Abstract
Purpose
Advanced stage ovarian clear cell carcinoma (OCCC) is unresponsive to conventional platinum-based chemotherapy. Frequent alterations in OCCC include deleterious mutations in the tumor suppressor ARID1A and activating mutations in the PI3K subunit PIK3CA. In this study, we aimed to identify currently unknown mutated kinases in OCCC patients and test druggability of downstream affected pathways in OCCC models.
Experimental Design
In a large set of OCCC patients (n=124), the human kinome (518 kinases) and additional cancer related genes were sequenced and copy number alterations were determined. Genetically characterized OCCC cell lines (n=17) and OCCC patient-derived xenografts (n=3) were used for drug testing of ERBB tyrosine kinase inhibitors erlotinib and lapatinib, the PARP inhibitor olaparib and the mTORC1/2 inhibitor AZD8055.
Results
We identified several putative driver mutations in kinases at low frequency that were not previously annotated in OCCC. Combining mutations and copy number alterations, 91% of all tumors are affected in the PI3K/AKT/mTOR pathway, the MAPK pathway or the ERBB family of receptor tyrosine kinases and 82% in the DNA repair pathway. Strong p-S6 staining in OCCC patients suggests high mTORC1/2 activity. We consistently found that the majority of OCCC cell lines are especially sensitive to mTORC1/2 inhibition by AZD8055 and not towards drugs targeting ERBB family of receptor tyrosine kinases or DNA repair signaling. We subsequently demonstrated the efficacy of mTORC1/2 inhibition in all our unique OCCC patient-derived xenograft models.
Conclusions
These results propose mTORC1/2 inhibition as an effective treatment strategy in OCCC.
Keywords: kinome sequencing, ovarian clear cell carcinoma, copy number alterations, mTORC1/2, PDX models
Introduction
In the United States, ovarian cancer is the fifth-leading cause of cancer deaths in women (1). Ovarian clear cell carcinoma (OCCC) is the second most common subtype of epithelial ovarian cancer. The majority of OCCC patients are diagnosed at an early stage (57-81% at stage I/II) and have better overall survival compared to stage matched high-grade serous (HGS) ovarian cancer, the most common subtype of ovarian cancer. In contrast, OCCC patients diagnosed at late stage respond poorly to standard platinum-based chemotherapy compared to late stage HGS ovarian carcinoma patients (2). In recent years, genetic studies in relatively small patient groups have revealed the mutational landscape in OCCC. The SWI-SNF chromatin remodeling complex DNA binding AT-rich interactive domain 1A gene (ARID1A) has been shown to be deleteriously mutated in 40-57% of OCCC patients, the highest percentage found in any cancer (3, 4). Loss of ARID1A protein, being a key component of the complex, may affect the expression of many genes (5). Activation of the PI3K/AKT/mTOR pathway, implicated in survival, protein synthesis and proliferation, is another major player in OCCC. PIK3CA, encoding the catalytic domain of PI3K, contains activating mutations in 30-40% of OCCC patients, whereas expression of the PI3K antagonist PTEN is diminished in 40% of OCCC patients (4, 6, 7). Furthermore, mutations in the oncogene KRAS and the tumor suppressor gene TP53 have been identified in 4.7%-14% and 10%-15% of OCCC patients, respectively (1, 4, 6, 8, 9). In addition to mutational aberrations, copy number alterations (CNA) have been found in OCCC tumor samples in the proto oncogene ZNF217, tumor suppressor genes, cyclin dependent kinase inhibitors CDKN2A and CDKN2B and the membrane receptor oncogene MET (10–12).
The identification of the most frequently mutated genes ARID1A and PIK3CA may lead to new therapeutic strategies. In particular, the effects of ARID1A loss are being investigated and vulnerabilities in ARID1A mutant cancers are being identified. Synthetic lethal interactions have recently been demonstrated in ARID1A mutant OCCC cancer cell lines by shRNA mediated inhibition of ARID1B, a homolog of ARID1A, as well as chemical inhibition of the histone H3 methyltransferase EZH2 and histone deacetylase HDAC6 (13–15). PI3K signaling-mediated tumor addiction through the well-studied hypermorphic mutant forms of PIK3CA (E545* and H1047*) was studied extensively in multiple cancer types including OCCC. Recent translational research in OCCC cell lines demonstrated sensitivity to PI3K/mTOR dual inhibition and AKT inhibition, although PIK3CA mutations did not predict sensitivity to these inhibitors (16, 17).
In the present study, we aimed to identify novel targetable mutations by means of high-coverage sequencing of all protein kinase genes, referred to as the kinome, and of a subgroup of cancer-related genes in a large set of OCCC. In addition, we determined copy number gains and losses in kinases and other genes of OCCC tumors using high-coverage single nucleotide polymorphism (SNP) arrays. To detect kinase mutations and CNA at both high and low frequency, we used a large cohort of 124 untreated primary OCCC tumors and most of the available OCCC cell lines (n=17). Finally, we functionally validated several candidate targets in OCCC cell lines and unique OCCC patient-derived xenograft (PDX) models. Our results indicate mTORC1/2 inhibition as an approach to guide future development of therapeutic strategies for OCCC.
Methods
Sample collection
Primary tumor samples from 124 OCCC patients and 47 paired control blood samples were prospectively collected from Belgium, Germany, Norway, Poland, The Netherlands, UK and USA. All patients gave written informed consent for samples to be collected and the corresponding ethical review boards approved the study. Tumor samples had to contain ≥40% tumor cells, of which ≥70% was OCCC, as determined by experienced gynecologic oncology pathologists. We obtained 17 human OCCC cell lines: TOV21G (ATCC, USA); RMG1, RMG2, OVMANA, OVTOKO and HAC2 (JCRB Cell Bank, Japan); JHOC5 (RIKEN Cell Bank, Japan); OVCA429 (Cell Biolabs, USA); OVSAYO, TUOC1, KK, OVAS, SMOV2 and KOC7C (Dr. Hiroaki Itamochi, Tottori University School of Medicine, Tottori, Japan); ES2 (Dr. Els Berns, Erasmus MC, Rotterdam, The Netherlands); TAYA (Dr. Yasushi Saga, Jichi Medical University, Yakushiji, Shimotsuke-shi, Tochigi, Japan) and OV207 (Dr. Vijayalakshmi Shridhar, Mayo Clinic, Rochester, MN, USA). All cells were maintained in RPMI supplemented with 10% fetal calf serum. All the cell lines were tested by STR profiling and tested as mycoplasma free. All cells were kept in culture for a maximum of 50 passages.
Kinome sequencing
Library construction, exome capture and sequencing
From 124 primary fresh frozen OCCC tumors and 47 paired controls, 3 µg DNA was prepared for sequencing using the following steps. Genomic DNA was sheared to produce 300 bp fragments (Covaris S220 USA); using SureSelect Target enrichment & Human Kinome Kit (Agilent technologies®, USA) kinase exons were tagged and captured; using biotinylated RNA library baits and streptavidin beads, exons were amplified and loaded on a HiSeq2500 Illumina sequencer using paired-end sequencing according to manufacturer’s protocols. The SureSelect Human Kinome Kit captures exons from 518 kinases, 13 diglyceride kinases, 18 PI3K domain and regulatory component genes and 48 cancer related genes (Supplementary Table 1). After sequencing, raw data was mapped to the human reference sequence NCBI build 37 (hg19) and processed according to our sequencing pipeline (Supplementary Fig. 1). Genome Analysis Toolkit (GATK, version 1.0.5069) was used for indel re-alignment and base quality recalibration on BAM files. See supplementary methods for further details on kinome sequencing.
SNP array
SNP genotyping, quality control
Genome-wide SNP genotyping was performed with HumanOmniExpressExome-8BeadChip (Illumina, USA) containing >900K SNPs, including >273K functional exomic markers to determine CNA in 108 primary OCCC tumors and 17 OCCC cell lines. DNA sample processing, hybridization, labeling, scanning and data extraction was performed according to Illumina infinium 2 protocol. Illumina GenomeStudio software was used for primary sample assessment and SNP call rate quality control of SNP intensity output files. See supplementary methods for further details on SNP array analysis.
In vitro inhibitor screening
The 17 OCCC cell lines were plated in 384 well plates with 250-2000 cells/well using an automatic cell dispenser. Cell lines were incubated with varying concentrations of AZD8055 (Axon Medchem, The Netherlands), gefitinib (Selleckchem, USA), lapatinib (Selleckchem, USA) or olaparib (Axon Medchem, The Netherlands) for 6 days. Cell viability was subsequently monitored using CellTiter-Blue cell viability assay (Promega, USA). Due to large cell density dependent variability in drug sensitivity, for each cell line the lowest and highest IC50 were removed. The cell lines CAPAN1, OE19, LOVO, OVCAR3, OVCAR4 and SKOV3 were exposed to AZD8055 to serve as sensitive and resistant controls.
