Summary
Patterns of species richness are commonly linked to life history strategies. In diatoms, an exceptionally diverse lineage of photosynthetic heterokonts important for global photosynthesis and burial of atmospheric carbon, lineages with different locomotory and reproductive traits differ dramatically in species richness, but any potential association between life history strategy and diversification has not been tested in a phylogenetic framework.
We constructed a time‐calibrated, 11‐gene, 1151‐taxon phylogeny of diatoms – the most inclusive diatom species tree to date. We used this phylogeny, together with a comprehensive inventory of first–last occurrences of Cenozoic fossil diatoms, to estimate ranges of expected species richness, diversification and its variation through time and across lineages.
Diversification rates varied with life history traits. Although anisogamous lineages diversified faster than oogamous ones, this increase was restricted to a nested clade with active motility in the vegetative cells.
We propose that the evolution of motility in vegetative cells, following an earlier transition from oogamy to anisogamy, facilitated outcrossing and improved utilization of habitat complexity, ultimately leading to enhanced opportunity for adaptive divergence across a variety of novel habitats. Together, these contributed to a species radiation that gave rise to the majority of present‐day diatom diversity.
Keywords: anisogamy, diatoms, diversification, life history, motility, oogamy
Introduction
Patterns of species richness across time, space and clades are commonly linked to evolutionary innovations and ecological opportunities that are thought to influence the rates of speciation and extinction. Changes in life history can affect developmental or reproductive strategies and have consequences for reproductive success and fitness, whereas traits related to locomotion—which are tightly linked to life history in many taxa—can improve dispersal abilities and facilitate range expansions, range shifts or colonizations of previously unavailable habitats. As a result, transitions in life history (e.g. selfing or outcrossing), locomotion (e.g. sedentary or mobile) or their interactions are often correlated with shifts in species diversification (e.g. Leclère et al., 2009; Goldberg et al., 2010; Ikeda et al., 2012). Numerous life history traits have been linked to net diversification (birth − death, r = b − d) in vascular plants. These include features associated with longevity (annual vs perennial, Drummond et al., 2012), seed dispersal (Leslie et al., 2013; Beaulieu & O'Meara, 2016) and mechanisms that promote outcrossing, such as self‐incompatibility (Goldberg et al., 2010) and heterostyly (de Vos et al., 2014). Similar patterns are also evident in animals. In insects, for example, the evolution of complete metamorphosis (Rainford et al., 2014), as well as its subsequent reduction in some groups (Cieslak et al., 2014), have been associated with increases in net diversification (but see Condamine et al., 2016). In hydrozoans, two alternative life cycles, one with and one without a mobile life history stage, have been maintained by species selection related to lower extinction rates in lineages with a medusa or medusoid free‐swimming stage that facilitates dispersal (Leclère et al., 2009). Owing to its role in dispersal, mating and interactions with environmental conditions in general, locomotion has also been linked with diversification. By facilitating long‐range dispersal, the presence and properties of motile life history stages can increase gene flow across populations and, in turn, reduce rates of speciation (e.g. Palumbi, 1992, 1994 and references therein). On the other hand, limited locomotory abilities could restrict gene flow between divergent and potentially locally adapted populations, thereby increasing rates of speciation (Duda & Rolán, 2005; Ikeda et al., 2012). Such interactions among life history, motility and ecology are common across the tree of life and are especially important in microbial eukaryotes (protists) in which the ability of self‐propelled locomotion often changes with alternating life history stages (Hoek et al., 1995).
One such interaction has played out in the evolutionary history of diatoms (Bacillariophyta), an ecologically, functionally and morphologically diverse clade of stramenopile algae (Andersen, 2004). Diatoms are thought to be ancestrally planktonic and oogamous, with nonmotile vegetative cells suspended in a dilute environment, necessitating motile flagellated gametes to reproduce successfully (Drebes, 1977; Round et al., 1990; Chepurnov et al., 2004). However, flagellated male gametes were lost at least twice: once in the common ancestor of the pennate diatoms – a large clade of predominantly benthic species whose gametes are behaviorally dimorphic and move via pseudopodia (Davidovich et al., 2010; Sato et al., 2011; Kaczmarska et al., 2017), and once in Ardissonea crystallina – a member of a benthic marine clade referred to as toxariids (Davidovich et al., 2017). Active, directed motility in vegetative cells evolved subsequently in a clade nested within pennate diatoms (raphid pennate diatoms; Harper, 1977; Round et al., 1990). Motility of these cells is enabled by a longitudinal slit through the cell wall called a raphe, which is lined with actin–myosin protein complexes (Poulsen et al., 1999) that move the cell across a substrate by displacing strands of extracellular mucilaginous secretions (Harper, 1977; Round et al., 1990). Diatoms outside this lineage can use these secretions for attachment and, in some cases, movement (Pickett‐Heaps et al., 1986, 1991; Kooistra et al., 2003), but the range, velocity and responsiveness of cells with raphe‐enabled motility are unmatched by other types of motility found in nonraphid diatoms (Consalvey et al., 2004). In all, three combinations of mode of sexual reproduction and locomotion of vegetative cells are known in diatoms: oogamous and nonmotile lineages (the paraphyletic group of ‘centric’ diatoms, excluding anisogamous toxariids), anisogamous and nonmotile lineages (the paraphyletic group of ‘araphid’ pennate diatoms, including the anisogamous toxariids), and anisogamous and motile lineages (the clade of raphid pennate diatoms) (Round et al., 1990; Theriot et al., 2015; Parks et al., 2017).
Comparisons between diatom groups with different combinations of reproductive and locomotory traits reveal drastic differences in species richness, with anisogamous diatoms far outnumbering oogamous diatoms and, within the former, lineages with motile vegetative cells (specifically, the raphid pennates) far outnumbering lineages with nonmotile vegetative cells (Guiry & Guiry, 2017; Kociolek et al., 2017). This apparent disparity in species richness and rates of diversification was first investigated by James Small (e.g. Small, 1945a,1945b, 1950), whose work on the geological duration of fossil taxa revealed differences in species turnover between diatoms with radially vs bilaterally symmetrical cell walls (Small, 1950; see also Van Valen, 1973). Small's results were framed in terms of cell symmetry, the primary basis of diatom classification at the time, but the division between ‘centric’ and ‘pennate’ also largely mirrors the split between oogamous and anisogamous diatoms. An added layer of complexity comes from common in vitro observations of selfing (homothally) in centric diatoms and the prevalence of outcrossing (heterothally) in pennates (Chepurnov et al., 2004 and references therein). As a result, Small's early findings of differences in diversification dynamics between centric and pennate diatoms also apply to variation in the mode of sexual reproduction and outcrossing: oogamous diatoms (generally capable of selfing in vitro) diversified more slowly than anisogamous diatoms (generally incapable of selfing in vitro).
These considerations suggest that life history—specifically the mode of sexual reproduction—is one of the key factors driving the observed disparity in species richness across diatom groups. However, directed motility of vegetative cells within the anisogamous lineage (raphid diatoms) conferred many ecological advantages as well, including the ability to quickly respond to changes in light and nutrient availability, diurnal and tidal migrations, and pheromonal movements (Palmer & Round, 1967; Sato et al., 2011; Cohn et al., 2015; Bondoc et al., 2016a,2016b). The aggregated effect of these benefits suggests an alternative hypothesis—namely, that the primary driver of species richness in raphid pennate diatoms was active motility of vegetative cells, the evolution of which fundamentally altered both inter‐ and intraspecies interactions (Drebes, 1977; Harper, 1977; Kingston, 1999), enabled the colonization of novel habitats (Palmer & Round, 1967; Sims et al., 2006) and allowed them to better exploit habitat heterogeneity (Consalvey et al., 2004; Cohn et al., 2015). Such a scenario would imply that Small's discovery of faster diversification of anisogamous diatoms is a result of faster diversification in the nested lineage of actively motile raphid pennate diatoms, analogous to the existence of an important unconsidered (or ‘hidden’) trait (Beaulieu & O'Meara, 2016) – in this case, a novel means of locomotion within a larger clade with a derived mode of sexual reproduction.
