Abstract
In this study, proteins were extracted from tomato seeds, the main by-product of tomato processing. The incubation for 138.62 min coupled with 3% alcalase was observed to be optimum to produce a tomato seed protein hydrolysate (TSPH) with the highest antioxidant properties. Under these conditions, predicted TSPH activities were 62.99% scavenging of DPPH radicals and 54.81% reduction of phosphomolybdate. Separation of TSPH by ultrafiltration provided three fractions (UF1–UF3) of which, UF3 (< 3 kDa) showed the strongest activity (73.15% DPPH scavenging and 60.1% phosphomolybdate reduction). UF3 was further separated by RP-HPLC into sub-fractions F1–F6. Biological testing found that F2 and F4 were the most active in scavenging DPPH radicals (60.36 and 21.23%) and reducing phosphomolybdate (57.3 and 48.0%). LC–ESI–MS/MS analysis showed that the higher activity of F2 might be explained by the presence of more peptides that contained tyrosine and histidine, known to enhance antioxidant activity through hydrogen or electron transfer. In the simulated gastrointestinal digestion test, peptides in F2 were more resistant compared to those in F4. These findings indicate that peptide fraction F2 might be more useful in the formulation of functional foods because of its greater antioxidant activity and resistance to digestion.
Keywords: Antioxidant peptides, Ultrafiltration, LC–ESI–MS/MS, Gastrointestinal digestion
Introduction
Oxidative stress is one the physiological modifications associated with the development of conditions such as cardiovascular, neuronal, digestive and cancerous diseases (Sarmadi and Ismail 2010). Antioxidant compounds taken as part of food products or in the form of supplement have been proven useful in the reduction of oxidative stress. Phenolic compounds are the most known antioxidant molecules, but in recent years, peptides derived from food proteins have been found to also possess antioxidant properties. These peptides often released from the initial protein sequence through enzymatic digestion or fermentation are generally recognized as been safe (Shahidi and Zhong 2010). In addition to their potential unique biological effects in body, they also possess nutritive values (Shahidi and Zhong 2010). The structure of a peptide plays an important role its antioxidant activity and such, the presence of some specific amino acids, the overall hydrophobic, imidazole (i.e. histidine), sulfur (cysteine and methionine), and hydroxyl groups are essential for the activities of peptides (Rajapakse et al. 2005; Ren et al. 2008). The mechanism by which antioxidant peptides regulate physiological pathways include direct radical scavenging, metal chelation, and regulation of the activity of enzymes. In endothelial cells for example, they can stimulate hemoxygenase-1 (HO-1), ferritin (an iron binding protein), or promote the synthesis of glutathione, a potent intracellular antioxidant peptide (Erdmann et al. 2008).
The usefulness of a bioactive peptide assayed using in vitro tests depends on its ability to be transferred into a biological system, which in turn is dependent on whether or not it can resist gastric and intestinal digestions. This can be best evaluated using an animal model, however simulated in vitro models are often used for preliminary investigation, thereby allowing the selection of the best candidate for further investigations. Over the past decades, simulated gastrointestinal assays have been developed to study chemical and structural changes that may occur to compounds after entering the body. These models take into account the effect of pH, ionic strength of the digestive fluid, concentration of bile acids, and the activity of enzymes (Delgado et al. 2011). Upon being absorbed, a peptide can be transported and reach a specific receptors to perform a specific function (Segura-Campos et al. 2011). One of the best method to investigate the fate of peptides and proteins in high performance chromatograph coupled to tandem mass spectrometry (LC–MS/MS). The technique is sensitive and accurate to identify peptides and any of their modification. For example, oxidation of methionine and tryptophan chains or fission of disulfide bands is detectable and identifiable with this technique (Panchaud et al. 2012). Tomatoes are one of the main agricultural products that are widely used in the food industry, home and restaurants. Tomato pomace, mainly composed of peel and seeds is the main by-product of tomato processing (Sogi et al. 2002). This pomace, composed of approximately 60% seeds is often used as livestock feed, soil amendment, or dumped in landfills (Sogi et al. 2005). Seeds, separated from the pomace can be used to produce proteins with unique functional properties. Defatted tomato seed meal samples containing 39.7–41.4% proteins were reported to possess higher water absorption capacity compared to soy protein isolates although their foaming and emulsification properties were lower (Shao et al. 2014). In a related work, fermentation of tomato seeds meals with Bacillus subtilis yielded hydrolysates with antioxidant and antibacterial properties (Moayedi et al. 2016). These works and others in the literature have focussed on the pomace or seeds. Meanwhile, extracting the proteins will provide an opportunity to investigate their functionalities or use them as a source of bioactive peptides. The objective of this study was to extract proteins from tomato seeds and then optimize conditions for the production of a hydrolysate with the best antioxidant activities followed by separation and identification of peptides. A second objective was to subject the most active fractions to simulated gastrointestinal digestion and investigate possible structural changes.
Materials and methods
Materials
Tomato seeds (Solanum lycopersicum) were purchased from Deland Company, Golestan, Iran. Alcalase 2.4 L [2.4 (Anson unit)/mL enzyme] from Bacillus licheniformis was purchased from Novozymes Co., Denmark. Pepsin from porcine gastric mucosa and Pancreatin from porcine pancreas were purchased from Sigma-Aldrich, (Milwaukee, WI, USA). Amicon Ultra filter centrifugal tubes (3, 10 kDa) were purchased from Millipore Sigma, USA. All chemicals used were of analytical grade.
Protein extraction
Tomato seeds were sun dried at 25–30 °C for 4 days. Major parts of the skins were removed using a 1 mm sieve. Skins remaining on the sieve were separated from seeds with a blowing fan then grounded with a pilot blender. The meal was prepared by defatting ground seeds with n-hexane (1:4 w/v) overnight. The dried defatted meal (20 g) was extracted for 1 h with NaCl solution 1.5% (10:1) in a stirred glass vessel at pH 11.5 that was kept constant by addition of NaOH (0.5 N). The slurry was centrifuged at 6000×g for 20 min (Combi 514R, Hanil, South Korea). The supernatant was collected and the pH was adjusted to the isoelectric point (4.0) using 0.5 M HCl. The protein (i.e. precipitate) was separated by centrifugation at 6000×g for 20 min and freeze dried to yield 9.53 g of isolate, representing a 47.7% extraction efficiency. The isolate contained 67.15% proteins.
