Abstract
RATIONALE:
There is continued interest in exploring new analytical technologies for the detection and quantitation of DNA adducts, biomarkers which provide direct evidence of exposure and genetic damage in cells. With the goal of reducing clean-up steps and improving sample throughput, a Differential Mobility Spectrometry/Mass Spectrometry (DMS/MS) platform has been introduced for adduct analysis.
METHODS:
A DMS/MS platform has been utilized for the analysis of dG-ABP, the deoxyguanosine adduct of the bladder carcinogen 4-aminobiphenyl (4-ABP). After optimization of the DMS parameters, each sample was analyzed in just 30 s following a simple protein precipitation step of the digested DNA.
RESULTS:
A detection limit of one modification in 10^6 nucleosides has been achieved using only 2 μg of DNA. A brief comparison (quantitative and qualitative) with liquid chromatography/mass spectrometry is also presented highlighting the advantages of using the DMS/MS method as a high-throughput platform.
CONCLUSIONS:
The data presented demonstrate the successful application of a DMS/MS/MS platform for the rapid quantitation of DNA adducts using, as a model analyte, the deoxyguanosine adduct of the bladder carcinogen 4-aminobiphenyl.
Exposure to harmful chemicals can cause these chemicals or their electrophilic metabolites to bind covalently to DNA forming DNA adducts. Failure of the cells to repair the chemical modifications can cause accumulation of these adducts leading to mutations which may have severe biological consequences. The linkage between DNA adduct formation and carcinogenicity has been suggested by several studies[1–3] and DNA adducts provide the direct evidence of exposure and genetic damage in cells. The genotoxic damage can be monitored by quantifying the level of these adducts which may also serve as a biomarker for health risk assessment.
Over the past decades, significant efforts have been made towards the analysis of DNA adducts. Different analytical methods of varying sensitivity such as nuclear magnetic resonance, circular dichroism, fluorescence spectroscopy, laser-induced fluorescence, immunoassay, electrochemical detection and capillary electrophoresis with laser-induced fluorescence detection have been employed to monitor the level of DNA adducts.[4–10] Notably, 32P-postlabeling[11] has been widely used for the detection of DNA adducts because of its high sensitivity. However, this method is criticized as being insufficiently specific and internal standards are not used to account for analyte losses during sample processing. Over the past several years, mass spectrometry based methods with the ability to provide structural information have been accepted as the platform of choice for DNA adduct analysis. Our laboratory[12,13] and those of several other investigators[14–16] have successfully applied high-performance/tandem mass spectrometry (HPLC/MS/MS) for the analysis of DNA adducts with high sensitivity and reliability.
There remains continued interest in exploring new analytical technologies for the detection and quantitation of DNA adducts, in particular with the goal of reducing clean-up steps and improving sample throughput to enable larger-scale population screening for the analysis of the public health impact of chemical exposures. Differential Mobility Spectrometry/Mass Spectrometry, which has been an area of considerable activity in our laboratory in recent years, has proved to be effective in a variety of applications, including analysis of peptides and drugs of forensic interest[17,18] and investigation of its capabilities for DNA adduct analysis is a logical extension of these efforts.
Since its inception in early 1990s,[19] Differential Mobility Spectrometry (DMS) has been used as a rapid gas-phase ion separation/filtration technique. Although DMS has been primarily used for separations, it has been demonstrated that its unique capability allows pre-filtering of ion species prior to mass analysis resulting in reduced noise and enhanced sensitivity.[20] Two different types of differential mobility ion filters are commonly used: the cylindrical FAIMS (field-asymmetric waveform ion mobility spectrometry) and the planar DMS. Both of them are continuous ion-filtering devices and utilize high-frequency asymmetric waveforms to separate ions. Schneider et al. have pointed out that FAIMS provides the potential advantage of ion focusing at atmospheric pressure because of radio-frequency (RF) fields and analyzer curvature, whereas DMS provides faster ion transit times and offers a transparent mode of operation which allows all ions to be transmitted without discrimination.[20]
It has been well established that addition of modifiers, mainly small organic molecules to the transport gas, enhances DMS separations.[17] In the presence of modifiers, the differential mobility separations are best described by the cluster-decluster model.[21] A number of different modifiers have been used, from alcohols like 2-propanol, 2-butanol, cyclopentanol, and others, which are most effective for positive ions, to halogenated compounds like 1,2,3-trichloropropane and methylene chloride for negative ions.[22] In a recent publication, Hall et al. have reported on the use of ethyl acetate as a modifier in forensic applications.[18] Alternating RF voltages, known as the Separation Voltage (SV), are applied across the planar DMS electrodes. There are two types of fields applied in a differential mobility filter: high field and low field. It is believed that during the low-field portion of the asymmetric wave, analyte ions form clusters with the neutral gas modifiers and, during the high-field portion, the ionneutral complex dissociates changing the effective crosssectional area for ion transmission. This change in the effective cross-sectional area causes differences in flight times of ions leading to improved separations. A counterbalancing DC voltage known as the compensation voltage (CV) is applied to transmit the analyte of interest.
CV scans at varying SVs with different potential modifiers introduced at various rates in the transport gas are normally performed in order to identify the optimal parameters for DMS ion transmission. Once the peak values are identified and set as the DMS parameters, ions of interest can be separated/filtered through the DMS cell in milliseconds which makes this method very suitable for coupling to mass spectrometers. In practice, DMS provides an extra dimension of orthogonal separation/filtration without adding to the overall analysis time. DMS is especially useful in combination with mass spectrometry because it is less correlated with mass[23] than ion mobility (IMS).[24,25]
With the stated aim to improve sample throughput by eliminating the time-consuming chromatographic analysis step and possibly by-passing, or at least reducing, sample clean-up steps, we report here on the application of DMS-MS/MS toward the analysis of DNA adducts. N-(2-Deoxyguanosine-8-yl)-4-ABP (dG-ABP), the deoxyguanosine adduct of the bladder carcinogen 4-aminobiphenyl (4-ABP), found in cigarette smoke, paints, food colors, hair dyes and fumes from heated oils and fuels, is used as a model compound in the present study.[26–29] A comparison of the DMS/MS approach with the results obtained using our current LC/MS/MS protocol is presented.
