Abstract
DNA methylation analysis of cervical scrapes using FAM19A4 and mir124‐2 genes has shown a good clinical performance in detecting cervical cancer and advanced CIN lesions in need of treatment in HPV‐positive women. To date, longitudinal data on the cancer risk of methylation test‐negative women are lacking. In our study, we assessed the longitudinal outcome of FAM19A4/mir124‐2 methylation analysis in an HPV‐positive screening cohort with 14 years of follow‐up. Archived HPV‐positive cervical scrapes of 1,040 women (age 29–61 years), who were enrolled in the POBASCAM screening trial (ISRCTN20781131) were tested for FAM19A4/mir124‐2 methylation. By linkage with the nationwide network and registry of histo‐ and cytopathology in the Netherlands (PALGA), 35 cervical cancers were identified during 14 years of follow‐up comprising three screens (baseline, and after 5 and 10 years). The baseline scrape of 36.1% (n = 375) women tested positive for FAM19A4/mir124‐2 methylation, including 24 women with cervical cancer in follow‐up, and 30.6% (n = 318) had abnormal cytology (threshold borderline dyskaryosis or ASCUS), including 14 women with cervical cancer in follow‐up. Within screening round capability of FAM19A4/mir124‐2 methylation to detect cervical cancer was 100% (11/11, 95% CI: 71.5–100). Kaplan–Meier estimate of 14‐year cumulative cervical cancer incidence was 1.7% (95% CI: 0.66–3.0) among baseline methylation‐negative and 2.4% (95% CI: 1.4–3.6) among baseline cytology‐negative women (risk difference: 0.71% [95% CI: 0.16–1.4]). In conclusion, a negative FAM19A4/mir124‐2 methylation test provides a low cervical cancer risk in HPV‐positive women of 30 years and older. FAM19A4/mir124‐2 methylation testing merits consideration as an objective triage test in HPV‐based cervical screening programs.
Keywords: cervical screening, HPV testing, triage, DNA methylation, cancer risk, cytology
Short abstract
What's new?
While HPV testing is increasingly being used for cervical‐cancer screening, there is a problem with this approach: Most HPV infections won't progress to (pre)malignant disease, which results in a significant number of unnecessary colposcopy referrals and over‐diagnoses. A better triage test is needed to discern which HPV+ women have clinically relevant disease. In this longitudinal study, the authors found that a methylation test may provide adequate predictive power. Low cervical‐cancer incidence after a negative FAM19A4/mir124‐2 methylation test among HPV+ women supports use of this methylation assay as safe, objective triage tool.
Abbreviations
- FAM19A4
Family with sequence similarity 19 (chemokine (C‐C)‐motif)‐like), member A4
- mir124‐2
microRNA 124‐2
- HPV
human papillomavirus
- POBASCAM
Population‐based screening trial Amsterdam
- PALGA
nationwide network and registry of histo‐ and cytopathology in the Netherlands
- ASC‐US
Atypical squamous cells of undetermined significance
- CIN
cervical intraepithelial neoplasia
- LSIL
low‐grade squamous intraepithelial lesion
- SCC
squamous cell carcinoma
- AC
adenocarcinoma
- PCR
Polymerase chain reaction
- qMSP
quantitative methylation‐specific PCR
- Cq
quantification cycle
- ACTB
β‐actin
- BMD
borderline or mild dyskaryosis
- 95% CI
95% confidence interval
- SD
standard deviation.
Introduction
Several randomized controlled trials have demonstrated that human papillomavirus (HPV)‐based cervical cancer screening is more effective than cytology‐based screening in preventing cervical cancer.1, 2, 3 As a consequence, primary HPV‐based cervical screening has been implemented in some countries and is under consideration in several other ones. However, since most HPV infections will not give rise to (pre)malignant disease, an important concern related to the adoption of this approach is the increased number of unnecessary colposcopy referrals and over‐diagnoses. This side‐effect can be tackled by applying an adequate triage test for HPV‐positive women to discern those with clinically relevant disease. Various studies have evaluated triage strategies for HPV‐positive women in screening cohorts, including virus‐ and host cell‐based strategies, such as HPV16/18 genotyping,4, 5 HPV E7 mRNA analysis,6, 7 cytology,8, 9 p16/ki67 dual staining,10, 11 epigenetic changes in the host and/or viral genome,12, 13 and combinations thereof.8, 9 At present, reflex cytology testing, which has been adopted in the Dutch HPV‐based screening program, is considered an appropriate triage test,1, 4, 8, 9, 14 although a short‐term repeat cytology is needed to assure a sufficiently low risk of cervical cancer for triage test‐negative women. Furthermore, cytology is subjective and prior knowledge of HPV presence, as is the case in the setting of primary HPV screening, will likely increase the false‐positivity rate.15 This problem could be overcome by using an objective triage test for HPV‐positive women.