Hydrolysis of proteins and optimization
The isolated protein was suspended in potassium phosphate buffer solution (pH 8) at a ratio of 1:5 (w/v). The alcalase at concentrations between 1 and 3% enzyme–substrate (E/S) ratios was added to the suspension and incubated at 50 °C for 30–180 min. All reactions were performed in a shaking incubator (Vision, Scientific co, LTD, Korea) with constant agitation (200 rpm). At the end of the incubation, the enzyme was inactivated by heating the mixture in a water bath at 85 °C for 20 min (Zhang et al. 2014). The mixture’s temperature was decreased using ice-bath, and centrifuged at 6700×g for 20 min at 10 °C to remove undigested proteins and denatured enzyme. The supernatant (i.e. hydrolysate) was dried using the freeze dryer (Operun-FDB5503, South Korea). The degree of hydrolysis was determined as trichloroacetic acid (TCA) solubility index and expressed as nitrogen soluble in 10% TCA. Five milliliters of hydrolysate was added to 5 mL of 10% TCA, strongly mixed, and then centrifuged for 20 min at 10 °C. Nitrogen content of the supernatant and the initial solution were determined and used to TCA solubility as reported in the literature (Hoyle and Merritt 1994).
The Minitab software (version 18; Minitab Inc., State College, PA, USA) was used to optimize the production of the protein hydrolysate with best radical scavenging activity. Response surface methodology (RSM) with central composite design and two variables (time and enzyme/substrate ratio), five replicates at the central point, without block and alpha = 1 was considered for optimization that led to thirteen hydrolysate treatments with the temperature set at 50 °C.
Evaluation of antioxidant activity
DPPH radical scavenging
Quadratic relationships obtained from RSM data were used to select the time (138.62 min) and 3% alcalase to produce tomato seed proteins hydrolysate (TSPH) at 50 °C. The radical scavenging activity of this hydrolysate was determined using 2,2-diphenyl-1-picrylhydrazyl; DPPH radical (DPPH) radical assay as reported in a previous work (Yen and Wu 1999). The TSPH sample prepared in 99.5% ethanol (1 mg/mL) were mixed 1:1 (v/v) with 0.1 mM DPPH dissolved in the same solvent. Test tubes (triplicate) were vigorously stirred for 2 min and kept in the dark at room temperature for 10 min. The absorbance was measured at 517 nm usinga spectrophotometer (UV/VIS T80 PG Instruments, UK) to determine their ability to quench the DPPH radical which is translated by a decrease in absorbance. It should be noted that, in the control sample ethanol was used instead of hydrolyzed proteins. The radical scavenging activity was calculated by following equation:
Ammonium phosphomolybdate assay
One hundred microliters of TSPH in distilled water (1 mg/mL) was mixed with 600 µL phosphomolybdate reagent (0.6 M sulfuric acid, 28 mM sodium phosphate and 4 mM ammonium molybdate). Test tubes were covered with aluminum foil and incubated in the water bath at 95 °C for 90 min. The mixtures were then allowed to reach room temperature and the absorbance was recorded at 765 nm. Blank was run using the same procedure but containing an equal volume of solvent instead of the protein hydrolysate. In this assay, a higher absorbance indicates a higher reducing power (Jan et al. 2013).
Ultrafiltration separation of hydrolyzed proteins
The hydrolysate was separated using 15 mL Amicon Ultra Centrifugal regenerated cellulose membranes (Merck Inc, Chicago, USA) with 3 or 10 kDa molecular weight cut off. Three fractions corresponding to molecular weight above 10 kDa (UF1), between 3 and 10 kDa (UF2), and below 3 kDa (UF3) were obtained and freeze dried. The antioxidant activities of UF1–UF3 were determined as described above using DPPH and phosphomolybdate assays.
High performance liquid chromatographic separation
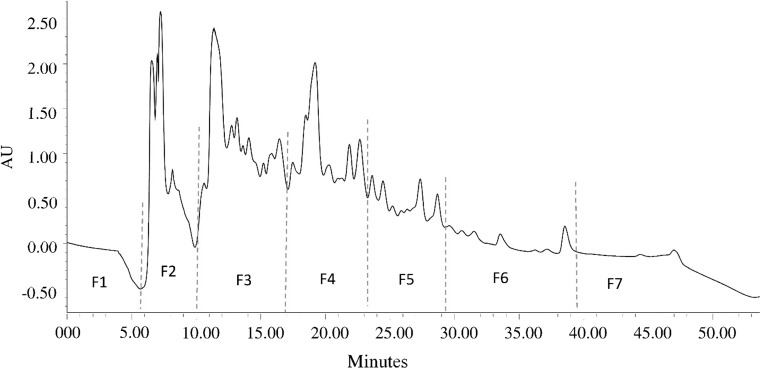
The ultrafiltration fraction UF3 was the most active. To concentrate bioactive peptides into a specific sub-fraction, a reverse phase preparative HPLC system was use to further fractionate UF3 based on a reported procedure (Vanvi and Tsopmo 2016). The system included 1525 binary pump, 2998 photodiode array detector (set at 280 nm), 2707 auto-sampler maintained at 8 °C, and fraction collector III from Waters (Montreal, QC, Canada). A Waters prep XBridge BEH column C18; 130 Å, 10 µm, 19 × 150 mm was used to separate peptide fractions in UF3. RP-HPLC separating is based on relative hydrophobicity of amino acid residues and the tendency of peptide chains to partition into the stationary phase of the column. Injection volumes were 500 µL (1 mg/mL), and mobile phases acetic acid 0.01% in water (A) and acetic acid 0.01% in methanol (B). Peptides were eluted with a linear gradient from 5 to 40% solvent B over 50 min at flow rate of 4 mL/min. The eluates were detected at 280 nm and grouped into 7 fractions according to the chromatogram 0–6 min (F1), 6–10 min (F2), 10–16 min (F3), 16–23 min (F4), 23–28 min (F5), 28–37 min (F6), 37–50 min (F7). The solvent in each fraction was removed under vacuum at 45 °C using Buchi Rotavapor R-215, sample suspended in water, freeze dried. The antioxidant activities of F1–F6 were also determined using DPPH and phosphomolybdate assays.
