Abstract
Skeletal muscle has a well-organized tissue structure comprised of aligned myofibers and an encasing extracellular matrix (ECM) sheath or lamina, within which reside satellite cells. We hypothesize that the organization of skeletal muscle tissues in culture can affect both the structure of the deposited ECM and the differentiation potential of developing myotubes. Further, we posit that cellular and ECM cues can be a strong determinant of myoblast fusion and morphology in 3D tissue culture environments. To test these, we utilized a thermoresponsive nanofabricated substratum (TNFS) to engineer anisotropic sheets of myoblasts which could then be transferred and stacked into multilayered tissues. Within such engineered tissues, we found that myoblasts rapidly sense topography and deposit structurally organized ECM proteins. Further, the initial tissue structure was found to exert significant control over myoblast fusion and eventual myotube organization. These results highlight the importance of ECM structure on myoblast fusion and organization, and provide insights into substrate-mediated control of myotube formation in the development of novel, more effective, engineered skeletal muscle tissues.
Introduction
The skeletal muscle microenvironment comprises local physical, chemical, and biological stimuli surrounding cells that often dictate or regulate cell function. These microenvironments include the extracellular matrix (ECM), the structural organization of which is central to muscle tissue development 1. Based on this understanding, efforts to recreate the structural cues of the ECM within controlled, in vitro 3D environments to aid cellular development now form a substantive literature 2–8. The majority of studies utilizing engineered 3D skeletal muscle tissues rely on the use of exogenous scaffold material, which distorts the cell-matrix ratio present in native skeletal muscle 9–12. Consequentially, analysis of the interaction between cells and their surrounding matrix, as well as the impact this interaction has on tissue development, is confounded by reliance on non-physiological models.
To avoid the presence of substantial exogenous ECM materials, and so more closely model the cell dense nature of the native skeletal musculature, thermoresponsive polymers incorporated onto cell culture surfaces can be used to detach intact monolayers of cells as sheets 13,14. This method preserves cell-deposited ECM and morphology when these detached sheets are transferred to new culture environments 15. In doing so, this approach allows for preservation of cell-cell and cell-ECM connections critical for maintaining correct tissue organization.
Despite the advantages of this system, typical cell sheet engineering utilizes substrates lacking topographical cues, thus limiting the organization of cells and subsequently leading to the generation of randomly organized tissues 13,15,16. This, in turn limits investigation of the effect of specific microenvironments on cell fate and function, and prevents accurate recapitulation of ECM architectures, such as the endomysium 17, when generating in vitro engineered skeletal muscle tissues. To address these limitations, we recently developed a platform utilizing nanotopographical cues to align myoblast monolayers, as well as a thermoresponsive release layer, termed thermoresponsive nanofabricated substratum (TNFS) 18. Nanopatterned cell sheets can be transferred from the TNFS and continue to develop with consistent alignment cues even when stacked into multilayered tissues. Although this phenomenon is useful for skeletal muscle tissue engineering purposes, it is not yet known how transferred cell sheets retain the structural signals imparted on them by the nanopatterned layers from which they were released. In this study, we demonstrate that our TNFS stacking method allows for transfer of organized cell-deposited ECM, which provides alignment cues and prevents cell sheet reorganization after detachment and transfer (Fig. 1). Further, we highlight that, due to the end-to-end nature of myoblast fusion to form myotubes 19, multilayered, aligned myoblast tissues are able to form structurally organized myotube cultures from myoblasts in TNFS-mediated cell sheets. Our results highlight the importance of ECM structure on myoblast fusion and organization, and provide insights into substrate-mediated control of myotube formation in the development of novel, more effective, engineered skeletal muscle tissues.
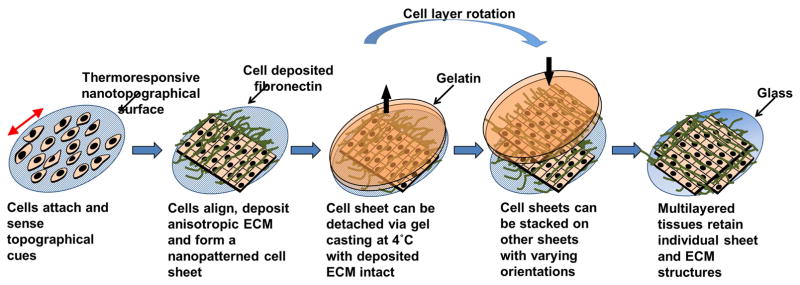
Figure 1. Engineering structurally organized single and multi-layered skeletal muscle tissue.
Using a thermoresponsive nanofabricated substrata (TNFS) and the gel casting method for engineering and transferring anisotropic cell and ECM monolayers, cells are cultured on TNFS for consistent orientation of cellular alignment (red double pointed arrow indicates substrate orientation). Cells deposit anisotropic ECM (green fibers) and form confluent cell sheets within 48 hours. Following casting in gelatin cell sheets are subjected to reduced temperature to trigger thermoresponse of polymer coated substrate and removed from culture dish following gelatin solidification (black arrow indicates direction of casted cell layer). Coherent cell sheets are stacked on other cell sheets with varying orientations and deposited on a new culture surface for experimental conditions, tissue bilayers with anisotropic cell and ECM orientation were cultured in growth or differentiation conditions for up to seven days maintaining anisotropy over the duration of experimental period.
