Abstract
Aberrant precursor mRNA splicing plays a pivotal role in liver diseases. However, roles of splicing regulators in alcoholic liver disease are unknown. Herein, we investigated a splicing regulator, Slu7, in the development of alcoholic steatohepatitis. Adenovirus-mediated alteration of hepatic Slu7 expression in mice pair fed either with or without (as control) ethanol in their diet was used. Knockdown of hepatic Slu7 by adenovirus-Slu7shRNA treatment ameliorated inflammation and attenuated liver injury in mice after ethanol administration. Mechanistically, reducing liver Slu7 expression increased the expression of sirtuin 1 (SIRT1) full-length and repressed the splicing of SIRT1 into SIRT1-ΔExon8 isoform in ethanol-fed mice. Knockdown of hepatic Slu7 in the ethanol-fed mice also ameliorated splicing of lipin-1 and serine/arginine-rich splicing factor 3 (Srsf3). In concordance with ameliorated splicing of SIRT1, lipin-1, and Srsf3, knockdown of hepatic Slu7 inhibited the activity of NF-κB, normalized iron and zinc homeostasis, reduced oxidative stress, and attenuated liver damage in ethanol-fed mice. In addition, hepatic Slu7 was significantly elevated in patients with alcoholic steatohepatitis. Our present study illustrates a novel role of Slu7 in alcoholic liver injury and suggests that dysregulated Slu7 may contribute to the pathogenesis of human alcoholic steatohepatitis.
Alcoholic steatohepatitis is a prevalent early form of alcoholic liver disease (ALD), which can progress to fibrosis, cirrhosis, hepatocellular carcinoma, and liver failure in clinical patients.1 Despite intensive studies over the past decades, the pathogenic mechanisms of alcoholic steatohepatitis remain incompletely understood, and patients with alcoholic steatohepatitis lack effective treatments.
Alternative splicing of precursor mRNA (pre-mRNA) is a vital step in gene expression that eliminates the introns and ligates the exons to form mature mRNAs that can be translated into proteins.2, 3 Defects in pre-mRNA splicing have been implicated in various human diseases, including liver diseases.3 In recent years, aberrant mRNAs and dysregulated splicing regulators in liver diseases are emerging as targets for new therapeutic interventions.3 However, alternative pre-mRNA splicing is an underappreciated mechanism in the pathogenesis of ALD.
The pre-mRNA splicing is catalyzed in two steps by spliceosome, a large ribonucleoprotein complex.4, 5, 6, 7 The splicing regulator Slu7 (human homolog hSlu7) participates in the selection of the 3′ site during the second step of splicing.4, 5, 6, 7, 8, 9 Human homolog hSlu7 protein has three functional domains, a zinc-knuckle motif, a lysine-rich region, and a nuclear localization signal, that play distinctive roles in determining its subcellular localization.8, 9 Slu7 is the only second step splicing protein with a potential zinc-binding domain in the form of a zinc-knuckle motif, suggesting the potential involvement of Slu7 in regulation of metabolism of metals, such as zinc and iron.4, 5, 6, 7, 8, 9, 10, 11
Slu7 not only functions as a splicing regulator but also plays an essential coregulatory role for maintaining liver functions.8, 9, 10, 11 In liver, Slu7 participates in the regulation of splicing and expression of multiple genes implicated in lipid metabolism, inflammation, development, and progression of fibrosis and cirrhosis.11 Defective Slu7 is likely to contribute to metabolic diseases, such as nonalcoholic fatty liver diseases, because experimental knockdown of Slu7 disrupted the gene expression involved in lipogenesis and impaired physiological metabolic responses in mouse liver.11
Sirtuin (SIRT) 1, an NAD+-dependent class III protein deacetylase, is an essential player in controlling the pathways of inflammatory responses and lipid metabolism.12 SIRT1 displays functional diversity, with its deacetylation activity fine-tuned via multiple complicated mechanisms.12 Emerging evidence demonstrates that alternative splicing of SIRT1 pre-mRNA produces several variants, such as SIRT1 full length (SIRT1-FL) and SIRT1-ΔExon8.13, 14, 15 SIRT1-ΔExon8 isoform lacks exon 8 because of in-frame splicing between exons 7 and 9 of SIRT1-FL RNA.13, 14 Because exon 8 encodes the deacetylase domain of SIRT1, the exclusion of exon 8 markedly attenuates SIRT1-ΔExon8 deacetylase activity.13, 14
Abnormality of SIRT1 and its signaling has been implicated in the pathogenesis of alcoholic steatosis/steatohepatitis.15 However, whether and how ethanol exposure influences alternative splicing of SIRT1 pre-mRNA and contributes to liver damage are currently unknown. Interestingly, SIRT1 is a target gene of Slu7 in cultured human hepatic cells, suggesting that Slu7 may regulate SIRT1 splicing.11
Lipin-1 regulates lipid metabolism and inflammation process via functioning as a phosphatidic acid phosphohydrolase enzyme and as a transcriptional coregulator.16 Lipin-1 pre-mRNA splicing produces two isoforms, lipin-1α and lipin-1β.16, 17 Lipin-1α is generated through skipping exon 7 from lipin-1β.17 Dysregulated lipin-1 and its pre-mRNA splicing have been implicated in metabolic diseases, including obesity.17, 18 Ethanol exposure inhibits lipin-1 pre-mRNA splicing by inhibiting lipin-1α generation, inducing the ratio of Lpin1 β/α in the livers of mice and of patients with alcoholic hepatitis.19, 20 More important, aberrant SIRT1 signaling is associated with disruption of lipin-1 pre-mRNA splicing by ethanol in mice.19
Serine/arginine-rich splicing factor 3 (Srsf3; alias SRp20) belongs to the highly conserved family of SR proteins.21 Srsf3 pre-mRNA alternative splicing generates a full-length isoform lacking exon 4 (Iso1) and an alternative isoform including exon 4 (Iso2).21, 22, 23 Slu7 regulates Srsf3 pre-mRNA splicing.11 Furthermore, errors in Slu7-Srsf3 axis contribute to abnormality of liver functions in rodents and humans.11, 21, 22, 23
We tested a hypothesis in the present study that dysregulated expression of liver Slu7 might be pivotal in the development of alcoholic steatohepatitis. Using adenovirus (Ad)–mediated gene alteration of hepatic Slu7 expression in a chronic-plus-binge ethanol feeding mouse model,24 we demonstrate, for the first time, that knockdown of Slu7 ameliorates the ethanol-induced inflammation and attenuates alcoholic liver injury in mice via modifying pre-mRNA splicing of SIRT1, lipin-1, and Srsf3.
Materials and Methods
Reagents, Plasmids, and Antibodies
Most supplies and chemicals were purchased from Sigma Chemical (St. Louis, MO), Schleicher and Schuell (Keene, NH), Gibco-BRL (Grand Island, NY), and DuPont NEN Research Products (Boston, MA). Murine vectors expressing Ad–green fluorescent protein (GFP; control), Ad-Slu7 [adenoviral type 5 (dE1/E3); promoter, cytomegalovirus], Ad-shRNA (control), and Ad-shSlu7 [adenoviral type 5 (dE1/E3); promoter, U6] were custom designed and obtained from Vector BioLabs, Inc. (Malvern, PA). Antibodies against Slu7, Srsf3, nuclear factor of activated T cells 4 (NFATc4), and acetylated forkhead box protein O 1 were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX). Acetylated NF-κB, NF-κB, and SIRT1 antibodies were purchased from Cell Signaling (Danvers, MA).
Animal Studies
Hepatocyte-specific SIRT1 knockout mice were kindly gifted by Dr. Xiaoling Li (National Institute of Environmental Health Sciences/NIH, Research Triangle Park, NC).19 A chronic-plus-binge mouse model (a single binge after chronic ethanol consumption) was used.24 Mice were fed with liquid diets providing 1 kcal/mL, which were freshly prepared daily from powder with Lieber-DeCarli formulation (Frenchtown, NJ). Female SIRT1 knockout and wild-type mice (10 to 12 weeks old) were divided into pair-fed dietary groups: ethanol and control (without ethanol). During the first 10 days, diet in the ethanol groups contained 5% ethanol, whereas the control mice were pair fed with same calories of dextrin maltose. At day 11, mice were gavaged either a single dose of ethanol [5 g/kg body weight, 31.5% ethanol (ethanol group)] or an isocaloric dose of dextrin maltose (control group). The mice were euthanized, and blood and tissue samples were collected 9 hours after gavage.
In vivo Ad-mediated gene transfer experiments were performed, as described previously.25, 26 During the 10 days of the chronic-plus-binge ethanol feeding period, overexpression of Slu7 or knockdown of Slu7 in female C57BL/6J mice was accomplished via retro-orbital injection of Ad-GFP, Ad-Slu7, Ad-shRNA, and Ad-shSlu7 (0.5 to 1.0 × 109 active viral particles in 200 μL phosphate-buffered saline) to mice twice on days 1 and 5. On the final day, mice were sacrificed, and tissue (liver and/or serum) samples were harvested. All of the animal experiments were approved by the Institutional Animal Care and Use Committee at Northeast Ohio Medical University.
Ad-Mediated Gene Transfer in Cultured Cells
Mouse AML-12 hepatocytes (ATCC, Manassas, VA) were cultured in Dulbecco’s modified Eagle’s medium/F12 medium supplemented with 100 μg/mL streptomycin, 10% fetal bovine serum, 0.1 μmol/L dexamethasone, 63 μg/mL penicillin G, and insulin-transferrin-selenium (Gibco-BRL). They were seeded in 6-well plates for 24 hours. On the day of transfection, cells were washed and switched to reduced serum medium (Invitrogen, Carlsbad, CA) and then infected with Ad-GFP, Ad-Slu7, Ad-shRNA, and Ad-shSlu7 for 36 to 48 hours.27
Western Blot Analysis
Western blot analyses were performed on liver tissue extract with 10% Bio-Rad MiniPROTEIN Tetra Cell system and then transferred to polyvinylidene difluoride membranes with Bio-Rad Blotting Module (Bio-Rad Laboratories, Hercules, CA).27, 28 The signals obtained for liver protein extracts were normalized by using polyclonal rabbit anti-actin β antibody (Sigma, Ronkonkoma, NY). Quantification and analysis of multiple protein bands were performed using AlphaView software version 3.4 (ProteinSimple, San Jose, CA).