Simulated gastrointestinal digestion
The experiment was done to simulate gastric and small intestinal digestion and describe in a previous procedure (Minekus et al. 2014). The two HPLC fractions F2 and F4 with the highest antioxidant activities were used for this section. The simulated gastric fluid (SGF, pH 2.0) contained the corresponding electrolytes KCl (6.9 mM), KH2PO4 (0.9 mM), NaHCO3 (25 mM), NaCl (47.2 mM), MgCl (H2O)6 (0.1 mM) and (NH4)2CO3 (0.5 mM). Five milligrams of sample were added to 4 mL of SGF, followed by 0.2 mL porcine pepsin (2000 U/mL, in SGF), and 25 µL CaCl2 (0.075 mM) and 975 mL water. The mixture was adjusted to pH 3.0 with 1 M HCl, incubated for 2 h at 37 °C with shaking at 120 rpm and then centrifuged for 20 min at 12,000×g to collect the supernatant. Five milliliters of this supernatant was mixed with 4 mL of simulated intestinal fluid (SIF, pH 7.0) made of KCl (6.8 mM), KH2PO4 (0.8 mM), NaHCO3 (85 mM), NaCl (38.4 mM) and MgCl (H2O)6 (0.33 mM), and then 0.2 mL pancreatin (100 U/mL, in SIF), 25 µL bile salts (10 mM) and 975 µL water. The solution was adjusted to pH 7.0 with 1 M NaOH, incubated (2 h, 37 °C, 120 rpm) and was centrifuged for 20 min at 12,000×g to collect and freeze dry supernatants. HPLC profiles of fractions F2 and F4 were compared before and after gastrointestinal digestions to investigate possible degradation of peptide molecules. Twenty microliters of each digest were injected into a Waters XBridge™ BEH, column (C18; 130 A, 5 µm, 4.6 × 150 mm) at a concentration of 1 mg/mL and separated with linear gradients of acetic acid 0.01% in water (A), and acetic acid 0.01% in methanol (B). F2 was analyzed with the following with a linear gradient of solvent B (10–30%) over 30 min and F4 with a linear gradient of solvent B (25–55%) over 30 min at rate 1 mL/min for 30 min The eluates were detected at 280 nm. Areas under the curve of peaks were used to calculate degradations.
Mass spectroscopy
Samples were analysed by nano LC/MSMS at Quebec Genomics Center (Sainte-Foy, QC, Canada). For each injection, 1 µg of peptide samples were injected and separated by online reversed-phase nanoscale capillary liquid chromatography (nanoLC) and analyzed by electrospray mass spectrometry (ESI–MS/MS). Experiments were performed with a Dionex UltiMate 3000 nanoRSLC chromatography system (Thermo Fisher Scientific/Dionex Softron GmbH, Germering, Germany) connected to an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) equipped with a nanoelectrospray ion source. The acquisition was done over 60 min. Mass spectra were acquired using a data dependent acquisition mode using Thermo XCalibur software version 3.0.63. Full scan mass spectra were acquired from 350 to 1800 m/z. Each MS scan was followed by acquisition of fragmentation MS/MS spectra of the most intense ions for a total cycle time of 3 s (top speed mode). The selected ions (including single charged ion) were isolated using the quadrupole analyzer in a window of 1.6 m/z and fragmented by higher energy Collision-induced Dissociation (HCD) with 35% of collision energy. All MS/MS peak lists (MGF files) were then analyzed using Mascot (Matrix Science, London, UK; version 2.5.1). The Search was limited to the CP_SolanumLycopersicum_ci_4081_UP000004994_20170830 database assuming nonspecific enzyme digestion. The Scaffold software (version 4.7.3, Proteome Software Inc, Portland, OR) was used to validate MS/MS (Nesvizhskii et al. 2003).
Statistical analysis
Minitab (version 18; Minitab Inc., State College, PA, US All) was used to optimize the production condition of hydrolysate protein. RSM with central composite design and two variables (time and enzyme/substrate ratio), five replicates at the central point, without block and alpha = 1 was considered for this test that led to produce 13 hydrolysate treatments. Chemical experiments were performed in triplicates. Data are presented as mean SD (Standard Deviation). The experimental results were analyzed based on response surface method. Comparisons were performed with the Duncan test the significant level set at p < 0.05. Microsoft Excel 2013 was used to plot the curves.