AZD8055 in vivo administration
All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Groningen (Groningen, the Netherlands) and carried out in accordance with the approved guideline “code of practice: animal experiments in cancer research” (Netherlands Inspectorate for Health Protection, Commodities and Veterinary Public Health, 1999). See supplementary methods for further details on AZD8055 in vivo administration.
Additional methods
K-means consensus clustering of kinome mutations and CNA, mRNA level determination, flow cytometry, MTT assays, long-term proliferation assays, Western blotting, immunohistochemical analysis, data visualization and statistical analysis are described in the supplementary methods.
Results
Kinome sequencing analysis
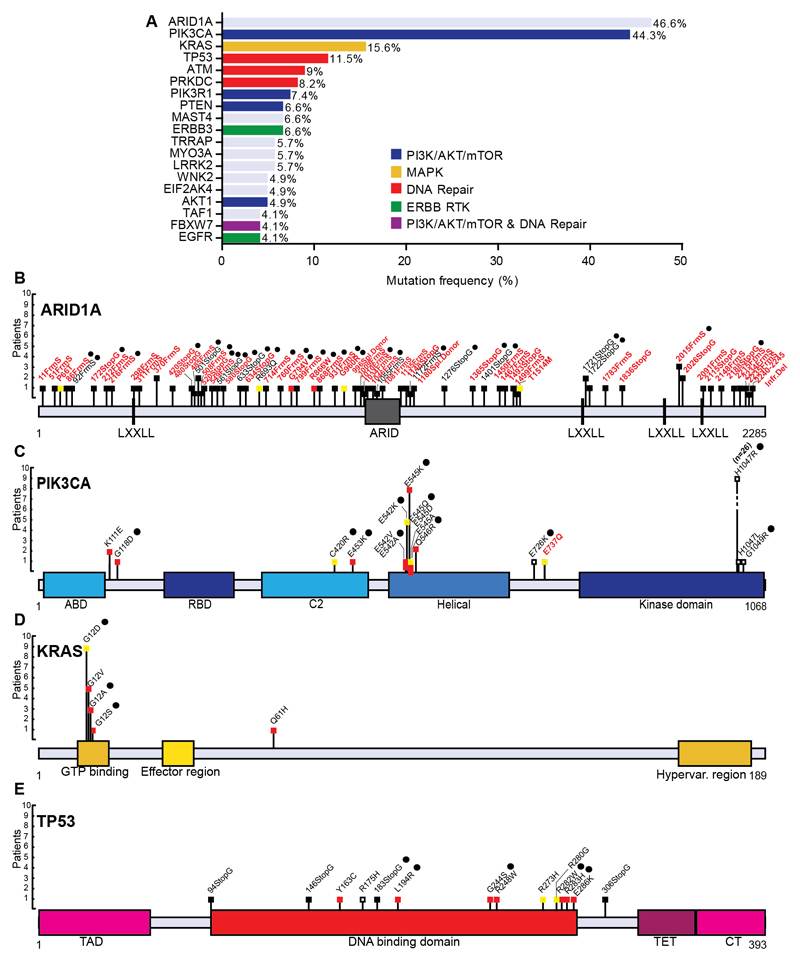
Across all OCCC tumors (n=124) and OCCC cell lines (n=17), 95.9% of all bases had >20 read coverage for variant calling. The mean coverage depth for aligned reads was 99.3x. On average 1.17% of the sequenced genes were mutated per patient. Genes with a high mutation frequency (>4%) across all OCCC tumors are shown in Figure 1A. Re-sequencing of these 19 genes using Haloplex confirmed 227 out of the 234 mutations (97%) that were originally identified by kinome sequencing using the same tumor DNA. The majority of genes with a high mutation frequency are implicated in well-known cancer related pathways like the PI3K/AKT/mTOR pathway (PIK3CA, PTEN, PIK3R1 and AKT1), MAPK signaling transduction pathway (KRAS), DNA repair pathway (TP53, ATM and PRKDC), ERBB family of receptor tyrosine kinase genes (ERBB3 and EGFR) and chromatin remodeling genes (ARID1A). The frequencies of previously described mutated genes observed in our study were in agreement with such frequencies reported earlier in smaller studies of OCCC (Fig. 1A) (3, 4, 6, 8, 9).
Figure 1. Most frequent OCCC mutations.
(A) Frequently mutated genes in OCCC as identified by kinome sequencing in 122 OCCC tumors using a 4% cutoff. ARID1A mutations (n=54 tumors) were revealed using haloplex sequencing on 116 OCCC tumors. Mutated genes involved in PI3K (PIK3CA, PIK3R1, PTEN, AKT1 and FBXW7, n=54, n=9, n=8, n=6, n=5 tumors, respectively), MAPK (KRAS, n=19 tumors), or DNA repair signaling (TP53, ATM, PRKDC and FBXW7, n=14, n=11, n=10 and n=5 tumors, respectively) and ERBB family of receptor tyrosine kinases (ERBB3 and EGFR, n=8 and n=5 tumors) are among the frequently mutated genes. Schematics of identified mutations in known OCCC mutated genes (B) ARID1A, (C) PIK3CA, (D) KRAS and (E) TP53. Mutation marks are shown in black (truncating), red (SIFT and PolyPhen damaging prediction), yellow (SIFT or PolyPhen damaging prediction) or white (SIFT and PolyPhen benign prediction). Mutation effects are indicated with a black spot when paired control was available and written in black (previously described mutation) or red (novel mutations).
ARID1A mutations were analyzed using Haloplex sequencing only. Out of 54 ARID1A mutant tumors, 18 tumors contained homozygous frameshift or stop-gain mutations, 13 tumors harbored more than one heterozygous mutation, while 23 tumors contained a single heterozygous frameshift or stop-gain mutation (Fig. 1B). The identified proportion of each type of mutation in ARID1A matches those reported in earlier studies (3, 4). ARID1A mutant and wild type tumors did not show differential mutation incidence in kinome genes (7.4 vs. 6.6 mutations per tumor on average).
Statistical binomial univariate testing of all 851 identified kinome mutations revealed 11 significantly mutated genes (p<0.05) relative to background mutations. These predicted oncogenic drivers were PIK3CA, KRAS, TP53, PTEN, AKT1, PIK3R1, FBXW7, ERBB3, ATM, CHEK2 and MYO3A (Supplementary Table 3). PIK3CA and KRAS exhibited mutations in established hotspot sites in aa Q542, Q545 and H1047 in PIK3CA and G12 in KRAS, while TP53 mutations were distributed across the TP53 DNA binding domain (Fig. 1C-E).
Interestingly, we identified mutations in 3 genes not previously described in OCCC (AKT1, PIK3R1 and ERBB3) and with an established role in PI3K/AKT/mTOR pathway activation. AKT1 missense mutations were identified in 6 tumors (4.9%). AKT1 is one of the key components of the PI3K/AKT/mTOR cascade. The PH domain of AKT1 interacts with its kinase domain and maintains the protein in a closed and inactive state (18). One candidate somatic mutation, D323N, was located in the kinase domain. The mutations R25H, L52R (occurring in 3 tumors) and W80R (both described in COSMIC) were all located in the AKT1 PH domain (Supplementary Fig. 2A). PIK3R1 was mutated in 9 tumors (7.3%), of which 7 tumors carried mutations in the inter-SH2-1 SH2-2 domain of the protein (aa 429-623). This domain binds PIK3CA and is required for the inhibitory role of PIK3R1 on PIK3CA (19). Two of the inter-SH2 domain mutations are described in COSMIC, E439* and T576* in-frame deletions. In addition, we identified 6 novel inter-SH2 domain mutations; two somatic missense (aa N453D and T471S, together in one tumor), two somatic in-frame deletions (aa QF455* and RE461*), one candidate somatic frameshift (starting in aa 582) and one candidate somatic stop-gain mutation (aa 571) (Supplementary Fig. 2B). In ERBB3 we identified 10 missense mutations across 8 distinct tumors (5.7%). At the cell membrane ERBB3 can dimerize with other ERBB family members and regulate downstream kinase signaling. Six mutations were located across the extracellular domains of ERBB3 and two could be identified as somatic. The D297Y mutation has been described as a hotspot location in ovarian and colorectal cancer (20). Furthermore, 3 mutations were in the intracellular C-terminal domain and one in the kinase domain (Supplementary Fig. 2C). Of the other significantly mutated genes, F-Box and WD Repeat Domain Containing 7 (FBXW7) has been described in PI3K regulation, DNA repair and mutations are mainly found in its WD repeats (21). ATM Serine/Threonine Kinase (ATM) and Checkpoint kinase 2 (CHEK2) are designated as DNA repair genes whereas Myosin IIIA (MYO3A) is an actin-dependent motor protein. Mutations in FBXW7 and ATM were previously described in OCCC (Supplementary Fig. 2D-G) (22).