A candidate, objective triage tool involves DNA hypermethylation analysis of promoter regions of certain host cell genes involved in cervical carcinogenesis.12, 16, 17, 18, 19, 20 Cervical cancer development after a persistent infection with high‐risk HPV is driven by additional host cell changes such as altered DNA methylation.12, 17, 20, 21 In earlier work, we have shown that methylation assays targeting FAM19A4 and/or mir124‐2 genes have competitive performance against other triage options.22, 23, 24, 25, 26 In cross‐sectional and short‐term clinical follow‐up studies among both cervical screening and gynecologic outpatient populations, FAM19A4 methylation analysis has shown a similar sensitivity for cervical intraepithelial neoplasia lesions or worse (CIN3+) as compared to cytology.22, 24 Methylation levels of these genes in cervical scrapes increase with the severity of underlying cervical lesion.22, 25 Of interest, FAM19A4 methylation analysis detects (virtually) all cervical carcinomas, and has proven to be more sensitive than cytology for the detection of CIN3 lesions with a longer duration of existence.27 In fact, FAM19A4 methylation analysis tested positive in all cervical scrapes of women with CIN3 lesions with a duration of the associated HPV infection of >5 years.22 Those lesions are characterized by a ‘cancer‐like’ (epi)genetic profile and have therefore been considered as advanced precancers.22, 28 Based on this feature, triage testing by methylation analysis has been proposed to confer a high reassurance against short‐term risk of cervical cancer.12 However, longitudinal data on the low cancer risk of methylation test‐negative women are lacking. Here, we assessed cross‐sectional (within screening round) performance of FAM19A4/mir124‐2 methylation‐based triage, and long‐term incidence of cervical cancer (cancer risk) among HPV‐positive women tested by the FAM19A4/mir124‐2 methylation assay within a cohort of women participating in the POBASCAM randomized controlled screening trial with 14 years of follow‐up, and compared figures to cytology.
Materials and Methods
Study population
The study design of the POBASCAM trial (Trial registration ID: NTR218; ISRCTN20781131), a population‐based randomized controlled trial for implementation of high‐risk HPV testing in cervical screening, has been published before.3, 29, 30 In brief, women aged 29–61 were included from January 1999 to September 2002. They were randomized (1:1) to either the intervention group (cytology and HPV co‐testing; n = 22,420) or control group (cytology with blinded HPV testing; n = 22,518). Women with a double‐negative HPV and cytology test (intervention group), or negative cytology test (control group) were referred to routine screening (interval 5 years). In both study arms, women with a cytology result of moderate dyskaryosis or worse (>ASC‐US/LSIL) were immediately referred to colposcopy. Remaining women [i.e. HPV‐positive with normal or borderline or mild dyskaryosis (ASC‐US/LSIL) cytology (intervention group), or women with ASC‐US/LSIL cytology (control group)] underwent follow‐up co‐testing (intervention group) or cytology testing (control group) at 6 and/or 18 months and were referred for colposcopy in case the HPV test was positive or cytology showed ASC‐US/LSIL (intervention group), or when repeat cytology result was ≥ASC‐US/LSIL (control group). At the second screening round after 5 years, participants in both study groups were managed according to the protocol of the intervention group. At the third screening round after 10 years, participants in both study groups were managed according to the protocol of the control group. Histological follow‐up data were tracked through the nationwide network and registry of histopathology and cytopathology (PALGA).31
FAM19A4/mir124‐2 methylation analysis in baseline samples
For the present study, we included all HPV‐positive women who were diagnosed with cervical cancer during 14 years of follow‐up (intervention arm n = 18; control arm n = 27), and all remaining HPV‐positive women of the control arm (n = 1149). Of 1,040 HPV‐positive women, sufficient left‐over material of baseline cervical scrapes was available and valid FAM19A4/mir124‐2 methylation test results were obtained. DNA from cervical scrapes was isolated using the Nucleo‐Spin 96 Tissue kit (Macherey‐Nagel, Duren, Germany) and a Microlab Star robotic system (Hamilton, Planegg, Germany) according to manufacturers’ protocol.32 Extracted DNA was subjected to bisulphite treatment using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA) as described previously.33, 34 Bisulphite‐converted DNA was used as template for DNA methylation analysis for FAM19A4 and mir124‐2 genes using a prototype version of the QIAsure Methylation Test (Qiagen, Hilden, Germany) and Rotorgene PCR‐system (Qiagen). All samples had a quantification cycle (Cq) value for ACTB <26.4 to assure adequate sample quality. ΔCq values are calculated per sample as the difference between the Cq value of the FAM19A4 or mir124‐2 targets and the Cq value of the reference (ACTB). This ΔCq is a relative quantitative value for the promoter methylation level of the FAM19A4 or mir124‐2 gene. For normalization, also the ΔCq value of a calibrator sample is calculated and subtracted from the ΔCq of the FAM19A4 or mir124‐2 targets resulting in a ΔΔCq value.35 The calibrator is a standardized low‐copy plasmid DNA sample with known copy number of the three targets (i.e., FAM19A4, mir124‐2, and ACTB). The methylation result of a sample was scored positive if 1 or both markers had ΔΔCq value below a specified threshold. Thresholds were determined performing an optimization procedure in a training cohort of HPV‐positive women by maximizing the CIN3+ sensitivity at a given, predefined specificity of 70%, and next validated in an independent HPV‐positive screening cohort.32
FAM19A4/mir124‐2 methylation testing in follow‐up samples
Whenever available, repeat cervical scrapes of women diagnosed with cervical cancer within the first screening round were tested for FAM19A4/mir124‐2 methylation (n = 11 scrapes; n = 9 women). In addition, of women with cervical cancer diagnosis during the second or third screening round, available cervical scrapes collected in the second screening round (n = 9 scrapes, n = 9 women) were subjected to FAM19A4/mir124‐2 methylation testing for longitudinal clinical performance evaluation. No cervical scrapes from the third screening round were available for testing.