We tested these hypotheses using a combination of phylogenetic and paleontological data, combining a large dataset of first–last occurrences of marine Cenozoic fossil diatoms with an 11‐gene phylogeny for 1151 diverse diatom taxa, which represents the most comprehensive analysis of the diatom phylogeny to date. We found that, although anisogamous diatoms diversified faster than oogamous diatoms, the higher rates were largely restricted to the nested clade of actively motile species. We propose that the evolution of directed motility enabled improved utilization of habitat complexity, colonization of novel habitats and more frequent or efficient sexual reproduction, ultimately resulting in increased genetic diversity, greater potential for evolutionary change and accelerated species diversification in the lineage of raphid pennate diatoms.
Materials and Methods
Phylogenetic dataset assembly
We compiled data for 11 genes that exhaust the set of markers with substantial representation in publicly available databases (Sorhannus & Fox, 2012; Theriot et al., 2015; Ruck et al., 2016). These included two nuclear rRNA genes (18S and 28S rRNA), seven plastid genes (16S rRNA, atpB, psaA, psaB, psbA, psbC and rbcL) and two mitochondrial genes (cob and coxI) (Supporting Information Table S1). The combined data from these genes encompasses as much diatom diversity as possible, whilst maximizing the power to resolve both old and recent divergences (Theriot et al., 2015). After removing environmental sequences, which are commonly identified to the genus level only, we used Usearch (Edgar, 2010) to identify and remove duplicate accessions (Table S1). We then aligned the sequences and performed several rounds of filtering to remove misidentified sequences, taxonomic duplicates and unnamed species that were identical (or nearly so) to a named sister taxon (based on zero or near‐zero branch lengths). Decisions regarding the removal of misidentified sequences were made on the basis of the manual inspection of maximum likelihood (ML) trees built with RAxML (Stamatakis, 2014). When an accession appeared correctly aligned, but was reconstructed phylogenetically far from its expected position (i.e. it fell outside the expected genus or family), we flagged it as misidentified and removed it from the alignment. These decisions were made in consultation with the primary literature to ensure that an accession in question had not been recently transferred to a different taxonomic group or had been previously known to have an unexpected phylogenetic placement compared with that suggested by its name. These checks were important because many diatom genera are not monophyletic and can comprise distant relatives that are taxonomically united on the basis of convergent (Alverson et al., 2007; Ruck & Theriot, 2011) or plesiomorphic (Ruck et al., 2016) characters. For taxonomic duplicates, we removed the accession with fewer genes, and for cases in which a pair of taxonomic duplicates had different gene complements, we combined their data into a single accession. Finally, we dropped all accessions represented by a single gene that was not 18S or rbcL, the two genes with the highest taxon occupancy in the dataset. Preliminary tree searches showed that these types of singletons were often inconsistently or erroneously placed phylogenetically. To enable the estimation of branch lengths at the split between diatoms and Parmales (Bolidophyceae), we included nuclear ribosomal RNA and plastid gene data for 13 additional outgroups from the stramenopile classes Chrysophyceae, Phaeophyceae, Xanthophyceae, Raphidophyceae, Eustigmatophyceae and Pelagophyceae. Data files, phylogenies and scripts are available from a Zenodo online repository: https://doi.org/10.5281/zenodo.583628.
Multiple sequence alignment, model partitioning and phylogenetic inference
Ribosomal RNA genes (16S, 18S and 28S) were aligned on the basis of covariance models of secondary structure (Cannone et al., 2002; Nawrocki et al., 2009). For the nuclear 18S and plastid 16S genes, we used covariance models for Eukarya and Bacteria, respectively, and removed sequences shorter than 250 nucleotides in length. For the 28S gene fragment, we used a covariance model developed from aligned full‐length 28S sequences from stramenopiles (Nakov et al., 2014). We masked secondary structure alignments to remove sites with low probability of positional homology based on the covariance models (Nawrocki et al., 2009). Protein‐coding genes were aligned with Mafft v.6.864b (Katoh & Standley, 2013) and adjusted manually to resolve gaps caused by incomplete codons. Single‐gene alignments were then concatenated into a single 11‐gene alignment.
We split the 11‐gene alignment into seven partitions (one for the combined rRNA (n = 1 partition), one per codon position for the combined plastid genes (n = 3 partitions) and one per codon position for the combined mitochondrial genes (n = 3 partitions) (Table S2)) and identified the best nucleotide substitution model using Iq‐tree v.1.4.1 (Nguyen et al., 2015). Tree searches were performed with Iq‐tree using the above partitioning scheme and substitution models and the edge‐proportional model to accommodate between‐partition differences in evolutionary rate. We performed 100 ML optimizations with default settings, except for the strength of perturbation of the nearest‐neighbor interchange (Iq‐tree option: ‐pers, default = 0.5), which we varied between 0.3 and 0.6 with 0.05‐unit increments. Support for the inferred relationships was assessed with 103 UltraFast bootstrap replicates (Minh et al., 2013). The interpretation of the UltraFast bootstrap values differs from that of a standard nonparametric bootstrap resampling in that only nodes with UltraFast bootstrap support ≥95% are considered to be strongly supported.
Time calibration
We time calibrated the phylograms using treePL (Sanderson, 2002; Smith & O'Meara, 2012) and 38 calibration points taken from first appearances of diatom lineages in the fossil record (Table S3). Most first‐appearance data came from the primary literature (Gersonde & Harwood, 1990; Harwood & Gersonde, 1990; Harwood & Nikolaev, 1995; Sims et al., 2006; Harwood et al., 2007), and many have been used previously (Medlin, 2015). However, given the size of our phylogeny and the importance of having calibrations distributed throughout the tree and covered time span, we also compiled first‐appearance records from the Neptune/Chronos database (Lazarus, 1994) which includes data from the Deep Sea Drilling Project (DSDP) and Ocean Drilling Project (ODP).
To construct calibration bounds, we followed a procedure that uses the difference between the first occurrences of two sister lineages (ghost lineage time), or the difference between the earliest and second earliest occurrence of a lineage (penultimate distance time) (Norris et al., 2015). For example, the first occurrences in the fossil record of the sister taxa Aulacoseira (Archepyrgis) and Melosira are 115 million yr ago (Ma) and 125 Ma, respectively (Table S3), resulting in a ghost lineage time of 10 million yr (Myr). Following Norris et al. (2015), a prior distribution for this node can be constructed using a lognormal distribution with a mean set to half the ghost lineage time (5 Myr in this case), a standard deviation of 1.814 Myr and an offset given by the earlier of the two first occurrences (Melosira, 125 Ma in this case). We used the 5% and 95% quantiles of this distribution as the minimum and maximum bounds for the age of the calibrated node in treePL. We used minimum age bounds only when there was not enough information to calculate the ghost lineage or penultimate distance time. We obtained a total of 21 calibrations with minimum and maximum bounds (including the most recent common ancestor (MRCA) of diatoms + Parmales and MRCA of all diatoms) and 17 calibrations with a minimum bound only. The majority of calibrations were within centric diatoms (19), with 11 in the araphid grade and six in the raphid clade.
To find the optimal rate‐smoothing parameter for the penalized likelihood time calibration (Sanderson, 2002), we used a random subsample and replace cross‐validation procedure (Smith & O'Meara, 2012) with a range of tested values for the smoothing parameter between 102 and 10−5. Cross‐validation showed that a value of 10−3 provided the best smoothing, indicating substantial evolutionary rate variation across the phylogeny, as expected for a phylogenetic tree of this size and age. We used the calibrations described in Table S3 and the rate‐smoothing procedure described above to time calibrate the best tree and all bootstrap phylogenies for downstream analyses.