Results and discussion
Optimization of hydrolysis conditions of proteins
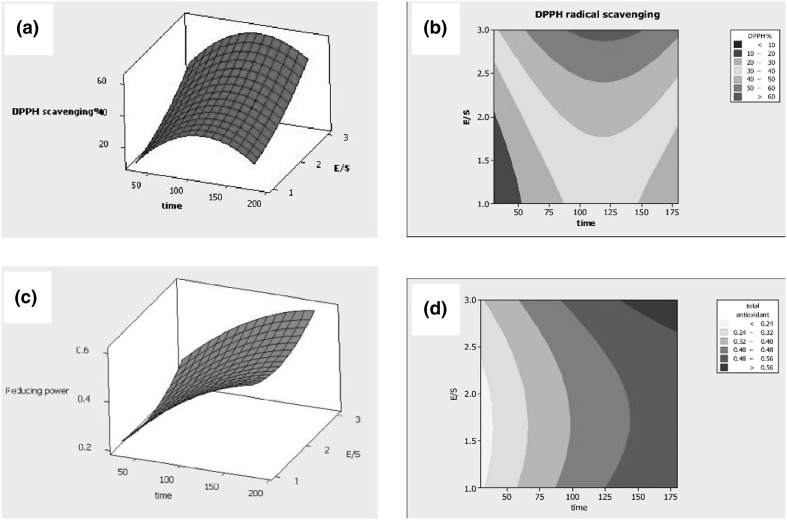
The response surface methodology (RSM) was used to optimize conditions to obtain a hydrolysate with the best antioxidant activities (DPPH and phosphomolybdate) with time and enzyme/substrate ratio as variables while the temperature was maintained constant (50 °C). Figure 1 indicated that increasing hydrolysis time and enzyme had an enhancing effect on free radical scavenging activity. The highest antioxidant power was achieved by applying a defined temperature of 50 °C, 3% enzyme and incubation for 138.62 min. The activity tomato seed protein hydrolysate (TPSH) under these conditions reached 62.99% for DPPH and 54.81% for phosphomolybdate. Coefficients of determination (R2), 83.15 for DPPH and 84.65 for phosphomolybdate showed that regression equations accurately explained the effect of variables (Table 1). These relationships, Eqs. 1 (DPPH) and 2 (phosphomolybdate) showed the quadratic link of variables with the response. The index of lack of fitness for phosphomolybdate reduction was not significant and indicated that the ability of this linear regression to predict the appropriate response. The DPPH radical scavenging capacity of TPSH in this study is in similar to that of tomato seed meals (61.4%) fermented with B. subtilis (Moayedi et al. 2016). In that study however, proteins were not extracted prior to fermentation and there was likely more contribution from non-peptides compared to data from this work.
| 1 |
| 2 |
Fig. 1.
Response surfaces and contour plots for the effect of variables: DPPH radical scavenging activity as a function of different hydrolysis conditions time (min) (a) and enzyme ratio (%) (b); phosphomulybdate reduction as a function of different hydrolysis time (min) (c) and enzyme ratio (%) (d)
Table 1.
ANOVA table for response as affected by independent variables during optimization experiments
| Phosphomolybdate reduction assay | DPPH radical scavenging | |||
|---|---|---|---|---|
| Regression coefficient | p value | Regression coefficient | p value | |
| Regression | 0.238 | 0.000 | − 9.544 | 0.000 |
| Time (X1) | 0.004 | 0.001 | 0.714 | 0.114 |
| Enzyme ratio (X2) | − 0.179 | 0.237 | − 4.629 | 0.003 |
| X1·X2 | − 1.033 | 0.010 | 0.010 | 0.007 |
| − 9.339 | 0.213 | − 0.003 | 0.011 | |
| 0.055 | 0.188 | 4.644 | 0.389 | |
| Lack of fit | 0.12 | 0.54 | ||
| R2 | 84.65 | 83.15 | 0.000 | |
Fractionation of TSPH by ultrafiltration and evaluation of antioxidant activity
Ultrafiltration is a fast and simple method often used to fractionate peptide mixtures based on similarity of molecular weights at both the laboratory and industrial scales (Castel et al. 2012). TSPH produced under optimum conditions had a trichloroacetic acid (TCA) solubility index of 51%. The TCA is one of the most significant criterion for characterizing the extent of protein hydrolysis. The TSPH was separated into three fractions: UF1 (> 10 kDa), UF2 (3–10 kDa) and UF3 (< 3 kDa). Antioxidant activities were determined based on abilities to scavenge DPPH radicals and reduce phosphomolybdate. The following data (DPPH, phosphomolybdate) expressed as IC50 were obtained for UF1 (1.15 ± 0.58 and 0.980 ± 0.2), UF2 (0.498 ± 0.5 and 0.532 ± 0.36), UF3 (0.336 ± 0.2 and 0.387 ± 0.23), and the whole hydrolysate (0.830 ± 0.44 and 0.490 ± 0.41). UF3 had the lowest IC50 (p < 0.05) in both assays and this might be because sequences of its peptides are the smallest. There are literature data indicated that the lower the molecular weight of peptides the greater their free radical scavenging activities in DPHH and other radical assays (Ranathunga et al. 2006; Chi et al. 2014). One possible explanation is the higher steric hindrance of larger molecules in the DPPH assay. Meanwhile, depending on the radical, medium size peptides can have better scavenging activities as reported for human peptide fraction (> 3 kDa) in the peroxyl radical scavenging activity (Tsopmo et al. 2009). Low molecular weight peptides are also physiologically important because they are carriers that might allow their intact absorption through intestinal barrier so that they can exert for example exert their antioxidant effects. Amongst the three fractions only UF3 had lower IC50 (p < 0.05) compared to the whole TPSH hydrolysate and other ultra-filtered fractions, therefore UF3 is a good candidate for further investigation. Other studies have reported that ultrafiltration enhanced the activity of fractions. For example, Foh et al. (2010) showed that the ferric reducing power of the ultrafiltration fraction with molecular weight less than 1000 Da significantly was higher than whole hydrolysate, it also showed an excellent DPPH radical-scavenging activity (Foh et al. 2010). Chi et al. (2014) reported ultra-filtrate fraction from monkfish muscle protein hydrolysate with the lowest molecular weight (MW < 1 kDa) possibly contained more effective antioxidant peptides than the other fractions at the same concentrations (Chi et al. 2014).