Results
Seeded myoblasts rapidly sense nanotopographic substrate cues and deposit aligned ECM shortly after attachment
Skeletal muscle myoblasts possess an innate ability to respond quickly to changes in mechanical cues 20. Such capacity is essential to enable dynamic manipulation of internal tension to accurately and reliably respond to changes in external load. In conventional cell cultures, the structural ECM niche is lost, leading to the development of non-physiological branching in developing myotubes, and arrest at early developmental stages, characterized by poor myofibril alignment, centralized nuclei, and weak contractility 21–24. Provision of physiological guidance cues and/or representative strains have proven efficacious at enhancing the development of skeletal muscle in vitro, and we have previously demonstrated that myoblasts grown on TNFS align within 24 hours to form an anisotropic cell sheet 18. However, the time course of morphological alignment, as well as deposition and structure of ECM, has not yet been investigated. To better characterize cellular responses to topographic substrate cues, we therefore sought to assess cytoskeletal organization and ECM deposition in myoblasts immediately after cell plating.
We specifically examined the cellular morphology and structure of deposited ECM on TNFS and controls at four timepoints. We found that cells were able to attach to TNFS within 15 minutes but remained spherical in shape, similar to cells in suspension. However, the cells remained firmly attached even after washing prior to fixation and extensions of the cell body indicating adhesion were apparent even during bright field microscopic analysis. Within 1 hour, cells began to elongate in parallel with the direction of the underlying topography, with a confluent, anisotropic sheet formed within 4 hours. By 24 hours, cells were tightly packed in a confluent monolayer and cell borders were not readily apparent. The overall dynamics of cell spreading on nanopatterned surfaces were similar to that of the unpatterned controls.
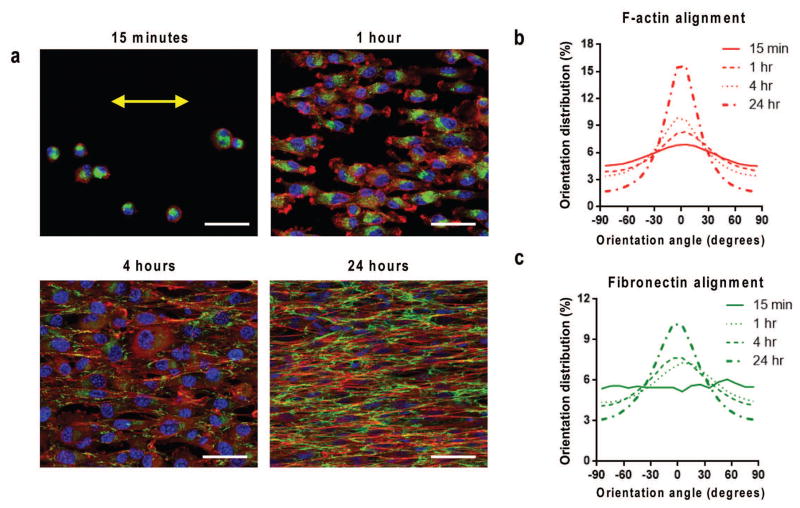
Subsequent immunostaining and confocal microscopy imaging revealed that cytoskeletal F-actin filaments were primarily located on the periphery of cells at 15 minutes and were predominantly concentrated at the leading edges of the cells aligned to the topography after 1 hour (Fig. 2a). Clearly filamentous cytoskeletal structure was evident within 4 hours of seeding, well aligned to the nanotopographical cues, and continued to develop throughout culture (Fig. 2a). Quantification of the alignment of the cytoskeleton demonstrated a time-dependent increase in F-actin alignment peaking within 5 degrees of the major axis of alignment (Fig. 2b). Our analysis showed that after only 15 minutes the alignment was randomly distributed, while following 24 hours of exposure to the nanotopographical cues the alignment of F-actin within 5 degrees of the major axis increased by 190%.
Figure 2. Rapid anisotropic cell attachment, cytoskeletal and ECM alignment.
(a) Confocal microscope image of F-actin (red), fibronectin (green), and nuclei (blue) showing TNFS-seeded C2C12 myoblasts at varied time intervals following seeding on nanotopographical substrates, demonstrating the rapid detection and alignment due to nanotopographical cues. Time course quantitative analysis of (b) cytoskeletal alignment and (c) cell-deposited ECM alignment of cells relative to major axis. Substrate nanopattern orientation is along the horizontal axis. Scale bars: 50μm.
As we were interested in the presence and structure of the ECM during anisotropic sheet formation on the TNFS, we also analyzed cell-deposited fibronectin through immunostaining and confocal imaging. Within 15 minutes, the presence of fibronectin was apparent within the body of the attached cells, but was not present on the remainder of the TNFS (Fig. 2a). As cell morphology began to elongate at 1 hour, the fibronectin was still predominately located within the cell body. After 4 hours of culture, fibronectin was primarily around the periphery of the cells and began to assume an aligned structure (Fig. 2a). Within 24 hours, the cell-deposited fibronectin had a well-defined anisotropic, web-like structure characteristic of fibronectin and reminiscent of a cell-deposited basal lamina (Fig. 2a). Control (unpatterned) surfaces also had cell-deposited fibronectin; however, it was predominately an isotropic web-like structure (Fig. 3c). Quantification of the alignment of cell-deposited fibronectin on nanopatterned surfaces demonstrated a time-dependent increase in alignment similar to that seen in the cytoskeleton with fibronectin alignment within 5 degrees of the major axis of alignment increased by 226% (Fig. 2c). These results demonstrate that cells rapidly attach to the TNFS, sense nanotopographical cues and elongate in the direction of these cues, and deposit ECM proteins (fibronectin) aligned to the nanotopographical cues.
