Abstract
Previous metabolic studies have demonstrated that leishmania parasites are able to synthesise proline from glutamic acid and threonine from aspartic acid. The first committed step in both biosynthetic pathways involves an amino acid kinase, either a glutamate 5‐kinase (G5K; http://www.chem.qmul.ac.uk/iubmb/enzyme/EC2/7/2/11.html) or an aspartokinase (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC2/7/2/4.html). Bioinformatic analysis of multiple leishmania genomes identifies a single amino acid‐kinase gene (LdBPK 262740.1) variously annotated as either a putative glutamate or aspartate kinase. To establish the catalytic function of this Leishmania donovani gene product, we have determined the physical and kinetic properties of the recombinant enzyme purified from Escherichia coli. The findings indicate that the enzyme is a bona fide G5K with no activity as an aspartokinase. Tetrameric G5K displays kinetic behaviour similar to its bacterial orthologues and is allosterically regulated by proline, the end product of the pathway. The structure‐activity relationships of proline analogues as inhibitors are broadly similar to the bacterial enzyme. However, unlike G5K from E. coli, leishmania G5K lacks a C‐terminal PUA (pseudouridine synthase and archaeosine transglycosylase) domain and does not undergo higher oligomerisation in the presence of proline. Gene replacement studies are suggestive, but not conclusive that G5K is essential.
Enzymes
Glutamate 5‐kinase (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC2/7/2/11.html); aspartokinase (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC2/7/2/4.html).
Keywords: biochemical pathway, drug target, inhibitors, Leishmania, proline biosynthesis
Abbreviations
- DDR
double drug‐resistant line
- G5K
glutamate 5‐kinase
- P5CS
Δ1–pyrroline‐5‐carboxylate synthase
- PUA domain
pseudouridine synthase and archaeosine transglycosylase domain
- RCWT
wild‐type leishmania‐overexpressing G5K
- ROS
reactive oxygen species
- SDR
single drug‐resistant line
Introduction
Leishmania parasites are the causative agents of visceral, cutaneous and mucocutaneous leishmaniasis. The life‐threatening visceral form is responsible for 30 000 to 50 000 deaths per annum and the incidence of the disfiguring cutaneous form is estimated at 700 000 to 1 300 000 new cases per annum (https://www.dndi.org/diseases-projects/leishmaniasis/). Treatment options are limited: there are no efficacious vaccines and currently available drugs are expensive, difficult to administer and toxic 1, 2, 3. The disease is transmitted between mammalian hosts by the bite of a female sand fly. During their life cycle, the parasites undergo remarkable morphological and biochemical changes, ranging from motile flagellated promastigote forms living in the alkaline midgut of the sand‐fly vector to sessile amastigotes growing in the acidic parasitophorous vacuole of the host macrophage 4. Not only do leishmania have to adapt to environmental changes in pH, temperature and osmotic tension but also to changes in nutrient availability 5. In addition to intermittent blood feeds, the female sand fly also feeds on sugars (mainly sucrose) either from aphid honeydew or from plants. Thus, leishmania parasites live in a sugar‐ and amino acid‐rich environment in the insect midgut and preferentially use simple sugars as a source of energy. Under glucose‐limiting conditions, leishmania promastigotes can also use proline as an energy source 6, 7. Some bloodsucking insects utilise abundant proline in their haemolymph as an energy source for flight 8, 9, 10 where proline concentrations can be as high as 60 mm; however, nothing is known about proline levels in the case of sand flies. In contrast to the promastigote, leishmania amastigotes live in a glucose‐poor environment and utilise fatty acids for energy production and amino acids for gluconeogenesis 5, 11, 12, 13.
Metabolic labelling studies with either [14C] or [13C] glucose indicate that label can be incorporated into glycine, serine, proline, or threonine in amino acid‐deficient medium 6, 14. [U13C] Aspartate is also readily incorporated into threonine suggesting the presence of the beta‐aspartate pathway to threonine 14. Glutamate and aspartate are of interest as the first committed step in the respective biosynthetic pathways involves an amino acid kinase, either glutamate 5‐kinase (G5K, http://www.chem.qmul.ac.uk/iubmb/enzyme/EC2/7/2/11.html) for proline synthesis, or aspartokinase (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC2/7/2/4.html) for threonine synthesis (Fig. 1). Both of these pathways are absent in the related parasite Trypanosoma brucei where threonine constitutes a major source of acetate for lipid biosynthesis 15, 16, 17 and proline is a major energy source in insect procyclic forms 18, 19, 20. Although bioinformatic analyses of leishmania genomes have identified suitable candidate genes for all of the remaining pathway steps in the biosynthesis of proline and threonine 21, only one putative amino acid kinase can be identified (e.g. LinJ.26.2740, LmJF.26.2710, and LdBPK_262740.1). These genes are annotated as putative G5Ks in GeneDB, but their enzymatic function has not been characterised. In addition, they have also been proposed as possible aspartokinases 14, 22. The aim of this study was to characterise the putative G5K from Leishmania donovani and assess its possible role in proline and/or threonine biosynthesis.
Figure 1.
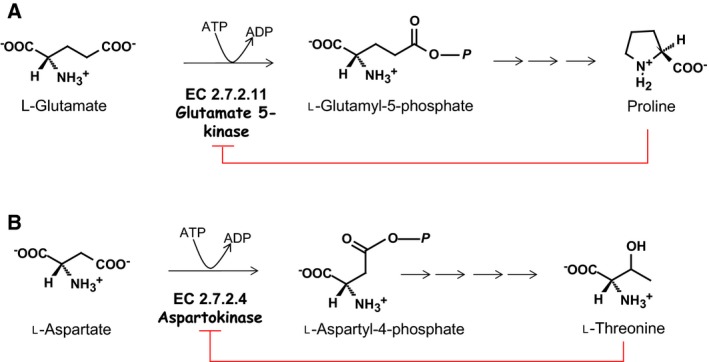
Proline and threonine biosynthetic pathways from glutamate and aspartate. Leishmania is predicted to synthesise proline from glutamate by the same pathway found in bacteria, comprising of three enzymes: γ‐glutamyl kinase (G5K, http://www.chem.qmul.ac.uk/iubmb/enzyme/EC2/7/2/11.html), γ‐glutamyl phosphate reductase (GPR, http://www.chem.qmul.ac.uk/iubmb/enzyme/EC1/2/1/41.html) and Δ1‐pyrroline‐5‐carboxylate reductase (P5C, http://www.chem.qmul.ac.uk/iubmb/enzyme/EC1/5/1/2.html). Biosynthesis of threonine is predicted to start with conversion of aspartate into l‐aspartyl‐4‐phosphate by aspartokinase (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC2/7/2/4.html) followed by aspartate‐semialdehyde dehydrogenase (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC1/2/1/11.html), homoserine dehydrogenase (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC1/1/1/3.html), homoserine kinase (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC2/7/1/39.html) and threonine synthase (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC4/2/3/1.html).
Results
Sequence analysis of G5Ks
The construction of phylogenetic trees spanning both higher eukaryotes to lower prokaryotes (Fig. 2) was built based on their amino sequence and evolutionary distances were calculated by Poisson correction method within mega7 programme. Leishmania G5Ks (e.g. LmjF26.2710, LinJ.26.2740, and LdBPK_262740.1) are located on a clade closer to bacterial and lower eukaryotes compared to higher eukaryotes. A comparison of G5K sequences from Escherichia coli, human and L. donovani (Fig. 3) illustrates some shared homology in relation to residues interacting with nucleotides, glutamate as well as putative binding motifs for ATP, the conserved G5K domain and leucine zipper. Of note, only E. coli contains the C‐terminal PUA (pseudouridine synthase and archaeosine transglycosylase) domain, which is present in some bacteria but absent in the Leishmania equivalent. The PUA domain is potentially involved in RNA binding but its exact function is still unknown 8, 9. The situation is different in humans and other higher eukaryotes in that G5K is part of a bifunctional enzyme (Δ1–pyrroline‐5‐carboxylate synthase, P5CS). Here, the kinase domain at the N‐terminus is fused with a glutamate‐5‐semi‐aldehyde dehydrogenase (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC1/2/1/41.html) domain at the C‐terminus. As with Leishmania the PUA domain is absent. It has been shown that in both bacteria and plants that proline biosynthesis is regulated by proline exerting feedback inhibition of G5K or the equivalent kinase domain of P5CS respectively 10.
Figure 2.