Diversification rate at predefined clades
Although the sample of 1151 taxa in our dataset encompasses a broad sample of diatom diversity and considerably expands upon previous efforts to reconstruct the diatom phylogeny, our species count still falls well short of the total estimated number of diatom species (see below). To accommodate unsampled diversity in our models, we pruned the phylogeny to include just one representative per genus (Fig. 1a–c), such that each tip was an unresolved polytomy representing the entire species diversity within that genus (N = 234 genera; ‘genus‐level’ analysis). This strategy breaks down when genera are nonmonophyletic, and so we also performed analyses with genera grouped into more inclusive clades (N = 45 clades; ‘clade‐level’ analysis).
Figure 1.
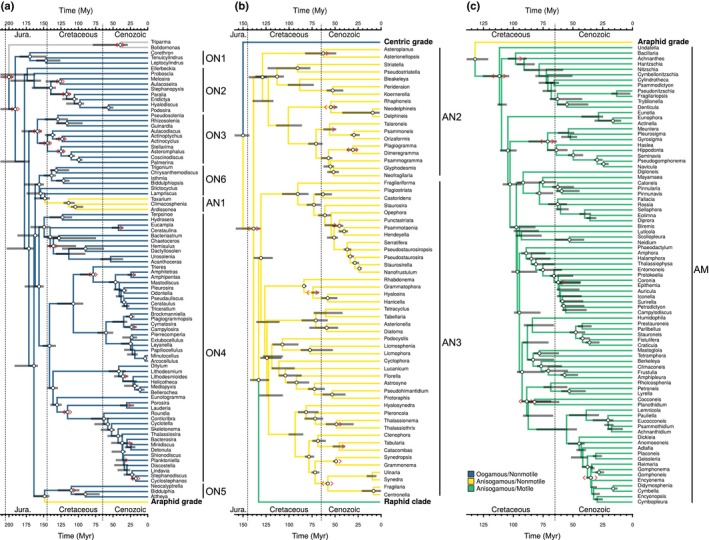
Time‐calibrated phylogeny of diatoms based on 11 genes with 1151 diatoms and six Parmales pruned down to one taxon per genus (N = 234). The highest scoring tree, based on 100 maximum likelihood optimizations, is shown. In (a–c), branches are colored according to life history and motility. White circles at nodes indicate UltraFast bootstrap support ≥95%. Gray bars denote the 5% and 95% quantiles of the distribution of node ages across bootstrap replicates in millions of years (Myr). Red angular brackets denote calibrations, single ‘>’ for a minimum bound only and double ‘< >' for minimum and maximum bounds. Clade labels for oogamous nonmotile (ON), anisogamous nonmotile (AN) and anisogamous motile (AM) lineages match the groups in Figs 2(c) and 3(b–d).
We calculated net diversification rates using (1) the crown or stem age of clades across bootstrap trees, (2) the total number of species per lineage (see below), and (3) an estimate of the diatom‐wide relative extinction rate (or ‘extinction fraction’, ε = d/b; see below). We also calculated the 95% confidence interval for the expected size of a clade given its crown or stem age (Magallón & Sanderson, 2001).
The total number of diatom species is unknown, and recent approximations vary by orders of magnitude, from 2 × 104 (Guiry, 2012) to 105 (Mann & Vanormelingen, 2013), indicating that our phylogenetic hypothesis includes, at best, 5% of all diatom species. We therefore calculated the net diversification rate across a range of diversity estimates: (1) N Total = 11 958 species, the AlgaeBase total number of taxonomically accepted names for all genera represented in the phylogeny (Guiry & Guiry, 2017); (2) N Total = 10 569 species, the DiatomBase total number of taxonomically accepted names for extant species in all genera represented in the phylogeny (Kociolek et al., 2017); (3) N Total = 20 000 species (Guiry, 2012); (4) N Total = 30 000 species, the lower estimate of Mann & Vanormelingen (2013); and (5) N Total = 100 000 species, the upper estimate of Mann & Vanormelingen (2013).
Standing diversity for analyses at the genus and clade levels was taken from both AlgaeBase and DiatomBase. We report results that used data from DiatomBase because it provides an estimate for the number of extant species, which is the quantity of interest for diversification models (Magallón & Sanderson, 2001). However, a comparison of extant diversity per genus from DiatomBase with total diversity per genus from AlgaeBase showed similar numbers, and both datasets provided qualitatively similar results.
Our calculations of net diversification required a fixed value for the relative extinction rate (Magallón & Sanderson, 2001). We followed the method of Foote (2000), as implemented in the R package paleotree v.2.6 (Bapst, 2012), to obtain an empirical estimate of relative extinction. This approach used the first and last occurrence of fossil species binned into discrete, nonoverlapping time intervals to calculate the instantaneous rates of speciation and extinction. Fossil data were taken from the Barron Diatom Catalog of first and last occurrences of Cenozoic diatoms (Lazarus et al., 2014). To obtain a single estimate of relative extinction for the entire set of fossils spanning the Cenozoic, we binned the data into discrete time intervals, calculated rates of speciation and extinction, and then averaged over time intervals. To assess the effect of interval duration on the estimates of relative extinction and downstream analyses, we repeated the calculations with bin durations of 0.5, 1.0, 2.5 and 5.0 Myr.
Discrete shifts and temporal variation in diversification
To complement the analysis based on predefined taxonomic ranks, we used a rank‐free approach to identify discrete shifts in diversification rate that uses a stepwise Akaike information criterion (AIC) procedure (Medusa; Alfaro et al., 2009; Brown et al., 2016). The addition of a discrete shift to the Medusa diversification model increases the number of model parameters, and so more complex models were retained only when the improvement in AIC was greater than a threshold derived from the size of the phylogeny (N = 234, ΔAICc = 6.7; ‘genus‐level’ analysis). To account for discordance between taxonomy and phylogeny, we repeated the analysis using a clade‐level phylogeny (N = 45, ΔAICc = 2.3; ‘clade‐level’ analysis). We restricted the algorithm to test only birth–death models and allowed shifts in diversification at both stems and nodes of the phylogeny.
To summarize Medusa results and account for uncertainty in divergence times across bootstrap trees, we focused on breakpoints detected in ≥50% or ≥75% of bootstrap trees. We also calculated temporal trends of net diversification (r = birth − death), net turnover rate (τ = birth + death sensu Beaulieu & O'Meara (2016)) and relative extinction (ε = death/birth). We split the phylogenies into 1‐Myr intervals and obtained summary statistics for parameters of all branches intersecting an interval (Tank et al., 2015).
Results
Time‐calibrated phylogeny of diatoms
Consistent with previous multigene‐based (e.g. Theriot et al., 2015) and transcriptome‐based (Parks et al., 2017) phylogenies, we reconstructed the centric diatoms as a grade of five large clades (Figs 1a, S1). A clade of Corethrales + Leptocylindrales was sister to all other diatoms, and a clade of the multipolar diatoms Attheya and Biddulphia was sister to the clade of pennate diatoms (Figs 1a, S1). The diatom crown age was estimated at 190.4 Ma (mean across bootstrap trees), placing the origin of diatoms near the Triassic–Jurassic boundary (Fig. 1a). Origins of genus‐level diversity within the grade of centric diatoms largely pre‐dated the Cenozoic, with notable exceptions within Eupodiscales, Cymatosirales, Lithodesmiales and Thalassiosirales (Fig. 1a). These results suggested both temporal and lineage‐specific variation in the origination of genera, whereby genera within the radial centric grade diversified much earlier than their counterparts in the polar centric grade (Fig. 1a). The pennate and raphid lineages also had their origins in the Cretaceous, with genus originations distributed more evenly throughout the Cretaceous and Cenozoic (Fig. 1b,c).
