Abstract
Objective(s):
This study was designed to determine the relationship of Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli isolates in multispecies biofilms and their individual phenotypic characters in biofilm consortia.
Materials and Methods:
The subject isolates were recovered from different food samples and identified on the basis of growth on differential and selective media. Tube methods, Congo-red agar method, and scanning electron microscopy (SEM) were used to study biofilms phenotypes. The hydrophobicity of the strains was evaluated by the adhesion to apolar solvent.
Results:
The results showed that E. coli dominated the pre-biofilm stage. It has been observed that E. coli adopted biofilm life much before S. aureus and P. aeruginosa. However, after adopting biofilm lifestyle, slowly and gradually, P. aeruginosa dominated the consortia and dispersed other stakeholders. The subject isolates of P. aeruginosa produce cis-2-decanoic acid to disperse or inhibit S. aureus and E. coli biofilms. Gas-chromatography and mass spectrometry results showed that cis-2-decanoic was higher in the co-culture condition and increased at late log-phase or at stationary phase. Although majority of S. aureus were unable to compete with P. aeruginosa, however, a minor population competed, survived, and persisted in biofilm consortia as small colony variants. The survivors showed higher expression of sigB and sarA genes. P. aeruginosa showed comparatively higher hydrophobic surface properties.
Conclusion:
Comparative analysis showed that cell surface hydrophobicity, growth rate, and small colony variants (SCVs) are correlated in biofilm consortia of the P. aeruginosa, S. aureus, and E. coli.
Key Words: Biofilms, Escherichia coli, Hydrophobicity, Pseudomonas aeruginosa, Small colony variants, Staphylococcus aureus
Introduction
In the natural environment, bacteria exhibit remarkably diverse and complex social cooperation and coordination. Majority of bacteria have the ability to switch from unicellular to multicellular lifestyle such as microbial colonies and biofilms (1, 2). The biofilms are bacterial aggregates adherent to each other and/or to a surface embedded in self-produced extracellular polymeric substances (3). These highly organized communities comprise a high level of morphotypes and phenotypic and genotypic heterogeneity where inter- and intra-species interaction and cooperation is a common phenomenon (4). In biofilm consortia, bacteria suspend their metabolism and cover themselves in a shell made of polysaccharides and proteins (5), which protects biofilm indwellers from the toxic effects of antibacterial agents. Most of the antibiotics target actively growing bacteria and are unable to penetrate protective layer of biofilms (5). In biofilm structure, coordination and cooperation take place via inter and intraspecies exchange of metabolites, signaling molecules, genetic material, and defensive compounds (4). More often diffusible signal molecules or cell-density related (quorum sensing - QS) molecules are released to communicate and induce expression of certain genes in neighboring cells (6). According to Atkinson and Williams (7), bacteria are not limited to communication within their own species but are capable of ‘listening in’ and ‘broadcasting to’ unrelated species to intercept messages and coerce cohabitants into behavioral modifications, either for the good of the population or for the benefit of one species over another. It is highly evident that several species of bacteria coexist and interact in multispecies biofilm consortia and all of the indwellers coordinate for survival (8). In settings where multiple species coexist, their interactions often are mediated through extracellular compounds (4). Development in one microbe can be influenced by small molecules secreted by other species (9). As the organisms adhere to a surface, they keep signaling to one another. Once they sense a quorum, genes are up-regulated and sticky exo-polysaccharides are produced that ‘glue’ the bacteria together (4, 5). It has been reported that Escherichia coli interacts with other bacteria, e.g. pseudomonas and staphylococci and is able to form multi-species biofilms (5). Similarly, Pseudomonas aeruginosa PAO1 has been reported to facilitate Staphylococcus aureus biofilm formation when both are grown in coculture biofilms (10). In the process, eDNA facilitates interspecies interaction between P. aeruginosa and S. aureus (4, 11). It has also been reported that exoproducts of P. aeruginosa recovered from cystic fibrosis patients stimulate S. aureus biofilms (12). Similarly, peptidoglycan shed by S. aureus was found to stimulate the production of virulence factors in P. aeruginosa (12, 13). Likewise, P. aeruginosa can greatly increase the ability of E. coli to persist and grow in aquatic environments as well as in biofilm formation (14). These three pathogens S. aureus, P. aeruginosa, and E. coli are well known for their versatility and pathogenicity. The study of physiology and behavior of multispecies biofilm of foodborne pathogens in food processing environments may provide the necessary information to prevent and reduce the contamination of food products. Therefore, it is necessary to understand the inter-species and intra-species interactions, i.e. population structure, cell surface charges, physiology, and function within a biofilm. Although, S. aureus, P. aeruginosa, and E. coli are associated with the hospital as well as community-acquired infections, to the best of our knowledge, nothing is yet known about the formation of a mixed-species biofilm composed of these pathogens. The aim of this study was to characterize the inter-species interaction of these pathogens in biofilm consortia.
Materials and Methods
In the present study, a total of seventeen biofilm-producing and five non-biofilm-producing strains of P. aeruginosa, E. coli, and S. aureus were used. The subject strains were isolated from different food samples. These isolates were identified on the basis of typical morphology by gram staining and growth on differential and selective media, e.g. Cetrimide Agar (Merck, Germany) for P. aeruginosa, Eosin methylene blue (EMB, Oxoid) and MacConkey agar (Oxoid) for E. coli, and Baird–Parker agar (Oxoid) with egg yolk-tellurite (Sigma) for S. aureus. Coagulase and DNase tests were used for confirmation of S. aureus and oxidase test was performed for confirmation of P. aeruginosa and E. coli. E. coli (ATCC8739), S. aureus (ATCC25923), and P. aeruginosa (ATCC9027) were used as positive control for identification.
Phenotypic characterization of slime-producing bacteria
Biofilm formation was initially confirmed by the Congo red agar method as described earlier (15-17). Briefly, BHI agar (Oxoid) plates containing 50 g/l sucrose and 0.8 g/l Congo red were prepared and streaked with strains and incubated aerobically for 24–48 hr at 37 °C. Positive results were indicated by black colonies with dry crystalline appearance. Weak slime producers usually remained pink, though occasional darkening at the center of colonies was observed.
Biofilm assay
A qualitative assessment of biofilm formation on glass slides was evaluated as described earlier by Mirani and Jamil (18). As it was difficult to study all of the seventeen isolates in different combinations, the isolates were randomly selected by using Microsoft Excel random number (RAND) generator from the group of subject isolates of this study. Briefly, two-inch pieces of glass slides were submerged in BHI broth (Oxoid) containing 0.1 ml of the 4 hr young culture of subject isolates of P. aeruginosa, E. coli, and S. aureus in a combination of randomly selected isolates and incubated at 37 °C for 48 hr to 96 hr.
P. aeruginosa + E. coli + S. aureus
P. aeruginosa + E. coli
P. aeruginosa + S. aureus
E. coli + S. aureus
All isolates in pure form
After incubation, glass slides were washed with phosphate buffer saline (pH 7.0) to remove unbound cells and debris; films were fixed with acetic acid for 15 min and stained with 3% crystal violet. Each experiment was repeated three times.
Quantification of biofilms
Biofilm formation was quantified by the addition of 2 × 200 μl of 95% ethanol as described previously (19) and absorbance was recorded with a spectrophotometer (Nicollet Evolution 300 BB) at 563 nm wavelength.
Enumeration of biofilm population
After maturation of the biofilm, the glass slides (4 mm) were gently washed three times with phosphate-buffered saline (PBS) to remove debris. After washing, glass slides were transferred to sterile 5 ml tubes containing 3 ml PBS and vortexed at 3000 rpm for 2 min to separate cells from biofilms. After vortexing, the extracted bacteria were enumerated using agar dilution plating technique. To perform it, 10 fold serial dilutions (1/10, 1/100, and 1/1000) were made from each sample containing the dislodged bacteria and 10 ml were seeded to calculate an accurate count of the biofilm population. Each experiment was performed in triplicate.
Evaluation of colony variance during biofilm development and detection of persister cells
The emergence of colony variants associated with biofilms of subject isolates was studied and these variants were enumerated, as described in a previous study (20). Biofilm biomass was harvested from a glass slide, resuspended in saline (total volume of 1 ml), homogenized for 30 sec to disrupt cell clusters by vigorous shaking, serially diluted, and plated on tryptone soy agar (Oxoid) and Baird–Parker agar (Oxoid), Cetrimide agar, and EMB agar plates. For the determination of stability of the colony variants, well-isolated colonies were sub-cultured on tryptone soy agar and incubated for 24 hr. This was repeated six times, and reversion with respect to colony size and biochemical reactions was monitored as described elsewhere (21). The experiments were performed in duplicate. The persister cells obtained were characterized for stability, oxidase, hemolysis, catalase production, clumping factor, coagulase production, and DNase production by the method of Bayston et al (22). The drop plate method described by researchers (23) was followed to count CFUs.