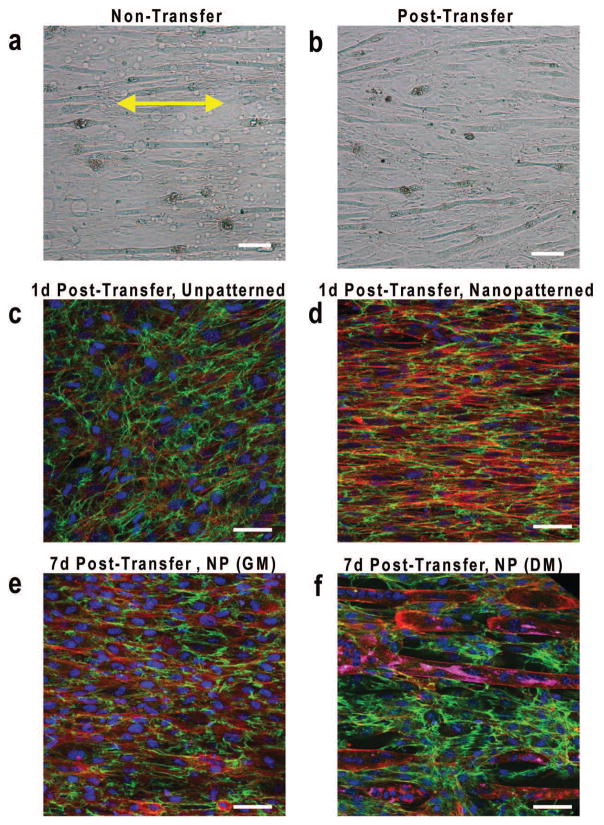
Figure 3. Myoblasts cultured on nanopatterned substrates align and secrete ECM in response to nanotopographical cues.
Bright field microscope images of myoblast monolayers differentiated for 7 days while (a) on TNFS or (b) transferred to a flat glass surface with formation of aligned myotubes. Yellow double-sided arrows indicate initial substrate nanopattern direction. Scale bar, 300 um. Confocal microscope images of F-actin (red), fibronectin (green), and nuclei (blue) staining showing gel-casted and transferred (c) unpatternend and (d) nanopatterned myoblast bilayer sheets 24 hours after transfer onto a flat glass surface. Confocal microscope images of nanopatterned bilayer sheets including MY-32 (magenta) following 7 days in (e) quiescent growth conditions and (f) differentiation conditions. Substrate nanopattern orientation is along the horizontal axis. Scale bars: 50 um.
Cell-deposited ECM transferred with the cell sheet maintains structural anisotropy and regulates myoblast fusion
To determine the effects of engineered nanotopographical cues versus cell-generated orientation cues on myoblast fusion, we differentiated myoblasts under two topographical conditions: maintaining cells on nanopatterned surfaces as well as removing the cell sheet and differentiating the cell sheet on a flat surface. Myoblasts in both conditions fused along the direction of alignment of the nanopattern on the TNFS, with myotubes remaining on the TNFS appearing to have a small degree of increased alignment (Fig. 3a, b). However, sheets of differentiated myotubes were unable to be transferred from the TNFS, requiring that myotube studies be conducted using undifferentiated nanopatterned cell sheets with differentiation following transfer. The effect of cell-deposited ECM as an alignment cue was observed utilizing the TNFS gel casting method by transferring nanopatterned myoblast cell sheets and unpatterned myoblast cell sheets to a flat surface. Following immunofluorescent imaging unpatterned cell sheets were observed to generate random alignment in contrast to the clearly aligned cytoskeletal and ECM present in nanopatterned cell sheets (Fig. 3c, d).
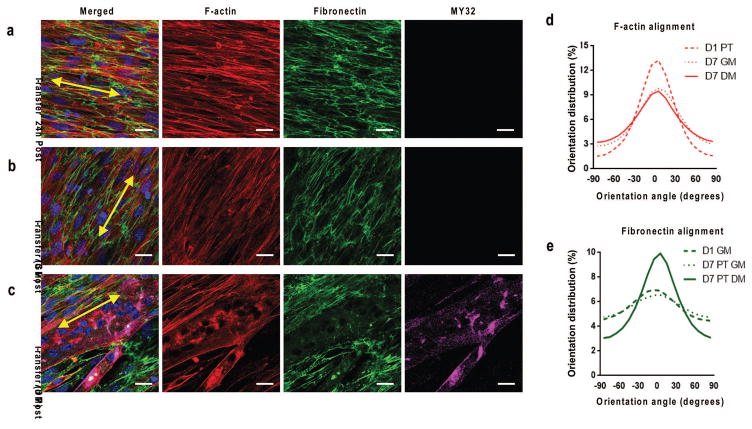
Imaging of transferred, nanopatterned cell sheets showed that cytoskeletal structure and cell-deposited ECM both maintained parallel alignment (Fig. 4a), similar to that of nanopatterned cell sheets immediately before transfer on TNFS (Fig. 2). This indicates that the transfer process does not significantly disrupt either cell morphology or ECM structure. TNFS-transferred cell sheets are then maintained in either a quiescent growth state or using a differentiation protocol with F-actin alignment maintaining overall structural anisotropy (Fig. 4b, c) in both conditions. These alignment differences were highlighted in the quantification of F-actin architecture for samples transferred to a flat surface (Fig. 4c). F-actin alignment declined progressively following transfer (Fig. 4d), presumably due to loss of nanotopographical cues guiding continued cell growth. Contrasting this, in transferred and differentiated nanopatterned cell sheets, the alignment of ECM, as measured by fibronectin alignment, increased with culture time (Fig. 4e). This time-dependent increase likely resulted from cell-mediated remodeling to allow for myotube formation, as the increase was seen only in cell sheets cultured in the differentiation protocol. This indicates that the cell-secreted ECM and induced cellular structure imparted by the TNFS are sufficient to dictate myotube formation with specified alignment, without continued culture on the nanotopographical substrate. This also further indicates that myoblasts actively remodel transferred ECM to allow for myotube formation along the original alignment of the nanopatterned cell sheet.