Pathway analysis of kinome mutations
We also investigated the pathways in which genes with a high mutation frequency are involved. To this end, all low mutation frequency genes (<4%) present in PI3K/AKT/mTOR, MAPK and DNA repair pathway genes, the ERBB family of receptor tyrosine kinases and mutations in ARID1A were mapped per tumor in order to determine mutational spectra across these signaling pathways in OCCC. One or more mutations were found in PI3K/AKT/mTOR-related genes in 76 patients (62.3%), in MAPK related genes in 22 patients (18%), in the ERBB family of receptor tyrosine kinases in 18 patients (14.8%) and in DNA repair pathway genes in 46 patients (37.7%). The co-occurrence of mutations in both ARID1A and PIK3CA in this large set of OCCC tumors was in accordance with the findings from a previous, smaller study (Supplementary Fig. 3A) (4, 23). PIK3R1 mutations were mutually exclusive with PIK3CA mutant tumors (p=0.043) and a trend was observed for PTEN with PIK3CA mutant tumors (p=0.0756). In general, TP53 mutant tumors (including pure OCCCs) were mutually exclusive with both ARID1A and PIK3CA mutant tumors (p=0.0031), in agreement with previous literature (5). Surprisingly, co-occurrence of TP53 and ARID1A or PIK3CA mutations was observed in a few cases. In total 108 out of 122 tumors (89%) comprised one or more mutations in genes belonging to the PI3K/AKT/mTOR pathway, MAPK pathway, DNA repair pathway and ERBB family of receptor tyrosine kinases (Supplementary Fig. 3A).
Evidently, there is a large overlap in tumors that were both ARID1A mutant and harbored a mutation in DNA repair genes or PI3K/AKT/mTOR, MAPK and ERBB family of receptor tyrosine kinase genes (Supplementary Fig. 3B). Subdividing tumors into ARID1A heterozygous-mutated and homozygous-mutated tumors did not change the overlap in mutations in PI3K/AKT/mTOR, MAPK, DNA repair, or ERBB family of receptor tyrosine kinase genes in both groups (data not shown). ARID1A wild type tumors more frequently contained mutations in DNA repair genes in addition to mutations in PI3K/AKT/mTOR, MAPK or the ERBB family of receptor tyrosine kinase genes as compared to ARID1A mutant tumors (49% vs. 26%, p=0.0385).
CNA analysis
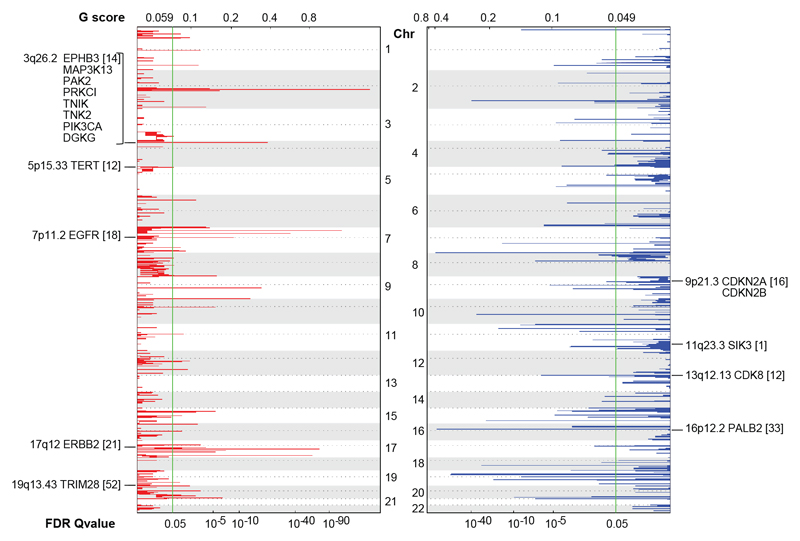
In 108 SNP genotyped primary OCCC tumors, GISTIC analysis annotated 324 significantly amplified genes located in 48 focal regions. We identified amplification of ZNF217 (20q13.20), a transcriptional regulator previously described in OCCC (11), in 29 tumors (27%, p=0.014). Using GISTIC analysis, we subsequently found multiple kinases and other cancer-related genes among the significantly amplified genes (Fig. 2).
Figure 2. Kinase CNA in OCCC.
Significant CNA in kinases across 108 OCCC tumors as determined by GISTIC analysis. All kinases and cancer related genes from the kinome sequencing gene panel that were focally significantly amplified (red) or deleted (blue) are indicated along the chromosomes vertically. Chromosomal location and total amount of tumors (between brackets) harboring the event are annotated with each gene name. The false-discovery rate (FDR) 0.05 threshold, indicated by the green line, and G-score are shown along the horizontal axis.
One recurrently amplified region (3q26.2) contained multiple kinases; PIK3CA, EPHB3, MAPK3K13, PAK2, PRKCI, TNIK, TNK2 and DGKG (14 tumors, 13%, p=0.046). The cancer-related gene TERT (5p15.33), included in kinome sequencing, was amplified in 12 tumors (11%, p=0.044). EGFR (7p11.2) emerged as the most significant recurrently amplified kinase; it was present in 18 OCCC tumors (17%, p<0.0001). Other recurrently amplified kinases included ERBB2 (17q12), which was amplified in 21 tumors (19%, p=0.002), and the chromatin-associated and transcriptional control-related kinase TRIM28 (19q13.43), which was amplified in 52 tumors (48%, p=0.0035).
Furthermore, a total of 118 significantly deleted genes were identified in 62 focally deleted regions. Two significantly deleted kinases could be identified: SIK3 (11q23.3) and the transcriptional repressor CDK8 (13q12.13) (Fig. 2). In addition, 3 known cancer related genes included in kinome sequencing were significantly deleted. The cell cycle regulators CDKN2A and CDKN2B (both located on 9p21.3) were deleted in 16 tumors (15%, p=0.024) and PALB2 (16p12.2), an essential chaperone of BRCA2, was deleted in 33 tumors (31%, p<0.0001).
A separate GISTIC analysis was implemented to compare ARID1A mutant tumors (n=45) with ARID1A wild type (n=63) tumors (Supplementary Fig. 4A). TRIM28 amplification and CDK8 deletion were significantly retained in only ARID1A mutant tumors (p<0.0001 and p=0.0067, respectively), whereas EGFR amplification was significantly retained in only ARID1A wild type tumors (p=0.0054) (Supplementary Fig. 4B-C).
Integration of kinome mutations and CNA
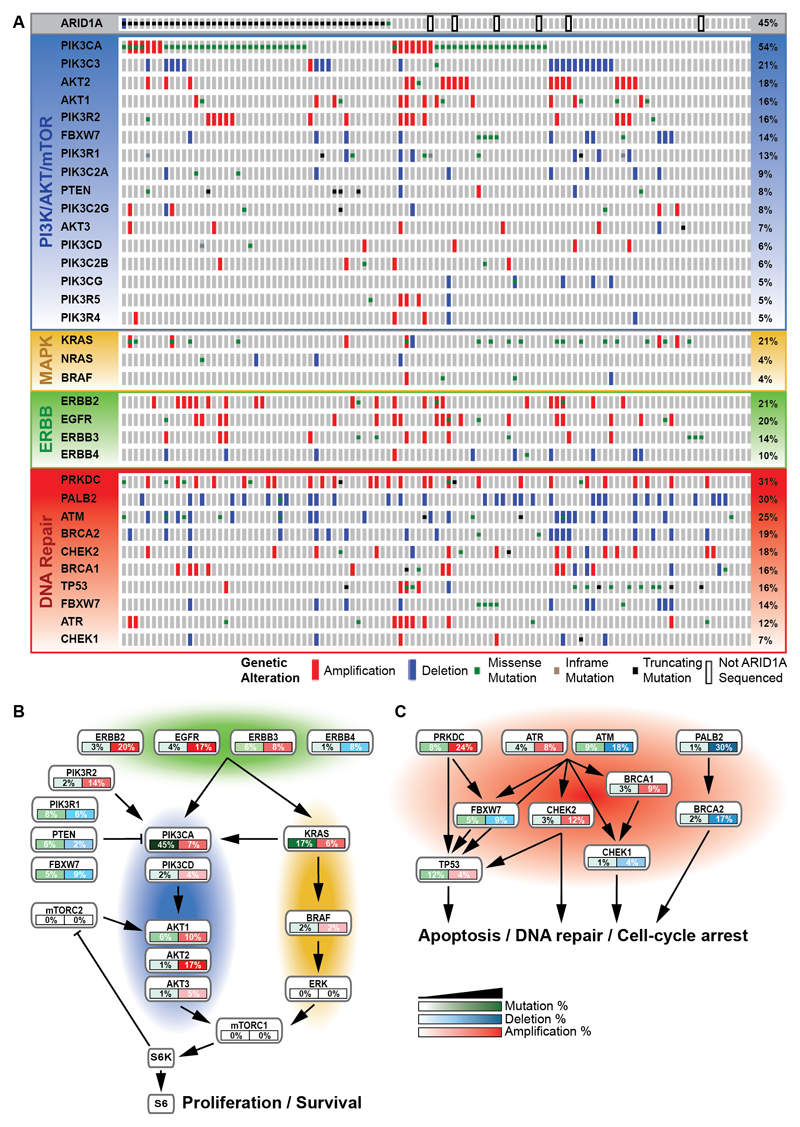
Both kinome sequencing and SNP data were available for 106 tumors. After merging all mutations and CNA events in the PI3K/AKT/mTOR, MAPK and DNA repair pathway and ERBB family of receptor tyrosine kinases, we identified at least one event in 103 of 106 tumors analyzed (97%) (Fig. 3A).
