Abstract
Antituberculosis drug‐induced adverse drug reactions (ATD‐ADRs) are increasing globally, and it is key to identify candidate ATD‐ADRs loci for clinical management. We prospectively enrolled 1,235 highly suspicious tuberculosis (TB) inpatients to investigate the profiles and genetic risk factors of ATD‐ADRs in the liver, kidneys, and blood. Overall, 644 subjects were eligible and genotyped for seven polymorphisms in drug‐metabolizing enzymes and transporter genes. Clinical follow‐up and blood analysis were performed regularly. We found that a notable rate of ATD‐ADRs (incidence: 16.5%, drug intervention rate: 10.4%), mainly involving hepatotoxicity (10.6%) and leukopenia (3.3%) in western China. CYP2D6 rs1135840 and NUDT15 rs116855232 increased the risks of hepatotoxicity and leukopenia with an odds ratio of 2.52 and 4.97, respectively. Both variants showed excellent negative predictive values (93.7% and 98.1%, respectively) but moderate sensitivities (72.7% and 52.4%, respectively). These data provide new insight into ATD‐ADRs in the Chinese population and may offer future leads for diagnosis and treatment.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Antituberculosis drug‐induced adverse drug reactions (ATD‐ADRs) are becoming increasingly common globally. Nevertheless, there is currently a paucity of information regarding the characteristics and risk factors underlying common and severe ATD‐ADRs, which are worthwhile to explore in clinical settings.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ The study investigated the overall profiles and genetic risk factors of ATD‐ADRs in the liver, kidneys, and blood of a large prospective tuberculosis cohort in western China.
WHAT THIS STUDY ADDS TO OUR KNOWLEDGE
☑ This study describes notable profiles of ATD‐ADRs in the liver, kidneys, and blood in patients from western China, with a significant impact on subsequent therapeutic orientation. This study also provides a novel evidence that CYP2D6 rs1135840 and NUDT15 rs116855232 are susceptibility loci for the development of hepatotoxicity and leukopenia, respectively.
HOW THIS MIGHT CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE
☑ Rs1135840 and rs116855232 are potential pharmacogenetic markers for hepatotoxicity and leukopenia in TB patients. Regular blood monitoring and pharmacogenetic analyses are useful for identifying patients at elevated risk of ATD‐ADRs during individualized TB treatment.
Tuberculosis (TB) remains a major global health concern.1 Although short‐course strategies have been successfully implemented, TB management remains associated with serious issues, such as an increasing prevalence of multidrug‐resistant Mycobacterium tuberculosis (MTB), disease relapse in elderly and immunocompromised patients, and the surge in antituberculosis drug‐induced adverse drug reactions (ATD‐ADRs). Long‐term antituberculosis (anti‐TB) combination regimens could lead to various types and levels of adverse drug reactions (ADRs), which could subsequently lead to therapy modification or discontinuation, the emergence of drug resistance,2 and even treatment failure or mortality.3 Antituberculosis drug‐induced hepatotoxicity (ATDH) is the most prevalent and serious ADR, with occurrence rates of 4.8–36% in different areas, and could cause liver failure and mortality in up to 15% and 67% of patients, respectively.4, 5, 6, 7 In addition to the leading ATD‐ADRs of hepatotoxicity, there are other severe ATD‐ADRs, such as nephrotoxicity and hematotoxicity, that are less frequent but are still lasting or fatal. These ADRs also pose considerable challenges to clinicians but have not yet been well elucidated.5, 8 More in‐depth knowledge of the characteristics and associated risk factors of ATD‐ADRs is urgently needed to predict and prevent ADRs, particularly among vulnerable populations.
Genomic variants are increasingly recognized as pivotal factors influencing the interindividual variability of drug effectiveness or the risk for ADRs.9, 10 The interrogation of genetic variants and their predisposition to ADRs has developed into clinical pharmacogenomics (PGx), which is widely used to screen patients who are at risk for ADRs from therapies for cancer and other diseases. PGx has particular value in determining an individual's combined medications.10, 11, 12, 13, 14 A preemptive PGx‐oriented approach has been proposed to achieve the goal of personalized medicine for many diseases and has also demonstrated significant potential in anti‐TB treatment.13, 14 Recently, N‐acetyl transferase 2 (NAT2) genotype testing has been proposed for isoniazid (INH) daily dose optimization by the Pharmacogenomics Knowledgebase (pharmaGKB) database15 and the China Precision Medicine Clinical Research and Application Association.16 NAT2 slow acetylator genotypes are designated biomarkers for the development of ATDH.17 Jung et al. and Azuma et al.18, 19 demonstrated that an NAT2 genotype‐guided regimen achieved therapeutic concentrations of INH, significantly prevented the occurrence of ATDH, and reduced early treatment failure from 38% to 15%, emphasizing the significance of pharmacogenetically oriented therapy in TB clinical practice. In addition to NAT2, polymorphisms in other metabolizing enzyme genes, such as cytochrome P450 2E1 and glutathione S‐transferase M1, and certain HLA alleles have been reported to affect patients' susceptibility to ATDH;20 however, robust and definite conclusions cannot be drawn due to controversial results from previous studies. The roles of other genes involved in drug disposition and drug response signaling pathways, such as drug transporter genes, are still seldom investigated. In addition, little is known about the genetic factors of nephrotoxicity and hematotoxicity during anti‐TB chemotherapy. There is currently a paucity of information regarding the genetic risk markers underlying common or severe ATD‐ADRs, which should be explored in clinical settings.
China ranks third among all high‐TB endemic countries, accounting for 8.6% of the global tuberculosis incidence,1 and most TB cases are reported in central and western China.21 Thus, identifying the features and genetic risk factors of ADRs during anti‐TB chemotherapy in the Chinese TB population is necessary. The objective of this prospective study was to determine the profiles of ATD‐ADRs in the liver, kidneys, and blood of patients in western China and to explore candidate genetic biomarkers to predict common ADRs in TB management.