EXPERIMENTAL
Chemicals and standards
Caution: 4-nitrobiphenyl and its derivatives are carcinogenic and should be handled using the appropriate personal protective equipment.
Calf-thymus DNA, nuclease p1 from Penicillium citrinium, deoxyribonuclease 1 (DNase I) type 2 from bovine pancreas, alkaline phosphatase from Escherichia coli (type IIIs), ethanol, and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). Snake venom phosphodiesterase was purchased from USB Corporation (Cleveland, OH, USA). N-(2-Deoxyguanosine-8-yl)-4-ABP (dG-ABP) was purchased from Toronto Research Chemicals (Toronto, ON, Canada). dG-ABP-d9 was previously synthesized and characterized in our laboratory[30] and introduced simultaneously with the dG-ABP analyte at the same fixed compensation voltage. Formic acid solution was purchased from Sigma-Aldrich Chemical Co.. HPLC grade water and methanol were purchased from Fisher Scientific (Fair Lawn, NJ, USA).
Modification of calf-thymus DNA with 4-nitrobiphenyl
The synthesis of ABP-modified calf thymus DNA was previously reported.[30] Briefly, 200 mg of 4-nitrobiphenyl (Toronto Research Chemicals (Toronto, ON, Canada)) was reduced to N-hydroxy-4-aminobiphenyl in the presence of 50 mg 5% Pd/C at 0°C. Next, 3.8 μg of N-hydroxy-4-aminobiphenyl dissolved in argon-purged ethanol was reacted overnight with mild agitation at room temperature with 3.6 mg of calf thymus DNA dissolved in 5.0 mL of 10 mM sodium citrate buffer, pH 5. The aqueous DNA reaction mixture was extracted three times with 5.5 mL distilled phenol (saturated with TE buffer: 40 mL phenol + 10 mL TE buffer) followed by extraction with 5.0 mL of n-butanol three times. The DNA was then precipitated with five volumes of ice-cold ethanol and air-dried. The DNA was redissolved in TE buffer to give approximately1 mg/mL concentration.
DNA quantitation, enzymatic digestion and protein precipitation
DNA quantitation was performed using a Quant-IT™ doublestranded (ds) DNA BR Assay kit with a Qubit fluorometer (Invitrogen Corp., Carlsbad, CA, USA). Aliquots containing 2 μg DNA (dissolved in 5mM Tris-Cl/10 mM ZnCl2) were removed for digestion and analysis for each sample point.
Calf thymus DNA was hydrolyzed according to a method previously described.[31] Samples were incubated at 98°C for 3–5 min and chilled in the freezer down to room temperature. Then 0.3 units of nuclease P1 (0.3 units μL−1 solution of 5 mM Tris-Cl, pH 7.4) and 3.1 KU of DNase I (1 μg μL−1 solution in 5 mM Tris/10 mM MgCl2, pH 7.4) were added per μg of DNA and incubated in a water bath maintained at 37°C. After 5 h, 0.003 units of phosphodiesterase (100 ng μL−1 in 5 mM Tris/10 mM MgCl2, pH 7.4), and 0.002 units of alkaline phosphatase per μg of DNA were added and the mixture was further incubated at 37°C for 18 h. Protein precipitation was carried out by adding five volumes of ice-cold ethanol and centrifuging at 10 000 rpm for 15 min. The samples were dried down and stored at —80°C until analysis.
Instrumentation
The fundamental goals of the proposed approach to the analysis of DNA adducts by DMS/MS was tested using two different planar DMS systems, one attached to a 3D ion trap (LCQ Classic; Thermo Finnigan, San Jose, CA, USA) and the second interfaced to an API 3000 triple quadrupole mass spectrometer (AB SCIEX, Framingham, MA, USA). The DMS filters varied only in terms of their dimensions and this comparison provided an assessment of the general effect and applicability of the DMS configuration to such analyses.
DMS/ion trap mass spectrometer
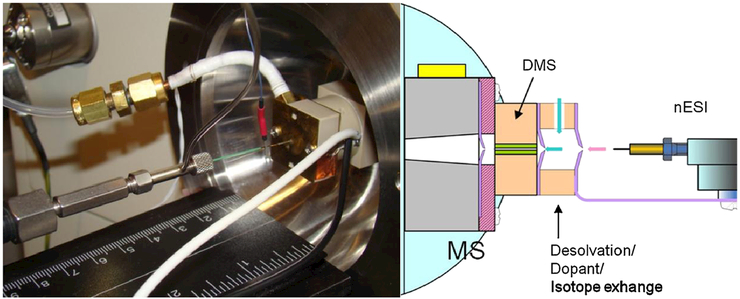
A planar DMS system developed by Sionex Corporation (now defunct) with a filter gap 0.5 mm high × 3.0 mm wide 10.0 mm long was used for the work (Fig. 1) which sat on the heated capillary of an LCQ Classic mass spectrometer (Thermo-Finnigan). Sionex Expert software was used to set the DMS parameters. The SV could be set at zero or set or scanned in the range from 500 to 1500 V and the CV could be set or scanned from —43 to +15 V. Although the system in use here is no longer commercially available, the underlying electronics technology has been described,[32] and successor commercial instrumentation (SelexION, AB SCIEX) has become commercially available.