Accelerated diversification in diatoms with actively motile vegetative cells
The analysis of Cenozoic marine diatom fossils showed that both speciation (birth, b) and extinction (death, d) decreased during the first 20 Myr of the Cenozoic (Fig. 2a). The last 40–45 Myr were marked by a slow increase in turnover (b + d) that first peaked at c. 35 Ma and then again towards the recent (Fig. 2a). These results were in agreement with recent paleontological studies based on similar data (Lazarus et al., 2014; Cermeño et al., 2015).
Figure 2.
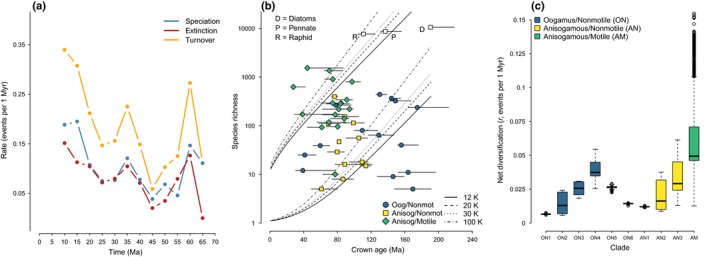
Diatom diversification based on fossil and phylogenetic data at predefined clades. (a) Rates of speciation (birth, b), extinction (death, d) and species turnover (τ = b + d) based on first–last occurrence data for Cenozoic diatoms from the Barron Diatom Catalog. (b) Confidence intervals (95%) for species richness of predefined diatom clades, based on crown ages, standing diversity and a relative extinction (ε = d/b) of ε = 0.751. Line types correspond to different approximations of total diatom diversity (see the Materials and Methods section). Species richness from DiatomBase is plotted against crown age (million years ago, Ma) and color coded by the combination of reproductive mode and locomotion. Error bars span the minimum and maximum crown ages of clades across bootstrap trees in million years (Myr). The clades of all diatoms (D), pennate diatoms (P) and raphid diatoms (R) are shown as open squares. Clades younger than 10 Myr or having fewer than five species are omitted. (c) Net diversification rates (r = b–d), averaged over bootstrap phylogenies and pooled based on the most inclusive monophyletic groups with a certain combination of reproductive and locomotory traits. Lines, boxes and whiskers represent medians, inter‐quartile range and quartile values + 1.5 × quartile values, respectively. See labels in Fig. 1 for contents of oogamous nonmotile (ON), anisogamous nonmotile (AN) and anisogamous motile (AM) groups. Oog, oogamous; Anisog, anisogamous; Nonmot, nonmotile. See Supporting Information Fig. S4 for an identical analysis with AlgaeBase instead of DiatomBase species richness.
Using these data, we calculated that Cenozoic marine fossil diatoms diversified at a mean rate of r = 0.029 (SD = 0.007) net speciation events per 1 Myr, with a relatively high mean relative extinction rate of ε = 0.751 (SD = 0.081; Table 1). These values were averages of calculations performed with the fossil data binned into discrete, nonoverlapping time intervals with durations of 0.5, 1.0, 2.5 and 5.0 Myr (Fig. S2). Comparison of results obtained with different interval durations indicated that the estimates of relative extinction varied with the granularity of the fossil data, being highest for 5‐Myr intervals and lowest for 0.5‐Myr intervals (Fig. S2). The most pronounced downstream effect of relative extinction calculated with different interval durations was noticed for young clades inferred to have diversified at a high rate (Fig. S3). For such clades, relative extinction calculated with a bin size of 5 Myr provided the most conservative values for net diversification (Fig. S3). Although this variation had a minor effect on downstream calculations of net diversification (Fig. S3), we nonetheless performed all downstream analyses with relative extinction averaged over the estimates obtained from the four different bin sizes (Table 1; Fig. S2).
Table 1.
Instantaneous rates of speciation (birth, b) and extinction (death, d) based on first–last occurrence data of Cenozoic marine diatoms summarized over time
| Median | Mean | SD | |
|---|---|---|---|
| Speciation (b) | 0.093 | 0.097 | 0.009 |
| Extinction (d) | 0.086 | 0.085 | 0.005 |
| Net diversification (r = b − d) | 0.029 | 0.027 | 0.007 |
| Relative extinction (ε = d/b) | 0.731 | 0.751 | 0.081 |
| Net turnover (τ = d + b) | 0.193 | 0.190 | 0.030 |
The fossil data were binned into discrete, nonoverlapping intervals with durations of 0.5, 1.0, 2.5 and 5.0 million yr (Myr). Summary statistics were calculated for each granularity, and the reported values are averages of the median parameter estimates across bin sizes. See Supporting Information Fig. S2 for more details on parameter estimates at different granularities.
Using the above estimate of relative extinction averaged through time and across bin sizes (ε = 0.751), the inferred crown age of diatoms (mean = 190.4 Ma; Fig. 1) and different approximations of total diatom diversity, we calculated a minimum net diversification of r = 0.044 events per 1 Myr per lineage (SD = 0.00033) for N Total = 20 000 species and a maximum of r = 0.052 (SD = 0.00039) for N Total = 100 000 species. The 95% confidence limits on expected species richness under the least conservative scenario (N Total = 100 000 species) indicated that the expected diversity of a clade with a crown age of 50 Myr could range from two to 218 species.
Although the numbers of oogamous/nonmotile genera (ON; number of genera N G = 86), anisogamous/nonmotile genera (AN; N G = 61) and anisogamous/motile genera (AM, raphid; N G = 85) included in our analyses were comparable (Fig. 1), at the species level, the number of described AM species (DiatomBase number of species, N S = 9533) far exceeds the numbers of described ON (N S = 1871) and AN (N S = 759) diatoms (Figs 2b, 3). This unbalanced distribution of species‐level diversity was evident in the diversity‐by‐age plot (Fig. 2b). The raphid pennate (AM) lineage as a whole was more diverse than expected, or, at least, at the upper bound of expected diversity for N Total = 100 000 species (Fig. 2b). The broader pennate diatom lineage (AN + AM) was also more diverse than expected when the assumed total diatom diversity was ≤ 20 000 species (Fig. 2b).
Figure 3.
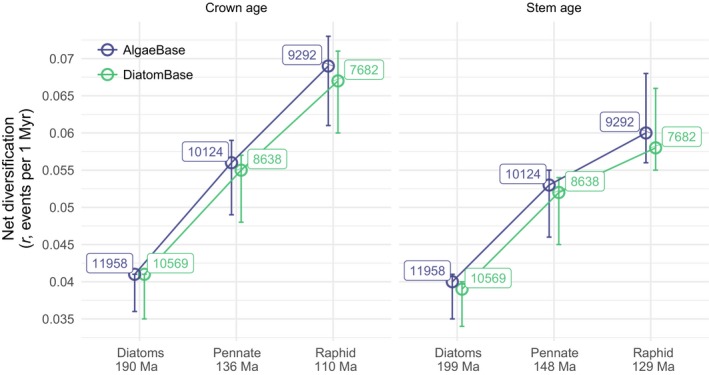
Net diversification (birth − death, r = b − d, events per million years) estimated from known standing diversity and crown or stem ages for all diatoms (ON + AN + AM), the bilaterally symmetrical clade (pennate diatoms, AN + AM) and the actively motile clade (raphid diatoms, AM). Shown are the mean values (circles) and minimum–maximum range (error bars) across bootstrap phylogenies. All calculations used relative extinction ε = 0.751 estimated from Cenozoic fossil data (Table 1). Numbers next to the points indicate the total number of species in the respective databases. Numbers below the clade names are the estimates of crown and stem ages in million years ago (Ma, Fig. 1). ON, oogamous nonmotile; AN, anisogamous nonmotile; AM, anisogamous motile.