Data and statistical analysis
All methylation testing was performed blinded for cyto‐ and histopathology outcomes, and data were matched afterwards. Follow‐up data were collected until July 2013,36 at which point women had had the opportunity of three rounds of five‐year screening each (i.e., three screens; at baseline, and after 5 and 10 years). Analyses were performed in SPSS Statistics for Windows version 22.
We determined the number of individuals who tested positive at baseline as a proportion of the number submitted for testing, and stratified for outcome (i.e., with or without cancer diagnosis during 14‐years of follow‐up). Cytology was labeled positive if the result was borderline dyskaryosis or worse (i.e., atypical squamous cells of undetermined significance (ASC‐US) or low grade squamous intraepithelial lesions (LSIL) or worse, and labeled negative otherwise. Six‐ and 18‐month follow‐up cervical scrapes were used to determine repeatability (or test–retest reliability), that is the relation between measurement outcome within a short period of time. Second screen scrapes were used to determine measurement outcome in relation to time to diagnosis of cervical cancer.
We used Kaplan‐Meier methods to estimate the cumulative incidence (risk) of histological outcome of cervical cancer up to 14 years of follow‐up. Time to event was defined as the number of years between the baseline test and the date of histologic diagnosis. Events occurring after 14 years were censored because they were likely to be detected at the fourth screen after baseline. In case of an interrupting event (e.g., uterus extirpation), time to event was censored at the date of the interrupting event. If no screening test result had been reported at the second and the third screen (n = 120) the time to event was censored after 4 years. If no screening test results had been reported at the third screen (n = 125), the time to event was censored 9 years after the study entry date. Women without event were censored after 14 years. Given the randomized controlled trial design, cancer risks in intervention and control arm were considered equal. Therefore, cases in control and intervention groups were pooled in order to evaluate highest number of cervical cancers diagnosed in the POBASCAM trial. As we do not expect that the relation between cytology and methylation results is different in the intervention arm and the control arm, the control arm was considered to provide sufficient number of women without cancer for the analyses. Cumulative cancer incidences were corrected for pooling, by dividing the cumulative incidence by 2. Separate estimates of corrected cumulative cancer incidence were reported for methylation and cytology results among HPV‐positive women. We assessed risk difference between baseline HPV‐positive, methylation‐negative and baseline HPV‐positive, cytology‐negative women. We constructed 95% confidence intervals (95% CI) for the risk difference via bootstrap in R version 3.2.5.
Results
Study population
A flowchart of the study population is presented in Figure 1. Of the 1040 HPV‐positive women included in the study cohort, 35 women were diagnosed with cervical cancer over a period of 14 years (three screening rounds), that is, 26 squamous cell carcinoma (SCC) and 9 adenocarcinoma (AC). The women with cancer had a mean age of 41 years (range, 29–59). The mean age of the HPV‐positive women without cervical cancer was 35 years (range, 29–61).
Figure 1.
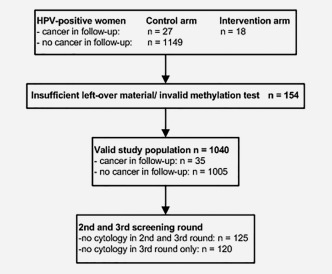
Flowchart of the study population.
Table 1 reports baseline methylation and cytology results overall and stratified for outcome. A total of 375 (36.1%) women tested positive for FAM19A4/mir124‐2 methylation at baseline, among whom 24 women with a cervical cancer diagnosis over a period of 14 years, and 665 women tested methylation‐negative at baseline, among whom 11 women with a cervical cancer diagnosis over a period of 14 years were identified. For cytology, 318 (30.6%) women had a baseline positive test (threshold borderline dyskaryosis or ASC‐US), among whom 14 women with a cervical cancer diagnosis over a period of 14 years, and 722 women were cytology‐negative at baseline, among whom 21 women with a cervical cancer diagnosis over a period of 14 years.
Table 1.
Baseline methylation and cytology results
| A. | ||||
|---|---|---|---|---|
| Total cohort | ||||
| Methylation | Cytology | |||
| − | + | Total | ||
| MM− | 543 | 122 | 665 | |
| MM+ | 179 | 196 | 375 | |
| Total | 722 | 318 | 1040 | |
| B. | ||||
|---|---|---|---|---|
| Women with cancer in follow‐up | ||||
| Methylation | Cytology | |||
| − | + | Total | ||
| MM− | 11 | 0 | 11 | |
| MM+ | 10 | 14 | 24 | |
| Total | 21 | 14 | 35 | |
| Women with cancer in follow‐up | ||||
|---|---|---|---|---|
| Methylation | Cytology | |||
| − | + | Total | ||
| MM− | 532 | 122 | 654 | |
| MM+ | 169 | 182 | 351 | |
| total | 701 | 304 | 1005 | |
Number of women positive or negative for methylation and cytology; (A) overall and (B) stratified for outcome (i.e., with or without cervical cancer diagnosis during 14‐years of follow‐up).
