Abstract
Amyotrophic lateral sclerosis type 4 (ALS4) is a rare, early-onset, autosomal dominant form of ALS, characterized by slow disease progression and sparing of respiratory musculature. Dominant, gain-of-function mutations in the senataxin gene (SETX) cause ALS4, but the mechanistic basis for motor neuron toxicity is unknown. SETX is a RNA-binding protein with a highly conserved helicase domain, but does not possess a low-complexity domain, making it unique among ALS-linked disease proteins. We derived ALS4 mouse models by expressing two different senataxin gene mutations (R2136H and L389S) via transgenesis and knock-in gene-targeting. Both approaches yielded SETX mutant mice that develop neuromuscular phenotypes and motor neuron degeneration. Neuropathological characterization of SETX mice revealed nuclear clearing of TDP-43, accompanied by TDP-43 cytosolic mislocalization, consistent with the hallmark pathology observed in human ALS patients. Post-mortem material from ALS4 patients exhibited TDP-43 mislocalization in spinal cord motor neurons, and motor neurons from SETX ALS4 mice displayed enhanced stress granule formation. Immunostaining analysis for nucleocytoplasmic transport proteins Ran and RanGAP1 uncovered nuclear membrane abnormalities in motor neurons of SETX ALS4 mice, and nuclear import was delayed in SETX ALS4 cortical neurons, indicative of impaired nucleocytoplasmic trafficking. SETX ALS4 mice thus recapitulated ALS disease phenotypes in association with TDP-43 mislocalization, and provided insight into the basis for TDP-43 histopathology, linking SETX dysfunction to common pathways of ALS motor neuron degeneration.
Introduction
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease characterized by the rapid and progressive loss of motor neurons in the brain and spinal cord, with sparing of sensory neurons. This motor neuron demise leads to muscle atrophy, muscle weakness, fasciculations, spasticity, and death within three to five years [31]. Overall, ALS shows familial inheritance in about 10% of cases, while the remaining 90% of patients are likely explained by a combination of environmental and genetic factors, with heritability estimated at about 50% [1, 63]. Although a number of risk factors and toxins have been implicated [11], germline mutations remain the only proven cause of ALS, with four genes (C9orf72, SOD1, TARDBP, and FUS) accounting for >50% of FALS [16]. From the total list of ~20 ALS disease genes, three themes emerge as relevant to motor neuron health: RNA metabolism, protein quality control, and cytoskeletal/axonal transport.
ALS4 is a rare, childhood or adolescent-onset autosomal dominant form of ALS, that is characterized by very slow disease progression and sparing of the respiratory musculature (OMIM: 602433). ALS4 patients exhibit some typical clinical features of ALS, including limb weakness, severe muscle wasting, and pyramidal signs [13, 48]. The mean age of onset of ALS4 is 17 years however, although the range is broad (5 – 63 years) [48]. ALS4 is unique among dominantly inherited forms of ALS in that patient life expectancy is normal; there is an absence of bulbar involvement; and there is symmetrical distribution of distal atrophy and weakness [48]. These features thus make ALS4 distinct from classical ALS. Only one ALS4 autopsy report has been published, demonstrating atrophic spinal cords with marked loss of anterior horn cells and degeneration of corticospinal tracts, as well as loss of neurons in the dorsal root ganglia and degeneration of the posterior columns in two patients [48]. Ubiquitin-positive cellular aggregates were not evident in neuronal cytoplasm; however, ubiquitin-positive axonal spheroids were present in the grey matter of the cord, dorsal root entry zones, and peripheral nerves [48].
In 2004, we identified the senataxin gene (SETX) as the cause of ALS4 by detecting a c.1166T>C mutation (L389S substitution) in all 49 affected members of a single large extended family [14]. While ALS4 is very rare, we reported unrelated ALS4 pedigrees with two different SETX single amino acid substitution mutations, R2136H and T3I. Subsequent studies identified additional unrelated ALS4 families with the L389S mutation, confirming the pathogenicity of this mutation [4, 51]. Recently, a SETX N264S substitution was reported in an adult-onset female patient presenting with lower motor neuron impairment. Other SETX mutations have also been reported in patients with motor neuron disease, but these are isolated cases without evidence for genetic transmission [58]. Interestingly, a different neurodegenerative disorder, known as ataxia with oculomotor apraxia type 2 (AOA2), is caused by recessive loss-of-function of SETX, with >80 different mutations identified to date (OMIM: 606002). Unlike related recessive ataxias, such as ataxia-telangiectasia [34], AOA2 patients do not display an increased risk of cancer [39]. As AOA2 carriers express reduced levels of SETX protein but do not develop ALS, SETX ALS4 mutations are presumed to be acting through a gain-of-function (GOF) mechanism, though the basis for this GOF toxicity remains to be defined.
SETX is a 2677 amino acid (aa) length protein that contains a 500 aa domain that is conserved in superfamily 1 helicase proteins [24], with homology to the budding yeast protein Sen1p. The SETX gene is ubiquitously expressed [14, 44], and a number of functions have been attributed to SETX, based upon studies of Sen1p, including roles in: (i) RNA polymerase II (RNAP II) transcription termination [55]; (ii) resolution of RNA/DNA hybrids(so-called R-Loops) [52]; (iii) processing of non-coding RNAs and mRNA [55]; (iv) nuclear exosome recruitment [9, 49]; and (v) formation of replication stress foci during S/G2-phase [49, 67]. Other roles for SETX have been proposed, such as the regulation of circadian rhythm genes, PER and CRY [46]. Based upon proteomics analysis, SETX is likely to play a role in transcription regulation [67], but may not regulate transcription termination in higher eukaryotes [32]. Significant effort has gone into testing the role of SETX in R-loop resolution, but Setx knock-out mice display no defects in this process [65]. Recent work suggests that ALS4 cells contain fewer R-loops, and this pathological alteration impairs proper gene expression regulation [26]. SETX may directly interact with Exosc9 [9, 49], and appears to mediate recruitment of the nuclear exosome complex to newly transcribed RNAs. SETX may also facilitate degradation of stalled late-replicating RNAs, as SETX localizes to sites of collision between DNA polymerase I and the RNA polymerase II [49, 67]. Finally, it should be noted that SETX is maintained in cells at very low levels, with a recent proteomics study finding SETX in the lowest abundance category (i.e. <500 molecules/cell) [7].
One of the very first mouse models developed to study a neurodegenerative disease was a transgenic model that featured expression of disease-causing SOD1 mutations identified in ALS1 patients. These SOD1 mice accurately recapitulate ALS1 disease phenotypes [27], and emerged as the mammalian model of choice for studying ALS pathobiology in general and for the testing of potential therapies for sporadic ALS. However, in 2006, the discovery of mislocalization of the RNA binding protein TDP-43 in the brains and spinal cords of ALS patients led to the realization that virtually all sporadic ALS patients and most familial ALS patients, except for SOD1 ALS1 patients, exhibit nuclear clearing and cytosolic aggregation of TDP-43 in their remaining upper and lower motor neurons [29, 54]. With the identification of ALS disease-causing mutations in the TDP-43 gene, and discovery of disease-linked mutations in different RNA binding proteins as a common cause of familial ALS [16], the ALS field sought to generate additional ALS mouse models in the hopes of recapitulating relevant neuromuscular disease phenotypes that would be accompanied by this hallmark TDP-43 mislocalization histopathology, but despite numerous efforts, this has not been realized [56].
To determine the mechanistic basis for SETX gain-of-function motor neuron toxicity, we developed ALS4 mouse models by standard transgenesis of the SETX R2136H mutation, and gene targeting to “knock-in” the highly penetrant SETX L389S mutation. We found that both PrP-SETX-R2136H and Setx-L389S+/− mice develop a slowly progressive motor phenotype, with motor neuron degeneration. When we immunostained lumbar spinal cord sections from SETX mutant mice, we observed nuclear clearing of TDP-43, accompanied by TDP-43 cytosolic mislocalization, which was reminiscent of the hallmark pathology observed in human ALS patients. Analysis of post-mortem material from two ALS4 patients established that TDP-43 mislocalization is a pathological feature of ALS4 motor neuron disease, and immunostaining analysis for Ran and RanGAP1 revealed frequent nuclear membrane abnormalities in motor neurons of ALS4 mice, suggesting that impaired nucleocytoplasmic trafficking may contribute to the TDP-43 mislocalization. Direct examination of nuclear import of a NLS-NES-eGFP reporter in primary cortical neurons from ALS4 mice indicated reduced uptake of the reporter into the nucleus, consistent with defective nucleocytoplasmic transport. Our findings indicate that these SETX ALS4 mouse models may connect SETX dysfunction to common pathways of ALS motor neuron degeneration.
Materials & Methods
Generation of ALS4 mouse models
Model 1 (PrP-SETX-R2136H)
We obtained a full-length SETX cDNA (RZPD (Berlin, Germany) and cloned the wild type (wt) sequence into a mouse prion promoter (PrP) expression cassette [61]. The R2136H mutation (c.6407G>A) was introduced by PCR amplifying a small amplicon from patient derived cDNA, and cloning it into the final PrP expression cassette. Both wt and R2136H coding sequences were verified by DNA sequencing prior to male pronuclear injections. Five wt and two R2136H mutant Tg lines were thus established. Based on the results of TaqMan qPCR analysis SETX transgene expression in mouse brain, two SETX wt and two R2136H Tg lines were maintained based upon comparable levels of transgene expression.
Model 2 (SETX-L389S+/−)
For the knock-in model, a targeting construct was generated by PCR amplification from murine BAC clone DNA (426D2). The Right-arm was cloned into a holding vector and subjected to site-directed mutagenesis to introduce the L389S substitution. Both targeting arms were then cloned into a standard targeting vector backbone (4317G9). Integrity of the ALS4 targeting construct was validated by DNA sequencing. Following ES cell electroporation and G418 selection, three Setx-L389S clones were found to be correctly integrated as determined by long-range PCR and Southern blot. A single clone was used for chimera development by injection into 3.5 day-old mouse blastocysts. Several chimeric mice displaying chimeric coat color from 45% to 95% were generated and two of these produced gene targeted F1 offspring. Neo targeting cassette removal was mediated by breeding our F1 mice with homozygous CMV-Cre “deleter” mice. Neo cassette ‘excision’ was validated by PCR and genotyping is achieved by PCR amplification across the remaining LoxP repeat which detects a 322-bp product compared to 287-bp for the wt allele.