Scanning electron microscopy (SEM)
Scanning electron microscopy (SEM) was done to analyze the production of extracellular matrix material and cell morphology as described earlier (18). Biofilm slides were divided into 4 mm sections and washed with distilled water to remove debris, and negatively stained with 0.2% uranyl acetate for 30 sec. These 4 mm slide sections showed the presence of biofilm material when examined directly in a GOEL-JEM-1200 EX II Electron Microscope.
Bacterial hydrophobicity assay
The hydrophobicity of strains was evaluated by the microbial adhesion to solvent test as described in a study (24). It consisted of evaluating the affinity of the cells towards apolar solvents (hexadecane). For the experiment, bacterial cells were harvested by centrifugation at 8500 g for 5 min and resuspended to ABS 578 nm in 0.01 M potassium phosphate buffer (pH 7.0). This bacterial suspension was mixed with a solvent in a ratio of 1:6 (0.4/2.4 v/v) by vortexing for 3 min to make an emulsion. The mixture was then left for 30 min until the separation of the two phases. Aqueous phase absorbance was measured (ABS 2) and the percentage of adhesion was expressed as %adhesion= (1–ABS 2/ ABS 1) × 100.
Extracellular fatty acid (Cis-2-decenoic acid) extraction and methylation
Fatty acids extraction was achieved by a method as described in a study (25). 50-ml samples of cell cultures of different groups were collected at different time intervals and centrifuged at 10,000 rpm for 15 min to remove cells and debris to collect the cell-free supernatant. The extracellular fatty acid in the supernatant was extracted with 50 ml of chloroform/ methanol mixture (2:1, v/v). The cell pellets were re-suspended in 10 ml distilled water and mixed with 20 mL chloroform/ methanol mixture (2:1, v/v). Cis-2-decenoic acid 100 μl/l was added as the internal standard. The chloroform layer was evaporated under a nitrogen stream and the dried fatty acid sample was used for the analysis of extracellular fatty acid. Separated fatty acids were methylated to prepare fatty acid methyl esters as described by Mirani and Jamil (26).
GC-MS analysis The GC-MS analysis for fatty acid methyl esters was performed as described previously by Mirani and Jamil (26) on an Agilent 6890 N Gas Chromatographic instrument coupled with an Agilent MS-5975 inert XL Mass Selective Detector and an Agilent auto-sampler 7683-B Injector (Agilent technologies, Little Fall, NY, USA). A capillary column HP-5MS (5% phenyl methylsiloxane) with a dimension of 60 m x 0.25 mm i.d × 0.25 µm film thickness (Agilent technologies, Palo, Alto, CA, USA) was used for the separation of fatty acid methyl esters. The initial temperature was 150 ºC, which was maintained for 2 min, raised to 240 ºC at the rate of 3 ºC/min and kept at 230 ºC for 10 min. The split ratio was 1:50 and helium was used as a carrier gas with the flow rate of 0.8 ml/min. The injector and detector were 240 and 270 ºC, respectively. The mass spectrometer was operated in the electron impact (EI) mode at 70 eV and scan range of 50–800 m/z. Library search identification of compounds from mass spectra was optimized and tested by matching test spectra against reference spectra in the NIST mass spectral database. Each experiment was repeated three times.
Calculation and standard analysis
Peak identification of analyzed extracellular fatty acids was covered by the comparisons with retention time and mass-spectra of known standards. Standard methyl esters of Cis-2-decenoic acid (100 μl/l) were used for the confirmation of GC-MS library results. All samples were used in duplicate, analyzed three times, and reported as n=2×3.
Polymerase chain reaction
The SCCmec elements (I–V) and mecA gene were identified as previously described (27, 28). Expression of icaA, sigB, and sarA genes was studied by using specific primers reported earlier (29-31). Total RNA was recovered from exponentially growing cells in tryptone soy broth (OD at 578 nm) using a dedicated kit (Qiagen Rneasy Mini, Qiagen, Hilden, Germany) and stored at −20 °C. DNA was removed from RNA extractions using DNase according to the manufacturer’s instructions and RNA concentration was quantified using a spectrophotometer (Evolution 300 BB, Thermoelectro Corporation, Madison, WI, USA). One µg of RNA was used per reverse transcriptase-PCR (One-step RT-PCR Kit, Qiagen) together with gene-specific primers. Moreover, P. aeruginosa and E. coli were reconfirmed by using specific primers targeting 16s ribosomal DNA (rDNA) and fimA, respectively, as described by Spilker et al. (32) and Naravaneni and Jamil (33) (supplement 1). E. coli (ATCC8739), S. aureus (ATCC25923) and P. aeruginosa (ATCC9027) were used as positive control for identification.
Statistical analysis
Data analysis was accomplished by using SPSS version 17.0 for frequencies. Data presented in result tables are mean of three independent experiments.
Results
Co-culture of biofilm positive P. aeruginosa, S. aureus and E. coli
P. aeruginosa, S. aureus, and E. coli are among the most prevalent foodborne pathogens. This study was designed to investigate the interspecies interactions of these pathogens in multi-species biofilms. For this study, seventeen biofilm positive isolates of each of P. aeruginosa, S. aureus, and E. coli were cocultured in TSB broth in different combinations. Initially, the individual characteristics of these isolates were studied in mono-species biofilm consortia. After adaptation to biofilm mode of life, all of these isolates were found to be more hydrophobic as compared to planktonic or wild-type isolates and cell surface hydrophobicity increased with incubation time (Table 1). This is a common character of biofilm positive isolates studied. Comparative analysis showed that P. aeruginosa possessed more hydrophobic surfaces among three subject isolates studied. Similarly, S. aureus showed more hydrophobic surface properties as compared to E. coli (Table 1). The other common character was the correlation of cell surface hydrophobicity and small colony variants or persister (metabolically inactive) or viable but not culturable population. This population increases with age of biofilm consortia. The old biofilm consortia (i.e. 48 to 96 hr) were found to be dominated by metabolically inactive and more hydrophobic cells (Table 1). These phenotypes were not recovered at pre-biofilm or at planktonic stage (Table 1 to 5). This phenotypic heterogeneity was a common character of S. aureus (100%) and P. aeruginosa (76.4%) biofilm consortia. However, in E. coli 41.1% of biofilm consortia have shown this phenotypic heterogeneity. Viable plate count showed that CFU count dropped one logarithmic step after adopting biofilm mode of life in the entire subject isolates (Table 1). Population analysis assay showed that at pre-biofilm stage E. coli population dominated the multi-species biofilm consortia and consistently over-grew P. aeruginosa and S. aureus (Table 2). Results indicated that subject isolates of S. aureus and E. coli produced different amounts of biofilm but had similar timing of biofilm peaks and declines. Although, P. aeruginosa takes more time, comparatively, to enter from planktonic to biofilm mode of lifestyle, after adopting biofilm lifestyle P. aeruginosa showed dominance over S. aureus and E. coli (Table 2). Biofilm mass study by the crystal violet method indicates that average P. aeruginosa biomass was greater than that of S. aureus and E. coli. P. aeruginosa formed highly organized biofilms with inter-connected cells and continuous surface coverage (Figures 1a, b & c). Similar properties were observed in S. aureus monoculture biofilms (Figures 1f & g). Interestingly, after adaptation of biofilm mode of life, a morphological change in P. aeruginosa has been noticed (Figures 1b &c). The cells covered with extracellular matrix material showed a bricks like appearance. Conversely, E. coli showed scant biofilms with very small, discontinuous micro-colonies in monoculture and in co-culture conditions (Figures 1d & e).
Table 1.