Figure 1.
Left: Actual DMS system sitting on the heated capillary of the ion trap mass spectrometer. Right: graphical representation of the DMS system.
Electrospray ionization (ESI) was performed using coated a 10 μm PicoTip emitter from New Objective (Woburn, MA, USA). The syringe was connected to the emitter tip using a 150 μm (i.d.) capillary tubing and the ESI voltage was applied to the union at the liquid-liquid junction between the capillary and the tip. Samples were introduced at a flow rate of 300 nL/min using a syringe pump from Harvard Apparatus (Holliston, MA, USA). The desolvation gas (ultra-high purity nitrogen) was introduced at a flow rate of approximately 100 cc/min into the desolvation region at a temperature of 100°C. The vacuum drag of the mass spectrometer was measured to be 600 cc/min. External air flow of approximately 500 cc/min also merged in with the desolvation gas into the DMS system. Ethyl acetate modifier was introduced into the desolvation region along with the nitrogen gas at a final concentration of approximately 1.5% v/v, calibrated by a weight loss measurement. The electrospray was held at 2 kV throughout the analysis.
DMS/triple quadrupole mass spectrometer
A prototype DMS API 3000 triple quadrupole mass spectrometer (AB SCIEX, Concord, Ontario. Canada) which has the removable DMS filter online in front of the first quadrupole was used for this study. The dimensions of the DMS were 1 mm × 10 mm × 15 mm. A modified version of the software Analyst version 1.5 which included parameters for SV and CV was used. The SV could be varied from 0 to 5000 V and the CV could be varied from −100 to +100 V. Electrospray was performed using a stainless steel 30 μmi.d. emitter from Proxeon (Odense, Denmark). Samples were introduced at a flow rate of 400 nL/min using a syringe pump from Harvard Apparatus. Electrospray was held constant at 3500 V throughout the analysis. Modifiers were introduced into the curtain gas (nitrogen, 1.1 L/min, 600 cc/min vacuum drag, 500 cc/min curtain gas outflow) using a second syringe pump from Harvard Apparatus for introduction of the modifier at the rate of 0.6% v/v.
RESULTS AND DISCUSSION
The first step in the protocol of DNA adduct analysis involves digestion of the DNA to mononucleosides for the liberation of DNA adducts. Once the digestion is complete, the sample is subjected to a protein-precipitation step and the supernatant is dried down. As shown in the left hand portion of the flow chart in Fig. 2, after the protein precipitation, solid-phase extraction (SPE) is required before LC/MS analysis in order to remove residual matrix components. This is typically accomplished by trapping of the analyte in the stationary phase and eluting it with high percentage of suitable organic solvent. SPE of DNA adducts is challenging because of the potential for analyte losses during the cleanup, given the relative proportions of typical adduct levels (pg) to the smallest available SPE bed masses (mg). It has also been pointed out that the polypropylene frit material from the SPE cartridges can leach out and cause ion suppression problems during the analysis.[33] These factors alone suggested the consideration of DMS/MS as an alternative option that may at least reduce some of the limitations imposed by the traditional LC/MS protocol.
Figure 2.
Comparison of the workflow for the analysis of DNA adducts by LC/MS and DMS/MS.
Detection of dG-ABP in calf thymus DNA using DMS/ion trap MS
While LC/MS has proved to be highly effective in meeting many of the requirements (e.g., sensitivity, sample quantity) for the analysis of DNA adducts, it is clear from the above discussion that this protocol is time-consuming and can create a bottleneck when analysis of multiple samples is involved. Given our previously demonstrated success of the utilization of DMS/MS for the rapid analysis of forensic samples,[20,34] we set as a goal the reduction of the overall analysis time by eliminating at least some of the steps associated with the LC/MS protocol. In accordance with this objective we sought to determine whether dG-ABP could be detected by DMS/MS in a DNA digest immediately after protein precipitation as indicated in the right hand portion of Fig. 2. To establish the feasibility of this goal, the initial investigation was conducted using a DMS filter interfaced to an LCQ Classic ion trap. For this purpose, 100 pg of dGABP was spiked into 2 μg of a calf thymus DNA digest and the sample was reconstituted to a total volume of 100 μL (70% MeOH/30% H2O/0.1% CH2O2 v/v/v) which was best accommodated by the volume of the connecting tubings to facilitate the electrospray and infused at a flow rate of 300 nL/min. Data were acquired under two different conditions: (i) DMS-transparent and (ii) DMS-on, ethyl acetate modifier. In DMS-transparent mode, both SV and CV are set to zero so that no DMS filtration occurs. For the DMS-on condition, the separation voltage (SV) was set at 1500 V and the compensation voltage (CV) was scanned from −43 V to +15 V. After the CV scan had been performed, the precursor ion mass (m/z 435) for the analyte dG-ABP was extracted from the total ion chromatogram. The CV corresponding to the apex of the extracted ion chromatogram was established as the CV for selective dG-ABP transmission in this system and introduction into the ion trap, thereby preventing unwanted matrix ions from getting into the trap. DMS-transparent mode transmits ions non-selectively, with residual losses due to the presence of the interface which were measured by Schneider et al.[18] to be on the order of 25% for a curtain-gas inlet.
In Fig. 3(A), it can be seen that when the DMS system is in transparent mode, i.e., no electric field applied, and without the use of modifier, the signal for the analyte of interest, dG-ABP (m/z 435), is obscured by background ions. Some of the largest background ion peaks are much broader than expected for small molecular ions and so may be due to multiply charged residual protein ion contamination. Heavy ions of this type are effectively separated by DMS with modifiers due to the large size differential between the protein or fragment and the modifier, and because positive compensation voltage values are characteristic of very large molecules.