Abbreviations: MM− = negative for FAM19A4/mir124‐2 methylation; MM+ = positive for FAM19A4/mir124‐2 methylation; − = normal cytology; + = cytology‐positive (≥ borderline dyskaryosis or ASC‐US).
FAM19A4/mir124‐2 methylation analysis stratified for histotype and time to diagnosis of cervical cancer
Table 2 reports data of FAM19A4/mir124‐2 methylation analysis and cytology at baseline stratified by cancer histotype and screening round wherein the cervical cancer was diagnosed. All cervical scrapes of women diagnosed with cervical cancer, both SCC and AC, within the first screening round were FAM19A4/mir124‐2 methylation‐positive. Table 3 reports methylation and cytology findings over time for the subset of women diagnosed with cervical cancer, of whom follow‐up scrapes collected closer to the date of cancer diagnosis were available. Figure 2 shows corresponding levels of methylation for FAM19A4 (Fig. 2 a) and mir124‐2 (Fig. 2 b) over time. All 6‐ and/or 18‐month follow‐up scrapes of nine women with cancer detected in the first screening round scored methylation‐positive, with stable or slightly increased levels over baseline scrape, supporting high repeatability of FAM19A4/mir124‐2 methylation analysis. Longitudinal data for cancers detected in second or third screening round support an increased frequency of FAM19A4/mir124‐2 methylation positivity in scrapes taken closer to the time of cervical cancer diagnosis, and confirm a cross‐sectional (within screening round) capability of FAM19A4/mir124‐2 methylation to identify cervical cancer of 100% (11/11; 95% CI:71.5–100). In general, the methylation levels between baseline and follow‐up cervical scrapes of these women increased and were highest closest to the time of cancer diagnosis.
Table 2.
Methylation and cytology results at baseline stratified for cancer histotype and time of cancer diagnosis
| Diagnosed within first round (i.e., up to 4 years from baseline) | Diagnosed within second round (i.e., 5–9 years from baseline) | Diagnosed within third round (i.e., 10–14 years from baseline) | ||||
|---|---|---|---|---|---|---|
| MM+ | Cyt+ | MM+ | Cyt+ | MM+ | Cyt+ | |
| SCC | 9/9 (100%) | 7/9 (77.7%) | 8/11 (72.7%) | 4/11 (36.4%) | 2/6 (33.3%) | 1/6 (16.7%) |
| AC | 3/3 (100%) | 2/3 (66.7%) | 1/3 (33.3%) | 0/3 (0%) | 1/3 (33.3%) | 0/3 (0%) |
| Total | 12/12 (100%) | 9/12 (75.0%) | 9/14 (64.3%) | 4/14 (28.6%) | 3/9 (33.3%) | 1/9 (11.1%) |
Abbreviations: AC= adenocarcinoma; SCC= squamous cell carcinoma; MM+ = positive for FAM19A4/mir124‐2 methylation; Cyt+ = cytology‐positive (≥ borderline dyskaryosis or ASC‐US).
Table 3.
FAM19A4/mir124‐2 methylation findings over time
| Patient ID | Histotype | Baseline | Follow‐up 6 months | Follow‐up 18 months | Second round | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cyto | Meth | Cyto | Meth | Cyto | Meth | Cyto | Meth | Cancer detected in round | ||
| Case 1 | SCC | Normal | Pos | BMD | Pos | BMD | Pos | 1 | ||
| Case 2 | AC | BMD | Pos | BMD | Pos | 1 | ||||
| Case 3 | SCC | BMD | Pos | BMD | Pos | >BMD | Pos | 1 | ||
| Case 4 | AC | BMD | Pos | >BMD | Pos | 1 | ||||
| Case 5 | SCC | BMD | Pos | >BMD | Pos | 1 | ||||
| Case 6 | SCC | BMD | Pos | >BMD | Pos | 1 | ||||
| Case 7 | SCC | >BMD | Pos | >BMD | Pos | 1 | ||||
| Case 8 | SCC | Normal | Pos | >BMD* | Pos | 1 | ||||
| Case 9 | AC | BMD | Pos | >BMD | Pos | 1 | ||||
| Case 10 | AC | Normal | Neg | Normal | Pos | 2 | ||||
| Case 11 | SCC | Normal | Neg | BMD | Pos | 2 | ||||
| Case 12 | AC | Normal | Pos | >BMD | Pos | 2 | ||||
| Case 13 | SCC | Normal | Pos | >BMD | Pos | 2 | ||||
| Case 14 | SCC | >BMD | Pos | >BMD | Pos | 2 | ||||
| Case 15 | AC | Normal | Neg | BMD | Pos | 3 | ||||
| Case 16 | SCC | Normal | Neg | Normal | Neg | 3 | ||||
| Case 17 | SCC | Normal | Pos | Normal | Pos | 3 | ||||
| Case 18 | SCC | >BMD | Pos | Normal | Pos | 3 | ||||
Methylation and cytology results of scrapes collected at baseline and follow‐up (i.e., 6 months, 18 months and/or second screening round) of women diagnosed with cervical cancer, ranked by screening round wherein the cancer was diagnosed (last column).