Characterization of Mono-Specie Biofilms of Pseudomonas aeruginosa (P.A), Escherichia coli (E.C) and Staphylococcus aureus (S.A). Cell surface hydrophobicity, Planktonic population and SCVs or persister cells population at pre and post biofilm stage
| Biofilm optical density (OD578a) |
Cell surface hydrophobicity |
Pre-biofilm population at 24 hr | Biofilm population at 48 hr |
SCVs or persister cell population of biofilms | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P.A | E.C | S.A | Pre-Biofilm Stage at 24 hr | Biofilm Stage at 48 hr |
|||||||||||||||||||
| 24 hr | 48 hr | 24 hr | 48 hr | 24 hr | 48 hr | P.A | E.C | S.A | P.A | E.C | S.A | P.A | E.C | S.A | P.A | E.C | S.A | P.A | E.C | S.A | |||
| 0.33 | 0.82 | 0.13 | 0.55 | 0.39 | 0.66 | 0.41 | 0.46 | 0.51 | 0.96 | 0.57 | 0.95 | 1x105 | 1x105 | 1x105 | 1x104 | 1x103 | 1x103 | 1x102 | <1cfu | 1x101 | |||
| 0.41 | 0.89 | 0.15 | 0.59 | 0.08 | 0.92 | 0.63 | 0.45 | 0.55 | 0.99 | 0.58 | 0.99 | 1x105 | 1x106 | 1x105 | 1x103 | 1x103 | 1x103 | 1x102 | <1cfu | 1x101 | |||
| 0.25 | 0.88 | 0.15 | 0.57 | 0.23 | 0.89 | 0.67 | 0.46 | 0.57 | 0.93 | 0.53 | 0.95 | 1x105 | 1x106 | 1x105 | 1x103 | 1x103 | 1x104 | 1x102 | <1cfu | 1x103 | |||
| 0.32 | 0.78 | 0.17 | 0.59 | 0.27 | 0.73 | 0.77 | 0.53 | 0.55 | 0.94 | 0.54 | 0.91 | 1x105 | 1x106 | 1x105 | 1x103 | 1x104 | 1x104 | 1x101 | <1cfu | 1x103 | |||
| 0.24 | 0.99 | 0.34 | 1.13 | 0.17 | 0.86 | 0.72 | 0.62 | 0.59 | 0.92 | 0.90 | 0.91 | 1x105 | 1x106 | 1x105 | 1x103 | 1x104 | 1x104 | 1x102 | 1x101 | 1x103 | |||
| 0.42 | 1.11 | 0.33 | 0.98 | 0.21 | 0.60 | 0.73 | 0.61 | 0.63 | 0.91 | 0.85 | 0.99 | 1x105 | 1x106 | 1x105 | 1x103 | 1x103 | 1x104 | 1x102 | 1x101 | 1x103 | |||
| 0.36 | 0.91 | 0.37 | 0.97 | 0.22 | 0.79 | 0.67 | 0.56 | 0.61 | 0.89 | 0.83 | 0.91 | 1x105 | 1x105 | 1x105 | 1x103 | 1x103 | 1x104 | 1x102 | <1cfu | 1x102 | |||
| 0.33 | 0.52 | 0.46 | 0.51 | 0.35 | 0.49 | 0.59 | 0.52 | 0.57 | 0.68 | 0.62 | 0.92 | 1x105 | 1x105 | 1x105 | 1x103 | 1x104 | 1x104 | <1cfu | <1cfu | 1x102 | |||
| 0.39 | 0.88 | 0.37 | 0.73 | 0.33 | 0.45 | 0.62 | 0.51 | 0.68 | 0.90 | 0.77 | 0.91 | 1x105 | 1x106 | 1x105 | 1x103 | 1x104 | 1x104 | 1x102 | 1x102 | 1x102 | |||
| 0.26 | 0.88 | 0.45 | 0.98 | 0.55 | 0.87 | 0.74 | 0.47 | 0.61 | 0.92 | 0.86 | 0.91 | 1x105 | 1x105 | 1x105 | 1x103 | 1x104 | 1x103 | 1x102 | 1x102 | 1x102 | |||
| 0.34 | 0.56 | 0.44 | 0.63 | 0.54 | 0.81 | 0.77 | 0.43 | 0.45 | 0.85 | 0.64 | 0.59 | 1x105 | 1x106 | 1x105 | 1x103 | 1x104 | 1x103 | 1x102 | <1cfu | 1x101 | |||
| 0.27 | 0.79 | 0.49 | 1.07 | 0.55 | 0.86 | 0.72 | 0.62 | 0.35 | 0.87 | 0.81 | 0.55 | 1x105 | 1x105 | 1x105 | 1x103 | 1x103 | 1x103 | <1cfu | <1cfu | 1x101 | |||
| 0.33 | 0.97 | 0.41 | 0.52 | 0.25 | 0.81 | 0.64 | 0.63 | 0.45 | 0.84 | 0.72 | 0.56 | 1x105 | 1x106 | 1x105 | 1x103 | 1x104 | 1x104 | 1x102 | <1cfu | 1x101 | |||
| 0.43 | 0.98 | 0.41 | 0.53 | 0.09 | 0.85 | 0.68 | 0.64 | 0.59 | 0.83 | 0.73 | 0.87 | 1x105 | 1x106 | 1x105 | 1x103 | 1x104 | 1x103 | 1x102 | 1x101 | 1x102 | |||
| 0.41 | 0.95 | 0.47 | 0.55 | 0.08 | 0.97 | 0.66 | 0.66 | 0.51 | 0.94 | 0.71 | 0.97 | 1x105 | 1x106 | 1x105 | 1x103 | 1x104 | 1x103 | <1cfu | 1x102 | 1x103 | |||
| 0.32 | 0.62 | 0.45 | 1.04 | 0.23 | 0.52 | 0.49 | 0.64 | 0.44 | 0.76 | 0.91 | 0.99 | 1x105 | 1x105 | 1x105 | 1x103 | 1x103 | 1x104 | <1cfu | 1x102 | 1x103 | |||
| 0.33 | 0.61 | 0.49 | 1.04 | 0.27 | 0.79 | 0.43 | 0.65 | 0.47 | 0.73 | 0.88 | 0.82 | 1x105 | 1x105 | 1x105 | 1x103 | 1x103 | 1x103 | 1x102 | <1cfu | 1x102 | |||
Table 2.
Characteristics of Multi-specie Biofilms of Escherichia coli, Staphylococcus aureus and Pseudomonas aeruginosa, Cell surface hydrophobicity, Planktonic population and SCVs or persister cells population at pre and post biofilm stage
| Combinations | S # | Biofilm OD578a | Cell surface hydrophobicity |
Pre-biofilm population after 18 hr incubation |
Biofilm population after 48 hr of incubation |
SCVs or persister cell population of biofilms |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18 hr | 24 hr | 48 hr | E. coli | S. aureus | P. aeruginosa | E. coli | S. aureus | P. aeruginosa | E. coli | S. aureus | P. aeruginosa | |||
| Biofilm positive isolates in multi-specie biofilms | 1 | 0.93 | 0.45 | 0.68 | 0.77 | 1x105 | 1x104 | 1x102 | 1x102 | <10CFU | 1x103 | 1x101 | <10CFU | 1x102 |
| 2 | 0.92 | 0.33 | 0.66 | 0.71 | 1x105 | 1x104 | 1x102 | 1x102 | <10CFU | 1x103 | 1x101 | <10CFU | 1x102 | |
| 3 | 0.89 | 0.41 | 0.73 | 0.82 | 1x105 | 1x104 | 1x103 | 1x102 | 1x103 | 1x103 | 1x101 | <10CFU | 1x102 | |
| 4 | 0.97 | 0.44 | 0.77 | 0.87 | 1x105 | 1x104 | 1x103 | <10CFU | 1x103 | 1x103 | <10CFU | 1x101 | 1x102 | |
| 5 | 0.99 | 0.22 | 0.71 | 0.89 | 1x104 | 1x104 | 1x103 | 1x102 | <10CFU | 1x103 | 1x101 | <10CFU | 1x102 | |
| 6 | 1.01 | 0.24 | 0.73 | 0.94 | 1x105 | 1x104 | 1x103 | 1x102 | <10CFU | 1x103 | <10CFU | <10CFU | 1x102 | |
| 7 | 1.03 | 0.26 | 0.71 | 0.91 | 1x105 | 1x105 | 1x103 | <1CFU | 1x103 | 1x103 | <10CFU | <10CFU | 1x102 | |
| 8 | 0.92 | 0.34 | 0.67 | 0.77 | 1x104 | 1x105 | 1x103 | 1x103 | <10CFU | 1x103 | <10CFU | <10CFU | 1x102 | |
| 9 | 1.04 | 0.27 | 0.69 | 0.93 | 1x105 | 1x104 | 1x103 | 1x103 | <10CFU | 1x103 | <10CFU | <10CFU | 1x102 | |
| Biofilm Negative Isolates in multi-specie biofilms | 1 | BND | 0.22 | 0.68 | 0.51 | 1x105 | 1x104 | 1x104 | BND | BND | BND | BND | BND | BND |
| 2 | BND | 0.19 | 0.66 | 0.52 | 1x105 | 1x105 | 1x104 | BND | BND | BND | BND | BND | BND | |
| 3 | 0.86 | 0.33 | 0.69 | 0.72 | 1x105 | 1x104 | 1x103 | 1x102 | <10CFU | 1x103 | <10CFU | <10CFU | 1x102 | |
| 4 | 0.88 | 0.37 | 0.41 | 0.74 | 1x105 | 1x104 | 1x103 | 1x102 | <10CFU | 1x103 | 1x101 | <10CFU | 1x101 | |
| 5 | BND | 0.31 | 0.44 | 0.46 | 1x105 | 1x104 | 1x104 | BND | BND | BND | BND | BND | BND | |
| 6 | BND | 0.28 | 0.43 | 0.51 | 1x105 | 1x104 | 1x104 | BND | BND | BND | BND | BND | BND | |
| 7 | BND | 0.27 | 0.42 | 0.55 | 1x105 | 1x104 | 1x104 | BND | BND | BND | BND | BND | BND | |
| 8 | BND | 0.22 | 0.34 | 0.52 | 1x105 | 1x104 | 1x104 | BND | BND | BND | BND | BND | BND | |
| 9 | BND | 0.18 | 0.33 | 0.47 | 1x105 | 1x104 | 1x103 | BND | BND | BND | BND | BND | BND | |
Figure 1.
