Summary
Pathogens secrete effector proteins to interfere with plant innate immunity, in which Ca2+/calmodulin (CaM) signalling plays key roles. Thus far, few effectors have been identified that directly interact with CaM for defence suppression. Here, we report that SFI5, an RXLR effector from Phytophthora infestans, suppresses microbe‐associated molecular pattern (MAMP)‐triggered immunity (MTI) by interacting with host CaMs.
We predicted the CaM‐binding site in SFI5 using in silico analysis. The interaction between SFI5 and CaM was tested by both in vitro and in vivo assays. MTI suppression by SFI5 and truncated variants were performed in a tomato protoplast system.
We found that both the predicted CaM‐binding site and the full‐length SFI5 protein interact with CaM in the presence of Ca2+. MTI responses, such as FRK1 upregulation, reactive oxygen species accumulation, and mitogen‐activated protein kinase activation were suppressed by truncated SFI5 proteins containing the C‐terminal CaM‐binding site but not by those without it. The plasma membrane localization of SFI5 and its ability to enhance infection were also perturbed by loss of the CaM‐binding site.
We conclude that CaM‐binding is required for localization and activity of SFI5. We propose that SFI5 suppresses plant immunity by interfering with immune signalling components after activation by CaMs.
Keywords: calcium, calmodulin, MAMP‐triggered immunity (MTI), RXLR effector, SFI5, tomato protoplast, virulence
Introduction
Plants rely on a multi‐layered immune system to combat potential pathogens. One important layer of inducible plant immune responses is the recognition by cell surface‐resident pattern recognition receptors (PRRs) of so‐called microbe‐associated molecular patterns (MAMPs), which are highly conserved molecules or structural components derived from microbes and indispensable for microbial fitness or lifestyle (Medzhitov & Janeway, 1997; Nürnberger & Brunner, 2002; Nürnberger et al., 2004; Ausubel, 2005; Boller & Felix, 2009; Macho & Zipfel, 2014). The signalling responses induced by PRR‐mediated perception of MAMPs is termed MAMP‐triggered immunity (MTI), which is accompanied by rapid Ca2+ influxes, accumulation of reactive oxygen species (ROS), activation of mitogen‐activated protein kinases (MAPKs), upregulation of immunity‐associated gene expression, synthesis of anti‐microbial proteins and callose deposition (Apel & Hirt, 2004; Macho & Zipfel, 2014; Bigeard et al., 2015; Lee et al., 2015).
One of the key mediators in plant immune responses is Ca2+/calmodulin (CaM) (Lecourieux et al., 2006; Cheval et al., 2013). CaM is a major Ca2+ sensor in eukaryotes and, after conformational change induced by Ca2+ binding to EF‐hand motifs, interacts with and regulates the function of diverse target proteins (McCormack et al., 2005). These include membrane receptors, protein kinases and transcription factors that are involved in plant development but also in plant immunity and adaptation to other stress conditions (Zhang et al., 2009; Reddy et al., 2011; Cho et al., 2016).
Microbes that can colonize plants have evolved multiple effectors that either prevent perception of MAMPs by membrane‐bound PRRs or interfere with cellular components of MTI signalling pathways (Boller & He, 2009; Bozkurt et al., 2012; Dou & Zhou, 2012; Giraldo & Valent, 2013; Doehlemann et al., 2014). The latter mainly involves effectors that translocate into host cells. For example, the bacterial pathogen Pseudomonas syringae secretes many effectors into plant cells using the type III secretion system, targeting various proteins to suppress MTI (Block & Alfano, 2011; Dou & Zhou, 2012; Fraiture & Brunner, 2014). Recently, one effector protein, HopE1, was shown to interact directly with CaM for MTI suppression (Guo et al., 2016). Oomycete pathogens, including downy mildews and Phytophthora species, secrete hundreds of candidate effectors that contain the RXLR motif shortly after the N‐terminal signal peptide (Tyler et al., 2006; Jiang et al., 2008; Haas et al., 2009), which is required for delivery into host cells (Whisson et al., 2007, 2016; Dou et al., 2008). Some of these effectors have been shown to target MTI components and suppress plant immunity (King et al., 2014; Whisson et al., 2016).
Phytophthora infestans is the causal agent of tomato and potato late blight disease, but it can also infect the model solanaceous plant Nicotiana benthamiana (Zheng et al., 2014; Whisson et al., 2016). Many RXLR effectors in P. infestans and other well‐known oomycete pathogens such as Phytophthora sojae and Hyaloperonospora arabidopsidis have been shown to suppress MTI at different steps of the signalling pathway, through different mechanisms and in different subcellular compartments. P. infestans effector AVR1 interacts with host exocyst component Sec5 to suppress CRN2‐induced cell death and PR1 secretion (Du et al., 2015). AVR3a represses the programmed cell death induced by the elicitor INF1 by targeting and stabilizing the host E3 ligase CMPG1 (Bos et al., 2006, 2010; Gilroy et al., 2011). PITG_03192 localizes to the host endoplasmic reticulum (ER), where it targets two NAC transcription factors, NTP1 and NTP2, and prevents their re‐localization into the nucleus upon MAMP application (McLellan et al., 2013). Pi04314 associates with different isoforms of host protein phosphatase type 1c (PP1c) and causes their re‐localization within the nucleus without affecting their biochemical activity. The PP1c isoforms were proposed to be susceptibility factors, utilized by Pi04314 to promote disease development by attenuating salicylic acid (SA) and jasmonate (JA) signalling (Boevink et al., 2016). PexRD2 interacts with host MAPK signalling to perturb both MTI and ETI (King et al., 2014). In P. sojae, Avr3b contains a Nudix hydrolase motif in the C‐terminal part of the effector domain and displays ADP‐ribose/NADH pyrophosphorylase enzymatic activity, which is dependent on the interaction with the plant cyclophilin CYP1 through a putative Glycine‐Proline motif (Dong et al., 2011; Kong et al., 2015). PsAvh23 suppresses defence gene expression by interacting with the ADA2 subunit of the histone acetyltransferase complex (Kong et al., 2017). In H. arabidopsidis, the nuclear‐localized effector, HaRxL44, was shown to associate with Mediator subunit 19a (MED19a), leading to proteasome‐dependent degradation of MED19a and the activation of jasmonate/ethylene (JA/ET)‐signalling to antagonize SA‐signalling and the activation of SA‐responsive genes (Caillaud et al., 2013).
In our previous study, we identified a range of P. infestans RXLR effectors, named Suppressor of early Flg22‐induced Immune response (SFI), that perturb the earliest signalling events of MTI in tomato protoplasts (Zheng et al., 2014). One of these, namely SFI5, localizes at the host plasma membrane, blocks the MAMP‐triggered activation of the MAP kinase cascade and facilitates P. infestans infection in N. benthamiana leaves. The mechanism underlying the mode of action of SFI5 in manipulating host immunity was not investigated.
In this study, we show that SFI5 contains a CaM‐binding motif at its C‐terminus. We demonstrate Ca2+‐dependent association between SFI5 and CaMs in both in vitro and in vivo assays. We provide strong correlative evidence that CaM‐association is required for the plasma membrane location and effector activity of SFI5 in tomato protoplasts and N. benthamiana leaves. We report for the first time that an effector association with CaM plays a critical role in suppression of immunity in the interactions between plants and filamentous pathogens.
Materials and Methods
Bioinformatics
The sequence of Phytophthora infestans SFI5 (PITG_13628) was analysed for CaM‐binding domains and canonical motifs according to the method described by Yap et al. (2000) and Mruk et al. (2014). Calmodulin sequences from Arabidopsis (Arabidopsis thaliana) and tomato (Solanum lycopersicum) were obtained from GenBank according to the descriptions of Day et al. (2002) and Zhao et al. (2013). Sequence alignment was performed by ClustalW program (Chenna et al., 2003).
Microbe and plant growth conditions
Escherichia coli DH5α and Rosetta™(DE3) (Merck, Germany) were routinely grown in Luria Bertani (LB) media with appropriate antibiotics at 37°C. Phytophthora infestans strain 88069 (Pieterse & Davidse, 1990) was grown on Rye Sucrose Agar at 19°C. Nicotiana benthamiana Domin (personal laboratory stock) and Solanum lycopersicum L. cv Moneymaker (Thompson & Morgan, Ipswich, Suffolk, UK) were grown under controlled conditions in a glasshouse as described previously (Zheng et al., 2014).
Gene cloning and constructs
Calmodulin genes were cloned by PCR amplification from tomato cDNAs with primer pairs shown in Supporting Information Table S1. The PCR amplicons were cloned using the Gateway® cloning recombination technology into the entry vector pDONR201 (Thermo Fisher Scientific, Darmstadt, Germany) and, subsequently, recombined into the expression vectors p2HAGW7 (N‐terminal HA‐tag) (Zheng et al., 2014) and p2FGW7 (C‐terminal GFP‐tag) or pB7WGF2 (N‐terminal GFP‐tag) (VIB, Belgium; Karimi et al., 2005), or pDEST15 (N‐terminal GST‐tag) and pDEST17 (N‐terminal His‐tag) for protein expression in E. coli (Thermo Fisher Scientific). The SFI5 gene without the region encoding the signal peptide (28–241 aa) was amplified by PCR, digested with XmnI and SalI restriction endonucleases and ligated into the vector pMAL‐p5x (N‐terminal MBP‐tag) (New England Biolabs, Frankfurt, Germany). Positive clones were confirmed by sequencing.
Truncated SFI5 genes without fragments encoding N‐ or C‐terminal domains were amplified by PCR using specific primers (Table S1), cloned into the entry vector pDONR201 (Thermo Fisher Scientific) and subsequently recombined into the expression vectors p2HAGW7 and p2FGW7 (Life Technologies, Darmstadt, Germany). Site‐directed mutagenesis was performed following the instruction manual of the QuikChange® II XL Site‐Directed Mutagenesis Kit (Agilent Technologies, Waldbronn, Germany) using the p2HAGW7‐SFI5 63–241 aa or p2FGW7‐SFI5 63–241 aa plasmid constructs as PCR template. Primers used for mutagenesis are listed in Table S1. All constructs were verified by sequencing.
