872 | 60.1701 |
| GSE18105 | 94 | 17 | 3.5782 | 0.2301 | 3.7274 | 0.1259 |
| GSE32323 | 17 | 17 | 3.6589 | 0.1686 | 3.7274 | 0.1259 |
| GSE33114 | 89 | 7 | 13.8090 | 9.1097 | 33.8167 | 15.1097 |
| GSE21510 | 123 | 25 | 3.5938 | 0.2735 | 3.9052 | 0.1615 |
| GSE22598 | 17 | 17 | 3.6589 | 0.1731 | 3.7274 | 0.1259 |
| GSE37364 | 43 | 38 | 61.3837 | 33.1756 | 106.4921 | 32.7381 |
| TCGA | 456 | 41 | 23.8587 | 20.8016 | 88.0016 | 42.7183 |
Abbreviations: GEO, Gene Expression Omnibus; TCGA, the Cancer Genome Atlas.
Table 2.
Analysis of overall survival in datasets obtained from GEO and TCGA
| Accession | No of samples | No of high expression | No of low expression | HR | 95% CI | P-value |
|---|---|---|---|---|---|---|
| GSE17536 | 175 | 88 | 87 | 2.087 | 1.284–3.391 | 0.003 |
| GSE17537 | 54 | 27 | 27 | 1.126 | 0.457–2.771 | 0.797 |
| GSE17538 | 229 | 115 | 114 | 1.482 | 0.974–2.255 | 0.066 |
| GSE29621 | 65 | 33 | 32 | 1.804 | 0.796–4.085 | 0.157 |
| GSE33114 | 89 | 45 | 44 | 1.049 | 0.405–2.72 | 0.921 |
| GSE38832 | 119 | 60 | 59 | 2.109 | 0.946–4.702 | 0.068 |
| GSE72970 | 88 | 44 | 44 | 1.716 | 1.048–2.81 | 0.032 |
| TCGA | 453 | 227 | 226 | 1.144 | 0.77–1.7 | 0.505 |
Abbreviations: GEO, Gene Expression Omnibus; TCGA, the Cancer Genome Atlas.
Figure 2.
Meta-analysis based on GEO and TCGA datasets.
Notes: (A) Forest plots to assess the expression of LINC00996 in tumor tissues and non-tumor tissues. (B) Forest plots to assess the effect of downregulated LINC00996 expression on OS in patients with CRC. Weights are from random effects analysis.
Abbreviations: GEO, Gene Expression Omnibus; OS, overall survival; SMD, standard mean difference; TCGA, the Cancer Genome Atlas, CRC, colorectal cancer.
Relationship between LINC00996 and clinicopathologic characteristics of CRC
We assessed relationships between LINC00996 and clinicopathological characteristics of CRC in TCGA, the result showed that the expression level of LINC00996 was lower in tumor tissues (23.8587±20.8016), compared with normal tissues (88.0016±42.7183; P<0.001). In addition, in the remote metastasis group, LINC00996 expression was also significantly downregulated (16.9839±19.0155), compared with the group without remote metastasis (26.1234±28.1145; P=0.002). Additionally, combined with GEO and TCGA data, meta-analysis revealed that the depletion of LINC00996 was associated with poorer OS in CRC patients (HR =1.39, 95% CI: 1.10–1.69, P<0.001, fixed) (Figure 2B). However, LINC00996 was not associated with gender, lymphatic invasion, tumor size, lymph node metastasis, and pathological stage. Clinicopathological characteristics are presented in Table 3.
Table 3.