Figure 3.
Full scan mass spectra of 100 pg dG-ABP in 2 μg calf thymus DNA recorded on the ion trap under two different conditions: (A) DMS-transparent (i.e., DMS field not applied), no modifier and (B) DMS-on (separation and compensation voltage fixed), ethyl acetate modifier.
When the DMS is turned on with ethyl acetate modifier introduced via the drift gas, the dG-ABP adduct can be selectively introduced into the trap with a significant reduction in background chemical noise (Fig. 3(B)). The reduction of these species can be attributed to the change in DMS CV caused by introduction of the modifier into the drift gas consistent with phenomena previously observed.[35] In addition, the increase in signal intensity of dG-ABP from 3.25 × 103 to 1.93 × 104 along with the indicated noise reduction are significant and demonstrate the usefulness of DMS to remove matrix ion interferences and improve sensitivity by selectively filling the trap. In the DMS-transparent mode there is premature saturation of the trap due to the presence of a multitude of matrix ions. This results in loss of resolution and peak broadening. However, selective DM filtration provides optimal conditions for filling the trap with the precursor ion of dG-ABP and retention of mass resolution.[36] The dG-ABP adduct detected in these preliminary experiments corresponded to approximately 4 modifications in 105 nucleosides based on the use of 2 μg DNA for the analysis.
Quantitation of dG-ABP in calf thymus DNA by DMS/triple quadrupole MS
Triple quadrupole mass spectrometers, which can operate in the multiple reaction monitoring (MRM) mode, are widely used for quantitation purposes. In the case of complex matrices, ion suppression during the ESI process and possibility of interferences necessitate the use of a clean-up before MS analysis. In the present application addressed here, DMS functions as a post-electrospray ion separation/filtration technique, analogous to that of an HPLC method. Given the successful demonstration of the capability of the DMS to filter out the matrix interferences when combined with an ion trap, we examined next the use of a DMS/triple quadrupole platform to explore the possibilities of quantitation of DNA adducts in a matrix of digested DNA after a simple protein-precipitation step. Samples were introduced into the DMS/MS/MS platform under nanoelectrospray conditions.
Prior to sample analysis, the instrument parameters for the DMS/MS were optimized by infusing 25 pg/μL dG-ABP in 70% MeOH/30% H2O/0.1% CH2O2 v/v/v. The instrument was operated in the MRM mode and the transition from the precursor ion [M + H]+ of dG-ABP (to the product ion [M + H-116]+ (m/z 435 → 319) was monitored at unit resolution in both Q1 and Q3. The electrospray was set at 3500 V. Declustering potential (DP) was set at 53 V, focusing potential (FP) at 190 V, entrance potential (EP) at 10 V, focusing lens 1 (IQ1) at −10.5 V, prefilter (ST) at −18.7 V, collision energy (CE) at 19 V and collision cell exit potential (CXP) at 18.8 V, respectively.
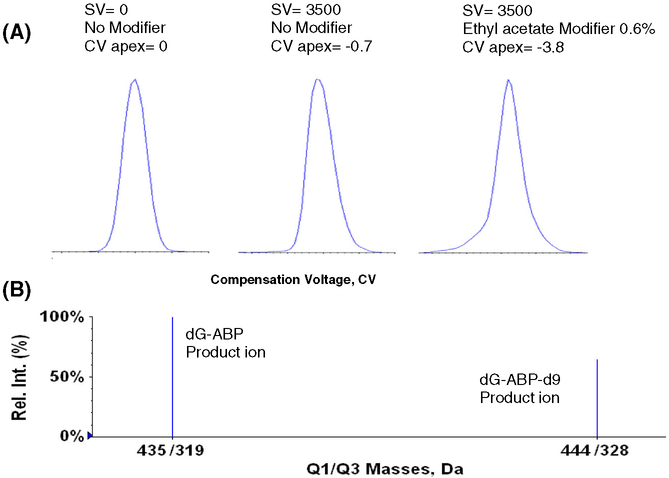
For the DMS analysis of dG-ABP adducts, two modifiers:(i) isopropanol and (ii) ethyl acetate, were screened at different modifier concentrations and at different separation voltages (SV). For the final analysis, ethyl acetate was established as the modifier of choice and was introduced at 0.6% of the curtain gas (v/v). With the separation voltage (SV) set at 3500 V, the compensation voltage (CV) was scanned from −30 V to +10 V. As shown in Fig. 4, in the presence of 0.6% ethyl acetate modifier the precursor ion of dG-ABP was shifted from a CV = 0 to a value of CV = −3.8 V and this value was used for all subsequent analyses. Figure 3 indicated that this also corresponds to a suppression of matrix ions.
Figure 4.
(A) Normalized extracted ion profile showing CV shifts for dG-ABP with and without ethyl acetate modifier at SV values of 0 and 3500 V and (B) MRM transitions monitored for dG-ABP and internal standard.