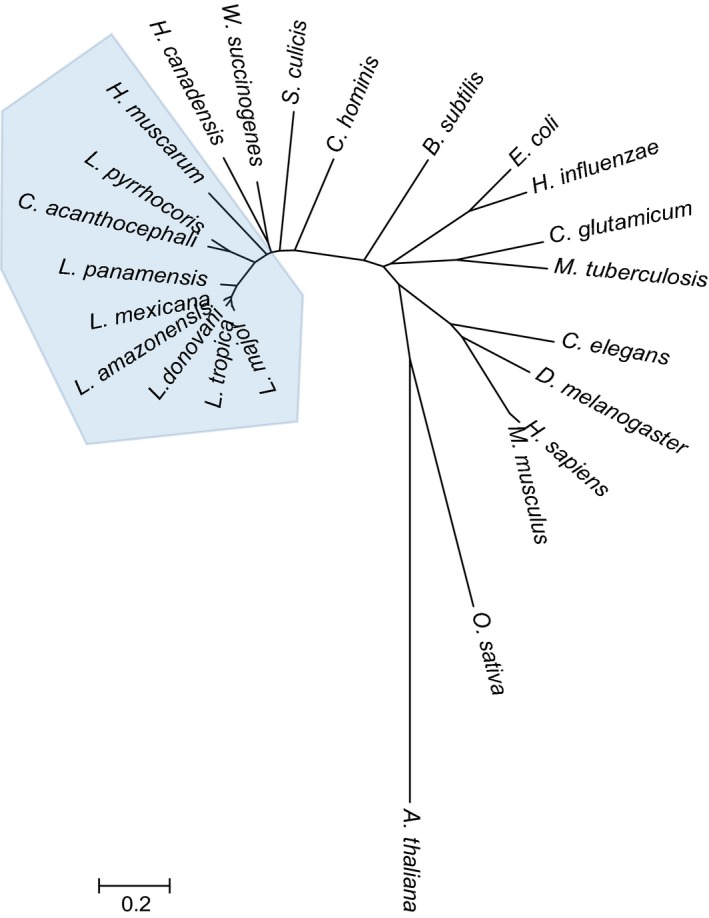
Phylogenetic relationship of G5K orthologues. The phylogenetic tree was constructed as described in the Experimental procedures. The full‐length sequence data were obtained from GenBank/EMBL databases under the following accession numbers: http://www.ncbi.nlm.nih.gov/protein/AEE32297.1 for Arabidopsis thaliana P5CS1; http://www.ncbi.nlm.nih.gov/protein/XP_015621839.1 for Oryza sativa P5CS; http://www.ncbi.nlm.nih.gov/protein/NP_001246877.1 for Drosophila melanogaster P5CS; http://www.ncbi.nlm.nih.gov/protein/CAA64224.1 for Homo sapiens P5CS; P5CS; http://www.ncbi.nlm.nih.gov/protein/NP_062672.2 for Mus musculus P5CS; http://www.ncbi.nlm.nih.gov/protein/CAC35828.2 for Caenorhabditis elegans P5CS; http://www.ncbi.nlm.nih.gov/protein/NP_216955.1 for Mycobacterium tuberculosis G5K; http://www.ncbi.nlm.nih.gov/protein/AAC44174.1 for Corynebacterium glutamicum G5K; http://www.ncbi.nlm.nih.gov/protein/AAC22560.1 for Haemophilus influenzae G5K; http://www.ncbi.nlm.nih.gov/protein/NP_414777.1 for Escherichia coli G5K; http://www.ncbi.nlm.nih.gov/protein/CAB13740.1 for Bacillus subtilis G5K; http://www.ncbi.nlm.nih.gov/protein/WP_012108545.1 for Campylobacter hominis G5K; http://www.ncbi.nlm.nih.gov/protein/EPY34138.1 for Strigomonas culicis G5K; http://www.ncbi.nlm.nih.gov/protein/WP_011138443.1 for Wolinella succinogenes G5K; http://www.ncbi.nlm.nih.gov/protein/EFR48368.1 Helicobacter canadensis G5K; http://www.ncbi.nlm.nih.gov/protein/AGT02536.1 for Herpetomonasmuscarum G5K; http://www.ncbi.nlm.nih.gov/protein/KPA74038.1 for Leptomonas pyrrhocoris G5K; http://www.ncbi.nlm.nih.gov/protein/AGT02656.1 for Crithidia acanthocephali G5K; http://www.ncbi.nlm.nih.gov/protein/AIN99380.1 for Leishmania panamensis G5K; http://www.ncbi.nlm.nih.gov/protein/AKK31239.1 for Leishmania mexicana G5K; http://www.ncbi.nlm.nih.gov/protein/AKK31245.1 for Leishmania amazonensis G5K; http://www.ncbi.nlm.nih.gov/protein/AKK31242.1 for Leishmania donovani G5K; http://www.ncbi.nlm.nih.gov/protein/AKK31236.1 for Leishmania tropica G5K; http://www.ncbi.nlm.nih.gov/protein/CAJ05678.1 for Leishmania major G5K. The trypanosomatid clade is highlighted in blue.
Figure 3.
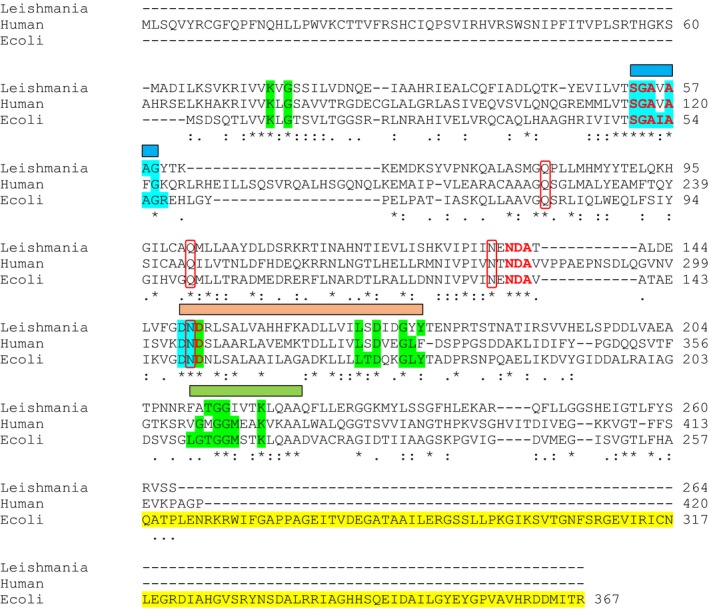
Multiple alignment and key functional residues in G5K orthologues. The amino acid sequence of Leishmania donovani G5K was compared to the human (excluding the C‐terminal PC5S domain) and Escherichia coli homologues. The amino acid sequences were aligned using muscle (http://www.ebi.ac.uk/Tools/msa/muscle/). Identical amino acid residues are highlighted*. Conserved residues which interact with the nucleotide (green), glutamate (red), interact with both (blue) and contain the PUA domain (yellow) are highlighted. Residues involved in linking the two catalytic centres of each dimer (red boxes). Binding motifs for ATP (blue rectangle), conserved G5K domain (green rectangle) and leucine zipper (peach rectangle) are also highlighted. Data from 23, 31, 80. All highlighted residues are identical in species causing mucocutaneous, cutaneous or visceral forms of leishmaniasis (L. donovani shows between 86 and 100% identity with L. guyanensis, L. panamensis, L. braziliensis, L. mexicana, L. tropica, Leishmania major and Leishmania infantum).
Cloning, expression and purification of recombinant LdG5K
The gene‐encoding LdG5K (0.792 kb) was amplified by PCR from L. donovani genomic DNA and cloned into a modified pGEX expression vector. After purification and on‐column proteolytic cleavage of the GST tag a single band was released (yield ~ 1 mg·L−1 of culture) with a Mr of ~ 29 kDa by SDS/PAGE (Fig. 4A). The theoretical Mr was calculated from the amino acid sequence to be 29.033 kDa. Confirmation was undertaken by tryptic analysis of the isolated protein determined by MALDI‐TOF mass spectrometry with 91% coverage and a mass of 29.10 kDa. The molecular weight of the native LdG5K was determined by size exclusion to be approximately 105 kDa consistent with a tetrameric quaternary structure (Fig. 4B).
Figure 4.
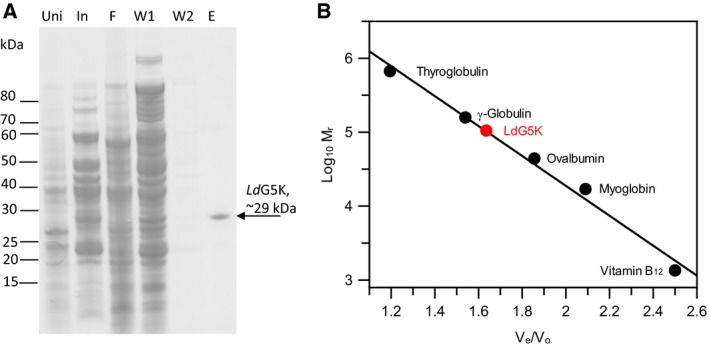
Purification and physical properties of recombinant LdG5K. (A) SDS/PAGE analysis of purification scheme using GST‐tagged LdG5K with on‐column cleavage resulting in the release of recombinant protein. Uni, uninduced sample; In, induced sample; F, flow through; W1 and W2, column wash 1 and 2; E, elution after on‐column cleavage with Prescission Protease. Molecular weight standards (kDa) highlighted on the left hand side of the gel. (B) Oligomeric structure of native LdG5K by size exclusion chromatography. Standard proteins and their molecular weights (Da) are as follows: Thyroglobulin (bovine), 670 000; γ globulin (bovine), 158 000; Ovalbumin (chicken), 44 000; Myoglobin (horse), 17 000; Vitamin B12, 1350. LdG5K, glutamate 5‐kinase.