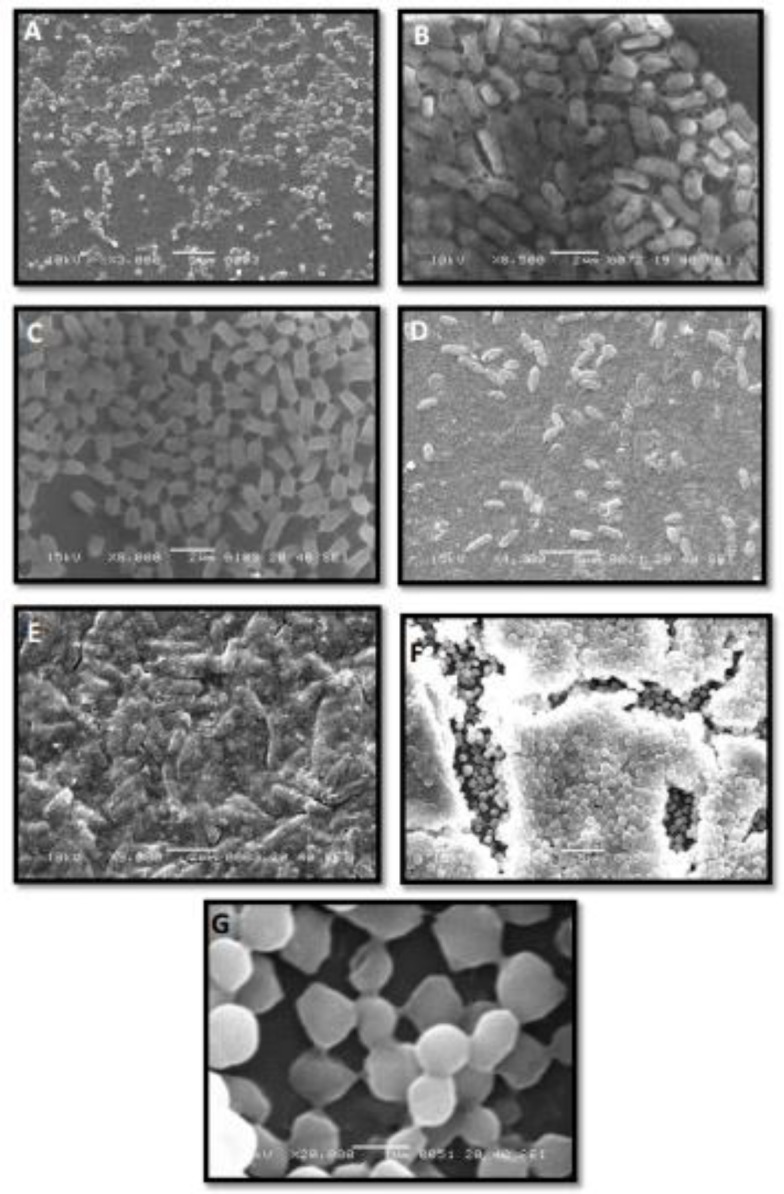
(A) Pseudomonas aeruginosa biofilm negative control, (B) P. aeruginosa biofilms, cells surrounded with the extracellular matrix material and showing brick-like appearance. (C) Highly adherent small colony variants of P. aeruginosa. (D) Biofilm negative Escherichia coli. (E) Biofilm consortia of E. coli. (F) Biofilm consortia of Staphylococcus aureus, cells surrounded with matrix material, showing multilayered structure. (F) Highly adherent small colony variants of S. aureus
Co-culture of S. aureus and P. aeruginosa
Out of five, three co-cultured combinations showed biofilm formation. Surprisingly, P. aeruginosa dominated S. aureus at the pre-biofilm stage as well as in biofilm consortia (Table 4). Population analysis profile of biofilm consortia showed only P. aeruginosa. Moreover, when biofilm negative isolates were co-cultured, out of five only one showed biofilm formations and this was also dominated by P. aeruginosa (Table 4). Co-culture of biofilm positive P. aeruginosa with biofilm negative S. aureus did not show any impact on P. aeruginosa biofilms. However, biofilm negative P. aeruginosa with biofilm positive S. aureus resulted in inhibition of S. aureus biofilm formation. Surprisingly, biofilm negative P. aeruginosa adopted biofilm mode of life after inhibition of S. aureus biofilm formation. On Congo-red agar, parallel inoculation of P. aeruginosa and biofilm positive S. aureus showed biofilm positive S. aureus growth without blackening. This suggests inhibition of polysaccharide production of S. aureus, which is a pre-requisite for S. aureus adhesion and biofilm formation (Table 2 & 4).
Table 4.
Characteristics of Duo-Specie Biofilms of Pseudomonas aeruginosa and Staphylococcus aureus Cell surface hydrophobicity, Planktonic population and SCVs or persister cells population at pre and post biofilm stage
| Combination | S # | Biofilm OD578a | Cell surface hydrophobicity |
Pre-biofilm population after 24 hr of incubation |
Biofilm population after 48 hr of incubation |
SCVs or persister cell population of biofilms |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| At 18 hr | At 24 hr | At 48 hr | P. aeruginosa | S. aureus | P. aeruginosa | S. aureus | P. aeruginosa | S. aureus | |||
| Biofilm positive isolates | 1 | 0.97 | 0.23 | 0.49 | 0.88 | 1x104 | 1x105 | 1x103 | 1x101 | 1x102 | <10CFU |
| 2 | 0.99 | 0.21 | 0.63 | 0.87 | 1x104 | 1x105 | 1x103 | <10CFU | 1x102 | <10CFU | |
| 3 | 0.93 | 0.21 | 0.47 | 0.81 | 1x104 | 1x105 | 1x103 | <10CFU | 1x102 | <10CFU | |
| 4 | 0.98 | 0.28 | 0.57 | 0.88 | 1x104 | 1x104 | 1x102 | <10CFU | 1x102 | <10CFU | |
| 5 | 0.97 | 0.31 | 0.61 | 0.82 | 1x104 | 1x105 | 1x102 | <10CFU | 1x102 | <10CFU | |
| Biofilm negative isolates | 1 | BND | 0.19 | 0.27 | 0.28 | 1x104 | 1x105 | BND | BND | BND | BND |
| 2 | BND | 0.18 | 0.27 | 0.31 | 1x104 | 1x105 | BND | BND | BND | BND | |
| 3 | 0.61 | 0.31 | 0.39 | 0.57 | 1x104 | 1x105 | 1x102 | <10CFU | <10CFU | <1CFU | |
| 4 | BND | 0.13 | 0.21 | 0.25 | 1x104 | 1x105 | BND | BND | BND | BND | |
| 5 | BND | 0.21 | 0.19 | 0.35 | 1x104 | 1x105 | BND | BND | BND | BND | |
| Biofilm positive S. aureus with biofilm negative P. aeruginosa |
1 | BND | 0.19 | 0.26 | 0.33 | 1x105 | 1X103 | BND | BND | BND | BND |
| 2 | BND | 0.21 | 0.28 | 0.31 | 1x104 | 1X104 | BND | BND | BND | BND | |
| 3 | 0.58 | 0.26 | 0.47 | 0.66 | 1x105 | 1X104 | 1x103 | <10CFU | <10CFU | <10CFU | |
| 4 | 0.66 | 0.19 | 0.49 | 0.73 | 1x105 | 1X103 | 1x103 | <10CFU | <10CFU | <10CFU | |
| 5 | 0.63 | 0.23 | 0.53 | 0.79 | 1x104 | 1X103 | 1x103 | <10CFU | <10CFU | <10CFU | |
| Biofilm negative S. aureus with biofilm positive P. aeruginosa |
1 | 0.66 | 0.22 | 0.56 | 0.77 | 1x104 | 1X104 | 1X103 | <10CFU | 1x102 | <10CFU |
| 2 | 0.81 | 0.22 | 0.49 | 0.81 | 1x104 | 1X104 | 1X103 | <10CFU | 1x102 | <10CFU | |
| 3 | 0.89 | 0.21 | 0.53 | 0.83 | 1x104 | 1X104 | 1X103 | <10CFU | <10CFU | <10CFU | |
| 4 | 0.82 | 0.23 | 0.52 | 0.84 | 1x104 | 1X104 | 1X103 | 1x102 | 1x102 | <10CFU | |
| 5 | 0.88 | 0.25 | 0.55 | 0.82 | 1x104 | 1X104 | 1X103 | 1x102 | 1x102 | <10CFU | |
Co-culture of S. aureus with E. coli
Co-culture of S. aureus with E. coli showed that the E. coli dominated pre-biofilm stage (Table 2 & 3). However, in biofilm consortia, S. aureus overcame E. coli and occupied more space. Like P. aeruginosa, S. aureus also showed organized and inter-connected cells. The aged biofilm consortia was also dominated by SCVs of S. aureus (Tables 2 & 3).