Expression of LINC00996 and clinicopathological parameters in CRC obtained from TCGA
| Clinicopathological features | n | LINC00996 relative expression
|
t | P-value |
|---|---|---|---|---|
| Mean ± SD | ||||
| Tissue | −0.954 | <0.001 | ||
| Tumor | 456 | 23.8587±20.8016 | ||
| Normal | 41 | 88.0016±42.7183 | ||
| Age | 0.067 | 0.947 | ||
| <60 | 124 | 25.1532±25.1862 | ||
| ≥60 | 326 | 24.9601±28.01607 | ||
| Gender | −0.439 | 0.661 | ||
| Male | 237 | 25.5485±30.0389 | ||
| Female | 213 | 24.4178±23.7941 | ||
| Lymphatic invasion | 1.012 | 0.312 | ||
| Yes | 161 | 23.1553±22.8174 | ||
| No | 246 | 25.9309±29.4945 | ||
| AJCC pathologic T | F=0.103a | 0.902 | ||
| Tis | 1 | 37 | ||
| T1 | 11 | 25.9091±23.0150 | ||
| T2–T4 | 438 | 24.9635±27.3828 | ||
| AJCC pathologic N | 1.075 | 0.283 | ||
| N0 NX | 265 | 26.1660±28.3905 | ||
| N1–2 | 185 | 23.3622±25.4817 | ||
| AJCC pathologic M | 3.25 | 0.002 | ||
| M0 MX | 381 | 26.1234±28.1145 | ||
| M1 | 62 | 16.9839±19.0155 | ||
| Pathologic stage | 0.861 | 0.39 | ||
| I–II | 250 | 25.8720±28.2844 | ||
| III–IV | 191 | 23.6073±26.1344 |
Note:
One-way analysis of variance analysis was performed.
Abbreviations: AJCC, American Joint Committee on Cancer; TCGA, the Cancer Genome Atlas; CRC, colorectal cancer.
Identification of LINC00996 related genes in CRC
To comprehend the potential mechanisms of LINC00996 involved in CRC, three independent programs were performed to identify genes related to LINC00996: Multi Experiment Matrix,19 RBPDB,20 and TANRIC.21 The first collects LINC00996 related genes in Multi Experiment Matrix, a web program that identifies LINC00996 related genes by gathering several hundreds of publicly available gene expression data sets from the ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) database. A total of 2,267 genes were collected when the score was set at 10−8. The second algorithm screens for LINC00996 related genes in RBPDB, which is a collection of experimental observations of RNA-binding sites. Here, 26 genes were screened. The third algorithm searches for LINC00996 related genes using TANRIC, an open-access web resource for interactive exploration of lncRNAs in cancer. A total of 348 genes related to LINC00996 were selected by setting the absolute value at 10−8. We recorded the overlapping genes more than two out of three programs. Eventually, 142 genes were noted as LINC00996 related genes in CRC.
GO enrichment, KEGG pathway analysis, and protein–protein interaction (PPI) network
Selecting Homo sapiens as the background of listed target genes in DAVID, we obtained the GO term annotations. The results of the GO analysis are summarized in Table 4 and Figure 3, and the top five enriched items are listed according to P-values. There were 35 significant annotations of the biological process (P<0.05). Some of them were involved in adaptive immune response, B cell receptor signaling pathway, G-protein coupled purinergic nucleotide receptor signaling pathway, regulation of immune response, etc. With regard to the cellular component of GO analysis, six significant terms were observed (P<0.05) and some of them correlated with the integral component of plasma membrane, external side of plasma membrane, plasma membrane, integral component of membrane, etc. In the molecular function of GO analysis, seven significant terms were obtained (P<0.05) and some of them were contributed to G-protein coupled purinergic nucleotide receptor activity, receptor activity, GTPase activator activity, transmembrane signaling receptor activity, etc.
Table 4.