RESULTS
Patient characteristics
The baseline characteristics of the TB patients are summarized in Table 1. A total of 644 active TB patients was ultimately eligible, of whom 61.03% were men and 84.47% were Han Chinese. The average age was 42.25 years, and the mean body mass index (BMI) was 19.2 kg/m2. Male patients were older and had higher smoking and drinking status than female patients. Four‐fifths of the TB patients had involved pulmonary tuberculosis (PTB), and 51.87% had previously received ATDs. Most patients (92.7%) suffered from local infectious symptoms, such as cough and sputum upon admission, followed by fever (52.64%) and poor appetite (37.58%). Most patients with chest images had infiltration and effusion (40.22%) or fibrosis and calcification (39.11%). The positive rates for TB‐DNA and an interferon gamma release assay (TB‐IGRA), which were 54.28% and 61.64% respectively, were considerably higher than those of MTB cultures and microscope smears (41.23% and 30.51%). Significant differences were observed in the proportion of PTB cases and the positive smear results between males and females. Quantitative laboratory traits of the participants at baseline, peak, and valley during anti‐TB therapy are also shown in Supplementary Table S1. Overall, most laboratory indicators in the ADR patients differed between the patients with and without ADRs. The patients with ADRs had a worsening presentation in laboratory examinations during anti‐TB therapy, while the presentation of those without ADRs did not change significantly.
Table 1.
The clinical features of TB patients
| Clinical features | All patients (n = 644) | Gender | ||
|---|---|---|---|---|
| Male (n = 393) | Female (n = 251) | P | ||
| Ethnicity (Han/Others) | 544/100 | 333/60 | 211/40 | 0.365 |
| Age, mean ± SD (years) | 42.25 ± 19.56 | 43.64 ± 20.42 | 40.08 ± 17.97 | 0.021 |
| BMI (kg/m2) | 19.20 ± 4.04 | 19.30 ± 5.32 | 18.42 ± 4.88 | 0.244 |
| Smoking, Yes / (Ever + No) | 254 (39.44) | 242 (61.58) | 12 (4.78) | < 0.001 |
| Drinking, Yes / (Ever + No) | 302 (46.89) | 271 (68.96) | 31 (12.35) | < 0.001 |
| TB status, n (%) | 0.495 | |||
| De novo | 310 (48.13) | 194 (49.36) | 116 (46.22) | |
| Retreated | 334 (51.87) | 199 (50.64) | 135 (63.78) | |
| TB subtype, n (%) | 0.039 | |||
| PTB | 329 (51.09) | 216 (54.96) | 113 (45.02) | |
| EPTB | 101 (15.68) | 54 (13.74) | 47 (18.73) | |
| PTB & EPTB | 214 (33.23) | 123 (31.30) | 91 (36.25) | |
| General symptoms, n (%) | ||||
| Fever | 339 (52.64) | 212 (53.94) | 127 (50.60) | 0.407 |
| Weight loss | 223 (34.63) | 144 (36.64) | 79 (31.47) | 0.179 |
| Night sweat | 188 (29.19) | 114 (29.01) | 74 (29.48) | 0.897 |
| Poor appetite | 242 (37.58) | 154 (39.19) | 88 (35.06) | 0.292 |
| Fatigue | 174 (27.02) | 112 (28.50) | 62 (24.70) | 0.290 |
| Local infectious symptoms, n (%) | 0.063 | |||
| Presence | 597 (92.70) | 358 (91.09) | 239 (95.22) | |
| Absence | 47 (7.30) | 35 (8.91) | 12 (4.78) | |
| Chest image of PTB (n = 271), n (%) | 0.697 | |||
| Normal | 1 (0.37) | 1 (0.54) | — | |
| Infiltration and effusion | 109 (40.22) | 74 (40.22) | 35 (40.23) | |
| Caseation and cavitation | 55 (20.30) | 40 (21.74) | 15 (17.24) | |
| Fibrosis and calcification | 106 (39.11) | 69 (37.50) | 37 (42.53) | |
| Microbiological results | ||||
| Positive culture, n (%) | 47/114 (41.23) | 32/71 (45.07) | 15/41 (34.88) | 0.248 |
| Positive smear, n (%) | 198/593 (30.51) | 136/362 (37.57) | 62/231 (26.84) | 0.007 |
| Positive TB‐DNA, n (%) | 222/409 (54.28) | 140/248 (56.45) | 82/161 (50.93) | 0.274 |
| Positive TB‐IGRA, n (%) | 98/159 (61.64) | 68/106 (64.15) | 32/53 (60.38) | 0.818 |
TB, tuberculosis; PTB, pulmonary tuberculosis; EPTB, extrapulmonary tuberculosis.
Profiles of ATD‐ADRs in the liver, blood, and kidneys
As shown in Table 2 and Table S2, the overall incidence rate of ADRs among the patients was 16.5% (106/644), and the main ATD‐ADRs included ATDH (10.6%) and leukopenia (3.3%). Sixty‐eight patients were considered cases of ATDH, all of which presented alterations in hepatic enzymes and 21 of which developed symptomatic hepatitis. ATDH cases were graded as mild (48.5%, 33/68), moderate (27.9%, 19/68), and severe (23.6%, 16/68) (Table S3), and age and gender were similar between the three groups (P = 0.723 and 0.895, respectively). Thirteen patients experienced uncommon hyperbilirubinemia (2.0%). Among hematological ADRs, leukopenia was common (3.3%, 21/644), followed by thrombocytopenia (2.5%, 16/644), whereas no cases of anemia were observed. Regarding ADRs in the kidneys, acute (AKI) or chronic (CKD) were rare—only 10 and 12 patients suffered from AKI or CKD, respectively. Females were at a greater risk of developing leukopenia and had a reduced risk for AKI (P = 0.002, 0.037, respectively), but given the limited number of samples, these results should be interpreted with particular caution.
Table 2.