Calibration curve of dG-ABP
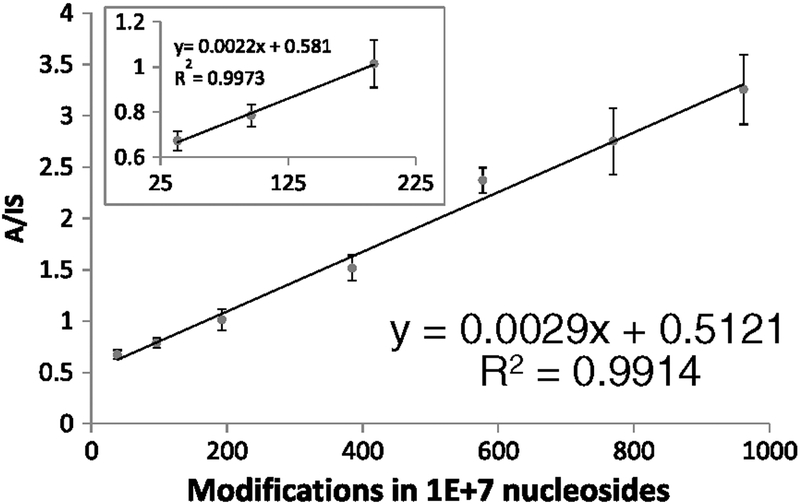
The calibration curve for dG-ABP was prepared in matrix-matched calf thymus DNA. Two micrograms of DNA was used for each point. Isotopically labeled dG-ABP-d9 was used as the internal standard. For the internal standard, the transition from the precursor ion [M + H]+ of dG-ABP-d9 to the product ion (m/z 444 → 328) [M + H-116]+ at unit resolution was monitored. A seven-point calibration curve across a concentration range [10–250 pg] was generated in triplicate and is presented in Fig. 5.
Figure 5.
Calibration curve of dG-ABP in calf thymus DNA. The error bars represent standard deviations of triplicate analyses and the inset a fit to the lowest part of the data.
The data was processed using Microsoft Excel. The Y-axis represents the ratio of the product ion intensities of dG-ABP and dG-ABP-d9 and the X-axis represents the DNA modifications present in the form of dG-ABP per 107 nucleosides analyzed. A linear regression analysis was performed in Excel for the seven points used to get a R2 value of 0.9914. To verify the linearity at the lower portion of the calibration curve, a linear regression analysis was performed on the first three points (Fig. 5, inset) which gave a R2 value of 0.9973, very much in line with that of the complete curve. The limit of detection (LOD) for the analysis was determined to be 10 dG-ABP adducts in 107 nucleosides with the limit of quantification (LOQ) represented by the lowest point in the calibration curve that was determined to be 38 modifications in 107 nucleosides.
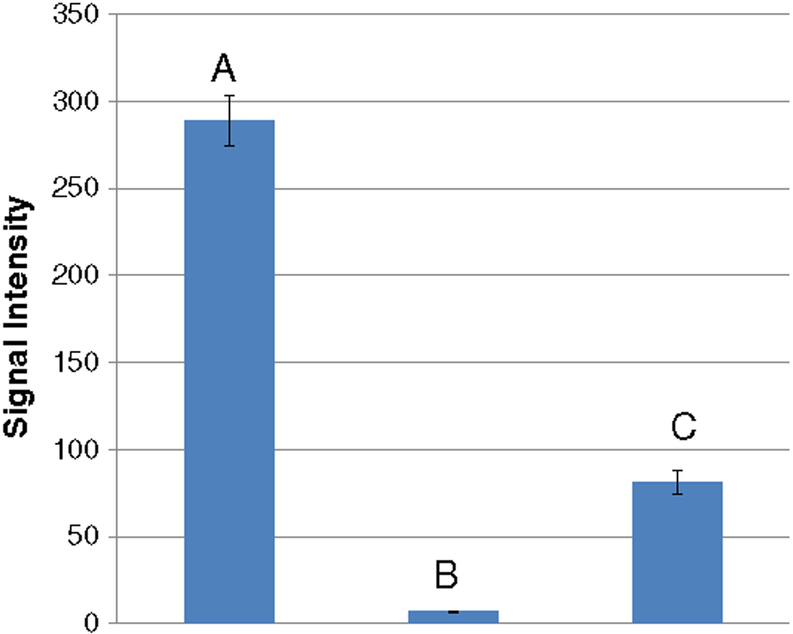
To assess the ability of DMS to selectively filter background contaminants, a blank sample of the calf thymus DNA digest was examined in the DMS-transparent mode and, as shown in bar A of Fig. 6, the matrix components produced a strong signal coincident with that of the MRM transition of the adduct. The bulk of this matrix interference was subsequently removed by turning on the DMS filter (bar B, Fig. 6), which reduced the background ion intensity by almost 20-fold. For comparison, the signal for the LOD is shown in Fig. 6 (bar C). This signal corresponds to a mass detection limit of 5.8 femtomoles based on the time over which the data were acquired during the infusion of the specific sample, corresponding to the analysis of 2 μg of DNA.
Figure 6.
Removal of interference using DMS. The signal from blank matrix CT (calf-thymus) DNA was removed in DMS-on mode to produce a linear calibration curve using ethyl acetate as the modifier: signals from (A) blank calf thymus DNA in the DMS-transparent mode, (B) blank calf thymus DNA in DMS-on mode, (C) 5.8 femtomoles of the analyte in 2 μg calf thymus DNA in DMS-on mode.
It should be noted that an attempt to generate a calibration curve for dG-ABP in the DMS-transparent mode using the same sample was unsuccessful. While MRM signals could be seen for analyte and the internal standard, the ratios were skewed throughout the seven points indicative of matrix ion interferences. Linear regression analysis performed in MS Excel yielded a R2 value of 0.6437 (graph not shown). Clearly, the ability of the DMS to remove the interferences and produce a linear calibration curve for an analyte which is present at the ppm level in a complex matrix is significant and this opens the possibility of using DMS/MS/MS as a high-throughput quantitative platform for adduct quantitation.
To further demonstrate the accuracy and precision of the calibration curve obtained in the DMS-on mode, two quality control points were evaluated in triplicate over a period of 2 days (Table 1). The values were calculated from the calibration curve based on the observed analyte to internal standard ratio.