The GO analysis of predicted target genes of LINC00996
| GO ID | Term | Count | P-value |
|---|---|---|---|
| Biological process | |||
| 0002250 | Adaptive immune response | 11 | 9.77×10−8 |
| 0050853 | B cell receptor signaling pathway | 8 | 1.01×10−7 |
| 0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway | 5 | 2.27×10−6 |
| 0050776 | Regulation of immune response | 9 | 3.93×10−5 |
| 0006955 | Immune response | 13 | 4.45×10−5 |
| Cellular component | |||
| 0005887 | Integral component of plasma membrane | 33 | 3.19×10−9 |
| 0009897 | External side of plasma membrane | 13 | 4.10×10−8 |
| 0005886 | Plasma membrane | 58 | 5.85×10−8 |
| 0016021 | Integral component of membrane | 53 | 2.67×10−3 |
| 0005622 | Intracellular | 18 | 1.38×10−3 |
| Molecular function | |||
| 0045028 | G-protein coupled purinergic nucleotide receptor activity | 5 | 1.75×10−6 |
| 0004872 | Receptor activity | 9 | 9.91×10−5 |
| 0005096 | GTPase activator activity | 8 | 2.59×10−3 |
| 0004888 | Transmembrane signaling receptor activity | 7 | 3.04×10−3 |
| 0005244 | Voltage-gated ion channel activity | 3 | 1.90×10−3 |
Notes: In the GO analysis of predicted target genes of LINC00996 in three databases, there were 35 available biological processes, six cellular components, and seven molecular functions (P<0.05). In this table, the top five terms of the GO analysis are presented.
Abbreviation: GO, gene ontology.
Figure 3.
GO enrichment analysis of LINC00996 related genes in CRC.
Note: LINC00996 related genes are divided into three functional groups: biological process, cellular component, and molecular function.
Abbreviations: CRC, colorectal cancer; GO, gene ontology.
