Abstract
β-Glucocerebrosidase (GCase) mutations cause Gaucher’s disease and are a high risk factor in Parkinson’s disease. The implementation of a small molecule modulator is a strategy to restore proper folding and lysosome delivery of degradation-prone mutant GCase. Here, we present a potent quinazoline modulator, JZ-4109, which stabilizes wild-type and N370S mutant GCase and increases GCase abundance in patient-derived fibroblast cells. We then developed a covalent modification strategy using a lysine targeted inactivator (JZ-5029) for in vitro mechanistic studies. By using native top-down mass spectrometry, we located two potentially covalently modified lysines. We obtained the first crystal structure, at 2.2 Å resolution, of a GCase with a noniminosugar modulator covalently bound, and were able to identify the exact lysine residue modified (Lys346) and reveal an allosteric binding site. GCase dimerization was induced by our modulator binding, which was observed by native mass spectrometry, its crystal structure, and size exclusion chromatography with a multiangle light scattering detector. Finally, the dimer form was confirmed by negative staining transmission electron microscopy studies. Our newly discovered allosteric site and observed GCase dimerization provide a new mechanistic insight into GCase and its noniminosugar modulators and facilitate the rational design of novel GCase modulators for Gaucher’s disease and Parkinson’s disease.
Graphical abstract

INTRODUCTION
Gaucher’s disease, the most common lysosomal storage disease, is caused by a recessively inherited deficiency in β-glucocerebrosidase (GCase)1 and subsequent accumulation of toxic lipid substrates. In recent years, GCase mutations were also found as a major risk factor for Parkinson’s disease (PD) and dementia with Lewy bodies (DLB).2–6 Our previous research in neurons indicated that the accumulation of glucosylceramide (GluCer), the substrate of GCase, promotes the formation of α-synuclein (α-syn) oligomers, which are considered toxic in PD.7 Previous work from our group and others have shown that α-syn aggregation can be induced by reduction of GCase activity in cells or animal models, suggesting a close link between α-syn metabolism and GCase activity.8–11 Enhancement of GCase activity is thought to be a potential therapeutic strategy for GCase-associated synucleinopathies, including PD.12,13 Many of the GCase mutations are missense mutations that result in single amino acid substitutions of these enzymes.14 Most of these mutants, including the prevalent Gaucher mutation N370S, are still functional.15 However, as a consequence of misfolding, mutant GCase can accumulate in the endoplasmic reticulum (ER), eventually being targeted for proteasome-mediated breakdown.
A therapeutic approach consists of restoring the proper folding and lysosome delivery of degradation-prone mutant enzymes using small molecules as pharmacological chaperones (PCs).15 Previous studies have shown that iminosugars increase the cellular activity of the N370S mutant form of GCase, as well as of the wild-type enzyme.15,16 Nevertheless, iminosugars tend to have poor selectivity and have relatively short half-lives in cells.17 Several different scaffolds of noniminosugar modulators (e.g., 1 and 2, Figure 1a) have been reported as GCase PCs since 2007.18–22
Figure 1.
Structure and activity of GCase modulators. (a) Chemical structures of noniminosugar pharmacological chaperones. (b) Dose-response curves for GCase modulator JZ-4109 and JZ-5029, and IFG. (c) Thermal melting curves of GCase with different concentrations of modulator JZ-4109 (increase in Tm relative to a DMSO control shown). (d) Western blot of fibroblast cell lysate after 3 days of compound treatments with DMSO as a control. (e) Normalization of GCase signal to GAPDH signal for each sample calculated from Western blots. (f) GCase activity assay for cell lysate with 4MU-β-Glc substrate after 3 days of treatment. (g) Time-dependent enzyme activity decrease with the treatment of inactivator JZ-5029. Each data point was collected in triplicate, and error bars represent the mean values ± standard deviation (SD). The data shown are representative of three independent experiments.
Our study indicated that one of these noniminosugar GCase modulators was effective at reducing α-syn inclusions, as well as associated downstream toxicity, in induced pluripotent stem cell (iPSC)-derived human midbrain dopamine (DA) neurons, demonstrating a therapeutic effect in PD.23 However, the binding site and the mechanism of action of these noniminosugar modulators are not known, which hinders further drug discovery efforts related to GCase modulation as a potential treatment for GD and PD.
We have developed a series of nanomolar potency GCase modulators,24 and here, we describe the identification of the binding site and mechanism of these quinazoline modulators by incorporation of an electrophilic group into one of the modulators, which led to covalent modification of a lysine residue close to the ligand. The binding site was characterized by native mass spectrometry and X-ray crystallography. We also used transmission electron microscopy (TEM) to confirm the GCase dimer conformation induced by the ligand, which supports its mechanism of action and a new approach to GCase stabilization. These results provide an important foundation for the future design of GCase modulators as novel therapeutics for synucleinopathies.
RESULTS
Discovery of Potent Quinazoline Modulators
To develop novel potent modulators of GCase and use them to elucidate their mechanism of action, a systematic structure– activity relationship (SAR) study on quinazoline derivatives was carried out. We identified a series of nanomolar potency noniminosugar GCase modulators,24 such as JZ-4109 (3) (Figure 1a and Supporting Information Scheme 1), with an IC50 value of 8 nM against wild-type recombinant GCase in comparison with 112 nM for isofagomine (IFG), a known iminosugar inhibitor (Figure 1b). Similar to other quinazoline inhibitors, JZ-4109 exhibited linear mixed inhibition, with an increase in Km and decrease in Vmax values upon increasing inhibitor concentrations (Supporting Information Figure S1). Compound JZ-4109 also possesses the ability to protect wild-type recombinant GCase from thermal denaturation in a dose-dependent manner with an approximate 8° C maximum thermal shift (Figure 1c).
Cellular Assays for Modulator JZ-4109
To investigate how JZ-4109 affects wild-type GCase enzyme activity and protein abundance in cells, we treated fibroblast cells from a healthy control with JZ-4109 and IFG, each at 0.2 and 2 µM for 3 days. We used an immunoblot to measure GCase abundance in cells and also measured enzyme activity in the whole cell lysate. We found that both the GCase protein abundance (Figure 1d and Figure 1e) and GCase activity significantly increased as a result of JZ-4109 treatment (Figure 1f), suggesting that JZ-4109 stabilizes GCase and thus increases enzyme activity in cells. We did not observe an increase in GBA mRNA upon JZ-4109 treatment demonstrating that the increase in protein abundance is not due to increased expression (Supporting Information Figure S2).