The characteristics of adverse drug reactions from TB patients
| Adverse drug reactions | All TB patients | Age | Age‐based | Gender | |||
|---|---|---|---|---|---|---|---|
| Mean ± SD | P | Old /Young | P | M/F | P | ||
| Thrombocytopenia | |||||||
| Presence | 16 | 42.81 ± 20.94 | 0.908 | 6/10 | 0.964 | 11/5 | 0.521 |
| Absence | 628 | 42.24 ± 19.55 | 232/396 | 382/246 | |||
| Leukopenia | |||||||
| Presence | 21 | 45.81 ± 21.05 | 0.397 | 8/13 | 0.912 | 6/15 | 0.002 |
| Absence | 623 | 42.13 ± 19.52 | 230/393 | 387/236 | |||
| Hyperbilirubinemia | |||||||
| Presence | 13 | 43.46 ± 18.14 | 0.822 | 3/10 | 0.295 | 10/3 | 0.235 |
| Absence | 631 | 42.23 ± 19.61 | 235/396 | 383/248 | |||
| ATDH | |||||||
| Presence | 68 | 42.66 ± 18.94 | 0.855 | 24/44 | 0.764 | 42/26 | 0.895 |
| Absence | 576 | 42.20 ± 19.65 | 214/362 | 351/225 | |||
| AKI | |||||||
| Presence | 10 | 55.70 ± 25.21 | 0.028 | 5/5 | 0.397 | 9/1 | 0.037 |
| Absence | 634 | 42.04 ± 19.41 | 233/401 | 384/250 | |||
| CKD | |||||||
| Presence | 12 | 54.25 ± 22.48 | 0.032 | 6/6 | 0.345 | 8/4 | 0.773 |
| Absence | 632 | 42.02 ± 19.45 | 400/232 | 385/247 | |||
Old ≥50 years and Young <50 years; M/F, Male/Female; ATDH, antituberculosis drug‐induced hepatotoxicity; AKI, acute kidney injury; CKD, chronic kidney damage. Significant associations are denoted in bold.
Although most of these 106 ADR events were mild (36.8%) and moderate (53.8%), the severe events, which accounted for 9.4% of the cases, should not be ignored (Table S2). A total of 67 ADR patients required therapeutic intervention, with ATDH and leukopenia being the most important reasons for intervention. ATDs were withdrawn or replaced from 43.4% of the ADR patients (46/106) and were then rechallenged later in most cases, with other drugs administered to 23.6% of the patients (25/106) to relieve symptoms. On eight occasions, severe hepatitis resulted in hospitalization, and INH, rifampicin (RIF), and pyrazinamide (PZA) were discontinued in two instances (one concurrent severe ATDH and AKI, one concomitant ATDH and leukopenia) and were not restarted in either.
Associations and effect evaluations of candidate single nucleotide polymorphisms (SNPs) on ATD‐ADRs
SNP distributions in the TB cases
Genetic variants and their characteristics are listed in Table S4. Seven successfully genotyped SNPs conformed to Hardy–Weinberg equilibrium (P > 0.05 for all). Compared with male patients, female patients had a higher proportion of the mutant T allele‐based genotype in NUDT15 rs116855232 (P = 0.029) (Table S5). There were no nominally significant differences in age and gender among the different genotypes for other SNPs (all P > 0.05).
Potential pharmacogenomic predictors of ATD‐ADRs
Details regarding the correlations among the genotypes and ADRs are presented in Table 3. Rs1135840 and rs116855232 were potential pharmacogenetic markers for identifying individuals at elevated risk of hepatotoxicity and leukopenia during anti‐TB therapy. TB patients with the GG genotype of rs1135840 experienced a higher risk (2.52‐fold) of ATDH than those with an A allele‐containing genotype (adjusted P = 0.009). Patients with rs1135840 GG showed a marginally higher alanine aminotransferase (ALT) value (P = 0.042, Table S6), but rs1135840 was not associated with ATDH severity. Regarding rs116855232, T allele carriers were more likely to suffer from leukopenia, exhibiting a significantly increased risk (odds ratio (OR) = 4.97; 95% confidence interval (CI) = 2.06–11.97, adjusted P = 0.003, Table 3). Sixteen of the 644 TB patients were identified with a homozygous TT mutation in rs116855232, and the TT genotype‐related risk of leukopenia was markedly higher in leukopenia (OR (95% CI) = 7.82 (2.05–29.87), adjusted P = 0.018). No significant differences among the frequencies of the other five SNPs were obtained between the ADR cases and the controls (all P > 0.05 before correction).
Table 3.