Figure 3.
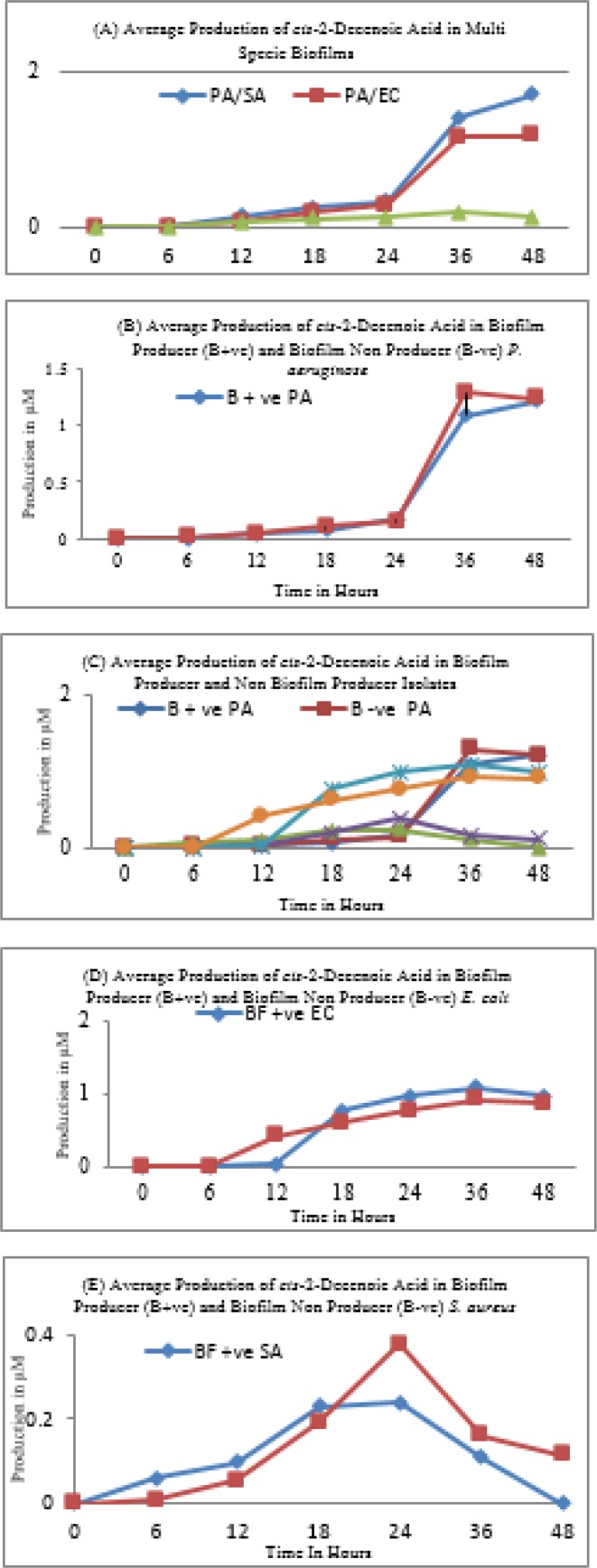
Production and impact of cis-2-Decenoic acid on biofilm. PA= Pseudomonas aeruginosa, SA=Staphylococcus aureus, EC=Escherichia coli
Table 3.
Characteristics of duel specie biofilms of Escherichia coli and Staphylococcus aureus Cell surface hydrophobicity, Planktonic population and SCVs or persister cells population at pre and post biofilm stage
| Combination | S # | Biofilm OD578a | Cell surface hydrophobicity |
Pre-biofilm population after 24 hr of incubation |
Biofilm population after 48 hr of incubation |
SCVs or persister cell population of biofilms |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| At 18 hr | At 24 hr | At 48 hr | E. coli | S. aureus | E. coli | S. aureus | E. coli | S. aureus | |||
| Biofilm positive Isolates | 1 | 0.89 | 0.33 | 0.61 | 0.77 | 1x105 | 1x104 | 1x102 | 1x103 | 1x101 | 1x102 |
| 2 | 0.83 | 0.41 | 0.63 | 0.74 | 1x105 | 1x104 | 1x101 | 1x103 | 1x101 | 1x102 | |
| 3 | 0.92 | 0.44 | 0.56 | 0.82 | 1x105 | 1x104 | 1x102 | 1x102 | <10CFU | 1x102 | |
| 4 | 0.99 | 0.42 | 0.55 | 0.93 | 1x105 | 1x105 | 1x102 | 1x102 | <10CFU | 1x103 | |
| 5 | 0.93 | 0.26 | 0.62 | 0.87 | 1x104 | 1x104 | 1x102 | 1x102 | <10CFU | 1x102 | |
| Biofilm negative isolates | 1 | BND | 0.22 | 0.33 | 0.51 | 1x105 | 1x105 | BND | BND | BND | BND |
| 2 | BND | 0.18 | 0.21 | 0.44 | 1x105 | 1x104 | BND | BND | BND | BND | |
| 3 | BND | 0.17 | 0.22 | 0.28 | 1x105 | 1x104 | BND | BND | BND | BND | |
| 4 | BND | 0.21 | 0.23 | 0.27 | 1x105 | 1x104 | BND | BND | BND | BND | |
| 5 | BND | 0.23 | 0.34 | 0.47 | 1x105 | 1x105 | BND | BND | BND | BND | |
| Biofilm positive S. aureus with biofilm negative E. coli |
1 | 0.66 | 0.22 | 0.49 | 0.73 | 1x105 | 1x104 | <10CFU | 1x103 | <10CFU | 1x102 |
| 2 | 0.72 | 0.21 | 0.44 | 0.72 | 1x105 | 1x104 | <10CFU | 1x103 | <10CFU | 1x101 | |
| 3 | 0.81 | 0.19 | 0.47 | 0.71 | 1x105 | 1x104 | 1x101 | 1x103 | <10CFU | 1x101 | |
| 4 | 0.79 | 0.32 | 0.66 | 0.69 | 1x105 | 1x104 | 1x101 | 1x103 | <10CFU | 1x101 | |
| 5 | 0.77 | 0.33 | 0.61 | 0.69 | 1x105 | 1x104 | <1CFU | 1x103 | <10CFU | 1x101 | |
| Biofilm negative S. aureus with biofilm positive E. coli |
1 | 0.66 | 0.45 | 0.59 | 0.77 | 1x105 | 1x104 | 1x102 | 1x103 | 1x101 | <10CFU |
| 2 | 0.71 | 0.43 | 0.61 | 0.78 | 1x105 | 1x104 | 1x102 | <10CFU | <10CFU | <10CFU | |
| 3 | 0.73 | 0.42 | 0.63 | 0.76 | 1x104 | 1x104 | 1x103 | <10CFU | <10CFU | <10CFU | |
| 4 | 0.77 | 0.41 | 0.62 | 0.69 | 1x105 | 1x104 | 1x103 | 1x101 | <10CFU | <10CFU | |
| 5 | 0.69 | 0.39 | 0.64 | 0.71 | 1x105 | 1x104 | 1x102 | 1x101 | <10CFU | <10CFU | |
Co-culture of P. aeruginosa with E. coli
In this combination, it was again noticed that E. coli dominated pre-biofilm stage (Table 5). However, after adoption of biofilm mode of life P. aeruginosa took over E. coli and slowly dominated the whole consortium. Although P. aeruginosa adopted biofilm mode of life later than E. coli, slowly it took over and occupied more space (Table 5).