Lipid Peroxidation Assays
The lipid peroxidation end product, malondialdehyde (MDA), in liver was measured on the basis of the formation of thiobarbituric reactive substances and expressed as the extent of MDA production.28
Assessment of Hepatic Lipid Accumulation
Liver tissues were homogenized in ice-cold phosphate-buffered saline. Total lipids were extracted with chloroform/methanol. Liver triglyceride and cholesterol contents were measured with BioVision assay kits (BioVision, Milpitas, CA), as described previously.27, 28
MPO Activity Assay
Liver myeloperoxidase (MPO) activity was measured using assay kits (Abcam, Cambridge, MA), as previously reported.28
Histopathological Analysis
Liver tissues were fixed in 10% formalin and embedded in paraffin.27, 28 Liver sections were stained with hematoxylin and eosin.
Serum Biochemical Assays
Serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were measured by using the MaxDiscovery ALT and AST Assay Kits (Bio Scientific, Austin, TX). Serum triglyceride and cholesterol were measured by using the Infinity Cholesterol and Triglyceride Reagents (Thermo Fisher Scientific, Waltham, MA). All analyses were performed using a SpectraMax i3× microplate reader (Molecular Devices, Sunnyvale, CA).
Real-Time Quantitative PCR Analysis
Liver total RNA was prepared using Trizol extraction method (Invitrogen). Real-time quantitative PCR was performed in an iCycler Spectrofluorometric thermal cycler (Bio-Rad Laboratories).27, 28 The relative amount of target mRNA was calculated using the comparative threshold cycle (CT) method by normalizing target mRNA CT values to those for glyceraldehyde-3-phosphate dehydrogenase. Gene-specific primer sets were either purchased or designed using Harvard's PrimerBank (http://pga.mgh.harvard.edu/primerbank, last accessed May 5, 2018) from Qiagen SuperArray Bioscience (Frederick, MD) (Table 1).
Table 1.
Primers Used in This Study
| Primer name | Direction | Sequence |
|---|---|---|
| Human GAPDH | Forward | 5′-AGGGCTGCTTTTAACTCTGGT-3′ |
| Reverse | 5′-CCCCACTTGATTTTGGAGGGA-3′ | |
| Human SLU7 | Forward | 5′-AAGAGGGATCGGTGGAATGG-3′ |
| Reverse | 5′-GGCTTTCAATGTTCGTTTTGCC-3′ | |
| Human SRSF3-ISO1 | Forward | 5′-GTGGCTGCCGTGTAAGAGTGG-3′ |
| Reverse | 5′-TCTCTTCTCCTATCTCTAGAAAG-3′ | |
| Human SRSF3-ISO2 | Forward | 5′-CTGACGACTGGCCAGCCTGG-3′ |
| Reverse | 5′-GTGGCTGCCGTGTAAGAGTGG-3′ | |
| E-selectin | Forward | 5′-AGCAGAGTTTCACGTTGCAGG-3′ |
| Reverse | 5′-TGGCGCAGATAAGGCTTCA-3′ | |
| GAPDH | Forward | 5′-TGACCTCAACTACATGGTCTACA-3′ |
| Reverse | 5′-CTTCCCATTCTCGGCCTTG-3′ | |
| iNOS | Forward | 5′-GTTCTCAGCCCAACAATACAAGA-3′ |
| Reverse | 5′-GTGGACGGGTCGATGTCAC-3′ | |
| IL-1β | Forward | 5′-TCGCTCAGGGTCACAAGAAA-3′ |
| Reverse | 5′-CATCAGAGGCAAGGAGGAAAAC-3′ | |
| IL-6 | Forward | 5′-TAGTCCTTCCTACCCCAATTTCC-3′ |
| Reverse | 5′-TTGGTCCTTAGCCACTCCTTC-3′ | |
| ICAM-1 | Forward | 5′-CAATTTCTCATGCCGCACAG-3′ |
| Reverse | 5′-AGCTGGAAGATCGAAAGTCCG-3′ | |
| Lipin-1α | Forward | 5′-GGTCCCCCAGCCCCAGTCCTT-3′ |
| Reverse | 5′-GCAGCCTGTGGCAATTCA-3′ | |
| Lipin-1ß | Forward | 5′-CAGCCTGGTAGATTGCCAGA-3′ |
| Reverse | 5′-GCAGCCTGTGGCAATTCA-3′ | |
| Ly6G | Forward | 5′-CTTCTCTGATGGATTTTGCGTTG-3′ |
| Reverse | 5′-AGTAGTGGGGCAGATGGGAAG-3′ | |
| MCP-1 | Forward | 5′-TTAAAAACCTGGATCGGAACCAA-3′ |
| Reverse | 5′-GCATTAGCTTCAGATTTACGGGT-3′ | |
| MIP-1α | Forward | 5′-TGAGAGTCTTGGAGGCAGCGA-3′ |
| Reverse | 5′-TGTGGCTACTTGGCAGCAAACA-3′ | |
| MIP-1β | Forward | 5′-AACACCATGAAGCTCTGCGT-3′ |
| Reverse | 5′-AGAAACAGCAGGAAGTGGGA-3′ | |
| Slu7 | Forward | 5′-AGTTAGTGCGACACCCATGAC-3′ |
| Reverse | 5′-CAGGAGCATTGCCCAGTTT-3′ | |
| Sirt1-full | Forward | 5′-TGACCGATGGACTCCTCACT-3′ |
| Reverse | 5′-ATTGTTCGAGGATCGGTGCC-3′ | |
| Sirt1-Δ8 | Forward | 5′-TAGCACTAATTCCAAGTAATC-3′ |
| Reverse | 5′-TTATGATTTGTCTGATGGATAGTTT-3′ | |
| Srsf3-Iso1 | Forward | 5′-GTGGCTGCCGTGTAAGAGTGG-3′ |
| Reverse | 5′-TCTCTTCTCCTATCTCTAGAAAG-3′ | |
| Srsf3-Iso2 | Forward | 5′-GTGGCTGCCGTGTAAGAGTGG-3′ |
| Reverse | 5′-CTGACGACTGGCCAGCCTGG-3′ | |
| TNF-α | Forward | 5′-AGGCTGCCCCGACTACGT-3′ |
| Reverse | 5′-GACTTTCTCCTGGTATGAGATAGCAAA-3′ | |
| VCAM-1 | Forward | 5′-TGAACCCAAACAGAGGCAGAGT-3′ |
| Reverse | 5′-GGTATCCCATCACTTGAGCAGG-3′ |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ICAM-1, intercellular adhesion molecule-1; iNOS, inducible nitric oxide synthase; ISO1/Iso1, full-length isoform lacking exon 4; ISO2/Iso2, alternative isoform including exon 4; MCP-1, monocyte chemoattractant protein-1; MIP, macrophage inflammatory protein; Sirt, sirtuin; SRSF3/Srsf3, serine/arginine-rich splicing factor 3; TNF-α, tumor necrosis factor-α; VCAM1, vascular cell adhesion molecule-1.
Assessment of Liver Mineral Status
Liver iron concentrations were determined using the Iron Assay Kit (Abcam).28 Mineral contents in the liver tissues were analyzed by inductively coupled plasma mass spectroscopy at the Center for Translational Biomedical Research, North Carolina Research Campus (University of North Carolina at Greensboro).
RT-PCR Analysis
Separate reactions for inclusion and exclusion isoforms were performed to evaluate splicing. Real-time quantitative RT-PCR was performed with primers specific for mouse Sirt1 exon 7 to exon 9.13, 14 PCR products were visualized on a UV transilluminator using 2% agarose gel electrophoresis and 200 ng/mL ethidium bromide. Sirt1 splice-variant–specific primer sequences were as follows: Sirt1-Exon 7, 5′-GATGAAGTTGACCTCCTCATTG-3′ (forward); and Sirt1-Exon 9, 5′-TTATCATTTGTCTGATGGATAGTTT-3′ (reverse). Results represent at least three independent experiments.
Human Alcoholic Steatohepatitis Samples and Normal Human Livers
Normal human liver and alcoholic steatohepatitis (with fibrosis) samples were obtained, as described previously,29 from the Liver Tissue Procurement and Distribution System at University of Minnesota, which obtained informed consent before sample collection and distribution. The inclusion criteria were also described previously.29 Alcoholic steatohepatitis was diagnosed on the basis of liver histology and the history of alcohol drinking. Normal healthy liver samples were also obtained from the Liver Tissue Procurement and Distribution System, collected from the part of donor livers that was not used for transplantation. The use of human tissue samples was approved by the institutional review board at Northeast Ohio Medical University.
Statistical Analysis
Statistical analysis was performed using one-way analysis of variance test, followed by the post-hoc Bonferroni test or a two-tailed unpaired t-test (GraphPad Prism, San Diego, CA). All values are expressed as means ± SEM. Differences were considered statistically significant at P < 0.05.
Results
Ad-Mediated Hepatic Slu7 Knockdown Attenuates Liver Injury in the Ethanol-Fed Mice
The role of Slu7 in experimental alcoholic steatohepatitis was examined using in vivo gain-of-function or loss-of-function strategies in mice.23 Hepatic Slu7 mRNA abundance was reduced significantly by approximately 30% in the mice injected with Ad-shSlu7 in comparison with mice injected with Ad-GFP (Supplemental Figure S1A). Hepatic Slu7 mRNA was increased by approximately 1.8-fold in mice injected with Ad-Slu7 compared with control mice (Supplemental Figure S1B). Chronic-plus-binge ethanol feeding induced liver injury but had no statistically significant effect on the body weights of mice treated with Ad-Slu7 or Ad-shSlu7 (Tables 2 and 3).
Table 2.
Selected Parameters in Slu7 Knockdown Mice after Ethanol Feeding
| Parameters | Ad-shRNA | Ad-shRNA + ethanol | Ad-shSlu7 | Ad-shSlu7 + ethanol |
|---|---|---|---|---|
| Starting body, g | 20.16 ± 0.4 | 20.53 ± 0.3 | 19.96 ± 0.4 | 20.54 ± 0.2 |
| Final body, g | 23.32 ± 0.3 | 21.61 ± 0.5 | 22.92 ± 0.4 | 21.77 ± 0.35 |
| Liver, g | 1.02 ± 0.02 | 1.00 ± 0.03 | 0.93 ± 0.02 | 0.95 ± 0.03 |
| Liver/body, % | 4.3 ± 0.05 | 4.6 ± 0.09∗ | 4.0 ± 0.05 | 4.4 ± 0.09∗ |
| Liver cholesterol, mg/mg protein | 1.04 ± 0.04 | 0.98 ± 0.2 | 1.02 ± 0.03 | 0.99 ± 0.1 |
| Serum | ||||
| Triglycerides, mg/dL | 40.68 ± 1.62 | 67.73 ± 2.68∗ | 34.61 ± 2.17 | 63.05 ± 2.55∗ |
| Cholesterol, mg/dL | 87.52 ± 7.7 | 111.63 ± 4.0∗ | 99.37 ± 6.5 | 115.54 ± 4.0∗ |
| β-Hydroxybutyrate, mmol/L | 0.20 ± 0.02 | 1.74 ± 0.3∗ | 0.20 ± 0.01 | 0.9 ± 0.1∗† |
Female C57BL/6J mice, aged 10 to 12 weeks, were divided into four groups, as follows: pair-fed Ad-shRNA control; ethanol-fed Ad-shRNA; pair-fed Ad-shSlu7 control; and ethanol-fed Ad-shSlu7. Data are expressed as means ± SEM. n = 5 to 12 animals.