Figure 3. Mutation and CNA distribution.
(A) Nonsynonymous mutation distribution in genes involved in the frequently mutated PI3K/AKT/mTOR (blue) and MAPK pathway (yellow), the ERBB family of receptor tyrosine kinases (green) and DNA repair pathway (red) as well as ARID1A and PALB2 are shown with OncoPrint. CNA in each mutated gene are added. The 106 OCCC tumors that were both kinome sequenced and SNP arrayed are shown on the horizontal axis ordered on total event frequency in the subsequently altered pathways. (B) As determined by kinome sequencing and SNP array analysis, the interacting ERBB family of receptor tyrosine kinases, PI3K/AKT/mTOR and MAPK pathway and (C) DNA repair pathway are commonly altered. These alterations are defined by mutations and CNA.
Amplification incidence of >10% was found in the PI3K/AKT/mTOR-related genes AKT2 (17%), PIK3R2 (14.2%), PIK3CA (12.3%), AKT1 (10.4%), the ERBB family of receptor tyrosine kinases ERBB2 (19.8%) and EGFR (17%), and the DNA repair genes PRKDC (24%) and CHEK2 (12.3%). AKT2, PIK3R2, ERBB2, EGFR and CHEK2 primarily contained amplifications, from which only ERBB2 (n=2) and EGFR (n=1) presented tumors that were both amplified and mutated. PIK3CA was mostly mutated, yet 4 PIK3CA mutant tumors carried both an amplification and a mutation. Amplifications and mutations never co-occurred in AKT1 and PRKDC.
Deletion incidence of >10% was observed in the PI3K/AKT/mTOR-related gene PIK3C3 (18.9%) and the DNA repair genes PALB2 (30.2%), ATM (17.9%) and BRCA2 (17%). PIK3C3, PALB2 and BRCA2 primarily harbored amplifications, while ATM amplifications and mutations co-occurred in 3 tumors.
We hypothesized that the alterations in genes described in Figure 3A can be added up to promote aberrant pathway signaling. Mutations or CNA occurred in PI3K/AKT/mTOR in 84.9% of tumors, in MAPK pathway in 27.4% of tumors and in ERBB receptor family of kinases in 42.5% of tumors. Combined mutations and CNA in PI3K/AKT/mTOR and MAPK pathway genes and the ERBB family of receptor tyrosine kinases indicated that 91% of all tumors were affected (Fig. 3B), while the DNA repair pathway was affected in 82% of all tumors (Fig. 3C).
Kinome profile reveals tumor clusters with differential survival
Clinical data was available for a subset of patients (n=70) (Supplementary Table 4). Disease-specific survival analysis revealed that ARID1A, PIK3CA or ARID1A plus PIK3CA alterations were not related to survival (Supplementary Fig. 5A-C). Kinome mutations and CNA events of all 106 tumors were integrated, and the tumors were grouped using K-means consensus clustering. Maximum cluster number was set at 8, since more clusters only marginally decreased friction (Supplementary Fig. 5D-F). Most tumors grouped together in cluster 1 (n=53), 3 (n=27) or 5 (n=13). A trend for worse disease-specific survival was demonstrated with cluster 3 compared with all other clusters (p=0.0638) (Supplementary Fig. 5G-H), which became highly significant upon selection for advanced stage OCCC patients (p<0.001, n=31) (Supplementary Fig. 5I). Grouping of mutation and CNA status for each cluster did not reveal unique genes in cluster 3 (Supplementary Fig. 5J-K).
Kinome profile based inhibitor screen identifies mTORC1/2 inhibition susceptibility
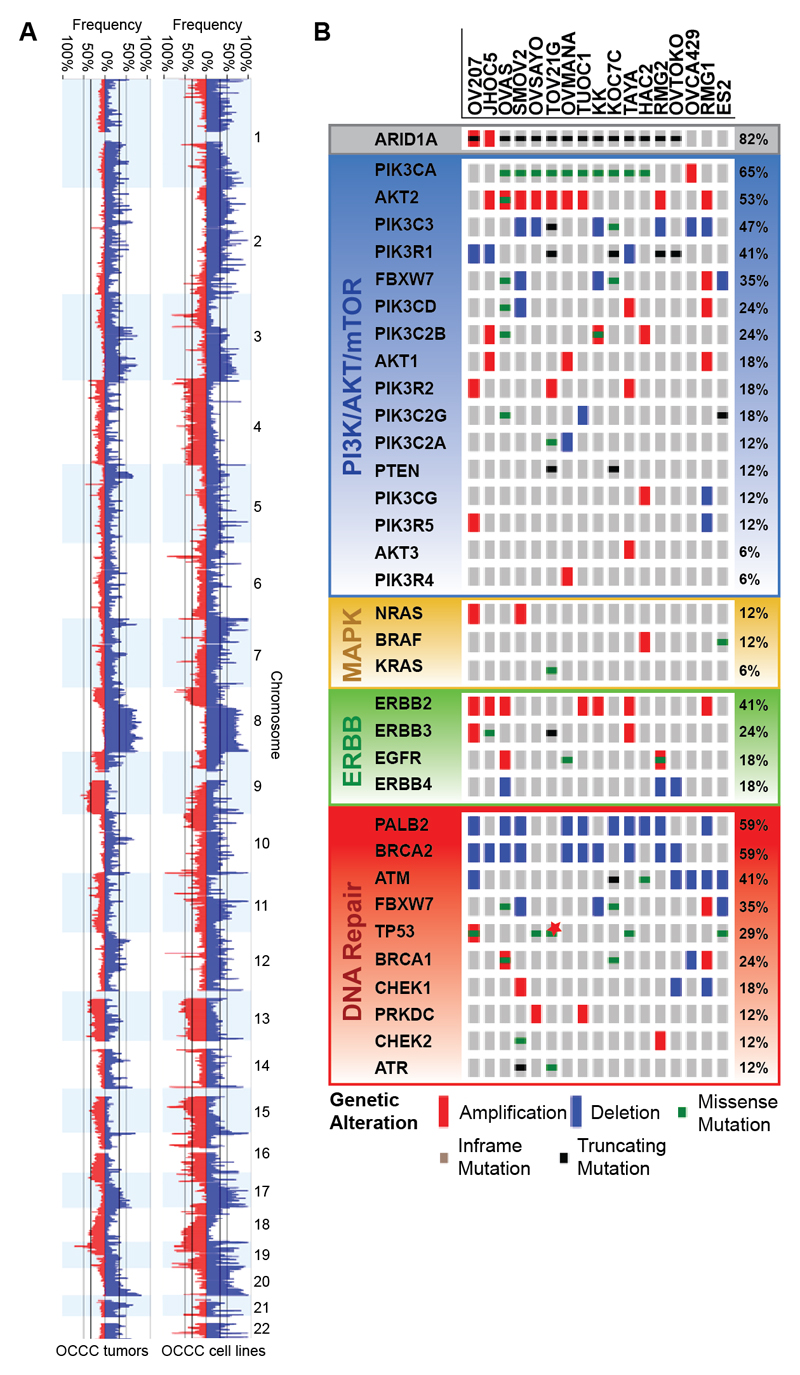
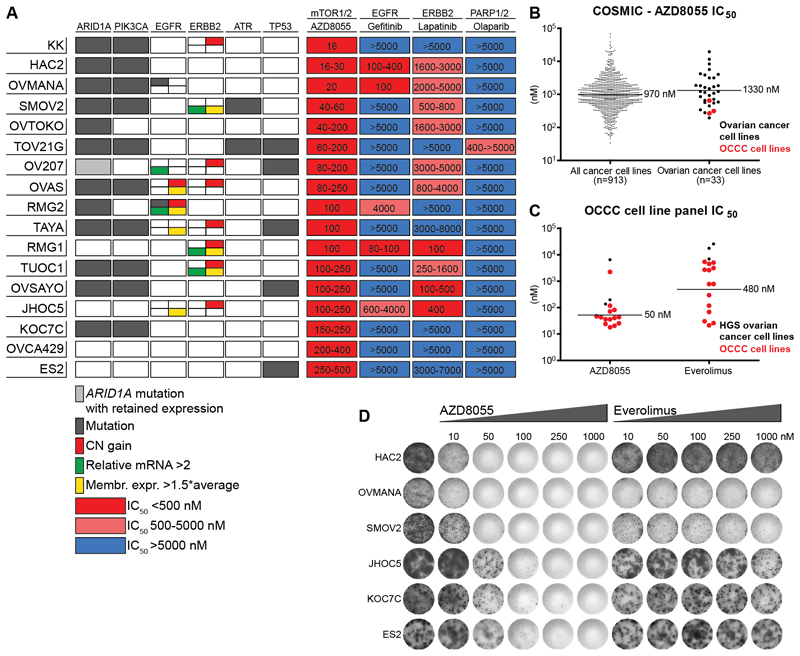
Kinome sequencing and SNP data of 17 OCCC cell lines presented similar alteration frequencies in PI3K/AKT/mTOR, MAPK and DNA repair pathway genes and the ERBB family of receptor tyrosine kinases as those observed in OCCC patients (Fig. 4A-B). However, an overrepresentation of ARID1A and TP53 mutations and an underrepresentation of KRAS mutations were found. Due to the resemblance of OCCC patient alterations with those in the cell line panel, we decided to screen for kinase inhibition vulnerabilities of the PI3K/AKT/mTOR and MAPK pathway downstream targets mTORC1/2 and the ERBB receptor tyrosine kinases EGFR and ERBB2. In addition, the DNA repair pathway was targeted. High-throughput drug sensitivity testing revealed nanomolar range efficacy against the mTORC1/2 inhibitor AZD8055 in all 17 OCCC cell lines tested (Fig. 5A). Sensitivity appeared to be irrespective of PIK3CA or ARID1A mutation status, suggesting that alterations upstream of mTORC1/2 may explain the comprehensive AZD8055 susceptibility. COSMICs cancer cell line drug screening data (n=913 cell lines) with AZD8055 (Cancerrxgene.org) and temsirolimus (targeting mTORC1) indicate that OCCC cell lines are among the most sensitive ovarian cancer cell lines (Fig. 5B and Supplementary Fig. 6A).