Biofilm inhibition assay and cis-2-decenoic acid production
Supernatants of all of the subject isolates of P. aeruginosa had a pronounced effect on the S. aureus and E. coli at the pre-biofilm stage. All biofilm producing isolates of S. aureus and E. coli were unable to adopt biofilm mode of life in the presence of cell-free supernatants of P. aeruginosa (Figures 2a, c, d, e & f). A similar inhibitory effect was noticed when heat-killed dead cells of P. aeruginosa were applied. Moreover, this cell-free extract was comparatively ineffective against established biofilms of S. aureus. Interestingly in established biofilms, the small colony variants of S. aureus were not affected by the presence of heat-killed cells or cell-free extract of P. aeruginosa and persisted for a long time and remained adherent. Contrary to this, cell-free extract of P. aeruginosa dispersed established biofilms of E. coli by killing or lysis of target cells. The results of Gas chromatography and mass spectrometry (GC-MS) revealed that cell-free extract of P. aeruginosa contains a sustainable amount of cis-2-decenoic acid (Figure 3). Comparative analysis showed that biofilm non-producer isolates produce more cis-2-decenoic acid and the highest production was achieved at late log phase or stationary phase (Figures 3b, d & e). The highest production of cis-2-decenoic was noticed when P. aeruginosa and S. aureus were grown in combination (Figure 3a). Similarly, E. coli and P. aeruginosa showed a higher quantity of cis-2-decenoic as compared to the E. coli and S. aureus combination (Figure 3a). Moreover, highest quantity of extracellular cis-2-decenoic fatty acid was recovered from P. aeruginosa and lowest quantity was recovered from S. aureus (Figure 3a). Moreover, cis-2-decenoic non-producer P. aeruginosa has no impact on E. coli and S. aureus growth either in planktonic or biofilm stage.
Figure 2.
Biofilm inhibition Assay. (A & F) On Congo-red gar plate, parallel growth of (PA) Pseudomonas aeruginosa and (SA) Staphylococcus aureus, showed that S. aureus is unable to produce biofilm in the presence of P. aeruginosa. However, the control streaked away from P. aeruginosa showed blackening, which suggests biofilm formation. (B) Biofilm positive S. aureus on Congo-red agar (C) Application of (DC) dead cells and (LC) live cells of P. aeruginosa showed inhibition of biofilm formation of S. aureus on Congo-red agar. (D) Application of heat-killed cells of P. aeruginosa showed inhibition of biofilm formation of S. aureus on Congo-red agar. (E) In Congo-red broth S. aureus (S) was unable to adapt biofilm mode of growth in the presence of P. aeruginosa (P). P=P. aeruginosa, S=S. aureus. P+S=P. aeruginosa + S. aureus
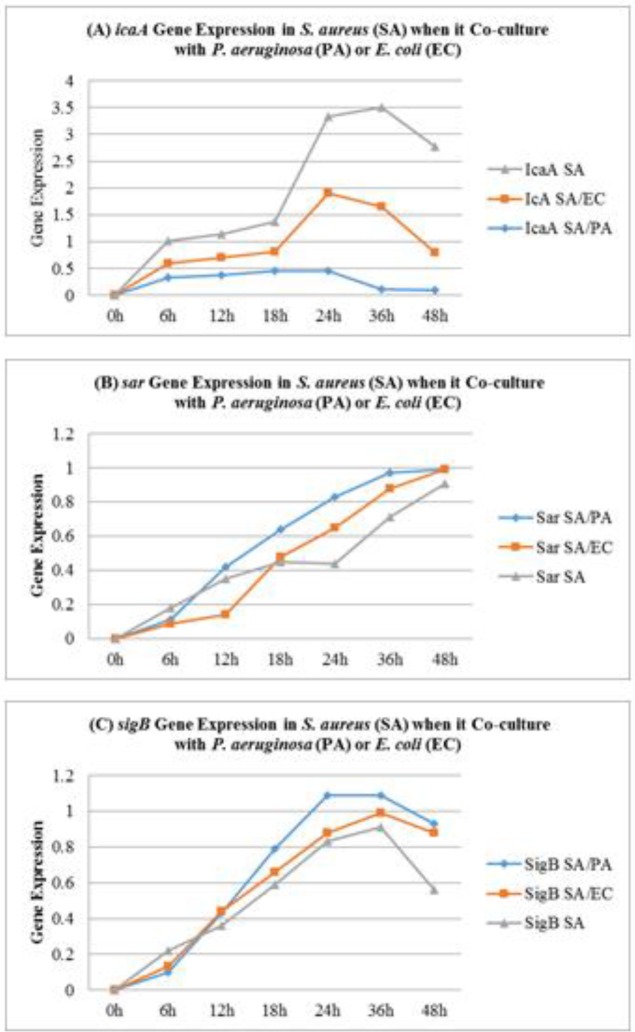
S. aureus icaA, Sar, and SigB gene expression in Co-culture conditions
In co-culture with P. aeruginosa, a significant reduction was noticed in the icaA gene, which is considered to be responsible for biofilm formation in S. aureus. The icaA ene was drastically influenced by the presence of P. aeruginosa and >10 fold reduction was noticed in the expression of this gene by RT-PCR. The presence of E. coli also influences the icaA gene expression in S. aureus. However, S. aureus isolates survived in the presence of P. aeruginosa and exhibited the highest activity of sigB and sarA. Similarly, the presence of E. coli also augments sigB and sarA gene expression in S. aureus (Figures 4a to 4c).
Figure 4.
Impact of Pseudomonas aeruginosa (PA) and Escherichia coli on icaA, sar and sigB gene expression of Staphylococcus aureus (SA) in co-culture conditions
Discussion
In nature, competition between bacterial strains and species appears to be a common phenomenon. According to a study (34), bacteria appear to attack back when they are attacked. It has been reported that stress factors, e.g. nutrient depletion, overcrowding, and invaders, induce bacteria to release their own antibiotics or toxins for self-defense (35). These antibiotics or toxins may also work as a signaling molecule for other bacteria (36). Consequently, other species or genera residing in the same niche become alert and defend themselves, sometimes by adopting the biofilm mode of life (37). It is well known that most of the antibiotics or other antibacterial molecules are unable to penetrate in biofilm consortia (38, 39). In a natural environment, biofilms mostly consist of complex and multiple communities (1). In this study, we attempted to explore multi-species biofilm formation, biofilm dispersion, and planktonic interaction in P. aeruginosa, S. aureus, and E. coli in a single assay. It has been noticed that at the pre-biofilm stage, the entire range of subject isolates exhibited hydrophilic surface properties. It suggests that this is a common character of monoculture as well as co-culture. However, after the adoption of biofilm mode of life, the surface properties of subject isolates were changed from hydrophilic to hydrophobic. According to Kochkodan et al. (40) and Giaouris et al. (41), cell surface hydrophobicity is a major factor for bacterial adhesion and biofilm formation. This is supported by Krasowska and Sigler (42). The other significant character of the subject isolates of P. aeruginosa and S. aureus is the appearance of metabolically inactive phenotypes i.e. small colony variants SCVs. These phenotypes dominate the biofilm consortia both in monoculture and co-culture state. The SCVs takeover metabolically active population as biofilm consortia becomes older. Interestingly, the hydro-phobicity of biofilm consortium increases as the population of SCVs increases. These characters were the common property of S. aureus and P. aeruginosa and in agreement with our previous study of S. aureus and MRSA biofilms (43). According to Kahl et al. (44), although the SCVs are metabolically inactive and most of the genes are non-functional but genes for biofilm formation and adhesion are up-regulated. Contrary to this, our study showed that icaA, sigB and sarA gene expression was more reduced or even arrested in SCVs of S. aureus (45). Interestingly, in present study, P. aeruginosa seems to augment sarA and sigB expression and diminish icaA gene expression in co-cultured isolates of S. aureus. The same phenomenon was noticed when S. aureus was co-cultured with E. coli. It has been noticed that in biofilm consortia, majority of S. aureus were dispersed or killed by the presence of P. aeruginosa and only minor population was able to survive. These survivors seem to adopt ica independent biofilm mode of life because the icaA gene expression was reduced or arrested in the presence of P. aeruginosa or E. coli. This was confirmed by the sarA and sigB gene expression studies. It is reported that sarA is significant for S. aureus biofilm development (46). These results indicate that in the conditions where icaA gene is absent or non-functional, sarA mediates biofilm formation in Staphylococci. Similarly, sigB plays a crucial role in the survival of S. aureus in stress environment (47). In a recent study, Tuchscherr et al. (47), reported that sigB enables S. aureus to switch from planktonic to small colony variants. It is reported that SCVs have a major role in stability and persistence of S. aureus and P. aeruginosa biofilms (45, 48). Comparative analysis of co-culture population showed that E. coli dominated pre-biofilm population and adopted biofilm mode of life before S. aureus and P. aeruginosa. This might be due to comparatively shorter generation time of E. coli and weaker competitive nature of S. aureus. Accordingly, Culotti and Packman (14) reported that co-culturing of P. aeruginosa augment the ability of E. coli to persist and grow in aquatic environment. Chu et al. (49) suggested that indole produced by E. coli arrests the quorum singling program and other virulence programs of P. aeruginosa. This supports E. coli to survive and dominate the P. aeruginosa and S. aureus in co-culture environment. Contrary to this, Sadowska et al. (50) reported that co-culturing of E. coli has no effect on the growth of S. aureus. This scenario depicted the dominance of E. coli at planktonic stage. However, once E. coli population reaches the decline stages and is unable to produce extracellular metabolites, P. aeruginosa and S. aureus proliferate and take advantage. Furthermore, after adoption of biofilm state S. aureus and P. aeruginosa occupy more space and