P < 0.05 versus pair-fed Ad-shRNA controls.
P < 0.05 versus ethanol-fed Ad-shRNA.
Table 3.
Selected Parameters in Slu7 Overexpression Mice after Ethanol Feeding
| Parameters | Ad-GFP | Ad-GFP + ethanol | Ad-Slu7 | Ad-Slu7 + ethanol |
|---|---|---|---|---|
| Starting body, g | 20.13 ± 0.3 | 20.73 ± 0.3 | 20.46 ± 0.3 | 20.20 ± 0.3 |
| Final body, g | 22.4 ± 0.2 | 20.93 ± 0.46 | 22.11 ± 0.7 | 20.46 ± 0.5 |
| Liver, g | 0.93 ± 0.06 | 1.04 ± 0.01 | 0.97 ± 0.03 | 0.99 ± 0.02 |
| Liver/body, % | 4.0 ± 0.003 | 5.1 ± 0.001∗ | 4.0 ± 0.001 | 5.0 ± 0.001∗ |
| Liver cholesterol, mg/mg protein | 1.16 ± 0.09 | 1.33 ± 0.04 | 1.36 ± 0.08 | 1.27 ± 0.05 |
| Serum | ||||
| Triglycerides, mg/dL | 43.43 ± 3.21 | 78.19 ± 3.66∗ | 35.42 ± 2.49 | 79.41 ± 4.04∗ |
| Cholesterol, mg/dL | 73.2 ± 10.95 | 130.96 ± 6.41∗ | 98.4 ± 6.01 | 94.41 ± 8.11 |
| β-Hydroxybutyrate, mmol/L | 0.25 ± 0.03 | 1.26 ± 0.1∗ | 0.24 ± 0.02 | 1.83 ± 0.1∗† |
Female C57BL/6J mice, aged 10 to 12 weeks, were divided into four groups, as follows: pair-fed Ad-GFP control; ethanol-fed Ad-GFP; pair-fed Ad-Slu7 control; and ethanol-fed Ad-Slu7. Data are expressed as means ± SEM. n = 5 to 12 animals.
P < 0.05 versus pair-fed Ad-GFP controls.
P < 0.05 versus ethanol-fed Ad-GFP.
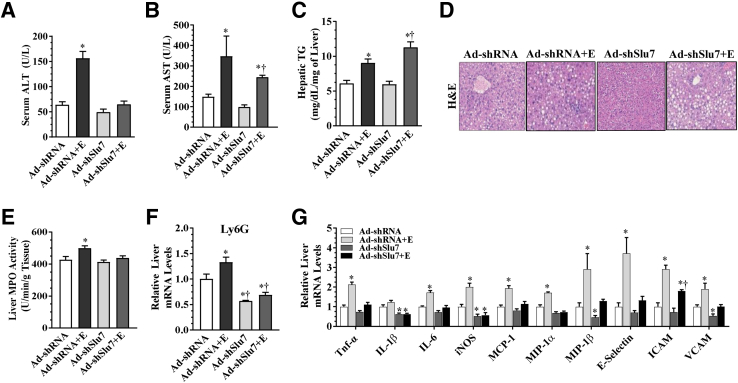
Ethanol feeding caused steatohepatitis with significantly higher levels of hepatic triglyceride, accumulation of lipid droplets in liver, and elevated levels of ALT and AST, compared with the pair-fed control mice (Figure 1, A–D, and Supplemental Figure S2, A–D). Remarkably, Ad-mediated Slu7 knockdown largely normalized the ethanol-associated elevation of serum ALT and AST but slightly exacerbated the ethanol-induced steatosis in mice (Figure 1, A–D). On the contrary, Ad-mediated overexpression of Slu7 augmented serum ALT and AST levels but mildly reduced hepatic lipid accumulation in the livers of ethanol-fed mice (Supplemental Figure S2, A–D). There were no significant differences among these groups in hepatic cholesterol content (Tables 2 and 3). Slu7 knockdown significantly augmented ethanol-elevated serum levels of β-hydroxybutyrate, whereas the elevated serum β-hydroxybutyrate in the ethanol-fed mice was attenuated by Slu7 overexpression (Tables 2 and 3). Collectively, these results demonstrate that hepatic Slu7 knockdown attenuates liver injury in mice that received ethanol.
Figure 1.
Ad-mediated hepatic Slu7 knockdown attenuates liver injury but slightly aggravates steatosis in mice after ethanol administration. Female C57BL/6J mice were pair fed either a control diet or an ethanol (E)–containing diet for 10 days, followed by single gavage of ethanol. During the 10-day chronic-plus-binge ethanol feeding period, Ad-shSlu7 or Ad-shRNA control (0.5 to 1.0 × 109 active viral particles in 200 μL of phosphate-buffered saline) was given to mice twice on days 1 and 5. A: Serum alanine aminotransferase (ALT). B: Serum aspartate aminotransferase (AST). C: Hepatic triglyceride (TG) content. D: Hematoxylin and eosin (H&E) staining of liver sections. E: Liver myeloperoxidase (MPO) activity. F: Relative liver mRNA levels of Ly6G. G: Relative liver mRNA levels of tumor necrosis factor-α (TNF-α), IL-1β, IL-6, inducible nitric oxide synthase (iNOS), monocyte chemoattractant protein (MCP)-1, macrophage inflammatory protein (MIP)-1α, MIP-1β, E-selectin, intercellular adhesion molecule (ICAM), and vascular cell adhesion molecule (VCAM). Data are expressed as means ± SEM (A–C and E–G). n = 5 to 12 mice. ∗P < 0.05 versus pair-fed Ad-shRNA controls; †P < 0.05 versus ethanol-fed Ad-shRNA. Original magnification, ×40 (D).
Slu7 Knockdown Ameliorates Hepatic Inflammation in the Ethanol-Fed Mice
Ethanol feeding caused neutrophilic inflammation in mice compared with pair-fed control mice, as revealed by increased hepatic MPO activity, and induced mRNA expression of Ly6G, a marker of neutrophilic inflammation (Figure 1, E and F). Knockdown of Slu7 abolished the ability of ethanol to increase hepatic neutrophilic inflammation in mice by attenuating hepatic MPO activity and reducing Ly6G gene expression (Figure 1, E and F). Congruently, Slu7 knockdown largely abolished the ethanol-mediated increases in mRNA abundances of a panel of hepatic inflammation mediators in mice, including IL-1β, tumor necrosis factor-α, IL-6, inducible nitric oxide synthase, macrophage inflammatory protein-1α, macrophage inflammatory protein-1β, monocyte chemoattractant protein-1, E-selectin, vascular cell adhesion molecule, and intercellular adhesion molecule (Figure 1G). Conversely, in comparison with pair-fed control mice, hepatic overexpression of Slu7 provoked inflammation response with elevated hepatic MPO activity in the ethanol-fed mice (Supplemental Figure S2E). Taken together, the results clearly demonstrate that knockdown of Slu7 ameliorates ethanol-provoked hepatic inflammation in mice.
Slu7 Promotes Production of SIRT1-ΔExon8 in Cultured AML-12 Hepatocytes
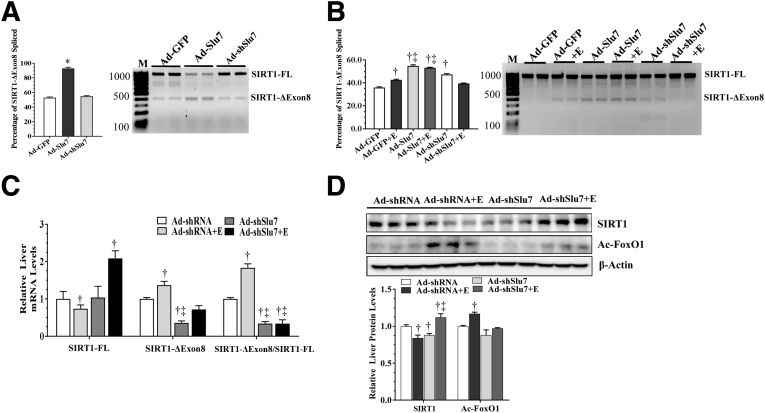
It was investigated whether Slu7 was involved in regulating SIRT1 pre-mRNA splicing by infecting mouse AML-12 hepatocytes with Ad-GFP, Ad-Slu7, and Ad-shSlu7, and the levels of SIRT1-FL and SIRT1-ΔExon8 were measured in these cells (Supplemental Figure S3). Overexpression of Slu7 in AML-12 cells markedly stimulated SIRT1 exon 8 skipping, as indicated by higher levels of SIRT1-ΔExon8 and lower levels of SIRT1-FL mRNA (Figure 2A). Conversely, Slu7 knockdown in AML-12 cells did not induce SIRT1 exon 8 skipping (Figure 2A). These results indicate that SIRT1 pre-mRNA undergoes alternative splicing via, at least in part, Slu7-mediated mechanisms in cultured AML-12 hepatocytes.
Figure 2.
Ad-mediated knockdown of hepatic Slu7 regulates hepatic sirtuin (SIRT) 1 pre-mRNA splicing in mice after ethanol administration. A: Mouse AML-12 hepatocytes were infected with Ad-GFP, Ad-Slu7, and Ad-shSlu7. Relative abundance of SIRT1-ΔExon8 or SIRT1 full length (SIRT1-FL). B: Female C57BL/6J mice were pair fed either a control diet or an ethanol (E)–containing diet for 10 days, followed by single gavage of ethanol. During the 10-day chronic-plus-binge ethanol feeding period, Ad-shSlu7 or Ad-shRNA control (0.5 to 1.0 × 109 active viral particles in 200 μL of phosphate-buffered saline) was given to mice twice on days 1 and 5. Relative abundance of SIRT1-ΔExon8 or SIRT1-FL. C: Relative liver mRNA levels of SIRT1-FL and SIRT1-ΔExon8 and ratio of SIRT1-ΔExon8/SIRT1-FL. D: Western blot analysis of liver SIRT1 and acetylated (Ac) forkhead box protein O (FoxO) 1 (on separate gels). Data are expressed as means ± SEM. n = 3 replications (A and B); n = 5 to 12 mice (C and D). ∗P < 0.05 versus Ad-GFP controls; †P < 0.05 versus pair-fed Ad-shRNA controls; ‡P < 0.05 versus ethanol-fed Ad-shRNA. M, markers.