In vitro pull‐down assay
pDEST17 vector expressing His‐SlCaM6 and pMAL‐p5x vector expressing MBP‐SFI5 (28–241 aa) were transformed into Rosetta™(DE3) competent cells, respectively. Positive colonies were cultivated in LB medium at 37°C with 200 rpm shaking until the culture reached an OD600 of 0.6, followed by addition of 0.5 mM isopropyl β‐d‐thiogalactopyranoside (IPTG) for protein induction. Upon induction, bacterial cultures were transferred to 28°C with 200 rpm shaking for 2 h before harvesting. His‐ and MBP‐fusion proteins were purified by HisTrap (HisTrap FF Crude; GE Healthcare, Freiburg, Germany) and MBPTrap (MBPTrap HP; GE Healthcare) columns, respectively, following the user manuals. For pull‐down assay, the same amount (5 mmol) of His‐SlCaM6 and MBP‐SFI5 were mixed together with 5 mM CaCl2 or 5 mM EGTA in 500 μl PBS buffer and incubated at 4°C for 1 h. Both protein mixtures were then incubated with 10 μl Ni‐NTA resin at 4°C for 30 min, followed by the removal of the supernatant and extensive wash (eight times) with wash buffer (250 mM This‐HCl, 500 mM NaCl, 25% (v/v) glycerol, 5 mM β‐mercaptoethanol, and 10 mM imidazole. pH 8.0). The binding proteins were eluted with 1 M imidazole and mixed with SDS sample buffer for SDS‐PAGE. Purified His‐SlCaM6 and MBP‐SFI5 proteins also were loaded for SDS‐PAGE as controls.
Transfection of tomato protoplasts
Tomato mesophyll protoplasts were isolated and transfected as described previously (Fraiture et al., 2014; Zheng et al., 2014).
Immunoprecipitation and mass spectrometry
The transformed protoplasts were harvested by centrifugation at 100 g for 1 min and the pellet was re‐suspended in 1 ml of immunoprecipitation (IP) buffer containing 50 mM HEPES (pH 7.4), 150 mM NaCl, 0.1% Triton X‐100, 1 mM EDTA, 1 mM DTT, 1× phosphatase inhibitor cocktail (PhosphoSTOP, Roche) and 1× protease inhibitor cocktail. Total protein was released by sonication and the cell debris was removed by centrifugation. The HA‐tagged proteins were immunoprecipitated from lysates by incubation with 20 μl of anti‐HA antibody‐coupled beads (anti‐HA affinity matrix, Roche) for 3–6 h while gently shaking at 4°C. Afterwards, the beads were washed three times with 1 ml of washing buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 0.2% Triton X‐100, 1× phosphatase inhibitor cocktail and 1× protease inhibitor cocktail). For elution, 50 μl SDS loading buffer without DTT was added to the beads, followed by boiling at 95°C for 10 min. The immunoprecipitated proteins were then further analysed by immunoblotting or LC‐MS/MS (tandem).
SDS‐PAGE and immunoblotting
Proteins were separated by 13.5% SDS‐PAGE, blotted and incubated with appropriate antibodies (Zheng et al., 2014). The antibody dilutions were: anti‐phospho‐p44/42 MAPK antibody (Cell Signalling Technology Europe BV, Leiden, Netherlands), 1 : 1000 in 5% BSA‐TBST; anti‐GFP (Acris), 1 : 5000 in 5% BSA‐TBST; anti‐HA (Sigma), 1 : 1000 in 5% BSA‐TBST; anti‐GST (Sigma), 1 : 7000 in 5% BSA‐TBST; anti‐MBP (Cell Signalling Technology), 1 : 10 000 in 5% BSA‐TBST; anti‐His (Abcam, Cambridge, UK), 1 : 5000 in 5% BSA‐TBST; anti‐rabbit IgG alkaline phosphatase (Sigma), 1 : 3000 in TBST; anti‐goat IgG alkaline phosphatase (Sigma), 1 : 3000 in TBST; anti‐mouse IgG alkaline phosphatase (Sigma), 1 : 3000 in TBST; anti‐goat IgG horseradish peroxidase (Sigma), 1 : 10 000 in TBST; anti‐mouse IgG horseradish peroxidase (Sigma), 1 : 10 000 in TBST. Immunodetection was performed with a NBT/BCIP solution (AppliChem, Darmstadt, Germany) or with the ECL Western blotting system (GE Healthcare).
Tris‐glycine native‐PAGE
pDEST15 vector expressing GST‐SlCaM6 was transformed into Rosetta™(DE3) competent cells, and the fusion protein was induced and purified with GSTrap column (GSTrap FF column, GE Healthcare) following the user manual. Native polyacrylamide gel electrophoresis (Native‐PAGE) was carried out as described previously (Niepmann & Zheng, 2006; Arndt et al., 2012). Equal volumes of 133 μM synthetic peptide and 50 μM GST‐ or His‐ SlCaM6 were mixed and incubated for 1 h at 4°C. Peptide–protein interactions were analysed on 12% tris‐glycine native gels (30 mM Tris‐HCl pH 7.5, 190 mM glycine, 5 mM DTT). NativeMark Unstained Protein Standard (Life Technologies) was used as a protein marker for the analysis. The electrophoresis was performed with the Mini‐PROTEAN Tetra Cell apparatus (BioRad) in native running buffer (25 mM Tris base, 192 mM Glycine) at a low current (10 mA) at 4°C. The proteins on the gel were visualized by Coomassie blue staining (Blakesley & Boezi, 1977).
1‐Anilinonaphthalene‐8‐sulfonate assay
1‐Anilinonaphthalene‐8‐sulfonate (ANS) (Sigma) was dissolved in ethanol at a stock concentration of 10 mM. The assays were performed as described by Chinpongpanich et al. (2011) with minor modifications. ANS (100 μM) and 1 μM purified recombinant GST‐SlCaM6 were incubated in the reaction buffer containing 20 mM Tris‐HCl (pH 7.5), 100 mM NaCl, and 1 mM CaCl2 for 15 min before addition of 0–100 μM synthetic peptide. Fluorescence was measured in a 96‐well plate reader (MWG) with λex = 360 nm and λem = 460 nm.
Luciferase reporter gene assays
Luciferase reporter gene assay was conducted following the method of Zheng et al. (2014). Protoplasts were treated with/without flg22 to a final concentration of 500 nM. The luminescence reflecting the luciferase activity was measured at different time‐points using a Mithras LB 940 luminometer (Berthold, Bad Wildbad, Germany). Luciferase activity (+/−flg22) was normalized by measuring β‐glucuronidase (GUS) activity upon lysis with an equal volume of CCLR solution and incubation of 10 μl protoplast extract with 90 μl MUG substrate.
MAP kinase activation assay
The MAP kinase activation was detected as described previously (Zheng et al., 2014). Two hundred microliters of transformed tomato protoplasts untreated or treated with 500 nM flg22 for 0 and 20 min were harvested for immunodetection by anti‐phospho‐p44/42 MAPK antibody.
Oxidative burst assay
Tomato protoplasts were used to measure reactive oxygen species (ROS) production using a luminol‐based assay (Halter et al., 2014). One hundred to 200 μl of transformed protoplast samples were incubated in W1 buffer without MES (0.5M mannitol, 20 mM KCl) at 20–22°C in the dark for 6–8 h. Before measurement, protoplasts were harvested by centrifugation at 100 g for 1 min and resuspended in 100 μl W5 buffer (18.4 g l−1 CaCl2 × 2H2O, 1.0 g l−1 glucose, 9.0 g l−1 NaCl, 0.4 g l−1 KCl) containing 200 μM luminol L‐012 (Wako Chemicals) and 20 μg ml−1 horseradish peroxidase and incubated for an additional 30 min in the dark. After treatment with 500 nM flg22, luminescence was recorded for 30 min using a Mithras LB 940 multiplate reader (Berthold).
Infection assay
Phytophthora infestans infection assay was performed as described previously (Zheng et al., 2014). Agrobacterium tumefaciens carrying GFP‐SFI5 deletion mutant constructs were infiltrated into N. benthamiana leaves, which were then inoculated by P. infestans strain 88069 1 d after A. tumefaciens infiltration. Lesion sizes were determined and photographed at 7 d post‐infection.
Confocal fluorescence microscopy
Nicotiana benthamiana leaf cells of wild‐type (WT), GFP‐LTi6b or mOrange‐LTi6b PM marker‐expressing transgenic lines (Kurup et al., 2005; Yang et al., 2016) were infiltrated with A. tumefaciens containing expression constructs at OD600 of 0.01. Cells transiently expressing SFI5 fusion constructs were imaged 48 h post‐infiltration on Zeiss 710 or Nikon A1R confocal microscopes. GFP was excited with 488 nm light and the emissions were detected between 500 and 530 nm. mRFP and mOrange were excited with 561 nm light and emissions were detected between 600 and 630 nm or 580 and 610 nm, respectively. Cells displaying a low intensity of GFP fluorescence were imaged to minimize artefacts of overexpression. For co‐expression of the GFP‐SFI5 constructs with the cytoplasmic mRFP control construct, A. tumefaciens suspensions were pre‐mixed before infiltration.
Images were processed with propriety confocal software or ImageJ as required. Figures were constructed with Adobe Photoshop and Adobe Illustrator.