disperse E. coli from biofilm consortia. It was noticed that as P. aeruginosa population increases in biofilm consortia the population of E. coli decreases. Same phenomenon was noticed for P. aeruginosa against S. aureus. This might be due to cell surface hydrophobicity. It was noticed that more hydrophobicity produces strong biofilms, persist for a long time and difficult to disperse (43). Cell surface hydrophobicity assays revealed that S. aureus and P. aeruginosa were more hydrophobic as compared to E. coli. The other property of S. aureus and P. aeruginosa which help them to persist for long time is the switch to small colony variants. These metabolically inactive phenotypes persist for long time and increases survival of S. aureus and P. aeruginosa under stress conditions (51). On the other hand, E. coli are unable to switch to small colony variants, so they are unable to withstand toxic effect of antibacterial metabolites of S. aureus or P. aeruginosa and are gradually dispersed from co-cultured consortia. Moreover, the findings of Estrela et al. (52) and Davies and Marques (53) suggested that P. aeruginosa producing diffusible signaling molecules e.g. cis-2-dodecenoic acid and cis-2-decenoic acid which are found to be effective in biofilm dispersion of not only P. aeruginosa, but also Streptococcus mutans, E. coli, K. pneumoniae, Proteus mirabilis, Streptococcus pyogenes, Bacillus subtilis, S. aureus, and the yeast, Candida albicans. In the present study, it was noticed that P. aeruginosa produces cis-2-decanoic acid and the production of this compound increases in co-culture state either with S. aureus or E. coli. This suggests that strong competitive behavior and dominance of P. aeruginosa. Consequently, E. coli and S. aureus are unable to adopt biofilm mode of life in the presence of P. aeruginosa. Similarly, P. aeruginosa occupy more space and it disperses S. aureus population when it grows with S. aureus. This is also supported by the findings of Filkins et al. (10) and Kim et al. (54). They have suggested that in co-culture P. aeruginosa reduces S. aureus viability by producing 2-heptyl-4-hydroxyquinoline N-oxide (HQNO) and siderophores (10). In three species biofilms, the P. aeruginosa dominate the consortia. Although, E. coli dominates the pre-biofilm stage but is unable to proliferate in three species biofilms, perhaps due to the presence of P. aeruginosa, which was evidently harmful for other members of three species biofilm consortia. This antagonistic effect is probably caused by extracellular metabolites of P. aeruginosa e.g. cis-2-decanoic acid. Moreover, cell-free extract as well as dead cells of P. aeruginosa were found to inhibit S. aureus and E. coli biofilm formation and also disperse pre-established biofilms. This suggests the antagonistic or antibacterial nature of compounds of P. aeruginosa. As we used heat-killed cells in this study, it suggests this compound is cell bound and heat stable. On the other hand cell-free extract also disperse biofilm indicating the extracellular activity of P. aeruginosa. Accordingly, Qin et al. (55) suggested that P. aeruginosa extracellular products are important microbial competition factors that overcome and disperse Staphylococcal biofilms. However, S. aureus or E. coli cell-free culture extract had no effect on P. aeruginosa or on each other’s biofilms or on planktonic growth.
Conclusion
The present study indicates that P. aeruginosa and S. aureus exhibited hydrophobic surface properties and are comparatively slow growing. Whereas subject isolates of E. coli exhibited hydrophilic surface properties and were comparatively fast growing. Due to hydrophobic surface properties and slow growth rate, P. aeruginosa and S. aureus showed strong biofilm formation and persistence. Among all three subject organisms, P. aeruginosa is comparatively more hydrophobic, slow growing, and adopts biofilm mode of life pretty late; in this category, S. aureus comes next. Moreover, other hydrophobic isolates, e.g. P. aeruginosa and S. aureus are more likely to switch to small colony variants. Biofilm consortia dominated with small colony variants persist for a long time and are difficult to disperse. With these properties, P. aeruginosa dominates and antagonizes other counterparts in biofilm consortia and also produces cis-2-decanoic acid to inhibit or disperse biofilms of other organisms. Although, majorities of the S. aureus were unable to compete with P. aeruginosa, however, a minor population did compete, survive, and persist in biofilm consortia as small colony variants. The survivors showed higher expression of sigB and sarA genes. These results suggest that cell surface hydrophobicity is directly proportional to biofilm formation and SCVs population and inversely proportional to the growth rate of the subject isolates.
Acknowledgment
We are thankful to Mr M Yousf Khan of the University of Karachi, Karachi, Pakistan for providing scanning electron microscopy facilities for this study.
Conflicts of Interest
All authors confirm that they have no conflicts of interest.
References
- 1.Berlanga M, Guerrero R. Living together in biofilms: the microbial cell factory and its biotechnological implications. Microb Cell Fact. 2016;15:165–175. doi: 10.1186/s12934-016-0569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Bodman SB, Willey JM, Diggle SP. Cell-cell communication in bacteria: united we stand. J Bacteriol. 2008;190:4377–4391. doi: 10.1128/JB.00486-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flemming H, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nature Rev Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 4.Giaouris E, Heir E, Desvaux M, Hébraud M, Møretrø T, Langsrud S, et al. Intra- and inter-species interactions within biofilms of important foodborne bacterial pathogens. Front Microbiol. 2015;6:841–866. doi: 10.3389/fmicb.2015.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Limoli D, Jones C, Wozniak D. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol Spectr. 2015;3:1–30. doi: 10.1128/microbiolspec.MB-0011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li HY, Tian X. Quorum sensing and bacterial social interactions in biofilms. Sensors. 2012;12:2519–2538. doi: 10.3390/s120302519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atkinson S, Williams P. Quorum sensing and social networking in the microbial world. J R Soc Interface. 2009;6:959–978. doi: 10.1098/rsif.2009.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shank EA, Kolter R. New developments in microbial interspecies signalling. Curr Opin Microbiol. 2009;12:205–214. doi: 10.1016/j.mib.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filkins LM, Graber JA, Olson DG, Dolben EL, Lynd LR, Bhuju S, et al. Co culture of Staphylococcus aureus with Pseudomonas aeruginosa drives S aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J Bacteriol. 2015;197:2252–2264. doi: 10.1128/JB.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hotterbeekx A, Singh SK, Goossens H, Kumar SM. In vivo and in vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front Cell Infect Microbiol. 2017;7:106–118. doi: 10.3389/fcimb.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orazi G, O’Toole GA. Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis infection. MBio. 2017;4:e00873–17. doi: 10.1128/mBio.00873-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci USA. 2013;110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Culotti A, Packman AI. Pseudomonas aeruginosa promotes Escherichia coli biofilm formation in nutrient-limited medium. PLoS One. 2014;9:e107186. doi: 10.1371/journal.pone.0107186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HS, Park HD. Ginger extract inhibits biofilm formation by Pseudomonas aeruginosa PA14. PLoS One. 2013;8:e76106. doi: 10.1371/journal.pone.0076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaiser TD, Pereira EM, Dos Santos KR, Maciel EL, Schuenck RP, Nunes AP. Modification of the congo red agar method to detect biofilm production by Staphylococcus epidermidis. Diagn Microbiol Infect Dis. 2013;75:235–239. doi: 10.1016/j.diagmicrobio.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Reichhardt C, Jacobson AN, Maher MC, Uang J, McCrate OA, Eckart M, Cegelski L. Congo red interactions with curli-producing E coli and native curli amyloid fibres. PLoS One. 2015;10:e0140388. doi: 10.1371/journal.pone.0140388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirani ZA, Jamil N. Effect of sub-lethal doses of vancomycin and oxacillin on biofilm formation by vancomycin intermediate resistant Staphylococcus aureus. J Basic Microbiol. 2011;51:191–195. doi: 10.1002/jobm.201000221. [DOI] [PubMed] [Google Scholar]