To further investigate the cellular function of modulator JZ-4109, we used a series of cellular tests. First, as impaired GCase ER-maturation is one effect of Gaucher’s disease associated mutants, we used Endo H and PNGase F assays to check whether treatment with modulator JZ-4109 affected GCase maturation. The results revealed that the increase in GCase signal after treatment was post-ER with no increase observed in ER-retained GCase which indicates that upon compound treatment GCase is effectively trafficked in cells and does not increase protein load in the ER, which could lead to ER stress which is not observed after treatment (Figure 2a, Supporting Information Figure S3). Second, as GCase is a lysosomal enzyme, we investigated lysosomal GCase abundance by lysosomal enrichment. The results showed that after treatment with modulator JZ-4109, there was an improvement of GCase levels and enzyme activity in lysosomal enrichment fractions (Figure 2b, Figure 2c, and Figure 2d). Third, we were interested in whether modulator JZ-4109 could stabilize mutant GCase in cells. We used homozygous N370S mutant fibroblast cells from a type-1 Gaucher’s disease patient. After JZ-4109 treatment, we found that both the protein abundance and activity of GCase was significantly increased. (Figure 2e and Figure 2f). These cellular experiments support the promising effect of modulator JZ-4109 on cellular GCase activity and drug discovery.
Figure 2.
Cellular assays for modulator JZ-4109 treatment. (a) Endo H and PNGase F assays for wild-type fibroblast after compound treatment. Modulator JZ-4109 (2 µM), IFG (2 µM), n = 3. (b) Lysosomal enrichment assay for HEK cells after JZ-4109 (2 µM) and IFG (2 µM) treatment. (c, d) statistical analysis for GCase in the lysosomal fraction (C, GCase only; D, GCase normalized with Lamp1). Single tail test, n = 3. (e) Western blot and Endo H assay of N370S fibroblast cells for GCase after JZ-4109 (0.2 and 2 µM) and IFG (0.2 and 2 µM) treatment, n = 3. (f) Normalized (to DMSO control) GCase activity in N370S fibroblast cells after JZ-4109 and IFG treatment. n = 3.
Covalent Modification of GCase with JZ-5029
To explore the modulator binding site, we developed a covalent modification strategy by attaching a carbon linker to the phenyl ring of the 2,3-dihydro-1H-indene in JZ-4109, terminating in an electrophilic N-hydroxysuccinimide (NHS) ester so that it could covalently attach to the protein (JZ-5029, 4, Figure 1a and Supporting Information Scheme S2). With a covalently modified protein, we could identify the binding site by proteomics and crystallography. Compound JZ-5029 had an IC50 value of 56 nM against wild-type GCase in a 4MU-β-Glc enzyme activity assay (Figure 1b). When JZ-5029 was incubated with wild-type GCase at pH 7.0, the enzyme activity decreased in a time-dependent manner (Figure 1g). After 4 h the inactive enzyme was dialyzed with the same buffer, and no enzyme activity returned. To demonstrate that JZ-5029 and JZ-4109 occupy the same binding site on GCase, we conducted an in vitro competition assay. In the presence of JZ-4109, the covalent modification rate by JZ-5029 was slowed down in a dose-dependent manner confirming that JZ-4109 competes with JZ-5029 for the binding site (Supporting Information Figure S4).
Native Mass Spectrometry of Covalently Modified GCase by JZ-5029
To confirm the presence of the modulator-bound GCase, we first performed native mass spectrometry (nMS). Interestingly, the intact spectrum of a species with twice the molecular weight of the GCase monomer was abundantly observed (Figure 3a), exhibiting a distribution of major peaks clustered around 126.6 kDa and differing in mass by 162 ± 1 Da, consistent with variable numbers of hexose subunits of monomer glycosylation. A minor monomeric species from solution was also observed (Figure 3b, top), with a distribution of masses consistent with the unmodified GCase. However, the monomer ejected from the gas-phase isolated dimer appeared to have modulator nearly completely bound, having an observed mass increase of 501 Da (corresponding to the expected JZ-5029 fragment bound) when compared to the monomer generated from solution (Figure 3b, bottom). Isolation of the ejected monomer and activation to induce peptide backbone fragmentation resulted in extensive fragment coverage from the C-terminus (Figure 3c), including four fragment ions that allowed for the localization of the modulator to a binding site in the TIM barrel domain of the protein (likely either Lys321 or Lys346).
Figure 3.
Multitiered native mass spectrometry analysis of JZ-5029-modified GCase. (a) Intact native mass spectrum of the GCase dimer bound to quinazoline modulator JZ-5029 with deconvoluted neutral masses in the inset. Multiple peaks correspond to variable glycosylation. (b) Top: spectrum of the 15+ monomer species directly measured from solution; Bottom: spectrum of the 15+ monomer resulting from gas-phase isolation of the dimer and ejection of the monomer using collisional activation. (c) Graphical fragment ion map with blue flags indicating ions fragmented from the monomer that have been matched to within 10 ppm of those predicted from the GCase sequence. Only residues 276–497 are shown; no N-terminal fragments were observed. The orange markers correspond to the two potential modulator binding sites (+500.2576 Da).
Crystal Structure of GCase Modified by JZ-5029
To further understand the mechanism of the quinazoline modulators and confirm our finding, we crystallized JZ-5029-modified GCase and resolved the X-ray crystal data to 2.2 Å resolution (Supporting Information Table S1). The crystals contain four monomers of GCase in the asymmetric unit (Figure 4a). The ligand binding site is located at the dimer interface near the catalytic pocket (Figure 4b and Supporting Information Figure S5). The ligand contains a chiral carbon at position 2 of the 2,3-dihydro-1H-indene ring; on the basis of the electron density map, the (R)-enantiomer is bound at two of the four molecules of the asymmetric unit (chains A and D), and the (S)-enantiomer is bound to the protomers (chains B and C) of the other two molecules. The general binding modes of the enantiomers are similar, except that the 2,3-dihydro-1H-indene moiety is flipped by 180° and the octynyl linker adopts a slightly different conformation (Figure 4c). The ligand is covalently attached to Lys346 of loop 2 (Supporting Information Figure 6), confirming our proteomics study. The nitrogen atom at position 1 of the quinazoline ring forms a H-bond with the carboxylic acid group of the catalytic Glu235 residue. This nitrogen was found to be the most important nitrogen in the quinazoline scaffold from the previous SAR study.19 The NH group connecting the quinazoline and the 2,3 dihydro-1H-indene ring forms another H-bond with Ser345. According to our SAR study (to be published in another journal), methylation of this NH group diminishes the inhibitory activity of JZ-4109, suggesting that the key contribution of the H-bond interaction is with Ser345. The quinazoline ring has a strong π–π stacking interaction with the phenyl ring of Tyr313, and the 3-pyridinyl ring has a π–amide interaction at a distance of 3.5 Å with Gln284. Phe347 adopted two conformations in the crystal, indicating its flexible movement upon ligand binding. It is notable that the phenyl ring of Phe347 in chain A lies between two 2,3-dihydro-1H-indene moieties of two enantiomers and forms a π–π stacking interaction with the two dihydroindene rings (Figure 4c and Supporting Information Figure S5). This π–π interaction was predicted by our SAR study of quinazoline modulators, and showed an important contribution to the inhibitory activity of GCase modulators. The 2,3-dihydro-1H-indene and carbon linker of the ligand show additional hydrophobic interactions with Phe316, Phe347, and Trp348 in the binding pocket, and with Leu241, Gly243, and Tyr244 from the other chain. These interactions stabilize the GCase dimer conformation.