SNP associated with adverse drug reactions from TB patients
| Genotypes | Thrombocytopenia (n = 16) | Leukopenia (n = 21) | Hyperbilirubinemia (n = 13) | ATDH (n = 68) | AKI (n = 10) | CKD (n = 12) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |
| ABCB1 rs1045642 | ||||||||||||
| AG+GG (n = 538) | 0.85 (0.24‐3.04) | 0.806 | 0.19 (0.34‐4.11) | 0.781 | 1.09 (0.24‐4.97) | 0.915 | 1.02 (0.52‐2.02) | 0.947 | 1.79 (0.22‐14.25) | 0.579 | 0.58 (0.16‐2.19) | 0.428 |
| AA (n = 106) | ||||||||||||
| ABCB1 rs1055302 | ||||||||||||
| CT+TT (n = 294) | 1.20 (0.44‐3.23) | 0.724 | 1.61 (0.67‐3.88) | 0.282 | 2.73 (0.83‐8.96) | 0.085 | 1.07 (0.64‐1.76) | 0.806 | 1.19 (0.34‐4.16) | 0.781 | 0.85 (0.27‐2.70) | 0.780 |
| CC (n = 350) | ||||||||||||
| CYP2D6 rs1135840 | ||||||||||||
| GG (n = 344) | 1.46 (0.52‐4.06) | 0.469 | 0.64 (0.27‐1.54) | 0.316 | 1.01 (0.34‐3.04) | 0.985 | 2.52 (1.43‐4.44) | 0.009* | 0.86 (0.25‐3.01) | 0.819 | 0.61 (0.19‐1.95) | 0.403 |
| CG+CC (n = 298) | ||||||||||||
| CYP2C19 rs3758580 | ||||||||||||
| CT+TT (n = 335) | 0.54 (0.20‐1.51) | 0.236 | 1.23 (0.51‐2.97) | 0.638 | 1.86 (0.55‐6.24) | 0.308 | 1.11 (0.67‐1.84) | 0.686 | 3.74 (0.79‐17.77) | 0.075 | 1.29 (0.41‐4.12) | 0.663 |
| CC (n = 308) | ||||||||||||
| CYP2C19 rs4244285 | ||||||||||||
| GG (n = 310) | 1.82 (0.65‐5.07) | 0.244 | 0.80 (0.33‐1.93) | 0.623 | 0.67 (0.22‐2.06) | 0.481 | 0.89 (0.54‐1.48) | 0.657 | 0.27 (0.06‐1.26) | 0.073 | 0.77 (0.24‐2.44) | 0.651 |
| AG+AA (n = 334) | ||||||||||||
| CYP2C19 rs4986894 | ||||||||||||
| TT (n = 310) | 1.82 (0.65‐5.07) | 0.244 | 0.80 (0.33‐1.93) | 0.623 | 0.67 (0.22‐2.06) | 0.481 | 0.89 (0.54‐1.48) | 0.657 | 0.27 (0.06‐1.26) | 0.073 | 0.77 (0.24‐2.44) | 0.651 |
| CT+CC (n = 334) | ||||||||||||
| NUDT15 rs116855232 | ||||||||||||
| CT+TT (n = 124) | 1.41 (0.45‐4.45) | 0.568 | 4.97 (2.06‐11.97) | 0.003* | 0.75 (0.17‐3.47) | 0.713 | 0.89 (0.46‐1.71) | 0.73 | 1.82 (0.46‐7.13) | 0.290 | 1.41 (0.38‐5.28) | 0.709 |
| CC (n = 520) | ||||||||||||
P value has been adjusted for age, gender and BMI.
Considering the low frequencies of some minor genotypes, seven SNPs were stratified based on the dominant or recessive model.
Notably, the rs116855232 TT genotype‐related risk is markedly higher (OR = 7.82, 95% CI = 2.05–29.87, adjusted P = 0.018) in leukopenia.
*Significant associations denoted in bold were performed with Bonferroni corrections.
Performances of significant SNP screening for ATDH and leukopenia
We evaluated the predictive capacities of the significant SNPs CYP2D6 rs1135840 and NUDT15 rs116855232 (Table 4). Because predictive biomarkers are used to exclude patients at risk for developing ADRs, negative predictive value (NPV) and sensitivity become the most important indicators in the analysis.30 Rs1135840 had good capabilities for these parameters, with an NPV of 93.7% and a sensitivity of 72.7%. The performance of rs116855232 in predicting ATD‐induced leukopenia was moderate, with an NPV of 98.1% but a sensitivity of only 52.4%. Both SNPs showed poor positive predictive values (PPVs).
Table 4.
Performance of pharmacogenetic screening for ATDH and leukopenia from TB patients
| Phenotype | rs1135840*GG |
Predictive parameters percent (95% CI) |
Phenotype | rs116855232* TT+CT |
Predictive parameters percent (95% CI) |
||||
|---|---|---|---|---|---|---|---|---|---|
| Pos | Neg | Total | Pos | Neg | Total | ||||
| ATDH |
Sen: 72.7 (61.0‐82.0) Spe: 48.6 (44.6‐52.7) PPV: 14.0 (10.7‐18.0) NPV: 93.7 (90.7‐96.2) Accuracy: 51.1 (47.2‐54.9) |
Leukopenia |
Sen: 52.4 (32.4‐71.7) Spe: 81.9 (78.6‐84.7) PPV: 8.9 (5.0‐15.2) NPV: 98.1 (96.5‐99.0) Accuracy: 80.9 (77.7‐83.8) |
||||||
| Presence | 48 | 18 | 66 | Presence | 11 | 10 | 21 | ||
| Absence | 296 | 280 | 576 | Absence | 113 | 510 | 623 | ||
| Total | 344 | 298 | 642 | Total | 124 | 520 | 644 | ||
Pos, positve; Neg, negative; Sen, sensitivity; Spe, specificity; PPV, positive predictive value; NPV, negative predictive value.
DISCUSSION
Globally, ADR has been a major contributor toward anti‐TB treatment discontinuation or failure, particularly in developing countries such as China. In the present study, ATD‐ADRs noticeably and substantially impacted anti‐TB treatment, with an incidence of 16.5% and an intervention rate of 10.4%; fortunately, most were reversible after timely interference, findings that were comparable to other studies from China.31, 32, 33 In our study, ATDH, the most frequent and serious ATD‐ADR, was moderate (10.6%) compared with the data in the literature on hospitalized patients (4.8–36%) because of different definitions used, heterogeneous ethnic origins, various sample sizes, and even potential confounding causes of hepatotoxicity. Advanced age and female gender are common risk factors for the occurrence and severity of ATDH; however, significance was not achieved in the present study. The indicators of an unhealthy lifestyle, such as drinking, smoking, and staying up late, are more frequent in younger generations than in the elderly, which may increase younger individuals' predisposition to hepatotoxicity.
Leukopenia and thrombocytopenia are increasing and unfavorable among Chinese TB patients. Despite the threat that they pose to patients, these events have been overlooked in studies from other regions of China.31, 32, 33 A recent retrospective study included a relatively smaller sample of patients from northern China.33 Two prospective reports,31, 32 which included outpatients and inpatients, came from the Centers for Disease Control and Prevention (CDC); however, since 2010 TB diagnosis and treatment have been transferred from local CDCs to specialized hospitals in China because of the better surveillance of ADRs in hospitals. Our prospective study included 644 TB inpatients from West China Hospital, the largest medical center in western China, and adopted a very strict ADR criterion and causality assessment. This is what differentiates the present study from other similar studies and makes the conclusions of the present study more persuasive and representative. Our findings also provide a basis for patient management, particularly in female patients, who develop hematological toxicity during TB therapy. The current study also evaluated other ADRs, including hyperbilirubinemia, AKI, and CKD, which were not fully understood in a few previous studies.34, 35 AKI and CKD caused by ATDs are rare but severe and have been reported concomitant with hepatitis in 10% of patients.7 As an absolute number, such a rate is substantial and is a cause for concern in the high TB‐burden regions of Asia and South Africa. This concomitance must be considered during the reintroduction to the drug or intermittent administration.