Results
SFI5 binds CaM in vitro in the presence of Ca2+
Phytophthora infestans SFI5 is a 241‐amino acid protein bearing the typical signature of RXLR effectors with an N‐terminal signal peptide, followed by a sequence (Ala28 to Arg62) containing the RXLR motif and a predicted C‐terminal effector domain of 178 amino acid residues (Phe63 to Arg241). Sequence analysis in the Calmodulin Target Database (Yap et al., 2000) revealed a putative CaM‐binding site located at the C‐terminal end of SFI5 including 18 amino acids between Pro222 and Leu239 (Fig. 1a). A genome‐wide analysis has revealed six CaM genes in tomato, which encode four isoforms – SlCaM1, SlCaM2, SlCaM3/4/5 and SlCaM6 – sharing 91–99% sequence identity (Zhao et al., 2013) (Fig. S1). We cloned SlCaM1, SlCaM3/4/5 and SlCaM6 but not SlCaM2 from a tomato cDNA library.
Figure 1.
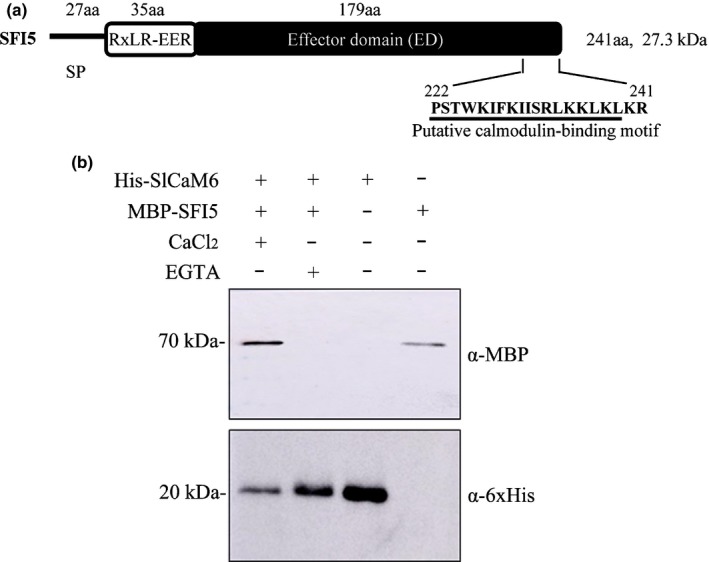
SFI5 binds calmodulin (CaM) in the presence of Ca2+. (a) Schematic representation of the SFI5 effector, showing the predicted amino acid (aa) length of the signal peptide (SP), host‐translocation motif‐containing domain (RxLR‐EER) and effector domain (ED). The putative CaM‐binding motif is underlined. The CaM‐binding motif was predicted in the Calmodulin Target Database (Yap et al., 2000). (b) Pull‐down assay of SFI5 and SlCaM6. MBP‐tagged SFI5 (MBP‐SFI5) and 6xHis‐tagged SlCaM6 (His‐SlCaM6) were mixed with 5 mM CaCl2 or 5 mM EGTA in PBS buffer and incubated for 1 h at 4°C. The mixtures were then incubated with Ni‐NTA agarose beads for 30 min, followed by extensive wash with wash buffer of Ni‐NTA beads. The bound proteins were eluted with 1 M imidazole and mixed with SDS sample buffer for SDS‐PAGE. Gel‐separated proteins were transferred to nitrocellulose membranes and immunodetected with anti‐MBP and anti‐6xHis antibodies, respectively. Purified MBP‐SFI5 and His‐SlCaM6 were used as controls.
We performed a pull‐down assay to analyse in vitro the interaction between SFI5 and CaM. The purified His‐SlCaM6 and MBP‐SFI5 proteins were expressed in E. coli and purified prior to interaction studies using Ni‐NTA affinity chromatography in the presence of Ca2+ or the Ca2+ chelator EGTA. As shown in Fig. 1(b), in the presence of Ca2+, both a 70 kDa band corresponding to MBP‐SFI5 and a 20 kDa band corresponding to His‐SlCaM6 were immunodetected in the Ni‐NTA eluate (Lane 1 of Fig. 1b). In the presence of EGTA, the 70 kDa MBP‐SFI5 was not detected (Lane 2 of Fig. 1b). Immunodetection of the purified His‐SlCaM6 and MBP‐SFI5 controls in lanes 3 and 4 of Fig. 1(b), confirmed the identity of the bands in lanes 1 and 2. Thus, we conclude that SFI5 interacts with SlCaM6 in vitro in a Ca2+‐dependent manner.
A C‐terminal 17‐aa peptide of SFI5 binds CaM in vitro in the presence of Ca2+
The predicted CaM‐binding site at the C‐terminus of SFI5 does not contain a canonical CaM‐binding motif (Mruk et al., 2014). However, a helical wheel projection of the 18‐amino acid stretch (Pro222 to Leu239) showed a basic amphipathic structure with one side enriched in positively charged residues (Lys226, Lys229, Arg233, Lys236) and the opposite side rich in hydrophobic residues (Trp225, Ile227, Phe228, Ile231), which resembles typical CaM recognition and binding motifs (Fig. 2a). Peptides corresponding to the predicted CaM‐binding site of SFI5 and with amino acid mutations, which led to different net charges, were synthesized for CaM‐binding analysis (Figs 2b, S2).
Figure 2.
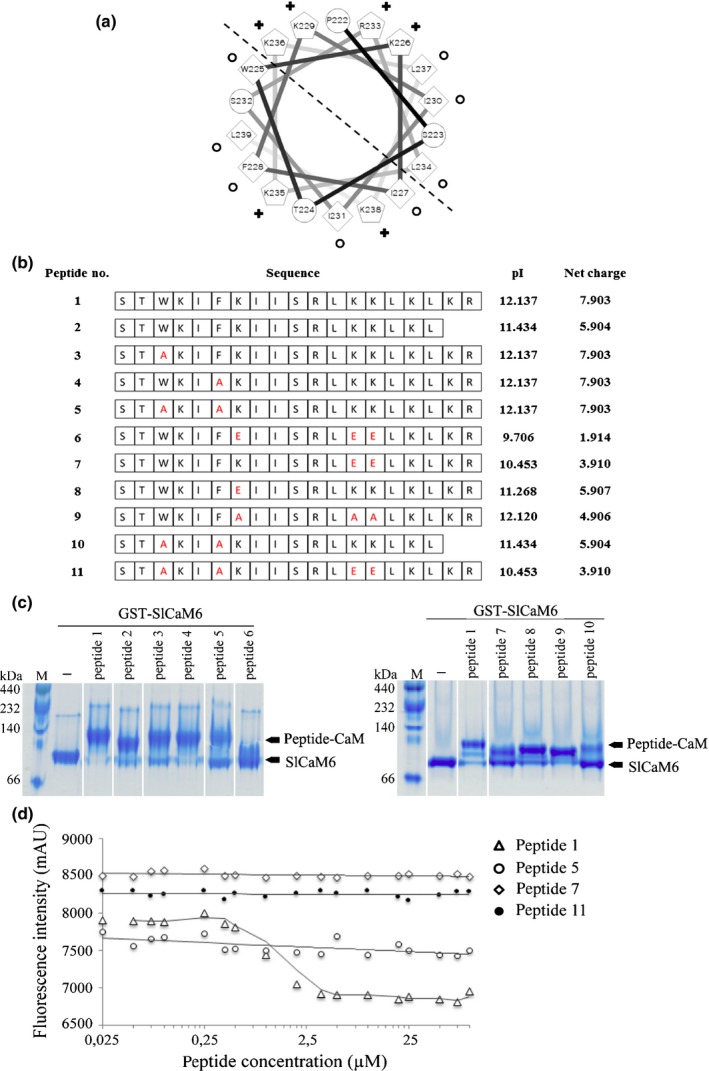
In vitro interaction between C‐terminal 17‐aa domain of SFI5 and GST‐SlCaM6. (a) Helical wheel projection of the predicted 18‐amino acid calmodulin (CaM)‐binding domain of SFI5 (Pro222 to Leu239). Hydrophobic and potentially positively charged residues are marked with circles and crosses, respectively. The dashed line divides the amphipathic helix into the hydrophobic side and hydrophilic side. Numbers refer to amino acid positions in SFI5 protein. (b) Synthetic peptides corresponding to the predicted CaM‐binding site with various amino acid mutations. Peptide 1 represents the last 19 aa at the C‐terminus of SFI5. Peptides 2–11 are truncated or mutated versions of peptide 1, in which the substituted amino acids are presented in red. The pI and net charge (at pH7.0) of each peptide were calculated by Editseq (Lasergene v.8; dnastar). (c) CaM mobility shift on tris/glycine native gels. Purified GST‐SlCaM6 (50 μM) was incubated with each synthetic peptide (133 μM) in the presence of 5 mM CaCl2. Samples were separated on tris/glycine native gels followed by Coomasssie blue staining. Arrows indicate the position of bands representing free GST‐SlCaM6 (SlCaM6) and GST‐SlCaM6‐peptide complex (Peptide‐CaM). (d) 1‐Anilinonaphthalene‐8‐sulfonate (ANS) fluorescence competition assay. GST‐SlCaM6 (1 μM), ANS (100 μM) in the buffer containing 20 mM Tris‐HCl pH7.5, 100 mM NaCl, 1 mM CaCl2 were incubated with increasing concentration of selected peptides for 30 min, and kinetic changes of fluorescence was monitored at an excitation wavelength of 360 nm (λex) and an emission wavelength of 460 nm (λem). Data points represent the mean ± SE of three technical replicates from three independent assays.