Figure 4.
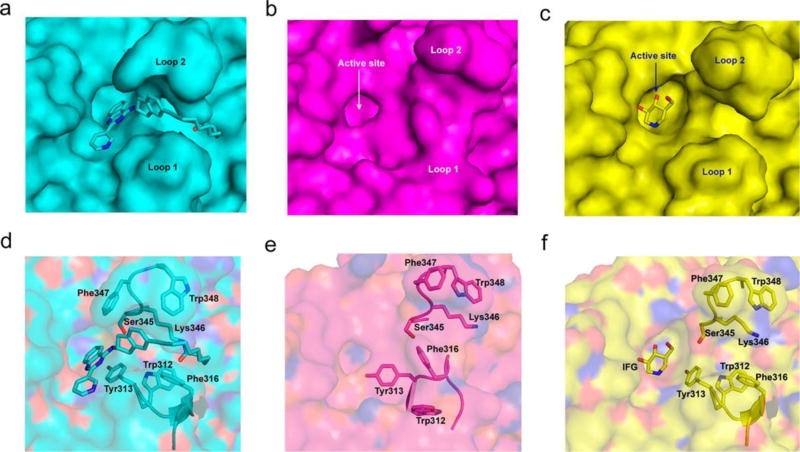
Crystal structure of JZ-5029-modified GCase. (a) Crystallographic unit cell of JZ-5029-modified GCase. (b) Location of the JZ-5029 binding site in the GCase dimer interface. (c) Interaction diagram of JZ-5029 (in cyan, the modified Lys346 is also presented) with GCase (Chain A, yellow; Chain B, pink). (d) Loop 1, loop 2, and loop 3 conformation changes in JZ-5029-modified GCase (cyan) compared to apo-GCase (mauve, pdb: 1OGS), and IFG bound GCase (yellow, pdb: 2NSX).
This ligand binding induces critical movements for two loops at the mouth of the active site, loop 1 (311–319) and loop 2 (342–354) in all four of the monomers (Figure 4d). The arrangement of loop 1 and loop 2 exposes an allosteric site for quinazoline modulators (Figure 5a), compared to apo-GCase (Figure 5b) and IFG-bound GCase (Figure 5c). Loop 1 adopts an α-helical conformation, which was also observed in the IFG binding crystal structure25,26 (Figure 4d). Loop 2 has a compacted conformation (Figure 5d) compared to apo-GCase (Figure 5e), but similar to the IFG binding conformation (Figure 5f). The movement of Phe347 partially covers the entrance of the active site (Figure 5a, 5d), suggesting the movement of Phe347 may manipulate the enzyme activity by controlling the substrate access to the active site.
Figure 5.
Surface representation and residue conformation of loop 1 and loop 2 in GCase. Surface representation of (a) JZ-5029-modified GCase (cyan), (b) apo-GCase (purple, pdb: 1OGS), (c) IFG-bound GCase (yellow, pdb: 2NSX). The residue movement of loop 1 and loop 2 of (d) JZ-5029-modified GCase (cyan), (e) apo-GCase (purple, pdb: 1OGS), (f) IFG-bound GCase (yellow, pdb: 2NSX).
Quinazoline Modulator-Induced GCase Dimerization
In the native mass spectrum, we observed that JZ-5029-modified GCase was in a dimer form. Here, we confirm an unusual dimer form present in our ligand-bound GCase crystal unit compared with the reported apo- and iminosugar-bound GCase structures. Chain A of the JZ-5029-modified dimer is twisted at an angle (Figure 6a) that exposes the active site (indicated in red in Figure 6b) when chain B is superimposed with apo-GCase. To test if this dimer form exists in solution, we analyzed solution samples with a size exclusion gel chromatography system equipped with a multiangle light scattering (SEC-MALS) detector. At pH 5.0, the wild-type GCase monomer was the predominant peak (black, 13.4 min, 62 kDa) (Figure 6c). When treated with JZ-4109 (0.25 to 2 equiv), the monomer peak shifted to an earlier time (10.2 min) with twice the monomer molecular weight (124 kDa) in a dose-dependent manner (Figure 6c). The dimer form was the major peak (71%) with JZ-5029-modified GCase, with only a small amount of the monomer (Figure 6d). Under the same conditions, when wild-type GCase was treated with IFG (0.5 to 10 equiv), only the monomer peak was observed, and elution time was delayed in a dose-dependent manner (Figure 6e) without formation of the dimer peak, suggesting a large conformation changing of GCase protein. To further understand the relationship between dimer formation and GCase modulating activity, we selected a set of compounds with an IC50 value range between 5 and 300 nM from our publication24 and evaluated their effects on GCase dimerization using SEC (Supporting Information Table S2). Using identical conditions, we found that all of the selected modulators stabilize GCase dimerization to varying degrees. Further examination revealed that the degree to which modulators induced GCase dimer formation strongly correlated with their enzyme activity (Supporting Information Figure S7).
Figure 6.