Follow‐up at 36 months instead of 18 months.
Abbreviations: AC = adenocarcinoma; BMD = borderline or mild dyskaryosis; cyto = cytology; meth = methylation; SCC= squamous cell carcinoma.
Figure 2.
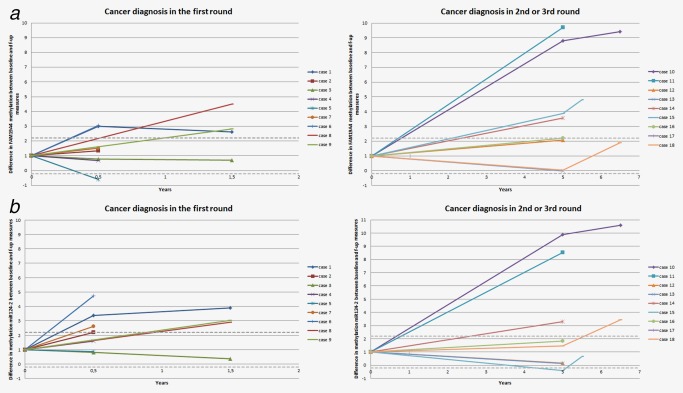
FAM19A4 (a) and mir124‐2 (b) methylation levels over time. Relative difference in FAM19A4 (a) and mir124‐2 (b) methylation levels between baseline and follow‐up cervical scrapes of women with cancer diagnosed in the first screening round (left panel), or in the second or third screening round (right panel). Baseline methylation levels are set to 1, and the difference over baseline level at the different time points is plotted. The grey dotted lines represent 2xSD (upper) or −2xSD (lower) of a repeat analysis. Samples within these lines are considered to have similar methylation level as compared to baseline. SD = standard deviation. [Color figure can be viewed at http://wileyonlinelibrary.com]
Long‐term incidence of cervical cancer in HPV‐positive women according to baseline FAM19A4/mir124‐2 methylation and cytology results
The corrected cumulative incidence of cervical cancer up to three screens corresponding to 14 years of follow‐up, among HPV‐positive women stratified for methylation or cytology status at baseline, is depicted in Figure 3. After the third screening round, the corrected cumulative cervical cancer incidence was 5.5% (95% CI: 3.6–7.7) for methylation‐positive and 4.1% (95% CI: 2.1–6.3) for cytology‐positive women. Cumulative incidence was 2.4% (95% CI: 1.4–3.6) for cytology‐negative and 1.7% (95% CI: 0.66–3.0) for methylation‐negative women. The risk difference between HPV‐positive, methylation‐negative and HPV‐positive, cytology‐negative women was 0.71% (95% CI: 0.16–1.4).
Figure 3.
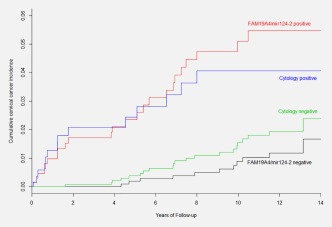
Fourteen‐year cumulative incidence of cervical cancer among HPV‐positive women stratified by FAM19A4/mir124‐2 methylation or cytology test result at baseline. Kaplan‐Meier cancer incidence curves for HPV‐positive women testing FAM19A4/mir124‐2 methylation‐positive or methylation‐negative compared to HPV‐positive women testing cytology‐positive (threshold borderline dyskaryosis or ASC‐US) or cytology‐negative at baseline. [Color figure can be viewed at http://wileyonlinelibrary.com]
Discussion
Our study provides a long‐term evaluation of the triage capacity of a FAM19A4/mir124‐2 methylation test in a screening cohort of HPV‐positive women of 30 years and older. FAM19A4/mir124‐2 methylation analysis on cervical scrapes exhibited a very high cross‐sectional (within screening round) sensitivity for cervical cancer, both SCC and AC, among HPV‐positive women. This translated into a low 14‐year cancer risk among HPV‐positive, methylation‐negative women of 1.7% (95% CI: 0.66–3.0). The low cervical cancer risk conferred by a negative test supports the use of this DNA methylation test as a safe triage alternative in HPV‐based screening programs.
In this HPV‐positive screening cohort, the cross‐sectional, within one screening round sensitivity of the methylation marker panel was 100% and better than cytology in all evaluated screening rounds. The longitudinal sensitivity for cervical cancer37 decreased with a longer time between the baseline cervical scrape and the moment of cervical cancer diagnosis. However, in screening practice where women will be retested every 3–5 years, a 100% cross‐sectional sensitivity will likely translate into a high reassurance against cervical cancer of primary HPV‐testing combined with methylation‐based triage. The longitudinal outcome data show a lower 14‐year cervical cancer risk among baseline FAM19A4/mir124‐2 methylation‐negative women as compared to baseline cytology‐negative women in this HPV‐positive screening cohort (1.7% and 2.4%, respectively; risk difference: 0.71% (95% CI: 0.16–1.4). It is instructive to compare methylation testing to cytology, an approach often proposed to triage HPV‐positive women. A methylation‐based triage assay may offer several advantages over cytology: it is objective, directly applicable to the DNA isolates that are generated during most HPV‐based screening workflows, has a high repeatability as shown herein, allows higher throughput, could be adapted to intermediate and low resource settings, and it can be directly applied to self‐collected samples.