Gene expression, siRNA knock-down & SETX expression constructs
RT-PCR Analysis
Total brain and spinal cord RNA samples were isolated using the Trizol method (Life Technologies). Quantification of mRNA was performed using an Applied Biosystems 7500 Real Time (RT) Sequence Detection System. ABI Assays-on-Demand with TaqMan-based probes for human SETX (Hs00981138_m1), mouse Setx (Mm00616677_m1) and Gapdh (Mm99999915_g1) were show to be highly specific with little to no cross reactivity. As the different primer-probe sets had similar amplification kinetics, we were able to use the comparative Ct (cycle threshold) method to detect gene expression as described previously [38]. The no-RT controls showed Ct values at least six cycles higher than plus-RT amplification.
SETX siRNA knock-down
For siRNA knockdown experiments, HeLa cells were transiently transfected for 48 hrs with a predesigned Silencer Select siRNA to SETX (s22951) or a Scramble control (si-CRL), using RNAiMAX™ Reagent (Invitrogen). Unless otherwise stated, siRNA was used at a final concentration of 10 nM.
Expression constructs
Details of the cloning and PCR amplification of the 1xFlag-Tagged SETX construct were outlined in our 2006 publication [15]. The TARDBP expression construct (Myc-TDP43-HA) was generated by cloning the human TDP-43 coding sequence into the Xho I restriction site of the pCI vector (Promega). Xho I and Myc tag sequences were added 5′-prime to the forward TDP-43 primer. Similarly, Xho I and HA tag sequences were added 5′-prime to the reverse TDP-43 primer. Primer sequences are available upon request. The Flag-hnRNP H expression construct was purchased from GeneCopoeia. The TDP-43-mGFP and FUS-mGFP Lenti vectors were purchased from Origene™. The mGFP (scientific name mTagGFP), provides the same brightness of fluorescence as eGFP, but is significantly more pH stable. All constructs generated by our group were validated by standard DNA Sanger sequencing prior to use.
Primary cortical neuron culture and Propidium Iodide staining
Primary cortical neurons (PCNs) were cultured from dissociated cortex of postnatal day-0 to day-2 (P0–P2) C57BL/6J pups as described previously [66]. PCNs were maintained until treatment by removing half of the CM and replacing it with fresh CM on day 3 (where day 0 is the day of cell seeding), day 5, and every second day after day 5. Propidium iodide (PI) stain is membrane impermeable and therefore does not enter viable cells with intact membranes. To perform this assay, we removed media from each sample being careful not to dislodge adherent cells, and did not rinse with PBS. We then incubated in replacement media at 37°C containing PI (1:3000) and Hoechst dye (1:10,000) for 15 min before imaging under a Zeiss 780 LSM confocal microscope at 10X. Briefly 3 images were taken per well and each experiment was performed in at least triplicate.
Primary Cerebellar Granule Neuron (CGN) culture
CGN’s were cultured from dissociated cerebella from 7–8 day-old mice, as previously described [18]. After complete dissociation of the tissue, cells were pelleted at 2000 rpm for 3.5 min in a swinging bucket centrifuge. The pellet was resuspended in 9 ml of Neurobasal A media supplemented with 125 μl L-Glutamine, 125 μl Pen-Strep, and 25 mM KCl at a density of 1×106 cells/ml. At day 14, neurons were subjected to potassium withdrawal, as described [18].
Primary motor neuron culture
Mixed ventral horn cultures were prepared as previously described [40]. Briefly, primary embryonic motor neurons were isolated on E13. Ventral horns were dissected from individual embryos and dissociated by 10 min incubation with trypsin (final concentration 0.025%, Type XII-S, Sigma Aldrich, Paisley, UK) and three trituration steps in 400 μl L-15, 50 μl 4% bovine serum albumin (BSA, Sigma Aldrich,) in L-15 and 50 μl DNase (1 μg/ml, Sigma Aldrich,). 1 ml 4% BSA was added to the cell suspension to form a cushion and cells were centrifuged at 239xg at RT for 5 min. Cell pellets were resuspended and then plated onto glass coverslips (precoated with polyornithine and laminin for at least 2 hrs each) and maintained in neurobasal medium, supplemented with 1% penicillin-streptomycin (50 units/ml penicillin, 50 μg/ml streptomycin), 2% B27 supplement, 2% horse serum, 0.05% 50 mM β-mercaptoethanol, 0.5 mM L-glutamine (Invitrogen), 0.1 ng/ml brain-derived neurotrophic factor, 0.1 ng/ml glial-derived neurotrophic factor and 0.5 ng/ml ciliary neurotrophic factor (Peprotech).
Immunoblotting and Immunoprecipitation
Western Blot Analysis
Protein lysates from whole brain, spinal cord tissue or cell line extracts were prepared as previously described [53]. We loaded 30–50 μg of homogenized proteins per lane, and after running 3%–8% Tris-Acetate gels (Invitrogen), samples were transferred to PVDF membranes (Millipore), which were blocked in 3% milk in PBS at RT for 1 hr. Membranes were incubated with an anti-SETX Ab (A301-105A, Bethyl), anti-Flag Ab (F1804, Sigma), anti-hnRNP H Ab (AbCam), anti-Cleaved Caspase-3 Ab (9664S, Cell Signalling) or β-actin (ab8226, AbCam) in PBS-T with 3% BSA at 4°C overnight. The primary antibody was visualized with horseradish-peroxidase conjugated anti-rabbit or anti-mouse IgG (Santa Cruz) at 1:5,000 dilution and enhanced chemiluminescence (Amersham). Densitometry analysis was performed using the NIH ImageJ software application and normalized to β-actin signal intensity.
Immunoprecipitation
Cells were rinsed twice with ice-cold PBS and lysed in ice-cold lysis buffer (25mM HEPES-KOH pH 7.4, 150mM NaCL, 5mM EDTA, 1% Triton X-10040 mM) supplemented with one tablet of EDTA-free protease inhibitors (#11873580001 from Roche) per 10 mL of lysis buffer. The soluble fractions from cell lysates were isolated by centrifugation at 12,000 rpm for 11 min in a microfuge. Protein lysates were quantified using Pierce BCA Protein Assay Kit (Thermo Scientific) following the manufactures protocol. For immunoprecipitations, primary antibodies were incubated with Dynabeads® (Invitrogen) overnight, and then washed with sterile PBS. Antibodies bound to Dynabeads were then incubated with lysates with rotation for 2 hours at 4°C. Immunoprecipitates were washed three times with lysis buffer. Immunoprecipitated proteins were denatured by the addition of 20 μl of sample buffer and heated for 10 min at 70°C, resolved by SDS-PAGE, and analyzed via Western blot analysis.
Behavioral phenotyping
All studies and procedures were approved by the University of Washington and University of California, San Diego, Institutional Animal Care and Use Committees (IACUC). All mice used in this study were maintained on the C57BL/6J common laboratory strain background, and we used sex-balanced and age-matched cohorts, with cohort size based upon the outcome of power analysis for normal distributions. Unless otherwise stated, mice were maintained on a 14-hr light/10-hr dark cycle as standard procedure, and housed in cages consisting of no more than four mice. Behavioral testing was performed by research technicians blinded to the genotype in all cases during the daytime.
Accelerating rotarod analysis
Mice were trained on the rotarod (Economex, Columbia Instruments) at day 0 by allowing them to run freely for up to 5 min. On days 1–4, mice were placed on the rotarod for comparative testing as follows: at time zero the rotarod was initiated at a speed of four rotations per min. Thereafter, the rotarod accelerates at a rate of 0.1 rotations per second, and as each mouse falls, time is recorded in seconds. Two additional trials of this routine were undertaken for each mouse for a maximum of 300 sec duration, providing at least five minutes of rest between trials. The average time per day was recorded for the three runs and also averaged for each mouse over the four days.
Neurological function - Composite phenotype scoring
We previously developed a protocol for the rapid and sensitive quantification of disease severity in neurodegenerative mouse models [28]. The composite phenotype score was performed as previously described for each of: Ledge test; Hind limb clasping, Gait; and Kyphosis [28]. At least six sex-matched mice in genotype group at 4, 6 and 12 months were tested.
Cell culture & immunocytochemistry
HEK293A, HEK293T, and HeLa cells were obtained from the American Type Culture Collection (ATCC) and ThermoFisher Scientific respectively, and grown in DMEM media with 10% FBS. Transfections were performed with Lipofectamine 2000, according to manufacturer’s instructions (Invitrogen). For purposes of authentication, HEK293A, HEK293T, and HeLa cells were tested by STR profile and mycoplasma analysis by the SCRM Stem Cell Genomics Core prior to use.
At 7 DIV, motor neuron cultures were fixed in 4% PFA for 20 minutes at RT and blocked and permeabilized for 1 hr in 5% serum in PBS-0.1% Triton X-100 (Sigma). Cells were incubated at 4°C overnight with the following primary antibodies: β-III tubulin clone TUJ1 (1:500, Biolegend), activated caspase 3 (1:400, New England Biolabs), G3BP (1:500, Abcam), and fragile X mental retardation protein (FMRP; 1:500, Abcam). Primary antibodies were detected using Alexa Fluor 488 or Alexa Fluor 568 secondary antibodies (Thermo Fisher Scientific) and nuclei were stained with DAPI (Sigma). Images were acquired using a Zeiss LSM 510 confocal microscope and Leica DMR fluorescence microscope. In order to activate apoptosis, cultures were treated with staurosporine (1 μM for 16 hrs). Motor neurons were identified by staining with β-III tubulin and by morphology. To determine the number of positively stained motor neurons for activated caspase 3, cells were counted if co-stained with β-III tubulin and caspase 3 and if the activated caspase 3 co-localized with the nuclear DAPI stain. Stress granules were induced by sodium arsenite treatment (0.5 mM for 90 min). Motor neurons were identified as positive for stress granules (G3BP) if three or more puncta were identified within the cytoplasm. Cell counts were performed in a blind manner. Cells were also stained for FMRP to confirm presence of stress granules.
Mouse histology, immunohistochemistry, and stereological counts
To preserve tissue, cardiac perfusion with PFA was undertaken [17], followed by embedding in paraffin wax, and 8 μm thick sections mounted on poly-L-lysine-coated slides (Thermo Scientific) were Nissl- or hematoxylin and eosin-stained according to standard methods. Alternately, Cryo-sections were generated by OCT-embedding – then 10-μm frozen sections were cut and mounted on poly-D-lysine-coated slides (Thermo Scientific). Immunostaining was performed using Elite plus kits (Vector laboratories). For double immunofluorescence, secondary Alexa Fluor-conjugated antibodies (1:1000, Molecular Probes, Invitrogen) were used. For motor neuron counts, perfused lumbar spinal cords from 3 to 4 animals per genotype were serially sectioned, and every 6th section (30 μm) was analyzed. Sections were mounted, dried, incubated overnight in 1:1 ethanol/chloroform to de-fat the sections, stained for 10 min in warm 0.1 % cresyl violet, dehydrated and coverslipped. Large neurons greater than 30 μm in diameter (as assessed using the integrated morphometry analysis package in Metamorph 7.7, Molecular Devices, Wokingham, UK) in the anterior horn of the lumbar spinal cord were counted in 15 sections. For immunostaining with Ran antibody (1:200, Cat# 610341, Fisher Scientific), antigen retrieval using R-Universal Epitope Recovery Buffer (Cat # 62719-10, Electron Microscopy Science) was performed. Briefly, free floating 25 μm-thick L4 spinal cord sections were placed in a cooker for 40 min (20 min for the cooker to reach 90–100°C + 20 min for buffer incubation). After 40 min, we let the sections cool for 20 min before immunohistochemistry. RanGAP1 antibody (1:200, Cat# sc-25630, Santa Cruz) was used without antigen retrieval.