Notably, the present genetic analysis indicated that CYP2D6 rs1135840 and NUDT15 rs116855232 were potential pharmacogenomic predictors of developing ATDH and leukopenia. The CYP2D6 gene encodes an important hepatic phase I drug‐metabolizing enzyme involved in the metabolism of up to 25% of medications.36 Lee et al. reported that rs1135840 GG is associated with higher norclozapine/clozapine serum drug levels.37 Zhang et al. found that CpG hypermethylation of CYP2D6 and rs1135840 increased the risk of ATDH in a small Chinese population.38 We determined that the rs1135840 GG genotype was associated with an increased risk of ATDH, which was in accordance with Zhang et al.'s findings.38 Previous significant SNPs that have been reported to be directly associated with ATDH are not in linkage disequilibrium with rs1135840 and are unlikely to be confounding factors for the association of rs1135840 with ATDH (Table S7). Located in exon 9 of the CYP2D6 gene, rs1135840 encodes a Ser/Thr missense substitution and is a part of the extremely common decreased‐functional haplotype CYP2D6*10. This variant could result in interindividual variability of CYP2D6 enzymatic activity or expression. Previous studies and our results suggest that the CYP2D6 gene may play a role in participating in the metabolism of ATDs or forming covalent adducts with ATDs, similar to the effect of some other general CYP enzymes on ATDs39, 40; however, this possibility should be verified in functional assays.
The NUDT15 enzyme catalyzes the hydrolysis of oxidatively damaged nucleotides to prevent them from being incorporated into DNA, thereby reducing DNA damage and the risk of apoptosis.41, 42 NUDT15 rs116855232 C>T (p.Arg139Cys), which is characteristic of Asian and Native American populations, was observed in 10.8% of our study population and seemed to make a significant contribution to the development of leukopenia. In TB patients, the presence of the NUDT15*T allele would confer a 4.97‐fold increased risk for leukopenia, and the TT genotype‐related risk is much higher (7.82‐fold). A well‐established association of NUDT15 C>T with thiopurine‐related leukopenia has been confirmed among different populations, endowing the homozygous TT with a remarkable up to 35‐fold increase in leukopenia risk.41, 42, 43, 44 Functional experiments have demonstrated that the loss‐of‐function mutation NUDT15 (rs116855232 TT) causes increased cell apoptosis and decreased cell viability, as well as low tolerance to 6‐MP treatment.41 In clinical practice in China, prior to azathioprine/6‐mercaptopurine initiation, patients are currently required to test NUDT15 rs116855232 C>T in advance, and a dose reduction of at least 50% is recommended for those with homozygous NUDT15 variants. These findings compellingly point to NUDT15 rs116855232 as a pharmacogenetic marker for drug‐induced leukopenia, particularly in patients with Asian or Native American ancestry.
Genetic risk assessment is useful to exclude ADRs for their high NPV.30 The NPVs of 93.7% and 98.1% obtained in our study are significant as screening tests, although well‐designed prospective studies are warranted to confirm and generalize as appropriate. The PPVs are low for identifying persons with ATDH and leukopenia, which is likely due to the relatively high frequency of the risk genotypes in the study population (∼53% for CYP2D6 and 19% for NUDT15) compared with the low incidence of these ADRs (∼10.6% for ATDH and 3.3% for leukopenia).30 These results also reflect that gene determinants are not sufficient to trigger ATD‐ADRs, and combinatorial indicators including genetic risk variants and nongenetic risk factors may be more powerful in predicting ATD‐ADRs.20 Genetic assessment and regular blood monitoring are helpful to identify patients with a high ATD‐ADR risk. A series of measures can be taken to prevent ATD‐ADRs in these patients, including the use of lower initial doses, dose reduction, and coadministration of protective agents. Prompt withdrawal of the offending medication and regimen changes remain the most critical strategy applied in clinical practice.45
Our study has several limitations. First, given the small number of patients with ADRs, multicenter studies are warranted to confirm the findings. Second, a combination of drug‐related factors, such as drug dosage, therapeutic drug monitoring, and genetic and nongenetic risk factors, should be incorporated into algorithms to improve predictive capability over that of genetic tests alone. Third, we assessed only the ADRs induced by standard multidrug regimens, and the specific effects of single drugs on ADRs, as well as the effects of high or toxic doses of ATDs in patients with extensively drug‐resistant tuberculosis and tuberculous meningitis should be investigated further. Furthermore, some patients took TB medicines for more than 8 months, which was longer than our follow‐up period; consequently, ATD‐ADRs among different courses of treatment still need to be studied intensively.
In summary, western China had a marked epidemic of ATD‐ADRs, significantly impacting subsequent therapeutic orientation. Regular blood examinations are suggested to closely monitor adverse events. We identified CYP2D6 rs1135840 and NUDT15 rs116855232 as genetic risk factors for the development of ATDH and leukopenia, highlighting the importance of pharmacogenetic analyses during individualized TB treatment.