In terms of the KEGG pathway analysis, there were 23 signaling pathways that were statistically significant (P<0.05). As shown in Table 5, all 23 enriched items are listed according to corrected P-values. The signaling pathways were mainly enriched in the JAK-STAT signaling pathway, cell adhesion molecules, NF-κB signaling pathway, HIF-1 signaling pathway, toll-like receptor (TLR) signaling pathway, PI3K-AKT signaling pathway, etc.
Table 5.
Pathway analysis of the predicted target genes of LINC00996
| Title | Count | P-value | Genes |
|---|---|---|---|
| Hematopoietic cell lineage | 7 | 5.10×10−6 | ITGA4 CD19 MS4A1 CD2 CD38 CD1C IL7R |
| Primary immunodeficiency | 5 | 2.22×10−5 | BTK CD19 CD79A IL7R PTPRC |
| Cytokine–cytokine receptor interaction | 8 | 2.05×10−4 | CCR7 IL2RB TNFRSF17 IL10RA CSF2RB IL7R IL18R1 CD27 |
| B cell receptor signaling pathway | 5 | 2.50×10−4 | BTK PRKCB CD19 CD79A PIK3CG |
| JAK-STAT signaling pathway | 6 | 5.93×10−4 | STAT4 IL2RB PIK3CG IL10RA CSF2RB IL7R |
| Leukocyte transendothelial migration | 5 | 1.52×10−3 | ITGA4 PRKCB PIK3CG CYBB RASSF5 |
| Cell adhesion molecules (CAMs) | 5 | 3.41×10−3 | SELL ITGA4 HLA-DOA CD2 PTPRC |
| NF-κB signaling pathway | 4 | 6.07×10−3 | BTK PRKCB ATM LY96 |
| Transcriptional misregulation in cancer | 5 | 6.78×10−3 | MEF2C CCR7 ATM IL2RB CCNT2 |
| Intestinal immune network for IgA production | 3 | 1.08×10−2 | ITGA4 HLA-DOA TNFRSF17 |
| Toxoplasmosis | 4 | 1.08×10−2 | HLA-DOA LY96 PIK3CG IL10RA |
| Non-small-cell lung cancer | 3 | 1.29×10−2 | PRKCB PIK3CG RASSF5 |
| Inflammatory bowel disease (IBD) | 3 | 1.88×10−2 | HLA-DOA STAT4 IL18R1 |
| Oxytocin signaling pathway | 4 | 2.23×10−2 | MEF2C PRKCB CD38 PIK3CG |
| Leishmaniasis | 3 | 2.23×10−2 | ITGA4 HLA-DOA PRKCB |