At a lower phylogenetic scale, only lineages within the actively motile raphid pennate (AM) clade had higher than expected species richness, whereas the species richness of clades within the grades of AN and ON diatoms either was within or below the bounds of expected species richness (Fig. 2b). The estimated diversification rate of raphid (AM) diatoms as a whole was 1.66 times greater than that of all diatoms (Fig. 3), and the diversification rate of AM diatoms was significantly higher than that of any of the AN and ON lineages, with rate ratios ranging between 1.6 and 10.7 (Tukey post hoc tests, P adj < 0.001, Fig. 2c). These comparisons were obtained by pooling and averaging diversification rate estimates of clades grouped into the most inclusive monophyletic groups with a particular combination of life history and locomotory traits. For example, for the AM lineages that together form a single monophyletic group (raphid pennate diatoms), we averaged over all AM clades, whereas, as the AN and ON lineages are paraphyletic, we pooled clade‐level parameters into the most inclusive monophyletic groups (e.g. Corethrales + Leptocylindrales; Fig. 1). Results using stem rather than crown clade ages (Fig. 3) and alternative sources for the number of species, i.e. AlgaeBase instead of DiatomBase, agreed with these results (Figs 3, S4).
Discrete shifts and temporal trends in diatom diversification
In the genus‐level analysis with DiatomBase species richness data, Medusa identified 23 rate shifts that occurred in at least 5% of the bootstrap trees. Of these, 13 were present in ≥50% and 10 were detected in ≥75% of trees (Fig. 4a). Most shifts occurred within the past c. 70 Myr, and upward shifts tended to be more recent, consistent with the Cenozoic increase in diatom diversity as a whole (Rabosky & Sorhannus, 2009; Lazarus et al., 2014; Cermeño et al., 2015), as well as many lineages of both ON (Alverson, 2014) and AM (Edwards, 1991) diatoms (Fig. 4a). Breakpoints at deeper internal branches were detected in Thalassiosirales (ON, upward shift), deeper within a clade of multipolar diatoms (ON, downward shift) and in two clades of AN diatoms (downward shifts, Fig. 4a). Two phylogenetically deep upward shifts coincided with the transitions from oogamy to anisogamy in the ancestor of pennate diatoms (frequency = 0.70, magnitude = 0.007) and with the evolution of active motility in the ancestor of raphid pennate diatoms (frequency = 0.69, magnitude = 0.014, Fig. 4a).
Figure 4.
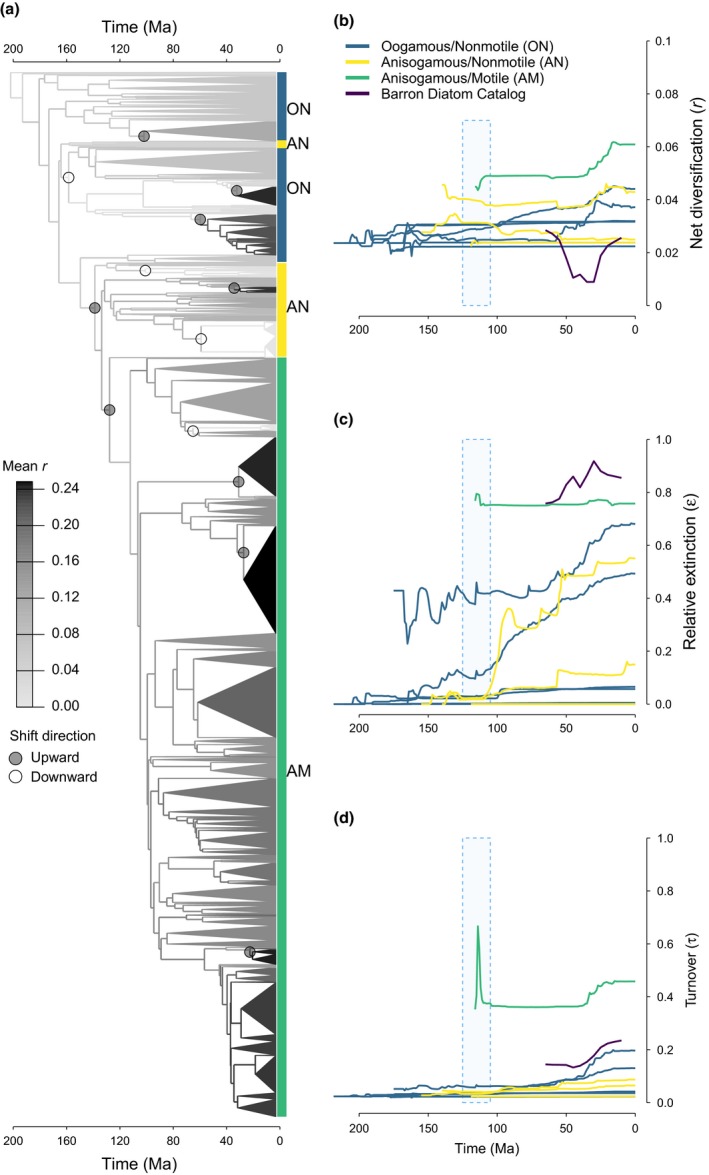
Discrete shifts and temporal trends of diversification across diatoms. (a) Medusa reconstruction of rates of (birth − death, r = b − d, events per million years) across the genus‐level phylogeny of diatoms + Parmales. Triangle size is proportional to the number of species per genus (from DiatomBase), and the shading corresponds to the estimated net diversification rate. Rate shifts detected in ≥50% of trees are shown. (b–d) Temporal trends in net diversification, relative extinction (ε = d/b) and turnover (τ = b + d) as estimated by the genus‐level Medusa analysis. Trend lines were calculated by slicing the bootstrap phylogenies into 1 million yr (Myr) intervals and averaging the branch‐associated parameters for each slice across trees. Trendlines are shown for the most inclusive clades with a combination of reproductive and locomotory traits, and match the groups in Fig. 2(c) and the clade labels in Fig. 1. Rates estimated from the Cenozoic marine fossil record (bin size, 5 Myr) were smoothed using nonparametric local polynomial regression (LOESS), but are otherwise not different from Fig. 2(a). The timing of the evolution of active motility (the range of crown age estimates for raphid pennates across bootstrap phylogenies) is shown as a blue rectangle. See Supporting Information Fig. S5 for an identical analysis with AlgaeBase instead of DiatomBase species richness. Ma, million years ago.
Averaged branch‐associated parameters binned into 1‐Myr intervals showed increases in the rates of net diversification, relative extinction and species turnover after the emergence of actively motile raphid pennate diatoms (Fig. 4b–d). Temporal trends in lineages with different life history strategies reinforced the finding of faster diversification in actively motile raphid diatoms (AM) compared with their nonmotile counterparts (ON and AN, Fig. 4b). This difference in diversification, however, was not caused by higher rates of speciation among raphid diatoms against a background of similar extinction rates, or the reverse, lower rates of extinction with comparable speciation. The difference instead was a result of increases in both the birth of new lineages and their extinction (turnover = birth + death, Fig. 4d). Accordingly, a higher fraction of species per unit time went extinct among AM lineages than in the ON and AN grades (Fig. 4c). We found qualitatively similar results when using species richness data from AlgaeBase instead of DiatomBase, and when we collapsed the phylogeny down to the 45 predefined clades used in the previous analysis (Figs 3, S4, S5).
Discussion
Diversity patterns are frequently associated with evolutionary innovations and ecological opportunities that are thought to alter the rates at which species are born and lost through time. Transitions in locomotory and life history strategies are important traits that may underlie major shifts in diversification (Leclère et al., 2009; Goldberg et al., 2010; Ikeda et al., 2012; Cieslak et al., 2014; de Vos et al., 2014). Changes in life history may also drive shifts in locomotion (or vice versa) to optimally match these traits, which are often linked in microbial eukaryotes (Hoek et al., 1995).