Figure 4. Transferred nanopatterned myoblast sheets remodel cytoskeletal and ECM structure depending on culture condition.
Confocal microscope image of F-actin(red), fibronectin (green), MY-32 (magenta) and nuclei (blue) staining to visualize structural organization of transferred myoblasts under (b) growth conditions and (c) differentiation conditions. Sheets cultured under growth conditions did not stain positive for myotubes. Yellow double-sided arrows indicate initial substrate nanopattern direction. Quantitative analysis of distribution of (d) cytoskeletal alignment and (e) cell-deposited ECM alignment under varied culture conditions of transferred nanopatterned cell sheets relative to major axis. Scale bars: 20μm.
Orientational control of bilayered tissues enhance myotube formation
In previous work we demonstrated that nanopatterned cell sheets can be stacked into multilayered tissues while maintaining overall sheet alignment and stacked tissue architecture 18. Based on the results, we hypothesized that transferred, anisotropic cell sheets would provide sufficient alignment cues to allow for maintenance of cell morphology and enhance formation of aligned myotubes in culture.
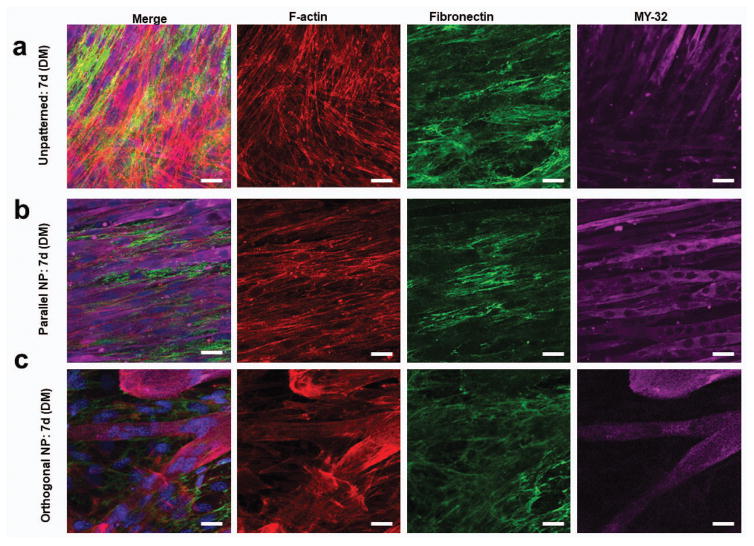
To determine the durability of the effects imparted by engineered nanotopographical cues on cell-sheet morphology, we investigated stacking cell layers in both parallel and orthogonal geometries using the TNFS-mediated cell sheet engineering technique and compared these results to unpatterned cell sheet bilayers. The initial morphology of orthogonal bilayers was similar to that seen in stacked unpatterned cell sheets, with crosshatching seen in myotubes as well as F-actin and fibronectin networks (Fig. 5a, c). Conversely, parallel bilayers demonstrated consistent and aligned myotubes with faintly visible myotubes in different planes of view seen in all samples, with consistently aligned F-actin and fibronectin networks (Fig. 5b).
Figure 5. Stacked myoblast bilayers maintain cytoskeletal and ECM structure long term under quiescent conditions.
Confocal microscope image of F-actin (red), fibronectin (green), MY-32 (magenta) and nuclei (blue) staining to visualize structural organization of transferred myoblasts under differentiation conditions 7 days after transfer of (a) unpatterned, (b) parallel, and (c) orthogonal bilayers. Scale bars: 20μm.
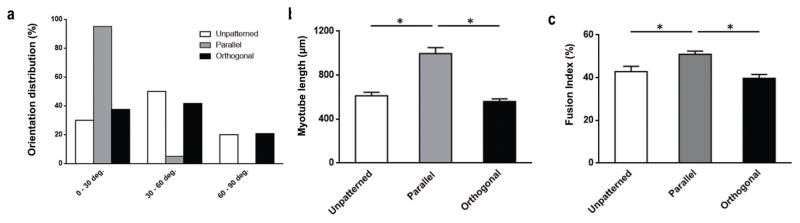
Due to the highly organized structure of skeletal muscle tissue, any engineering approach seeking to replicate this organization must, in addition to providing the proper developmental cues for myotube maturation, enable durable cellular alignment. Such enhanced and durable alignment is seen in the parallel bilayers generated by the gel casting method (>30% aligned with the major axis and 100% within 30 degrees) in contrast to both unpatterned and orthogonal bilayers (<15% aligned with the major axis and <50% within 30 degrees) (Fig. 6a). In addition to enhanced alignment, the morphology of the parallel bilayers more closely resembled in vivo skeletal muscle tissue, with longer myotubes (Fig. 6b) having an increased cell fusion index (Fig. 6c).
Figure 6. Nanopatterned cell sheet myotubes have greater alignment and maturity compared to unpatterned cell sheet myotubes.
(a) Percentage of myotubes oriented along major axis (graph is ±90 degrees from center point of each condition’s distribution) of cell sheet following 7 days of differentiation as bilayer cultures. (b) Average length of myotubes following 7 days of culture in conditions as indicated, no significant difference seen between unpatterned and orthogonal conditions. (c) Myotube fusion index percentage, calculated as the ratio of the nuclei number in myotubes with two or more nuclei versus the total number of nuclei observed in differentiated bilayer cultures in conditions as indicated, no significant difference seen between unpatterned and orthogonal conditions. Significance denoted by bars with star over conditions with p < 0.05. Error bars are SEM.