Biochemical characterisation and determination of the kinetic parameters
Previous enzymatic activity assays have used fixed time‐point assays measuring product formation, either ADP 23 or γ‐glutamyl phosphate converted into γ‐glutamyl hydroxamate 24. A spectrophotometric assay previously described for trypanothione synthetase activity 25 was adapted to follow LdG5K activity with glutamate, aspartate or other amino acid analogues. The assay measured ADP formation by coupling the reaction to pyruvate kinase/lactate dehydrogenase and monitoring oxidation of NADH at 340 nm (Fig. 5A). The kinase exhibited typical hyperbolic kinetics with respect to l‐glutamate and ATP (Fig. 5B,C) with k cat and K m determined to be: 12.8 ± 0.3 s−1 and 10 ± 0.7 mm; 11.7 ± 0.3 s−1 and 0.6 ± 0.07 mm, respectively. Activation of G5K by Mg2+ shows high substrate inhibition with k cat 11.6 ± 0.8 s−1, K a 2.0 ± 0.4 mm and K i of 43 ± 7 mm (Fig. 5D). These parameters are broadly similar to those for bacterial glutamate kinases (Table 1). Several analogues of ATP and l‐glutamate were tested as alternative substrates. GTP, CTP or UTP did not replace ATP as phosphate donor as reported for G5K from Thermotoga maritima 26. Glutaric acid showed 29% of the activity with glutamate as substrate, whereas aminovaleric acid and 2‐amino‐4‐phosphonobutyric acid were inactive. No activity whatsoever could be detected over a range of aspartate concentrations (1–200 mm). l‐citrulline and d,l‐ornithine at 1 mm concentration did not inhibit the enzymatic conversion of l‐glutamate. These data indicate that the enzyme is a G5K, with no functional activity as an aspartokinase.
Figure 5.
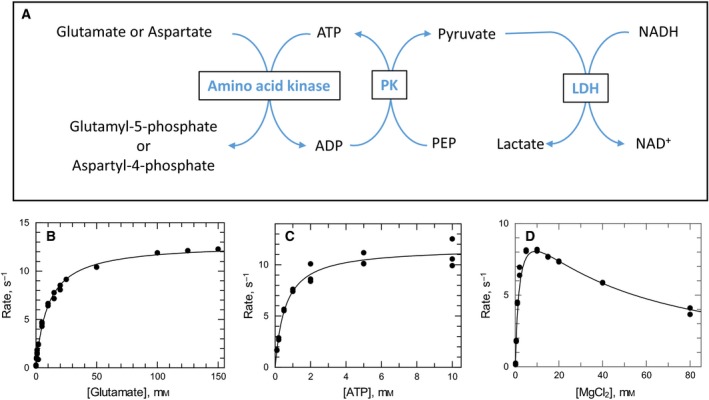
LdG5K activity relationship with varying substrates. In the standard coupled assay monitoring the oxidation of NADH (A), the enzyme concentration was fixed at 0.75 μg with varying concentrations of the following: (B) l‐glutamate range 0–150 mm with fixed 5 mm ATP and 5 mm MgCl2 concentrations; (C) ATP range 0–10 mm with fixed 5 mm MgCl2, and Na Glutamate, 150 mm concentration; (D) MgCl2 range 0–80 mm with fixed 5 mm ATP and 150 mm Na Glutamate concentration.
Table 1.
Comparison of G5K kinetic parameters with other species
| Substrate/Metal ion | Parameter | Units | Leishmania donovani | Escherichia coli a | Thermotoga maritima b | ||
|---|---|---|---|---|---|---|---|
| WT | G5K domain | ||||||
| ATP |
|
mm | 0.6 ± 0.07 | 1.6 | 1.6 | 3.3 | |
| k cat | s−1 | 11.7 ± 0.3 | 85.6 | 33.9 | 52 | ||
| k cat/ | m −1·s−1 | 19 500 | 53 600 | 21 200 | 15 800 | ||
| Glutamate |
|
mm | 10 ± 0.7 | 91 | 227 | 23 | |
| k cat | s−1 | 12.8 ± 0.3 | 109 | 48.8 | 51 | ||
| k cat/ | m −1·s−1 | 1280 | 1200 | 215 | 2200 | ||
| Magnesium |
|
mm | 2.0 ± 0.4 | 4.3 | 0.11 | – | |
|
|
mm | 43 ± 7 | ~ 15 | 4.8 | – | ||
| k cat | s−1 | 11.6 ± 0.8 | 77.2 | 21.7 | – | ||
The activity of LdG5K on varying concentrations of ATP, glutamate and free magnesium was determined by Pyruvate Kinase/Lactate Dehydrogenase coupling system. Free Mg was fitted to high substrate inhibition equation as described in the Experimental procedures. Data calculated from aPérez‐Arellano et al. 81 for the full‐length protein (WT) and for the protein lacking the PUA domain (G5K domain) and bPérez‐Arellano and Cervera 26.
Effect of proline on LdG5K activity and its oligomeric state
In the absence of proline, the substrate glutamate obeys hyperbolic kinetics (Fig. 5). However, the kinetic behaviour with glutamate as variable substrate becomes sigmoidal in the presence of increasing concentrations of proline (Fig. 6A). Relative to the kinetic parameters in the absence of proline, Hill slope increases to a maximum of 3.0, S 0.5 increases linearly, whereas V max decreases with increasing proline concentrations (Fig. 6B). This behaviour is similar to that reported for E. coli G5K 27. The effect of excess proline (10 mm) on the oligomeric state of LdG5K was examined as studies undertaken for the E. coli enzyme resulted in aggregation of tetramers into decamers 23. This was not the case for LdG5K, with no change in the oligomeric state being detected by size exclusion (Fig. 6C). This supports the conclusion that higher oligomerisation of E. coli G5K requires a PUA domain and that allosteric kinetic behaviour is conferred solely by the G5K domain 23.
Figure 6.
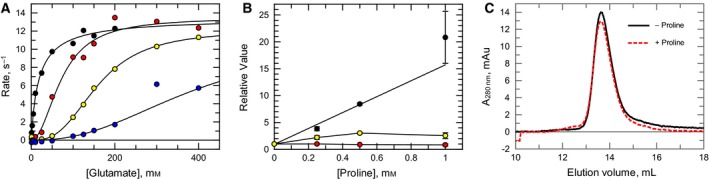
Effect of l‐proline on kinetic behaviour of LdG5K with l‐glutamate and influence on the oligomeric state. (A) The effect of varying l‐glutamate concentration on LdG5K activity in the absence (black) or presence of fixed concentrations (mm) of l‐proline (0.25, red; 0.5 yellow; and 1, blue). (B) Effect of proline on kinetic parameters: S 0.5 (black); k cat (red); Hill slope (yellow). Values (±standard error of the fit to the Hill equation) are expressed relative to parameters in the absence of proline (S 0.5 19.0 ± 2.9 mm; k cat 13.5 ± 0.6 s−1; Hill slope 0.94 ± 0.09). (C) Elution profiles of LdG5K incubated in the presence (black line) and absence (red dotted line) of 10 mm l‐proline.
Inhibition of LdG5K by proline analogues
A panel of l‐proline analogues were surveyed for their inhibitory activity against LdG5K at a fixed concentration of 1 mm. Of these, d‐proline was inactive while l‐proline and three other compounds inhibited activity by more than 30% (Table 2). l‐proline was the most effective with an IC50 value of 0.39 ± 0.08 mm, followed by 3,4‐dehydro‐l‐proline, l‐azetidine‐2‐carboxylic acid and l‐4‐thiazolidine carboxylic acid. Methylation of either the α‐carbon or the imino nitrogen or esterification of the α‐carboxylate abolished the inhibitory activity of l‐proline. Similarly expansion of the pyrrolidine ring to a piperidine or various substitutions on the pyrrolidine ring were not tolerated (Table 2).
Table 2.
Inhibition of recombinant LdG5K by proline analogues compared with Escherichia coli G5K. IC50 values where measured if LdG5K activity was < 70% in the presence of 1 mm proline analogue. The IC50 values were measured as described in the Experimental procedures by measuring the enzyme velocity as a function of inhibitor concentration and the data were fitted using a four‐parameter IC50 equation. NT, not tested
| Proline analogue | LdG5K activity (%) with 1 mm | LdG5K IC50 (mm) | EcG5K IC50 (mm)a | |
|---|---|---|---|---|
| l‐proline |
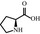
|
4 | 0.39 ± 0.08 | 0.15 |
| d‐proline |
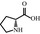
|
103 | – | NT |
| 3,4‐dehydro‐d,l‐proline |
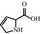
|
9 | 0.49 ± 0.02 | 0.16 |
| l‐azetidine‐2‐carboxylic acid |
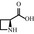
|
25 | 0.73 ± 0.07 | 1.38 |
| l‐4‐thioproline |
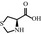
|
66 | 1.8 ± 0.2 | 1.0 |
| l‐pipecolinic acid |
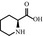
|
100 | – | NT |
| d‐pipecolinic acid |
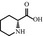
|
100 | – | NT |
| N‐methyl‐l‐proline |

|
100 | – | NT |
| l‐proline methyl ester |
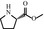
|
100 | – | 3.5 |
| α‐methyl‐l‐proline |
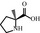
|
82 | – | 4.7 |
| Cis‐3‐hydroxy‐l‐proline |

|
100 | – | NT |
| Cis‐4‐hydroxy‐l‐proline |
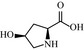
|
87 | – | 1.22 |
| Trans‐4‐hydroxy‐l‐proline |
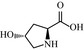
|
89 | – | 22.6 |
| N‐acetyl‐l‐proline |
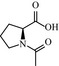
|
93 | – | > 100 |
| l‐trans‐pyrrolidine‐2,4‐dicarboxylic acid |
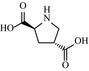
|
84 | – | NT |
| l‐prolinamide |
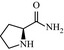
|
96 | – | NT |
| Trans‐1‐acetyl‐4‐hydroxy‐l‐proline |
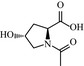
|
100 | – | NT |
aData from Pérez‐Arellano et al. 23.