Knockdown of Slu7 Inhibits SIRT1 Pre-mRNA Splicing in Mice after Ethanol Administration
To further gain mechanistic insights into the roles of Slu7-SIRT1 axis in the development of alcoholic steatohepatitis, the effects of Slu7 were examined on SIRT1 pre-mRNA splicing in ethanol-fed mice. Ethanol-fed mice had mildly, but significantly, higher levels of SIRT1-ΔExon8 compared with pair-fed control mice (Figure 2B). In comparison to control mice, Slu7 overexpression increased SIRT1-ΔExon8 levels in mice fed with or without ethanol (Figure 2B). Conversely, Slu7 knockdown normalized SIRT1-ΔExon8 elevation in the ethanol-fed mice to the levels of control mice (Figure 2B). The relative levels of SIRT1-ΔExon8 were paradoxically slightly enhanced by Slu7 knockdown compared with control mice, implying that SIRT1 splicing may be regulated differently in cultured hepatocytes and in animals (Figure 2B).
It was examined whether SIRT1 mRNA isoforms were regulated by altered Slu7 expression in the livers of ethanol-fed mice. Not surprisingly, the livers of ethanol-fed mice had lower levels of SIRT1-FL mRNA and, congruently, higher SIRT1-ΔExon8 mRNA levels compared with pair-fed control mice (Figure 2C). The ratio of hepatic SIRT1-ΔExon8/SIRT1-FL was higher in the ethanol-fed mice than in control mice (Figure 2C). Knockdown of Slu7 substantially increased SIRT1-FL mRNA and decreased SIRT1-ΔExon8 levels in the ethanol-fed mice compared with control mice (Figure 2C). Accordingly, Slu7 knockdown markedly reduced the ratio of SIRT1-ΔExon8/SIRT1-FL in mice fed with or without ethanol to the levels lower than control mice (Figure 2C).
It was further determined whether Slu7 knockdown altered SIRT1 protein expression and its deacetylase activities in the livers of ethanol-fed mice. Consistent with our previous findings,30 ethanol feeding to mice significantly decreased the protein levels of hepatic SIRT1 compared with control animals (Figure 2D). However, the protein levels of hepatic SIRT1 were equal to or higher than the control levels on Slu7 knockdown in the ethanol-fed mice (Figure 2D). Furthermore, Slu7 knockdown abolished the ethanol-induced hyperacetylation of forkhead box protein O 1, which is a known target of SIRT1, indicating that hepatic SIRT1 enzymatic activity was restored by hepatic Slu7 knockdown in ethanol-fed mice (Figure 2D). Conversely, in comparison with pair-fed control mice, Slu7 overexpression induced SIRT1 exon 8 exclusion, increased the ratio of SIRT1-ΔExon8/SIRT1-FL, and enhanced levels of forkhead box protein O 1 acetylation in the livers of ethanol-fed mice (Supplemental Figure S4). Altogether, the results demonstrate that knockdown of Slu7 inhibits SIRT1 pre-mRNA splicing in mice after ethanol administration.
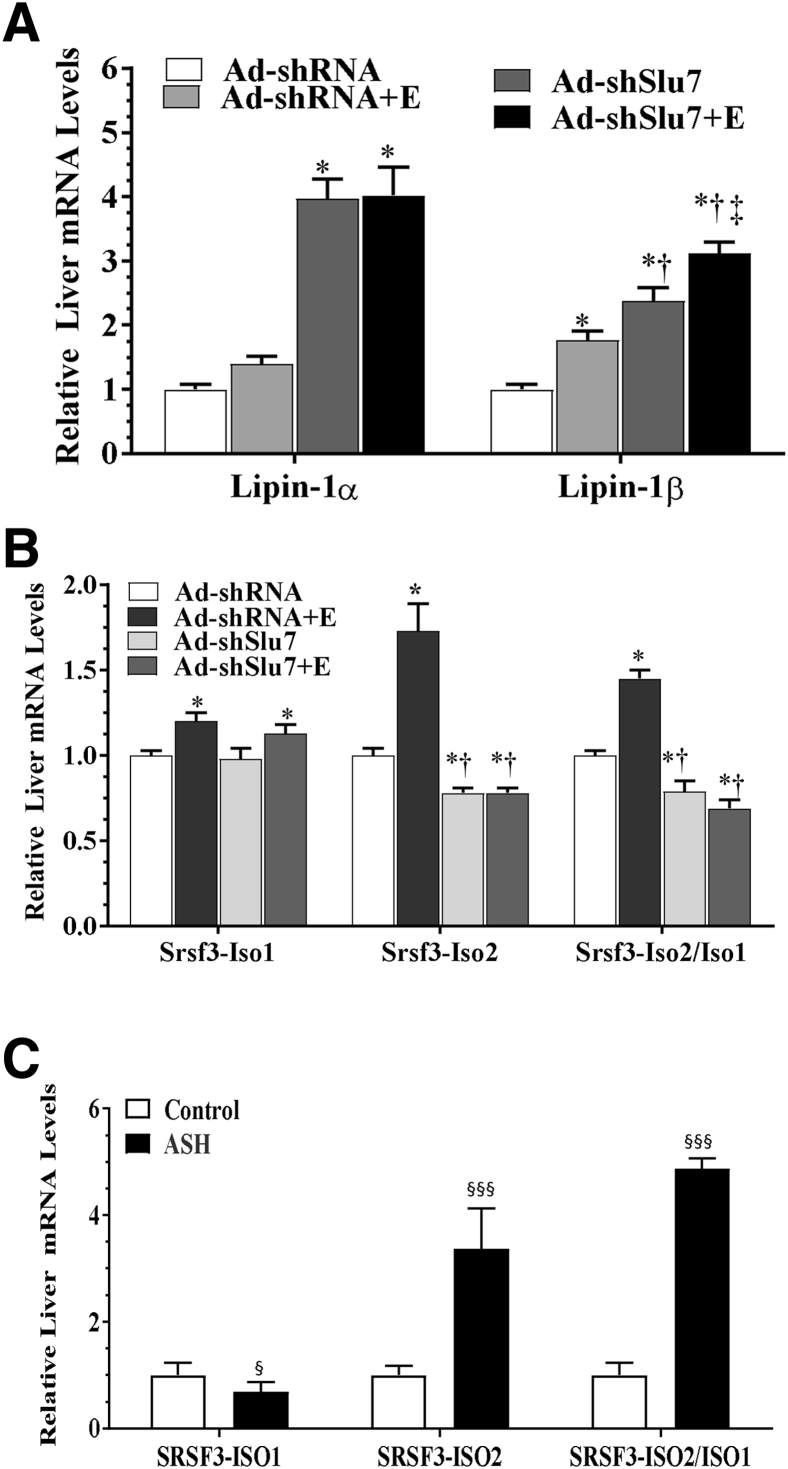
Regulation of Alternative Splicing of Lipin-1 by Reduced Expression of Slu7 in Ethanol-Fed Mice
It was investigated whether stimulated lipin-1 splicing contributed to the hepatic improvement in ethanol-fed mice on Slu7 knockdown.19, 20 Ethanol feeding to mice significantly increased mRNA expression level of lipin-1β without affecting lipin-1α mRNA levels (Figure 3A). However, in comparison with control mice, knockdown of Slu7 drastically increased lipin-1α mRNA levels by nearly threefold in the livers of mice fed with or without ethanol (Figure 3A). Intriguingly, lipin-1β mRNA levels were higher than those in control mice, and Slu7 knockdown further increased lipin-1β mRNA levels in the ethanol-fed mice (Figure 3A). These results demonstrate that knockdown of Slu7 increases the production of lipin-1α in ethanol-fed mice, suggesting that Slu7 may stimulate lipin-1 exon 7 skipping.
Figure 3.
Ad-mediated knockdown of hepatic Slu7 regulates hepatic lipin-1 and serine/arginine-rich splicing factor (Srsf) 3 splicing in ethanol-fed mice. Female C57BL/6J mice were pair fed either a control diet or an ethanol (E)–containing diet for 10 days, followed by single gavage of ethanol. During the 10-day chronic-plus-binge ethanol feeding period, Ad-shSlu7 or Ad-shRNA control (0.5 to 1.0 × 109 active viral particles in 200 μL of phosphate-buffered saline) was given to mice twice on days 1 and 5. A: Relative hepatic mRNA levels of lipin-1, lipin-1α, lipin-1β, and lipin-1α/β. B: Relative liver mRNA levels of Srsf3, Srsf3–full-length isoform lacking exon 4 (Iso1), Srsf3–alternative isoform including exon 4 (Iso2), and ratio of Srsf3-Iso2/Iso1. C: Real-time PCR analyses of liver tissues from normal livers and patients with alcoholic steatohepatitis (ASH). Data are expressed as means ± SEM. n = 5 to 12 mice (A and B); n = 7 (C, normal liver); n = 5 (C, ASH). ∗P < 0.05 versus pair-fed Ad-shRNA controls; †P < 0.05 versus ethanol-fed Ad-shRNA; ‡P < 0.05 versus pair-fed Ad-shSlu7; §P < 0.05, §§§P < 0.001 versus normal livers.
Knockdown of Slu7 Ameliorates Pre-mRNA Srsf3 Splicing in Ethanol-Fed Mice
The influence of Slu7 on Srsf3 pre-mRNA splicing in ethanol-administrated mice was investigated. Feeding ethanol markedly increased Srsf3-Iso2 and significantly elevated the ratio of Srsf3-Iso2/Iso1 compared with the pair-fed control mice (Figure 3B). Strikingly, knockdown of Slu7 reduced the Srsf3-Iso2/Iso1 ratio in the ethanol-fed mice to levels lower than the controls (Figure 3B). These results demonstrated that knockdown of Slu7 in mice ameliorated ethanol-induced disruption of hepatic Srsf3 splicing.