Discussion
Previous research demonstrates the feasibility of engineering multilayered tissues stacked in specific alignment configurations that are maintained after stacking and subsequent cell culture 14,18. While maintained tissue structure is often advantageous for creating multilayered tissues with complex architectures, and allows for more robust structure-function analysis, we sought to illuminate how tissue architecture is maintained following the removal of cultured cells from the nanotopographic cues provided by the TNFS system. We found that cells rapidly sense the underlying nanotopographical cues, alter their morphology, and then deposit ECM along the periphery of the cell, creating an anisotropic, confluent cell sheet within 24 hours of culture. The nanopatterned sheets could then be transferred and stacked as multilayered tissues with largely intact ECM structure that does not reorganize during quiescent conditions, but is remodeled by differentiating cells.
Previous studies have utilized numerous methods to induce alignment of myoblasts and skeletal muscle tissues, including microfabricated posts 9, micropatterned 2D surfaces 25 and 3D microchannels 26. Although these approaches were able to induce global alignment of myoblasts, these approaches rely on constraining of cell morphology for structural organization. As a result, there is a distinct lack of uniformity of the degree of alignment within the tissues, with a decrease of cellular alignment away from the constrained edges 25. The use of nanotopography, which does not constrain cell morphology, allows for uniform tissue alignment without the reliance of any pillar, edge, or wall, and subsequently allows for scalable, organized tissues. Additionally, the use of nanofabrication techniques such as CFL allows for more complex tissue organizations in 2D, with the subsequent gel casting and stacking approach allowing for a hierarchal control of 3D tissue structure.
The use of nanotopographical cues also allows for the rapid deposition of anisotropic ECM, likely forming a structurally organized basal lamina, which is then transferred as part of the nanopatterned cell sheet. This basal lamina provides continuous directional cues to overlying cells, and so prevents reorganization and migration of myoblasts after transfer off of the TNFS. However, under differentiation conditions, myoblasts actively migrate and remodel their microenvironment in order to fuse to form nascent myotubes, expressing matrix metalloproteinases both during in vitro studies of myoblast fusion as well as in vivo studies of muscle regeneration 27,28. Additionally, ECM composition is another important factor in satellite cell behavior, with a healthy ECM a prerequisite for myogenesis in mature muscle tissue 29,30. In line with these studies, we observed the removal and remodeling of deposited ECM during the myoblast fusion process for both 2D and 3D tissues. Myoblasts appeared to be aligned to a greater degree when maintained on the TNFS, presumably due to topographical cues imparted by the substrate. These results demonstrate that cell-generated ECM can affect the formation and alignment of myotubes. A different group has utilized micropatterned, aligned myoblast sheets in a similar manner to engineer multilayered, aligned myoblast tissues 31. However, their stacked myoblast sheets ultimately reorganized to match the orientation of the top myoblast sheet, consistently generating only aligned, multilayered tissues after 24h of culture. In contrast to these findings, our myoblast sheets maintained orientation of the individual layers, as seen in these and previous experiments 18. Takahashi et al., hypothesize that myoblast fluidity through the cells sheets is responsible for the reorganization. Other studies have shown that ECM architecture influences 3D migration, with increasing ECM porosity correlating with increased migration speed 32,33. It is quite possible that the utilization of nanotopography, as well as the dense, aligned cell-deposited ECM, restricts such movement of myoblasts within our sheets, possibly due to the deposited ECM structure 34. Further, we observed that myoblasts within cell sheets actively remodel their microenvironment during differentiation and then migrate between cell sheets in order to form myotubes, indicating that a more remodeling-active myoblast phenotype can migrate between sheets, possibly only after reorganizing the ECM. Finally, aligning myoblasts in a singular direction during differentiation facilitates greater fusion of cultured myoblasts and elongation of the resulting myotubes.
During in vivo regeneration of damaged muscle tissues, satellite cells migrate to the site of injury and, in addition to fusing with resident myotubes, differentiate to form multiple small myotubes within the basal lamina sheath of the original myofiber 17. Many studies have focused on the chemotactic response of satellite cells and myoblasts to form myotubes, but few studies have examined the specific role of microenvironmental structure on myoblast biology. As satellite cells have been well documented to mobilize from up or downstream of the site of injury 35 and as the muscle basal lamina survives various types of tissue injury and is believed to play a critical role in regeneration 30,36 our studies indicate the structure of the basal lamina can be an important determinant of myotube alignment during differentiation in the absence of chemotactic signals. Further, other studies have shown the importance of cytoskeletal remodeling and structure in myoblast fusion 17.
In conjunction with these studies, our work could inform future strategies for engineering skeletal muscle tissues for repair, holding promise for both tissue transfer techniques and design parameters for in vitro or in vivo tissue scaffolds. Specifically, the use of structurally organized, 3D myoblast tissues may hold promise to improve tissue patch or tube techniques for transplantation to sites of injured tissue. For example, it has previously been shown that a 3-dimensional microenvironment can enhance myogenesis of myoblasts over 2D culture, with a further improvement once the cells are aligned 9. Additionally, the use of a 3D supporting scaffold allows for improved integration of transplanted satellite cells over direct cell injection 37. Finally, functional 3D tissue constructs have also been shown to restore active force generation in conditions such as volumetric muscle loss 38. As a scaffold inherently reduces cell-cell connections, the use of a scaffold-free approach towards engineering 3D, structurally organized skeletal muscle tissues could further improve integration in a transplantation model, while also limiting potential host-biomaterials interactions. This could allow for improved therapies for clinical regeneration of skeletal muscle, such as for treatment of volumetric muscle loss. Additionally, generation of biomimetically aligned tissue layers holds promise for the examination of disease phenotypes that operate on tissue scales 39. This study further supports the concept that imparting alignment through initial topographic cues induces cytoskeletal and cell secreted-ECM alignment, which then dictates the orientation of myotube formation without need for maintenance of extrinsic cues.