Genotypic analysis of WT and KO cells
To assess the possible role of G5K in the leishmania life cycle, we sought to generate a proline auxotroph by gene replacement. For a nonessential gene this is normally straightforward requiring two rounds of gene replacement with drug selectable markers to produce a null mutant as shown in the schematic diagram (Fig. 7A). In the case of an essential gene, gene duplication takes place, sometimes involving partial or complete chromosomal duplication; alternatively, the gene of interest is retained at the correct locus and the drug selectable marker is inserted elsewhere in the genome. Such genomic rearrangements are suggestive, but not conclusive evidence that a gene is essential. Addition of a ‘rescue copy’ of the gene of interest either expressed on a plasmid or targeted to a different genomic locus (e.g. ribosomal DNA locus) frequently allows replacement of both chromosomal copies, although this can also fail for technical reasons or the locus being refractory to genetic manipulation 28.
Figure 7.
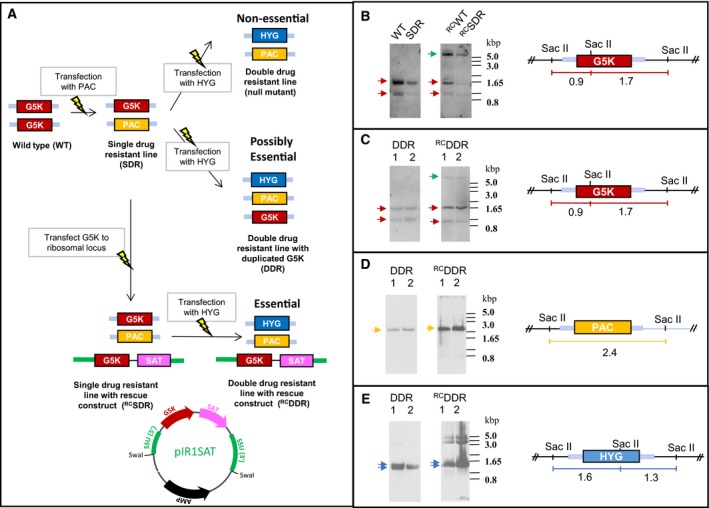
Assessing the essentiality of G5K. (A) Schematic representation of the gene replacement strategy and possible outcomes. The light blue bars represent the 5′‐ and 3′‐ UTRs flanking the G5K locus used for gene replacement by homologous recombination. A map of the rescue construct pIR1SAT_LdG5K is included showing the Swa I restriction sites used to linearise the construct targeted for insertion into the (ectopic) ribosomal DNA locus. (B–E) Genotypic analysis of transgenic clones for WT, SDR, DDR and their corresponding rescue constructs (RCWT,RCSDR and RCDDR). gDNA samples were digested with Sac II and Southern blots prepared as described in Experimental procedures. Blots were sequentially probed and stripped with the following: LdG5K (B, C); puromycin N‐acetyl transferase (D) and hygromycin phosphotransferase (E). Coloured arrows indicate that gene replacement had occurred at the G5K chromosomal locus. Presence of the rescue construct in WT, SDR and DDR clones is highlighted by a green arrow.
It is important to note that, although leishmania are aneuploid, the G5K‐bearing chromosome 26 is diploid in all species 29, 30. Restriction enzyme mapping confirmed that G5K is present as a single copy per haploid genome in the LdBOB strain used here. Single drug‐resistant lines (SDR) were obtained by transfection with the PAC gene replacement construct followed by selection for puromycin resistance in either WT leishmania or in WT cells previously transfected with the overexpression rescue cassette for G5K (RCWT). Resistant lines were obtained bearing the correct replacement of one G5K allele to produce a SDR and RCSDR clonal line (Fig. 7B). Attempts to delete the second allele by transfection with the HYG construct in the SDR clone to produce a double knockout lacking G5K were unsuccessful (Fig. 7C). Although Southern blot analysis confirmed that PAC and HYG had been correctly integrated at the G5K locus (Fig. 7D,E), a copy of the endogenous G5K was also retained at the correct genomic locus (Fig. 7C). Transfection of the clone bearing the ectopic rescue construct (RCDDR) with HYG also failed to generate a chromosomal null mutant (Fig. 7C). The detection of two higher bands (5–4 kbp range) with the HYG marker as well as a doublet band potentially representing the correct size 1.6 and 1.3 kb fragments indicates correct insertion at the G5K locus and elsewhere in the genome (Fig. 7E). There was little difference in growth rates for WT, DDR and the equivalent rescue clonal lines with doubling times of 8.41 ± 0.07; 8.54 ± 0.05; 9.09 ± 0.03 and 9.85 ± 0.01 h respectively. To establish that the overexpressing lines were producing functional protein that would act as a rescue when both allelic copies were deleted, extracts of WT and RCWT lines were assayed for G5K activity. In the RCWT extract a net G5K activity of 34.7 ± 0.9 nmol·min−1·mg−1 was obtained against a background ATPase activity of 12.9 ± 1.8 nmol·min−1·mg−1. No net G5K activity could be detected for the WT extract due to the high background ATPase activity. However, addition of recombinant LdG5K to the WT extracts was readily measurable indicating that the extracts did not contain an inhibitor of the reaction. Thus, the failure to create a chromosomal null mutant cannot be ascribed to failure of G5K expression, but could be due to other technical reasons. Together, the results obtained here are suggestive, but not conclusive that G5K is essential for growth and survival of the parasite.
Discussion
This study provides definitive evidence that the protein encoded by LdBPK_262740.1 is a bone fide G5K (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC2/7/2/11.html) and not an aspartate kinase (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC2/7/2/4.html). The kinetic parameters are broadly equivalent to the few prokaryotic G5Ks that have been studied in any detail (Table 1). Catalytic processes for microbial G5K's and plant P5CS enzymes are regulated through proline feedback inhibition. Proline, the end product of the pathway, induces the parasite enzyme to switch from hyperbolic to sigmoidal behaviour with respect to glutamate, with changes in the S 0.5 for glutamate, Hill coefficient and k cat similar to those reported for E. coli G5K 27. However, unlike the E. coli enzyme 23 the tetrameric structure of LdG5K is not affected by proline possibly due to the lack of a PUA domain. In E. coli G5K the addition of proline shifts the oligomeric state from tetramer (a dimer of dimers in crystallographic studies 31) to a possible dodecamer (trimer of tetramers) at infinite proline concentration 23. Deletion of the PUA domain (absent in LdG5K) abolishes this aggregation, but not the feedback inhibition by proline. Molecular modelling and site‐directed mutagenesis studies on the E. coli and Campylobacter jejuni G5Ks predict a proline‐binding pocket adjacent to the substrate binding site involving Asn34 and Asp 137 23, residues that are conserved in LdG5K (Fig. 3). The structure‐activity relationship of proline analogues determined here agree well with, and extend, the previous study 23 and emphasise the specificity of this binding pocket. Based on the estimated intracellular concentrations of glutamate (14 mm) and proline (3 mm) of promastigotes grown in amino acid replete medium 32, our studies predict that the enzyme will be essentially inactive under these nutrient‐rich conditions (Fig. 6A). Thus, the regulatory parameters determined here appear to be physiologically relevant to the intact cell, supporting the observation that biosynthesis only occurs under conditions of amino acid starvation 14. Unlike the long isoform of mammalian P5CS 33, which is involved in arginine biosynthesis, citrulline and ornithine do not inhibit LdG5K. This is consistent with the fact that leishmania are auxotrophic for arginine when cultured in the presence of glucose 32 and lack genes for two key pathway enzymes linking proline biosynthesis with ornithine / arginine metabolism (ornithine aminotransferase and ornithine deiminase).
Metabolism of proline in leishmania has similarities and differences with its trypanosomatid cousins, the African trypanosome, Trypanosoma brucei and the South American trypanosome, Trypanosoma cruzi. While all three species are capable of using proline as an energy source, converting proline into glutamate and subsequently into tricarboxylic acid intermediates 6, 18, 34, 35, metabolic labelling studies show that only leishmania is capable of synthesising proline de novo from either glutamate or glucose 14, 18, 34, 36. There is an absolute requirement for proline to support growth of T. brucei procyclic forms 18, whereas this is not the case for several Leishmania spp. 32. Given the lack of bioinformatic or biochemical evidence for alternative routes for proline biosynthesis from ornithine (via ornithine δ‐aminotransferase or ornithine cyclodeaminase) 37 this suggests that the G5K pathway is necessary and sufficient for growth of promastigotes in media lacking proline.
In addition to proline's role as a protein building block, as a major energy source and as a source of nitrogen, this imino acid has been implicated in response to various forms of cellular stress. Intracellular accumulation of proline is a common phenomenon in response to environmental stress in bacteria, protozoa, algae, plant and marine invertebrates (for a review see 36, 38). In other organisms proline has been shown to function as an osmoprotectant, as a small molecule chaperone preventing protein aggregation, as a metal chelator and as a scavenger of reactive oxygen species (ROS), specifically the hydroxyl radical and singlet oxygen 38. Proline is also proposed to help maintain the GSH thiol redox balance by scavenging ROS and by stabilising key antioxidant enzymes.