The major strengths of the current study are its setting nested within a population‐based screening program, its large size, the long follow‐up period (14 years), and the wide age range (29–61 years). A limitation to our study was the post‐hoc design and the relatively limited number of cervical cancers. Women were managed according to cytology either or not in combination with HPV‐testing, but not according to methylation findings. The presented incidence estimates of cervical cancer were tracked through the nationwide histopathology and cytopathology registry PALGA,31 which does not contain information on gynecological procedures. Although we were unable to assess how many cancer cases may be missed because women did not comply with the colposcopy advice, we expect that this will not markedly affect our estimates given that PALGA covers (almost) all pathology diagnoses in the Netherlands and therefore potentially missed cancers should have been registered in the following rounds when they become symptomatic. Another aspect is that within the POBASCAM trial cytology and HPV testing were performed without knowledge of the other test result. It is known that guided cytological screening performed with prior knowledge of HPV status results in an improved detection of cervical (pre)cancer at the cost of a loss in specificity.15 Accordingly, sensitivity figures of cytology triage might be underestimated in our study, and the observed risk difference between methylation‐negative women and cytology‐negative women might be slightly overestimated. This bias of HPV knowledge does not apply to the FAM19A4/mir124‐2 methylation test, given its objective nature. Methylation analysis of FAM19A4 and mir124‐2 has previously been evaluated in various clinical settings, including cervical cancer screening populations, non‐attendees and gynecologic outpatient populations. In these studies, high clinical sensitivity and specificity were obtained and therefore this marker panel is currently further validated in clinical cohorts from different European countries (Valid‐screen project 666800). Other host‐cell methylation markers include single markers or marker combinations of JAM3, TERT, C13ORF18, EPB41L3, ANKRD18CP, LHX8, DLX1, ITGA4, RXFP3, SOX17, ZNF671, GHSR, SST, ZIC1, ASTN1, SOX1, and DCC.19, 38, 39, 40, 41, 42 Although these markers have shown promising results so far, they have been studied less extensively and only cross‐sectionally, as compared to the FAM19A4/mir124‐2 methylation test. In order to find the best possible triage markers in various settings, the next step in biomarker development43 warrants marker comparisons and prospective screening/intervention studies.
In summary, our data show that a negative FAM19A4/mir124‐2 methylation test provides a low cervical cancer risk in HPV‐positive women of 30 years and older. Therefore, FAM19A4/mir124‐2 methylation testing merits consideration as a safe and objective, alternative triage test in HPV‐based cervical cancer screening programs.
Conflict of Interest
CJLMM, PJFS, RDMS, and DAMH have minority stake in Self‐screen B.V., a spin‐off company of VU University Medical Center Amsterdam of which CJLMM is director since September 2017. Self‐screen B.V holds patents related to the work and has developed and manufactured the FAM19A4/mir124‐2 methylation assay, which is licensed to Qiagen (QIAsure Methylation Test). CJLMM has participated in the sponsored speaker's bureau of Merck and Qiagen, and served occasionally on the scientific advisory board of Qiagen and Merck. CJLMM has a very small number of shares of Qiagen, has occasionally been consultant for Qiagen and until April 2016 was a minority shareholder of Diassay B.V. PJFS has been on the speaker's bureau of Roche, Gen‐Probe, Qiagen and Seegene. He is consultant for Crucell Holland B.V. DAMH has been on the speaker's bureau of Qiagen and serves occasionally on the scientific advisory board of Pfizer and Bristol‐Meyer Squibb. JB has received consultancy fees from Roche, GlaxoSmithKline, and Merck, and received travel support from DDL. All fees were collected by his employer. All other authors declare that they have no conflicts of interest.
Acknowledgments
We thank A. Splunter, M. Bogaarts and S. Mongera for their excellent technical assistance.