GCase dimer conformation and SEC-MALS chromatogram. (a) Comparison of JZ-5029-modified GCase dimer (pink) conformation to apo-GCase dimer (cyan, pdb: 1OGS); aligned by chain B: (b) Left, compound JZ-5029 (tan) modified GCase dimer surface representation with exposed active site (indicated in red); Right, predicted apo-GCase dimer surface representation (cyan, pdb: 1OGS). (c) SEC-MALS chromatogram of wild-type GCase (black) and a mixture of JZ-4109 (4.12, 8.25, 16.5, and 33.0 µM) and GCase (16.5 µM) with the calculated molar masses (g/mol) dotted on the chromatogram. (d) SEC-MALS chromatogram of wild-type GCase (black) and JZ-5029-modified (red) GCase with the calculated molar masses (g/mol) dotted on the chromatogram. (e) SEC-MALS chromatogram of wild-type GCase (black) and a mixture of IFG (8.25, 16.5, 33.0, and 165 µM) and GCase (16.5 µM) with the calculated molar masses (g/mol) dotted on the chromatogram
Characterization of JZ-5029-Modified GCase by TEM
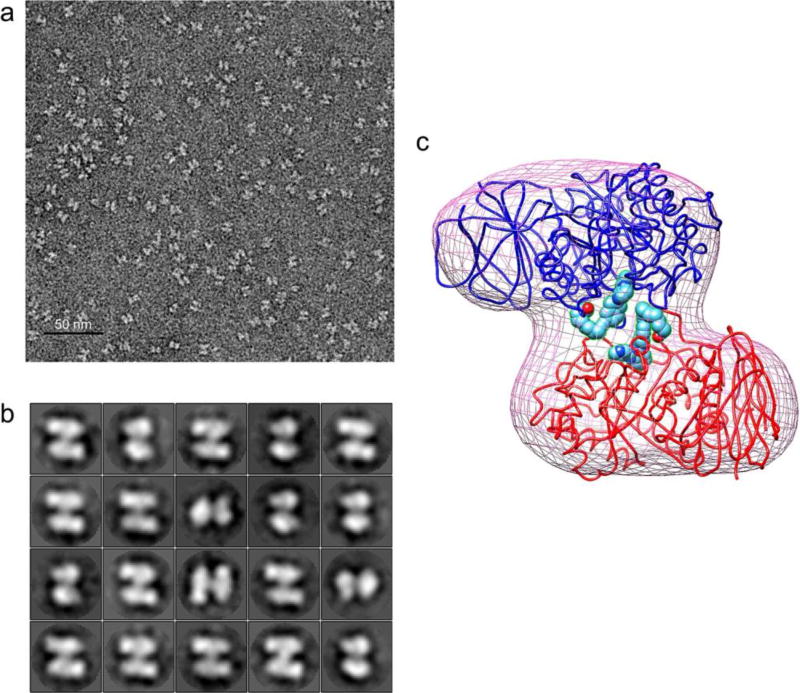
To characterize the dimer form of JZ-5029-modified GCase, the dimer fractions from SEC were collected and visualized using negative staining transmission electron microscopy (TEM). As shown in Figure 7a, a butterfly shaped dimer form is easily discernible in the TEM micrographs. The average dimensions of the particles are about 10 nm × 10 nm. The particles were selected, and the images were analyzed by two-dimensional reference-free classification (Figure 7b). Because we have obtained high resolution X-ray structural data for the JZ-5029-modified GCase, we were able to compare it with the 3D model generated with the TEM data. Here, we show that the JZ-5029-modified dimer fit into the 3D TEM model (Figure 7c).
Figure 7.
TEM results for JZ-5029-modified GCase dimer. (a) Representative negative staining TEM micrograph of the JZ-5029-modified GCase dimer. (b) Representative reference free class averages of JZ-5029-modified GCase dimer. (c) Docking of JZ-5029-modified GCase dimer (blue, Chain A; red, Chain B; spheres, JZ-5029) into 3D TEM model (purple mesh)
DISCUSSION
Preclinical studies have reported that GCase pharmacological chaperones (PCs) can affect α-synuclein processing.27 The first PC to enter clinical trials was isofagomine (IFG), which failed to show general clinical effects.28 Consequently, more effort was invested in the development of noniminosugar GCase modulators that had greater selectivity and drug-like properties than the iminosugar compounds. A series of nanomolar- potency quinazoline modulators was developed from our systematic SAR study.24 Among them, JZ-4109 (3, Figure 1a) was one of the most potent modulators as an in vitro enzyme inhibitor (Figure 1b), and had a similar PC effect as IFG in the thermal shift (Figure 1c) and wild-type and N370S patient-derived fibroblast cell assays (Figure 1d,e). Further cell assays confirmed that JZ-4109 increased the mature lysosome GCase levels and enhanced the enzyme activity (Figure 1f), highlighting the potential application of these quinazoline modulators in amplification of the wild-type and mutant GCase enzyme activity.
To explore the binding site of these noniminosugar modulators, we attempted to obtain a crystal structure with JZ-4109 bound to GCase by either incubation of JZ-4109 with GCase or by soaking it into the apo-GCase crystal. However, we were unable to crystallize the complex, either because of the limited solubility of JZ-4109, an unknown action mechanism, or an inaccessible binding site in the crystal. Consequently, we focused on developing a covalent modulator to modify GCase, in which the ligand would become part of the protein, resulting in the complete occupation of the binding site with the ligand. The primary amino group of lysine residues is a highly reactive nucleophile located on most protein surfaces, which can be modified by many diverse chemicals.29,30 N-Hydroxysuccinimide (NHS) esters have been used to modify lysine residues in numerous biological systems without denaturation of the protein.31 The phenyl ring of the 2,3-dihydro-1H-indene moiety of compound 3 was modified by installation of a long carbon linker terminating in an NHS ester; this compound, JZ-5029 (4), had comparable activity to that of JZ-4109, but produced time-dependent inactivation of GCase (Figure 1g).
With native mass spectrometry, we observed GCase was covalently modified by our compound (JZ-5029) and formed a dimer as the major species (Figure 3a). The mass shift of the modified GCase monomer, compared to the wild-type GCase monomer, was consistent with the ligand molecular weight, indicating complete and selective modification of the GCase dimer by JZ-5029 (Figure 1b). Further fragmentation of the protein allowed us to locate the modified fragment in the peptide 316–358 (Figure 3c), which included two lysine residues (Lys321 or Lys346). Because of the low signal/noise ratio of the peptide with further fragmentation, we were unable to identify exactly which lysine was modified by mass spectrometry.
Having obtained GCase covalently modified with JZ-5029, we rescreened crystallization conditions and were able to crystallize the modified enzyme, which was resolved to 2.2 Å resolution (Figure 4). As we suspected, the ligand covalently modified Lys346 (Figure 4c), one of two lysines identified by native top-down mass spectrometry. Covalent attachment of JZ-5029 induced large conformation changes of loop 1 and loop 2 compared to apo-GCase (Figure 4d). The important rearrangement of loop 1 and loop 2 exposes an allosteric site for quinazoline modulators (Figure 5a). The loop 1 conformation was changed to an α-helix, which was also found to be a key effect of IFG PC activity.25,26 Tyr313 in loop 1 had been found to exist in multiple conformations in different GCase crystals. The major π–π interaction between the ligand quinazoline ring and Tyr313 stabilizes the loop 1 conformation, and the H-bond with Ser345 locks the conformation of loop 2 into a compacted shape. Both of these interactions contribute to the PC activity of GCase modulators. The residue movement on loop 2 contributes to the enzyme activity, suggesting that this binding site may also adapt to the activators, namely, noninhibitory GCase modulators. We also observed that the ligand binding site was located at the GCase dimer interface (Figure 4b). Additional hydrophobic interactions of Phe347 on chain A with another monomer strengthened the binding of the ligand (Figure 4c), supporting the notion that the compound binds at the dimer interface, thereby stabilizing the dimer form. These hydrophobic interactions between the ligand and both monomers contribute to the potent inhibitory activity of JZ-4109. Using SEC-MALS (Figure 6), we observed that GCase was stabilized as a dimer by JZ-4109 (Figure 6c) as well as a series of other quinazoline modulators, and through covalent modification by JZ-5029 (Figure 6d), whereas the iminosugar inhibitor IFG did not induce any dimer form (Figure 6e). The strong correlation between induced GCase dimer and compound enzyme activity demonstrated that the ability of dimer forming by GCase modulators contributed to their enzyme activity. The delay of the IFG elution peak may be due to the changing of GCase conformation induced by IFG binding. These data suggest that our quinazoline modulators act by a mechanism different from that of the iminosugars.