| Neuroactive ligand-receptor interaction | 5 | 2.51×10−2 | P2RY14 P2RY8 P2RY13 P2RY10 CYSLTR1 |
| Influenza A | 4 | 2.82×10−2 | HLA-DOA PRKCB TLR7 PIK3CG |
| Fc gamma R-mediated phagocytosis | 3 | 3.46×10−2 | PRKCB PIK3CG PTPRC |
| AGE-RAGE signaling pathway in diabetic complications | 3 | 3.91×10−2 | PRKCB PIK3CG CYBB |
| Epstein–Barr virus infection | 4 | 3.91×10−2 | CD19 CD38 PIK3CG IL10RA |
| HIF-1 signaling pathway | 3 | 3.91×10−2 | PRKCB PIK3CG CYBB |
| Toll-like receptor signaling pathway | 3 | 4.04×10−2 | LY96 TLR7 PIK3CG |
| PI3K-AKT signaling pathway | 5 | 4.12×10−2 | ITGA4 CD19 IL2RB PIK3CG IL7R |
Note: The pathway analysis was performed in Kyoto Encyclopedia of Genes and Genomes database, and there were 23 available pathways that were significant (P<0.05).
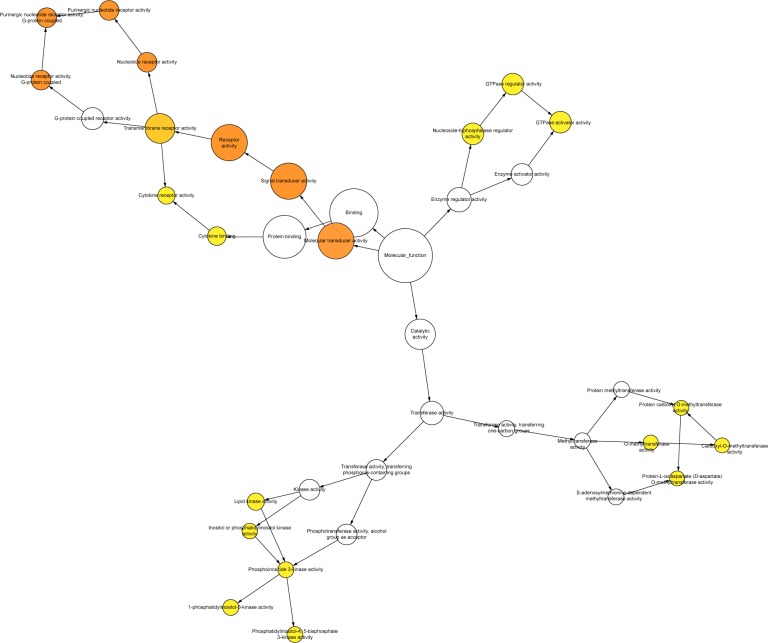
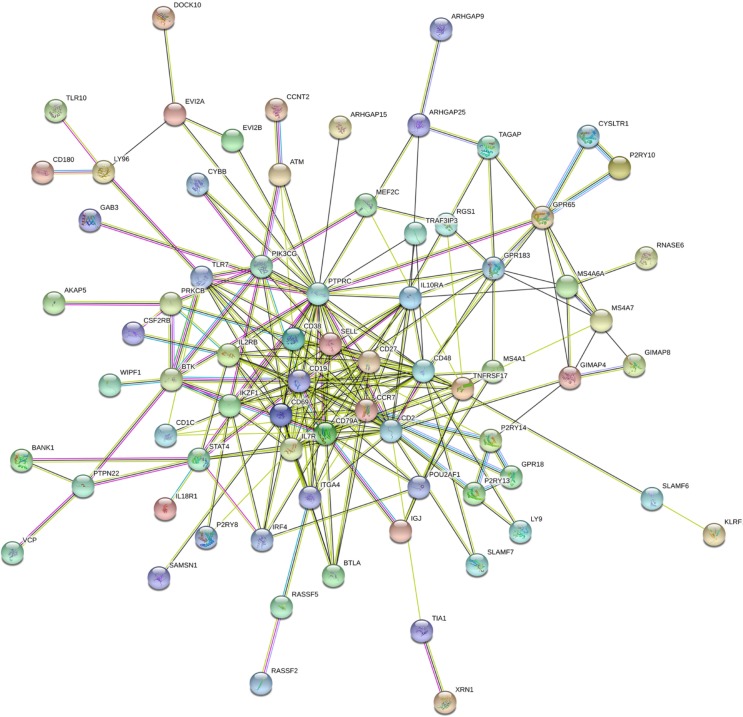
BINGO analysis of the overlapping 142 genes was conducted. The networks of BINGO analysis, including three parts (biological process, cellular component, and molecular function), are presented in Figures 4–6, respectively. Furthermore, hub genes analysis was conducted by the protein–protein interaction, and the results are presented in Figure 7. In the network, several genes demonstrated higher connectivity, including CD19, CD2, CD48, GPR183, and PTPRC.
Figure 4.
Network in BINGO analysis of the LINC00996 related genes of biological process.
Figure 5.
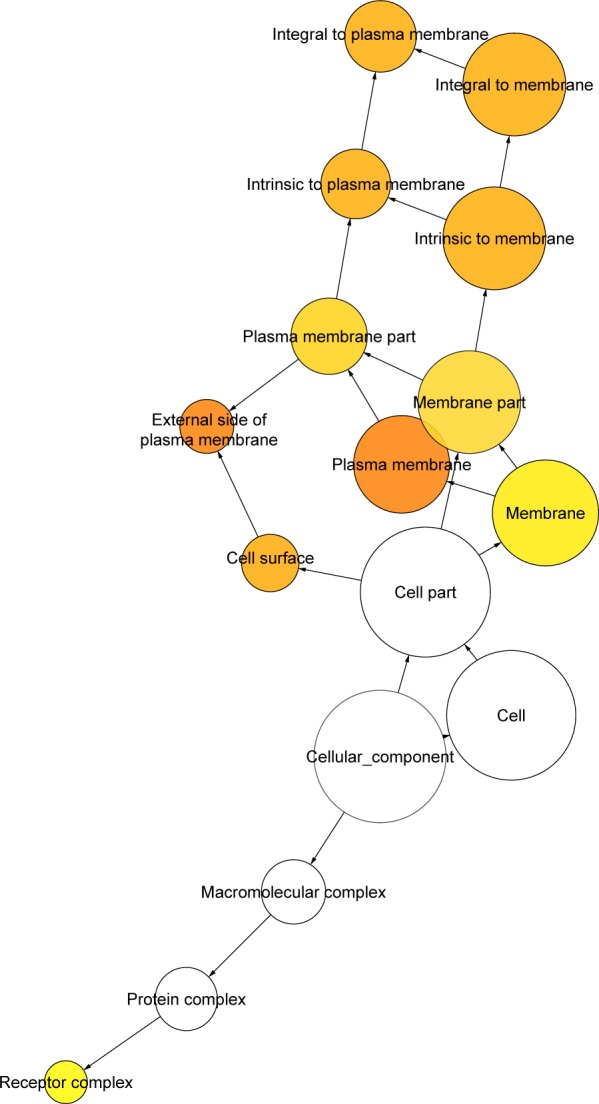
Network in BINGO analysis of the LINC00996 related genes of cellular component.
Figure 6.
Network in BINGO analysis of the LINC00996 related genes of molecular function.
Figure 7.
Protein–protein interactions of the LINC00996 related genes.