Figure 4. OCCC cell line CNA and mutation distribution.
(A) Chr 1-22 CNA (amplifications in blue, deletions in red) in OCCC cell lines (n=17) and tumors (n=108) depicted from Nexus Copy Number. (B) OCCC cell lines nonsynonymous mutation distribution in genes involved in the frequently mutated PI3K/AKT/mTOR (blue) and MAPK pathway (yellow), ERBB family of receptor tyrosine kinases (green) and DNA repair (red) pathway as well as ARID1A and PALB2 are shown with OncoPrint. CNA in each mutated gene are added. The 17 OCCC cell lines that were both kinome sequenced and SNP arrayed are shown on the horizontal axis ordered on total event frequency in the subsequently altered pathways. JHOC5 and OV207 are ARID1A mutant but retain ARID1A expression. The genes AKT2, PIK3C3, PIK3CD, PIK3C2B, PIK3R2, PIK3C2G, PIK3C2A, PIK3CG, AKT3, PIK3R4, ERBB4, PALB2 and CHEK1 were not sequenced in JHOC5, HAC2 and OVCA429, *TP53 mutation in TOV21G was detected just above threshold.
Figure 5. OCCC cell line panel inhibitor screening.
(A) Schematic representation of mutation status, mRNA level, copy number gain, membrane receptor expression and a heatmap of inhibitor IC50 in 17 OCCC cell lines. For each cell line horizontally: mutation status of the genes ARID1A, PIK3CA, EGFR, ATR and TP53 are indicated in grey, EGFR and ERBB2 mRNA level relative to GAPDH >2 are indicated in green, copy number gain is indicated in red, EGFR and ERBB2 membrane expression relative to average mean fluorescence intensity >2 are indicated in yellow. Cell line IC50 for AZD8055 (mTORC1/2), gefitinib (EGFR), lapatinib (ERBB2) and olaparib (PARP1/2) are shown horizontally, and cell lines are ordered on AZD8055 sensitivity vertically. (B) IC50 of the mTORC1/2 inhibitor AZD8055 from COSMICs (Cancerxgene.org) drug screening database in all cancer cell lines vs. ovarian cancer cell lines, horizontal lines indicate geometric mean. (C) AZD8055 and everolimus IC50 determined for 14 OCCC cell lines (ES2, KOC7C, SMOV2, JHOC5, RMG1, OVMANA, HAC2, OV207, OVTOKO, TOV21G, OVAS, OVCA429, TUOC1 and RMG2) by MTT assay. HGS ovarian cancer cell lines PEA1, PEO14 and OVCAR3 were used as a resistant control, horizontal lines indicate geometric mean of only the OCCC cell lines. Data is derived from n≥2 experiments. (D) Long-term proliferation assay after exposure to increasing concentrations of AZD8055 and everolimus. Results are representative of n=3 experiments.
MTT assay-based IC50 determination in 14 out of 17 OCCC cell lines demonstrated higher susceptibility to AZD8055 and the mTORC1 inhibitor everolimus in comparison to 3 HGS ovarian cancer cell lines (Fig. 5C). In contrast to AZD8055, proliferation inhibition by everolimus displayed a plateau phase in a wide concentration range and a large variation in sensitivity among OCCC cell lines (Fig. 5D and Supplementary Fig. 6B). Moreover, everolimus treatment resulted in increased p-AKT473 levels, while AZD8055 and MLN0128, another mTORC1/2 inhibitor, reduced this upregulation (Supplementary Fig. 6C). AZD8055 and dactolisib, an mTORC1/2-PI3K inhibitor for which OCCC cell lines are highly sensitive (Supplementary Fig. 6A), strongly reduced p-AKT473 and to a lesser extent p-AKT308 at high concentrations (Supplementary Fig. 6D). The absence of PARP cleavage indicated that everolimus, AZD8055 and dactolisib do not induce apoptosis in an in vitro setting (Supplementary Fig. 6D). OCCC cell lines presented a large IC50 variation for upstream inhibitors of the PI3K/AKT/mTOR and MAPK pathway, e.g. PI3K inhibitor GDC0941 and MEK inhibitor selumetinib, compared with AZD8055 and dactolisib (Supplementary Fig. 6E). Long-term proliferation inhibition by dactolisib was stronger compared with AZD8055 (Supplementary Fig. 6F). Limited sensitivity was observed towards the EGFR inhibitor gefitinib and ERBB2/EGFR inhibitor lapatinib. EGFR mutations were observed in two cell lines, but only OVMANA (R705K) displayed gefitinib sensitivity. In cells with elevated levels of ERBB2 mRNA and high ERBB2 membrane expression (RMG1, SMOV2 and TUOC1), lapatinib sensitivity was observed (Fig. 5A). However, some of the tested cell lines without EGFR mutations or elevated ERBB2 levels also displayed sensitivity to gefitinib and lapatinib, indicating involvement of alternative mechanisms. The PARP1 inhibitor olaparib showed some efficacy against ATR mutant TOV21G cells.
In conclusion, OCCC cell lines exhibit exquisite sensitivity to mTORC1/2 inhibitors, whereas limited sensitivity was observed towards ERBB receptor tyrosine kinase and DNA repair inhibitors despite the high aberration frequency in those pathways.