SRSF3-ISO2/ISO1 ratio was substantially increased in liver sample from patients with alcoholic steatohepatitis in comparison with healthy controls (Figure 3C).29 These results indicated clinical relevance of aberrant SRSF3 pre-mRNA splicing in pathogenesis of ALD.
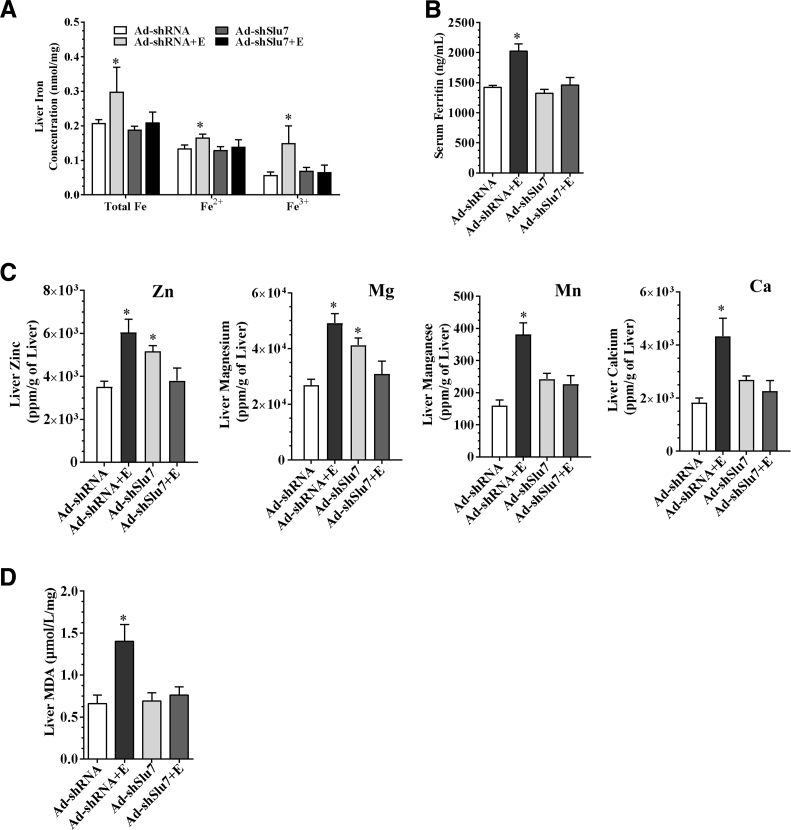
Knockdown of Slu7 Ameliorates the Disruptions in Metal Homeostasis and Attenuates Oxidative Stress in the Liver of Ethanol-Fed Mice
The involvement of metals in the mechanisms of alleviating liver injury on Slu7 knockdown was investigated in the ethanol-fed mice. Ethanol administration to mice significantly increased the total concentrations of iron, ferrous, and ferric, compared with the pair-fed control mice (Figure 4A). However, knockdown of Slu7 normalized the ethanol-elevated iron, ferrous, and ferric concentrations in the livers of mice (Figure 4A). Accordingly, although ethanol feeding significantly increased circulating levels of ferritin, which is a marker of iron storage,31 serum ferritin levels were attenuated on Slu7 knockdown in mice (Figure 4B). Surprisingly, genetic ablation of hepatocyte-specific SIRT1 completely blocked the ability of chronic-plus-binge ethanol feeding to induce hepatic accumulation of total iron and ferrous in mice, suggesting that ethanol-mediated hepatic iron overload may be independent of hepatocyte-specific SIRT1 (Supplemental Figure S5A).
Figure 4.
Slu7 knockdown modulates hepatic iron and zinc homeostasis and reduces oxidative stress in mice. Female C57BL/6J mice were pair fed either a control diet or an ethanol (E)–containing diet for 10 days, followed by single gavage of ethanol. During the 10-day chronic-plus-binge ethanol feeding period, Ad-shSlu7 or Ad-shRNA control (0.5 to 1.0 × 109 active viral particles in 200 μL of phosphate-buffered saline) was given to mice twice on days 1 and 5. A: Liver total iron (Fe), ferrous (Fe2+), ferric (Fe3+) concentrations. B: Serum ferritin concentrations. C: Liver zinc (Zn), magnesium (Mg), manganese (Mn), and calcium (Ca). D: Liver malondialdehyde (MDA) contents. Data are expressed as means ± SEM. n = 5 to 12 mice. ∗P < 0.05 versus pair-fed Ad-shRNA controls.
Furthermore, although metals, such as zinc, magnesium, manganese, and calcium, were significantly increased in the ethanol-fed mice, Slu7 knockdown attenuated concentrations of these metals in the livers of ethanol-fed mice (Figure 4C). In contrast, Slu7 overexpression was associated with augmented hepatic iron and ferrous contents and serum ferritin levels in mice fed with or without ethanol (Supplemental Figure S5, B and C).
Abnormal iron metabolism and hepatic iron overload are major causes of ethanol-induced reactive oxygen species generation and inflammation in association with development of alcoholic liver injury.28, 31, 32 In comparison with pair-fed control mice, ethanol feeding to mice significantly increased hepatic levels of MDA (Figure 4D). Slu7 knockdown completely normalized the ethanol-induced MDA elevation in mice (Figure 4D). Slu7 overexpression had the opposite effect on the MDA levels, as demonstrated by higher hepatic MDA levels in ethanol-fed mice (Supplemental Figure S5D). These data demonstrate that knocking down Slu7 normalizes metals, such as iron and zinc homeostasis, and attenuates the hepatic oxidative stress in mice after ethanol administration.
Knockdown of Slu7 Attenuates the Activities of NF-κB and NFATc4 in Ethanol-Fed Mice
The observations that Slu7 knockdown ameliorated ethanol-induced hepatic inflammation suggested involvement of decreased activities of hepatic NF-κB and NFATc4 via optimizing SIRT1–lipin-1 signaling in these mice.15, 19, 20
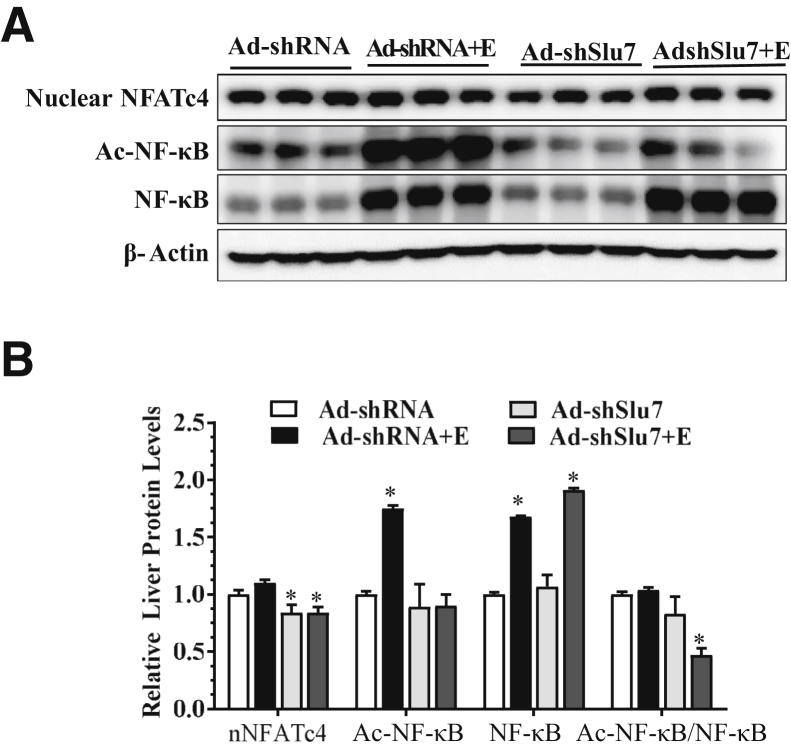
Ethanol feeding to mice increased NF-κB activity, as revealed by enhanced acetylated NF-κB levels in comparison to pair-fed control mice (Figure 5). However, the ability of ethanol to induce NF-κB hyperacetylation was largely abolished on Slu7 knockdown in mice (Figure 5). Hepatic Slu7 overexpression had the opposite effect on NF-κB activity (Supplemental Figure S6).
Figure 5.
Slu7 knockdown attenuates hepatic NF-κB and nuclear factor of activated T cells 4 (NFATc4) activities in the ethanol administrated mice. Female C57BL/6J mice were pair fed either a control diet or an ethanol (E)–containing diet for 10 days, followed by single gavage of ethanol. During the 10-day chronic-plus-binge ethanol feeding period, Ad-shSlu7 or Ad-shRNA control (0.5 to 1.0 × 109 active viral particles in 200 μL of phosphate-buffered saline) was given to mice twice on days 1 and 5. A: Representative Western blot analysis of liver nuclear NFATc4, acetylated (Ac) NF-κB, and NF-κB (on separate gels). The β-actin blot is the same as shown in Figure 2D. B: Relative liver protein levels of liver nNFATc4, Ac-NF-κB, and NF-κB. Data are expressed as means ± SEM (B). n = 5 to 12 mice. ∗P < 0.05 versus pair-fed Ad-shRNA controls.
Nuclear accumulation of NFATc4 was slightly increased in the ethanol-fed mice than pair-fed control mice (Figure 5). However, Slu7 knockdown reduced the nuclear protein levels of NFATc4 to lower than control levels in mice fed with or without ethanol (Figure 5). Collectively, these results demonstrate that Slu7 knockdown attenuates both NF-κB and NFATc4 and subsequently reduces inflammation in the livers of ethanol-fed mice.
Up-Regulation of Hepatic Slu7 in Ethanol-Fed Mice and in Patients with Alcoholic Steatohepatitis
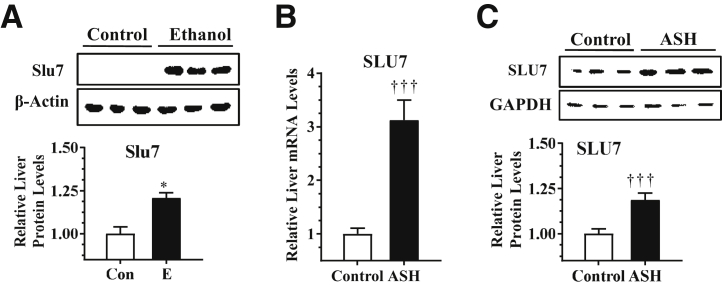
The protein expression levels of hepatic Slu7 in mice were examined using the chronic-plus-binge ethanol feeding protocol.24 The protein levels of hepatic Slu7 were significantly higher in the ethanol-fed mice than pair-fed control mice (Figure 6A).