These possible roles for proline have not been extensively studied in the trypanosomatids. Proline, along with glutamate and alanine, have been implicated in responses to osmotic stress 39, 40 and this may be of biological relevance to these parasites’ survival in the insect gut. In T. cruzi proline is reported to modulate resistance to ROS and resistance to trypanocidal drugs through a novel d,l‐proline transporter 41. T. cruzi also possesses an intracellular proline racemase and a secreted proline racemase that acts as B‐cell mitogen modulating the immune response 42. However, the leishmania genome does not contain a proline racemase.
One specific role for proline in leishmania is in response to purine starvation 43. Leishmania are purine auxotrophs and lack the ability to synthesise purines de novo. Thus, they are entirely dependent on an exogenous supply from the extracellular medium. Purine starvation not only markedly upregulates purine transporters and purine salvage enzymes but also upregulates proline biosynthetic pathway and downregulates proline degradation, resulting in a 6‐ to 10‐fold increase in intracellular proline concentration 43. The role of proline in this stress response is not understood, but could act to augment the increased levels of proteins involved in protein folding/protein stability, as well as key trypanothione‐dependent antioxidant enzymes observed 43.
One key question is why do Leishmania spp. have this de novo biosynthetic pathway, whereas T. brucei and T. cruzi do not? One possibility concerns the different parasite locations in their mammalian hosts. T. brucei resides extracellularly in the blood, lymph and interstitial fluid, whereas T. cruzi resides intracellularly in the cytosol of many different cell types in many different tissues. Thus, both of these organisms should have ready access to proline and therefore have no requirement for de novo biosynthesis. In contrast, leishmania parasites reside within an acidic phagolysosomal compartment in macrophages – the parasitophorous vacuole (PV). Information on the availability of nutrients in the PV is lacking and can only be inferred from the nutritional requirements of the parasites. Thus, purines, haem, vitamins, iron and essential amino acids must be available in the PV in sufficient amounts to sustain growth 44. Studies with Salmonella auxotrophs suggest that the macrophage acidic compartment in which they reside is nutrient poor for purine, pyrimidine, aromatic amino acids, histidine and proline 45, 46, 47. Notably, Salmonella enterica lacking G5K (ProB) are defective for survival in vitro in macrophages and in vivo in mice 46. A related study in Mycobacterium tuberculosis showed that a proline auxotroph (proC, Δ1‐pyrolline 5‐carboxylate reductase) displays attenuated virulence both in bone marrow‐derived macrophages and in mice 48. While both of these studies indicate that proline is limiting in these specialised macrophage phagolysosomal compartments, it may not apply to the leishmania PV, which is known to be in contact with the external medium via endocytosis 49.
To assess the possible role of G5K in the leishmania life cycle, we sought to generate a proline auxotroph by gene replacement. Despite the presence of proline in the medium that should serve as a nutritional rescue, it was not possible to create a G5K null mutant. Instead, both drug selection markers were correctly inserted at the G5K locus, with retention of an additional copy of G5K. This result is suggestive, but not conclusive, that G5K is essential. Additional evidence of essentiality can be obtained using a genetic rescue strategy that should allow direct gene replacement without additional chromosomal alterations. In our case, multiple attempts to create a chromosomal G5K null with an episomal rescue construct in the ribosomal locus were unsuccessful even in the presence of proline (0.35 mm) in the growth medium. Further work is required to assess whether G5K has an additional structural or enzymatic function that remains to be elucidated.
A second possible unique function relates to a class of drugs used to treat leishmaniasis, namely, the pentavalent antimonials, sodium stibogluconate (pentostam) and meglumine antimonate (glucantime). These metalloid‐sugar complexes are thought to be activated by reduction of SbV to the trivalent (SbIII) form in the host macrophage, the intracellular amastigote or possibly both 3. The mode of action of SbIII is not fully understood, but is likely to include inhibition of key dithiol antioxidant proteins such as trypanothione reductase and tryparedoxin peroxidase with perturbation of thiol‐redox homeostasis and thiol‐buffering capacity 50, 51, 52, 53. Resistance to antimonial compounds in Bihar State, India is now so widespread that antimonials are no longer recommended as front‐line treatment. One theory why this has occurred in India, but not in other geographic regions, relates to the metalloid, arsenic. The hypothesis is that the human population living in parts of Bihar are exposed to high levels of arsenic in drinking water and, in infected patients, arsenic selects for parasite resistance to trivalent arsenic and concomitant resistance to antimonials 54. Evidence to support this theory has been obtained in vitro 55, 56 and in animal models 57, although a retrospective clinical epidemiological study proved to be insufficiently powered to provide a definitive answer to this question 58. It is noteworthy that arsenic‐contaminated soil has a harmful effect on plant growth through perturbation of numerous metabolic pathways and other biological functions 59 and antimony‐contaminated soil induces similar adverse effects on growth 60. Strikingly, plants exposed to either inorganic arsenic or antimony respond by increasing their intracellular proline content 61, 62, 63. In leishmania, no changes in expression of G5K, glutamate‐5‐semi‐aldehyde dehydrogenase or pyrroline‐5‐carboxylate reductase have been noted in whole‐genome sequencing, proteomics or gene expression studies comparing drug‐sensitive and drug‐resistant parasites 64, 65, 66, 67, 68. However, a metabolomic study noted elevated glutamate and proline in antimony‐resistant cells compared with susceptible Leishmania infantum promastigotes 69. The mechanism by which glutamate or proline convey resistance is not understood: both amino acids are precursors for the antioxidant metabolites glutathione and trypanothione and proline could act as a scavenger of ROS induced by SbIII 70. Whether these changes in amino acid content occur in antimony‐resistant clinical isolates has not been studied.
In conclusion, this study confirms the key putative enzyme in proline biosynthesis is indeed a classical G5K. G5K lacks aspartokinase activity leaving the question of how parasites synthesise threonine unanswered. Further work is required to establish the functional roles of proline in this parasite. In particular, is G5K a potential drug target in the intracellular amastigote stage of the life cycle?
Experimental procedures
Chemicals and reagents
Chemicals and reagents used in this study were all of the highest purity which are commercially available. All substrates, cofactors and enzymes were products from Sigma Aldrich (Gillingham, UK).
Leishmania donovani promastigote cell culture maintenance
Leishmania donovani LdBOB promastigotes (derived from MHOM/SD/62/1S‐CL2D) were routinely passaged in modified M199 medium with 10% fetal calf serum as previously described 71.
Cloning and generation of transgenic cell lines
All constructs made were prepared and sequenced for electroporation using QIAprep Miniprep Plasmid Kit (Qiagen, Venlo, the Netherlands). Primers used in this study to generate the overexpression and knockout constructs (Table 3) are based on the GeneDB sequences for L. infantum G5K (LinJ.26.2740).
Table 3.
Primers used for knockdown, knockout and rescue constructs. Upper case letters refer to gene sequences and enzyme restriction sites are underlined
| Primer name | Sequence |
|---|---|
| ORF‐BamHI forward | 5′‐ggatccaATGGCGGACATCTTGAAG‐3′ |
| ORF‐BamHI reverse | 5′‐ggatccTCAAGATGAAACTCTCGA‐3′ |
| 5′UTR‐NotI forward | 5′‐ataagaatgcggccgcCGCTAATGATTACACTAC‐3′ |
| 5′UTR‐HindIII/PmeI reverse | 5′‐gtttaaacttacggaccgtcaagcttGCTTGATTCCGCGTGGAC‐3′ |
| 3′UTR‐PmeI/BamHI forward | 5′‐gacggtccgtaagtttaaacggatccAGCGCTTAGGGATGTCAC‐3′ |
| 3′UTR‐NotI reverse | 5′‐ataagtaagcggccgcGTTCGATACCGTTTTGAGA‐3′ |
| G5K‐BamHI forward | 5′‐gcgcggatccATGGCGGACATCTTGAAGT‐3′ |
| G5K‐EcoRI forward | 5′‐gcgcgaattcTCAAGATGAAACTCTCGAAT‐3′ |
For the gene replacement cassettes, the 5′ and 3′ UTRs directly adjacent to the ORF were amplified using 5′UTR‐NotI forward, 5′UTR‐HindIII/PmeI reverse and 3′UTR‐Pme I/BamHI forward, 3′UTR‐NotI reverse respectively. Using the following conditions: 95 °C for 1 min; 55 °C for 2.5 min; 72 °C for 3 min for 30 cycles, PCR products were used together in a further overlapping extension PCR to yield a product containing the 5′‐UTR linked to the 3′‐UTR via a short HindIII – BamHI linker region with a NotI site at each end. (Table 3). This product was subsequently ligated into the NotI site of pGEM‐5Zf(+) vector (Promega, Southampton, UK). The selectable drug resistance genes puromycin N‐acetyl transferase (PAC) and hygromycin phosphotransferase (HYG) were introduced into this vector via the HindIII and BamHI within the linker region cloning sites and sequenced.