Complex formation between GST‐SlCaM6 and synthetic peptides was shown by gel shift experiments on tris‐glycine native gels. All peptides were mixed with recombinant GST‐SlCaM6 in the presence of Ca2+. As shown in Fig. 2(c), mixing peptide 1 with GST‐SlCaM6 resulted in a band shift to a higher molecular weight compared to the GST‐SlCaM6 control, indicating complex formation between the predicted CaM‐binding motif and SlCaM6. Peptide 2, in which the last two amino acid residues (Lys240‐Arg241) of peptide 1 were removed, was able to bind SlCaM6 with nearly the same efficacy as peptide 1, although its net charge at pH 7 decreased from + 7.9 to + 5.9 (Fig. 2b). This suggests that these two residues (Lys240‐Arg241) are not crucial for CaM association. Replacing two hydrophobic residues, Trp225 and Phe228, with Ala in peptides 3 and 4, respectively, did not severely affect the complex formation with SlCaM6. However, replacing both Trp225 and Phe228 with Ala significantly reduced the binding affinity of peptide 5 and peptide 10 to SlCaM6, with c. 50–75% of the SlCaM6 remaining in an unbound state. The three lysine residues Lys229, Lys235 and Lys236 were predicted by computer modelling to be important for direct interaction with CaM (Yap et al., 2000; Mruk et al., 2014). Replacement of these lysine residues with the negatively charged residue Glu caused a drastic reduction of the net charge of peptide 6 and abolished the interaction with SlCaM6. The replacement of Lys235 and Lys236 only with Glu (peptide 7) or Lys229 only with Glu (peptide 8) had less effect on the net charge and resulted in an increase of peptide binding to SlCaM6 (Fig. 2b,c). Interestingly, replacing the three lysine residues with alanine in peptide 9 also caused less reduction of the net charge and did not affect peptide binding to SlCaM6.
The gel shift assay also was performed with His‐tagged SlCaM6 and additional synthetic peptides with mutations were tested (Fig. S2). The replacement of both Trp225 and Phe228 with His significantly reduced, but did not abolish the binding of peptide 12 to SlCaM6. No binding was detected when Trp225 and Phe228 were replaced with Glu (peptide 13), or in the quadruple mutant, in which Trp225, Phe228, Lys235 and Lys236 were replaced by Glu (peptide 14). The results with His‐SlCaM6 are highly consistent with those of the assay with GST‐tagged SlCaM6. In summary, the net charge of the C‐terminal helix of SFI5 is a crucial factor for CaM‐binding and Trp225, Phe228, Lys235 and Lys236 are important residues for the interaction.
To underpin the importance of the two hydrophobic residues (Trp225 and Phe228) and the two basic residues (Lys235 and Lys236) for CaM‐binding, we carried out CaM competition assays with four synthetic peptides (1, 5, 7 and 11) and 1‐Anilinonaphthalene‐8‐sulfonate (ANS), a compound with high affinity to CaM which is fluorescent upon binding. The kinetics of ANS fluorescence change was monitored in the presence of increasing concentrations of different peptides. As shown in Fig. 2(d), the fluorescence curve rapidly declined with increasing concentration of peptide 1 (IC50 = 1 μM), indicating that competition occurred between ANS and the peptide 1 for SlCaM6‐binding. By contrast, ANS fluorescence decreased smoothly or remained unchanged with increasing concentrations of peptide 5, 7 and 11 (the last a quadruple mutant of Trp225, Phe228, Lys235 and Lys236). This result confirmed the importance of the mutated amino acids within the CaM‐binding site of SFI5.
The CaM binding motif of SFI5 is necessary for the interaction with tomato CaMs in vivo
In order to find out the host targets of SFI5 and confirm the results of the in vitro interaction between SFI5 and SlCaM6, we transiently expressed N‐terminal HA‐tagged SFI5 in tomato protoplasts. Total proteins of transgenic tomato protoplasts were isolated and immunoprecipitated by anti‐HA beads followed by LC‐MS/MS analysis. Through this approach, we identified a number of peptides that match tomato CaMs among the proteins isolated from protoplasts expressing HA‐SFI5 (Table S2). CaMs were not identified in immunoprecipitated material from protoplasts expressing HA‐SFI1, another MTI‐suppressing RXLR effector of P. infestans (Zheng et al., 2014) or transformed with the empty vector control (Table S2). In the next step, we performed pairwise interaction studies with SFI5 and SlCaM1, SlCaM3/4/5 and SlCaM6 in tomato protoplasts. GFP‐tagged SFI5 (GFP‐SFI5) and HA‐tagged CaMs were co‐expressed in tomato protoplasts. Total protein extracts from transformed protoplasts were prepared and immunoprecipitated with anti‐HA antibody‐coupled beads, followed by immunoblotting with anti‐GFP and anti‐HA antibodies, respectively. The result in Fig. 3(a) shows that GFP‐tagged SFI5 associated with all HA‐tagged CaMs without apparent specificity. To confirm whether the interaction between SFI5 and CaMs relies on the presence of Ca2+ and MAMP perception, we performed co‐immunoprecipitation assays with tomato protoplasts expressing HA‐SlCaM3/4/5 and GFP‐SFI5 treated with flg22, a CaM antagonist N‐(6‐aminohexyl)‐5‐chloro‐1‐naphthalene‐sulphonamide (W7) or Ca2+ channel blockers (LaCl3 and EDTA). The results in Fig. 3(b) show that the protein association was independent of the flg22 treatment but significantly inhibited by the presence of CaM antagonist and Ca2+ channel blockers.
Figure 3.
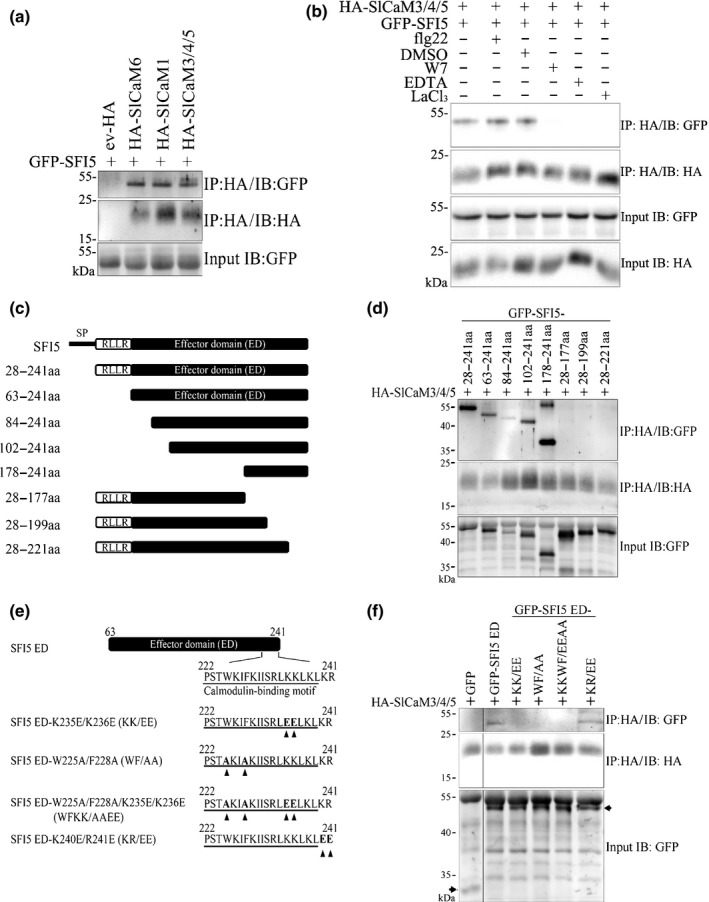
SFI5 interacts with tomato calmodulins (CaMs) in vivo. (a) Co‐immunoprecipitation analysis of transiently expressed GFP‐SFI5 and HA‐SlCaMs in tomato protoplasts. Extracted proteins (input) were subjected to immunoprecipitation (IP) with anti‐HA affinity matrix followed by immunobloting (IB) with anti‐HA antibody to detect the tomato CaMs and anti‐GFP antibody to detect the SFI5 fusion. (b) Co‐immunoprecipitation analysis of transiently expressed GFP‐SFI5 and HA‐SlCaM3/4/5 in tomato protoplasts upon different treatments. Protoplast samples were treated with 500 nM flg22, 0.75% DMSO (mock), 250 nM W7 (stock solution: 33 mM W7 dissolved in 99.5% DMSO) or 1 mM LaCl3 before the protoplasts were harvested for the IP with anti‐HA antibody. For EDTA treatment, total proteins were extracted with IP buffer containing 20 mM EDTA. (c) Schematic diagrams of SFI5 deletion mutants. Numbers indicate positions of amino acid (aa) residues based on the full‐length protein sequence. (d) Co‐immunoprecipitation analysis of transiently expressed GFP‐SFI5 deletion mutants and HA‐SlCaM3/4/5 in tomato protoplasts. (e) Schematic diagrams of site‐directed mutants of SFI5 effector domain (ED). Numbers indicate the amino acid positions based on the full‐length protein sequence. KK/EE, WF/AA, WFKK/AAEE and KR/EE correspond to amino acid exchanges of Lys235 and Lys236 with Glu, Trp225 and Phe228 with Ala, Lys235 and Lys236 with Glu and Trp225 and Phe228 with Ala, and Lys240 and Arg241 with Glu, respectively. Mutations are indicated by arrowheads (f) Co‐immunoprecipitation analysis of transiently expressed GFP‐SFI5 ED site‐directed mutants with HA‐SlCaM3/4/5 in tomato protoplasts. Bands corresponding to GFP and GFP‐tagged SFI5 ED point mutants are indicated by arrows. These results are representative of three replicates. GFP, green fluorescent protein.
We used the results gained from the in vitro binding assay to define the molecular determinants of the interaction between GFP‐SFI5 and SlCaM3/4/5 in tomato protoplasts. We generated a series of N‐terminal or C‐terminal deletion constructs of SFI5 and employed site‐directed mutagenesis to replace the four key residues (Trp225, Phe228, Lys235 and Lys236) in order to validate their role in the interaction between SFI5 and SlCaMs in vivo (Fig. 3c,e). After co‐immunoprecipitation and Western blot analysis, SlCaM3/4/5 interactions were detected with all N‐terminal deletion mutants of SFI5 including the shortest truncated protein corresponding to the last 63 amino acids (178–241 aa). By contrast, none of the three C‐terminal deletion variants, including the one lacking only the amphipathic helix of 19 amino acids (28–221 aa), was able to associate with HA‐SlCaM3/4/5 (Fig. 3d). The SFI5 variants carrying mutations in two hydrophobic residues (Trp225Ala/Phe228Ala – WF/AA), basic residues (Lys235Glu/Lys236Glu – KK/EE) and quadruple residues (Trp225Ala/Phe228Ala/Lys235Glu/Lys236Glu – WFKK/AAEE) failed to associate with HA‐SlCaM3 (Fig. 3e,f). As anticipated, the variant in which the last two amino acids (Lys240 and Arg241) were replaced by glutamic acid (KR/EE) did not affect CaM‐binding (Fig. 3e,f).