GCase has been suggested to exist as a dimer in vivo in spleen and fibroblast cells32 and also in a complex with LIMP-2 (GCase transporter).33 A recent study observed that GCase is in a monomer–dimer equilibration in vitro at higher concentrations (8 µM), and its activator, saposin C, can dissociate the GCase dimer to monomers.34 However, wild-type GCase is in equilibrium among several different forms, which makes it difficult to characterize a specific dimer conformation in the mixture. We found that inactivation of GCase by covalent ligand JZ-5029 produced a stable dimer form, which was separated and purified by size exclusion gel filtration chromatography. Because we were able to obtain the crystal structure of JZ-5029-modified GCase, we could use low resolution negative staining TEM to further characterize the dimer fraction of GCase in solution for comparison. The JZ-5029-modified dimer preferred a butterfly shape (Figure 7a and Figure 7b), which could be easily identified from their microscope images and allowed us to classify the images. Three dimer conformations were predicted to be stable in solution using the PISA server with the apo-GCase crystal structure.34 The authors of this paper predicted that the dimer form that has the catalytic site buried at the interface is the preferable conformation;34 this is consistent with the images from our TEM data.
CONCLUSIONS
In summary, we found that conversion of a potent non-iminosugar modulator JZ-4109 (3) of GCase to a covalent activator JZ-5029 (4) led to the modification and identification of a modified lysine and to the first crystal structure of a potent noniminosugar modulator bound to GCase. The ligand attaches to Lys346, a lysine residue at the interface of the GCase dimer, which stabilizes the dimer form in a way that exposes the active site. Further characterization of the dimer form with TEM supports a butterfly shaped dimer in solution as well as in the crystal. To the best of our knowledge, this is the first study that identifies a noniminosugar modulator allosteric binding site and a GCase dimer conformation. This novel structure-based design strategy exploits the potential binding pockets of the dimer interface to induce GCase dimerization and achieve high potency of a noniminosugar GCase modulator. It is expected that this identification of a novel allosteric binding pocket at the interface of the GCase dimer will facilitate future design of high potency noninhibitory GCase modulators for the ongoing drug discovery efforts in PD and related synucleinopathies. Moreover, this may be a general approach for stabilizing the active dimer form of GCase and related enzymes.
EXPERIMENTAL SECTION
Preparation of Modulators JZ-4109 (3) and JZ-5029 (4)
The detailed syntheses and characterization of modulators JZ-4109 (3) and JZ-5029 (4) can be found in the Supporting Information.
Compound Activity Assay with 4MU-β-Glc Substrate
The compounds in DMSO solution (0.5 µL/well) were transferred to a 96-well black microtiter plate (the final 12 concentrations were 0.76 nM to 1.56 µM). The enzyme solution (33.5 µL, 7.5 nM final concentration) was transferred to the wells. After 5 min of incubation at room temperature, the enzyme reaction was initiated by the addition of 33 µL/well 4MU-β-Glc (1.5 mM) solution. The reaction was terminated by the addition of 33 µL/well stop solution (1 M NaOH and 1 M glycine mixture, pH 10) after 30 min of incubation at 37 °C. The fluorescence was then measured in a Biotek Synergy H1 multimode plate reader with Ex = 365 nm and Em = 440 nm.
Thermodenaturation Experiment.19
This assay measures the change in the melting temperature of the recombinant wild-type GCase in the presence of different concentrations of the inhibitors. A mixture of wild-type GCase and SYPRO Orange (5000 stock concentration, Invitrogen, Carlsbad, CA) was added to a 96-well skirted thin-wall PCR plate (Bio-Rad, Hercules, CA) with final concentrations of 1 and 5 µM, respectively. Wild-type GCase and SYPRO Orange were diluted in 150 mM phosphate/citrate buffer at pH 4.8. A 12-point DMSO dilution series was made separately in a 96-well polypropylene plate (Thermo Fisher Scientific, Hudson, NH) for all analogues, with final concentrations ranging from 0.25 to 100 µM. 1 µL of each dilution point of each compound was transferred to the aforementioned wild-type GCase-SYPRO Orange mixture, with a final DMSO concentration of 2% (1 µL in 50 µL final volume). DMSO alone was also transferred to the PCR plate for each dilution series as a control sample. The plate was immediately centrifuged at 1000 rpm for 10 s and subsequently sealed with Optical-Quality Sealing Tape (Bio-Rad). The plate was then heated using an iQ5 real-time PCR detection system (Bio-Rad) from 20 to 95 °C with an increment of 1 °C and a ramping rate of 0.1°C/s. SYPRO Orange fluorescence was monitored by a CCD camera using excitation and emission wavelengths of 490 and 575 nm, respectively. Protein melting temperature (Tm) was obtained through an EXCEL-based DSF worksheet and GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA).
Cell Culture and Compound Treatment
Fibroblast cells (2132, male, healthy control) were cultured in fibroblast medium (500 mL) DMEM Medium (Life Tech), 15% heat-inactivated FBS (Life Tech), 200 mM l-glutamine (5 mL, Life Tech), and Pen Strep, (5 mL, Life Tech) at 37 °C and 5% CO2. The fibroblast cells were treated with compounds for 3 days. During the 3-day treatment, media was changed every day.
Endo H and PNGase F Assay
Endo H and PNGase F (New England Biolabs) were used to cleave glycan from protein. Denatured cell lysate (10 µg) was mixed with 2 µL Endo H or PNGase F, respectively, and incubated at 37 °C for 1 h. Western blots were run to examine the Endo H and PNGase F digestions. The post-ER from GCase does not respond to Endo H treatment (Endo H resistant) but does to PNGase F treatment. ER from GCase responds to both Endo H (Endo H sensitive) and PNGase F.