Discussion
The prognosis of CRC is fundamentally based on the clinical stage. However, some patients eventually die from cancer recurrence and metastasis following surgery, whereas other patients with a similar stage do not. This difference may be due to the different malignant potential of tumors classified in the same stage. In recent years, in order to prolong the survival time of CRC patients, numerous therapies have been applied in clinical managements, including interventional operation, radiotherapy, and combined therapy. However, the survival rate remains poor. Accumulating studies have reported that the molecular mechanisms play key roles in the carcinogenesis of CRC, and a large number of lncRNAs participate in the process.22–24 Therefore, it is of great significance to explore tumor related lncRNAs and their related signaling pathways, which will provide a sufficient theoretical basis to elucidate the potential mechanisms and to search for effective therapeutic targets for CRC patients.
In this paper, we investigated the roles of LINC00996 in CRC combined with the GEO and TCGA databases, which offer significant amounts of raw data. Our results indicated that LINC00996 is significantly decreased in CRC tissues compared with non-tumor tissues. In addition, we found that low level of LINC00996 is associated with remote metastasis. However, the loss of LINC00996 has a minimal effect on gender, lymphatic invasion, tumor size, lymph node metastasis, and pathological stage. We also discovered that patients with lower LINC00996 expression exhibited poor OS compared with patients with higher LINC00996 expression. Together, these findings revealed that the depletion of LINC00996 plays a crucial role in tumorigenesis and invasion in CRC patients.
No research on the molecule mechanism of LINC00996 in CRC has been reported. In order to explore the mechanism comprehensively, we attempted to predict the potential pathways whereby LINC00996 modulates the tumorigenesis of CRC. First, we identified 142 related genes by searching online biological databases. Next, we further performed GO and KEGG pathway analysis to identify biological functions enriched among those associated genes. We found that these genes were involved in various biological processes and molecular functions. The results of GO analysis demonstrate that LINC00996 is closely related to cellular membrane activities. In addition, pathway analysis revealed that some cell adhesion molecules may be regulated by LINC00996. The above findings suggest that the loss of LINC00996 is possibly involved in the process of tumor metastasis, which is consisted with our findings.
Interestingly, we also found NF-κB, JAK-STAT, HIF-1, TLR, and PI3K-AKT signaling pathways are significant pathways in CRC, as identified by the KEGG pathway analysis. It also has been established that they are involved in the process of tumorigenesis.25–29 Accordingly, we conjectured that LINC00996 represses the progression of CRC by regulating these pathways. In CRC, various molecular mechanisms involved in the carcinogenesis have been excavated. Aberrant activation of NF-κB signaling in CRC cell lines and tumor tissues could be involved in tumorigenesis, particularly in cell proliferation.30 Conversely, the inhibition of NF-κB induces apoptosis and suppresses epithelial–mesenchymal transition (EMT) in CRC cells.31,32 It has been reported that significantly increased phosphorylation levels of the JAK-STAT signaling pathway was observed in colorectal adenoma tissues as compared with the findings in normal colorectal tissue.33 Additionally, persistent JAK-STAT activation in colon cancer is reported to be associated with enhanced cell proliferation and tumor growth.34 In in vitro studies, suppressing JAK-STAT signaling attenuates CRC progression.35,36 Activation of HIF-1 signaling is considered as tumor-initiating factor, and it promotes EMT directly.37 Targeting HIF-1 signaling results in antiangiogenic effects in CRC.38 The TLR pathway drives cellular proliferation and interacts with PI3K-AKT and NF-κB signaling, and is associated with the upregulation of antiapoptotic genes.39 As a key pathogenetic signaling pathway of CRC, PI3K-AKT is directly involved in the carcinogenesis and progression of CRC through a variety of mechanisms.40,41
In our study, several potential limitations should be considered when interpreting the results. First, lncRNAs do not directly encode protein; they play a variety of roles via the mediation of mRNAs or microRNAs. Thus, other studies on the integrated analysis of lncRNA, mRNA, and microRNA expression profiles should be implemented to explore the comprehensive mechanism of LINC00996 in CRC. Second, our results are based on the bioinformatics analysis of GEO and TCGA data. Therefore, further experiments must be designed and performed to validate the contribution of LINC00996 in CRC.