Figure 6.
Ethanol (E) up-regulates hepatic Slu7 in mice and in patients with alcoholic steatohepatitis (ASH). A: Mice were pair fed either a control (Con) diet or an ethanol-containing diet for 10 days, followed by single gavage of ethanol. Western blot analysis of hepatic Slu7. B and C: Hepatic levels of SLU7 were determined in liver tissues from normal livers and patients with ASH. B: Relative hepatic mRNA levels of SLU7. C: Western blot analysis of hepatic SLU7. Data are expressed as means ± SEM (A–C). n = 7 (normal liver); n = 5 (ASH); n = 4 to 6 mice (A). ∗P < 0.05 versus pair-fed controls; †††P < 0.001 versus normal livers. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To further investigate whether SLU7 is involved in the pathogenesis of alcoholic steatohepatitis, the expression of hepatic SLU7 was examined in patients with alcoholic steatohepatitis.29 SLU7 mRNA level was increased by approximately 30% and protein level was enhanced by approximately 20% in liver samples from patients with alcoholic steatohepatitis compared with those from healthy individuals (Figure 6, B and C). The result demonstrated that hepatic SLU7 is up-regulated in ethanol-fed mice and in patients with alcoholic steatohepatitis.
Discussion
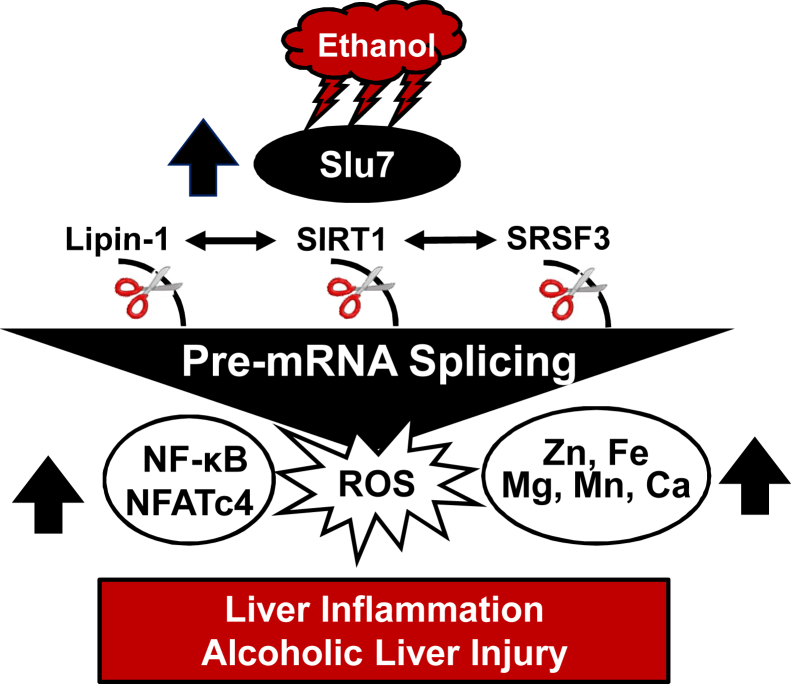
The present study unveiled a new role of splicing regulator Slu7 in the development of alcoholic steatohepatitis. Ad-mediated hepatic Slu7 knockdown ameliorated inflammation and attenuated ethanol-induced liver damage but slightly exacerbated steatosis in mice. Mechanistically, Slu7 knockdown inhibited SIRT1 pre-mRNA splicing by increasing SIRT1-FL and repressing the SIRT1-ΔExon8 in the livers of ethanol-fed mice. Concurrently, Slu7 knockdown in mice ameliorated the aberrant pre-mRNA splicing of lipin-1 and Srsf3 in response to ethanol challenge. Furthermore, Slu7 knockdown alleviated overload of metals, such as iron and zinc, accompanied by reducing hepatic oxidative stress, and diminished activities of NF-κB and NFATc4. This, in turn, limited inflammation in mice after ethanol administration. In addition, hepatic Slu7 was significantly up-regulated in patients with alcoholic steatohepatitis. Taken together, our findings demonstrated that knockdown of Slu7 ameliorated the ethanol-induced liver injury mainly via modifying alternative splicing of hepatic SIRT1, lipin-1, and Srsf3 in mice (Figure 7).
Figure 7.
Proposed role of Slu7 in alcoholic liver injury. Chronic-plus-binge ethanol feeding induces hepatic Slu7 expression. Elevated Slu7 subsequently disrupts sirtuin (SIRT) 1, lipin-1, and serine/arginine-rich splicing factor (Srsf) 3 pre-mRNA splicing and/or expression, which, in turn, activates NF-κB and nuclear factor of activated T cells 4 (NFATc4), disturbs metal homeostasis, enhances reactive oxygen species (ROS), increases hepatic inflammation, and promotes alcoholic liver injury.
Our present study has provided the first evidence that ethanol exposure induces hepatic SIRT1 pre-mRNA splicing in mice. The SIRT1 isoform, SIRT1-ΔExon8, displays distinct characteristics compared with SIRT1-FL.13, 14 For instance, SIRT1-ΔExon8 has minimal deacetylation activity.13, 14 Indeed, our results have demonstrated hyperacetylation of NF-κB and forkhead box protein O 1, two known indicators of SIRT1 deacetylase activity, in the livers of ethanol-fed mice, which suggests the involvement of SIRT1 splicing. SIRT1 deacetylase activity exerts anti-inflammatory effects by deacetylation of the Rel/p65 subunit of NF-κB at lysine residues and suppresses NF-κB activity.12, 15, 19 In this context, ethanol-mediated switch in the relative amounts of SIRT1-FL and SIRT1-ΔExon8 is likely to contribute to hyperacetylation of NF-κB and subsequently promote inflammation and liver injury in ethanol-fed mice.
The regulatory mechanisms of the ethanol-initiated and Slu7-mediated SIRT1 pre-mRNA alternative splicing remain to be clarified. It is suggested that the RNA-binding protein HuR binds to AT/T-rich domains around the exon 8 region to stimulate alternative splicing of SIRT1 pre-mRNA.14 Ethanol exposure perturbs HuR in macrophages.33 Slu7 up-regulates HuR in the livers of ethanol-fed mice (M. You, unpublished observation). The ethanol-mediated up-regulation of Slu7 may affect hepatic HuR, leading to SIRT1 pre-mRNA splicing.
It is intriguing that Slu7 knockdown in mice elevated SIRT1-FL and increased SIRT1 deacetylase activity but slightly aggravated steatosis in response to ethanol challenge. It is likely that the ethanol-mediated disruption of Slu7 contributes to development of steatosis independent of SIRT1 pre-mRNA splicing. Lipin-1β mRNA abundance was increased in the livers of ethanol-fed mice on Slu7 knockdown. Given that elevation of lipin-1β is associated with alcoholic steatosis,19, 34, 35 the exacerbated steatosis observed in ethanol-fed mice on Slu7 knockdown may be mediated, at least in part, by increased lipin-1β mRNA abundance. Furthermore, Slu7 knockdown diminished the ethanol-elevated plasma levels of β-hydroxybutyrate, an indicator of liver fatty acid oxidation. Thus, knockdown of Slu7 may suppress hepatic fatty acid oxidizing capacity in ethanol-fed mice, therefore exacerbating steatosis.
A surprising discovery was that Slu7 knockdown stimulated skipping of exon 7 in lipin-1β to generate lipin-1α but also increased lipin-1β mRNA abundance in the livers of ethanol-fed mice. Slu7 knockdown may stimulate lipin-1 exon 7 skipping to decrease lipin-1β and, simultaneously, enhance the stability of lipin-1β in response to ethanol challenge. These paradoxical effects may neutralize each other, leading to higher lipin-1β mRNA levels by Slu7 knockdown in ethanol-fed mice.
Ethanol-mediated elevation of Slu7 disrupted hepatic Srsf3 pre-mRNA slicing in mice and in patients with alcoholic steatohepatitis. Liver-specific ablation of Srsf3 in mice caused progressive liver damage, including steatosis, hepatitis, and fibrosis, similar to the progression of human liver disease.23 Dysregulation of the hepatic Slu7-Srsf3 axis during ethanol exposure may represent an important mechanism that contributes to progression of human alcoholic steatohepatitis, and it may show that SIRT1 participates in the ethanol-mediated disruption of Slu7-Srsf3 axis.
Aberrant metal homeostasis is implicated in the pathogenesis of ALD.28, 31 Slu7 protein is the only known second step splicing protein regulating iron and zinc metabolism.11 Conceivably, dysregulated Slu7 contributes to the elevation of hepatic metals and subsequent liver toxicity in mice after ethanol intake. SIRT1 is involved in regulation of metals, such as iron.29, 35, 36 It is logical to speculate that elevation of hepatic metals, particularly iron, in ethanol-fed mice may be regulated via Slu7-mediated SIRT1 pre-mRNA splicing. Surprisingly, current findings suggest that the ethanol-mediated hepatic iron overload is likely independent of liver-specific SIRT1. Genetic ablation of liver-specific SIRT1 also abolishes the ability of ethanol to generate SIRT1-ΔExon8 from SIRT1-FL in mice. Therefore, the possibility of Slu7-SIRT1-ΔExon8 axis connecting to iron overload in livers of ethanol-fed mice cannot be excluded. The causal role of SIRT1-ΔExon8 in the ethanol-mediated accumulation of metals needs to be further evaluated by performing ethanol feeding experiments in mice infected with Ad-SIRT1-ΔExon8 or Ad-SIRT1-ΔExon8shRNA.
The exact mechanism of hepatic Slu7 up-regulation after ethanol consumption remains to be determined. Furthermore, ethanol may regulate Slu7 distinctively in different hepatic cells, such as parenchymal (eg, hepatocytes) or nonparenchymal (eg, Kupffer) cells. Interestingly, although Slu7 was up-regulated in cultured hepatocytes exposed to ethanol, Kupffer cells isolated from ethanol-fed rats had lower Slu7 mRNA levels, suggesting that ethanol-regulated hepatic Slu7 might be mediated through cell-specific mechanisms (L. Nagy and M. You, unpublished observation). Further investigation is under way in our laboratory using hepatocyte- or myeloid cell–specific Slu7 knockout mice to identify cell specificity and signaling pathways responsible for altered Slu7 by ethanol.