- 19.O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 20.Allegrucci M, Sauer K. Characterization of colony morphology variants isolated from Streptococcus pneumonia biofilms. J Bacteriol. 2007;189:2030–2038. doi: 10.1128/JB.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 22.Bayston R, Ashraf W, Smith T. Triclosan resistance in methicillin-resistant Staphylococcus aureus expressed as small colony variants: a novel mode of evasion of susceptibility to antiseptics. J Antimicrob Chemother. 2007;59:848–853. doi: 10.1093/jac/dkm031. [DOI] [PubMed] [Google Scholar]
- 23.Chen CY, Nace GW, Irwin PL. A 6×6 drop plate method for simultaneous colony counting and MPN enumeration of Campylobacter jejuni, Listeria monocytogenes, and Escherichia coli. J Microbiol Meth. 2003;55:475–479. doi: 10.1016/s0167-7012(03)00194-5. [DOI] [PubMed] [Google Scholar]
- 24.Kouidhi B, Zmantar T, Hentati H, Bakhrouf A. Cell surface hydrophobicity, biofilm formation, adhesives properties and molecular detection of adhesins genes in Staphylococcus aureus associated to dental caries. Microb Pathog. 2010;49:14–22. doi: 10.1016/j.micpath.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Liu YZ, Yuan K, Ji CR. New method of extraction on the chemical components of Chinese medicinal plants extracting method by smashing of plant tissues (EMS) Henan Sc. 1993;11:2652–2681. [Google Scholar]
- 26.Mirani ZA, Jamil N. Role of extracellular fatty acids in vancomycin-induced biofilm formation by vancomycin-resistant Staphylococcus aureus. Pak J Pharm Sci. 2013;26:383–389. [PubMed] [Google Scholar]
- 27.Kilic A, Guclu AU, Senses Z, Bedir O, Aydogan H, Basustaoglu AC. Staphylococcal cassette chromosome mec(SCCmec) characterization and panton-valentine leukocidin gene occurrence for methicillin-resistant Staphylococcus aureus in Turkey, from 2003 to 2006. Antonie van Leeuwenhoek. 2008;94:607–614. doi: 10.1007/s10482-008-9278-3. [DOI] [PubMed] [Google Scholar]
- 28.Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuchscherr L, Bischoff M, Lattar SM, Llana MN, Pfortner H, Niemann S, et al. Sigma factor sigB is crucial to mediate Staphylococcus aureus adaptation during chronic infections. PLoS Pathog. 2015;11:e1004870. doi: 10.1371/journal.ppat.1004870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iqbal Z, Seleem MN, Hussain HI, Huang L, Hao H, Yuan Z. Comparative virulence studies and transcriptome analysis of Staphylococcus aureus strains isolated from animals. Sci Rep. 2016;6:35442–35453. doi: 10.1038/srep35442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nuryastuti T, van der Mei HC, Busscher HJ, Iravati S, Aman AT, Krom BP. Effect of cinnamon oil on icaA expression and biofilm formation by Staphylococcus epidermidis. Appl Environ Microbiol. 2009;75:6850–6855. doi: 10.1128/AEM.00875-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spilker T, Coenye T, Vandamme P, LiPuma JJ. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol. 2004;42:2074–2079. doi: 10.1128/JCM.42.5.2074-2079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naravaneni R, Jamil K. Rapid detection of food-borne pathogens by using molecular techniques. J Med Microbiol. 2005;54:51–54. doi: 10.1099/jmm.0.45687-0. [DOI] [PubMed] [Google Scholar]
- 34.Oliveira MN, Martinez-Garcia E, Xavier J, Durham WM, Kolter R, Kim W, et al. Biofilm formation as a response to ecological competition. PLoS Biol. 2015;13:e1002191. doi: 10.1371/journal.pbio.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cornforth DM, Foster KR. Antibiotics and the art of bacterial war. Proc Natl Acad Sci U S A. 2015;112:10827–10828. doi: 10.1073/pnas.1513608112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romero D, Traxler MF, Lopez D, Kolter R. Antibiotics as signal molecules. Chem Rev. 2011;111:5492–5505. doi: 10.1021/cr2000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davey ME, O’Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vega NM, Gore J. Collective antibiotic resistance: mechanisms and implications. Curr Opin Microbiol. 2014;21C:28–34. doi: 10.1016/j.mib.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beloin C, Renard S, Ghigo JM, Lebeaux D. Novel approaches to combat bacterial biofilms. Curr Opin Pharmacol. 2014;18C:61–68. doi: 10.1016/j.coph.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Kochkodan V, Tsarenko S, Potapchenko N, Kosinova V, Goncharuk V. Adhesion of microorganisms to polymer membranes: a photobactericidal effect of surface treatment with TiO2. Desalination. 2008;220:380–385. [Google Scholar]
- 41.Giaouris E, Chapot-Chartier MP, Briandet R. Surface physicochemical analysis of natural Lactococcus lactis strains reveals the existence of hydrophobic and low charged strains with altered adhesive properties. Int J Food Microbiol. 2009;131:2–9. doi: 10.1016/j.ijfoodmicro.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 42.Krasowska A, Sigler K. How microorganisms use hydrophobicity and what does this mean for human needs? Front Cell Infect Microbiol. 2014;4:112. doi: 10.3389/fcimb.2014.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirani ZA, Naz S, Khan F, Aziz M, Khan MN, Khan SI. Antibacterial fatty acids destabilize hydrophobic and multicellular aggregates of biofilm in S aureus. J Antibiot. 2017;70:115–121. doi: 10.1038/ja.2016.76. [DOI] [PubMed] [Google Scholar]
- 44.Kahl BC, Becker K, Löffler B. Clinical significance and pathogenesis of staphylococcal small colony variants in persistent infections. Clin Microbiol Rev. 2016;2:401–427. doi: 10.1128/CMR.00069-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mirani ZA, Aziz M, Khan SI. Small colony variants have a major role in stability and persistence of Staphylococcus aureus biofilms. J Antibiot. 2015;68:98–105. doi: 10.1038/ja.2014.115. [DOI] [PubMed] [Google Scholar]
- 46.Valle J, Toledo-Arana A, Berasain C, Ghigo JM, Amorena B, Penades JR, et al. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol. 2003;48:1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 47.Tuchscherr L, Loffler B. Staphylococcus aureus dynamically adapts global regulators and virulence factor expression in the course from acute to chronic infection. Curr Genet. 2016;62:15–17. doi: 10.1007/s00294-015-0503-0. [DOI] [PubMed] [Google Scholar]
- 48.Häussler S. Biofilm formation by the small colony variant phenotype of Pseudomonas aeruginosa. Environ Microbiol. 2004;4:546–551. doi: 10.1111/j.1462-2920.2004.00618.x. [DOI] [PubMed] [Google Scholar]
- 49.Chu W, Zere TR, Weber MM, Wood TK, Whiteley M, Hidalgo-Romano B, et al. Indole production promotes Escherichia coli mixed-culture growth with Pseudomonas aeruginosa by inhibiting quorum signalling. Appl Environ Microbiol. 2012;78:411–419. doi: 10.1128/AEM.06396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sadowska B, Walencka E, Wieckowska-Szakiel M, Różalska B. Bacteria competing with the adhesion and biofilm formation by Staphylococcus aureus. Folia Microbiol (Praha) 2010;55:497–501. doi: 10.1007/s12223-010-0082-x. [DOI] [PubMed] [Google Scholar]
- 51.Hoffman LR, Deziel E, D’Argenio DA, Lepine F, Emerson J, McNamara S, et al. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2006;103:19890–19895. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Estrela AB, Abraham WR. Combining biofilm-controlling compounds and antibiotics as a promising new way to control biofilm infections. Pharmaceuticals. 2010;3:1374–1393. doi: 10.3390/ph3051374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davies DG, Marques CN. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol. 2009;191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim S, Yoon Y, Choi KH. Pseudomonas aeruginosa DesB promotes Staphylococcus aureus growth inhibition in coculture by controlling the synthesis of HAQs. PLoS One. 2015;10:1–16. doi: 10.1371/journal.pone.0134624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qin Z, Yang Y, Qu D, Molin S, Tolker-Nielsen T. Pseudomonas aeruginosa extracellular products inhibit staphylococcal growth, and disrupt established biofilms produced by Staphylococcus epidermidis. Microbiol. 2009;155:2148–2156. doi: 10.1099/mic.0.028001-0. [DOI] [PubMed] [Google Scholar]