To generate a recovery construct expressing LdG5K, pIR1SAT_LdG5K, the G5K ORF was amplified from L. donovani genomic DNA using oeG5K forward and reverse primers (Table 3) and subsequently cloned into the integrating stable expression vector pIR1SAT 72. For recombinant expression in E. coli, LdG5K was amplified from the pIR1SAT_LdG5K construct, with G5K forward and reverse primers (Table 3), subsequently cloned into a modified pGEX‐6P‐1 vector (Amersham Biosciences, Buckinghamshire, UK) containing in‐frame N‐terminal His‐GST tag, using the BamHI and the EcoRI restriction sites. All the constructs generated were verified by DNA sequencing (http://www.dnaseq.co.uk).
Generation of L. donovani promastigote G5K knockout and overexpresser cells
DNA constructs were prepared for electroporation into LdBOB promastigotes and cloned with all procedures performed as previously described 73. Briefly, parasites were electroporated using Human T‐cell Nucleofactor with programme V‐033. Post‐transfection, cells were allowed to rest for 16–24 h prior to appropriate selection for PAC (20 μg·mL−1 puromycin) or HYG (50 μg·mL−1 hygromycin). Cloned cell lines were generated by limiting dilution and maintained in the presence of selective drugs. To generate WT and SDR lines overexpressing LdG5K, the pIR1SAT_LdG5K recovery construct was linearised by digestion with SwaI and electroporated into cells with selection for expression of streptothricin acetyltransferase (SAT) with 100 μg nourseothricin·mL−1.
Genotypic analysis
DNA was prepared for Southern blot analysis from WT and KO cells. WT DNA (5 μg) was digested various restriction and resolved on a 0.8% agarose gel. The DNA was subsequently transferred onto nitrocellulose and UV cross‐linked. The blots were hybridised with probes specific for G5K ORF and 5′ UTR. The probes were labelled with fluorescein–dUTP by DIG labelling kit (Roche, Welwyn Garden City, UK) and the blots processed as previously described 74.
Recombinant expression and purification of LdG5K
Recombinant pHis‐GST:LdG5K was expressed in E. coli strain C41(DE3) pLysS. Transformed cells were cultured in auto‐induction medium (LB medium supplemented with 0.5 g·L−1 glucose and 2 g·L−1 α‐lactose) plus 100 μg·mL−1 ampicillin at 37 °C with shaking at 200 r.p.m. for 2 h and the temperature was reduced to 18 °C overnight. Uninduced cells were grown in medium lacking glucose and α‐lactose. Soluble protein was purified from clarified lysate on glutathione (GST) resin. Briefly, cells were harvested by resuspending in lysis buffer [PBS/0.25 m NaCl/5% (v/v) glycerol] supplemented with EDTA‐free complete protease inhibitor cocktail (Roche) and lysed using a continuous cell disruptor (Constant Systems, Daventry, UK) at 30 000 psi. Clarified lysates (centrifuged at 45 000 g for 45 min, 4 °C) were applied to GST resin (GE Healthcare, Little Chalfont, UK), pre‐equilibrated in lysis buffer. The lysate was batch bound at 4 °C for 2 h with subsequent washing prior to on‐column cleavage by PreScission protease for 24 h according to manufacturer's instructions. Protein fractions were analysed by SDS/PAGE using a NuPAGE Novex 4–12% Bis‐Tris gel (Life Technologies, Paisley, UK) and visualised for purity by Coomassie Brilliant Blue staining. Prepared enzyme was buffer exchanged and concentrated using 3 MWCO filtration unit (Vivaspin 20; Millipore, Watford, UK) and stored in 50 mm HEPES pH 7.5 containing 20% (v/v) glycerol and 0.0005% (w/v) Na azide as snap‐frozen aliquots at −80 °C. The sequence and identity of the purified protein was verified by tryptic mass fingerprinting with 79% sequence coverage (Proteomic and Mass Spectrometry facility, University of Dundee). Proteins were analysed by SDS/PAGE using 4–15% Bis‐Tris NuPAGE polyacrylamide gels (Invitrogen, Life Sciences, Paisley, UK) and 1× MES running buffer according to manufacturer's instructions. Protein concentration was measured spectrophotometrically at 595 nm using the Bradford reagent (Bio‐Rad, Watford, UK) and BSA as a standard 75.
Analytical gel filtration
The native confirmation of LdG5K was confirmed by size exclusion chromatography using a Superdex 200 H/R 10/30 column fitted onto an AKTA FPLC system (both from GE Healthcare) with a buffer comprising 50 mm Tris‐HCl, pH 7.2 and 0.15 m NaCl running at 0.5 mL·min−1 at room temperature. Gel filtration protein standards (Bio‐Rad) used were as follows: vitamin B12 (1350 Da); horse myoglobin (17 000 Da); chicken ovalbumin (44 000 Da); bovine γ‐globulin (158 000 Da) and bovine thyroglobulin (670 000 Da). The void volume was 8.3 mL. The equation for estimating the molecular mass was derived from plots of V e/V o against log MW of the standards.
Kinetic characterisation of recombinant LdG5K and clarified L. donovani lysates
A continuous spectrophotometric enzyme assay was developed to follow the conversion of ATP into ADP using a coupled enzyme system utilising pyruvate kinase (PK) and lactate dehydrogenase (LDH; Fig. 4). Both coupling enzymes were present in excess and G5K activity was measured by the consumption of NADH at 340 nm. Standard assay conditions contained the following in a 1 mL reaction mix: 0.1 m imidazole, pH 7.4; 1 mm DTT; 5 mm ATP; 5 mm MgCl2; 0.1 mg·mL−1 BSA; 50 U PK/LDH; 1 mm phosphoenolpyruvate; 250 mm NADH; 0.75 μg LdG5K and the reaction was initiated with 100 mm Na glutamate. The assay components were pre‐equilibrated at 25 °C for 5 min prior to initiation with the substrate. When ATP was varied, 150 mm Na glutamate and 5 mm MgCl2 were kept in excess of ATP. When glutamate was varied 5 mm ATP and 5 mm MgCl2 were kept constant. Activity was calculated from the initial linear rate with the extinction coefficient of NADH, 6220 m −1·cm−1 76. All data were analysed by nonlinear regression using GraFit and fitted to the Michaelis–Menten equation, except for determining the free Mg concentration requirement which was fitted to a high substrate inhibition equation:
In the presence of proline the enzyme displayed sigmoidal kinetics with respect to glutamate and data were fitted by nonlinear regression to the Hill equation describing allosteric behaviour:
where n = Hill coefficient.
Log‐phase cultures of L. donovani WT and WT‐overexpressing G5K (1 × 107 cells·mL−1) were harvested by centrifugation (800 g, 10 min, 4 °C) and washed twice with ice‐cold PBS containing cOmplete™, EDTA‐free protease inhibitor cocktail (Roche, Basel, Switzerland). Clarified lysates were prepared as previously described 74. Briefly, cells were osmotically lysed with ice‐cold distilled water (containing cOmplete™, EDTA‐free protease inhibitors), frozen and thawed three times in liquid nitrogen (a biosafety procedure) and finally resuspended in an equal volume of 2× assay buffer (0.2 m imidazole, pH7.4; 2 mm DTT). The lysates were buffer exchanged into 1× assay buffer to remove cofactors and substrates prior to determining protein content.
Inhibitors
Initially single‐point assays of a set of proline analogues were tested at 1 mm concentration using the coupled assay described above. Any analogue which resulted in > 70% inhibition in LdG5K activity was retested under varying concentrations to determine their IC50 value. All measurements were done in triplicate.
Phylogenetic analysis
The phylogenetic tree was constructed from a highly conserved region of deduced G5K and P5CS amino acid sequences. The evolutionary history was inferred using the Neighbour‐Joining method 77. The optimal tree with the sum of branch length = 7.43079248 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method 78 and are in the units of the number of amino acid substitutions per site. The analysis involved 24 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 203 positions in the final dataset. Evolutionary analyses were conducted in mega7 79.
Conflicts of interest
The authors declare no real or perceived conflicts of interest.
Author contributions
AHF conceived the project and NS, HBO and AHF designed the experiments. NS performed the experiments. All authors analysed the data and contributed to the manuscript.
Acknowledgement
This work was supported by a grant from the Welcome Trust (079838).