Lysosomal Enrichment
Lysosomal enrichment kit (#89839, Thermo Fisher Scientific) and HEK cells were used to enrich the lysosome fraction. Cells were seeded in 10 cm dishes, and the cells were treated with JZ-4109, IFG, and DMSO (0.1% v/v) separately. After 3-day treatment, cells were harvested, using three confluent dishes for one sample. Cells were homogenized with a Dounce tissue grinder. OptiPrep Gradients (iodixanol-5,5′-[(2-hydroxy-1,3-propane-diyl)-bis(acetylamino)]bis[N,N′-bis(2,3dihydroxypropyl-2,4,6-triiodo-1,3-benzenecarboxamide) were prepared at 17%, 20%, 23%, 27%, and 30%. Cell lysate was loaded at the top of the gradients and was ultracentrifuged at 140,000g at 4 °C for 2 h. After centrifugation, the top layer was the lysosomal fraction, which was collected and used for Western blots.
Western Blotting
Triton (1%) lysis buffer was used to lyse the fibroblast cells after compound treatment, and then 20% SDS sample buffer was used to denature the proteins at 100 °C for 10 min. Bis-Tris gel (10%, Life Tech) was used for running gels and Trans-Blot Turbo PVDF kit (Bio-Rad) was used to transfer membranes. GCase antibody (Sigma-Aldrich) and GAPDH primary antibodies were incubated with the membranes overnight, then the incubated membranes were treated with the secondary antibody (Peroxidase-AffiniPure Goat Anti-Rabbit/Mouse IgG (H+L), Jackson Immunoresearch Lab) for 30 min. Chemidoc MP system (Bio-Rad) was used to scan the membranes and analyze the imaging.
Real Time Quantitative PCR of GBA1 and ER Stress Markers
Fibroblasts were seeded in 10 cm dishes, and the cells were treated with JZ-4109 (0.2 and 2 µM) or with DMSO. After 3 days of treatment, cells were harvested, and RNA was extracted using RNeasy mini kit (Qiagen, Germantown, MD). cDNA was synthesized from RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystem, Forest City, CA). qPCR analysis was performed using 7500 Fast Real-Time PCR system (Applied Biosystem). The primers used to measure mRNA levels are as followed: GBA1, ACCACAATGTCCGCCTACTC, GCTGCTTCTGGGTCTGTCA; BIP, CATCAAGTTCTTGCCGTTCA, ATGTCTTTGTTTGCCC-ACCT; CHOP, AGCGACAGAGCCAAAATCAG, TCTGCTTT-CAGGTGTGGTGA; ATF4, GTTCTCCAGCGACAAGGCTA, ATCCTGCTTGCTGTTGTTGG; GAPDH, ACAACTTTGGTA-TCGTGGAAGG, GCCATCACGCCACAGTTTC. Relative mRNA levels were calculated using the 2−ΔΔCT method and normalized GAPDH.
GCase Covalent Modification by Compound JZ-5029
JZ-5029 in DMSO (0.89 mM, 5 µL, 2 equiv) was added to wild-type GCase (22 µM, 95 µL, 1 equiv) in 0.1 M phosphate buffer (pH 7.0) with 1 mM taurocholate in one portion, immediately vortexed for 5 s, and incubated on ice. At indicated time points, the reaction solution (2 µL) was sampled and diluted (1:3125 dilution) into the assay buffer (50 mM citric acid, 176 mM K2HPO4, and 0.01% Tween-20 (v/v) at pH 5.9). After 4 h, the reaction solution was dialyzed by repeated (4×) buffer exchanges with 0.1 M phosphate buffer (pH 7.0) using Amicon Ultra 0.5 mL 10K centrifugal filters (Millipore). The enzyme was adjusted to 22 µM and aliquoted for activity. The diluted solutions were assayed with 4MU-β-Glc using the method detailed above.
A competition covalent modification assay with JZ-4109 was conducted using a method similar to the one described above. Wildtype GCase (22 µM) was pretreated with JZ-4109 (5.5, 11, and 22 µM) and with the same volume of DMSO as a blank control, and then treated with JZ-5029 (44 µM). At the indicated time points, the modification rate by JZ-5029 was monitored by enzyme activity assay.
Native Top-down Mass Spectrometry
Purified GCase samples were buffer exchanged with 30 kDa molecular weight cutoff filters (Millipore) into 100 mM ammonium acetate (pH 5.0). All samples were directly infused into a Q-Exactive HF mass spectrometer (Thermo Fisher Scientific), specially modified to detect large proteins and complexes.35 Mass spectra of the intact dimer and monomers were collected at a resolving power of 15,000 at 200 m/z. The spectrum of the GCase ejected monomer was acquired by first isolating the 24+ to 21+ charge states of the intact dimer (ranging from 5000 to 6200 m/z) using a quadrupole mass filter positioned behind the electrospray ion source and in front of the collision cell and Orbitrap mass analyzer. Using higher-energy collisional dissociation (HCD) in the collision cell, the monomer was ejected from the dimer in the gas phase. Fragment ions from dissociation of peptide backbone bonds in the monomer were produced by activating the dimer in the source region to eject the monomer, then applying HCD to ions of the isolated monomer species; fragmentation spectra were acquired at 120,000 resolving power at 200 m/z. Fragment ions were analyzed with ProSight Lite36 and mMass software.37
SEC-MALS-QELS
Molecular weights of the wild-type GCase (16.5 µM), mixture of GCase (16.5 µM) and JZ-4109 (4.125, 8.25, 16.5, and 33 µM), JZ-5029-modified GCase (16.5 µM), and a mixture of GCase and IFG (8.25, 16.5, 33, and 165 µM) were determined by conducting SEC-MALS experiments using an Agilent Technologies 1100 HPLC system (Agilent Technologies, Santa Clara, CA) equipped with a Dawn HeleosII 18-angle MALS light scattering detector, an Optilab T-rEX (refractometer with EXtended range) refractive index detector, a WyattQELS quasi-elastic (dynamic) light scattering (QELS) detector, and ASTRA software (all four from Wyatt Technology Europe GmbH, Dernbach, Germany). A total of 50 µL (1 mg/mL) of the protein samples in 100 mM sodium acetate, 100 mM sodium chloride, pH 5.0 buffer were injected and run on a TSKgel SuperSW3000 column (Tosoh Bioscience LLC, King of Prussia, PA) pre-equilibrated with the same buffer at a flow rate of 0.3 mL/min at 22 °C. Bovine serum albumin (BSA) (Sigma-Aldrich Corp., St. Louis, MO) was used as a control. The mixture of GCase (16.5 µM) and other selected compounds (33 µM) was tested under the same conditions detailed above.