METHODS
Study design and patient population
We consecutively recruited 1,235 highly suspicious TB patients at West China Hospital, western China, between December 2014 and November 2016. In total, 833 TB patients were confirmed by experienced respiratory physicians, based on typical TB symptoms, microbiological and/or radiological evidence of MTB, and response to ATDs. Patients with hepatitis, HIV infection, and renal, hematological, and cardiac diseases were excluded. Those with definite signs of aberrant liver and renal function and abnormal hematologic indicators before anti‐TB treatment were also excluded. All patients underwent short‐course chemotherapy consisting of at least oral INH (300–400 mg/day), RIF (450–600 mg/day), PZA (1,500–2,000 mg/day), and ethambutol (EMB, 750–900 mg/day) for the first 2 months, followed by INH, RIF, and EMB for at least 4 more months. If patients developed a definite ADR, their treatments were adjusted accordingly. Twenty‐nine patients were poorly adherent to the treatment or dropped out of the 8‐month follow‐up. Ultimately, 644 TB patients were enrolled in the prospective study.
Demographic and clinical data of the patients were obtained from medical records. Blood samples were collected from each participant for genotyping. Pretherapeutic laboratory tests and chest radiographs were performed in the clinical lab of West China Hospital. After ATDs were started, blood work, including analysis of biochemical parameters and hematological blood counts, and urine analysis were performed twice a month during the first 2 months and monthly in subsequent follow‐up appointments. The peak or valley values of laboratory indicators during the 8‐month follow‐up were recorded and used to assess ATD‐ADRs. Patients were regularly questioned about symptoms regarding ATD‐ADRs over the telephone or WeChat if possible. A flow diagram of the process used to enroll the participants is shown in Figure 1. This study was approved by the Committee on Human Research, Publications and Ethics, West China Hospital, Sichuan University (China), and written informed consent was obtained from each participant.
Figure 1.
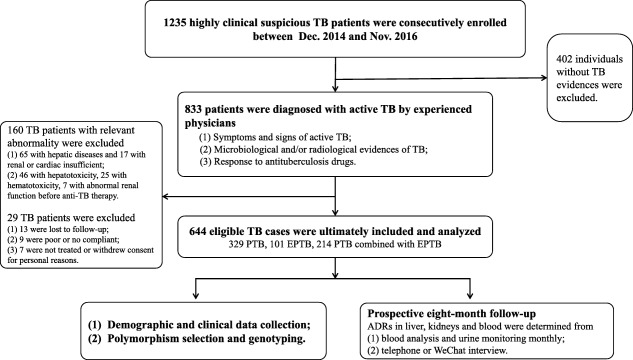
Flow chart of the study population. PTB, pulmonary tuberculosis; EPTB, extrapulmonary tuberculosis; anti‐TB, antituberculosis.
Definition of ATD‐ADRs and their severity
ATD‐ADRs in the liver, kidneys, and blood were strictly defined as grade 2 or 3 according to the National Institutes of Health and Common Toxicity Criteria for Adverse Events v. 4.0 (CTCAE v. 4.0)22 unless stated otherwise. Causality assessments of ATD‐ADRs were used to determine whether ADRs were present or not.23
ADRs in liver
(1) ATDH. Individuals who presented with ALT and aspartate aminotransferase (AST) levels ≥ 3 × the upper limit of normal (ULN) (120 IU/L) in the presence of hepatitis symptoms such as jaundice, nausea, vomiting, and abdominal pain and individuals presenting with ALT and AST levels ≥ 5 × the ULN (150 IU/L), with or without symptoms, were considered to have ATDH.22, 24 The severity of ATDH was classified as mild, moderate, or severe.22, 25 (2) Hyperbilirubinemia. An increase in total bilirubin ≥ 1.5 × the ULN (42 μmol/L) was defined as hyperbilirubinemia.22
Renal impairment
CKD was defined as the persistence of kidney injury (eGFR < 60 ml/min /1.73 m2 and presence of proteinuria) for more than 3 months.26 AKI was determined based on the presence of severe kidney injury within 48 h according to the consensus26 and the KDIGO AKI Guideline.27
Hematotoxicity
Anemia (hemoglobin concentration ≤ 80 × 1012/L), leukopenia (absolute counts ≤ 2.0 × 109/L), and thrombocytopenia (absolute counts ≤ 75 × 109/L) were considered as forms of hematological toxicity.22
ATD‐ADRs severity
ATD‐ADRs were graded as mild, moderate, and severe. ADRs were described as mild when they had little impact on TB management and when treatment was unnecessary. Moderate ADRs were designated when related symptoms and signs returned to normal upon the change/discontinuation of ATDs or when symptomatic treatment was initiated. Severe ADRs were those that led to prolonged hospital stays, required special care, or caused disability or death.28 For patients with moderate and severe ADRs, in addition to symptomatic treatment, TB medications were stopped if necessary and blood analyses were performed weekly until the blood indicators returned to normal levels.
Candidate polymorphism selection and genotyping
Key genes in the drug metabolic pathways and drug transport pathways, including CYP2D6, CYP2C19, ABCB1, and NUDT15, which were representative and highly polymorphic, were selected based on their biological functions and putative evidence for associations with ATD‐ADRs. CYP2D6 and CYP2C19 encode two important hepatic phase I drug‐metabolizing enzymes, and ABCB1 encodes P‐glycoprotein, a well‐known drug transporter protein, and these genes may be involved in metabolism/transport of ATDs (see Supplementary References 1–12). Additionally, NUDT15 encodes an important hydrolytic enzyme in the Nudix hydrolase superfamily. We selected candidate SNPs in these four genes by performing thorough searches of the dbSNP database and 1000 Genomes Project. SNPs were included if they were located in the promoter or exon region and had a minor allele frequency >0.05 among the Han Chinese in Beijing and if they had been implicated in the toxicity or ADRs of other drugs but rarely in that of anti‐TB drugs. Under the experimental conditions required for genotyping seven SNPs (CYP2D6 rs1135840; CYP2C19 rs3758580, rs4244285, rs4986894; ABCB1 rs1045642, rs1055302; NUDT15 rs116855232) were eventually included. Genomic DNA was extracted from peripheral blood and was genotyped using a commercial custom‐by‐design 48‐Plex SNPscan Kit, as described previously,29 which is based on patented SNP genotyping technology with a double‐ligation and multiplex fluorescence polymerase chain reaction. For quality control purposes, 30 random samples were genotyped in duplicate, with a concordance rate of 100%, and three samples with different genotypes for each SNP were randomly selected to confirm the genotyping results by direct sequencing.