References
- 1. Field MC, Horn D, Fairlamb AH, Ferguson MA, Gray DW, Read KD, De Rycker M, Torrie LS, Wyatt PG, Wyllie S et al (2017) Anti‐trypanosomatid drug discovery: an ongoing challenge and a continuing need. Nat Rev Microbiol 15, 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fairlamb AH, Gow NAR, Matthews KR & Waters AP (2016) Drug resistance in eukaryotic microorganisms. Nat Microbiol 1, e16092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Croft SL, Sundar S & Fairlamb AH (2006) Drug resistance in leishmaniasis. Clin Microbiol Rev 19, 111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stuart K, Brun R, Croft S, Fairlamb A, Gurtler RE, McKerrow J, Reed S & Tarleton R (2008) Kinetoplastids: related protozoan pathogens, different diseases. J Clin Invest 118, 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosenzweig D, Smith D, Opperdoes F, Stern S, Olafson RW & Zilberstein D (2008) Retooling Leishmania metabolism: from sand fly gut to human macrophage. FASEB J 22, 590–602. [DOI] [PubMed] [Google Scholar]
- 6. Krassner SM & Flory B (1972) Proline metabolism in Leishmania donovani promastigotes. J Protozool 19, 682–685. [DOI] [PubMed] [Google Scholar]
- 7. Ter Kuile BH & Opperdoes FR (1992) A chemostat study on proline uptake and metabolism of Leishmania donovani . J Protozool 39, 555–558. [DOI] [PubMed] [Google Scholar]
- 8. Bursell E (1978) Quantitative aspects of proline utilization during flight in tsetse flies. Physiol Entomol 3, 265–272. [Google Scholar]
- 9. Teulier L, Weber JM, Crevier J & Darveau CA (2016) Proline as a fuel for insect flight: enhancing carbohydrate oxidation in hymenopterans. Proc Biol Sci 283, e20160333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uchida K, Ohmori D, Yamakura F & Suzuki K (1990) Changes in free amino acid concentration in the hemolymph of the female Culex pipiens pallens (Diptera: Culicidae), after a blood meal. J Med Entomol 27, 302–308. [DOI] [PubMed] [Google Scholar]
- 11. Tsigankov P, Gherardini PF, Helmer‐Citterich M & Zilberstein D (2012) What has proteomics taught us about Leishmania development? Parasitology 139, 1146–1157. [DOI] [PubMed] [Google Scholar]
- 12. Naderer T, Ellis MA, Sernee MF, De Souza DP, Curtis J, Handman E & McConville MJ (2006) Virulence of Leishmania major in macrophages and mice requires the gluconeogenic enzyme fructose‐1,6‐bisphosphatase. Proc Natl Acad Sci USA 103, 5502–5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koslowsky DJ, Goringer HU, Morales TH & Stuart K (1992) In vitro guide RNA/m‐RNA chimera formation in Trypanosoma brucei RNA editing. Nature 356, 807–809. [DOI] [PubMed] [Google Scholar]
- 14. Saunders EC, Ng WW, Chambers JM, Ng M, Naderer T, Kromer JO, Likic VA & McConville MJ (2011) Isotopomer profiling of Leishmania mexicana promastigotes reveals important roles for succinate fermentation and aspartate uptake in tricarboxylic acid cycle (TCA) anaplerosis, glutamate synthesis, and growth. J Biol Chem 286, 27706–27717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Millerioux Y, Ebikeme C, Biran M, Morand P, Bouyssou G, Vincent IM, Mazet M, Riviere L, Franconi JM, Burchmore RJS et al (2013) The threonine degradation pathway of the Trypanosoma brucei procyclic form: the main carbon source for lipid biosynthesis is under metabolic control. Mol Microbiol 90, 114–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Weelden SWH, Van Hellemond JJ, Opperdoes FR & Tielens AGM (2005) New functions for parts of the Krebs cycle in procyclic Trypanosoma brucei, a cycle not operating as a cycle. J Biol Chem 280, 12451–12460. [DOI] [PubMed] [Google Scholar]
- 17. Cross GAM, Klein RA & Linstead DJ (1975) Utilization of amino acids by Trypanosoma brucei in culture: L‐threonine as a precursor for acetate. Parasitology 71, 311–326. [DOI] [PubMed] [Google Scholar]
- 18. Mantilla BS, Marchese L, Casas‐Sanchez A, Dyer NA, Ejeh N, Biran M, Bringaud F, Lehane MJ, Acosta‐Serrano A & Silber AM (2017) Proline metabolism is essential for Trypanosoma brucei brucei survival in the tsetse vector. PLoS Pathog 13, e1006158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ter Kuile BH (1997) Adaptation of metabolic enzyme activities of Trypanosoma brucei promastigotes to growth rate and carbon regimen. J Bacteriol 179, 4699–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Evans DA & Brown RC (1972) Utilization of glucose and proline by culture forms of Trypanosoma brucei . J Protozool 19, 686–690. [DOI] [PubMed] [Google Scholar]
- 21. Opperdoes FR & Coombs GH (2007) Metabolism of Leishmania: proven and predicted. Trends Parasitol 23, 149–158. [DOI] [PubMed] [Google Scholar]
- 22. Ong HB, Lee WS, Patterson S, Wyllie S & Fairlamb AH (2015) Homoserine and quorum‐sensing acyl homoserine lactones as alternative sources of threonine: a potential role for homoserine kinase in insect‐stage Trypanosoma brucei . Mol Microbiol 95, 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pérez‐Arellano I, Carmona‐Alvarez F, Gallego J & Cervera J (2010) Molecular mechanisms modulating glutamate kinase activity. Identification of the proline feedback inhibitor binding site. J Mol Biol 404, 890–901. [DOI] [PubMed] [Google Scholar]
- 24. Krishna RV & Leisinger T (1979) Biosynthesis of proline in Pseudomonas aeruginosa. Partial purification and characterization of γ‐glutamyl kinase. Biochem J 181, 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Torrie LS, Wyllie S, Spinks D, Oza SL, Thompson S, Harrison JR, Gilbert IH, Wyatt PG, Fairlamb AH & Frearson JA (2009) Chemical validation of trypanothione synthetase: a potential drug target for human trypanosomiasis. J Biol Chem 284, 36137–36145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pérez‐Arellano I & Cervera J (2010) Glutamate kinase from Thermotoga maritima: characterization of a thermophilic enzyme for proline biosynthesis. Extremophiles 14, 409–415. [DOI] [PubMed] [Google Scholar]
- 27. Pérez‐Arellano I, Rubio V & Cervera J (2006) Mapping active site residues in glutamate‐5‐kinase. The substrate glutamate and the feed‐back inhibitor proline bind at overlapping sites. FEBS Lett 580, 6247–6253. [DOI] [PubMed] [Google Scholar]
- 28. Jones NG, Catta‐Preta CMC, Lima APCA & Mottram JC (2018) Genetically validated drug targets in Leishmania: current knowledge and future prospects. ACS Infect Dis 4, 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lachaud L, Bourgeois N, Kuk N, Morelle C, Crobu L, Merlin G, Bastien P, Pages M & Sterkers Y (2014) Constitutive mosaic aneuploidy is a unique genetic feature widespread in the Leishmania genus. Microbes Infect 16, 61–66. [DOI] [PubMed] [Google Scholar]
- 30. Rogers MB, Hilley JD, Dickens NJ, Wilkes J, Bates PA, Depledge DP, Harris D, Her Y, Herzyk P, Imamura H et al (2011) Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania . Genome Res 21, 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marco‐Marin C, Gil‐Ortiz F, Pérez‐Arellano I, Cervera J, Fita I & Rubio V (2007) A novel two‐domain architecture within the amino acid kinase enzyme family revealed by the crystal structure of Escherichia coli glutamate 5‐kinase. J Mol Biol 367, 1431–1446. [DOI] [PubMed] [Google Scholar]
- 32. Nayak A, Akpunarlieva S, Barrett M & Burchmore R (2018) A defined medium for Leishmania culture allows definition of essential amino acids. Exp Parasitol 185, 39–52. [DOI] [PubMed] [Google Scholar]
- 33. Hu CA, Lin WW, Obie C & Valle D (1999) Molecular enzymology of mammalian Δ1‐pyrroline‐5‐carboxylate synthase. Alternative splice donor utilization generates isoforms with different sensitivity to ornithine inhibition. J Biol Chem 274, 6754–6762. [DOI] [PubMed] [Google Scholar]
- 34. Sylvester D & Krassner SM (1976) Proline metabolism in Trypanosoma cruzi epimastigotes. Comp Biochem Physiol B 55, 443–447. [DOI] [PubMed] [Google Scholar]
- 35. Ford WC & Bowman IB (1973) Metabolism of proline by the culture midgut form of Trypanosoma rhodesiense . Trans R Soc Trop Med Hyg 67, 257. [DOI] [PubMed] [Google Scholar]
- 36. Bringaud F, Barrett MP & Zilberstein D (2012) Multiple roles of proline transport and metabolism in trypanosomatids. Front Biosci (Landmark Ed) 17, 349–374. [DOI] [PubMed] [Google Scholar]
- 37. Fichman Y, Gerdes SY, Kovacs H, Szabados L, Zilberstein A & Csonka LN (2015) Evolution of proline biosynthesis: enzymology, bioinformatics, genetics, and transcriptional regulation. Biol Rev Camb Philos Soc 90, 1065–1099. [DOI] [PubMed] [Google Scholar]
- 38. Liang X, Zhang L, Natarajan SK & Becker DF (2013) Proline mechanisms of stress survival. Antioxid Redox Signal 19, 998–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vieira LL & Cabantchik ZI (1995) Amino acid uptake and intracellular accumulation in Leishmania major promastigotes are largely determined by an H+‐pump generated membrane potential. Mol Biochem Parasitol 75, 15–23. [DOI] [PubMed] [Google Scholar]
- 40. Inbar E, Schlisselberg D, Suter GM, Rentsch D & Zilberstein D (2013) A versatile proline/alanine transporter in the unicellular pathogen Leishmania donovani regulates amino acid homoeostasis and osmotic stress responses. Biochem J 449, 555–566. [DOI] [PubMed] [Google Scholar]