Table 1.
Evaluation of quality control points for dG-ABP calibration curve in the DMS-on mode
| Modifications in 107nudeosides | Average Analyte/Int. Std | Average precision % RSD | Average % accuracy |
|---|---|---|---|
| 58 | 0.67 | 5.8 | 98.5 |
| 346 | 1.62 | 1.6 | 106.5 |
Quantitation of dG-ABP in modified CT-DNA: Comparison of DMS/MS/MS and LC/MS/MS platforms
Over the past several years LC/MS/MS has established itself as the premier method for the analysis of DNA adducts. It was thus necessary to compare the DMS/MS quantitation approach with existing LC/MS methodology. For the analysis of dG-ABP, a validated method employing a small molecule microfluidic chip interfaced with an Agilent 6330 ion trap mass spectrometer has been recently published from our group[13] and used to assess the significance of the presence of this adduct in several chemoprevention studies.[37,38]
For this comparison, calf thymus DNA was reacted with NOH-4-ABP as described in the Experimental section. Internal standard was added to a 2 μg portion to account for any losses during sample preparation and the DNA was digested to the mononucleoside level. After protein precipitation, the supernatant was dried down and stored at −80°C until analysis. Just prior to analysis, the sample was reconstituted in 100 μL of 70% Me0H/30% H2O/0.1% CH2O2 v/v/v and vortexed for 1 min. The sample was introduced into the DMS/MS platform at the rate of 400 nL/min by infusion. For the analysis, the MS parameters set were the same as the ones used for the preparation of calibration curve. The product ions for dG-ABP and dG-ABP-d9 were monitored in the MRM mode and the adduct level was calculated based on the ratio observed.
Because preliminary analysis of the sample suggested the presence of a significant level of DNA adduction, three samples were prepared by diluting the modified DNA with a blank ct-DNA digest. Each sample was then analyzed in triplicate by both DMS/MS/MS and the previously validated LC/MS/MS method by two different individuals. The results are presented in Table 2 and show excellent quantitative agreement between the two platforms when data are compared at the same linear segments of their respective calibration curves. This is evidenced by the numbers shown for Samples 1 and 2 which are within 10% of each other. For Sample 3, the variation (176 ± 8 vs. 137 ± 13 per 107 normal nucleosides) may be attributed to the fact that the dG-ABP adduction level computed by LC/MS is based on the extrapolation of the calibration curve; the analyte to internal standard ratio observed for this point was well above the highest point in the LC/MS curve. Most importantly, however, the modifications computed by the two different methods are in the same order of magnitude and are reasonably close to each other. Given the nature of the complexity of the samples, the observed difference in adduct levels may also be a result of sample preparation steps.
Table 2.
Quantitation of dG-ABP in calf thymus DNA (# adducts in 107 normal nucleosides) by DMS/MS and LC/MS
| Sample | DMS/MS/MS | LC/MS/MS |
|---|---|---|
| 1 | 55±4 | 51±3 |
| 2 | 94±4 | 99±9 |
| 3 | 176±8 | 137±13 |
CONCLUSIONS
The data presented demonstrate the successful application of a DMS/MS/MS platform for the quantitation of DNA adducts using as a model analyte the deoxyguanosine adduct of the bladder carcinogen 4-aminobiphenyl. In the field of quantitative bioanalytical chemistry, the best way to evaluate a new system or procedure is to directly compare it with an existing validated method. Significantly, in this preliminary investigation there is strong coincidence of the results obtained by DMS/MS/MS with those produced in the analysis of the same samples using a validated LC/MS/MS protocol.
In comparing the results obtained by each method, it is acknowledged that our current microchip-based LC/MS/MS methodology involves a highly sensitive method with a LOQ of 2 adducts per 108 normal nucleosides, i.e. a 100-fold higher sensitivity. This comparison, however, should also be considered in the context of the information given in Fig. 2 which outlines the entire sample preparation schemes for the respective analyses. As shown in Fig. 2, after protein precipitation, the sample is lyophilized, purified by SPE, lyophilized for the second time and reconstituted in an appropriate solvent for LC/MS analysis. SPE and the second lyophilization procedures can consume well over an hour while the chromatography can take up to 20 min which does not include LC re-equilibration that can take an additional 3–5 min.
In contrast to the time-consuming LC/MS/MS protocol, in the current work, the lyophilized samples (after protein precipitation) were reconstituted and directly analyzed by DMS/MS. In DMS-on mode, the analyte of interest can be selectively introduced into the mass spectrometer by setting the CV to a unique voltage for that particular analyte which has allowed us to bypass the LC step completely. Once the parameters for DMS/MS analysis were set, data were acquired in just 30 s and, since DMS is a continuous method, subsequent samples can be introduced without the need for re-equilibration. It should be noted that the calibration curve using the DMS/MS/MS platform was completed in approximately 3 h as opposed to the nearly 24 h needed for the construction of the calibration curve by LC/MS/MS. The time savings presented by the use of DMS is significant and offers great potential for the improvement of sample throughput for the analysis of DNA adducts.