Materials and Methods
Fabrication of the thermoresponsive nanofabricated substratum (TNFS)
The thermoresponsive nanofabricated substratum were fabricated using previously published methods 18. Briefly, a UV-curable PUA mold was fabricated and used as the nanopatterned template for the poly(urethane acrylate)-poly(glycidyl methacrylate) (PUA-PGMA) substratum. Glass coverslips were primed with an adhesion promotor and allowed to dry. A previously mixed and degassed 1% GMA-PUA (w/v) prepolymer was aliquoted onto a PUA template consisting of 800 nm ridge and groove width and 500 nm deep parallel grooves and ridges; a dried coverslip was then placed primed-side down onto the prepolymer. The template-prepolymer-glass was cured under 365 nm UV light to initiate photopolymerization for 50 seconds. After polymerization, the PUA template was peeled off from the PUA-PGMA substratum using forceps. The substratum was then positioned to cover a 13mm diameter opening within a 35mm petri dish (Cell E&G) and using UV-curable NOA 83H (Norland Products) was adhered over the well and UV-cured overnight to finalize polymerization. To functionalize the surface of the nanofabricated substrate with a thermorsponsive polymer, amine-terminated poly(N-isopropylacrylamide) (PNIPAM) solution (1g/mL) was reacted with the PUA-PGMA substratum in a shaker at room temperature for 24 h at 55 rpm to allow for thermoresponsive functionalization. The TNFS was then washed with DI water to remove excess pNIPAM and sterilized with 294 nm UV light for 1 hour prior to use.
Cell Culture and Seeding on the TNFS
C2C12 mouse myoblasts were cultured in Dulbecco’s Modified Eagle medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Sigma) and 1% penicillin-streptomycin (Sigma) in an incubator at 37°C, 5% CO2. Cells were split at 80% confluency to prevent spontaneous differentiation and used within 7 passages of a thaw. To seed cells onto the TNFS, cells were split and seeded at a density of 1.25x105 cells/cm2 to form a confluent monolayer within 48 hours of culture for subsequent transfer. Cells were imaged with a bright-field microscope (Nikon TS100) during culture. Cell sheets were allowed to reach 100% confluency (24–48 hours in culture after seeding) before transfer onto treated glass or stacked on top of another cell sheet using our gel casting method. Once transferred, the single or bilayer tissues were cultured either in growth medium (GM, 10% FBS) to maintain quiescence or differentiation medium (2% horse serum, HS) to induce myotube fusion for up to 7 days after transfer. Tissues were subsequently fixed at 24 hours and 7 days after transfer.
Gel Casting Transfer and Stacking of Nanopatterned Cell Sheets
To transfer individual cell sheets or to engineer bilayer tissues, the gel casting method was used as previously described 18. Briefly, after cell sheets reached full confluency within 24–48 hours, the cells on the TNFS were rinsed and incubated with room temperature DPBS for up to 30 minutes to promote sheet detachment, at which point the DPBS was removed and melted (37°C) 7.5% wt/v gelatin was then added to the culture dish and incubated at 4°C for 15 minutes to allow for gelatin solidification. The solidified gelatin was then manually transferred with the adhered cell sheet to a new surface or transferred to another cell sheet cultured on a TNFS for multilayered stacked tissues. An orientation key was used to allow for orthogonal or aligned tissue stacking. The single and bilayer sheets were then transferred via the casted gelatin to a glass coverslip for subsequent culture and the gelatin was melted and removed. Using the gel casting method, complex, scaffold-free tissues with intact ECM microenvironments can be engineered to study aspects of myogenesis (Fig. 1).
Immunofluorescent Staining
Cells were washed with phosphate buffered saline (PBS, Sigma) and fixed in 4% paraformaldehyde (Sigma) for 15 min at room temperature (22°C). Fixed cells were then washed with DPBS, permeabilized and blocked with a solution of 5% bovine serum albumin (Sigma) and 0.25% Triton X-100 (Sigma) in PBS for 1 hour at room temperature, then washed with PBS. For fibronectin (produced in rabbit, Abcam) and myosin heavy chain staining (produced in mouse, Abcam), cells were incubated with the respective primary antibody at a dilution of 1:1000 (fibronectin) or 1:500 (MHC) in 1% BSA in PBS over night at 4°C. After primary incubation, cells and sheets were subsequently washed with DPBS and incubated with the respective fluorescently labeled secondary antibodies for 1 hour at 37°C. For F-actin staining, cells were incubated with AlexaFluor 488 conjugated phalloidin (Invitrogen) during the secondary staining step.
Quantitative Analysis of Cell Alignment
To assess alignment, immunofluorescent images were analyzed using an automated, modified pixel gradient-based MATLAB script as previously published 40. Briefly, images of phalloidin-stained cells were taken at three representative fields at 10X magnification and analyzed by being passed through a set of filters to calculate the gradient magnitude of each image pixel and generate an orientation vector. This orientation gradient data is then used to generate a histogram plot of overall cell alignment.