Targeting of mTORC1/2 is effective in PDX models
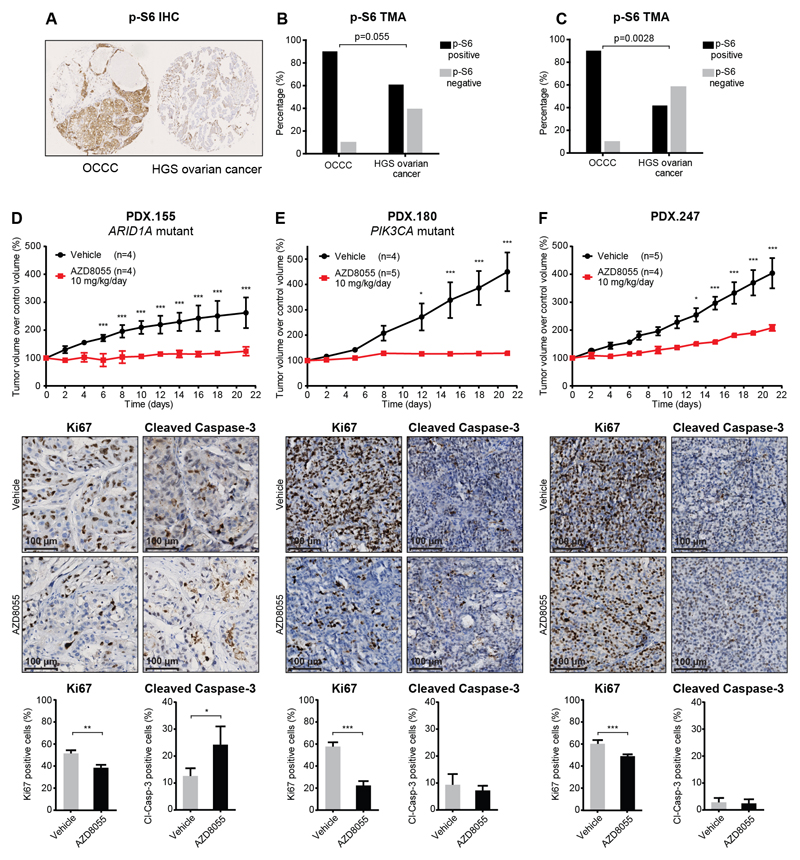
Considering the high susceptibility of OCCC cell lines to AZD8055, we validated mTORC1/2 activity in OCCC tumor samples. Immunostaining of the mTORC1/2 downstream target p-S6 was performed on two different tissue microarrays (TMAs) with primary tumor material from HGS ovarian cancer and OCCC (n=136 and n=83). OCCC tumors were more frequently p-S6 positive compared to HGS ovarian cancer in both datasets (p=0.053 and p=0.0028, respectively) (Fig. 6A-C). Subsequently, we tested AZD8055 efficacy in OCCC PDX-bearing NSG mice. Sequencing of 19 genes with the highest mutation frequency in OCCC patients showed mutations in ARID1A (1148* stop-gain), PTEN (H93R) and BRCA1 (V356A and 1533* stop-gain) in PDX.155, in PIK3CA (K111E, recurrent in OCCC tumors) and ATM (T1020I) in PDX.180 and none in PDX.247. All mutations were present in the sequenced F0 (patient tumor) F1 and F2 generation. GISTIC analysis indicated CNA across the PDX models that resemble CNA in OCCC patients (Supplementary Fig. 7A). Significant growth differences were observed between AZD8055 (10 mg/kg/day) and vehicle-treated mice in all 3 PDX models (Fig. 6D-F). AZD8055 treatment reduced tumor growth in PDX.247 and inhibited tumor growth in PDX.155 and PDX.180, which is reflected in a reduction of proliferation marker Ki67 positivity (Fig. 6D-F). Remarkably, an increased staining of the apoptosis marker Cleaved Caspase-3 was observed in PDX.155 after AZD8055 treatment (Fig. 6D-F). Phosphorylation of mTORC1/2 downstream target S6 was clearly detectable in the vehicle treated tumors but did not significantly change after AZD8055 treatment (Supplementary Fig. 7B-D).
Figure 6. p-S6 expression and mTORC1/2 inhibition in OCCC PDX models.
(A) Representation of p-S6 expression in an OCCC tumor (left) and HGS ovarian cancer tumor (right) as determined by IHC on TMA. (B) Percentage of positive p-S6 expression in primary OCCC tumors (positive n=10, negative n=1) and HGS ovarian cancer tumors (positive n=76, negative n=49), (C) and positive p-S6 expression in a second TMA of primary OCCC (positive n=10, negative n=1) and HGS ovarian cancer tumors (positive n=30, negative n=42), determined by Fisher’s exact test, two-tailed. (D) Tumor growth, Ki67 expression and Cleaved Caspase-3 positivity in PDX.155, (E) PDX.180 and (F) PDX.247 tumors following AZD8055 or vehicle treatment. PDX.155 vehicle mice were treated in F4 generation and AZD8055 mice were treated in F5 generation. Tumor volume is represented as percentage of initial tumor volume at start of treatment. Quantification of Ki67 and Cleaved Caspase-3 positivity in tumors from vehicle and AZD8055 treated mice, sacrificed on day 21, are shown below respective PDX model tumor growth charts. * indicates p<0.05, ** indicates p<0.01 and *** indicates p<0.001.
Discussion
Advanced stage OCCC patients respond poorly to standard platinum-based chemotherapy, indicating an unmet need for novel treatment strategies. Kinome sequencing of 518 kinases and 79 cancer-related genes and SNP array analysis of 108 OCCC tumors revealed mutations, amplifications and deletions in well-known genes such as ARID1A, PIK3CA, TP53 and KRAS and several newly identified genes. Collectively, alterations in genes from the PI3K/AKT/mTOR pathway, the MAPK pathway and the ERBB family of receptor tyrosine kinases were found in 91% of tumors. In line with these alterations, high mTOR signaling was frequently observed in OCCC and mTORC1/2 inhibition was highly effective in OCCC cell lines and PDX models. These results indicate that especially targeting mTOR1/2 could be an effective therapeutic approach for OCCC patients.
ARID1A was the most common mutated gene in our OCCC dataset, with a mutation rate of 46.6%. Overall, the mutation rates observed for ARID1A, PIK3CA, TP53 and KRAS matched earlier sequencing studies on smaller OCCC datasets (3, 4, 6, 8, 9, 24). Moreover, mutation incidences in PIK3CA, TP53 and KRAS correlated with the frequencies found in ‘pure OCCC’ tumors as described by Friedlander et al. (22), indicating that our sample cohort also contains pure OCCC tumors. ARID1A and PIK3CA mutations have been shown as early onset mutations in the development of OCCC (23, 25). Previous studies have shown that onset of oncogenesis regularly requires two or more mutational hits in a proto-oncogene or tumor suppressor gene, as was demonstrated in a de novo OCCC mouse model (26). Indeed, in our tumor cohort, 94% of the PIK3CA and ARID1A mutant tumors contained at least one additional mutation or CNA in PI3K/AKT/mTOR, MAPK or DNA repair pathway genes or the ERBB family of receptor tyrosine kinases, which was stage independent. Considering earlier reports and our observed OCCC mutation pattern, we hypothesize that a mutation in ARID1A or PIK3CA is accompanied by either a CNA or mutation in one of the aforementioned pathways to promote onset of OCCC. A limitation of the present study can be the imbalance in tumor samples and matched controls. Accordingly, we could designate a restricted number of mutations as somatic. Not all clinical data were available and therefore we may have underestimated the relation between kinome alteration status and clinicopathological characteristics or clinical outcome.
Genes mutated at low frequency (1-10%) are generally difficult to designate as oncogenic drivers. Therefore, we used a large OCCC dataset and binomial testing of observed kinome variants. Eleven genes were found to be significantly mutated relative to the background mutation rate, and some of these had not previously been described in OCCC. Validation of variants in these low-mutation-frequency genes across multiple cancer types using the COSMIC database enabled us to predict AKT1, PIK3R1, FBXW7, ERBB3, ATM, CHEK2 and MYO3A as novel drivers of OCCC (27). Functional effects of AKT1 and PIK3R1 mutations in these domains were described in previous research. For instance, mutations in the AKT1 PH-domain were shown to constitutively activate AKT1 (18). Likewise, mutations in the PIK3R1 inter SH1-SH2 domain resulted in loss of PIK3R1 function, thus generating PIK3CA hyperactivation (19). These studies support the assumption that AKT1 and PIK3R1 are oncogenic drivers in OCCC. Furthermore, AKT1, PIK3R1, FBXW7 and ERBB3 all contribute to mTOR signaling, while ATM, CHEK2 and FBXW7 are implicated in DNA repair signaling (19, 21, 28). MYO3A mutations are found infrequently in cancer, but are still implicated in resistance to trastuzumab in breast cancer (29).
In the present study, kinome sequencing and SNP array analysis revealed substantial percentages of EGFR and ERBB2 mutations or amplifications in OCCC tumors. EGFR amplifications (associated with endometriosis, a precursor of OCCC) and ERBB2 amplifications were previously demonstrated in OCCC by comparative genomic hybridization (30, 31). Furthermore, sensitivity towards the clinically available inhibitors gefitinib and lapatinib was shown in OCCC cell lines harboring EGFR or ERBB2 alterations and may be explored further in the clinic (32, 33). In previous studies, EGFR-targeting therapy such as gefitinib or erlotinib and EGFR-ERBB2 targeting therapy using lapatinib did not provide clinical benefit in pretreated ovarian cancer patients (34–36), but these studies did not focus on OCCC patients. Additionally, alterations in DNA repair pathway genes were identified in 82% of tumors, suggesting opportunities to target DNA repair cascades in OCCC in future research. Unfortunately, PARP1/2 inhibition by olaparib was not effective in our OCCC cell line panel, including BRCA1 mutant cell lines. Interestingly, inhibition of PARP1/2 by the PARP trapping agent talazoparib demonstrated efficacy in a subset of OCCC cell lines (37).