Statistical analysis
Univariate analysis was used to analyze categorical variables with the chi‐square test and continuous variables were analyzed with the Mann–Whitney U‐test. Associations between SNPs, TB characteristics, and ATD‐ADRs were evaluated using the unconditional logistic regression after adjusting for age, gender, and BMI. ORs and 95% CIs were used as measures of associations. Bonferroni corrections were used to adjust significant associations. The above‐mentioned analyses were performed using SPSS v. 22.0 (IBM, Armonk, NY). Performances of SNPs, including sensitivity, specificity, PPV, and NPV, were calculated according to the OpenEpi (Dean, A.G. et al. The OpenEpi Collection of Epidemiologic Calculators <http://www.openepi.com/Menu/OE_Menu.html> Accessed 4 April 2013). Statistical significance was set at 0.05.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
X.H., M.Z., H.B., X.C., and Y.Z. wrote the article; B.Y. designed the research; X.H., H.B., L.W., Y.C., L.D., Z.Z., W.P., T.L., and J.S. performed the research; X.H., M.Z., Y.L., and X.L. analyzed the data; M.Z. and X.C. contributed new reagents/analytical tools.
Supporting information
Supporting Information 1
Supporting Information 2
ACKNOWLEDGMENTS
Xuejiao Hu, Mei Zhang, and Hao Bai contributed equally to this work. This work was funded by grants from the National Natural Science Foundation of China (81672095) and the Projects of the Health and Family Plan in Sichuan Province (16ZD004). The authors also thank the Shanghai Genesky Bio‐Tech Genetic Core Lab for providing assistance with genotyping techniques.
Contributor Information
Xuerong Chen, Email: docbwy@126.com.
Yanhong Zhou, Email: docbwy@126.com.
Binwu Ying, Email: docbwy@126.com.
References
- 1. Global Tuberculosis Report 2016. Geneva: World Health Organization. <http://www.who.int/tb/publications/global_report/en/> (2016) Accessed 13 October 2016.
- 2. Sharma, S.K. et al Safety of 3 different reintroduction regimens of antituberculosis drugs after development of antituberculosis treatment‐induced hepatotoxicity. Clin. Infect. Dis. 50, 833–839 (2010). [DOI] [PubMed] [Google Scholar]
- 3. Munro, S.A. , Lewin, S.A. , Smith, H.J. , Engel, M.E. , Fretheim, A. & Volmink, J. Patient adherence to tuberculosis treatment: a systematic review of qualitative research. PLoS Med. 4, e238 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yamada, S. et al Genetic variation in carboxylesterase genes and susceptibility to isoniazid‐induced hepatotoxicity. Pharmacogenom. J. 10, 524–536 (2010). [DOI] [PubMed] [Google Scholar]
- 5. Fernandez‐Villar, A. et al Isoniazid hepatotoxicity among drug users: the role of hepatitis C. Clin. Infect. Dis. 36, 293–298 (2003). [DOI] [PubMed] [Google Scholar]
- 6. Forget, E.J. & Menzies, D. Adverse reactions to first‐line antituberculosis drugs. Expert Opin. Drug Saf. 5, 231–249 (2006). [DOI] [PubMed] [Google Scholar]
- 7. Kumar, R. et al Antituberculosis therapy‐induced acute liver failure: magnitude, profile, prognosis, and predictors of outcome. Hepatology 51, 1665–1674 (2010). [DOI] [PubMed] [Google Scholar]
- 8. Costiniuk, C.T. et al Acute renal failure and disseminated intravascular coagulation associated with rifampin in tuberculosis treatment. Int. J. Tuberc. Lung Dis. 15, 421 (2011). [PubMed] [Google Scholar]
- 9. Cheung, C.L. et al HLA‐B 38:02:01 predicts carbimazole/methimazole‐induced agranulocytosis. Clin. Pharmacol. Ther. 99, 555–561 (2016). [DOI] [PubMed] [Google Scholar]
- 10. Pirmohamed, M. , Aithal, G.P. , Behr, E. , Daly, A. & Roden, D. The phenotype standardization project: improving pharmacogenetic studies of serious adverse drug reactions. Clin. Pharmacol. Ther. 89, 784–785 (2011). [DOI] [PubMed] [Google Scholar]
- 11. Low, S.K. , Takahashi, A. , Mushiroda, T. & Kubo, M. Genome‐wide association study: a useful tool to identify common genetic variants associated with drug toxicity and efficacy in cancer pharmacogenomics. Clin. Cancer Res. 20, 2541–2552 (2014). [DOI] [PubMed] [Google Scholar]
- 12. Olson, J.E. et al Participant‐perceived understanding and perspectives on pharmacogenomics: the Mayo Clinic RIGHT protocol (Right Drug, Right Dose, Right Time). Genet. Med. 19, 819–825 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. O'Donnell, P.H. et al Pharmacogenomics‐based point‐of‐care clinical decision support significantly alters drug prescribing. Clin. Pharmacol. Ther. 11, 709 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma, Q. & Lu, A.Y. Pharmacogenetics, pharmacogenomics, and individualized medicine. Pharmacol. Rev. 63, 437–459 (2011). [DOI] [PubMed] [Google Scholar]
- 15. Whirl‐Carrillo, M. et al Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 92, 414–417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang, C. & Yao, S.K. Precision Medicine: A Compendium of Drug Therapy, 1st ed (People's Medical Publishing House, Beijing, 2016). [Google Scholar]
- 17. Matsumoto, T. , Ohno, M. & Azuma, J. Future of pharmacogenetics‐based therapy for tuberculosis. Pharmacogenomics 15, 601–607 (2014). [DOI] [PubMed] [Google Scholar]
- 18. Jung, J.A. et al A proposal for an individualized pharmacogenetic‐guided isoniazid dosage regimen for patients with tuberculosis. Drug. Des. Dev. Ther. 9, 5433–5438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Azuma, J. et al NAT2 genotype guided regimen reduces isoniazid‐induced liver injury and early treatment failure in the 6‐month four‐drug standard treatment of tuberculosis: a randomized controlled trial for pharmacogenetics‐based therapy. Eur. J. Clin. Pharmacol. 69, 1091–1101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Russmann, S. , Jetter, A. & Kullak‐Ublick, G.A. Pharmacogenetics of drug‐induced liver injury. Hepatology 52, 748–761 (2010). [DOI] [PubMed] [Google Scholar]
- 21. Wang, L. et al Tuberculosis prevalence in China, 1990‐2010; a longitudinal analysis of national survey data. Lancet 383, 2057–2064 (2014). [DOI] [PubMed] [Google Scholar]
- 22. National Cancer Institute, National Institutes of Health . Common Terminology Criteria for Adverse Events Version 4.0. <http://evs.nci.nih.gov/ftp1/CTCAE/About.html> (2009). Accessed May 2009.