TDP-43 DAB staining
Formalin-fixed, paraffin-embedded tissue blocks were obtained from cord L3–L5 lumbar section of 6 μM. Sections were deparaffinized, rehydrated, washed with PBS, and incubated in proteinase K solution (proteinase K was diluted in Tris buffer to the final concentration of 200μg/ml) for 1 h at 37 °C in a humidified chamber. The slides underwent a pretreatment, antigen retrieval step by placing the slides into HEIR buffer (pH = 6.0 or citric acid) at 80°C for 1-hr. The detection system was the peroxidase system, with diaminobenzidine (DAB) as the chromogen performed manually. In this procedure, TDP-43 was used as the primary Ab (ProteinTech, 12892-1-AP) at 1:100 dilution in TBS buffer with 3% Donkey serum. For each of the n=3 mice per genotype, we took five cord sections (separate by ~ 50 μM) for estimates of TDP-43 nuclear evacuation. The ubiquitin antibody used for human sample DAB staining was obtained from Life Technologies (MA1-10035), and used at 1:5000 dilution.
Fluorescence recovery after photobleaching (FRAP)
Shuttling marker lentivirus particle were produced by triple transfecting pMD2.G, R8.74 and s-GFP-L plasmids into HEK293T cells according to standard protocol. PCN’s were plated onto Thermo Scientific Nunc chambered coverglass slides (Cat# 155382, Lab-Tek). We did not replace regular NBAP media for the clear optical imaging media due to the risk of damaging neurons and affecting nuclear transport rate. PCNs were maintained at 37°C with 5% CO2 during FRAP analysis using a Zeiss LSM780 rear port microscope with Zen blue analysis software. We empirically established FRAP conditions such that > 85% of GFP nuclear signal was ablated, and neurons remaining viable. After five initial scans, photobleaching at 90% intensity in the 488 nm channel for 100 iterations was undertaken. During the recovery phase, we imaged every three seconds for 500 cycles, but would typically stop the experiment after 3–5 min of recovery time, as the s-GFP-L recovery rate was readily established by this time. We also monitored regions in the cytoplasm and the background as control to ensure bleaching did not bleed over into this reservoir of fluorophore. In this way, we determined the percentage fluorescent recovery per min for at least 50 neurons per pup and averaged these for each of n ≥ 3 pups per genotype.
Statistical analysis
All data were prepared for analysis with standard spread sheet software (Microsoft Excel). Statistical analysis was done using Microsoft Excel, Prism 4.0 (Graph Pad), or the VassarStats website <http://faculty.vassar.edu/lowry/VassarStats.html>. For ANOVA, if statistical significance (P < 0.05) was achieved, we performed Bonferroni post-hoc analysis to account for multiple comparisons. The level of significance (alpha) was always set at 0.05.
Results
Derivation and expression characterization of SETX ALS4 model mice
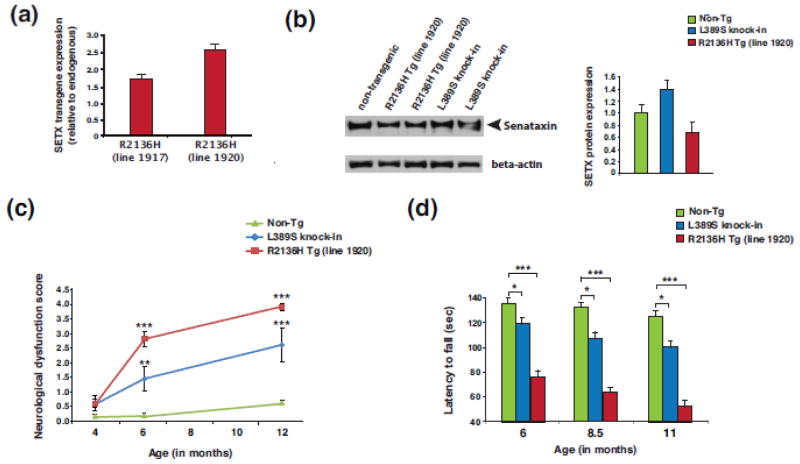
ALS4 is a very rare disorder, with only two SETX mutations (L389S and R2136H) defined as completely penetrant. To create a mouse model for ALS4, we chose to pursue two different strategies in parallel: 1) standard transgenesis, and 2) gene targeting of the murine Setx gene in embryonic stem cells. To derive transgenic mice, we cloned the human SETX cDNA into the mouse prion protein (PrP) promoter expression system [61], and introduced the R2136H mutation. We then performed injection into the male pronucleus of fertilized oocytes, using either normal PrP-SETX-wt or PRP-SETX-R2136H, and identified five PrP-SETX-wt founders and two PrP-SETX-R2136H founders, all of which were bred to generate multiple independent lines of SETX transgenic mice. RT-PCR analysis of transgene-positive mice from the two different PrP-SETX-R2136H lines (#1917 and #1920) indicated expression of SETX-R2136H at levels respectively ~1.5x and ~2.5x that of endogenous mouse SETX (Figure 1a). RT-PCR analysis of the five PrP-SETX-wt lines revealed two lines (#1406 and #1416) with comparable human SETX transgene expression (Online Resource Figure 1a), which were retained for use as controls. RT-PCR analysis of the endogenous mouse Setx gene in the five PrP-SETX-wt lines and two PrP-SETX-R2136H lines further indicated that human SETX transgene expression did not significantly alter endogenous mouse Setx gene expression at the RNA level (Online Resource Figure 1b). To introduce the SETX L389S mutation into the highly conserved endogenous mouse Setx gene, we performed gene targeting in embryonic stem cells using a standard approach, and after deriving mice that exhibited germ line transmission of the L389S mutation, we crossed these F1 progeny with CMV-Cre mice to remove the floxed selection cassette (Online Resource Figure 2). Resultant Setx-L389S+/− mice were thus obtained, and the colony expanded for further characterization.
Figure 1. Characterization of expression and motor function of SETX ALS4 mice.
(a) Human SETX transgene expression levels from whole brain RNAs were determined by Taqman qRT-PCR analysis for two month-old SETX-R2136H mice (lines #1917 and #1920). Expression level is shown relative to endogenous murine Setx, and normalized to Gapdh. n = 3 mice/genotype, n = 3 technical replicates.
(b) SETX immunoblot analysis of brainstem protein lysates for two-month old mice of the indicated genotype. Beta-actin served as the loading control. Densitometry quantification of SETX normalized to beta-actin is shown in the graph to the right, with non-transgenic (Non-Tg) arbitrarily set to 1.
(c) Composite neurological phenotype analysis of mice of the indicated age and genotype (n = 6 – 8 mice/genotype). **P < .01, ***P < .001; ANOVA with post-hoc Bonferroni test.
(d) We measured the average latency to fall from the accelerating rotarod for mice of the indicated age and genotype (n = 8 mice/genotype, 3 runs/day averaged over 4 days). The same cohorts of mice were used for the entire experiment. *P < .05, ***P < .001; ANOVA with post-hoc Bonferroni test. Error bars = s.e.m.
The SETX protein product is very large, consisting of 2,677 amino acids. To assure production of full-length SETX protein and to confirm relative expression levels, we performed immunoblot analysis of SETX. While we detected full-length SETX protein of ~303 kDa molecular mass in brain lysates from both PrP-SETX-R2136H and Setx-L389S+/− mice, we found that SETX protein expression levels in both PrP-SETX-R2136H and Setx-L389S+/− mice were comparable to endogenous mouse SETX levels in non-transgenic controls (Figure 1b), despite our expectation that PrP-SETX-R2136H mice would exhibit elevated SETX protein expression based upon the RT-PCR analysis (Figure 1a). To explore SETX protein level regulation, we transfected HeLa cells with an expression vector containing Flag-tagged SETX cDNA, and confirmed detection of an ~305 kDa protein upon immunoblotting with anti-Flag antibody (Online Resource Figure 3a). We then obtained and validated a human SETX siRNA, and transfected HeLa cells with either a scrambled control siRNA, SETX siRNA, or the Flag-SETX expression construct, and performed immunoblot analysis of SETX. The anti-SETX band observed in non-transfected and control siRNA-transfected HeLa cells was absent in SETX siRNA-transfected HeLa cells, and co-migrated with the anti-SETX band detected in Flag-SETX transfected HeLa cells (Online Resource Figure 3b), which is consistent with the minimal (<1%) difference in molecular mass between Flag-SETX and endogenous SETX. Furthermore, we repeatedly observed levels of SETX protein in Flag-SETX transfected HeLa cells that were comparable to endogenous levels of SETX protein in non-transfected and control siRNA-transfected cells (Online Resource Figure 3b). Taken together, immunoblot analysis of SETX protein expression in PrP-SETX transgenic mice and in SETX-transfected HeLa cells suggests that elevated levels of recombinant SETX protein are not easily maintained. Such regulation may be primarily post-translational as shown for the yeast orthologue Sen1p [20], or via auto-regulation, akin to what was shown for TDP-43 [5, 60], which prevents its expression from exceeding endogenous levels. Hence, the expression levels obtained by RT-PCR analysis of human-specific SETX in PrP-SETX-R2136H mice reflect the relative level of human transgenic SETX to mouse endogenous Setx (Figure 1a), such that ~60% of line 1917 SETX RNA expression is from the human SETX-R2136H transgene, and ~71% of line 1920 SETX RNA expression is from the human SETX-R2136H transgene.
SETX ALS4 mice develop motor phenotypes and motor neuron degeneration, and exhibit neurotoxicity
To evaluate ALS4 model mice for disease phenotypes, we performed a composite neurological screening examination [28], and noted the onset of motor abnormalities at 6 months of age in both PrP-SETX-R2136H and Setx-L389S+/− mice (Figure 1c). Neurological dysfunction was progressive, with both PrP-SETX-R2136H and Setx-L389S+/− mice displaying worsening motor function with age (Online Resource Figure 4a and Figure 1c). We also performed rotarod analysis on cohorts of non-transgenic, PrP-SETX-R2136H, and Setx-L389S+/− mice, and observed impaired motor coordination beginning at 6 months of age; the motor incoordination progressively worsened with age (Figure 1d). Behavioral analysis of PrP-SETX-wt mice did not reveal evidence of significant neurological impairment or motor incoordination upon rotarod analysis at 6 months of age, but indicated the onset of a mild motor deficit beginning at 8.5 months of age (Online Resource Figure 4b). Motor impairment in the PrP-SETX-wt mice remained modest in comparison to the PrP-SETX-R2136H mice, however (Online Resource Figure 4b).
ALS4 patients exhibit a combination of upper and lower motor neuron degeneration; however, lower motor neuron disease signs and symptoms are much more prominent [48]. To determine if ALS4 model mice exhibit lower motor neuron pathology, we compared lumbar spinal cord sections from 12 month-old non-transgenic, PrP-SETX-R2136H, and Setx-L389S+/− mice, and found no significant difference in the numbers of choline acetyltransferase (ChAT)-positive neurons between the three cohorts, though Setx-L389S+/− mice displayed a modest reduction (Online Resource Figure 5a–b). However, we did note that ventral horn motor neurons appeared to be degenerating in ALS4 model mice. Analysis of motor neuron area confirmed this impression, as ventral horn motor neurons from both PrP-SETX-R2136H and Setx-L389S+/− mice were significantly smaller in size in comparison to age- and littermate-matched control mice (Figure 2a). Within the posterior horn, staining patterns were normal for GFAP (glial fibrillary acidic protein), NeuN (neuronal nuclear antigen), and CC1 (an oligodendrocyte antigen), providing no detectable evidence for sensory neuron abnormality in ALS4 model mice. We then generated sections of spinal cord motor axons from the L5 root, measured the axon diameters, and counted axon numbers based upon diameter. This analysis of L5 spinal cord root motor axons revealed that Setx-L389S+/− mice suffer a significant decrease in the number of large caliber axons (i.e. > 9 μm in diameter), though PrP-SETX-R2136H mice retain large caliber axons at numbers that are comparable to their control littermates (Figure 2b–c). We also undertook a detailed analysis of the neuromuscular junctions in these same genotypes by staining gastrocnemius muscle for bungarotoxin and synaptophysin, and observed similar numbers of innervated skeletal muscle contacts, though Setx-L389S+/− mice displayed a modest increase in the number of innervated neuromuscular junctions (Online Resource Figure 5c).
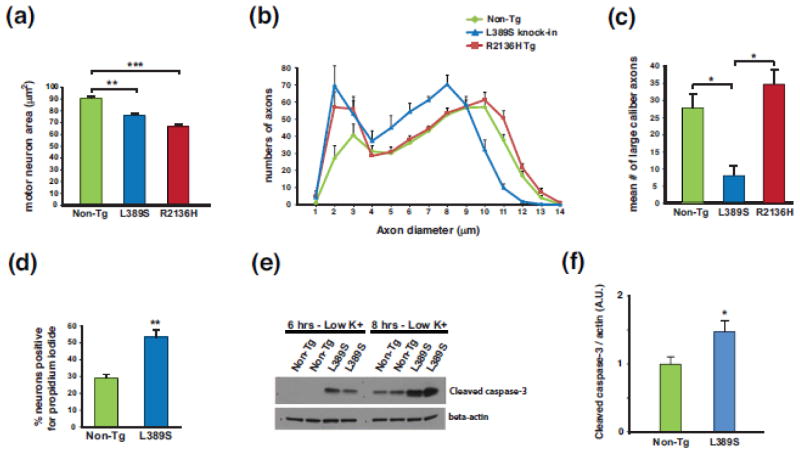
Figure 2. SETX ALS4 mice display motor neuron degeneration and neuron toxicity.
(a) We measured motor neuron area at the L5 level of the lumbar spinal cord in sections from 13 month-old mice of the indicated genotype (n = 3 mice/genotype; 100–120 motor neurons/genotype). **P < .01, ***P < .001; ANOVA with post-hoc Bonferroni test.
(b) We quantified axon numbers, measuring diameters, in the L5 motor root for mice of the indicated genotype (n = 3 mice/genotype; 800 – 850 axons counted/individual mouse).
(c) Comparison of number of large caliber (> 9 μm diameter) motor axons from (b). *P < .05; ANOVA with post-hoc Bonferroni test. Error bars = s.e.m.
(d) Quantification of propidium iodide-positive primary cortical neurons from SETX-L389S+/− mice and littermate control mice (n = 3 mice/genotype; 9 technical replicates/individual mouse). **P < .01, t-test.
(e) We cultured primary cerebellar granule neurons (CGNs) from SETX-L389S+/− mice and littermate control mice in media containing 25 mM KCl, subjected the CGNs to potassium withdrawal by switching them to 5 mM KCl for either 6 hrs or 8 hrs, and then performed immunoblot analysis of CGN protein lysates for cleaved caspase-3. Beta-actin served as the loading control.
(f) Quantification of cleaved caspase-3 level normalized to beta-actin for CGNs subjected to potassium withdrawal for 8 hrs (n = 9 mice/genotype). *P < .05, t-test.
Error bars = s.e.m.
To further evaluate the neurotoxicity of ALS4 SETX disease mutations, we cultured primary cortical neurons from Setx-L389S+/− mice and littermate control mice, and noted a nearly twofold increase in cell death in Setx-L389S+/− cortical neurons based upon a propidium iodide exclusion assay (Figure 2d). As C9orf72 ALS patients exhibit cerebellar neuropathology [2], we also cultured cerebellar granule neurons from Setx-L389S+/− mice and littermate control mice, and observed increased susceptibility of Setx-L389S+/− neurons to cell death upon potassium chloride withdrawal (Figure 2e–f), indicating that the SETX L389S mutation elicits neurotoxicity in primary neurons.
SETX ALS4 mice and human patients display TDP-43 mislocalization and histopathology
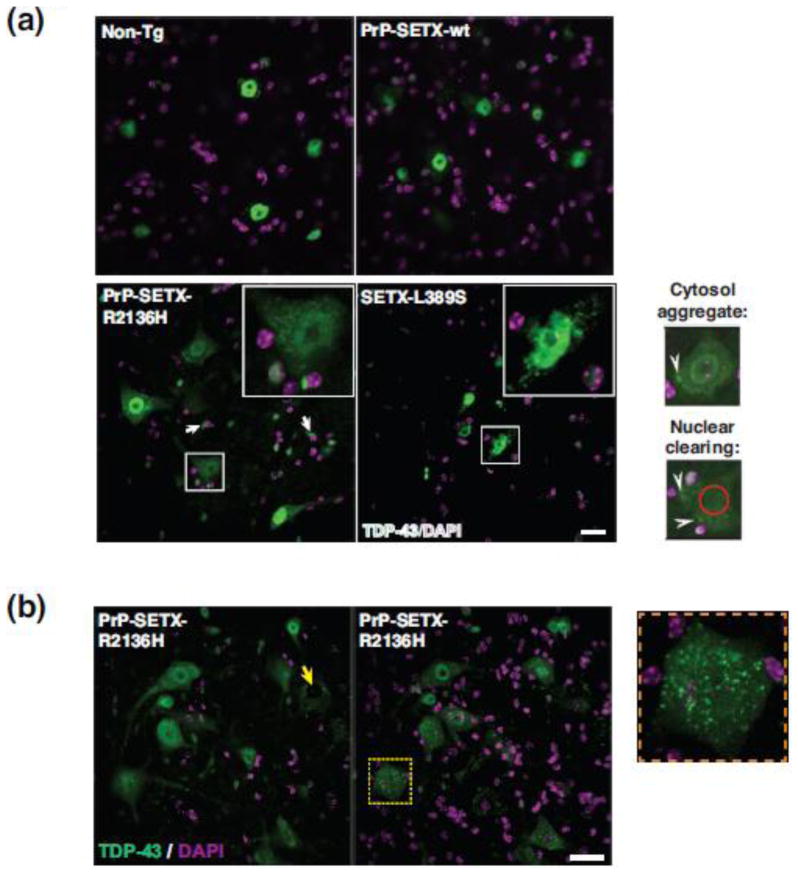
TDP-43 nuclear clearing and cytosolic aggregation have emerged as pathognomonic hallmarks of ALS motor neuron degeneration in sporadic and most familial forms of ALS [3, 45]. To determine if ALS4 mice exhibit such TDP-43 histopathology, we performed immunostaining for TDP-43 on lumbar spinal cord sections from 10 month-old mice, and observed TDP-43 mislocalization in spinal cord sections from both PrP-SETX-R2136H and Setx-L389S+/− mice, but not in cord sections from non-transgenic and PrP-SETX-wt controls (Figure 3a). TDP-43 mislocalization was prominent in motor neurons, but was also present in glia (Figure 3a). This TDP-43 histopathology consisted of the hallmark nuclear clearing and cytosolic mislocalization documented in human ALS patients (Figure 3a–b). To quantify the frequency of TDP-43 mislocalization, we performed an extensive analysis of TDP-43 nuclear clearing on L5 lumbar spinal cord sections (Figure 4a), and noted that ~10% of ventral horn motor neurons from PrP-SETX-R2136H and Setx-L389S+/− mice display TDP-43 mislocalization (Figure 4b). TDP-43 immunostaining analysis of brain sections of both PrP-SETX-R2136H and Setx-L389S+/− mice did not reveal any evidence of TDP-43 mislocalization in the cortex.
Figure 3. SETX ALS4 mice display hallmark ALS TDP-43 histopathology.
(a) We immunostained L5 lumbar spinal cord sections from 10 month-old mice of the indicated genotypes for TDP-43, and counterstained with DAPI. Non-Tg and PrP-SETX-wt mice displayed intense TDP-43 staining throughout motor neuron nuclei, but PrP-SETX-R2136H and SETX-L389S+/− mice exhibited reduced TDP-43 nuclear staining and cytosolic mislocalization in motor neurons (boxed). TDP-43 mislocalization was also present in glial cells (arrows). Representative examples of TDP-43 cytosolic aggregation and pronounced nuclear clearing are respectively indicated by white arrowheads and red circle in high power views of motor neurons (lower right). Scale bar = 20 μm
(b) We immunostained L5 lumbar spinal cord sections from 10 month-old PrP-SETX-R2136H mice for TDP-43, and counterstained with DAPI. Here we see representative examples of TDP-43 nuclear clearing (yellow arrow) and TDP-43 cytosolic mislocalization (boxed), which is also shown at higher power (right). Scale bar = 20 μm
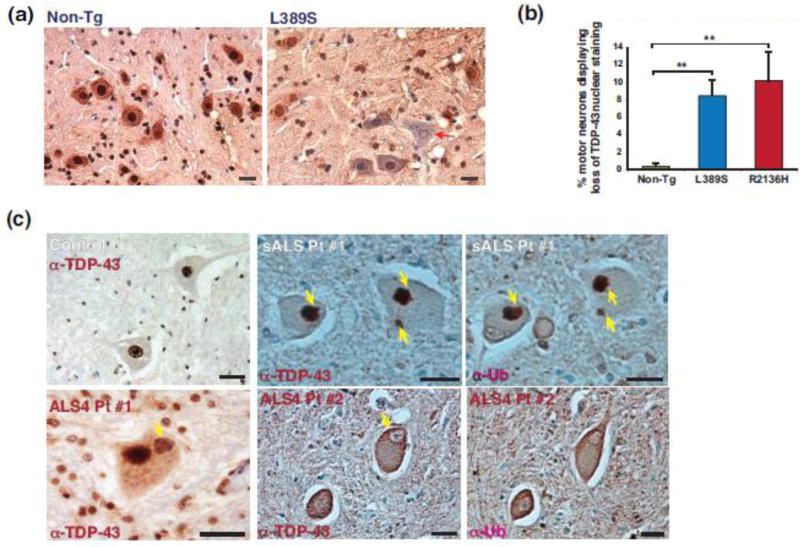
Figure 4. TDP-43 mislocalization and nuclear abnormality occur in SETX ALS4 mice and human ALS4 patients.
(a) We prepared L5 lumbar spinal cord sections from 10 month-old mice of the indicated genotypes, performed 3,3′-diaminobenzidine (DAB) staining of TDP-43, and counterstained with hematoxylin to assess TDP-43 mislocalization. Here we see a representative example of a SETX ALS4 mouse motor neuron that has lost TDP-43 nuclear staining. Scale bar = 20 μm
(b) Quantification of motor neurons exhibiting loss of TDP-43 nuclear staining from (A). n = 3 mice/genotype, 100 – 150 motor neurons/individual mouse. **P < .01; ANOVA with post-hoc Bonferroni test.
(c) We obtained L5 lumbar spinal cord sections from a control individual, a sporadic ALS (sALS) patient, and two different ALS4 patients (#1 and #2), and performed anti-TDP-43 and anti-ubiquitin (Ub) DAB staining. While the human control spinal cord sections displayed nuclear TDP-43 without cytosolic accumulation, we detected TDP-43 cytosolic aggregates in the sALS patient, and documented that these cytosolic aggregates stained positive for ubiquitin (yellow arrows). We also observed examples of TDP-43 mislocalized cytosolic accumulations in ALS4 (#1, yellow arrow). Similar to ALS4 mice, where we observed frequent examples of TDP-43 nuclear clearance, we found similar examples with Fisher hematoxylin staining in ALS4 spinal cord sections (#2, yellow arrow). Scale bars = 20 μm
ALS4 is very rare, with less than a dozen families ascertained around the world. To determine if human ALS4 patients exhibit TDP-43 mislocalization, we obtained post-mortem lumbar spinal cord from two related ALS4 patients, an age-matched control individual, and a sporadic ALS patient who had tested negative for mutations in common familial ALS genes. Upon TDP-43 immunostaining, we observed TDP-43 cytosolic aggregates in motor neurons from the lumbar cord of the sporadic ALS patient, as expected (Figure 4c). TDP-43 immunostaining of lumbar cord sections from the ALS4 patients revealed cytosolic aggregates accompanied by nuclear clearing in motor neurons (Figure 4c). These results thus demonstrate that TDP-43 mislocalization is a feature of ALS4 motor neuron disease in humans.
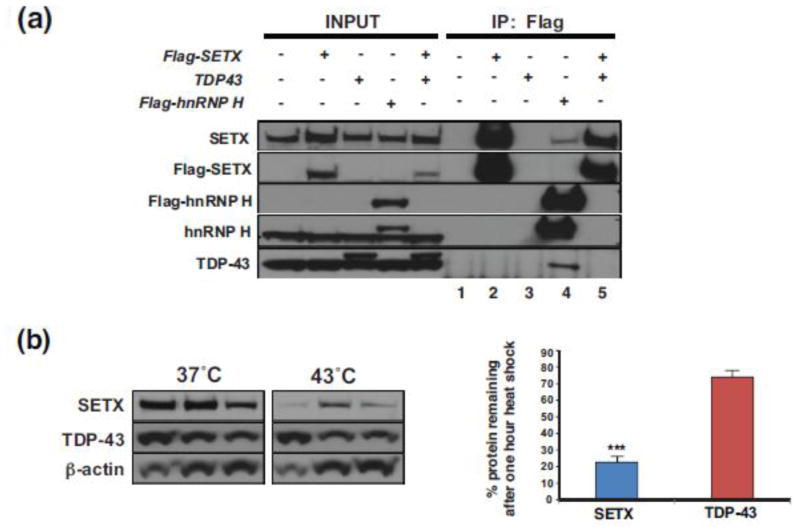
SETX protein does not interact with TDP-43 and is less stable than TDP-43
The presence of TDP-43 histopathology in ALS4 patients and model mice indicates that gain-of-function mutations in SETX, a RNA-binding protein with helicase activity, initiates a pathological process that alters TDP-43 function or localization. To ascertain the basis for the TDP-43 mislocalization in ALS4, we first considered the possibility that SETX and TDP-43 physically interact, as the TDP-43 interactome is enriched for ‘RNA splicing’ factors [22], and TDP-43 has been shown to regulate mRNA splicing [12, 33] and to suppress cryptic exon inclusion [37], while SETX has been implicated in RNA processing and RNA splicing [55]. Although previous TDP-43 proteomic studies did not identify SETX as an interactor [10, 22], detection of a TDP-43 – SETX interaction could be hindered by the low expression level of SETX. To determine if SETX and TDP-43 do physically interact, we performed co-immunoprecipitation (co-IP) assays in HEK293 cells by transfecting Flag-SETX or TDP-43 alone or in combination, and did not detect TDP-43 when we IP’d Flag-SETX, though we could detect TDP-43 when we IP’d Flag-tagged hnRNP H, a known interactor [22] (Figure 5a). We then inspected the amino acid sequence of SETX, as a number of ALS disease-causing genes encode RNA binding proteins that contain low complexity domains (LCDs), which often are the site of pathogenic mutation [30, 57]. Despite being a RNA-binding protein, our analysis did not reveal LCDs in the SETX amino acid coding region. To further assess the biophysical properties of SETX in comparison to the LCD-containing RNA-binding protein TDP-43, we subjected HeLa cells to a one hour heat shock at 43°C, and compared the stability of SETX to TDP-43 at baseline temperature (37°C) and after one hour heat shock at 43°C by immunoblot analysis (Figure 5b). While both SETX protein and TDP-43 protein are similarly stable at 37°C, we observed a marked reduction in SETX levels upon heat shock in comparison to TDP-43, which exhibited only a modest reduction in its protein levels (Figure 5b).
Figure 5. SETX protein does not interact with TDP-43 and is less stable than TDP-43.
(a) HEK293A cells were transfected with a Flag-SETX expression vector, Myc-TDP-43-HA expression vector, and/or a Flag-hnRNP H expression vector as indicated, and then subjected to immunoprecipitation (IP) with an anti-Flag antibody. Immunoblot analysis with the indicated antibodies was performed. Note that anti-TDP-43 immunoblotting of Flag-SETX IPs in lanes 2 and 5 does not detect TDP-43, but immunoblotting of the Flag hnRNP H IP (lane 4) detects both SETX and TDP-43, confirming the validity of the IP technique. No detectable proteins in lanes 1 and 3 rule out artefacts of the Flag IP technique, as these samples did not contain any Flag-tagged proteins.
(b) HEK293A cells were maintained at 37°C or raised to the heat shock temperature of 43°C for 1 hr. Immunoblot analysis of protein extracts after one hour at 37°C or 43°C (n = 3 separate cell cultures/treatment, as shown) indicate limited turnover of TDP-43, but marked reduction in SETX protein at the heat shock temperature of 43°C. Beta-actin served as the loading control. Densitometry quantification of SETX and TDP-43 protein levels normalized to beta-actin is shown in the graph to the right. ***P < .001, t-test. Error bars = s.e.m.
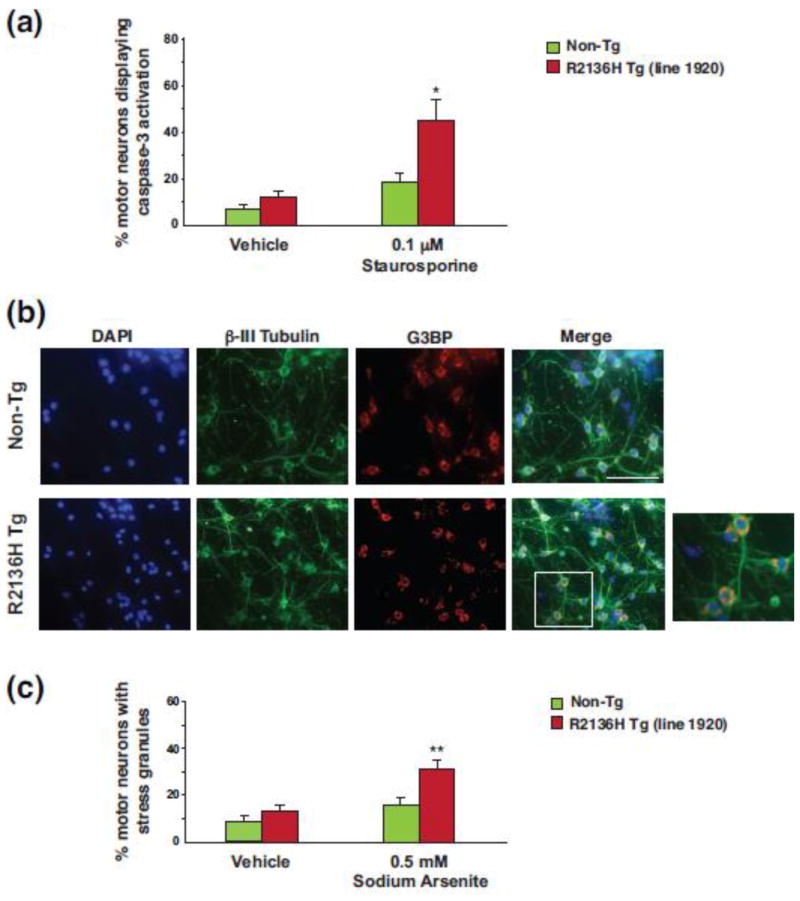
Motor neurons from ALS4 SETX mice are vulnerable to apoptotic activation and stress granule formation
To further evaluate the neurotoxicity of ALS4 SETX disease mutations, we cultured primary spinal cord motor neurons from embryonic day 13 (E13) PrP-SETX-R2136H transgenic mice, which were selected for these studies because of their more severe phenotype, and then treated the cultured E13 motor neurons with either staurosporine or vehicle. Immunostaining with an antibody directed against activated caspase-3 revealed a marked increase in apoptotic activation in E13 motor neurons from PrP-SETX-R2136H transgenic mice in comparison to E13 motor neurons from non-transgenic littermate control mice (Figure 6a). As a propensity to increased stress granule formation may promote TDP-43 stabilization and cytosolic aggregation [43], we again derived E13 motor neuron cultures and then exposed the motor neurons to sodium arsenite, an inducer of oxidative stress that leads to stress granule formation, after which we immunostained the motor neurons with antibodies against β-III tubulin and the Ras-GAP SH3-domain binding protein (G3BP) (Figure 6b). We observed a marked increase in stress granule formation in E13 motor neurons derived from PrP-SETX-R2136H transgenic mice (Figure 6c). To corroborate these findings, we immunostained primary motor neurons for fragile X mental retardation protein (FMRP), another component of stress granules, and confirmed enhanced stress granule formation in PrP-SETX-R2136H motor neurons (Online Resource Figure 6).
Figure 6. Motor neurons from SETX ALS4 mice exhibit apoptotic activation and increased formation of stress granules.
(a) Primary spinal cord motor neurons were prepared from embryonic day 13 (E13) mice of the indicated genotypes, treated with 0.1 μM staurosporine or vehicle (DMSO), immunostained for activated caspase-3, and counted as either caspase-3 positive or negative. *P < .05, t-test.
(b) Primary E13 spinal cord motor neurons were prepared from mice of the indicated genotypes, treated with 0.5 μM sodium arsenite or vehicle (water), and then immunostained for β-III tubulin and G3BP (stress granule marker). Inset shows enhanced stress granule formation in PrP-SETX-R2136H motor neurons. Scale bar = 10 μm
(c) Quantification of motor neurons displaying stress granule formation in (b). n = 3 mice/genotype, ≥ 300 motor neurons/genotype. **P < .01, t-test. Error bars = s.e.m.
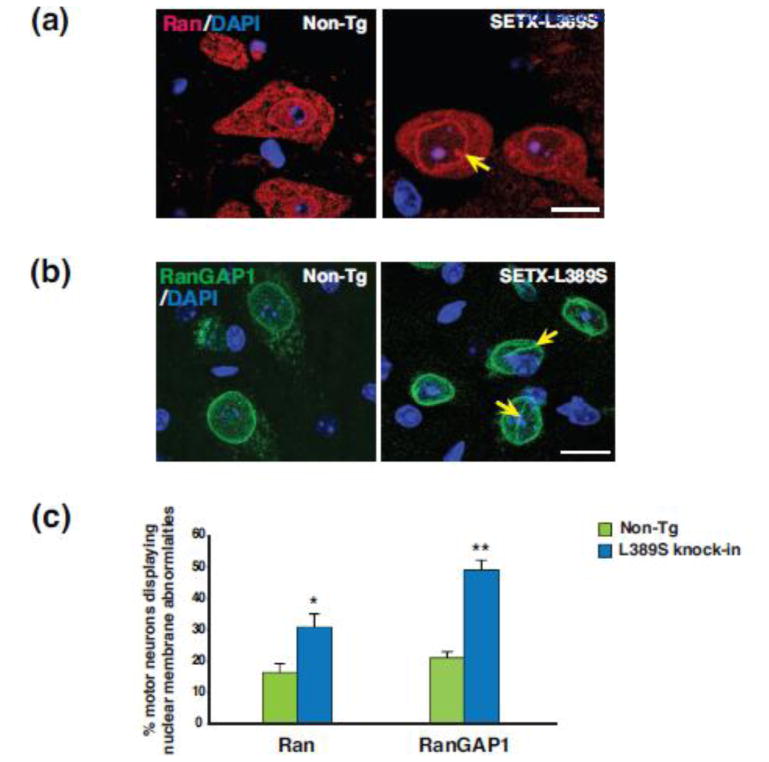
SETX ALS4 mice exhibit nuclear membrane defects and impaired nucleocytoplasmic transport
TDP-43 shuttles between the nucleus and cytosol [6]; as defects in nucleocytoplasmic shuttling have been observed as a central feature of ALS motor neuron disease in familial ALS [36, 68], we examined the nuclear membrane morphology of ventral horn motor neurons in Setx-L389S+/− ALS4 mice by immunostaining for the nucleocytoplasmic transport factors Ran and RanGAP1. This analysis revealed frequent instances of nuclear membrane irregularity in Setx-L389S+/− mice (Figure 7a–b), and when we quantified the frequency of nuclear membrane defects, we found a marked increase in the occurrence of nuclear membrane abnormalities in Setx-L389S+/− mice (Figure 7c).
Figure 7. SETX ALS4 mice exhibit nuclear membrane abnormalities.
(a) We immunostained L5 lumbar spinal cord sections from 11.5 month-old mice of the indicated genotypes for Ran, and counterstained with DAPI. The nuclear membrane was usually smooth without invagination in motor neurons from Non-Tg mice, but displayed frequent irregularities in SETX-L389S+/− mice, such as this invagination (yellow arrow). Scale bar = 20 μm
(b) We immunostained L5 lumbar spinal cord sections from 11.5 month-old mice of the indicated genotypes for RanGAP1, and counterstained with DAPI. The nuclear membrane outline was typically round and regular without invagination in motor neurons from Non-Tg mice, but displayed frequent abnormalities in SETX-L389S+/− mice, such as coffee bean-like invaginations (yellow arrows). Scale bar = 20 μm
(c) Quantification of nuclear membrane abnormalities in motor neurons from Non-Tg and SETX-L389S+/− mice in (a & b). n = 3 mice/genotype, 50 – 80 motor neurons/individual mouse. *P < .05, **P < .01, t-test.
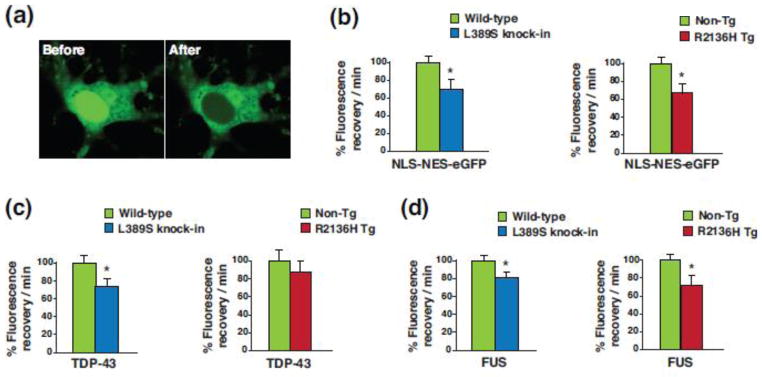
To further explore this phenomenon in ALS4, we obtained an eGFP expression construct that contains both a NLS and NES [64], and packaged this eGFP shuttling vector reporter (NLS-NES-eGFP) into lentivirus. We then cultured primary cortical neurons from non-transgenic, PrP-SETX-R2136H and Setx-L389S+/− mice, and found that upon cortical neuron transduction with NLS-NES-eGFP lentivirus, GFP fluorescence was readily detectable in both the nucleus and cytosol of transduced neurons (Figure 8a). To evaluate nucleocytoplasmic shuttling function in transduced neurons, we performed fluorescence recovery after photobleaching (FRAP) by selectively photobleaching the nucleus (Figure 8a), and then measuring the rate of fluorescence recovery in the nucleus. We documented significant reductions in the nuclear import rate of NLS-NES-eGFP for neurons from both PrP-SETX-R2136H and Setx-L389S+/− mice (Figure 8b). To determine if this nuclear shuttling defect affects the localization of ALS disease relevant proteins that are known to shuttle into and out of the nucleus, we repeated the FRAP assay, first with TPD-43-mGFP and then with FUS-mGFP. We observed a reduction in the nuclear import rate of TDP-43-mGFP in cortical neurons from PrP-SETX-R2136H and Setx-L389S+/− mice, but this reduction was significant for only Setx-L389S+/− neurons (Figure 8c). In the case of FUS-mGFP, nuclear import was significantly decreased for cortical neurons from both PrP-SETX-R2136H and Setx-L389S+/− mice (Figure 8d).
Figure 8. SETX ALS4 mice display defective nucleocytoplasmic transport.
(a) Primary cortical neurons from Non-Tg, PrP-SETX-R2136H, and SETX-L389S+/− mice were transduced with lentivirus containing a NLS-NES-eGFP shuttling reporter and subjected to photobleaching of the nucleus. Here we see the distribution of eGFP fluorescence in transduced cortical neurons before and after photobleaching of the nucleus.
(b) We performed FRAP of NLS-NES-eGFP transduced primary cortical neurons from mice of the indicated genotypes, and measured the recovery of eGFP signal in the nucleus over time (n = 4 mice/genotype). Fluorescence recovery/min was set to 100% for Wild-type or Non-Tg. *P < .05, t-test.
(c) We performed FRAP of TDP-43-mGFP transduced primary cortical neurons from mice of the indicated genotypes, and measured the recovery of mGFP signal in the nucleus over time (n = 3 mice/genotype). Fluorescence recovery/min was set to 100% for Wild-type or Non-Tg. *P < .05, t-test.
(d) We performed FRAP of FUS-mGFP transduced primary cortical neurons from mice of the indicated genotypes, and measured the recovery of mGFP signal in the nucleus over time (n = 3 mice/genotype). Fluorescence recovery/min was set to 100% for Wild-type or Non-Tg. *P < .05, t-test. Error bars = s.e.m.
Discussion
Over the last decade, dramatic insights into the genetic, molecular, and cellular basis of ALS have uncovered a number of pathways now believed to be of central importance for motor neuron health. Among these crucial pathways are RNA metabolism, proteostasis, and axonal transport [57]. We have focused our efforts on understanding how gain-of-function mutations in SETX selectively produce motor neuron dysfunction in ALS4, with the expectation that elucidation of SETX gain-of-function motor neuron toxicity might contribute to our understanding of ALS disease pathogenesis. Indeed, defining shared pathogenic mechanisms between different genetic forms of ALS and related conditions that impact RNA biology is a crucial goal for the field.
In this study, we sought to create a representative mouse model of ALS4 by expressing two different SETX gene mutations (R2136H and L389S) via two different, but complementary strategies: transgenesis and gene targeting. Both approaches yielded lines of SETX mutant mice that developed neuromuscular phenotypes and motor neuron degeneration, and characterization of the PrP-SETX-R2136H transgenic mice and Setx-L389S heterozygous knock-in mice yielded valuable insights into the nature of SETX motor neuron toxicity. When we measured the RNA expression levels of the PrP-SETX-R2136H transgene, we detected significant increases above endogenous SETX levels, especially in line #1920, but immunoblot analysis indicated that SETX protein levels were roughly equivalent between PrP-SETX-R2136H, Setx-L389S+/−, and non-transgenic control mice. This finding led us to examine SETX expression regulation, and indicated that SETX may be subject to some degree of auto-regulation, explaining why the cell does not tolerate SETX expression above endogenous levels. This is reminiscent of TDP-43, which has been shown to self-regulate its own expression rather tightly [5, 47]; for SETX, regulation appears to be at the protein level. When we evaluated motor function and neuropathology in ALS4 mice, we observed similar disease phenotypes, as both PrP-SETX-R2136H line 1920 and Setx-L389S+/− mice developed slowly progressive neurological defects, including clasping and impaired rotarod performance. Importantly, the observed disease phenotypes were only of a neuromuscular nature, as affected mice did not exhibit ataxia, seizures, tremors, or involuntary movement control phenotypes. Both lines of mice displayed a relatively normal lifespan with motor neuron degeneration but without a significant loss of motor neurons, which is reminiscent of the disease course of human ALS4 patients [48]. PrP-SETX-R2136H line 1920 mice exhibited a more severe neurological phenotype than age-matched Setx-L389S+/− mice, likely due to their higher level of mutant SETX expression (~70% of endogenous vs. ~50% of endogenous in the Setx L389S knock-in mice). However, only Setx-L389S+/− mice suffered a marked loss of large caliber axons at the L5 root of the spinal cord, which may reflect a greater propensity to neuropathy for the L389S mutation [51], though the very limited number of ALS4 patients precludes any definitive conclusions as to genotype-phenotype correlations. Furthermore, direct comparison of these two disease alleles in our ALS4 mouse models is complicated by the fact that expression regulation differs between the PrP-SETX-R2136H transgenic mice and Setx L389S knock-in mice.
Because ALS4 is very rare, scant pathological material from affected patients has been studied; hence, it was unknown whether TDP-43 histopathology would be a feature of ALS4 motor neuron disease. We thus performed TDP-43 immunostaining on ALS4 mice and documented nuclear clearing and cytosolic mislocalization of TDP-43 in ~10% of spinal cord motor neurons. This observation predicted that human ALS4 patients would exhibit TDP-43 histopathology, which we confirmed upon TDP-43 immunostaining analysis of two ALS4 patients. However, more importantly, detection of TDP-43 mislocalization in spinal cord motor neurons of PrP-SETX-R2136H and Setx-L389S+/− mice demonstrated that it is possible to recapitulate the hallmark TDP-43 histopathology seen in human ALS patients in mice. This is a significant finding, as considerable effort has been expended to create ALS mouse models that exhibit ALS disease-relevant motor phenotypes in combination with TDP-43 nuclear clearing and cytosolic mislocalization in motor neurons, and except for transgenic over-expression of FALS-linked ubiquilin-2 mutations [35], such mouse models have failed to yield neuromuscular disease phenotypes in combination with human ALS-like TDP-43 mislocalization [41, 59]. The absence of a suitable ALS rodent model with TDP-43 mislocalization has been problematic for advancing translational research on ALS. For example, one ALS therapy development approach seeks to modulate TDP-43 mislocalization, and one promising strategy for achieving this treatment goal is dosage reduction of ataxin-2, as decreased ataxin-2 suppresses TDP-43 toxicity in yeast and fly models [21]. Building on these observations, one group recently evaluated ataxin-2 dosage reduction by crossing TDP-43 transgenic mice with ataxin-2 knock-out mice and delivering ataxin-2 antisense oligonucleotides to TDP-43 transgenic mice, and reported amelioration of very aggressive, but non-ALS representative neurological dysfunction and neuropathology [8]. Hence, there is a need for ALS mouse models that will allow investigators to better evaluate candidate ALS-FTD therapies directed against TDP-43 histopathology.
SETX is a DNA/RNA helicase involved in the regulation of transcription termination, splicing, and RNA exosome-mediated transcript degradation [50, 52, 55], and thus is one of at least six RNA-binding proteins which can be mutated to produce dominantly inherited ALS. Despite being a RNA-binding protein, SETX differs from other RNA binding-proteins implicated in ALS, as it lacks a LCD. In support of SETX being distinct from LCD-containing RNA binding proteins, SETX is degraded upon heat shock. How then does ALS4 SETX gain-of-function motor neuron toxicity elicit TDP-43 mislocalization? The SETX helicase domain is unique in that it is present in only two other human proteins, RENT1 and IGHMBP2. RENT1 is an essential component of the RNA nonsense-mediated decay pathway [62], and Rent1 null mice show embryonic lethality [42]. Recessive mutations of the IGHMBP2 gene, however, cause a severe form of spinal muscle atrophy with respiratory distress (SMARD) [25], with obvious disease overlap with ALS4. The mechanistic basis for SMARD motor neuron disease in humans has remained mysterious, though studies of the nmd mouse model of SMARD have linked IGHMBP2 to protein translation at the level of tRNA regulation [19]. The availability of representative mouse models of ALS4 will similarly facilitate investigations into the molecular basis of SETX function in motor neurons and SETX dysfunction in motor neuron disease.
One proposed mechanism for TDP-43 mislocalization and partial colocalization with cytosolic granules is that impaired nucleocytoplasmic trafficking retards return of TDP-43 to the nucleus when motor neurons suffer cell stress. In this scenario, TDP-43 localizes to stress granules in the cytosol, but upon stress granule disassembly, TDP-43 is inhibited from reentering the nucleus. Studies of primary E13 motor neurons from ALS4 SETX mice revealed a propensity for stress granule formation, suggesting that SETX mutant protein expression may impact the regulation of this process. To directly examine nucleocytoplasmic trafficking, we employed FRAP in cortical neurons transduced with a NLS-NES-eGFP shuttling reporter, and documented impaired nuclear fluorescence recovery in neurons from PrP-SETX-R2136H and Setx-L389S+/− mice. We also observed impaired nuclear import of TDP-43 and FUS in neurons from ALS4 mice. These findings suggest that altered nucleocytoplasmic transport could be contributing to the TDP-43 mislocalization observed in SETX ALS4 mice, and imply that ALS4 may share pathogenic features with more common forms of familial ALS, including especially C9orf72 G4C2 repeat expansion ALS [23, 68]. If this turns out to be the case, then fundamentally important pathways of RNA regulation may be altered as a consequence of SETX gain-of-function mutations such that nucleocytoplasmic trafficking balance is no longer maintained. Determining the targets of SETX may thus shed light on cellular pathways and processes that are not only altered in ALS4, but also in sporadic ALS and more common familial forms of ALS. Indeed, a screen for modifiers of C9orf72 G4C2 58-repeat toxicity in Drosophila has identified the SETX fly orthologue as a potential suppressor [23]. Hence, further studies of SETX normal function and ALS4 pathobiology may reveal key points of intersection and shared disease pathways in sporadic and familial forms of ALS.
Supplementary Material
Acknowledgments
We are grateful to R. Palmiter (University of Washington) for providing the ES cell targeting vector (4317G9), B. Crain and L. Ostrow (Johns Hopkins Medical College) for providing ALS4 patient tissue samples, and A. Wörner (Max-Planck Institute of Biochemistry) for providing the NLS-NES-eGFP shuttling reporter construct. We wish to thank J.F. Ervin for technical assistance. This work was supported by funding from the US National Institutes of Health (R01 GM094384 [C.L.B.]), the Robert Packard Center for ALS Research at Johns Hopkins School of Medicine [A.R.L.S.], and the Motor Neurone Disease Association (MNDA) [B.M & L.G.].
References
- 1.Al-Chalabi A, Fang F, Hanby MF, Leigh PN, Shaw CE, Ye W, Rijsdijk F. An estimate of amyotrophic lateral sclerosis heritability using twin data. J Neurol Neurosurg Psychiatry. 2010;81:1324–1326. doi: 10.1136/jnnp.2010.207464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Sarraj S, King A, Troakes C, Smith B, Maekawa S, Bodi I, Rogelj B, Al-Chalabi A, Hortobagyi T, Shaw CE. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122:691–702. doi: 10.1007/s00401-011-0911-2. [DOI] [PubMed] [Google Scholar]
- 3.Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. S0006-291X(06)02318-7. [DOI] [PubMed] [Google Scholar]
- 4.Avemaria F, Lunetta C, Tarlarini C, Mosca L, Maestri E, Marocchi A, Melazzini M, Penco S, Corbo M. Mutation in the senataxin gene found in a patient affected by familial ALS with juvenile onset and slow progression. Amyotroph Lateral Scler. 2011;12:228–230. doi: 10.3109/17482968.2011.566930. [DOI] [PubMed] [Google Scholar]
- 5.Ayala YM, De Conti L, Avendano-Vazquez SE, Dhir A, Romano M, D’Ambrogio A, Tollervey J, Ule J, Baralle M, Buratti E, et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30:277–288. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayala YM, Zago P, D’Ambrogio A, Xu YF, Petrucelli L, Buratti E, Baralle FE. Structural determinants of the cellular localization and shuttling of TDP-43. J Cell Sci. 2008;121:3778–3785. doi: 10.1242/jcs.038950. [DOI] [PubMed] [Google Scholar]
- 7.Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. The quantitative proteome of a human cell line. Mol Syst Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker LA, Huang B, Bieri G, Ma R, Knowles DA, Jafar-Nejad P, Messing J, Kim HJ, Soriano A, Auburger G, et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017;544:367–371. doi: 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett CL, Chen Y, Vignali M, Lo RS, Mason AG, Unal A, Huq Saifee NP, Fields S, La Spada AR. Protein interaction analysis of senataxin and the ALS4 L389S mutant yields insights into senataxin post-translational modification and uncovers mutant-specific binding with a brain cytoplasmic RNA-encoded peptide. PLoS One. 2013;8:e78837. doi: 10.1371/journal.pone.0078837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blokhuis AM, Koppers M, Groen EJ, van den Heuvel DM, Dini Modigliani S, Anink JJ, Fumoto K, van Diggelen F, Snelting A, Sodaar P, et al. Comparative interactomics analysis of different ALS-associated proteins identifies converging molecular pathways. Acta Neuropathol. 2016;132:175–196. doi: 10.1007/s00401-016-1575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bozzoni V, Pansarasa O, Diamanti L, Nosari G, Cereda C, Ceroni M. Amyotrophic lateral sclerosis and environmental factors. Funct Neurol. 2016;31:7–19. doi: 10.11138/FNeur/2016.31.1.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chance PF, Rabin BA, Ryan SG, Ding Y, Scavina M, Crain B, Griffin JW, Cornblath DR. Linkage of the gene for an autosomal dominant form of juvenile amyotrophic lateral sclerosis to chromosome 9q34. Am J Hum Genet. 1998;62:633–640. doi: 10.1086/301769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YZ, Hashemi SH, Anderson SK, Huang Y, Moreira MC, Lynch DR, Glass IA, Chance PF, Bennett CL. Senataxin, the yeast Sen1p orthologue: characterization of a unique protein in which recessive mutations cause ataxia and dominant mutations cause motor neuron disease. Neurobiol Dis. 2006;23:97–108. doi: 10.1016/j.nbd.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Corcia P, Couratier P, Blasco H, Andres CR, Beltran S, Meininger V, Vourc’h P. Genetics of amyotrophic lateral sclerosis. Rev Neurol (Paris) 2017 doi: 10.1016/j.neurol.2017.03.030. [DOI] [PubMed] [Google Scholar]
- 17.Custer SK, Garden GA, Gill N, Rueb U, Libby RT, Schultz C, Guyenet SJ, Deller T, Westrum LE, Sopher BL, et al. Bergmann glia expression of polyglutamine-expanded ataxin-7 produces neurodegeneration by impairing glutamate transport. Nat Neurosci. 2006;9:1302–1311. doi: 10.1038/nn1750. [DOI] [PubMed] [Google Scholar]
- 18.Dastidar SG, Landrieu PM, D’Mello SR. FoxG1 promotes the survival of postmitotic neurons. J Neurosci. 2011;31:402–413. doi: 10.1523/JNEUROSCI.2897-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Planell-Saguer M, Schroeder DG, Rodicio MC, Cox GA, Mourelatos Z. Biochemical and genetic evidence for a role of IGHMBP2 in the translational machinery. Hum Mol Genet. 2009;18:2115–2126. doi: 10.1093/hmg/ddp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeMarini DJ, Papa FR, Swaminathan S, Ursic D, Rasmussen TP, Culbertson MR, Hochstrasser M. The yeast SEN3 gene encodes a regulatory subunit of the 26S proteasome complex required for ubiquitin-dependent protein degradation in vivo. Mol Cell Biol. 1995;15:6311–6321. doi: 10.1128/mcb.15.11.6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freibaum BD, Chitta RK, High AA, Taylor JP. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabaldon T, Koonin EV. Functional and evolutionary implications of gene orthology. Nat Rev Genet. 2013;14:360–366. doi: 10.1038/nrg3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grohmann K, Schuelke M, Diers A, Hoffmann K, Lucke B, Adams C, Bertini E, Leonhardt-Horti H, Muntoni F, Ouvrier R, et al. Mutations in the gene encoding immunoglobulin mu-binding protein 2 cause spinal muscular atrophy with respiratory distress type 1. Nat Genet. 2001;29:75–77. doi: 10.1038/ng703. [DOI] [PubMed] [Google Scholar]
- 26.Grunseich C, Wang IX, Watts JA, Burdick JT, Guber RD, Zhu Z, Bruzel A, Lanman T, Chen K, Schindler AB, et al. Senataxin Mutation Reveals How R-Loops Promote Transcription by Blocking DNA Methylation at Gene Promoters. Mol Cell. 2018;69:426–437. e427. doi: 10.1016/j.molcel.2017.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation [see comments] [published erratum appears in Science 1995 Jul 14;269(5221):149] Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 28.Guyenet SJ, Furrer SA, Damian VM, Baughan TD, La Spada AR, Garden GA. A simple composite phenotype scoring system for evaluating mouse models of cerebellar ataxia. J Vis Exp. 2010 doi: 10.3791/1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. ng.132. [DOI] [PubMed] [Google Scholar]
- 30.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiernan MC, Vucic S, Cheah BC, Turner MR, Eisen A, Hardiman O, Burrell JR, Zoing MC. Amyotrophic lateral sclerosis. Lancet. 2011;377:942–955. doi: 10.1016/S0140-6736(10)61156-7. [DOI] [PubMed] [Google Scholar]
- 32.Kuehner JN, Pearson EL, Moore C. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol. 2011;12:283–294. doi: 10.1038/nrm3098. nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling SC, Liang TY, Mazur C, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–1497. doi: 10.1038/nn.3230. nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lavin MF, Shiloh Y. The genetic defect in ataxia-telangiectasia. Annu Rev Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- 35.Le NT, Chang L, Kovlyagina I, Georgiou P, Safren N, Braunstein KE, Kvarta MD, Van Dyke AM, LeGates TA, Philips T, et al. Motor neuron disease, TDP-43 pathology, and memory deficits in mice expressing ALS-FTD-linked UBQLN2 mutations. Proc Natl Acad Sci U S A. 2016;113:E7580–E7589. doi: 10.1073/pnas.1608432113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, et al. C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell. 2016;167:774–788. e717. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ling JP, Pletnikova O, Troncoso JC, Wong PC. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015;349:650–655. doi: 10.1126/science.aab0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Lynch DR, Braastad CD, Nagan N. Ovarian failure in ataxia with oculomotor apraxia type 2. Am J Med Genet A. 2007;143:1775–1777. doi: 10.1002/ajmg.a.31816. [DOI] [PubMed] [Google Scholar]
- 40.Malik B, Nirmalananthan N, Bilsland LG, La Spada AR, Hanna MG, Schiavo G, Gallo JM, Greensmith L. Absence of disturbed axonal transport in spinal and bulbar muscular atrophy. Hum Mol Genet. 2011;20:1776–1786. doi: 10.1093/hmg/ddr061. ddr061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGoldrick P, Joyce PI, Fisher EM, Greensmith L. Rodent models of amyotrophic lateral sclerosis. Biochim Biophys Acta. 2013;1832:1421–1436. doi: 10.1016/j.bbadis.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 42.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 43.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreira MC, Klur S, Watanabe M, Nemeth AH, Le Ber I, Moniz JC, Tranchant C, Aubourg P, Tazir M, Schols L, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet. 2004;36:225–227. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- 45.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. 314/5796/130. [DOI] [PubMed] [Google Scholar]
- 46.Padmanabhan K, Robles MS, Westerling T, Weitz CJ. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science. 2012;337:599–602. doi: 10.1126/science.1221592. [DOI] [PubMed] [Google Scholar]
- 47.Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [pii] nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rabin BA, Griffin JW, Crain BJ, Scavina M, Chance PF, Cornblath DR. Autosomal dominant juvenile amyotrophic lateral sclerosis. Brain. 1999;122:1539–1550. doi: 10.1093/brain/122.8.1539. [DOI] [PubMed] [Google Scholar]
- 49.Richard P, Feng S, Manley JL. A SUMO-dependent interaction between Senataxin and the exosome, disrupted in the neurodegenerative disease AOA2, targets the exosome to sites of transcription-induced DNA damage. Genes Dev. 2013;27:2227–2232. doi: 10.1101/gad.224923.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269D. doi: 10.1101/gad.1792809. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rudnik-Schoneborn S, Arning L, Epplen JT, Zerres K. SETX gene mutation in a family diagnosed autosomal dominant proximal spinal muscular atrophy. Neuromuscul Disord. 2012;22:258–262. doi: 10.1016/j.nmd.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 52.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sopher BL, Thomas PS, Jr, LaFevre-Bernt MA, Holm IE, Wilke SA, Ware CB, Jin LW, Libby RT, Ellerby LM, La Spada AR. Androgen receptor YAC transgenic mice recapitulate SBMA motor neuronopathy and implicate VEGF164 in the motor neuron degeneration. Neuron. 2004;41:687–699. doi: 10.1016/s0896-6273(04)00082-0. [DOI] [PubMed] [Google Scholar]
- 54.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. doi:1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suraweera A, Lim Y, Woods R, Birrell GW, Nasim T, Becherel OJ, Lavin MF. Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum Mol Genet. 2009;18:3384–3396. doi: 10.1093/hmg/ddp278. [DOI] [PubMed] [Google Scholar]
- 56.Swarup V, Julien JP. ALS pathogenesis: Recent insights from genetics and mouse models. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:363–369. doi: 10.1016/j.pnpbp.2010.08.006. S0278-5846(10)00309-X. [DOI] [PubMed] [Google Scholar]
- 57.Taylor JP, Brown RH, Jr, Cleveland DW. Decoding ALS: from genes to mechanism. Nature. 2016;539:197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tripolszki K, Torok D, Goudenege D, Farkas K, Sulak A, Torok N, Engelhardt JI, Klivenyi P, Procaccio V, Nagy N, et al. High-throughput sequencing revealed a novel SETX mutation in a Hungarian patient with amyotrophic lateral sclerosis. Brain Behav. 2017;7:e00669. doi: 10.1002/brb3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Den Bosch L. Genetic rodent models of amyotrophic lateral sclerosis. J Biomed Biotechnol. 2011;2011:348765. doi: 10.1155/2011/348765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang IF, Chang HY, Hou SC, Liou GG, Way TD, James Shen CK. The self-interaction of native TDP-43 C terminus inhibits its degradation and contributes to early proteinopathies. Nat Commun. 2012;3:766. doi: 10.1038/ncomms1766. [DOI] [PubMed] [Google Scholar]
- 61.Wang J, Xu G, Slunt HH, Gonzales V, Coonfield M, Fromholt D, Copeland NG, Jenkins NA, Borchelt DR. Coincident thresholds of mutant protein for paralytic disease and protein aggregation caused by restrictively expressed superoxide dismutase cDNA. Neurobiol Dis. 2005;20:943–952. doi: 10.1016/j.nbd.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 62.Weng Y, Czaplinski K, Peltz SW. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wingo TS, Cutler DJ, Yarab N, Kelly CM, Glass JD. The heritability of amyotrophic lateral sclerosis in a clinically ascertained United States research registry. PLoS One. 2011;6:e27985. doi: 10.1371/journal.pone.0027985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Woerner AC, Frottin F, Hornburg D, Feng LR, Meissner F, Patra M, Tatzelt J, Mann M, Winklhofer KF, Hartl FU, et al. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science. 2016;351:173–176. doi: 10.1126/science.aad2033. [DOI] [PubMed] [Google Scholar]
- 65.Yeo AJ, Becherel OJ, Luff JE, Cullen JK, Wongsurawat T, Jenjaroenpoon P, Kuznetsov VA, McKinnon PJ, Lavin MF. R-loops in proliferating cells but not in the brain: implications for AOA2 and other autosomal recessive ataxias. PLoS One. 2014;9:e90219. doi: 10.1371/journal.pone.0090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Young JE, Garden GA, Martinez RA, Tanaka F, Sandoval CM, Smith AC, Sopher BL, Lin A, Fischbeck KH, Ellerby LM, et al. Polyglutamine-expanded androgen receptor truncation fragments activate a Bax-dependent apoptotic cascade mediated by DP5/Hrk. J Neurosci. 2009;29:1987–1997. doi: 10.1523/JNEUROSCI.4072-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yuce O, West SC. Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol Cell Biol. 2013;33:406–417. doi: 10.1128/MCB.01195-12. MCB.01195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.