Having established our initial goal of demonstrating the feasibility of rapid analysis of DNA adducts from a biological medium by DMS/MS/MS, we look forward to investigating the further optimization of this approach to bring the sensitivity of the analysis to levels comparable to those achieved by LC/MS/MS. It should be noted that in the current work utilizing the DMS/MS platform, we processed only 2 μg of DNA per analysis and, in terms of absolute sensitivity, we only utilized the equivalent of 4 ng of DNA during the 30 s period of data acquisition. These detection and quantitation levels are already within the range of modifications encountered in routine in vitro applications. Future efforts will focus on addressing ion suppression effects, likely introduced by contaminants or other impurities still present in the sample after the protein precipitation. Incorporation of an online sample enrichment/clean-up step prior to electrospray should help reduce the sample complexities encountered in DNA adduct analysis and it should also improve the sensitivity of the DMS/MS/MS platform to detect lower levels of modification. The added benefits of the DMS method can be further realized with more sensitive mass spectrometers such as the commercially available AB Sciex SelexION integrated on a 5500/6500 triple quadrupole. With improvements in sensitivity, DMS/MS/MS holds great potential for establishing itself as the high-throughput analytical platform for studies in the field of DNA adducts.
Acknowledgements
This work was supported by a grant RO1-CA69390 from NIH. SLC acknowledges support through NIH R01 AI101798 (A.J. Fornace Jr. PI). We would also like to thank Dr. Erkin Nazarov for his assistance with DMS-related issues.
REFERENCES
- [1].Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat. Rev. Cancer 2003, 3, 733. [DOI] [PubMed] [Google Scholar]
- [2].Hemminki K. DNA adducts, mutations and cancer. Carcinogenesis 1993, 14, 2007. [DOI] [PubMed] [Google Scholar]
- [3].Loechler EL. The role of adduct site-specific mutagenesis in understanding how carcinogen-DNA adducts cause mutations: perspective, prospects and problems. Carcinogenesis 1996,17, 895. [DOI] [PubMed] [Google Scholar]
- [4].Pradhan P, Jernstrom B, Seidel A, Norden B, Graslund A. Induced circular dichroism of benzo[a]pyrene-7,8-dihydrodiol 9,10-epoxide stereoisomers covalently bound to deoxyribooligonucleotides used to probe equilibrium distribution between groove binding and intercalative adduct conformations. Biochemistry 1998,37,4664. [DOI] [PubMed] [Google Scholar]
- [5].Vahakangas K, Trivers G, Rowe M, Harris CC. Benzo(a) pyrene diolepoxide-DNA adducts detected by synchronous fluorescence spectrophotometry. Environ. Health Perspect 1985, 62, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Poirier MC. Antisera specific for carcinogen-DNA adducts and carcinogen-modified DNA: applications for detection of xenobiotics in biological samples. Mutat. Res 1993, 288, 31. [DOI] [PubMed] [Google Scholar]
- [7].Wang P, Giese RW. Phosphate-specific fluorescence labeling with BO-IMI: reaction details. J. Chromatogr. A 1998, 809, 211. [DOI] [PubMed] [Google Scholar]
- [8].Bogdanov MB, Beal MF, McCabe DR, Griffin RM, Matson WR. A carbon column-based liquid chromatography electrochemical approach to routine 8-hydroxy-2’-deoxyguanosine measurements in urine and other biologic matrices: a one-year evaluation of methods. Free Radical Biol. Med 1999,27, 647. [DOI] [PubMed] [Google Scholar]
- [9].Rindgen D, Nakajima M, Wehrli S, Xu K, Blair IA. Covalent modifications to 2’-deoxyguanosine by 4-oxo-2-nonenal, a novel product of lipid peroxidation. Chem. Res. Toxicol 1999,12,1195. [DOI] [PubMed] [Google Scholar]
- [10].Tan WG, Carnelley TJ, Murphy P, Wang H, Lee J, Barker S, Weinfeld M, Le XC. Detection of DNA adducts of benzo[a]pyrene using immunoelectrophoresis with laser-induced fluorescence. J. Chromatogr. A 2001, 924, 377. [DOI] [PubMed] [Google Scholar]
- [11].Gupta RC, Reddy MV, Randerath K. 32P-postlabeling analysis of non-radioactive aromatic carcinogen-DNA adducts. Carcinogenesis 1982, 3, 1081. [DOI] [PubMed] [Google Scholar]
- [12].Glick J, Xiong W, Lin Y, Noronha AM, Wilds CJ, Vouros P. The influence of cytosine methylation on the chemoselectivity of benzo[a]pyrene diol epoxideoligonucleotide adducts determined using nanoLC/MS/MS. J. Mass Spectrom 2009, 44,1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Randall KL, Argoti D, Paonessa JD, Ding Y, Oaks Z, Zhang Y, Vouros P. An improved liquid chromatography-tandem mass spectrometry method for the quantification of 4-aminobiphenyl DNA adducts in urinary bladder cells and tissues. J. Chromatogr. A 2010,1217,4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Farmer PB, Singh R. Use of DNA adducts to identify human health risk from exposure to hazardous environmental pollutants: the increasing role of mass spectrometry in assessing biologically effective doses of genotoxic carcinogens. Mutat. Res 2008, 659, 68. [DOI] [PubMed] [Google Scholar]
- [15].Nauwelaers G, Bessette EE, Gu D, Tang Y, Rageul J, Fessard V, Yuan JM, Yu MC, Langouet S, Turesky RJ. DNA adduct formation of 4-aminobiphenyl and heterocyclic aromatic amines in human hepatocytes. Chem. Res. Toxicol 2011,24, 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Singh R, Farmer PB. Liquid chromatography-electrospray ionization-mass spectrometry: the future of DNA adduct detection. Carcinogenesis 2006, 27, 178. [DOI] [PubMed] [Google Scholar]