Mutations in mTOR have been detected in several tumor types including HGS ovarian cancer (38). Although mTOR mutations were not found in OCCC tumors, alterations in upstream pathways that promote mTORC1/2 activation were found in 91% of these tumors. In line with these results, abundant high p-S6 expression was observed in OCCC tumors compared to HGS ovarian cancer tumors. Moreover, OCCC cell lines demonstrated nanomolar range sensitivity towards the mTORC1/2 inhibitor AZD8055 and higher sensitivity compared with HGS ovarian cancer cell lines. To our knowledge we were the first to exploit the use of PDX models of OCCC. In vivo administration of AZD8055 resulted in a significant growth inhibitory effect in 3 OCCC PDX models, consisting of a PIK3CA mutant, an ARID1A mutant and a PIK3CA and ARID1A wild type model. These models reflect the genetic makeup of OCCC. Hence, we propose mTORC1/2 inhibition as a future treatment strategy in OCCC that should be explored with urgency. Additional inhibition of ARID1A synthetic lethal targets in combination with mTORC1/2 inhibition can be utilized in ARID1A mutant OCCC tumors (14, 15).
In the clinic, inhibition of mTORC1 with temsirolimus added to standard chemotherapy did not improve overall survival in newly diagnosed stage 3-4 OCCC patients as compared to historical controls (39). However, by blocking both mTORC1 and mTORC2, re-activation of PI3K and MAPK signaling via mTORC2 can be prevented in OCCC. This may contribute to a better response in OCCC patients. AZD8055 and another mTORC1/2 inhibitor, OSI-027, were tested in non-OCCC cancer types. Disappointingly, anti-tumor efficacy was only observed above maximum tolerated dose and both drugs are no longer in clinical development (40, 41). Based on our results, it is conceivable that tumor responses in OCCC patients can be attained at or below maximum tolerated dose with AZD8055 or OSI-027. A new generation of mTORC1/2 inhibitors, such as MLN0128, is now being evaluated in multiple phase II clinical trials (NCT02724020 and NCT02725268). Given the large variation in mutations in specific genes of the PI3K/AKT/mTOR and MAPK pathway between patients and the high variation in sensitivity towards PI3K and MEK inhibition in our OCCC cell lines, PI3K or MEK monotherapy tumor response in OCCC patients will presumably be limited. MLN0128 is now being combined with a PI3Kα inhibitor (MLN1117) in an ongoing phase II trial in advanced endometrial cancer (NCT02725268). Drugs targeting PI3K as well as mTORC1/2, e.g. dactolisib, DS-7423 and XL765, showed high toxicity in the clinical setting, despite promising efficacy in in vitro models such as ovarian cancer (16, 42–45). Based on our mutational analysis in OCCC patients, drug sensitivity screens and molecular analyses in OCCC cell lines, a strategy that combines inhibition of mTORC1/2 with PI3K or MEK will have the highest likelihood to improve clinical benefit for the majority of OCCC patients. However, the use of single target inhibitors of mTORC1/2, PI3K and MEK at suboptimal concentrations is probably crucial to maximize efficacy and minimize toxicities that were observed with PI3K-mTORC1/2 dual inhibitors. Overall, our findings set the stage to explore mTORC1/2 targeting phase II clinical trials in OCCC in the future.
Supplementary Material
Translational relevance.
Advanced stage ovarian clear cell carcinoma (OCCC) is less responsive to platinum-based chemotherapy compared to high-grade serous ovarian carcinoma. Our in-depth analyses of a large set of OCCC patients reveal numerous genomic alterations related to activation of mTORC1/2. High sensitivity, especially to inhibitors targeting both mTORC1 and mTORC2 is observed in a large OCCC cell line panel. Similar results are obtained in OCCC patient-derived xenografts, signifying the clinical implications of these genomic alterations. Targeting of mTORC1 in combination with standard chemotherapy did not improve overall survival in OCCC patients. Therefore, mTORC1/2 inhibitors, currently evaluated in phase II clinical trials, are proposed for the treatment of OCCC. Based on the mutational landscape in OCCC, our in vitro results and the toxicity observed with dual inhibitors of PI3K and mTORC1/2, future treatment combinations of mTORC1/2 inhibitors with either PI3K or MEK inhibitors could be considered to improve clinical benefit for OCCC patients.
Acknowledgements
We acknowledge Dr. Els Berns, Dr. Yasushi Saga and Dr. Vijayalakshmi Shridhar for their kind contribution of OCCC cell lines. We acknowledge the University of Cambridge, National Institute for Health Research Cambridge Biomedical Research Centre, Cambridge Experimental Cancer Medicine Centre and Hutchison Whampoa Limited for their support. The authors thank Jolanda A.L. Visser, Neeltje M. Kooi, Gerda de Vries and Fernanda X. Rosas Plaza for help with the PDX models. In addition, we acknowledge Anna Piskorz, Mercedes Jimenez-Linan and Karen Hosking. G. Bea A. Wisman and Steven de Jong are members of the EurOPDX Consortium.
Footnotes
Author contributions:
G.B.A.W., R.B., A.G.J.Z. and S.J. conceived and supervised the project. J.J.C. and K.B. performed the majority of experiments and data analysis with the assistance of G.B.A.W., R.S.N.F., T.T., H.K., G.J.M., E.M.H., A.M.C.G., E.W.D., D.W., R.J.C.K., and L.M.; H.I., A.K.L.R., M.J.B., H.B.S., I.G., E.N., J.B., E.I.B., J.K. and B.S. provided samples. J.J.C., G.B.A.W., A.G.J.Z and S.J. wrote the manuscript. All authors discussed the results and commented on the manuscript.
This research was supported by grants from the Dutch Cancer Society (KWF, RUG 2010-4833, 2011-5231 and 2012-5477), the Cock-Hadders foundation and Cancer Research UK (A15601).
The authors declare no potential conflicts of interest.
References
- 1.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anglesio MS, Carey MS, Kobel M, Mackay H, Huntsman DG, Vancouver Ovarian Clear Cell Symposium Speakers Clear cell carcinoma of the ovary: a report from the first Ovarian Clear Cell Symposium, June 24th, 2010. Gynecol Oncol. 2011;121:407–15. doi: 10.1016/j.ygyno.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–43. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones S, Wang TL, Shih I, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan B, Wang TL, Shih I. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011;71:6718–27. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo KT, Mao TL, Jones S, Veras E, Ayhan A, Wang TL, et al. Frequent activating mutations of PIK3CA in ovarian clear cell carcinoma. Am J Pathol. 2009;174:1597–601. doi: 10.2353/ajpath.2009.081000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashiguchi Y, Tsuda H, Inoue T, Berkowitz RS, Mok SC. PTEN expression in clear cell adenocarcinoma of the ovary. Gynecol Oncol. 2006;101:71–5. doi: 10.1016/j.ygyno.2005.09.047. [DOI] [PubMed] [Google Scholar]
- 8.Zannoni GF, Improta G, Chiarello G, Pettinato A, Petrillo M, Scollo P, et al. Mutational status of KRAS, NRAS, and BRAF in primary clear cell ovarian carcinoma. Virchows Arch. 2014;465:193–8. doi: 10.1007/s00428-014-1599-1. [DOI] [PubMed] [Google Scholar]
- 9.Ho ES, Lai CR, Hsieh YT, Chen JT, Lin AJ, Hung MH, et al. P53 Mutation is Infrequent in Clear Cell Carcinoma of the Ovary. Gynecol Oncol. 2001;80:189–93. doi: 10.1006/gyno.2000.6025. [DOI] [PubMed] [Google Scholar]
- 10.Kuo KT, Mao TL, Chen X, Feng Y, Nakayama K, Wang Y, et al. DNA copy numbers profiles in affinity-purified ovarian clear cell carcinoma. Clin Cancer Res. 2010;16:1997–2008. doi: 10.1158/1078-0432.CCR-09-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahman MT, Nakayama K, Rahman M, Katagiri H, Katagiri A, Ishibashi T, et al. Gene amplification of ZNF217 located at chr20q13.2 is associated with lymph node metastasis in ovarian clear cell carcinoma. Anticancer Res. 2012;32:3091–5. [PubMed] [Google Scholar]