From the in vitro and in vivo interaction assays, we concluded that SFI5 has a unique CaM‐binding site formed by a 17‐aa core region (Ser223 to Leu239) containing an alpha helical fold and amphipathic properties, in which the two hydrophobic residues (Trp225 and Phe228) and the two basic residues (Lys235 and Lys236) play critical roles in binding CaMs.
The CaM‐binding motif participates in the plasma membrane localization of SFI5
GFP‐SFI5 is located mainly at the plasma membrane (PM) in tomato protoplasts and N. benthamiana leaves, with very little fluorescence detectable in the cytoplasm (Zheng et al., 2014). To determine whether the CaM‐binding site is important for the intracellular localization of SFI5, full length SFI5 and N‐terminal or C‐terminal deletion variants or amino acid point mutants (Fig. 3c,e) were coexpressed with a PM marker, mOrange‐LTi6b, or cytoplasmic mRFP. Laser scanning confocal microscopy imaging shows that all three GFP‐tagged N‐terminal deletion forms of SFI5 used in this analysis (GFP‐SFI5 28–241 aa, GFP‐SFI5 63–241 aa and GFP‐SFI5 84–241 aa) were predominantly localized at the PM, as illustrated by the overlap of the GFP and mOrange fluorescence signals (Fig. 4), and the failure of the GFP fluorescence to overlap with free mRFP signal. This indicates that the N‐terminal region of SFI5 is not required for the plasma membrane localization. By contrast, the two GFP‐tagged C‐terminal deletion variants (GFP‐SFI5 28–199 aa and GFP‐SFI5 28–221 aa) and the quadruple point mutant (GFP‐SFI5 ED‐WFKK/AAEE) of SFI5 localized predominantly in the cytosol, with GFP fluorescent peaks representing trans‐vacuolar cytoplasmic strands, and a minimal overlap with mOrange‐LTi6b (Fig. 4). This suggests that the PM‐association of SFI5 is dependent on the C‐terminal CaM‐binding motif. Mutation of the last two amino acids (KR/EE) increased the cytoplasmic signal from the GFP fusion (GFP‐SFI5 ED KR/EE) but did not abolish the PM localization (Fig. S3). Co‐localization studies performed in tomato protoplasts using the aforementioned deletion and point mutants of SFI5 and the bacterial effector AvrPto, which is associated with the PM through the presence of an N‐terminal myristoylation site (Shan et al., 2000; He et al., 2006), mirrored the results obtained in N. benthamiana leaves (Fig. S4). Next, we investigated whether SFI5 and CaM interact at the PM. Co‐expression of mRFP‐SlCaM3/4/5 and GFP‐SFI5 28‐241 aa showed an absence of signal overlap, suggesting that if CaM binding is required for the PM localisation of SFI5, the interaction does not occur at the PM and that CaM likely dissociates from SFI5 before it locates at the PM (Fig. S5).
Figure 4.
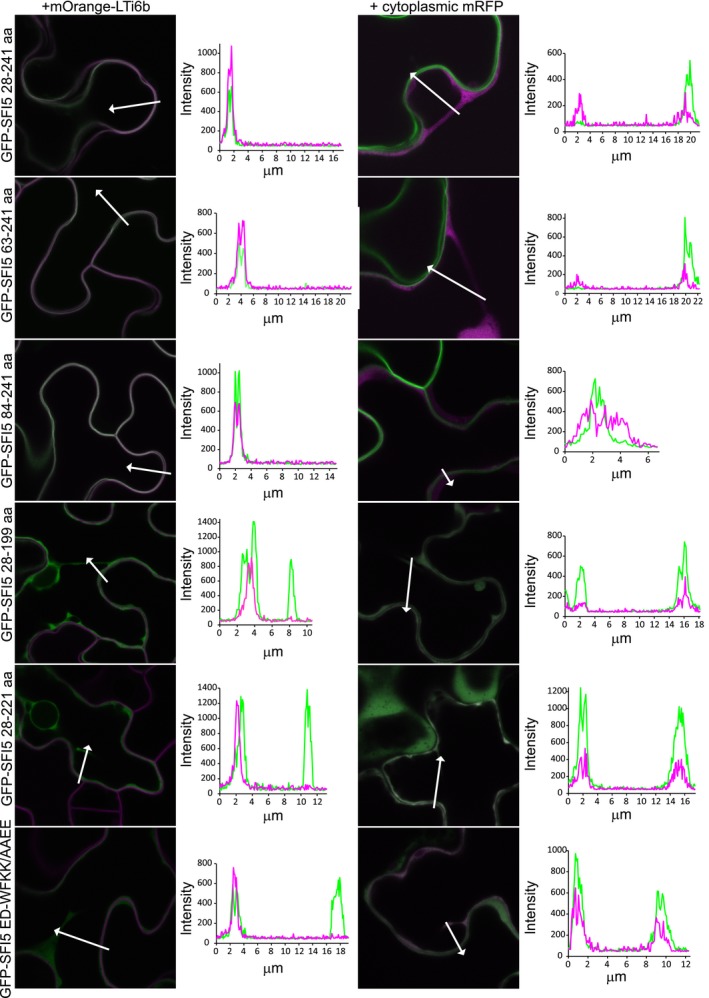
The calmodulin (CaM) binding motif of SFI5 is required for plasma membrane localization in Nicotiana benthamiana leaves. Single optical section images of GFP‐tagged SFI5 deletion and mutant forms (as indicated to the left of the images) coexpressed with the PM marker mOrange‐LTi6b or cytoplasmic mRFP (as indicated above the image columns). The fluorophores are shown in green for GFP and magenta for the mRFP or mOrange. The arrows indicate the lines drawn to generate the intensity profiles shown to the right of each image. Each profile was drawn such that both a cytoplasmic strand, or region of cytoplasm, and the plasma membrane were crossed. The lengths of the profiles are shown in the graphs in μm and the intensities are as generated by the confocal software. GFP, green fluorescent protein.
Both the C‐terminal CaM‐binding motif and N‐terminal region of SFI5 are required for effector activity
SFI5 is an effector that disturbs the earliest signalling events of MTI responses, including flg22‐dependent pFRK1‐Luc reporter gene expression, post‐translational activation of SlMPK1 and SlMPK3, and ROS production, and thereby promotes P. infestans infection in plant leaves (Zheng et al., 2014). To investigate the importance of the CaM‐binding site of SFI5 for disruption of MTI responses, HA‐tagged SFI5 deletion and point mutation variants were transiently expressed in tomato protoplasts (Fig. S6) and tested in a set of bio‐assays to measure early immune responses triggered by flg22.
First, we measured the impact of truncated and mutated SFI5 proteins on the induction of the pFRK1‐Luc reporter gene expression after flg22 treatment using the method described in Zheng et al. (2014). As shown in Fig. 5(a), expressing SFI5 C‐terminal deletion variants (HA‐SFI5 28‐177 aa, HA‐SFI5 28‐199 aa or HA‐SFI5 28–221 aa) failed to block flg22‐induced Luc activity in comparison to expressing SFI5 (HA‐SFI5 28–241 aa) or AvrPto. Point mutations in the CaM‐binding region (HA‐SFI5 ED‐KK/EE, HA‐SFI5 ED‐WF/AA or HA‐SFI5 ED‐WFKK/AAEE) also reduced the ability of SFI5 to block flg22‐triggered reporter gene activation, although the reduction was less than that of the deletion mutants. The HA‐SFI5 ED‐KR/EE mutant was as active as the positive controls (Fig. 5a). Interestingly, suppression of the flg22‐mediated reporter gene activation also was attenuated in tomato protoplasts expressing N‐terminal deletion constructs of SFI5 (HA‐SFI5 84–241 aa, HA‐SFI5 102–241 aa or HA‐SFI5 178–241 aa), except for the one lacking only the RXLR motif (HA‐SFI5 63‐241aa) which retained full suppression activity (Fig. 5a).
Figure 5.