Fractionation of UF3 by HPLC and evaluation of antioxidant activity
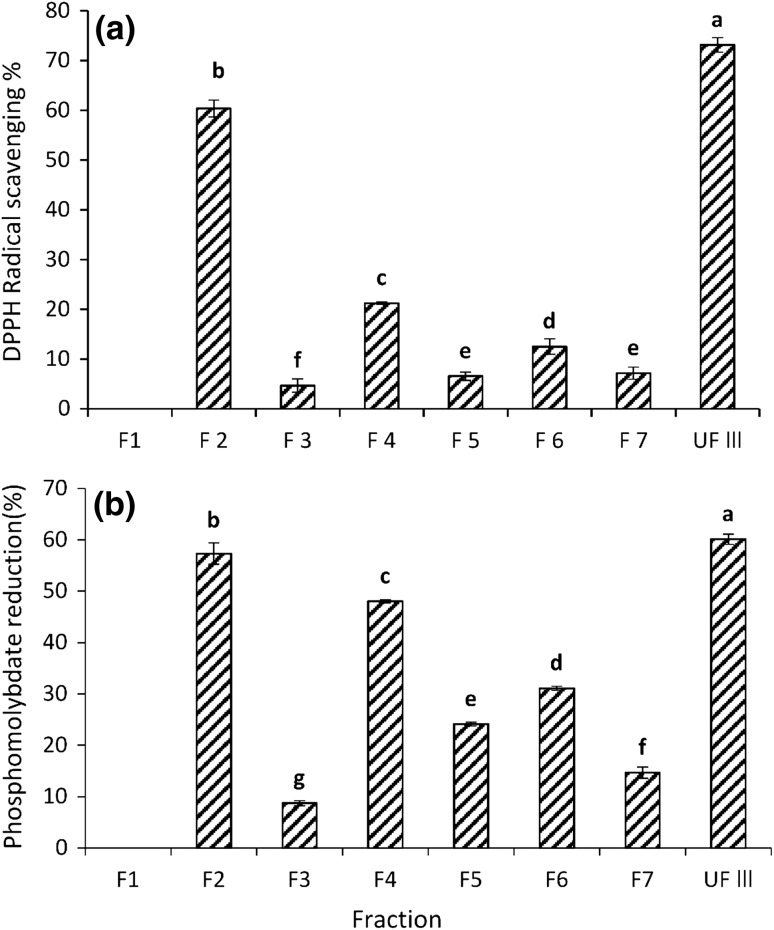
A reverse phase preparative HPLC system was used to further fractionate peptides present UF3 of MW less than 3 kDa, Eluates were pooled and combined into seven fractions (F1–F7) as shown in Fig. 2. The separation was performed on a C18 column and can therefore considered to be based on the hydrophobicity the peptides (Baakdah and Tsopmo 2016). Earlier fractions eluted with greater percentage of water should contain more hydrophilic peptides while those in later eluting fractions should be more hydrophobic because of increasing content of the organic solvent. Fractions were dried and assayed for antioxidant activities at 1 mg/mL concentrations. In the DPPH assay, fractions F2 and F4 had highest (p < 0.05) DPPH radical scavenging activities 60.36 and 21.23%, respectively (Fig. 3a). In the phosphomolybdate reducing test, both fractions also had the highest reducing activities 57.3% (F2) and 48% (F4) (Fig. 3b). It is clear that the HPLC separation of UF3 concentrated the activity in mostly two fractions. There was no relationship between possible increased of either DPPH or phosphomolybdate activities with increased hydrophobicity. Meanwhile in a recent study, the oxygen radical absorbance capacity (ORAC) of HPLC fractions of pepsin hydrolyzed oat proteins separated with the same column increased with increasing hydrophobicity (i.e. content of organic solvent) (Vanvi and Tsopmo 2016). There are several reasons that might explain why there was no correlation in the present work. They include the type of proteins from tomato seeds and oats, differences in assays (DPPH vs. ORAC), and proteases (alcalase vs. pepsin). Compared to UF3 (73.15% DPPH scavenging and 60.1% phosphomolybdate reducing activity at 1 mg/mL), the separation did not significantly enhance the antioxidant activity of fractions. The most active fraction F2 had comparable phosphomolybdate reducing activity but about 17.5% lower DPPH activity at this concentration (Fig. 3). A decrease in antioxidant activity was also reported after HPLC separation of hydrolyzed proteins by (Kim et al. 2007). In their study HPH III fraction from hoki (Johnius belengerii) frame protein hydrolysate that was obtained through separation by ion exchange chromatography showed 83.39% hydroxyl radical scavenging, while, further separation by RP-HPLC (Capcell Pak C18 UG-120 column) and RP-HPLC (Synchropak RPP-100 column) led to decrease in radical scavenging, respectively, below 80 and 30%. It could be due to sample limitations. The intensity of peaks in F2 and F4 (Fig. 2) were higher compared to those of in peaks in F5 and F6 but close to peaks in F3. Meanwhile, this observation is not a reflection of quantities, because the UV absorbance is related only to the presence of some functional groups. F2 is the most potent fraction and because it was eluted earlier with 90–70% of water, one would expect it to contain peptides with polar amino acids such as histidine or glutamine. However, for best activities the presence of hydrophobic amino acids and electron stabilizing amino acid such as tyrosine or tryptophan is also important (Rajapakse et al. 2005).
Fig. 2.
RP-HPLC chromatogram of the UF3 fraction (< 3 kDa) separated on a Waters prep XBridge™ BEH™ column C18 with gradient of 0.01% acetic acid in water and methanol. Fractions were monitored at 280 nm pooled into F1–F7
Fig. 3.
Antioxidant activity of peptide fractions obtained by RP-HPLC on C18 column and detected at 280 nm. a DPPH radical scavenging activity; b Phosphomulybdate reducing activity. Values are means of triplicate determinations (mean ± SD). Different letters represent significant differences
Identification peptides sequences by mass spectrometry
Tandem spectrometry (LC–MS/MS) was used for the identification of peptides in the two HPLC fractions with the highest antioxidant activities. The system was set to perform fragmentation on multiple charge ion peaks because this decrease the possibility of detecting non-peptide molecules. The sequences of peptides were obtained after the analysis of MS/MS peak lists using Mascot and X! Tandem, and statistical interpretation with the Scaffold software as described in the literature (Searle 2010). Several peptides were identified in both fractions (Table 2). A careful analysis showed that many peptides in F2 contained within their sequences amino acids such as tyrosine (Y), histidine (H), proline (P), serine (S), aspartate (D) and glutamate (E) which are known to enhance antioxidant activities of peptides (Hernández-Ledesma et al. 2005; Zhang et al. 2014). Examples of peptides in F2 are HTQHQFFHG, THPDVPGEPT, STTTKKHHPQYL and GVSLIRHVIQ. In F4, peptides such as YRQYPFQQ, SDLDPIRHK and QDRHQKIGQF (Table 2) also possessed amino acids that might be responsible its antioxidant activity, meanwhile, the number of these amino acids was less compared to those F2. This might explain the lower antioxidant activities of F4 relative to F2 although, the bioactivity of peptides is not just linked to the presence of specific amino acids. The location along the chain, the size of the peptide, and a certain degree of hydrophobicity are also important. The role of the above-mentioned amino acids in antioxidant activities is related to various properties. For example, tyrosine is a good hydrogen donor because of the presence of a phenolic group while, histidine is a good hydrogen donor as well but also has chelating and reducing properties due to the presence of an imidazole moiety (Shahidi and Zhong 2008; Darmawan et al. 2010). Hydrophobic amino acids also play important roles in antioxidant activity specifically, in lipid environments due to increased solubility that makes peptides close to oxidized species (Elias et al. 2008; Meshginfar et al. 2017).