Quantification of Myotube Alignment and Morphology
Confocal microscope images were taken at 20X magnification at randomly chosen locations per sample stained for MHC (minimum of n=3 per condition). The MHC channel was isolated from each image and subsequently analyzed using ImageJ. To quantify myotube morphology, a freehand trace of myotubes with a positive MHC signal was drawn per each identifiable myotube. An ellipse was fit to the freehand trace to determine overall orientation angle and length. At least 10 myotubes were traced per sample, yielding at least 30 myotube measurements per condition. Myotube lengths were averaged and statistical significance was characterized using a one-way ANOVA, with Tukey’s tests for multiple comparisons, with significance determined as a p-value <0.05. Myotube orientations were binned into 20 degree increments, with each myotube having one orientation and counted. The orientations were then plotted as a histogram, with the major axis of alignment set to 0 degrees, and the y-axis as a percent of myotube population per bin. Myotube fusion index was calculated as the ratio of nuclei present in myotubes with two or more nuclei over the total number of nuclei observed in the indicated conditions with significance determined as a p-value <0.05.
Acknowledgments
This work was supported by a National Institutes of Health Grants R01 NS094388 and R21 AR064395 (to DH Kim).
References
- 1.Thorsteinsdóttir S, Deries M, Cachaço AS, Bajanca F. The extracellular matrix dimension of skeletal muscle development. Developmental Biology. 2011;354(2):191–207. doi: 10.1016/j.ydbio.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Patterning proteins and cells using soft lithography. Biomaterials. 1999;20(23):2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 3.Kim D-H, Provenzano PP, Smith CL, Levchenko A. Matrix nanotopography as a regulator of cell function. The Journal of Cell Biology. 2012;197(3):351–360. doi: 10.1083/jcb.201108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lund AW, Yener B, Stegemann JP, Plopper GE. The Natural and Engineered 3D Microenvironment as a Regulatory Cue During Stem Cell Fate Determination. Tissue Engineering Part B: Reviews. 2009;15(3):371–380. doi: 10.1089/ten.teb.2009.0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nature Biotechnology. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 6.Ray A, Lee O, Win Z, Edwards RM, Alford PW, Kim D-H, Provenzano PP. Anisotropic forces from spatially constrained focal adhesions mediate contact guidance directed cell migration. Nature Communications. 2017;8:14923. doi: 10.1038/ncomms14923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevens MM, George JH. Exploring and Engineering the Cell Surface Interface. Science. 2005;310(5751):1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Wu Y, Guo B, Ma PX. Nanofiber Yarn/Hydrogel Core–Shell Scaffolds Mimicking Native Skeletal Muscle Tissue for Guiding 3D Myoblast Alignment, Elongation, and Differentiation. ACS Nano. 2015;9(9):9167–9179. doi: 10.1021/acsnano.5b03644. [DOI] [PubMed] [Google Scholar]
- 9.Bian W, Bursac N. Engineered skeletal muscle tissue networks with controllable architecture. Biomaterials. 2009;30(7):1401–1412. doi: 10.1016/j.biomaterials.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hosseini V, Ahadian S, Ostrovidov S, Camci-Unal G, Chen S, Kaji H, Ramalingam M, Khademhosseini A. Engineered Contractile Skeletal Muscle Tissue on a Microgrooved Methacrylated Gelatin Substrate. Tissue Engineering Part A. 2012;18(23–24):2453–2465. doi: 10.1089/ten.tea.2012.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khademhosseini A, Langer R. Microengineered hydrogels for tissue engineering. Biomaterials. 2007;28(34):5087–5092. doi: 10.1016/j.biomaterials.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 12.Nam K-H, Kim P, Wood DK, Kwon S, Provenzano PP, Kim D-H. Multiscale Cues Drive Collective Cell Migration. Scientific Reports. 2016;6:29749. doi: 10.1038/srep29749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ide T, Nishida K, Yamato M, Sumide T, Utsumi M, Nozaki T, Kikuchi A, Okano T, Tano Y. Structural characterization of bioengineered human corneal endothelial cell sheets fabricated on temperature-responsive culture dishes. Biomaterials. 2006;27(4):607–614. doi: 10.1016/j.biomaterials.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi H, Okano T. Cell Sheet-Based Tissue Engineering for Organizing Anisotropic Tissue Constructs Produced Using Microfabricated Thermoresponsive Substrates. Advanced Healthcare Materials. 2015;4(16):2388–2407. doi: 10.1002/adhm.201500194. [DOI] [PubMed] [Google Scholar]
- 15.Canavan HE, Cheng X, Graham DJ, Ratner BD, Castner DG. Surface Characterization of the Extracellular Matrix Remaining after Cell Detachment from a Thermoresponsive Polymer. Langmuir. 2005;21(5):1949–1955. doi: 10.1021/la048546c. [DOI] [PubMed] [Google Scholar]
- 16.Shimizu T, Yamato M, Isoi Y, Akutsu T, Setomaru T, Abe K, Kikuchi A, Umezu M, Okano T. Fabrication of Pulsatile Cardiac Tissue Grafts Using a Novel 3-Dimensional Cell Sheet Manipulation Technique and Temperature-Responsive Cell Culture Surfaces. Circulation Research. 2002;90(3):e40–e48. doi: 10.1161/hh0302.105722. [DOI] [PubMed] [Google Scholar]
- 17.Bischoff R. Interaction between Satellite Cells and Skeletal Muscle Fibers. Development. 1990;110(3):653-s-653. doi: 10.1242/dev.109.4.943. [DOI] [PubMed] [Google Scholar]