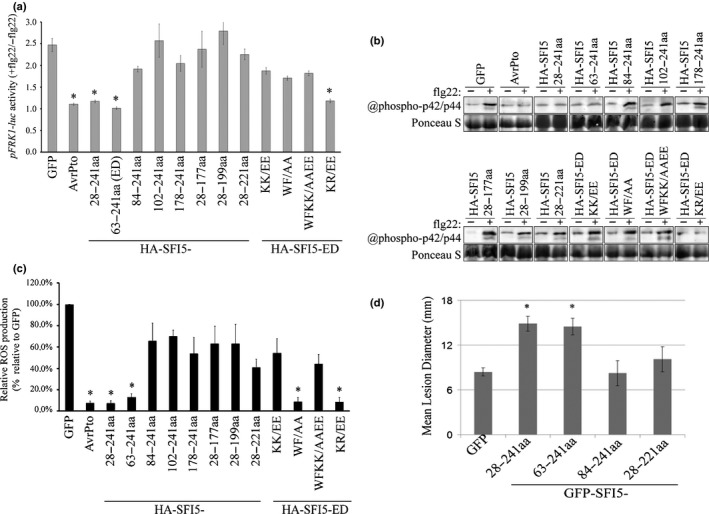
Both the calmodulin (CaM) binding motif and N‐terminal region of SFI5 are required for the suppression of flg22‐triggered early immune responses in tomato protoplasts and Phytophthora infestans infection in Nicotiana benthamiana. (a) Tomato protoplasts coexpressing HA‐tagged SFI5 deletion or point mutants with the two reporter genes pFRK1‐Luc and pUBQ10‐GUS were treated with or without flg22 (+/−flg22) and the pFRK1‐Luc activity was measured after 3 h. The promoter activity is calculated as the ratio of flg22‐induced luciferase activity relative to the untreated sample, which was normalized to the internal GUS activities (pFRK1‐Luc activity +flg22/−flg22). GFP and AvrPto served respectively as a negative and positive control for the suppression of pFRK1‐Luc activation by flg22. (b) Tomato protoplasts expressing HA‐tagged SFI5 deletion or point mutants were collected 20 min after flg22 treatment (+) or without flg22 treatment (−), and the phosphorylated mitogen‐activated protein kinases MAPK were detected by immunoblotting with the antibody raised against phosphorylated MAPK p44/p42. Ponceau S staining is shown as a loading control. (c) The oxidative burst in tomato protoplasts expressing HA‐tagged SFI5 deletion or point mutants is represented as the percentage of total photon counts measured between 6 and 20 min after flg22 treatment of the GFP control, which was set to 100%. GFP and AvrPto served respectively as negative and positive controls for the suppression of pFRK1‐Luc and MAPK activation and ROS burst by flg22. Data in (a, c) represent the mean ± SE from four independent experiments, for each of which three technical replicates were carried out. *, P < 0.05 (one‐way ANOVA followed by Dunnett's multiple comparison test). The results in (b) are representative of at least three independent experiments. (d) N‐terminal or C‐terminal deletion mutants of SFI5 were each transiently expressed via agro‐infiltration in one half of a N. benthamiana leaves and empty vector (GFP) was expressed in the other half. After 24 h, the infiltrated leaves were inoculated with P. infestans. Disease lesion diameters were measured 7 d post‐inoculation. Results are mean ± SE from three biological replicates, each of which used 24 leaves for inoculation per construct. Significant difference (*, P < 0.01) in lesion size compared to empty vector control was determined by one‐way ANOVA. GFP, green fluorescent protein.
Second, immunodetection of flg22‐dependent SlMPK1 and SlMPK3 activation using the p44/p42 antibody was performed with tomato protoplasts expressing the SFI5 deletion and point‐mutation constructs. The results shown in Fig. 5(b) are consistent with the pFRK1‐Luc induction assay.
Third, the results of the oxidative burst assays in tomato protoplasts expressing SFI5 deletion and point mutation constructs were largely in agreement with the results of pFRK1‐Luc induction and MAPK activation assays. Notably, the HA‐SFI5 mutant carrying a substitution for the two hydrophobic residues (HA‐SFI5 ED‐WF/AA) retained the ability to repress ROS production under the defined experimental conditions (Fig. 5c).
In order to further determine the role of the N‐terminal and CaM binding domains in the virulence function of SFI5, two GFP‐SFI5 variants with an N‐terminal deletion (63–241 aa and 84–241 aa) and one with a C‐terminal deletion (28–221 aa) were transiently expressed in N. benthamiana leaves followed by inoculation of P. infestans. As anticipated, both SFI5 (28–241 aa)‐expressing leaves and effector domain (63–241 aa)‐expressing leaves showed significantly larger lesion size compared to those on the GFP expressing control leaves at 7 d post‐pathogen inoculation. By contrast, leaves expressing two SFI5 deletion variants (84–241 aa and 28–221 aa) did not enhance the susceptibility to P. infestans infection since the lesion sizes were similar to those on control leaves (Fig. 5d). Altogether, these results (summarized in Table 1) indicate that there is a correlation between CaM binding, suppression of early MTI signalling and virulence function of SFI5. In addition, another domain of unknown function, located at the N‐terminal part of SFI5, after the RXLR motif and spanning c. 20 aa residues (Phe63‐Lys84), is required for the activity and function of SFI5 in the host cell.
Table 1.
Overview of in vivo Ca2+/calmodulin (CaM)‐binding, localization, and microbe‐associated molecular pattern (MAMP)‐triggered immunity (MTI)‐suppression of truncated or mutated SFI5
| SFI5 | Domain | CaM‐binding | PM‐localization | FRK1‐suppression | MAPK suppression | ROS suppression |
|---|---|---|---|---|---|---|
| 28–241 | + | + | + | + | + | |
| 63–241 | + | + | + | + | + | |
| 84–241 | + | + | − | − | − | |
| 102–241 | + | Not tested | − | − | − | |
| 178–241 | + | Not tested | − | − | − | |
| 28–177 | − | Not tested | − | − | − | |
| 28–199 | − | − | − | − | − | |
| 28–221 | − | − | − | − | − | |
| SFI5 ED | Mutation | CaM‐binding | PM‐localization | FRK1‐suppression | MAPK suppression | ROS suppression |
| WF/AA | ± | Not tested | − | − | + | |
| KK/EE | ± | Not tested | − | − | − | |
| WFKK/AAEE | − | − | − | − | − | |
| KR/EE | + | + | + | + | + |
MAPK, mitogen‐activated protein kinase; PM, plasma membrane; ROS, reactive oxygen species.
Discussion
Phytophthora infestans is an oomycete pathogen that infects the crop plants tomato and potato in nature, and the model plant Nicotiana benthamiana in the laboratory (Haas et al., 2009; King et al., 2014; Whisson et al., 2016). To colonize host plants, P. infestans secretes effector proteins, many of which contain an RxLR motif downstream of the N‐terminal signal peptide for translocation into the plant cell and a C‐terminal effector domain several of which have been shown to suppress plant immunity. So far, there are no studies showing that RXLR effectors suppress immunity by interacting with Ca2+ signalling components. Thus, SFI5 is the first example of an oomycete effector that suppresses microbe‐associated molecular patterns (MAMP)‐triggered immunity (MTI) responses by interacting with calmodulin (CaM).
The structure–function analysis presented here has accurately delimited the domain and residues of SFI5 that are involved in CaM binding. The C‐terminal 18 aa‐region of SFI5 forms an amphipathic α‐helix wheel with the segregation of basic and hydrophobic residues on opposite sides (Fig. 2a), characteristic of CaM‐binding domains (O'Neil & DeGrado, 1990; Meador et al., 1992; Crivici & Ikura, 1995). The CaM‐binding site of SFI5 appears to be noncanonical due to the weak similarity to any known CaM‐binding domains. To verify the CaM‐binding ability, peptides corresponding to the 18 aa‐region were tested for binding with SlCaM6 from tomato by tris‐glycine native gels (Fig. 2b,c). The results showed that the predicted CaM‐binding site of SFI5 is functional, which was confirmed further by in vitro and in vivo interaction assays with full‐length SFI5 and CaMs from tomato (Fig. 3a). The two hydrophobic residues (Trp225 and Phe228) are critical anchor residues but other hydrophobic residues at the same side of the α‐helix wheel (Ile227, Ile231 or Leu239) might also be important for the interaction with CaM. It is worth noting that mutation of the Trp225 and Phe228 residues did not dramatically affect the suppression of flg22‐triggered oxidative burst (Fig. 5c). The 1‐Anilinonaphthalene‐8‐sulfonate (ANS) fluorescence competition assay with peptide 5 (WF/AA) suggests abrogation of the interaction with CaM (Fig. 2d) but the gel shift assay showed a residual complex formation (Fig. 2c). It is therefore possible that replacement of the two strong hydrophobic amino acids with a weak hydrophobic residue (alanine) compromised, but did not completely abolish CaM‐binding activity of SFI5 in vivo, which was sufficient to suppress the oxidative burst. Additional Trp225/Phe228 mutant peptides, in which these two hydrophobic residues were converted into histidine or glutamate residues, did not bind to CaM (Fig. S2a,b). Dose–response assays with those SFI5 variants would help to better quantify CaM binding and their impact on the biological response. This would confirm the importance of CaM binding for the MTI‐suppressing activity of SFI5.
The molecular determinants of CaM that are required for the interaction with SFI5 are unknown. CaMs and CaMs‐like (CMLs) form a highly conserved Ca2+ sensor protein family, which is present in all eukaryotes. The 3D structure of CaM has the hallmark of a dumbbell shape with four EF‐hand Ca2+‐binding motifs organized in pairs and embedded in two globular domains separated by a long flexible helix. After Ca2+ binding, CaM undergoes conformational changes that expose its hydrophobic surfaces and subsequently interacts with a large array of proteins that are implicated in many cellular processes (Bouche et al., 2005; McCormack et al., 2005). It would be interesting to solve the crystal structure of SFI5 in complex with CaM, which could provide new insights on the structural variability of CaM/CaM‐binding protein interactions and on the mode of action of SFI5 in planta.
The CaM‐binding motif of SFI5 is required for the plasma membrane localization, MTI‐suppression and for promoting P. infestans colonization (Fig. 5; Table 1) but we did not observe CaM‐SFI5 co‐localization at the plasma membrane (Fig. S5). A simple and likely explanation for this is that CaM binding and activation may be required for SFI5 to interact subsequently with operative targets that are localized at the plasma membrane (PM). However, dissociation of CaM from SFI5 may be required for it to interact with the operative targets. In vivo Förster resonance energy transfer‐fluorescence lifetime imaging microscopy (FRET‐FLIM) would provide an alternative approach to study the interaction between SFI5 and CaM and perhaps provide information on the dynamics of their coupling. This technique has been successfully applied to study the spatiotemporal interaction dynamics of barley Mildew resistance Locus O (MLO) with its activator, CaM. It revealed that an increasing number of MLO/CaM complexes in the vicinity of the penetration sites was coincident with successful pathogen entry into host cells (Bhat et al., 2005).
Bioinformatic analyses do not predict the presence of a transmembrane or membrane‐anchoring domain in the sequence of SFI5. To further determine the importance and the role of CaM‐mediated PM‐localization for the SFI5 MTI‐suppressing activity, a PM anchor myristoylation site could be introduced into SFI5 and SFI5 variants deficient in CaM‐binding and the resulting proteins could be tested for their MTI‐suppressing activity. We have used this targeting approach to mis‐locate the RXLR effector SFI1 from the nucleus to the plasma membrane to thereby demonstrate that SFI1 must enter the nucleus to suppress MTI (Zheng et al., 2014). However, it has not been used to investigate whether simple restoration of PM localization would restore function.