- 41. Saye M, Miranda MR, di Girolamo F, de los Milagros CM & Pereira CA (2014) Proline modulates the Trypanosoma cruzi resistance to reactive oxygen species and drugs through a novel D, L‐proline transporter. J Parasitol Res 9, e92028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reina‐San‐Martin B, Degrave W, Rougeot C, Cosson A, Chamond N, Cordeiro‐Da‐Silva A, Arala‐Chaves M, Coutinho A & Minoprio P (2000) A B‐cell mitogen from a pathogenic trypanosome is a eukaryotic proline racemase. Nat Med 6, 890–897. [DOI] [PubMed] [Google Scholar]
- 43. Martin JL, Yates PA, Soysa R, Alfaro JF, Yang F, Burnum‐Johnson KE, Petyuk VA, Weitz KK, Camp DG, Smith RD et al (2014) Metabolic reprogramming during purine stress in the protozoan pathogen Leishmania donovani . PLoS Pathog 10, e1003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kun H, Moore A, Mascola L, Steurer F, Lawrence G, Kubak B, Radhakrishna S, Leiby D, Herron R, Mone T et al (2009) Transmission of Trypanosoma cruzi by heart transplantation. Clin Infect Dis 48, 1534–1540. [DOI] [PubMed] [Google Scholar]
- 45. Fields PI, Swanson RV, Haidaris CG & Heffron F (1986) Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA 83, 5189–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee EJ, Choi J & Groisman EA (2014) Control of a Salmonella virulence operon by proline‐charged tRNA(Pro). Proc Natl Acad Sci USA 111, 3140–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Steeb B, Claudi B, Burton NA, Tienz P, Schmidt A, Farhan H, Maze A & Bumann D (2013) Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS Pathog 9, e1003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smith DA, Parish T, Stoker NG & Bancroft GJ (2001) Characterization of auxotrophic mutants of Mycobacterium tuberculosis and their potential as vaccine candidates. Infect Immun 69, 1142–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Burchmore RJS & Barrett MP (2001) Life in vacuoles – nutrient acquisition by Leishmania amastigotes. Int J Parasitol 31, 1311–1320. [DOI] [PubMed] [Google Scholar]
- 50. Wyllie S, Mandal G, Singh N, Sundar S, Fairlamb AH & Chatterjee M (2010) Elevated levels of tryparedoxin peroxidase in antimony unresponsive Leishmania donovani field isolates. Mol Biochem Parasitol 173, 162–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wyllie S, Cunningham ML & Fairlamb AH (2004) Dual action of antimonial drugs on thiol redox metabolism in the human pathogen Leishmania donovani . J Biol Chem 279, 39925–39932. [DOI] [PubMed] [Google Scholar]
- 52. Cunningham ML & Fairlamb AH (1995) Trypanothione reductase from Leishmania donovani – purification, characterisation and inhibition by trivalent antimonials. Eur J Biochem 230, 460–468. [DOI] [PubMed] [Google Scholar]
- 53. Baiocco P, Colotti G, Franceschini S & Ilari A (2009) Molecular basis of antimony treatment in leishmaniasis. J Med Chem 52, 2603–2612. [DOI] [PubMed] [Google Scholar]
- 54. Perry MR, Wyllie S, Prajapati VK, Feldmann J, Sundar S, Boelaert M & Fairlamb AH (2011) Visceral leishmaniasis and arsenic: an ancient poison contributing to antimonial treatment failure in the Indian subcontinent? PLoS Negl Trop Dis 5, e1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Haimeur A & Ouellette M (1998) Gene amplification in Leishmania tarentolae selected for resistance to sodium stibogluconate. Antimicrob Agents Chemother 42, 1689–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dey S, Papadopoulou B, Haimer A, Roy G, Grondin K, Dou D, Rosen BP & Ouellette M (1994) High level arsenite resistance in Leishmania tarentolae is mediated by an active extrusion system. Mol Biochem Parasitol 67, 49–57. [DOI] [PubMed] [Google Scholar]
- 57. Perry MR, Wyllie S, Raab A, Feldmann J & Fairlamb AH (2013) Chronic exposure to arsenic in drinking water can lead to resistance to antimonial drugs in a mouse model of visceral leishmaniasis. Proc Natl Acad Sci USA 110, 19932–19937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Perry MR, Prajapati VK, Menten J, Raab A, Feldmann J, Chakraborti D, Sundar S, Fairlamb AH, Boelaert M & Picado A (2015) Arsenic exposure and outcomes of antimonial treatment in visceral leishmaniasis patients in Bihar, India: a retrospective cohort study. PLoS Negl Trop Dis 9, e0003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Abbas G, Murtaza B, Bibi I, Shahid M, Niazi NK, Khan MI, Amjad M & Hussain M (2018) Arsenic uptake, toxicity, detoxification, and speciation in plants: physiological, biochemical, and molecular aspects. Int J Environ Res Public Health 15, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vaculikova M, Vaculik M, Simkova L, Fialova I, Kochanova Z, Sedlakova B & Luxova M (2014) Influence of silicon on maize roots exposed to antimony – growth and antioxidative response. Plant Physiol Biochem 83, 279–284. [DOI] [PubMed] [Google Scholar]
- 61. Begum MC, Islam MS, Islam M, Amin R, Parvez MS & Kabir AH (2016) Biochemical and molecular responses underlying differential arsenic tolerance in rice (Oryza sativa L.). Plant Physiol Biochem 104, 266–277. [DOI] [PubMed] [Google Scholar]
- 62. Tripathi P, Singh RP, Sharma YK & Tripathi RD (2015) Arsenite stress variably stimulates pro‐oxidant enzymes, anatomical deformities, photosynthetic pigment reduction, and antioxidants in arsenic‐tolerant and sensitive rice seedlings. Environ Toxicol Chem 34, 1562–1571. [DOI] [PubMed] [Google Scholar]
- 63. Pavlik M, Pavlikova D, Staszkova L, Neuberg M, Kaliszova R, Szakova J & Tlustos P (2010) The effect of arsenic contamination on amino acids metabolism in Spinacia oleracea L. Ecotoxicol Environ Saf 73, 1309–1313. [DOI] [PubMed] [Google Scholar]
- 64. Leprohon P, Legare D & Ouellette M (2009) Intracellular localization of the ABCC proteins of Leishmania and their role in resistance to antimonials. Antimicrob Agents Chemother 53, 2646–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brotherton MC, Bourassa S, Leprohon P, Legare D, Poirier GG, Droit A & Ouellette M (2013) Proteomic and genomic analyses of antimony resistant Leishmania infantum mutant. J Parasitol Res 8, e81899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Decuypere S, Vanaerschot M, Brunker K, Imamura H, Muller S, Khanal B, Rijal S, Dujardin JC & Coombs GH (2012) Molecular mechanisms of drug resistance in natural Leishmania populations vary with genetic background. PLoS Negl Trop Dis 6, e1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Biyani N, Singh AK, Mandal S, Chawla B & Madhubala R (2011) Differential expression of proteins in antimony‐susceptible and ‐resistant isolates of Leishmania donovani . Mol Biochem Parasitol 179, 91–99. [DOI] [PubMed] [Google Scholar]
- 68. Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, Cotton JA, Hilley JD, De Doncker S, Maes I, Mottram JC et al (2011) Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res 21, 2143–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rojo D, Canuto GAB, Castilho‐Martins EA, Tavares MFM, Barbas C, Lopez‐Gonzalvez A & Rivas L (2015) A multiplatform metabolomic approach to the basis of antimonial action and resistance in Leishmania infantum . J Parasitol Res 10, e0130675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mehta A & Shaha C (2006) Mechanism of metalloid‐induced death in Leishmania spp.: role of iron, reactive oxygen species, Ca2+, and glutathione. Free Radic Biol Med 40, 1857–1868. [DOI] [PubMed] [Google Scholar]
- 71. Goyard S, Segawa H, Gordon J, Showalter M, Duncan R, Turco SJ & Beverley SM (2003) An in vitro system for developmental and genetic studies of Leishmania donovani phosphoglycans. Mol Biochem Parasitol 130, 31–42. [DOI] [PubMed] [Google Scholar]
- 72. Gaur U, Showalter M, Hickerson S, Dalvi R, Turco SJ, Wilson ME & Beverley SM (2009) Leishmania donovani lacking the Golgi GDP‐Man transporter LPG2 exhibit attenuated virulence in mammalian hosts. Exp Parasitol 122, 182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wyllie S, Patterson S & Fairlamb AH (2013) Assessing the essentiality of Leishmania donovani nitroreductase and its role in nitro drug activation. Antimicrob Agents Chemother 57, 901–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sienkiewicz N, Ong HB & Fairlamb AH (2010) Trypanosoma brucei pteridine reductase 1 is essential for survival in vitro and for virulence in mice. Mol Microbiol 77, 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 76. Bergmeyer HU (1975) New values for the molar extinction coefficients of NADH and NADPH for the use in routine laboratories. Z Klin Chem Klin Biochem 13, 507–508. [PubMed] [Google Scholar]
- 77. Saitou N & Nei M (1987) The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 78. Zuckerkandl E & Pauling L (1965) Molecules as documents of evolutionary history. J Theor Biol 8, 357–366. [DOI] [PubMed] [Google Scholar]
- 79. Kumar S, Stecher G & Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhuang GQ, Li B, Guo HY, Liu JL & Chen F (2011) Molecular cloning and characterization of P5CS gene from Jatropha curcas L. Afr J Biotechnol 10, 14803–14811. [Google Scholar]
- 81. Pérez‐Arellano I, Rubio V & Cervera J (2005) Dissection of Escherichia coli glutamate 5‐kinase: functional impact of the deletion of the PUA domain. FEBS Lett 579, 6903–6908. [DOI] [PubMed] [Google Scholar]