Crystallization, Data Collection, Structure Determination and Refinement
Wild-type GCase was covalently modified by JZ-5029 as described above and partially deglycosylated using Nglycosidase F (Glyko).38 The purified protein was used in initial crystallization trials employing a standard sparse matrix screen with approximately 1200 different conditions (proprietary initial screen and Qiagen). The crystallization hits were optimized further by systematically varying parameters critically influencing crystallization, such as temperature, protein concentration, drop ratio, and concentration of the components of the crystallization condition. The final conditions were also refined by systematically varying pH or precipitant concentrations as well as screening for additional compounds enhancing crystallization. The optimized condition contains 20% (w/v) PEG3350, 0.2 M (NH4)2SO4, 0.1 M NaOAc, pH 4.50, and 0.50% (w/v) lauryldimethylamine N-oxide (LDAO) at 20 °C. Crystals were flash frozen in liquid nitrogen in a reservoir solution containing 20% ethylene glycol. Diffraction data were measured at a temperature of 100 K. The crystallographic data were collected at the SWISS LIGHT SOURCE (SLS, Villigen, Switzerland) using cryogenic conditions.
The structure was solved by molecular replacement. A previously solved structure of GCase (1OGS.pdb) was used as a search model. Subsequent model building and refinement were performed according to standard protocols with the software packages CCP4 and COOT. Statistics of the final structure and the refinement process are listed in Supporting Information Table S1. Figures were generated using PyMOL (DeLano Scientific).
TEM Data Acquisition and Processing
A solution of JZ-5029-modified GCase dimer was applied to 400 mesh copper grids coated with ultrathin carbon film (Electron Microscopy Sciences). The grids were stained with 1% uranyl formate and imaged in a JEOL1400 TEM operating at 120 kV at a magnification of 40,540×. Ten tilted image pair micrographs were taken at 0° and 40°. Xmipp3 software was used for the particle picking, extraction and random conical tilt reconstruction of 5000 particles chosen from the micrographs to generate an initial 3D model.39 Leginon40 automated data collection was then used to collect 100 images from which ≈10,000 particles were picked using DoG Picker.41 Data was subsequently processed using RELION2 to generate a 3D model of JZ-5029 using 3D classification and 3D refinement subroutines.42 JZ-5029-modified GCase dimer from crystal structure was then docked in the model manually.
Statistics
Error bars represent standard error among at least three independent experiments with at least two replicates per condition. Statistical analysis was performed using Prism Software v 5.01 (GraphPad Software).
Supplementary Material
Acknowledgments
This work was supported by R01NS076054 (to D.K.). J.Z. and M.S. are also supported by a fellowship from Lysosomal Therapeutics Inc. (Cambridge, MA) and the Deutsche Forschungsgemeinschaft (Heisenberg Programme), respectively. O.S.S. is supported by a US National Science Foundation Graduate Research Fellowship (2014171659). N.L.K. acknowledges support from the W.M. Keck Foundation (DT061512) and the Neuroproteomics Center funded by the National Institute on Drug Abuse (P30 DA018310). We thank Dali Liu from Loyola University for his fruitful discussions and critical reading of the paper. We thank Habibi Goudarzi and Saman Shafaie from IMSERC at Northwestern University for their assistance with HRMS experiments, Chi-Hao Luan from Northwestern University’s High Throughput Analysis Laboratory for his assistance with thermal denaturation experiments, and Theint Aung and Arabela A. Grigorescu from Northwestern University’s Keck Biophysics Facility for their assistance with SEC-MALS experiments. This work made use of the IMSERC at Northwestern University, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF NNCI-1542205), the State of Illinois, and the International Institute for Nanotechnology (IIN). This research used resources of the Swiss Light Source (SLS Villigen, Switzerland). This work used resources of the Northwestern University Structural Biology Facility which is generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center.
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b13003.
Chemical compound information, source data and experiment procedures (PDF)
The authors declare no competing financial interest.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession number 5LVX.
References
- 1.Grabowski GA. Lancet. 2008;372:1263. doi: 10.1016/S0140-6736(08)61522-6. [DOI] [PubMed] [Google Scholar]
- 2.Aharon-Peretz J, Rosenbaum H, Gershoni-Baruch R. N. Engl. J. Med. 2004;351:1972. doi: 10.1056/NEJMoa033277. [DOI] [PubMed] [Google Scholar]
- 3.Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, Bar-Shira A, Berg D, Bras J, Brice A, Chen CM, Clark LN, Condroyer C, De Marco EV, Durr A, Eblan MJ, Fahn S, Farrer MJ, Fung HC, Gan-Or Z, Gasser T, Gershoni-Baruch R, Giladi N, Griffith A, Gurevich T, Januario C, Kropp P, Lang AE, Lee-Chen GJ, Lesage S, Marder K, Mata IF, Mirelman A, Mitsui J, Mizuta I, Nicoletti G, Oliveira C, Ottman R, Orr-Urtreger A, Pereira LV, Quattrone A, Rogaeva E, Rolfs A, Rosenbaum H, Rozenberg R, Samii A, Samaddar T, Schulte C, Sharma M, Singleton A, Spitz M, Tan EK, Tayebi N, Toda T, Troiano AR, Tsuji S, Wittstock M, Wolfsberg TG, Wu YR, Zabetian CP, Zhao Y, Ziegler SG. N. Engl. J. Med. 2009;361:1651. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schapira AHV, Olanow CW, Greenamyre JT, Bezard E. Lancet. 2014;384:545. doi: 10.1016/S0140-6736(14)61010-2. [DOI] [PubMed] [Google Scholar]