Although we provide strong correlative evidence that CaM‐binding is required for full function of SFI5, it is still unclear how SFI5 interferes with MTI signalling. One possibility is that SFI5 directly targets and inhibits the function of CaM or CMLs that regulate the activity of downstream CaM‐ and CML‐binding proteins which have a positive role on MTI. Large scale protein–protein interaction screens combined with the availability of genome and transcriptome resources have revealed a considerable number of putative CaM/CML‐binding proteins implicated in the regulation of plant immunity. Experimental evidence includes the finding that one member of the CaM‐binding transcription activator (CAMTA) family in Arabidopsis, AtCAMTA3 (also designated as AtSR1), was shown to suppress the expression of genes of the salicylic acid (SA) biosynthetic pathway thereby repressing SA‐dependent plant defence against bacteria and fungi (Galon et al., 2008; Du et al., 2009). Barley MLO, a PM‐resident protein, requires Ca2+/CaM association to compromise plant defence against powdery mildew (Kim et al., 2002). In addition, AtCaM7 was recently shown to interact and to co‐localize with ATP‐binding cassette (ABC) transporter PENETRATION 3 (PEN3) at the plasma membrane‐cytoplasm interface and may be involved in PEN3‐mediated nonhost resistance (Campe et al., 2016), whereas PEN3 focal accumulation was elicited upon MAMP perception or infection by adapted bacterial pathogens (Underwood & Somerville, 2013; Xin & He, 2013). However, the observation that functional SFI5 does not co‐localize with CaM at the PM potentially favours additional unknown proteins at the PM being targeted to suppress MTI.
There are few publications reporting an interaction of CaMs/CMLs with microbial effectors. Recently, HopE1 from P. syringae was discovered to interact with CaM and this interaction was required for further association with host microtubule‐associated protein 65 (MAP65). Upon association, MAP65 dissociates from the microtubule network which is thought to cause suppression of MAMP‐induced pathogenesis‐related protein secretion and enhanced susceptibility to bacterial infection (Guo et al., 2016). To date, it is unclear how HopE1 manipulates MAP65, but a conclusion of the authors was that CaM serves as a factor to activate HopE1 function in host cells. This is similar to our hypothesis that CaM is not the operative target of SFI5 but that SFI5 utilizes plant CaMs as positive regulators of its effector activity after translocation into the host cell. Given that SFI5 does not display specificity in binding distinct CaMs from tomato and Arabidopsis, it is possible that SFI5 association with CaM promotes the interaction with other CaM‐binding components. In this respect, the identification of potential additional targets might provide new findings on the mode of action SFI5.
The finding of a physical association of SFI5 with different CaM isoforms is the first direct link between an oomycete plant pathogen effector and components of Ca2+/CaM signalling in plants. Our current model predicts that SFI5 activation in host cells requires the association with CaM, in a Ca2+‐dependent manner, at the C‐terminal Pro222‐Leu239 alpha helix, triggering a conformational change of SFI5 conferring its ability to affect MAMP signal transduction pathways by manipulating one or several unknown membrane‐associated proteins, likely pattern recognition receptors and/or signalling components. Further molecular and biochemical studies are needed to dissect the specific mode of action of SFI5 in host cells and to unravel the molecular basis of the nonfunctionality of SFI5 in nonhost plants.
Author contributions
X.Z., N.W., P.C.B. and F.B. designed the experiments; X.Z., N.W., C.H., P.C.B. and H.M. performed the experiments; and X.Z., N.W., P.C.B., P.R.J.B., C.H. and F.B. wrote the manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Alignment of tomato and Arabidopsis CaMs.
Fig. S2 In vitro interaction between C‐terminal 17‐aa domain of SFI5 and His‐SlCaM6.
Fig. S3 The mutation KR/EE does not abolish GFP‐SFI5 ED plasma membrane localization in Nicotiana benthamiana leaves.
Fig. S4 The CaM binding motif of SFI5 is required for plasma membrane localization in tomato protoplasts.
Fig. S5 mRFP‐SlCaM3/4/5 and GFP‐SFI5 do not co‐localize in Nicotiana benthamiana leaves.
Fig. S6 Expression of HA‐tagged SFI5 deletion and site‐directed mutants in tomato protoplasts.
Table S1 List of primers
Table S2 Potential SFI5‐interacting proteins identified by LC‐MS/MS analysis
Acknowledgements
We acknowledge Jen Sheen (Massachusetts General Hospital and Harvard Medical School, Boston, USA) for providing the pFRK1‐Luc and AvrPto plasmid constructs. This work was supported by grants from the German Research Foundation (DFG: BR 3875/3‐1 to F.B.), China National Science Foundation (NSF 31701862 to X.Z.), the Biotechnology and Biological Sciences Research Council (BBSRC) (grants BB/G015244/1, BB/K018183/1, BB/L026880/1) and Scottish Government Rural and Environment Science and Analytical Services Division (RESAS). The funders had no role in study, design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Apel K, Hirt H. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology 55: 373–399. [DOI] [PubMed] [Google Scholar]
- Arndt C, Koristka S, Bartsch H, Bachmann M. 2012. Native polyacrylamide gels. Methods in Molecular Biology 869: 49–53. [DOI] [PubMed] [Google Scholar]
- Ausubel FM. 2005. Are innate immune signalling pathways in plants and animals conserved? Nature Immunology 6: 973–979. [DOI] [PubMed] [Google Scholar]
- Bhat RA, Miklis M, Schmelzer E, Schulze‐Lefert P, Panstruga R. 2005. Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proceedings of the National Academy of Sciences, USA 102: 3135–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H. 2015. Signalling mechanisms in pattern‐triggered immunity (PTI). Molecular Plant 8: 521–539. [DOI] [PubMed] [Google Scholar]
- Blakesley RW, Boezi JA. 1977. A new staining technique for proteins in polyacrylamide gels using Coomassie brilliant blue G250. Analytical Biochemistry 82: 580–582. [DOI] [PubMed] [Google Scholar]
- Block A, Alfano JR. 2011. Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Current Opinion in Microbiology 14: 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink PC, Wang X, McLellan H, He Q, Naqvi S, Armstrong MR, Zhang W, Hein I, Gilroy EM, Tian Z et al 2016. A Phytophthora infestans RXLR effector targets plant PP1c isoforms that promote late blight disease. Nature Communications 7: 10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. 2009. A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annual Review of Plant Biology 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Boller T, He SY. 2009. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JI, Armstrong MR, Gilroy EM, Boevink PC, Hein I, Taylor RM, Zhendong T, Engelhardt S, Vetukuri RR, Harrower B et al 2010. Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proceedings of the National Academy of Sciences, USA 107: 9909–9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JI, Kanneganti TD, Young C, Cakir C, Huitema E, Win J, Armstrong MR, Birch PR, Kamoun S. 2006. The C‐terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a‐mediated hypersensitivity and suppress INF1‐induced cell death in Nicotiana benthamiana . Plant Journal 48: 165–176. [DOI] [PubMed] [Google Scholar]
- Bouche N, Yellin A, Snedden WA, Fromm H. 2005. Plant‐specific calmodulin‐binding proteins. Annual Review of Plant Biology 56: 435–466. [DOI] [PubMed] [Google Scholar]
- Bozkurt TO, Schornack S, Banfield MJ, Kamoun S. 2012. Oomycetes, effectors, and all that jazz. Current Opinion in Plant Biology 15: 483–492. [DOI] [PubMed] [Google Scholar]
- Caillaud MC, Asai S, Rallapalli G, Piquerez S, Fabro G, Jones JD. 2013. A downy mildew effector attenuates salicylic acid‐triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biology 11: e1001732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campe R, Langenbach C, Leissing F, Popescu GV, Popescu SC, Goellner K, Beckers GJ, Conrath U. 2016. ABC transporter PEN3/PDR8/ABCG36 interacts with calmodulin that, like PEN3, is required for Arabidopsis nonhost resistance. New Phytologist 209: 294–306. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research 31: 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheval C, Aldon D, Galaud JP, Ranty B. 2013. Calcium/calmodulin‐mediated regulation of plant immunity. Biochimica et Biophysica Acta 1833: 1766–1771. [DOI] [PubMed] [Google Scholar]
- Chinpongpanich A, Wutipraditkul N, Thairat S, Buaboocha T. 2011. Biophysical characterization of calmodulin and calmodulin‐like proteins from rice, Oryza sativa L. Acta Biochimica et Biophysica Sinica 43: 867–876. [DOI] [PubMed] [Google Scholar]
- Cho KM, Nguyen HT, Kim SY, Shin JS, Cho DH, Hong SB, Shin JS, Ok SH. 2016. CML10, a variant of calmodulin, modulates ascorbic acid synthesis. New Phytologist 209: 664–678. [DOI] [PubMed] [Google Scholar]
- Crivici A, Ikura M. 1995. Molecular and structural basis of target recognition by calmodulin. Annual Review of Biophysics and Biomolecular Structure 24: 85–116. [DOI] [PubMed] [Google Scholar]
- Day IS, Reddy VS, Shad Ali G, Reddy AS. 2002. Analysis of EF‐hand‐containing proteins in Arabidopsis. Genome Biology 3: RESEARCH0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G, Requena N, Schaefer P, Brunner F, O'Connell R, Parker JE. 2014. Reprogramming of plant cells by filamentous plant‐colonizing microbes. New Phytologist 204: 803–814. [DOI] [PubMed] [Google Scholar]