It is intriguing that Ad-mediated partial depletion of hepatic Slu7 improved the liver functions in ethanol-fed mice. Loss of Slu7 was associated with disrupted liver functions and development of cirrhotic and preneoplastic liver diseases in rodents and humans.6, 7, 8, 9, 10, 11 Ad-mediated partial Slu7 knockdown may be necessary to obtain the optimal gene and protein expressions of Slu7 and subsequently prevent liver injury from ethanol consumption in mice. Future ethanol feeding studies using genetically modified models (eg, Slu7 heterozygous knockout mice or liver-specific Slu7 null mice) may provide better answers.
In summary, we identified the splicing regulator Slu7 as a specific and new target of ethanol and introduced a novel paradigm by which the Slu7-mediated pre-mRNA splicing events can be regulated by ethanol. More important, defective Slu7-SIRT1 signaling and mis-splicing of Srsf3 and lipin-1 were associated with inflammation and liver dysfunction in patients with ALD.19 Our findings highlighted the importance of accurate Slu7-mediated SIRT1, lipin-1, and Srsf3 pre-mRNA splicing for liver functions and the effects of deranged splicing machinery in human ALD. Further characterization of the mechanisms controlling Slu7 gene expression and its cellular targets may overcome the barrier of developing new nutritional or pharmacologic agents as therapeutic interventions in human alcoholic steatohepatitis.
Acknowledgments
We thank Dr. Xiaoling Li (National Institute of Environmental Health Sciences/NIH, Research Triangle Park, NC) for providing sirtuin 1 knockout mice.
Footnotes
Supported, in part, by NIH/National Institute on Alcohol Abuse and Alcoholism grants R01AA015951 and R01AA013623 (M.Y.), P50AA024333 (L.E.N.), and R21AA024946 (Y.Z.).
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2018.05.004.
Supplemental Data
Hepatic Slu7 mRNA and protein expression levels. Female C57/6J mice were pair fed either a control diet or an ethanol (E)–containing diet for 10 days, followed by single gavage of ethanol. During the 10-day chronic-plus-binge ethanol feeding period, Ad-GFP, Ad-Slu7, Ad-shSlu7, or Ad-shRNA control (0.5 to 1.0 × 109 active viral particles in 200 μL of phosphate-buffered saline) was given to mice twice on days 1 and 5. A: Relative liver mRNA levels of Slu7. B: Relative liver protein levels of Slu7. Data are expressed as means ± SEM. n = 5 to 12 mice. ∗P < 0.05 versus pair-fed Ad-GFP controls; †P < 0.05 versus ethanol-fed Ad-GFP; ‡P < 0.05 versus pair-fed Ad-Slu7.
Hepatic Slu7 overexpression exacerbates liver injury but slightly alleviates steatosis in mice after ethanol administration. Mice were fed ethanol (E) and treated as described in Supplemental Figure S1. A: Serum alanine aminotransferase (ALT). B: Serum aspartate aminotransferase (AST). C: Hepatic triglycerides (TGs) content. D: Hematoxylin and eosin (H&E) staining of liver sections. E: Liver myeloperoxidase (MPO) activity. Data are expressed as means ± SEM (A–C and E). n = 5 to 12 mice. ∗P < 0.05 versus pair-fed Ad-GFP controls; †P < 0.05 versus ethanol-fed Ad-GFP. Original magnification, ×40 (D).
The mRNA expression levels of Slu7 in AML-12 cells. Mouse AML-12 hepatocytes were infected with Ad-GFP, Ad-Slu7, and Ad-shSlu7. Relative mRNA levels of Slu7. Data are expressed as means ± SEM. n = 3 replications. ∗P < 0.05 versus Ad-GFP controls; †P < 0.05 versus Ad-Slu7.
Hepatic Slu7 overexpression impairs sirtuin (SIRT) 1 pre-mRNA splicing in mice after ethanol administration. Mice were fed ethanol (E) for 10 days, followed by single gavage of ethanol. During the 10-day chronic-plus-binge ethanol feeding period, Ad-GFP, Ad-Slu7, Ad-shSlu7, or Ad-shRNA control (0.5 to 1.0 × 109 active viral particles in 200 μL of phosphate-buffered saline) was given to mice twice on days 1 and 5. A: Relative liver mRNA levels of SIRT1 full length (FL) and SIRT1-ΔExon8 and ratio of SIRT1-FL/SIRT1-ΔExon8. B: Western blot analysis of liver acetylated (Ac) forkhead box protein O (FoxO) 1. Data are expressed as means ± SEM. n = 5 to 12 mice. ∗P < 0.05 versus pair-fed Ad-GFP controls; †P < 0.05 versus ethanol-fed Ad-GFP.
Hepatic Slu7 overexpression disrupts hepatic iron homeostasis in mice. A: Sirtuin 1 knockout (LKO) and wild-type (WT) mice were fed ethanol-containing and control diets using the chronic-plus-binge ethanol feeding protocol. Liver total iron (Fe), ferrous (Fe2+), and ferric (Fe3+) levels. B: Mice were fed ethanol (E) for 10 days, followed by single gavage of ethanol. During the 10-day chronic-plus-binge ethanol feeding period, Ad-GFP, Ad-Slu7, Ad-shSlu7, or Ad-shRNA control (0.5 to 1.0 × 109 active viral particles in 200 μL of phosphate-buffered saline) was given to mice twice on days 1 and 5. Liver Fe, Fe2+, and Fe3+ concentrations. C: Serum ferritin concentrations. D: Liver malondialdehyde (MDA). Data are expressed as means ± SEM. n = 5 to 12 mice. ∗P < 0.05 versus WT controls; †P < 0.05 versus pair-fed Ad-GFP control; ‡P < 0.05 versus ethanol-fed Ad-GFP.
Hepatic Slu7 overexpression activates hepatic NF-κB in ethanol-fed mice. Mice were fed ethanol (E) for 10 days, followed by single gavage of ethanol. During the 10-day chronic-plus-binge ethanol feeding period, Ad-GFP, Ad-Slu7, Ad-shSlu7, or Ad-shRNA control (0.5 to 1.0 × 109 active viral particles in 200 μL of phosphate-buffered saline) was given to mice twice on days 1 and 5. A: Western blot analysis of liver acetylated (Ac) NF-κB and NF-κB (on separate gels). The β-actin blot is the same as shown in Supplemental Figure S4B. B: Relative liver Ac NF-κB and NF-κB protein levels. Data are expressed as means ± SEM (B). n = 5 to 12 mice. ∗P < 0.05 versus pair-fed Ad-GFP controls; †P < 0.05 versus ethanol-fed Ad-GFP.
References
- 1.Farooq M.O., Bataller R. Pathogenesis and management of alcoholic liver disease. Dig Dis. 2016;34:347–355. doi: 10.1159/000444545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gallego-Paez L.M., Bordone M.C., Leote A.C., Saraiva-Agostinho N., Ascensão-Ferreira M., Barbosa-Morais N.L. Alternative splicing: the pledge, the turn, and the prestige: the key role of alternative splicing in human biological systems. Hum Genet. 2017;136:1015–1042. doi: 10.1007/s00439-017-1790-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berasain C., Goñi S., Castillo J., Latasa M.U., Prieto J., Avila M.A. Impairment of pre mRNA splicing in liver disease: mechanisms and consequences. World J Gastroenterol. 2010;16:3091–3102. doi: 10.3748/wjg.v16.i25.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chua K., Reed R. The RNA splicing factor hSlu7 is required for correct 3' splice-site choice. Nature. 1999;402:207–210. doi: 10.1038/46086. [DOI] [PubMed] [Google Scholar]
- 5.James S.A., Turner W., Schwer B. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA. 2011;8:1068–1077. doi: 10.1017/s1355838202022033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chua K., Reed R. Human step II splicing factor hSlu7 functions in restructuring the spliceosome between the catalytic steps of splicing. Genes Dev. 1999;13:841–850. doi: 10.1101/gad.13.7.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shomron N., Reznik M., Ast G. Splicing factor hSlu7 contains a unique functional domain required to retain the protein within the nucleus. Mol Biol Cell. 2004;15:3782–3795. doi: 10.1091/mbc.E04-02-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shomron N., Alberstein M., Reznik M., Ast G. Stress alters the subcellular distribution of hSlu7 and thus modulates alternative splicing. J Cell Sci. 2005;118:1151–1159. doi: 10.1242/jcs.01720. [DOI] [PubMed] [Google Scholar]
- 9.Castillo J., Goñi S., Latasa M.U., Perugorría M.J., Calvo A., Muntané J., Bioulac-Sage P., Balabaud C., Prieto J., Avila M.A., Berasain C. Amphiregulin induces the alternative splicing of p73 into its oncogenic isoform DeltaEx2p73 in human hepatocellular tumors. Gastroenterology. 2009;137:1805–1815. doi: 10.1053/j.gastro.2009.07.065. [DOI] [PubMed] [Google Scholar]
- 10.Urtasun R., Elizalde M., Azkona M., Latasa M.U., García-Irigoyen O., Uriarte I., Fernández-Barrena M.G., Vicent S., Alonso M.M., Muntané J., Prieto J., Ávila M.A., Berasain C. Splicing regulator SLU7 preserves survival of hepatocellular carcinoma cells and other solid tumors via oncogenic miR-17-92 cluster expression. Oncogene. 2016;35:4719–4729. doi: 10.1038/onc.2015.517. [DOI] [PubMed] [Google Scholar]
- 11.Elizalde M., Urtasun R., Azkona M., Latasa M.U., Goñi S., García-Irigoyen O., Uriarte I., Segura V., Collantes M., Di Scala M., Lujambio A., Prieto J., Ávila M.A., Berasain C. Splicing regulator SLU7 is essential for maintaining liver homeostasis. J Clin Invest. 2014;124:2909–2920. doi: 10.1172/JCI74382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang H.C., Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2014;25:138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch C.J., Shah Z.H., Allison S.J., Ahmed S.U., Ford J., Warnock L.J., Li H., Serrano M., Milner J. SIRT1 undergoes alternative splicing in a novel auto-regulatory loop with p53. PLoS One. 2010;5:e13502. doi: 10.1371/journal.pone.0013502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao W., Zhao J., Hou M., Wang Y., Zhang Y., Zhao X., Zhang C., Guo D. HuR and TIA1/TIAL1 are involved in regulation of alternative splicing of SIRT1 pre-mRNA. Int J Mol Sci. 2014;15:2946–2958. doi: 10.3390/ijms15022946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.You M., Jogasuria A., Taylor C., Wu J. Sirtuin 1 signaling and alcoholic fatty liver disease. Hepatobiliary Surg Nutr. 2015;4:88–100. doi: 10.3978/j.issn.2304-3881.2014.12.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han G.S., Carman G.M. Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J Biol Chem. 2010;285:14628–14638. doi: 10.1074/jbc.M110.117747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Péterfy M., Phan J., Reue K. Alternatively spliced lipin isoforms exhibit distinct expression pattern, subcellular localization, and role in adipogenesis. J Biol Chem. 2005;280:32883–32889. doi: 10.1074/jbc.M503885200. [DOI] [PubMed] [Google Scholar]