- 5.Sidransky E, Lopez G. Lancet Neurol. 2012;11:986. doi: 10.1016/S1474-4422(12)70190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin MK, Farrer MJ. Genome Med. 2014;6:48. doi: 10.1186/gm566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazzulli JR, Xu YH, Sun Y, Knight AL, McLean PJ, Caldwell GA, Sidransky E, Grabowski GA, Krainc D. Cell. 2011;146:37. doi: 10.1016/j.cell.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazzulli JR, Zunke F, Isacson O, Studer L, Krainc D. Proc. Natl. Acad. Sci. U. S. A. 2016;113:1931. doi: 10.1073/pnas.1520335113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu YH, Sun Y, Ran H, Quinn B, Witte D, Grabowski GA. Mol. Genet. Metab. 2011;102:436. doi: 10.1016/j.ymgme.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manning-Bog AB, Schule B, Langston JW. NeuroToxicology. 2009;30:1127. doi: 10.1016/j.neuro.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Osellame LD, Rahim AA, Hargreaves IP, Gegg ME, Richard-Londt A, Brandner S, Waddington SN, Schapira AH, Duchen MR. Cell Metab. 2013;17:941. doi: 10.1016/j.cmet.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sardi SP, Clarke J, Viel C, Chan M, Tamsett TJ, Treleaven CM, Bu J, Sweet L, Passini MA, Dodge JC, Yu WH, Sidman RL, Cheng SH, Shihabuddin LS. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3537. doi: 10.1073/pnas.1220464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sybertz E, Krainc DJ. Lipid Res. 2014;55:1996. doi: 10.1194/jlr.R047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hruska KS, LaMarca ME, Scott CR, Sidransky E. Hum. Mutat. 2008;29:567. doi: 10.1002/humu.20676. [DOI] [PubMed] [Google Scholar]
- 15.Sawkar AR, Cheng WC, Beutler E, Wong CH, Balch WE, Kelly JW. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15428. doi: 10.1073/pnas.192582899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawkar AR, Schmitz M, Zimmer KP, Reczek D, Edmunds T, Balch WE, Kelly JW. ACS Chem. Biol. 2006;1:235. doi: 10.1021/cb600187q. [DOI] [PubMed] [Google Scholar]
- 17.Butters TD, Dwek RA, Platt FM. Glycobiology. 2005;15:43R. doi: 10.1093/glycob/cwi076. [DOI] [PubMed] [Google Scholar]
- 18.Zheng W, Padia J, Urban DJ, Jadhav A, Goker-Alpan O, Simeonov A, Goldin E, Auld D, LaMarca ME, Inglese J, Austin CP, Sidransky E. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13192. doi: 10.1073/pnas.0705637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marugan JJ, Zheng W, Motabar O, Southall N, Goldin E, Westbroek W, Stubblefield BK, Sidransky E, Aungst RA, Lea WA, Simeonov A, Leister W, Austin CP. J. Med. Chem. 2011;54:1033. doi: 10.1021/jm1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marugan JJ, Huang W, Motabar O, Zheng W, Xiao J, Patnaik S, Southall N, Westbroek W, Lea WA, Simeonov A, Goldin E, Debernardi MA, Sidransky E. Med Chem Comm. 2012;3:56. doi: 10.1039/C1MD00200G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tropak MB, Kornhaber GJ, Rigat BA, Maegawa GH, Buttner JD, Blanchard JE, Murphy C, Tuske SJ, Coales SJ, Hamuro Y, Brown ED, Mahuran DJ. Chem Bio Chem. 2008;9:2650. doi: 10.1002/cbic.200800304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W, Zheng W, Urban DJ, Inglese J, Sidransky E, Austin CP, Thomas C. J. Bioorg. Med. Chem. Lett. 2007;17:5783. doi: 10.1016/j.bmcl.2007.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazzulli JR, Zunke F, Tsunemi T, Toker NJ, Jeon S, Burbulla LF, Patnaik S, Sidransky E, Marugan JJ, Sue CM, Krainc D. J. Neurosci. 2016;36:7693. doi: 10.1523/JNEUROSCI.0628-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng J, Chen L, Schwake M, Silverman RB, Krainc D. J. Med. Chem. 2016;59:8508–8520. doi: 10.1021/acs.jmedchem.6b00930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lieberman RL, D’Aquino JA, Ringe D, Petsko GA. Biochemistry. 2009;48:4816. doi: 10.1021/bi9002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieberman RL, Wustman BA, Huertas P, Powe AC, Jr, Pine CW, Khanna R, Schlossmacher MG, Ringe D, Petsko GA. Nat. Chem. Biol. 2007;3:101. doi: 10.1038/nchembio850. [DOI] [PubMed] [Google Scholar]
- 27.Cullen V, Sardi SP, Ng J, Xu YH, Sun Y, Tomlinson JJ, Kolodziej P, Kahn I, Saftig P, Woulfe J, Rochet JC, Glicksman MA, Cheng SH, Grabowski GA, Shihabuddin LS, Schlossmacher MG. Ann. Neurol. 2011;69:940. doi: 10.1002/ana.22400. [DOI] [PubMed] [Google Scholar]
- 28.Grabowski GA. Hematology Am. Soc. Hematol. Educ. Program. 2012;2012:13. doi: 10.1182/asheducation-2012.1.13. [DOI] [PubMed] [Google Scholar]
- 29.Choi S, Connelly S, Reixach N, Wilson IA, Kelly JW. Nat. Chem. Biol. 2010;6:133. doi: 10.1038/nchembio.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura T, Kawai Y, Kitamoto N, Osawa T, Kato Y. Chem. Res. Toxicol. 2009;22:536. doi: 10.1021/tx8003906. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Murria MJ, Valero ML, del Pino MMS. J. Proteomics. 2011;74:137. doi: 10.1016/j.jprot.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Mueller OT, Rosenberg A. J. Biol. Chem. 1977;252:825. [PubMed] [Google Scholar]
- 33.Reczek D, Schwake M, Schroder J, Hughes H, Blanz J, Jin XY, Brondyk W, Van Patten S, Edmunds T, Saftig P. Cell. 2007;131:770. doi: 10.1016/j.cell.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 34.Gruschus JM, Jiang Z, Yap TL, Hill SA, Grishaev A, Piszczek G, Sidransky E, Lee JC. Biochem. Biophys. Res. Commun. 2015;457:561. doi: 10.1016/j.bbrc.2015.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belov ME, Damoc E, Denisov E, Compton PD, Horning S, Makarov AA, Kelleher NL. Anal. Chem. 2013;85:11163. doi: 10.1021/ac4029328. [DOI] [PubMed] [Google Scholar]
- 36.Fellers RT, Greer JB, Early BP, Yu X, LeDuc RD, Kelleher NL, Thomas PM. Proteomics. 2015;15:1235. doi: 10.1002/pmic.201570050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strohalm M, Kavan D, Novak P, Volny M, Havlicek V. Anal. Chem. 2010;82:4648. doi: 10.1021/ac100818g. [DOI] [PubMed] [Google Scholar]
- 38.Dvir H, Harel M, McCarthy AA, Toker L, Silman I, Futerman AH, Sussman JL. EMBO Rep. 2003;4:704. doi: 10.1038/sj.embor.embor873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de la Rosa-Trevin JM, Oton J, Marabini R, Zaldivar A, Vargas J, Carazo JM, Sorzano CO. J. Struct. Biol. 2013;184:321. doi: 10.1016/j.jsb.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 40.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. J. Struct. Biol. 2005;151:41. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. J. Struct. Biol. 2009;166:205. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheres SH. J. Mol. Biol. 2012;415:406. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.