- Dong S, Yin W, Kong G, Yang X, Qutob D, Chen Q, Kale SD, Sui Y, Zhang Z, Dou D et al 2011. Phytophthora sojae avirulence effector Avr3b is a secreted NADH and ADP‐ribose pyrophosphorylase that modulates plant immunity. PLoS Pathogens 7: e1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou D, Kale SD, Wang X, Jiang RH, Bruce NA, Arredondo FD, Zhang X, Tyler BM. 2008. RXLR‐mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen‐encoded machinery. Plant Cell 20: 1930–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou D, Zhou JM. 2012. Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host & Microbe 12: 484–495. [DOI] [PubMed] [Google Scholar]
- Du L, Ali GS, Simons KA, Hou J, Yang T, Reddy AS, Poovaiah BW. 2009. Ca2+/calmodulin regulates salicylic‐acid‐mediated plant immunity. Nature 457: 1154–1158. [DOI] [PubMed] [Google Scholar]
- Du Y, Berg J, Govers F, Bouwmeester K. 2015. Immune activation mediated by the late blight resistance protein R1 requires nuclear localization of R1 and the effector AVR1. New Phytologist 207: 735–747. [DOI] [PubMed] [Google Scholar]
- Fraiture M, Brunner F. 2014. Killing two birds with one stone: trans‐kingdom suppression of PAMP/MAMP‐induced immunity by T3E from enteropathogenic bacteria. Frontiers in Microbiology 5: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraiture M, Zheng X, Brunner F. 2014. An Arabidopsis and tomato mesophyll protoplast system for fast identification of early MAMP‐triggered immunity‐suppressing effectors. Methods in Molecular Biology 1127: 213–230. [DOI] [PubMed] [Google Scholar]
- Galon Y, Nave R, Boyce JM, Nachmias D, Knight MR, Fromm H. 2008. Calmodulin‐binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Letters 582: 943–948. [DOI] [PubMed] [Google Scholar]
- Gilroy EM, Taylor RM, Hein I, Boevink P, Sadanandom A, Birch PR. 2011. CMPG1‐dependent cell death follows perception of diverse pathogen elicitors at the host plasma membrane and is suppressed by Phytophthora infestans RXLR effector AVR3a. New Phytologist 190: 653–666. [DOI] [PubMed] [Google Scholar]
- Giraldo MC, Valent B. 2013. Filamentous plant pathogen effectors in action. Nature Reviews Microbiology 11: 800–814. [DOI] [PubMed] [Google Scholar]
- Guo M, Kim P, Li G, Elowsky CG, Alfano JR. 2016. A bacterial effector co‐opts calmodulin to target the plant microtubule network. Cell Host & Microbe 19: 67–78. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Kamoun S, Zody MC, Jiang RH, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto‐Alalibo T et al 2009. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature 461: 393–398. [DOI] [PubMed] [Google Scholar]
- Halter T, Imkampe J, Mazzotta S, Wierzba M, Postel S, Bucherl C, Kiefer C, Stahl M, Chinchilla D, Wang X et al 2014. The leucine‐rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Current Biology 24: 134‐143. [DOI] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nürnberger T, Sheen J. 2006. Specific bacterial suppressors of MAMP signalling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125: 563–575. [DOI] [PubMed] [Google Scholar]
- Jiang RH, Tripathy S, Govers F, Tyler BM. 2008. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proceedings of the National Academy of Sciences, USA 105: 4874–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P. 2005. Modular cloning and expression of tagged fluorescent protein in plant cells. Trends in Plant Sciences 10: 103–105. [DOI] [PubMed] [Google Scholar]
- Kim MC, Panstruga R, Elliott C, Muller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze‐Lefert P. 2002. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416: 447–451. [DOI] [PubMed] [Google Scholar]
- King SR, McLellan H, Boevink PC, Armstrong MR, Bukharova T, Sukarta O, Win J, Kamoun S, Birch PR, Banfield MJ. 2014. Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKK epsilon to suppress plant immune signalling. Plant Cell 26: 1345–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Qiu X, Kang J, Wang Y, Chen H, Huang J, Qiu M, Zhao Y, Kong G, Ma Z. 2017. A Phytophthora effector manipulates host histone acetylation and reprograms defense gene expression to promote infection. Current Biology 27: 981–991. [DOI] [PubMed] [Google Scholar]
- Kong G, Zhao Y, Jing M, Huang J, Yang J, Xia Y, Kong L, Ye W, Xiong Q, Qiao Y et al 2015. The activation of Phytophthora effector Avr3b by plant cyclophilin is required for the nudix hydrolase activity of Avr3b. PLoS Pathogens 11: e1005139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurup S, Runions J, Kohler U, Laplaze L, Hodge S, Haseloff J. 2005. Marking cell lineages in living tissues. Plant Journal 42: 444–453. [DOI] [PubMed] [Google Scholar]
- Lecourieux D, Ranjeva R, Pugin A. 2006. Calcium in plant defence‐signalling pathways. New Phytologist 171: 249–269. [DOI] [PubMed] [Google Scholar]
- Lee J, Eschen‐Lippold L, Lassowskat I, Bottcher C, Scheel D. 2015. Cellular reprogramming through mitogen‐activated protein kinases. Frontiers in Plant Science 6: 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zipfel C. 2014. Plant PRRs and the activation of innate immune signalling. Molecular Cell 54: 263–272. [DOI] [PubMed] [Google Scholar]
- McCormack E, Tsai YC, Braam J. 2005. Handling calcium signalling: Arabidopsis CaMs and CMLs. Trends in Plant Science 10: 383–389. [DOI] [PubMed] [Google Scholar]
- McLellan H, Boevink PC, Armstrong MR, Pritchard L, Gomez S, Morales J, Whisson SC, Beynon JL, Birch PR. 2013. An RxLR effector from Phytophthora infestans prevents re‐localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathogens 9: e1003670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meador WE, Means AR, Quiocho FA. 1992. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin‐peptide complex. Science 257: 1251–1255. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA Jr. 1997. Innate immunity: the virtues of a nonclonal system of recognition. Cell 91: 295–298. [DOI] [PubMed] [Google Scholar]
- Mruk K, Farley BM, Ritacco AW, Kobertz WR. 2014. Calmodulation meta‐analysis: predicting calmodulin binding via canonical motif clustering. Journal of General Physiology 144: 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepmann M, Zheng J. 2006. Discontinuous native protein gel electrophoresis. Electrophoresis 27: 3949–3951. [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Brunner F. 2002. Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen‐associated molecular patterns. Current Opinion in Plant Biology 5: 318–324. [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Brunner F, Kemmerling B, Piater L. 2004. Innate immunity in plants and animals: striking similarities and obvious differences. Immunological Reviews 198: 249–266. [DOI] [PubMed] [Google Scholar]
- O'Neil KT, DeGrado WF. 1990. How calmodulin binds its targets: sequence independent recognition of amphiphilic α‐helices. Trends in Biochemical Sciences 15: 59–64. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Davidse LC. 1990. Attempted cloning of the Phytophthora infestans pyr‐gene for use in a homologous transformation system. Phytophthora Newsletter 16: 7–8. [Google Scholar]
- Reddy AS, Ali GS, Celesnik H, Day IS. 2011. Coping with stresses: roles of calcium‐ and calcium/calmodulin‐regulated gene expression. Plant Cell 23: 2010–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Thara VK, Martin GB, Zhou JM, Tang X. 2000. The pseudomonas AvrPto protein is differentially recognized by tomato and tobacco and is localized to the plant plasma membrane. Plant Cell 12: 2323–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL et al 2006. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313: 1261–1266. [DOI] [PubMed] [Google Scholar]
- Underwood W, Somerville SC. 2013. Perception of conserved pathogen elicitors at the plasma membrane leads to relocalization of the Arabidopsis PEN3 transporter. Proceedings of the National Academy of Sciences, USA 110: 12492–12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S et al 2007. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450: 115–118. [DOI] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Wang S, Birch PRJ. 2016. The cell biology of late blight disease. Current Opinion in Microbiology 34: 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XF, He SY. 2013. Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signalling in plants. Annual review of Phytopathology 51: 473–498. [DOI] [PubMed] [Google Scholar]
- Yang L, McLellan H, Naqvi S, He Q, Boevink PC, Armstrong M, Giuliani LM, Zhang W, Tian Z, Zhan J et al 2016. Potato NPH3/RPT2‐like protein StNRL1, targeted by a Phytophthora infestans RXLR effector, is a susceptibility factor. Plant Physiology 171: 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Kim J, Truong K, Sherman M, Yuan T, Ikura M. 2000. Calmodulin target database. Journal of Structural and Functional Genomics 1: 8–14. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhou RG, Gao YJ, Zheng SZ, Xu P, Zhang SQ, Sun DY. 2009. Molecular and genetic evidence for the key role of AtCaM3 in heat‐shock signal transduction in Arabidopsis. Plant Physiology 149: 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Liu W, Xu YP, Cao JY, Braam J, Cai XZ. 2013. Genome‐wide identification and functional analyses of calmodulin genes in Solanaceous species. BMC Plant Biology 13: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, McLellan H, Fraiture M, Liu X, Boevink PC, Gilroy EM, Chen Y, Kandel K, Sessa G, Birch PR et al 2014. Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22‐triggered immunity. PLoS Pathogens 10: e1004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Alignment of tomato and Arabidopsis CaMs.
Fig. S2 In vitro interaction between C‐terminal 17‐aa domain of SFI5 and His‐SlCaM6.
Fig. S3 The mutation KR/EE does not abolish GFP‐SFI5 ED plasma membrane localization in Nicotiana benthamiana leaves.
Fig. S4 The CaM binding motif of SFI5 is required for plasma membrane localization in tomato protoplasts.
Fig. S5 mRFP‐SlCaM3/4/5 and GFP‐SFI5 do not co‐localize in Nicotiana benthamiana leaves.
Fig. S6 Expression of HA‐tagged SFI5 deletion and site‐directed mutants in tomato protoplasts.
Table S1 List of primers
Table S2 Potential SFI5‐interacting proteins identified by LC‐MS/MS analysis