Conclusion
Based on our mining from GEO and TCGA datasets, our findings revealed that LINC00996 expression is downregulated in CRC tissues, and the depletion of LINC00996 has an unfavorable effect on the OS of CRC patients. Additionally, LINC00996 might repress tumorigenesis and metastasis via modulating tumor related signalings, such as JAK-STAT, NF-κB, HIF-1, TLR, and PI3K-AKT signaling pathways. However, further studies are essential to improve and validate our understanding of the roles of LINC00996 in CRC.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22(4):191–197. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jemal A, Murray T, Ward E, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005;55(1):10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 4.Sporn MB. The war on cancer. Lancet. 1996;347(9012):1377–1381. doi: 10.1016/s0140-6736(96)91015-6. [DOI] [PubMed] [Google Scholar]
- 5.Beltran M, Puig I, Peña C, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22(6):756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23(8):1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer MK, Niknafs YS, Malik R, et al. The landscape of long non-coding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei S, Zhao M, Wang X, Li Y, Wang K. PU.1 controls the expression of long noncoding RNA HOTAIRM1 during granulocytic differentiation. J Hematol Oncol. 2016;9(1):44. doi: 10.1186/s13045-016-0274-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garzon R, Volinia S, Papaioannou D, et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2014;111(52):18679–18684. doi: 10.1073/pnas.1422050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song H, Sun W, Ye G, et al. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225. doi: 10.1186/1479-5876-11-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng HT, Shi DB, Wang YW, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7(6):3174–3181. [PMC free article] [PubMed] [Google Scholar]
- 12.Lai MC, Yang Z, Zhou L, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29(3):1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 13.Nie FQ, Sun M, Yang JS, et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14(1):268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 14.Lu X, Zhou C, Li R, Deng Y, Zhao L, Zhai W. Long noncoding RNA AFAP1-AS1 promoted tumor growth and invasion in cholangiocarcinoma. Cell Physiol Biochem. 2017;42(1):222–230. doi: 10.1159/000477319. [DOI] [PubMed] [Google Scholar]
- 15.Dennis G, Sherman BT, Hosack DA, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 16.Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34(Web Server issue):W720–W724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adler P, Kolde R, Kull M, et al. Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods. Genome Biol. 2009;10(12):R139. doi: 10.1186/gb-2009-10-12-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res. 2011;39(Database issue):D301–D308. doi: 10.1093/nar/gkq1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Han L, Roebuck P, et al. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015;75(18):3728–3737. doi: 10.1158/0008-5472.CAN-15-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiang JF, Yin QF, Chen T, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24(5):513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p- mediated regulation of Wnt/β-catenin signaling. Mol Cancer. 2017;16(1):9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang JZ, Xu CL, Wu H, Shen SJ. lncRNA SNHG12 promotes cell growth and inhibits cell apoptosis in colorectal cancer cells. Braz J Med Biol Res. 2017;50(3):e6079. doi: 10.1590/1414-431X20176079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karin M, Cao Y, Greten FR, Li ZW, Zw L. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2(4):301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 26.Constantinescu SN, Girardot M, Pecquet C. Mining for JAK-STAT mutations in cancer. Trends Biochem Sci. 2008;33(3):122–131. doi: 10.1016/j.tibs.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Galanis A, Pappa A, Giannakakis A, Lanitis E, Dangaj D, Sandaltzopoulos R. Reactive oxygen species and HIF-1 signalling in cancer. Cancer Lett. 2008;266(1):12–20. doi: 10.1016/j.canlet.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 28.Kelly MG, Alvero AB, Chen R, et al. TLR-4 signaling promotes tumor growth and paclitaxel chemoresistance in ovarian cancer. Cancer Res. 2006;66(7):3859–3868. doi: 10.1158/0008-5472.CAN-05-3948. [DOI] [PubMed] [Google Scholar]