- [17].Levin DS, Miller RA, Nazarov EG, Vouros P. Rapid separation and quantitative analysis of peptides using a new nanoelectrospray-differential mobility spectrometer-mass spectrometer system. Anal. Chem 2006, 78, 5443. [DOI] [PubMed] [Google Scholar]
- [18].Hall AB, Coy SL, Nazarov EG, Vouros P. Rapid separation and characterization of cocaine and cocaine cutting agents by differential mobility spectrometry-mass spectrometry. J. Forensic Sci 2012, 57, 750. [DOI] [PubMed] [Google Scholar]
- [19].Buryakov IA, Krylov EV, Nazarov EG, Rasulev UK. A new method of separation of multi-atomic ions by mobility at atmospheric pressure using a high-frequency amplitude-asymmetric strong electric field. Int. J. Mass Spectrom. Ion Processes 1993, 128,143. [Google Scholar]
- [20].Schneider B, Covey T, Coy S, Krylov E, Nazarov E. Planar differential mobility spectrometer as a pre-filter for atmospheric pressure ionization mass spectrometry. Int. J. Mass Spectrom 2010, 298, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Krylov EV, Nazarov EG. Electric field dependence of the ion mobility. Int. J. Mass Spectrom 2009, 285, 149. [Google Scholar]
- [22].Coy SL, Krylov EV, Schneider BB, Covey TR,Brenner DJ, Tyburski JB, Patterson AD, Krausz KW, Fornace AJ, Nazarov EG. Detection of radiation-exposure biomarkers by differential mobility prefiltered mass spectrometry (DMS-MS). Int. J. Mass Spectrom 2010,291,108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shvartsburg AA, Tang K, Smith RD. Two-Dimensional Ion Mobility Analyses of Proteins and Peptides. Humana Press, New York, 2009, pp. 417–445. [DOI] [PubMed] [Google Scholar]
- [24].Dwivedi P, Wu P, Klopsch SJ, Puzon GJ, Xun L, Hill HH. Metabolic profiling by ion mobility mass spectrometry (IMMS). Metabolomics 2008, 4, 63. [Google Scholar]
- [25].McLean JA. The mass-mobility correlation redux: The conformational landscape of anhydrous biomolecules. J. Am. Soc. Mass Spectrom 2009, 20,1775. [DOI] [PubMed] [Google Scholar]
- [26].Garrigos MC, Reche F, Pernias K, Sanchez A, Jimenez A. Determination of some aromatic amines in finger-paints for children’s use by supercritical fluid extraction combined with gas chromatography. J. Chromatogr. A 1998, 819, 259. [DOI] [PubMed] [Google Scholar]
- [27].Oh SW, Kang MN, Cho CW, Lee MW. Detection of carcinogenic amines from dyestuffs or dyed substrates. Dyes and Pigments 1997, 33, 119. [Google Scholar]
- [28].Tokiwa H, Nakagawa R, Horikawa K. Mutagenic/carcinogenic agents in indoor pollutants; the dinitropyrenes generated by kerosene heaters and fuel gas and liquefied petroleum gas burners. Mutat. Res 1985,157, 39. [DOI] [PubMed] [Google Scholar]
- [29].Turesky RJ, Freeman JP, Holland RD, Nestorick DM,Miller DW, Ratnasinghe DL, Kadlubar FF. Identification of aminobiphenyl derivatives in commercial hair dyes. Chem. Res. Toxicol 2003,16,1162. [DOI] [PubMed] [Google Scholar]
- [30].Ricicki EM, Soglia JR, Teitel C, Kane R, Kadlubar F, Vouros P. Detection and quantification of N-(deoxyguanosin-8-yl)-4-aminobiphenyl adducts in human pancreas tissue using capillary liquid chromatography-microelectrospray mass spectrometry. Chem. Res. Toxicol 2005,18, 692. [DOI] [PubMed] [Google Scholar]
- [31].Gangl ET, Turesky RJ, Vouros P. Determination of in vitro- and in vivo-formed DNA adducts of 2-amino-3-methylimidazo[4,5-f]quinoline by capillary liquid chromatography/microelectrospray mass spectrometry. Chem. Res. Toxicol 1999, 12, 1019. [DOI] [PubMed] [Google Scholar]
- [32].Krylov EV, Coy SL, Vandermey J, Schneider BB, Covey TR,Nazarov EG. Selection and generation of waveforms for differential mobility spectrometry. Rev. Sci. Instrum 2010, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Goodenough AK, Schut HA, Turesky RJ. Novel LC-ESI/MS/MSn method for the characterization and quantification of 2’-deoxyguanosine adducts of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by 2-D linear quadrupole ion trap mass spectrometry. Chem. Res. Toxicol 2007, 20, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hall AB, Coy SL, Nazarov E, Vouros P. Development of rapid methodologies for the isolation and quantitation of drug metabolites by differential mobility spectrometry-mass spectrometry. Int. J. Ion Mobility Spectrom 2012, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Levin DS, Vouros P, Miller RA, Nazarov EG, Morris JC. Characterization of gas-phase molecular interactions on differential mobility ion behavior utilizing an electrospray ionization-differential mobility-mass spectrometer system. Anal. Chem 2006, 78, 96. [DOI] [PubMed] [Google Scholar]
- [36].Hall AB, Coy SL, Kafle A, Glick J, Nazarov E, Vouros P. Extending the dynamic range of the ion trap by differential mobility filtration. J. Am. Soc. Mass Spectrom 2013, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ding Y, Paonessa JD, Randall KL, Argoti D, Chen L, Vouros P, Zhang Y. Sulforaphane inhibits 4-aminobiphenyl-induced DNA damage in bladder cells and tissues. Carcinogenesis 2010, 31,1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Paonessa JD, Ding Y, Randall KL, Munday R, Argoti D, Vouros P, Zhang Y. Identification of an unintended consequence of Nrf2-directed cytoprotection against a key tobacco carcinogen plus a counteracting chemopreventive intervention. Cancer Res. 2011, 71, 3904. [DOI] [PMC free article] [PubMed] [Google Scholar]