Our results suggest that an interaction between motility and life history during diatom evolution may have promoted an increase in the rate of diversification of raphid pennate diatoms, a lineage that evolved a novel mechanism of directed motility in vegetative cells following the earlier loss of flagellated male gametes (Figs 2, 3, 4). At least 137 Ma (Fig. 1), the MRCA of pennate diatoms experienced a reduction in the morphological and behavioral differences between gametes, such that the typical egg‐and‐sperm oogamy present in the diatom ancestor, and largely preserved across the grade of ON (‘centric’) diatoms, was replaced with anisogamy. It is unclear what, if any, selective pressures led to the loss of the ancestral oogamous reproductive system. Oogamy is thought to be advantageous because a species can maximize the number of gametes and, as a result, successful copulation encounters by producing many small and highly motile male gametes, as well as zygote survivorship by producing one or a few large (and immobile) female gametes (Parker et al., 1972; Maynard Smith, 1978; Matsuda & Abrams, 1999; Bulmer & Parker, 2002; Togashi et al., 2012). In pennate diatoms, however, the flagellum of male gametes was lost, and dimorphism between gametes diminished, such that gametes in a copulation pair converged towards a similar, intermediate size (reviewed in Kaczmarska et al., 2013), but differed in their behavior (Drebes, 1977), with male gametes that moved by pseudopodia to search for a nonmotile female gamete (Davidovich et al., 2010, 2017; Sato et al., 2011; Kaczmarska et al., 2017). The evolution of the raphe and active motility in vegetative cells enabled gametangia, rather than gametes, to search for mating partners, promoting further reduction in sexual dimorphism and gamete mobility. An added benefit of raphe‐enabled locomotion is linked to the evolution of a system resembling internal fertilization, whereby copulation occurs between closely spaced cells protected by a mucilaginous ‘copulation envelope’ (Drebes, 1977; Round et al., 1990; Kaczmarska et al., 2013). Thus, in raphid pennate diatoms, the choice of a compatible mate precedes investment in meiosis, providing greater certainty of copulation and increasing the odds of the successful production of a viable zygote.
The novel combination of traits in raphid diatoms probably makes it easier to search for and find a genetically compatible mate. The diplontic life cycle of diatoms is characterized by long periods of asexual reproduction and short, periodic bouts of sexual reproduction. If vegetative cells cannot move, repeated vegetative divisions might lead to clonal patches of sibling cells, especially in benthic, nonmotile species that often live attached to substrates. This, in turn, might increase the chance of selfing or delay sexual reproduction until a nonclonal partner is encountered. The frequency of sexual reproduction and the rates of selfing or outcrossing in natural populations of diatoms are unknown and difficult to estimate. However, in culture, the majority of surveyed pennate diatoms are heterothallic, with mating occurring only between genetically compatible clones (Chepurnov & Mann, 2004; Chepurnov et al., 2004; but see, e.g., Davidovich et al., 2010, 2017). This led to the hypothesis that heterothally is the ancestral condition in pennates (Chepurnov et al., 2004). Moreover, when selfing has been observed and the progeny followed for a few generations thereafter, the cultured strains experienced inbreeding depression, resulting in reduced rates of gamete fusion and inviable zygotes (e.g. Chepurnov & Mann, 1999). It follows that selfing is probably rare in natural populations, and the various motility mechanisms that have evolved in different life history stages (gametes vs vegetative cells) might represent alternative strategies for increasing the frequency or efficiency of sex and outcrossing. The combination of life history and motility traits present in raphid pennate diatoms suggests that sexual reproduction is both more frequent and more efficient in this lineage compared with clades with nonmotile vegetative cells.
This interaction between locomotion and life history had lasting consequences for the ways in which raphid diatoms interact with conspecifics, heterospecifics and the environment. Raphe‐enabled motility relies on an actin–myosin system (Poulsen et al., 1999), and is directional, reversible and much faster than other forms of movement present in some nonraphid diatoms (Pickett‐Heaps et al., 1986, 1991; Kooistra et al., 2003). Active motility in vegetative cells enabled directed movement towards microhabitats with specific light, nutrient and temperature conditions (Cohn et al., 2015; Bondoc et al., 2016a), diurnal and tidal migrations (Palmer & Round, 1967), gravitactic behaviors (Frankenbach et al., 2014), predator avoidance (Kingston, 1999) and pheromonal migration (Sato et al., 2011; Gillard et al., 2013; Bondoc et al., 2016b; Moeys et al., 2016; Basu et al., 2017). This broad range of ecological benefits – unavailable to nonraphid diatoms – expanded the repertoire of habitats available for colonization, creating new opportunities for niche specialization. Active motility, and its tight association with substrate, might have lowered the overall rates of passive dispersal, reducing long‐range connectivity between populations, ultimately leading to greater isolation between local populations. Finally, the potentially higher frequency of sexual reproduction might have contributed to faster rates of adaptive divergence and the maintenance of reproductive isolation (Barraclough et al., 2003). Overall, the ability to move in response to both biotic and environmental stimuli appears to have provided greater potential for adaptive change and improved flexibility in dealing with habitat complexity in benthic habitats, where raphid diatoms thrive. Ultimately, the benefits of this novel locomotory trait might have contributed to the elevated diversification rate found in raphid pennate diatoms (Figs 2, 3, 4).
Although the myriad benefits of raphe‐enabled motility have been thoroughly characterized (Consalvey et al., 2004), these associations are nevertheless correlative and with a sample size of one (i.e. the gain of a raphe occurred just once in diatom evolution). This highlights a long‐standing challenge in comparative evolutionary biology. Namely, it is impossible to derive statistical support for an association between a trait and property, whether it is diversification or the evolution of another trait, when the focal trait evolved once (Maddison & FitzJohn, 2015; Beaulieu & O'Meara, 2016). Moreover, although it is likely that locomotion had a large effect on diatom evolution, diversification within raphid pennate diatoms was certainly influenced by other environmental, ecological and genetic factors. In this regard, the diversification of raphe‐bearing pennate diatoms resembles that of many other exceptionally diverse clades (e.g. flowering plants and insects), whose evolutionary success appears to be linked, at least superficially, to innovations that evolved only once. A more complete understanding of the diversification of diatoms, including raphid pennates, will need to account for the synergistic effects of other traits, biogeography and environmental changes, whose combined influence probably contributed to their diversification (Donoghue & Sanderson, 2015).
Author contributions
T.N., J.M.B. and A.J.A. designed the study. T.N. collected the data and performed the analyses. T.N., J.M.B. and A.J.A. wrote the manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Phylogeny of 1151 diatoms and 20 outgroups reconstructed using an 11‐gene dataset.
Fig. S2 Diatom diversification estimated from first–last occurrence data for Cenozoic fossils.
Fig. S3 Granularity of fossil data and its effect on the phylogenetic estimates of diversification.
Fig. S4 Diatom diversification at predefined ranks with diversity data from AlgaeBase.
Fig. S5 Discrete shifts and temporal trends of diversification across diatoms with diversity data from AlgaeBase.
Table S1 Sequence data for diatoms and Parmales
Table S2 Properties of the phylogenetic dataset, partitioning and model selection
Table S3 ;Minimum and maximum bounds for the calibration of internal nodes
Acknowledgements
We thank Jakub Witkowski (Uniwersytet Szczeciński, Poland) and Matt Ashworth (The University of Texas at Austin, TX, USA) for discussions on the diatom fossil record. Pat Kociolek (University of Colorado, Boulder, CO, USA) provided helpful information about diatom species numbers in AlgaeBase and DiatomBase. We thank Adam Siepielski (University of Arkansas, Fayetteville, AR, USA) and Edward Theriot (The University of Texas at Austin, TX, USA) for comments on an earlier version of the manuscript. This research used computational resources available through the Arkansas High Performance Computing Center, which was funded through multiple National Science Foundation (NSF) grants and the Arkansas Economic Development Commission. This work was supported by the NSF (grant no. DEB‐1353131 to AJA) and by a grant from the Simons Foundation (403249, A.J.A.).