- 23. World Health Organization . Uppsala Monitoring Centre. The use of the WHO‐UMC system for standardized case causality assessment. <https://www.who-umc.org/media/2768/standardised-case-causality-assessment.pdf> (2012). Accessed 17 April 2012.
- 24. Nahid, P. et al Executive Summary: Official American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America Clinical Practice Guidelines: Treatment of Drug‐Susceptible Tuberculosis. Clin. Infect. Dis. 63, 853–867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tostmann, A. , Boeree, M.J. , Aarnoutse, R.E. , de Lange, W.C. , van der Ven, A.J. & Dekhuijzen, R. Antituberculosis drug‐induced hepatotoxicity: concise up‐to‐date review. J. Gastroenterol. Hepatol. 23, 192–202 (2008). [DOI] [PubMed] [Google Scholar]
- 26. Mehta, R.L. et al Phenotype standardization for drug‐induced kidney disease. Kidney Int. 88, 226–234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kellum, J.A. & Lameire, N. Diagnosis, evaluation, and management of acute kidney injury: a KDIGO summary (Part 1). Crit. Care 17, 204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marra, F. et al Adverse drug reactions associated with first‐line anti‐tuberculosis drug regimens. Int. J. Tuberc. Lung Dis. 11, 868–875 (2007). [PubMed] [Google Scholar]
- 29. Yin, J. et al Interleukin 17A rs4711998 A>G polymorphism was associated with a decreased risk of esophageal cancer in a Chinese population. Dis. Esophagus 27, 87–92 (2014). [DOI] [PubMed] [Google Scholar]
- 30. Singer, J.B. et al A genome‐wide study identifies HLA alleles associated with lumiracoxib‐related liver injury. Nat. Genet. 42, 711–714 (2010). [DOI] [PubMed] [Google Scholar]
- 31. Lv, X. et al Adverse reactions due to directly observed treatment strategy therapy in Chinese tuberculosis patients: a prospective study. PLoS One 8, e65037 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang, T. et al Adverse events in treating smear‐positive tuberculosis patients in China. Int. J. Environ. Res. Public Health 13, E86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Han, X.Q. et al Prevalence and risk factors associated with adverse drug reactions among previously treated tuberculosis patients in China. Biomed. Environ. Sci. 30, 139–142 (2017). [DOI] [PubMed] [Google Scholar]
- 34. Kizilbash, Q. Successful management of acute interstitial nephritis in two cases of disseminated tuberculosis. Tuberculosis (Edinb). 101S, S135–S136 (2016). [DOI] [PubMed] [Google Scholar]
- 35. Muthukumar, T. , Jayakumar, M. , Fernando, E.M. & Muthusethupathi, M.A. Acute renal failure due to rifampicin: a study of 25 patients. Am. J. Kidney Dis. 40, 690–696 (2002). [DOI] [PubMed] [Google Scholar]
- 36. Owen, R.P. , Sangkuhl, K. , Klein, T.E. & Altman, R.B. Cytochrome P450 2D6. Pharmacogenet. Genomics 19, 559–562 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee, S.T. et al Association study of 27 annotated genes for clozapine pharmacogenetics: validation of preexisting studies and identification of a new candidate gene, ABCB1, for treatment response. J. Clin. Psychopharmacol. 32, 441–448 (2012). [DOI] [PubMed] [Google Scholar]
- 38. Zhang, J. et al Correlation of CpG island methylation of the cytochrome P450 2E1/2D6 genes with liver injury induced by anti‐tuberculosis drugs: a nested case‐control study. Int. J. Environ. Res. Public Health 13, E776 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sahu, R.K. , Singh, K. & Subodh, S. Adverse drug reactions to anti‐TB Drugs: pharmacogenomics perspective for identification of host genetic markers. Curr. Drug Metab. 16, 538–552 (2015). [DOI] [PubMed] [Google Scholar]
- 40. Metushi, I. et al Mechanism of isoniazid‐induced hepatotoxicity: then and now. Br. J. Clin. Pharmacol. 81, 1030–1036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang, S.K. et al A common missense variant in NUDT15 confers susceptibility to thiopurine‐induced leukopenia. Nat. Genet. 46, 1017–1020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang, J.J. et al Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J. Clin. Oncol. 33, 1235–1242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tanaka, Y. , Nakadate, H. , Kondoh, K. , Nakamura, K. , Koh, K. & Manabe, A. Interaction between NUDT15 and ABCC4 variants enhances intolerability of 6‐mercaptopurine in Japanese patients with childhood acute lymphoblastic leukemia. Pharmacogenom. J. 18, 12 (2017). [DOI] [PubMed] [Google Scholar]
- 44. Moriyama, T. et al NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat. Genet. 48, 367–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ramappa, V. & Aithal, G.P. Hepatotoxicity related to anti‐tuberculosis drugs: mechanisms and management. J. Clin. Exp. Hepatol. 3, 37–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information 1
Supporting Information 2