- 12.Yamashita Y, Akatsuka S, Shinjo K, Yatabe Y, Kobayashi H, Seko H, et al. Met is the most frequently amplified gene in endometriosis-associated ovarian clear cell adenocarcinoma and correlates with worsened prognosis. PLoS One. 2013;8:e57724. doi: 10.1371/journal.pone.0057724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helming KC, Wang X, Wilson BG, Vazquez F, Haswell JR, Manchester HE, et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat Med. 2014;20:251–4. doi: 10.1038/nm.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bitler BG, Aird KM, Garipov A, Li H, Amatangelo M, Kossenkov AV, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–8. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bitler BG, Wu S, Park PH, Hai Y, Aird KM, Wang Y, et al. ARID1A-mutated ovarian cancers depend on HDAC6 activity. Nat Cell Biol. 2017;19:962–73. doi: 10.1038/ncb3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oishi T, Itamochi H, Kudoh A, Nonaka M, Kato M, Nishimura M, et al. The PI3K/mTOR dual inhibitor NVP-BEZ235 reduces the growth of ovarian clear cell carcinoma. Oncol Rep. 2014;32:553–8. doi: 10.3892/or.2014.3268. [DOI] [PubMed] [Google Scholar]
- 17.Sasano T, Mabuchi S, Kuroda H, Kawano M, Matsumoto Y, Takahashi R, et al. Preclinical Efficacy for AKT Targeting in Clear Cell Carcinoma of the Ovary. Mol Cancer Res. 2015;13:795–806. doi: 10.1158/1541-7786.MCR-14-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parikh C, Janakiraman V, Wu WI, Foo CK, Kljavin NM, Chaudhuri S, et al. Disruption of PH-kinase domain interactions leads to oncogenic activation of AKT in human cancers. Proc Natl Acad Sci U S A. 2012;109:19368–73. doi: 10.1073/pnas.1204384109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaiswal BS, Kljavin NM, Stawiski EW, Chan E, Parikh C, Durinck S, et al. Oncogenic ERBB3 mutations in human cancers. Cancer Cell. 2013;23:603–17. doi: 10.1016/j.ccr.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y, Li G. Role of the ubiquitin ligase Fbw7 in cancer progression. Cancer Metastasis Rev. 2012;31:75–87. doi: 10.1007/s10555-011-9330-z. [DOI] [PubMed] [Google Scholar]
- 22.Friedlander ML, Russell K, Millis S, Gatalica Z, Bender R, Voss A. Molecular Profiling of Clear Cell Ovarian Cancers: Identifying Potential Treatment Targets for Clinical Trials. Int J Gynecol Cancer. 2016;26:648–54. doi: 10.1097/IGC.0000000000000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto S, Tsuda H, Takano M, Tamai S, Matsubara O. Loss of ARID1A protein expression occurs as an early event in ovarian clear-cell carcinoma development and frequently coexists with PIK3CA mutations. Mod Pathol. 2012;25:615–24. doi: 10.1038/modpathol.2011.189. [DOI] [PubMed] [Google Scholar]
- 24.Wang YK, Bashashati A, Anglesio MS, Cochrane DR, Grewal DS, Ha G, et al. Genomic consequences of aberrant DNA repair mechanisms stratify ovarian cancer histotypes. Nat Genet. 2017;49:856–65. doi: 10.1038/ng.3849. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto S, Tsuda H, Takano M, Iwaya K, Tamai S, Matsubara O. PIK3CA mutation is an early event in the development of endometriosis-associated ovarian clear cell adenocarcinoma. J Pathol. 2011;225:189–94. doi: 10.1002/path.2940. [DOI] [PubMed] [Google Scholar]
- 26.Chandler RL, Damrauer JS, Raab JR, Schisler JC, Wilkerson MD, Didion JP, et al. Coexistent ARID1A-PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling. Nat Commun. 2015;6:6118. doi: 10.1038/ncomms7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleuren ED, Zhang L, Wu J, Daly RJ. The kinome 'at large' in cancer. Nat Rev Cancer. 2016;16:83–98. doi: 10.1038/nrc.2015.18. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Q, Karnak D, Tan M, Lawrence TS, Morgan MA, Sun Y. FBXW7 Facilitates Nonhomologous End-Joining via K63-Linked Polyubiquitylation of XRCC4. Mol Cell. 2016;61:419–33. doi: 10.1016/j.molcel.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lapin V, Shirdel EA, Wei X, Mason JM, Jurisica I, Mak TW. Kinome-wide screening of HER2+ breast cancer cells for molecules that mediate cell proliferation or sensitize cells to trastuzumab therapy. Oncogenesis. 2014;3:e133. doi: 10.1038/oncsis.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamoto A, Sehouli J, Yanaihara N, Hirata Y, Braicu I, Kim BG, et al. Somatic copy number alterations associated with Japanese or endometriosis in ovarian clear cell adenocarcinoma. PLoS One. 2015;10:e0116977. doi: 10.1371/journal.pone.0116977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan DS, Iravani M, McCluggage WG, Lambros MB, Milanezi F, Mackay A, et al. Genomic analysis reveals the molecular heterogeneity of ovarian clear cell carcinomas. Clin Cancer Res. 2011;17:1521–34. doi: 10.1158/1078-0432.CCR-10-1688. [DOI] [PubMed] [Google Scholar]
- 32.Cohen MH, Williams GA, Sridhara R, Chen G, McGuinn WD, Jr, Morse D, et al. United States Food and Drug Administration Drug Approval summary: Gefitinib (ZD1839; Iressa) tablets. Clin Cancer Res. 2004;10:1212–8. doi: 10.1158/1078-0432.ccr-03-0564. [DOI] [PubMed] [Google Scholar]
- 33.Ryan Q, Ibrahim A, Cohen MH, Johnson J, Ko CW, Sridhara R, et al. FDA drug approval summary: lapatinib in combination with capecitabine for previously treated metastatic breast cancer that overexpresses HER-2. Oncologist. 2008;13:1114–9. doi: 10.1634/theoncologist.2008-0816. [DOI] [PubMed] [Google Scholar]
- 34.Posadas EM, Liel MS, Kwitkowski V, Minasian L, Godwin AK, Hussain MM, et al. A phase II and pharmacodynamic study of gefitinib in patients with refractory or recurrent epithelial ovarian cancer. Cancer. 2007;109:1323–30. doi: 10.1002/cncr.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blank SV, Christos P, Curtin JP, Goldman N, Runowicz CD, Sparano JA, et al. Erlotinib added to carboplatin and paclitaxel as first-line treatment of ovarian cancer: a phase II study based on surgical reassessment. Gynecol Oncol. 2010;119:451–6. doi: 10.1016/j.ygyno.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia AA, Sill MW, Lankes HA, Godwin AK, Mannel RS, Armstrong DK, et al. A phase II evaluation of lapatinib in the treatment of persistent or recurrent epithelial ovarian or primary peritoneal carcinoma: a gynecologic oncology group study. Gynecol Oncol. 2012;124:569–74. doi: 10.1016/j.ygyno.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkerson PM, Dedes KJ, Samartzis EP, Dedes I, Lambros MB, Natrajan R, et al. Preclinical evaluation of the PARP inhibitor BMN-673 for the treatment of ovarian clear cell cancer. Oncotarget. 2017;8:6057–66. doi: 10.18632/oncotarget.14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4:554–63. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farley JH, Brady WE, Fujiwara K, Nomura H, Yunokawa M, Tokunaga H, et al. A phase II evaluation of temsirolimus in combination with carboplatin and paclitaxel followed by temsirolimus consolidation as first-line therapy in the treatment of stage III-IV clear cell carcinoma of the ovary. JCO. 2016;34:5531. [Google Scholar]
- 40.Naing A, Aghajanian C, Raymond E, Olmos D, Schwartz G, Oelmann E, et al. Safety, tolerability, pharmacokinetics and pharmacodynamics of AZD8055 in advanced solid tumours and lymphoma. Br J Cancer. 2012;107:1093–9. doi: 10.1038/bjc.2012.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mateo J, Olmos D, Dumez H, Poondru S, Samberg NL, Barr S, et al. A first in man, dose-finding study of the mTORC1/mTORC2 inhibitor OSI-027 in patients with advanced solid malignancies. Br J Cancer. 2016;114:889–96. doi: 10.1038/bjc.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kashiyama T, Oda K, Ikeda Y, Shiose Y, Hirota Y, Inaba K, et al. Antitumor activity and induction of TP53-dependent apoptosis toward ovarian clear cell adenocarcinoma by the dual PI3K/mTOR inhibitor DS-7423. PLoS One. 2014;9:e87220. doi: 10.1371/journal.pone.0087220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu P, Laird AD, Du X, Wu J, Won KA, Yamaguchi K, et al. Characterization of the activity of the PI3K/mTOR inhibitor XL765 (SAR245409) in tumor models with diverse genetic alterations affecting the PI3K pathway. Mol Cancer Ther. 2014;13:1078–91. doi: 10.1158/1535-7163.MCT-13-0709. [DOI] [PubMed] [Google Scholar]
- 44.Fazio N, Buzzoni R, Baudin E, Antonuzzo L, Hubner RA, Lahner H, et al. A Phase II Study of BEZ235 in Patients with Everolimus-resistant, Advanced Pancreatic Neuroendocrine Tumours. Anticancer Res. 2016;36:713–9. [PMC free article] [PubMed] [Google Scholar]
- 45.Mehnert JM, Edelman G, Stein M, Camisa H, Lager J, Dedieu JF, et al. A phase I dose-escalation study of the safety and pharmacokinetics of a tablet formulation of voxtalisib, a phosphoinositide 3-kinase inhibitor, in patients with solid tumors. Invest New Drugs. 2018;36:36–44. doi: 10.1007/s10637-017-0467-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.