References
- 1. Ronco G, Dillner J, Elfström KM, et al. Efficacy of HPV‐based screening for prevention of invasive cervical cancer: follow‐up of four European randomised controlled trials. Lancet. 2013;39:1–9. [DOI] [PubMed] [Google Scholar]
- 2. Arbyn M, Ronco G, Anttila A, et al. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine. 2012;30: F88–99. [DOI] [PubMed] [Google Scholar]
- 3. Rijkaart DC, Berkhof J, Rozendaal L, et al. Human papillomavirus testing for the detection of high‐grade cervical intraepithelial neoplasia and cancer: final results of the POBASCAM randomised controlled trial. Lancet Oncol. 2012;13:78–88. [DOI] [PubMed] [Google Scholar]
- 4. Castle PE, Stoler MH, Wright TC, et al. Performance of carcinogenic human papillomavirus (HPV) testing and HPV16 or HPV18 genotyping for cervical cancer screening of women aged 25 years and older: a subanalysis of the ATHENA study. Lancet Oncol. 2011;12:880–90. [DOI] [PubMed] [Google Scholar]
- 5. Cox JT, Castle PE, Behrens CM, et al. Comparison of cervical cancer screening strategies incorporating different combinations of cytology, HPV testing, and genotyping for HPV 16/18: results from the ATHENA HPV study. Am J Obstet Gynecol. 2013;208:184.e1–.e11. [DOI] [PubMed] [Google Scholar]
- 6. Burger EA, Kornør H, Klemp M, et al. HPV mRNA tests for the detection of cervical intraepithelial neoplasia: a systematic review. Gynecol Oncol. 2011;120:430–8. [DOI] [PubMed] [Google Scholar]
- 7. Luttmer R, Berkhof J, Dijkstra MG, et al. Comparing triage algorithms using HPV DNA genotyping, HPV E7 mRNA detection and cytology in high‐risk HPV DNA‐positive women. J Clin Virol. 2015;67:59–66. [DOI] [PubMed] [Google Scholar]
- 8. Rijkaart DC, Berkhof J, van Kemenade FJ, et al. Evaluation of 14 triage strategies for HPV DNA‐positive women in population‐based cervical screening. Int J Cancer. 2012;130:602–10. [DOI] [PubMed] [Google Scholar]
- 9. Dijkstra M, van Niekerk D, Rijkaart D, et al. Primary hrHPV DNA testing in Cervical Cancer screening: how to manage screen positive women? A POBASCAM Trial sub study. Cancer Epidemiol Biomarkers Prev. 2014;23:55–63. [DOI] [PubMed] [Google Scholar]
- 10. Uijterwaal MH, Witte BI, Van Kemenade FJ, et al. Triaging borderline/mild dyskaryotic Pap cytology with p16/Ki‐67 dual‐stained cytology testing: cross‐sectional and longitudinal outcome study. Br J Cancer. 2014;110:1579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carozzi F, Gillio‐Tos A, Confortini M, et al. Risk of high‐grade cervical intraepithelial neoplasia during follow‐up in HPV‐positive women according to baseline p16‐INK4A results: a prospective analysis of a nested substudy of the NTCC randomised controlled trial. Lancet Oncol. 2013;14:168–76. [DOI] [PubMed] [Google Scholar]
- 12. Steenbergen RDM, Snijders PJF, Heideman DAM, et al. Clinical implications of (epi)genetic changes in HPV‐induced cervical precancerous lesions. Nat Rev Cancer. 2014;14:395–405. [DOI] [PubMed] [Google Scholar]
- 13. Tornesello ML, Buonaguro L, Giorgi‐Rossi P, et al. Viral and cellular biomarkers in the diagnosis of cervical intraepithelial neoplasia and cancer. Biomed Res Int. 2013;2013:1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Naucler P, Ryd W, Törnberg S, et al. Efficacy of HPV DNA testing with cytology triage and/or repeat HPV DNA testing in primary cervical cancer screening. J Natl Cancer Inst. 2009;101:88–99. [DOI] [PubMed] [Google Scholar]
- 15. Moriarty AT, Nayar R, Arnold T, et al. The Tahoe Study: bias in the interpretation of Papanicolaou test results when human papillomavirus status is known. Arch Pathol Lab Med. 2014;138:1182–5. [DOI] [PubMed] [Google Scholar]
- 16. Lorincz AT. Cancer diagnostic classifiers based on quantitative DNA methylation. Expert Rev Mol Diagn. 2014;14:293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wentzensen N, Sherman ME, Schiffman M, et al. Utility of methylation markers in cervical cancer early detection: appraisal of the state‐of‐the‐science. Gynecol Oncol. 2009;112:293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boers A, Bosgraaf RP, van Leeuwen RW, et al. DNA methylation analysis in self‐sampled brush material as a triage test in hrHPV‐positive women. Br J Cancer. 2014;111:1095–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boers A, Wang R, Van Leeuwen RW, et al. Discovery of new methylation markers to improve screening for cervical intraepithelial neoplasia grade 2/3. Clin Epigenet . 2016;8: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Steenbergen RDM, Ongenaert M, Snellenberg S, et al. Methylation‐specific digital karyotyping of HPV16E6E7 expressing human keratinocytes identifies novel methylation events in cervical carcinogenesis. J Pathol. 2013;231:53–62. [DOI] [PubMed] [Google Scholar]
- 21. Chen Y‐C, Huang R‐L, Huang Y‐K, Laio Y‐P, Su P‐H, Wang H‐C, Chang C‐C, Lin Y‐W, Yu M‐H, Chu T‐Y, Lai H‐C. Methylomics analysis identifies epigenetically silenced genes and implies an activation of β‐catenin signaling in cervical cancer. Int J Cancer. 2013;1–11. [DOI] [PubMed] [Google Scholar]