- 18.Jiao A, Trosper NE, Yang HS, Kim J, Tsui JH, Frankel SD, Murry CE, Kim D-H. Thermoresponsive Nanofabricated Substratum for the Engineering of Three-Dimensional Tissues with Layer-by-Layer Architectural Control. ACS Nano. 2014;8(5):4430–4439. doi: 10.1021/nn4063962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakelam MJ. The fusion of myoblasts. Biochemical Journal. 1985;228(1):1–12. doi: 10.1042/bj2280001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boonen KJM, Post MJ. The Muscle Stem Cell Niche: Regulation of Satellite Cells During Regeneration. Tissue Engineering Part B: Reviews. 2008;14(4):419–431. doi: 10.1089/ten.teb.2008.0045. [DOI] [PubMed] [Google Scholar]
- 21.Charrin S, Latil M, Soave S, Polesskaya A, Chrétien F, Boucheix C, Rubinstein E. Normal muscle regeneration requires tight control of muscle cell fusion by tetraspanins CD9 and CD81. Nature Communications. 2013;4:1674. doi: 10.1038/ncomms2675. [DOI] [PubMed] [Google Scholar]
- 22.Fear J. Observations on the fusion of chick embryo myoblasts in culture. Journal of Anatomy. 1977;124(Pt 2):437–444. [PMC free article] [PubMed] [Google Scholar]
- 23.Mackey AL, Magnan M, Chazaud B, Kjaer M. Human skeletal muscle fibroblasts stimulate in vitro myogenesis and in vivo muscle regeneration. The Journal of Physiology. 2017;595(15):5115–5127. doi: 10.1113/JP273997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salucci S, Baldassarri V, Falcieri E, Burattini S. α-Actinin involvement in Z-disk assembly during skeletal muscle C2C12 cells in vitro differentiation. Micron. 2015;68(Supplement C):47–53. doi: 10.1016/j.micron.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Junkin M, Leung SL, Whitman S, Gregorio CC, Wong PK. Cellular self-organization by autocatalytic alignment feedback. Journal of Cell Science. 2011;124(24):4213–4220. doi: 10.1242/jcs.088898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen Z, Guo S, Ye D, Chen J, Kang C, Qiu S, Lu D, Li Q, Xu K, Lv J, et al. Skeletal Muscle Regeneration on Protein-Grafted and Microchannel-Patterned Scaffold for Hypopharyngeal Tissue Engineering. BioMed Research International. 2013;2013:146953. doi: 10.1155/2013/146953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W, Pan H, Murray K, Jefferson BS, Li Y. Matrix Metalloproteinase-1 Promotes Muscle Cell Migration and Differentiation. The American Journal of Pathology. 2009;174(2):541–549. doi: 10.2353/ajpath.2009.080509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimowska M, Brzoska E, Swierczynska M, Streminska W, Moraczewski J. Distinct patterns of MMP-9 and MMP-2 activity in slow and fast twitch skeletal muscle regeneration in vivo. Int J Dev Biol. 2008;52(2–3):307–14. doi: 10.1387/ijdb.072331mz. [DOI] [PubMed] [Google Scholar]
- 29.Fuchs E, Tumbar T, Guasch G. Socializing with the Neighbors. Cell. 116(6):769–778. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 30.Lukjanenko L, Jung MJ, Hegde N, Perruisseau-Carrier C, Migliavacca E, Rozo M, Karaz S, Jacot G, Schmidt M, Li L, et al. Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nature Medicine. 2016;22:897. doi: 10.1038/nm.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi H, Shimizu T, Nakayama M, Yamato M, Okano T. The use of anisotropic cell sheets to control orientation during the self-organization of 3D muscle tissue. Biomaterials. 2013;34(30):7372–7380. doi: 10.1016/j.biomaterials.2013.06.033. [DOI] [PubMed] [Google Scholar]
- 32.Doyle AD, Petrie RJ, Kutys ML, Yamada KM. Dimensions in cell migration. Current Opinion in Cell Biology. 2013;25(5):642–649. doi: 10.1016/j.ceb.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wold K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. Journal of Cell Biology. 2013;201(7):1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doyle AD, Carvajal N, Jin A, Matsumoto K, Yamada KM. Local 3D matrix microenvironment regulates cell migration through spatiotemporal dynamics of contractility-dependent adhesions. Nature Communications. 2015;6:8720. doi: 10.1038/ncomms9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein-Ogus C, Harris JB. Preliminary observations of satellite cells in undamaged fibres of the rat soleus muscle assaulted by a snake-venom toxin. Cell and Tissue Research. 1983;230(3):671–776. doi: 10.1007/BF00216210. [DOI] [PubMed] [Google Scholar]
- 36.Carlson BM, Faulkner JA. The regeneration of skeletal muscle fibers following injury: a review. Med Sci Sports Exerc. 1983;15(3):187–98. [PubMed] [Google Scholar]
- 37.Boldrin L, Elvassore N, Malerba A, Flaibani M, Cimetta E, Piccoli M, Baroni MD, Gazzola MV, Messina C, Gamba P, et al. Satellite cells delivered by micro-patterned scaffolds: a new strategy for cell transplantation in muscle diseases. Tissue Eng. 2007;13(2):253–62. doi: 10.1089/ten.2006.0093. [DOI] [PubMed] [Google Scholar]
- 38.Quarta M, Cromie M, Chacon R, Blonigan J, Garcia V, Akimenko I, Hamer M, Paine P, Stok M, Shrager JB, et al. Bioengineered constructs combined with exercise enhance stem cell-mediated treatment of volumetric muscle loss. Nature Communications. 2017;8:15613. doi: 10.1038/ncomms15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macadangdang J, Guan X, Smith AS, Lucero R, Czerniecki S, Childers MK, Mack DL, Kim DH. Nanopatterned human iPSC-based model of a Dystrophin-null cardiomyopathic phenotype. Cell Mol Bioeng. 2015;8(3):320–332. doi: 10.1007/s12195-015-0413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D-H, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh K-Y, Tung L, Levchenko A. Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs. Proceedings of the National Academy of Sciences. 2010;107(2):565–570. doi: 10.1073/pnas.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]