Table 2.
Primary biological sequence information
| Peptide fraction | Amino acid sequence | Molecular weight | Predicted protein | Accession number |
|---|---|---|---|---|
| F2 | LIRHVIQSR | 1120.6826 | 12S seed storage protein CRA1 | K4CSI2 |
| F2 | SLSLPNFHPMPRL | 1507.7977 | 11S globulin seed storage protein 2 | K4CUT2 |
| F2 | GHSVIYVQ | 901.4659 | 11S globulin seed storage protein | K4CUT2 |
| F2 | GLLLPHYN | 925.5023 | 11S globulin subunit beta | K4CW41 |
| F2 | ASHGDFRIL | 1014.5251 | Vicilin precursor | B0JEU3 |
| F2 | GREQEREQEQEQEEGDVHYQ | 2502.0642 | Vicilin precursor | B0JEU3 |
| F2 | REQEQEQEEGDVHYQ | 1902.7981 | Vicilin precursor | B0JEU3 |
| F2 | DQSYFVAGPEHRQQ | 1660.7609 | Vicilin precursor | B0JEU3 |
| F2 | THPDVPGEPT | 1048.4821 | Oil body-associated protein 1A | K4CWY3 |
| F2 | ESDLDPIRHK | 1208.6156 | Vicilin protein At2g18540 | K4BB70 |
| F2 | STTTKKHHPQYL | 1439.7519 | Vicilin protein At2g18540 | K4BB70 |
| F2 | VAPDMEHPHGTPGHRHH | 1910.8715 | Peroxygenase | K4DHG6 |
| F2 | HTQHQFFHG | 1137.5094 | Oleosin 1 | K4C4M8 |
| F2 | SGHKIPAIGL | 991.5814 | Aldose reductase | K4CRY0 |
| F2 | PSYLNTPLL | 1016.5545 | 12S seed storage protein CRA1 | K4CSI2 |
| F2 | GVSLIRHVIQ | 1120.6720 | 12S seed storage protein CRA1 | K4CSI2 |
| F2 | VVRPPFSQ | 928.5133 | 12S seed storage protein CRA1 | K4CSI2 |
| F4 | LIRHVIQSR | 1120.6823 | 12S seed storage protein CRA1 | K4CSI2 |
| F4 | YRQYPFQQ | 1128.5349 | 1S globulin seed storage protein 2 | K4CUT2 |
| F4 | SLPNFHPMPR | 1210.5917 | 11S globulin seed storage protein 2 | K4CUT2 |
| F4 | NIGHPTRSDVYNPR | 1624.8070 | Legumin B | K4BDY2 |
| F4 | SPEFEEEQPHRP | 1480.6572 | 11S globulin subunit beta | K4CW41 |
| F4 | ASEEQIRAISEHASRS | 1769.8643 | Vicilin peptides 2–2 | B0JEU3 |
| F4 | THPDVPGEPT | 1048.4821 | Oil body-associated protein 1A | K4CWY3 |
| F4 | MEGPSHGVHPL | 1159.5437 | Oil body-associated protein 1A | K4CWY3 |
| F4 | ESDLDPIRHK | 1208.6156 | Vicilin protein At2g18540 | K4BB70 |
| F4 | KQVHPDIGIS | 1092.5924 | Histone H2B | K4D553 |
| F4 | AKIDWKETPQAH | 1422.7245 | 18.2 kDa class I heat shock protein | K4CRX4 |
| F4 | SDLDPIRHK | 1079.5728 | Vicilin | K4BB70 |
Simulated gastrointestinal digestion: Analysis by RP-HPLC
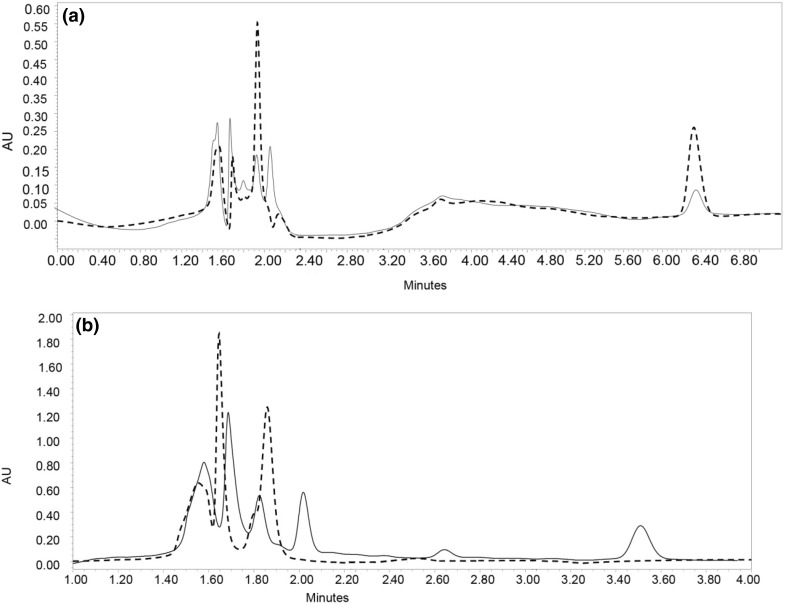
Fractions 2 and 4 were subjected to two step enzymatic hydrolysis with pepsin and pancreatin to evaluate the susceptibility of their peptides gastro-intestinal digestion. The behavior of peptides under these conditions might provide information on their potential presence in vivo and therefore, their potential physiological impact. Each fraction and its digests from the simulated digestion were analyzed by HPLC. Area under the curve of each peak relative to total areas of peaks were used to their relative abundance of peptides before and after digestion. Six main peaks were detected in F2, five of which, remained after the simulated gastrointestinal digestion (Fig. 4a). The relative percentage of areas under peaks 1 and 2 in F2 decreased from 44.9 to 13.1% and 37.5 to 6.0%, respectively after digestion and this might represent a partial digestion of some of peptides that were part of those peaks. For peaks 4 and 6, areas under the curve increased from 6.7 to 15.5% and 27.7 to 28.7%, respectively. The increase is likely due to the fact that the total number of peaks were five in digested F2 compared to six in the non-digested F2 but also because of reduced areas of other peaks. In the case of F4, six peaks were detected before digestion (Fig. 4b), but only three were present after treatment with pepsin and pancreatin. This showed that peptides in F4 are less resistant under the simulated digestion compared to those in F2. This finding indicate that peptide fraction F2 might be more useful in the formulation of functional foods because of its antioxidant activity. In addition, greater resistance to gastrointestinal digestion might make its peptides bioavailable, although, this remains to be tested in biologically relevant models. The net charge of ionizable group of amino acids in peptide chains affects their solubility and possibly their digestion (Marcolini et al. 2015). Fraction F2 were eluted early with more percentage of aqueous solvent and should there contained more ionizable amino acids as reflected by the identified peptides (Table 2). Other studies have reported that alcalase as a serine protease can cleave proteins and release peptides amino acid residues that are not available for active site of pepsin and pancreatin (Segura-Campos et al. 2011). In a related study, the simulated gastrointestinal digestion with pepsin and trypsin of amaranth antioxidant peptides reported minimal changes in activities and a slight increase in peptide peak areas especially in the early times of HPLC separation (Delgado et al. 2011). This is in agreement with data from this work where there changes in area of peaks after digestion.
Fig. 4.
HPLC chromatogram of peptide fractions. a Gastrointestinal digested F2 (dash line), non-digested F2 (black line). b Gastrointestinal digested F4 (dash line), non-digested F4 (black line)
Conclusion
The hydrolysis of tomato seed proteins with alcalase yielded a hydrolysate with good antioxidant activities. Although HPLC fractionation of this hydrolysate did not enhance its activity, it provided two fractions with antioxidant power of up to 95% of that the non-separated sample. Peptides identified in these fractions contain amino acids that make them suitable for evaluation in food systems because of the presence of hydrogen/electron donor and hydrophobic residues. Peptides in fraction F2 have the potential of being bioavailable because of their high stability simulated gastrointestinal digestion.
Acknowledgements
This work was supported by a grant from the National Science and Engineering Research Council of Canada No.: 371908 (AT), and a fellowship from the Ministry of Science, Research and Technology of the Islamic Republic of Iran to NM.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interests.
Contributor Information
Alireza Sadeghi Mahoonak, Phone: +98-911-377-9307, Email: sadeghiaz@gau.ac.ir.
Apollinaire Tsopmo, Phone: +1-613-520-2600, Email: apollinaire_tsopmo@carleton.ca.
References
- Baakdah MM, Tsopmo A. Identification of peptides, metal binding and lipid peroxidation activities of HPLC fractions of hydrolyzed oat bran proteins. J Food Sci Technol. 2016;53:3593–3601. doi: 10.1007/s13197-016-2341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel V, Andrich O, Netto FM, et al. Comparison between isoelectric precipitation and ultrafiltration processes to obtain Amaranth mantegazzianus protein concentrates at pilot plant scale. J Food Eng. 2012;112:288–295. doi: 10.1016/j.jfoodeng.2012.05.010. [DOI] [Google Scholar]
- Chi CF, Wang B, Deng YY, et al. Isolation and characterization of three antioxidant pentapeptides from protein hydrolysate of monkfish (Lophius litulon) muscle. Food Res Int. 2014;55:222–228. doi: 10.1016/j.foodres.2013.11.018. [DOI] [Google Scholar]
- Darmawan R, Bringe NA, de Mejia EG. Antioxidant capacity of alcalase hydrolysates and protein profiles of two conventional and seven low glycinin soybean cultivars. Plant Foods Hum Nutr. 2010;65:233–240. doi: 10.1007/s11130-010-0185-1. [DOI] [PubMed] [Google Scholar]
- Delgado MCO, Tironi VA, Añón M. Antioxidant activity of amaranth protein or their hydrolysates under simulated gastrointestinal digestion. LWT Food Sci Technol. 2011;44:1752–1760. doi: 10.1016/j.lwt.2011.04.002. [DOI] [Google Scholar]
- Elias RJ, Kellerby SS, Decker EA. Antioxidant activity of proteins and peptides. Crit Rev Food Sci Nutr. 2008;48:430–441. doi: 10.1080/10408390701425615. [DOI] [PubMed] [Google Scholar]
- Erdmann K, Cheung BWY, Schröder H. The possible roles of food-derived bioactive peptides in reducing the risk of cardiovascular disease. J Nutr Biochem. 2008;19:643–654. doi: 10.1016/j.jnutbio.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Foh MBK, Qixing J, Amadou I, Xia WS. Influence of ultrafiltration on antioxidant activity of tilapia (Oreochromis niloticus) protein hydrolysate. Adv J Food Sci Technol. 2010;2:227–235. [Google Scholar]
- Hernández-Ledesma B, Dávalos A, Bartolomé B, Amigo L. Preparation of antioxidant enzymatic hydrolysates from α-lactalbumin and β-lactoglobulln. Identification of active peptides by HPLC-MS/MS. J Agric Food Chem. 2005;53:588–593. doi: 10.1021/jf048626m. [DOI] [PubMed] [Google Scholar]
- Hoyle NT, Merritt JH. Quality of fish protein hydrolysates from herring (Clupea harengus) J Food Sci. 1994;59:76–79. doi: 10.1111/j.1365-2621.1994.tb06901.x. [DOI] [Google Scholar]
- Jan S, Khan MR, Rashid U, Bokhari J. Assessment of antioxidant potential, total phenolics and flavonoids of different solvent fractions of monotheca buxifolia fruit. Osong Public Heal Res Perspect. 2013;4:246–254. doi: 10.1016/j.phrp.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Je JY, Kim SK. Purification and characterization of antioxidant peptide from hoki (Johnius belengerii) frame protein by gastrointestinal digestion. J Nutr Biochem. 2007;18:31–38. doi: 10.1016/j.jnutbio.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Marcolini E, Babini E, Bordoni A, et al. Bioaccessibility of the bioactive peptide carnosine during in vitro digestion of cured beef meat. J Agric Food Chem. 2015;63:4973–4978. doi: 10.1021/acs.jafc.5b01157. [DOI] [PubMed] [Google Scholar]
- Meshginfar N, Sadeghi Mahoonak A, Ghorbani M, Aalami M. Effects of protein hydrolysate from sheep visceral on oxidative stability of soybean oil and chicken sausage. J Food Process Preserv. 2017;41:e12875. doi: 10.1111/jfpp.12875. [DOI] [Google Scholar]
- Minekus M, Alminger M, Alvito P, et al. A standardised static in vitro digestion method suitable for food—an international consensus. Food Funct. 2014;5:1113–1124. doi: 10.1039/C3FO60702J. [DOI] [PubMed] [Google Scholar]
- Moayedi A, Hashemi M, Safari M. Valorization of tomato waste proteins through production of antioxidant and antibacterial hydrolysates by proteolytic Bacillus subtilis: optimization of fermentation conditions. J Food Sci Technol. 2016;53:391–400. doi: 10.1007/s13197-015-1965-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Panchaud A, Affolter M, Kussmann M. Mass spectrometry for nutritional peptidomics: how to analyze food bioactives and their health effects. J Proteomics. 2012;75:3546–3559. doi: 10.1016/j.jprot.2011.12.022. [DOI] [PubMed] [Google Scholar]
- Rajapakse N, Mendis E, Byun H-G, Kim S-K. Purification and in vitro antioxidative effects of giant squid muscle peptides on free radical-mediated oxidative systems. J Nutr Biochem. 2005;16:562–569. doi: 10.1016/j.jnutbio.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Ranathunga S, Rajapakse N, Kim SK. Purification and characterization of antioxidative peptide derived from muscle of conger eel (Conger myriaster) Eur Food Res Technol. 2006;222:310–315. doi: 10.1007/s00217-005-0079-x. [DOI] [Google Scholar]
- Ren J, Zhao M, Shi J, et al. Purification and identification of antioxidant peptides from grass carp muscle hydrolysates by consecutive chromatography and electrospray ionization-mass spectrometry. Food Chem. 2008;108:727–736. doi: 10.1016/j.foodchem.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Sarmadi BH, Ismail A. Antioxidative peptides from food proteins: a review. Peptides. 2010;31:1949–1956. doi: 10.1016/j.peptides.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 2010;10:1265–1269. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- Segura-Campos M, Chel-Guerrero L, Betancur-Ancona D, Hernandez-Escalante VM. Bioavailability of bioactive peptides. Food Rev Int. 2011;27:213–226. doi: 10.1080/87559129.2011.563395. [DOI] [Google Scholar]
- Shahidi F, Zhong Y. Bioactive peptides. J AOAC Int. 2008;91:914–931. [PubMed] [Google Scholar]
- Shahidi F, Zhong Y. Novel antioxidants in food quality preservation and health promotion. Eur J Lipid Sci Technol. 2010;112:930–940. doi: 10.1002/ejlt.201000044. [DOI] [Google Scholar]
- Shao D, Atungulu GG, Pan Z, et al. Characteristics of isolation and functionality of protein from tomato pomace produced with different industrial processing methods. Food Bioprocess Technol. 2014;7:532–541. doi: 10.1007/s11947-013-1057-0. [DOI] [Google Scholar]
- Sogi DS, Arora MS, Garg SK, Bawa AS. Fractionation and electrophoresis of tomato waste seed proteins. Food Chem. 2002;76:449–454. doi: 10.1016/S0308-8146(01)00304-1. [DOI] [Google Scholar]
- Sogi DS, Bhatia R, Garg SK, Bawa AS. Biological evaluation of tomato waste seed meals and protein concentrate. Food Chem. 2005;89:53–56. doi: 10.1016/j.foodchem.2004.01.083. [DOI] [Google Scholar]
- Tsopmo A, Diehl-Jones BW, Aluko RE, et al. Tryptophan released from mother’s milk has antioxidant properties. Pediatr Res. 2009;66:614–618. doi: 10.1203/PDR.0b013e3181be9e7e. [DOI] [PubMed] [Google Scholar]
- Vanvi A, Tsopmo A. Pepsin digested oat bran proteins: separation, antioxidant activity, and identification of new peptides. J Chem. 2016;2016:1–8. doi: 10.1155/2016/8216378. [DOI] [Google Scholar]
- Yen G-C, Wu J-Y. Antioxidant and radical scavenging properties of extracts from Ganoderma tsugae. Food Chem. 1999;65:375–379. doi: 10.1016/S0308-8146(98)00239-8. [DOI] [Google Scholar]
- Zhang M, Mu TH, Sun MJ. Purification and identification of antioxidant peptides from sweet potato protein hydrolysates by Alcalase. J Funct Foods. 2014;7:191–200. doi: 10.1016/j.jff.2014.02.012. [DOI] [Google Scholar]