- 22. De Strooper LMA, Meijer CJLM, Berkhof J, et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev Res. 2014;7:1251–7. [DOI] [PubMed] [Google Scholar]
- 23. De Strooper LMA, Verhoef VMJ, Berkhof J, et al. Validation of the FAM19A4/mir124‐2 DNA methylation test for both lavage‐ and brush‐based self‐samples to detect cervical (pre)cancer in HPV‐positive women. Gynecol Oncol. 2016;141:341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luttmer R, De Strooper LMA, Berkhof J, et al. Comparing the performance of FAM19A4 methylation analysis, cytology and HPV16/18 genotyping for the detection of cervical (pre)cancer in high‐risk HPV‐positive women of a gynecologic outpatient population (COMETH study). Int J Cancer. 2016;1002:992–1002. [DOI] [PubMed] [Google Scholar]
- 25. Luttmer R, De Strooper LMA, Dijkstra MG, et al. FAM19A4 methylation analysis in self‐samples compared with cervical scrapes for detecting cervical (pre)cancer in HPV‐positive women. Br J Cancer. 2016;115:579–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luttmer R, De Strooper LMA, Steenbergen RDM, et al. Management of high‐risk HPV‐positive women for detection of cervical (pre)cancer. Expert Rev Mol Diagn. 2016;16:961–74. [DOI] [PubMed] [Google Scholar]
- 27. De Strooper LMA, Hesselink AT, Berkhof J, et al. Combined CADM1/MAL methylation and cytology testing for colposcopy triage of high‐risk HPV‐positive women. Cancer Epidemiol Biomarkers Prev. 2014;23:1933–7. [DOI] [PubMed] [Google Scholar]
- 28. Bierkens M, Hesselink AT, Meijer CJLM, et al. CADM1 and MAL promoter methylation levels in hrHPV‐positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer. 2013;133:1293–9. [DOI] [PubMed] [Google Scholar]
- 29. Bulkmans NWJ, Berkhof J, Rozendaal L, et al. Human papillomavirus DNA testing for the detection of cervical intraepithelial neoplasia grade 3 and cancer: 5‐year follow‐up of a randomised controlled implementation trial. Lancet. 2007;370:1764–72. [DOI] [PubMed] [Google Scholar]
- 30. Bulkmans NWJ, Rozendaal L, Snijders PJF, et al. POBASCAM, a population‐based randomized controlled trial for implementation of high‐risk HPV testing in cervical screening: design, methods and baseline data of 44,102 women. Int J Cancer. 2004;110:94–101. [DOI] [PubMed] [Google Scholar]
- 31. Casparie M, Tiebosch ATMG, Burger G, et al. Pathology databanking and biobanking in The Netherlands, a central role for PALGA, the nationwide histopathology and cytopathology data network and archive. Cell Oncol. 2007;29:19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hesselink AT, Heideman DAM, Steenbergen RDM, et al. Combined promoter methylation analysis of CADM1 and MAL: an objective triage tool for high‐risk human papillomavirus DNA‐positive women. Clin Cancer Res. 2011;17:2459–65. [DOI] [PubMed] [Google Scholar]
- 33. Overmeer RM, Henken FE, Snijders PJF, et al. Association between dense CADM1 promoter methylation and reduced protein expression in high‐grade CIN and cervical SCC. J Pathol. 2008;215:388–97. [DOI] [PubMed] [Google Scholar]
- 34. Overmeer RM, Henken FE, Bierkens M, et al. Repression of MAL tumour suppressor activity by promoter methylation during cervical carcinogenesis. J Pathol. 2009;219:327–36. [DOI] [PubMed] [Google Scholar]
- 35. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods. 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 36. Dijkstra MG, van Zummeren M, Rozendaal L, van Kemenade FJ, Helmerhorst TJM, Snijders PJF, Meijer CJLM, Berkhof J. Safety of extending screening intervals beyond five years in cervical screening programmes with testing for high risk human papillomavirus: 14 year follow‐up of population based randomised cohort in the Netherlands. BMJ. 2016;355 [DOI] [PubMed] [Google Scholar]
- 37. Elfström KM, Smelov V, Johansson ALV. Long term duration of protective effect for HPV negative women: follow‐up of primary HPV screening randomised controlled trial. BMJ. 2014;130:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eijsink JJH, Lendvai Á, Deregowski V, et al. A four‐gene methylation marker panel as triage test in high‐risk human papillomavirus positive patients. Int J Cancer. 2012;130:1861–9. [DOI] [PubMed] [Google Scholar]
- 39. Clarke MA, Luhn P, Gage JC, et al. Discovery and validation of candidate host DNA methylation markers for detection of cervical precancer and cancer. Int J Cancer. 2017;141:701–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eijsink JJH, Yang N, Lendvai A, et al. Detection of cervical neoplasia by DNA methylation analysis in cervico‐vaginal lavages, a feasibility study. Gynecol Oncol. 2011;120:280–3. [DOI] [PubMed] [Google Scholar]
- 41. Brentnall AR, Vasiljević N, Scibior‐Bentkowska D, et al. A DNA methylation classifier of cervical precancer based on human papillomavirus and human genes. Int J Cancer. 2014;135:1425–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schmitz M, Wunsch K, Hoyer H, et al. Performance of a methylation specific real‐time PCR assay as a triage test for HPV‐positive women. Clin Epigenetics. 2017;9:118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–61. [DOI] [PubMed] [Google Scholar]