- 18.Phan J., Reue K. Lipin, a lipodystrophy and obesity gene. Cell Metab. 2005;1:73–83. doi: 10.1016/j.cmet.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Yin H., Hu M., Liang X., Ajmo J.M., Li X., Bataller R., Odena G., Stevens S.M., Jr., You M. Deletion of SIRT1 from hepatocytes in mice disrupts lipin-1 signaling and aggravates alcoholic fatty liver. Gastroenterology. 2014;146:801–811. doi: 10.1053/j.gastro.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu M., Yin H., Mitra M.S., Liang X., Ajmo J.M., Nadra K., Chrast R., Finck B.N., You M. Hepatic-specific lipin-1 deficiency exacerbates experimental alcohol-induced steatohepatitis in mice. Hepatology. 2013;58:1953–1963. doi: 10.1002/hep.26589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sen S., Jumaa H., Webster N.J. Splicing factor SRSF3 is crucial for hepatocyte differentiation and metabolic function. Nat Commun. 2013;4:1336. doi: 10.1038/ncomms2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 23.Sen S., Langiewicz M., Jumaa H., Webster N.J.G. Deletion of serine/arginine-rich splicing factor 3 in hepatocytes predisposes to hepatocellular carcinoma in mice. Hepatology. 2015;61:171–183. doi: 10.1002/hep.27380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertola A., Mathews S., Ki S.H., Wang H., Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nat Protoc. 2013;8:627–637. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin H., Liang X., Jogasuria A., Davidson N.O., You M. miR-217 regulates ethanol-induced hepatic inflammation by disrupting sirtuin 1-lipin-1 signaling. Am J Pathol. 2015;185:1286–1296. doi: 10.1016/j.ajpath.2015.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Xu S., Giles A., Nakamura K., Lee J.W., Hou X., Donmez G., Li J., Luo Z., Walsh K., Guarente L., Zang M. Hepatic overexpression of SIRT1 in mice attenuates endoplasmic reticulum stress and insulin resistance in the liver. FASEB J. 2011;25:1664–1679. doi: 10.1096/fj.10-173492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai Y., Jogasuria A., Yin H., Xu M.J., Hu X., Wang J., Kim C., Wu J., Lee K., Gao B., You M. The detrimental role played by lipocalin-2 in alcoholic fatty liver in mice. Am J Pathol. 2016;186:2417–2428. doi: 10.1016/j.ajpath.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu X., Jogasuria A., Wang J., Kim C., Han Y., Shen H., Wu J., You M. MitoNEET deficiency alleviates experimental alcoholic steatohepatitis in mice by stimulating endocrine adiponectin-Fgf15 axis. J Biol Chem. 2016;291:22482–22495. doi: 10.1074/jbc.M116.737015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J., Xu Y., Li Y., Jadhav K., You M., Yin L., Zhang Y., Xu J., Xu Y., Li Y., Jadhav K., You M., Yin L., Zhang Y. Carboxylesterase 1 is regulated by hepatocyte nuclear factor 4α and protects against alcohol- and MCD diet-induced liver injury. Sci Rep. 2016;6:24277. doi: 10.1038/srep24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.You M., Liang X., Ajmo J.M., Ness G.C. Involvement of mammalian sirtuin 1 in the action of ethanol in the liver. Am J Physiol Gastrointest Liver Physiol. 2008;294:G892–G898. doi: 10.1152/ajpgi.00575.2007. [DOI] [PubMed] [Google Scholar]
- 31.Datz C., Müller E., Aigner E. Iron overload and non-alcoholic fatty liver disease. Minerva Endocrinol. 2017;42:173–183. doi: 10.23736/S0391-1977.16.02565-7. [DOI] [PubMed] [Google Scholar]
- 32.Harrison-Findik D.D. Role of alcohol in the regulation of iron metabolism. World J Gastroenterol. 2007;13:4925–4930. doi: 10.3748/wjg.v13.i37.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMullen M.R., Cocuzzi E., Hatzoglou M., Nagy L.E. Chronic ethanol exposure increases the binding of HuR to the TNFα 3′-untranslated region in macrophages. J Biol Chem. 2003;278:38333–38341. doi: 10.1074/jbc.M304566200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu M., Wang F., Li X., Rogers C.Q., Liang X., Finck B.N., Mitra M.S., Zhang R., Mitchell D.A., You M. Regulation of hepatic lipin-1 by ethanol: role of AMP-activated protein kinase/sterol regulatory element-binding protein 1 signaling in mice. Hepatology. 2012;55:437–446. doi: 10.1002/hep.24708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H., Shen F., Sherban A., Nocon A., Li Y., Wang H., Xu M.J., Rui X., Han J., Jiang B., Lee D., Li N., Keyhani-Nejad F., Fan J.G., Liu F., Kamat A., Musi N., Guarente L., Pacher P., Gao B., Zang M. DEP domain-containing mTOR-interacting protein suppresses lipogenesis and ameliorates hepatic steatosis and acute-on-chronic liver injury in alcoholic liver disease. Hepatology. 2018 doi: 10.1002/hep.29849. [Epub ahead of print] doi: 10.1002/hep.29849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno-Navarrete J.M., Moreno M., Ortega F., Sabater M., Xifra G., Ricart W., Fernández-Real J.M. CISD1 in association with obesity-associated dysfunctional adipogenesis in human visceral adipose tissue. Obesity (Silver Spring) 2016;24:139–147. doi: 10.1002/oby.21334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hepatic Slu7 mRNA and protein expression levels. Female C57/6J mice were pair fed either a control diet or an ethanol (E)–containing diet for 10 days, followed by single gavage of ethanol. During the 10-day chronic-plus-binge ethanol feeding period, Ad-GFP, Ad-Slu7, Ad-shSlu7, or Ad-shRNA control (0.5 to 1.0 × 109 active viral particles in 200 μL of phosphate-buffered saline) was given to mice twice on days 1 and 5. A: Relative liver mRNA levels of Slu7. B: Relative liver protein levels of Slu7. Data are expressed as means ± SEM. n = 5 to 12 mice. ∗P < 0.05 versus pair-fed Ad-GFP controls; †P < 0.05 versus ethanol-fed Ad-GFP; ‡P < 0.05 versus pair-fed Ad-Slu7.
Hepatic Slu7 overexpression exacerbates liver injury but slightly alleviates steatosis in mice after ethanol administration. Mice were fed ethanol (E) and treated as described in Supplemental Figure S1. A: Serum alanine aminotransferase (ALT). B: Serum aspartate aminotransferase (AST). C: Hepatic triglycerides (TGs) content. D: Hematoxylin and eosin (H&E) staining of liver sections. E: Liver myeloperoxidase (MPO) activity. Data are expressed as means ± SEM (A–C and E). n = 5 to 12 mice. ∗P < 0.05 versus pair-fed Ad-GFP controls; †P < 0.05 versus ethanol-fed Ad-GFP. Original magnification, ×40 (D).
The mRNA expression levels of Slu7 in AML-12 cells. Mouse AML-12 hepatocytes were infected with Ad-GFP, Ad-Slu7, and Ad-shSlu7. Relative mRNA levels of Slu7. Data are expressed as means ± SEM. n = 3 replications. ∗P < 0.05 versus Ad-GFP controls; †P < 0.05 versus Ad-Slu7.
Hepatic Slu7 overexpression impairs sirtuin (SIRT) 1 pre-mRNA splicing in mice after ethanol administration. Mice were fed ethanol (E) for 10 days, followed by single gavage of ethanol. During the 10-day chronic-plus-binge ethanol feeding period, Ad-GFP, Ad-Slu7, Ad-shSlu7, or Ad-shRNA control (0.5 to 1.0 × 109 active viral particles in 200 μL of phosphate-buffered saline) was given to mice twice on days 1 and 5. A: Relative liver mRNA levels of SIRT1 full length (FL) and SIRT1-ΔExon8 and ratio of SIRT1-FL/SIRT1-ΔExon8. B: Western blot analysis of liver acetylated (Ac) forkhead box protein O (FoxO) 1. Data are expressed as means ± SEM. n = 5 to 12 mice. ∗P < 0.05 versus pair-fed Ad-GFP controls; †P < 0.05 versus ethanol-fed Ad-GFP.
Hepatic Slu7 overexpression disrupts hepatic iron homeostasis in mice. A: Sirtuin 1 knockout (LKO) and wild-type (WT) mice were fed ethanol-containing and control diets using the chronic-plus-binge ethanol feeding protocol. Liver total iron (Fe), ferrous (Fe2+), and ferric (Fe3+) levels. B: Mice were fed ethanol (E) for 10 days, followed by single gavage of ethanol. During the 10-day chronic-plus-binge ethanol feeding period, Ad-GFP, Ad-Slu7, Ad-shSlu7, or Ad-shRNA control (0.5 to 1.0 × 109 active viral particles in 200 μL of phosphate-buffered saline) was given to mice twice on days 1 and 5. Liver Fe, Fe2+, and Fe3+ concentrations. C: Serum ferritin concentrations. D: Liver malondialdehyde (MDA). Data are expressed as means ± SEM. n = 5 to 12 mice. ∗P < 0.05 versus WT controls; †P < 0.05 versus pair-fed Ad-GFP control; ‡P < 0.05 versus ethanol-fed Ad-GFP.
Hepatic Slu7 overexpression activates hepatic NF-κB in ethanol-fed mice. Mice were fed ethanol (E) for 10 days, followed by single gavage of ethanol. During the 10-day chronic-plus-binge ethanol feeding period, Ad-GFP, Ad-Slu7, Ad-shSlu7, or Ad-shRNA control (0.5 to 1.0 × 109 active viral particles in 200 μL of phosphate-buffered saline) was given to mice twice on days 1 and 5. A: Western blot analysis of liver acetylated (Ac) NF-κB and NF-κB (on separate gels). The β-actin blot is the same as shown in Supplemental Figure S4B. B: Relative liver Ac NF-κB and NF-κB protein levels. Data are expressed as means ± SEM (B). n = 5 to 12 mice. ∗P < 0.05 versus pair-fed Ad-GFP controls; †P < 0.05 versus ethanol-fed Ad-GFP.