References
- Alfaro ME, Santini F, Brock C, Alamillo H, Dornburg A, Rabosky DL, Carnevale G, Harmon LJ. 2009. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proceedings of the National Academy of Sciences, USA 106: 13410–13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson AJ. 2014. Timing marine–freshwater transitions in the diatom order Thalassiosirales. Paleobiology 40: 91–101. [Google Scholar]
- Alverson AJ, Jansen RK, Theriot EC. 2007. Bridging the Rubicon: phylogenetic analysis reveals repeated colonizations of marine and fresh waters by thalassiosiroid diatoms. Molecular Phylogenetics and Evolution 45: 193–210. [DOI] [PubMed] [Google Scholar]
- Andersen RA. 2004. Biology and systematics of heterokont and haptophyte algae. American Journal of Botany 91: 1508–1522. [DOI] [PubMed] [Google Scholar]
- Bapst D. 2012. Paleotree: an R package for paleontological and phylogenetic analyses of evolution. Methods in Ecology and Evolution 3: 803–807. [Google Scholar]
- Barraclough TG, Birky CW Jr, Burt A. 2003. Diversification in sexual and asexual organisms. Evolution 57: 2166–2172. [DOI] [PubMed] [Google Scholar]
- Basu S, Patil S, Mapleson D, Russo MT, Vitale L, Fevola C, Maumus F, Casotti R, Mock T, Caccamo M et al 2017. Finding a partner in the ocean: molecular and evolutionary bases of the response to sexual cues in a planktonic diatom. New Phytologist 215: 140–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, O'Meara BC. 2016. Detecting hidden diversification shifts in models of trait‐dependent speciation and extinction. Systematic Biology 65: 583–601. [DOI] [PubMed] [Google Scholar]
- Bondoc KGV, Heuschele J, Gillard J, Vyverman W, Pohnert G. 2016a. Selective silicate‐directed motility in diatoms. Nature Communications 7: 10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondoc KGV, Lembke C, Vyverman W, Pohnert G. 2016b. Searching for a mate: pheromone‐directed movement of the benthic diatom Seminavis robusta . Microbial Ecology 72: 287–294. [DOI] [PubMed] [Google Scholar]
- Brown JW, FitzJohn RG, Alfaro ME, Harmon LJ. 2016. turboMEDUSA: Modelling evolutionary diversification using stepwise AIC. R package version 0.951. [WWW document] URL https://github.com/josephwb/turboMEDUSA [accessed 28 August 2017].
- Bulmer MG, Parker GA. 2002. The evolution of anisogamy: a game‐theoretic approach. Proceedings of the Royal Society of London B: Biological Sciences 269: 2381–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Müller KM et al 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermeño P, Falkowski PG, Romero OE, Schaller MF, Vallina SM. 2015. Continental erosion and the Cenozoic rise of marine diatoms. Proceedings of the National Academy of Sciences, USA 112: 4239–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepurnov V, Mann D. 1999. Variation in the sexual behaviour of Achnanthes longipes (Bacillariophyta). II. Inbred monoecious lineages. European Journal of Phycology 34: 1–11. [Google Scholar]
- Chepurnov VA, Mann DG. 2004. Auxosporulation of Licmophora communis (Bacillariophyta) and a review of mating systems and sexual reproduction in araphid pennate diatoms. Phycological Research 52: 1–12. [Google Scholar]
- Chepurnov VA, Mann DG, Sabbe K, Vyverman W. 2004. Experimental studies on sexual reproduction in diatoms. International Review of Cytology 237: 91–154. [DOI] [PubMed] [Google Scholar]
- Cieslak A, Fresneda J, Ribera I. 2014. Life‐history specialization was not an evolutionary dead‐end in Pyrenean cave beetles. Proceedings of the Royal Society of London B: Biological Sciences 281: 20132978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn SA, Halpin D, Hawley N, Ismail A, Kaplan Z, Kordes T, Kuhn J, Macke W, Marhaver K, Ness B et al 2015. Comparative analysis of light‐stimulated motility responses in three diatom species. Diatom Research 30: 213–225. [Google Scholar]
- Condamine FL, Clapham ME, Kergoat GJ. 2016. Global patterns of insect diversification: towards a reconciliation of fossil and molecular evidence? Scientific Reports 6: 19208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consalvey M, Paterson DM, Underwood GJC. 2004. The ups and downs of life in a benthic biofilm: migration of benthic diatoms. Diatom Research 19: 181–202. [Google Scholar]
- Davidovich NA, Davidovich OI, Podunay YA, Gastineau R, Kaczmarska I, Poulíčková A, Witkowski A. 2017. Ardissonea crystallina has a type of sexual reproduction that is unusual for centric diatoms. Scientific Reports 7: 14670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich NA, Kaczmarska I, Ehrman JM. 2010. Heterothallic and homothallic sexual reproduction in Tabularia fasciculata (Bacillariophyta). Fottea 10: 251–266. [Google Scholar]
- Donoghue MJ, Sanderson MJ. 2015. Confluence, synnovation, and depauperons in plant diversification. New Phytologist 207: 260–274. [DOI] [PubMed] [Google Scholar]
- Drebes G. 1977. Sexuality In: Werner D, ed. The biology of diatoms. Oxford, UK: Blackwell Scientific Publications, 250–283. [Google Scholar]
- Drummond CS, Eastwood RJ, Miotto STS, Hughes CE. 2012. Multiple continental radiations and correlates of diversification in Lupinus (Leguminosae): testing for key innovation with incomplete taxon sampling. Systematic Biology 61: 443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda TF Jr, Rolán E. 2005. Explosive radiation of Cape Verde Conus, a marine species flock. Molecular Ecology 14: 267–272. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Edwards AR. 1991. The Oamaru Diatomite. New Zealand Geological Survey paleontological bulletin, 64. DSIR Geology & Geophysics, Lower Hutt, New Zealand.
- Foote M. 2000. Origination and extinction components of taxonomic diversity: general problems In: Erwin DH, Wing SL, eds. Deep time: paleobiology's perspective. Lawrence, KS, USA: The Paleontological Society, 74–102. [Google Scholar]
- Frankenbach S, Pais C, Martinez M, Laviale M, Ezequiel J, Serôdio J. 2014. Evidence for gravitactic behaviour in benthic diatoms. European Journal of Phycology 49: 429–435. [Google Scholar]
- Gersonde R, Harwood DM. 1990. 25. Lower Cretaceous diatoms from ODP LEG 113 Site 693 (Weddel Sea). Part 1: Vegetative cells. In: Barker PF, Kenneth JP. et al, eds. Proceedings of the Ocean Drilling Program, Scientific Results 113: 365–402. [Google Scholar]
- Gillard J, Frenkel J, Devos V, Sabbe K, Paul C, Rempt M, Inzé D, Pohnert G, Vuylsteke M, Vyverman W. 2013. Metabolomics enables the structure elucidation of a diatom sex pheromone. Angewandte Chemie, International Edition 52: 854–857. [DOI] [PubMed] [Google Scholar]
- Goldberg EE, Kohn JR, Lande R, Robertson KA, Smith SA, Igić B. 2010. Species selection maintains self‐incompatibility. Science 330: 493–495. [DOI] [PubMed] [Google Scholar]
- Guiry MD. 2012. How many species of algae are there? Journal of Phycology 48: 1057–1063. [DOI] [PubMed] [Google Scholar]
- Guiry MD, Guiry GM. 2017. AlgaeBase. World‐wide electronic publication, National University of Ireland, Galway. [WWW document] URL http://www.algaebase.org [accessed 30 November 2017].
- Harper M. 1977. Movements In: Werner D, ed. Botanical monographs. The biology of diatoms. Berkeley, Los Angeles, CA, USA: University of California Press, 224–249. [Google Scholar]
- Harwood DM, Gersonde R. 1990. Lower Cretaceous diatoms from ODP LEG 113 Site 693 (Weddel Sea). Part 2: Resting spores, chysophycean cysts and endoskeletal dinoflagellate, and notes on the origin of diatoms. In: Barker PF, Kenneth JP. et al, eds. Proceedings of the Ocean Drilling Program, Scientific Results 113: 403–425. [Google Scholar]
- Harwood DM, Nikolaev VA. 1995. Cretaceous diatoms: morphology, taxonomy, biostratigraphy In: Babcock LE, Ausich WI, eds. Short courses in paleontology. Siliceous microfossils. Knoxville, TN, USA: The Paleontological Society, 81–106. [Google Scholar]
- Harwood DM, Nikolaev VA, Winter DM. 2007. Cretaceous records of diatom evolution, radiation, and expansion. Paleontological Society Papers 13: 33–59. [Google Scholar]
- Hoek C, Mann D, Jahns HM. 1995. Algae: an introduction to phycology. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Ikeda H, Nishikawa M, Sota T. 2012. Loss of flight promotes beetle diversification. Nature Communications 3: 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarska I, Gray BS Jr, Ehrman JM, Thaler M. 2017. Sexual reproduction in plagiogrammacean diatoms: first insights into the early pennates. PLoS ONE 12: e0181413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarska I, Poulíčková A, Sato S, Edlund MB, Idei M, Watanabe T, Mann DG. 2013. Proposals for a terminology for diatom sexual reproduction, auxospores and resting stages. Diatom Research 28: 263–294. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston MB. 1999. Wave effects on the vertical migration of two benthic microalgae: Hantzschia virgata var. intermedia and Euglena proxima . Estuaries and Coasts 22: 81–91. [Google Scholar]
- Kociolek JP, Balasubramanian K, Blanco S, Coste M, Ector L, Liu Y, Kulikovskiy M, Lundholm N, Ludwig T, Potapova M et al 2017. DiatomBase. [WWW document] URL http://www.diatombase.org [accessed 30 November 2017].
- Kooistra WH, De Stefano M, Mann DG, Salma N, Medlin LK. 2003. Phylogenetic position of Toxarium, a pennate‐like lineage within centric diatoms (Bacillariophyceae). Journal of Phycology 39: 185–197. [Google Scholar]
- Lazarus D. 1994. Neptune: a marine micropaleontology database. Mathematical Geology 26: 817–832. [Google Scholar]
- Lazarus D, Barron J, Renaudie J, Diver P, Türke A. 2014. Cenozoic planktonic marine diatom diversity and correlation to climate change. PLoS ONE 9: e84857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclère L, Schuchert P, Cruaud C, Couloux A, Manuel M. 2009. Molecular phylogenetics of Thecata (Hydrozoa, Cnidaria) reveals long‐term maintenance of life history traits despite high frequency of recent character changes. Systematic Biology 58: 509–526. [DOI] [PubMed] [Google Scholar]
- Leslie AB, Beaulieu JM, Crane PR, Donoghue MJ. 2013. Explaining the distribution of breeding and dispersal syndromes in conifers. Proceedings of the Royal Society of London B: Biological Sciences 280: 20131812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, FitzJohn RG. 2015. The unsolved challenge to phylogenetic correlation tests for categorical characters. Systematic Biology 64: 127–136. [DOI] [PubMed] [Google Scholar]
- Magallón S, Sanderson MJ. 2001. Absolute diversification rates in angiosperm clades. Evolution 55: 1762–1780. [DOI] [PubMed] [Google Scholar]
- Mann DG, Vanormelingen P. 2013. An inordinate fondness? The number, distributions, and origins of diatom species. Journal of Eukaryotic Microbiology 60: 414–420. [DOI] [PubMed] [Google Scholar]
- Matsuda H, Abrams PA. 1999. Why are equally sized gametes so rare? The instability of isogamy and the cost of anisogamy. Evolutionary Ecology Research 1: 769–784. [Google Scholar]
- Maynard Smith J. 1978. The evolution of sex. New York, NY, USA: Cambridge University Press Archive. [Google Scholar]
- Medlin L. 2015. A timescale for diatom evolution based on four molecular markers: reassessment of ghost lineages and major steps defining diatom evolution. Life and Environment 65: 219–238. [Google Scholar]
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30: 1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeys S, Frenkel J, Lembke C, Gillard JTF, Devos V, Van den Berge K, Bouillon B, Huysman MJJ, De Decker S, Scharf J et al 2016. A sex‐inducing pheromone triggers cell cycle arrest and mate attraction in the diatom Seminavis robusta . Scientific Reports 6: 19252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakov T, Ruck EC, Galachyants Y, Spaulding SA, Theriot EC. 2014. Molecular phylogeny of the Cymbellales (Bacillariophyceae, Heterokontophyta) with a comparison of models for accommodating rate variation across sites. Phycologia 53: 359–373. [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR. 2009. Infernal 1.0: inference of RNA alignments. Bioinformatics 25: 1335–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L‐T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ‐TREE: a fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris RW, Strope CL, McCandlish DM, Stoltzfus A. 2015. Bayesian priors for tree calibration: evaluating two new approaches based on fossil intervals. BioRxiv: 014340.
- Palmer JD, Round FE. 1967. Persistent, vertical‐migration rhythms in benthic microflora. VI. The tidal and diurnal nature of the rhythm in the diatom Hantzschia virgata . Biological Bulletin 132: 44–55. [Google Scholar]
- Palumbi SR. 1992. Marine speciation on a small planet. Trends in Ecology & Evolution 7: 114–118. [DOI] [PubMed] [Google Scholar]
- Palumbi SR. 1994. Genetic divergence, reproductive isolation, and marine speciation. Annual Review of Ecology and Systematics 25: 547–572. [Google Scholar]
- Parker GA, Baker RR, Smith V. 1972. The origin and evolution of gamete dimorphism and the male–female phenomenon. Journal of Theoretical Biology 36: 529–553. [DOI] [PubMed] [Google Scholar]
- Parks MB, Wickett NJ, Alverson AJ. 2017. Signal, uncertainty, and conflict in phylogenomic data for a diverse lineage of microbial eukaryotes (Diatoms, Bacillariophyta). Molecular Biology and Evolution 35: 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett‐Heaps J, Hill DRA, Blaze KL. 1991. Active gliding motility in an araphid marine diatom, Ardissonea (formerly Synedra) crystallina . Journal of Phycology 27: 718–725. [Google Scholar]
- Pickett‐Heaps JD, Hill DRA, Wetherbee R. 1986. Cellular movement in the centric diatom Odontella sinensis . Journal of Phycology 22: 334–339. [Google Scholar]
- Poulsen NC, Spector I, Spurck TP, Schultz TF, Wetherbee R. 1999. Diatom gliding is the result of an actin–myosin motility system. Cell Motility and the Cytoskeleton 44: 23–33. [DOI] [PubMed] [Google Scholar]
- Rabosky DL, Sorhannus U. 2009. Diversity dynamics of marine planktonic diatoms across the Cenozoic. Nature 457: 183–186. [DOI] [PubMed] [Google Scholar]
- Rainford JL, Hofreiter M, Nicholson DB, Mayhew PJ. 2014. Phylogenetic distribution of extant richness suggests metamorphosis is a key innovation driving diversification in insects. PLoS ONE 9: e109085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round FE, Crawford RM, Mann DG. 1990. Diatoms: biology and morphology of the genera. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Ruck EC, Nakov T, Alverson AJ, Theriot EC. 2016. Phylogeny, ecology, morphological evolution, and reclassification of the diatom orders Surirellales and Rhopalodiales. Molecular Phylogenetics and Evolution 103: 155–171. [DOI] [PubMed] [Google Scholar]
- Ruck EC, Theriot EC. 2011. Origin and evolution of the canal raphe system in diatoms. Protist 162: 723–737. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. 2002. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Molecular Biology and Evolution 19: 101–109. [DOI] [PubMed] [Google Scholar]
- Sato S, Beakes G, Idei M, Nagumo T, Mann DG. 2011. Novel sex cells and evidence for sex pheromones in diatoms. PLoS ONE 6: e26923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims PA, Mann DG, Medlin LK. 2006. Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia 45: 361–402. [Google Scholar]
- