In the accelerating and expanding field of research on genetic variation, it has become standard practice to work with a combination of datasets generated by multiple research groups at different times and by different methods. Synthesizing these data is important for genotype imputation, meta-analysis, and other applications, but may be difficult because alleles are typically observed and recorded on only one of the two DNA strands in genotyping and sequencing experiments. Different nomenclatures have arisen to designate strand orientation when reporting single nucleotide polymorphism (SNP) genotypes, but they are neither widely understood nor uniformly applied. Here we define the most common allele strand orientation nomenclatures and provide guidance in achieving strand consistency.
The majority of SNPs are ‘strand unambiguous’, such that genotypes called on different strands are readily identifiable (e.g., A/G alleles on one strand are T/C alleles on the opposite strand). However, determining strand orientation at ‘strand ambiguous’ SNPs is more complicated, where alleles are symmetrical across strands (A/T and C/G). It is assumed that all researchers, as a minimum for consistency, report the two alleles of a biallelic SNP on the same strand. It is the choice and the definition of which strand is used that leads to ambiguity. Generally, SNP alleles are reported for a single strand designated in one of four strand naming conventions: ‘probe/target’, ‘plus/minus’, ‘TOP/BOT’, and ‘forward/reverse’, defined as follows.
Probe/target
When SNPs are assayed with a site-specific probe, one of the two strands corresponds to (i.e., is collinear with) the probe sequence itself, and the other to the complementary genomic target sequence that flanks or spans the SNP site. Sometimes the probe strand is called the ‘design’ strand (in reference to assay design). Although the specifics vary between platforms, alternative alleles at a SNP site are often initially represented using the generic letter codes A and B. In the following, an italicized A refers to this generic allele designation and not to adenine. In Illumina annotation each SNP is defined with design allele nucleotides, and these occur on the same strand as the probe sequence; the order in which the alternative alleles are given specifies the generic A and B allele designations [1]. To illustrate, for a SNP defined as [T/G], the A allele is T and the B allele is G. In Affymetrix allele-specific hybridization technology, the letter codes A and B are assigned differently and could therefore occur on either the probe or target strand [2].
Plus (+)/minus (−)
In all human reference chromosomes, as for other eukaryotes [3], the plus (+) strand is defined as the strand with its 5′ end at the tip of the short arm [4,5] (Genome Reference Consortium, personal communication, March 27, 2012). SNP alleles reported on the same strand as the (+) strand are called ‘plus’ alleles and those on the (−) strand are called ‘minus’ alleles. Providing SNP alleles on the plus genomic strand is the convention in publicly available SNP datasets such as the HapMap (www.hapmap.org) and 1000 Genomes Projects (www.1000genomes.org).
Although the plus/minus designation is anchored at the telomeres of each chromosome, the orientation of intervening sequences may change between genome builds as gaps are filled in and sequences are refined. Thus when reporting plus/minus strand, one must specify a genome build. The fluid nature of plus/minus orientation has partly motivated the development of alternative nomenclatures.
Illumina TOP/BOT strand
The TOP/BOT strand naming convention, developed by Illumina and subsequently adopted by dbSNP, has been thoroughly defined elsewhere [1]. In brief, Illumina strand designation is determined by either the SNP alternative nucleotides or its flanking sequence. For unambiguous SNPs the TOP strand is defined as the one that contains an A nucleotide allele. The A is designated generically as allele A, whereas the alternative allele on the TOP strand is designated as allele B. For ambiguous SNPs the strand designation and allele A/B assignments are determined by flanking sequence in a similar manner. This strand definition is ‘local’ to a SNP in that alleles reported on the TOP strand for two neighboring SNPs may be on different physical strands of DNA [6]. Furthermore, the TOP/BOT strand definition is intended to be independent of any genome build or design strand. Another key feature of this naming system is that allele A for a TOP strand probe is the base pair complement of allele A for a BOT strand probe, such that the generic A/B genotype coding remains consistent regardless of which strand is probe or target. This nomenclature offers relative stability in the face of changing human genome assemblies and SNP databases.
Forward/reverse
The dbSNP resource of the US National Center for Biotechnology Information (NCBI) contains detailed information for each SNP in its database. Each refSNP (or ‘rs’) entry consists of one or more submitted SNP (or ‘ss’) records, each submitted by individual laboratories. Each dbSNP record shows a flanking DNA sequence, which is simply taken from the submission with the longest flanking sequence [6,7]. SNP alleles reported on the same strand as this exemplar sequence in dbSNP sequence are called ‘forward’ alleles. Conversely, alleles on the opposite strand are called ‘reverse’ alleles. Note that the dbSNP meaning of ‘forward’ is easily confused with (+) genomic strand, which has been referred to as the ‘forward’ strand by the HapMap project [8,9].
Achieving strand consistency
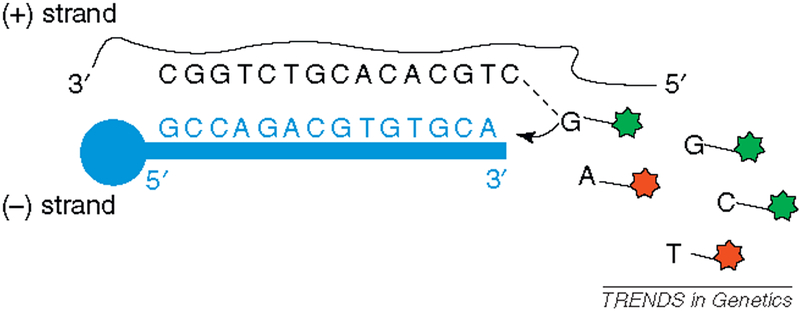
The most basic level of strand consistency requires only that genotypes are reported on the same DNA strand across datasets. At strand-unambiguous SNPs, discrepant nucleotides are sufficient to identify strand inconsistencies (e.g., A/C in one dataset and T/G in another). However, harmonizing strand-ambiguous SNPs requires converting allele calls to a specific strand, according to one of the strand naming conventions described above. Given a nucleotide sequence with a SNP and its flanking bases (e.g., CATCCC[A/C]TGCACA) one can determine whether the strand of that sequence is (i) plus or minus, by sequence matching with the genomic reference sequence; (ii) TOP or BOT, from the SNP itself or its flanking sequence [1]; and (iii) forward or reverse, from the ‘ss’ sequence record in dbSNP. Determination of probe or target strand requires additional information about assay design. In practice, genotyping assay vendors generally supply annotations that can be used to make strand conversions. Box 1 gives an example of how to interpret Illumina annotation to create a table of allele call conversions. Figure I shows a simplified schematic of the genotyping probe at this example SNP. However, SNP annotations are not infallible and further checks on strand consistency are useful. Commonly used checks are comparisons of minor allele frequency and patterns of linkage disequilibrium between the datasets to be harmonized [10,11].
Box 1. An example of allele conversion using Illumina annotation.
Here we use Illumina-provided annotation for an example SNP (rs216614) in Table I to derive a set of allele call conversions in Table II. In Table I, ‘SNP’ gives alternative alleles on the probe sequence strand, ‘IlmnStrand’ gives the TOP/BOT status of the probe sequence strand, ‘TopGenomicSeq’ gives the sequence surrounding the SNP on the TOP strand, ‘RefStrand’ gives the plus/minus status of the probe sequence strand, and ‘IlmnID’ encodes the correspondence between TOP/BOT and forward/reverse (dbSNP) strands. The ‘design’ alleles (on the probe sequence strand) are given directly by ‘SNP’ = [T/G] and, following the Illumina convention, the first nucleotide corresponds to allele A and the second to allele B. The TOP strand alleles are given in brackets in ‘TopGenomicSeq’. The ‘B_R’ in ‘IlmnID’ specifies that the dbSNP reverse strand corresponds with the BOT strand. The corresponding SNP assay is depicted in Figure I.
Figure I.
A simplified schematic of the SNP probe, where the probe sequence is in blue and the target sequence in black text. The ‘design’ alleles (T or G) are the fluorescently labeled nucleotides recruited to the allele probe in this two-color primer-extension assay. Adapted from materials available on the Illumina website (www.illumina.org).
Table I.
Excerpt from Illumina HumanOmni11-Quad_v1–0_C annotation file (build 37)
| IlmnID | Name | IlmnStrand | SNP | TopGenomicSeq | RefStrand |
|---|---|---|---|---|---|
| rs216614–131_B_R_1865662557 | rs216614 | BOT | [T/G] | …CATCCC[A/C]TGCACA… | - |
Table II.
rs216614 allele-mapping table
| AB | TOP | Design | Forward | Plus |
|---|---|---|---|---|
| A | A | T | A | A |
| B | C | G | C | C |
Our intent is not to advocate one allele nomenclature above all others because the universal adoption of one naming system is both unlikely and unnecessary. Instead, our aim is to explain the different nomenclatures and the need for precise documentation of allele designations for each dataset. Increased understanding and documentation will facilitate continued data sharing and collaboration within the genetics research community.
Acknowledgments
This work was supported in part by the following National Institutes of Health grants: GENEVA Coordinating Center (U01 HG004446); GARNET Coordinating Center (U01 HG005157); Center for Inherited Disease Research (U01HG004438, NIH contract numbers HHSN268200782096C and HHSN268201100011I); and Broad Center for Genotyping and Analysis (U01HG04424).
References
- 1.Illumina Inc. (2006) ‘TOP/BOT’ strand and ‘A/B’ allele (Technical Note). http://www.illumina.com/documents/products/technotes/technote_topbot.pdf
- 2.Affymetrix Inc. (2012) Affymetrix genotyping glossary. http://www.affymetrix.com/support/help/genotyping_glossary/index.affx
- 3.Cherry JM et al. (1998) SGD: Saccharomyces genome database. Nucleic Acids Res. 26, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunham I et al. (1999) The DNA sequence of human chromosome 22. Nature 402, 489–495 [DOI] [PubMed] [Google Scholar]
- 5.Cartwright RA and Graur D (2011) The multiple personalities of Watson and Crick strands. Biol. Direct 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Center for Biotechnology Information (2005) Sequence formatting in dbSNP reports. http://www.ncbi.nlm.nih.gov/books/NBK44414
- 7.Kitts AK and Sherry S (2002) The single nucleotide polymorphism database (dbSNP) of nucleotide sequence variation In The NCBI Handbook (McEntyre J and Ostell J, eds), National Center for Biotechnology Information; (Chap. 5) In: http://www.ncbi.nlm.nih.gov/books/NBK21101/) [Google Scholar]
- 8.Frazer KA et al. (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altshuler DM et al. (2010) Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Browning SR (2009–2011) Strand-switching utility for BEAGLE. http://faculty.washington.edu/sguy/beagle/strand_switching/strand_switching.html
- 11.Howie B and Marchini J (2009–2012) IMPUTE2 strand alignment options. http://mathgen.stats.ox.ac.uk/impute/strand_alignment_options.html