- 29.Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, González-Barón M. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004;30(2):193–204. doi: 10.1016/j.ctrv.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 30.de Simone V, Franzè E, Ronchetti G, et al. Th17-type cytokines, IL-6 and TNF-α synergistically activate STAT3 and NF-kB to promote colorectal cancer cell growth. Oncogene. 2015;34(27):3493–3503. doi: 10.1038/onc.2014.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan X, Shen H, Jiang H, Hu D, Wang J, Wu X. External Qi of Yan Xin Qigong inhibits activation of Akt, Erk1/2 and NF-κB and induces cell cycle arrest and apoptosis in colorectal cancer cells. Cell Physiol Biochem. 2013;31(1):113–122. doi: 10.1159/000343354. [DOI] [PubMed] [Google Scholar]
- 32.Feng M, Feng J, Chen W, et al. Lipocalin2 suppresses metastasis of colorectal cancer by attenuating NF-κB-dependent activation of snail and epithelial mesenchymal transition. Mol Cancer. 2016;15(1):77. doi: 10.1186/s12943-016-0564-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchiyama T, Takahashi H, Endo H, et al. Role of the long form leptin receptor and of the STAT3 signaling pathway in colorectal cancer progression. Int J Oncol. 2011;39(4):935–940. doi: 10.3892/ijo.2011.1105. [DOI] [PubMed] [Google Scholar]
- 34.Corvinus FM, Orth C, Moriggl R, et al. Persistent STAT3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia. 2005;7(6):545–555. doi: 10.1593/neo.04571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Jin H, Xu R, Mei Q, Fan D. Triptolide downregulates Rac1 and the JAK/STAT3 pathway and inhibits colitis-related colon cancer progression. Exp Mol Med. 2009;41(10):717–727. doi: 10.3858/emm.2009.41.10.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong H, Du W, Zhang YJ, et al. Trichostatin A, a histone deacetylase inhibitor, suppresses JAK2/STAT3 signaling via inducing the promoter-associated histone acetylation of SOCS1 and SOCS3 in human colorectal cancer cells. Mol Carcinog. 2012;51(2):174–184. doi: 10.1002/mc.20777. [DOI] [PubMed] [Google Scholar]
- 37.Zhang W, Shi X, Peng Y, et al. HIF-1α promotes epithelial-mesenchymal transition and metastasis through direct regulation of ZEB1 in colorectal cancer. PLoS One. 2015;10(6):e0129603. doi: 10.1371/journal.pone.0129603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagaraju GP, Park W, Wen J, et al. Antiangiogenic effects of ganetespib in colorectal cancer mediated through inhibition of HIF-1α and STAT-3. Angiogenesis. 2013;16(4):903–917. doi: 10.1007/s10456-013-9364-7. [DOI] [PubMed] [Google Scholar]
- 39.Liu YD, Ji CB, Li SB, et al. Toll-like receptor 2 stimulation promotes colorectal cancer cell growth via PI3K/Akt and NF-κB signaling pathways. Int Immunopharmacol. 2018;59:375–383. doi: 10.1016/j.intimp.2018.04.033. [DOI] [PubMed] [Google Scholar]
- 40.Sun K, Wang S, He J, et al. NCOA5 promotes proliferation, migration and invasion of colorectal cancer cells via activation of PI3K/AKT pathway. Oncotarget. 2017;8(64):107932–107946. doi: 10.18632/oncotarget.22429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou R, Shao Z, Liu J, et al. COPS5 and LASP1 synergistically interact to downregulate 14-3-3σ expression and promote colorectal cancer progression via activating PI3K/AKT pathway. Int J Cancer. 2018;142(9):1853–1864. doi: 10.1002/ijc.31206. [DOI] [PubMed] [Google Scholar]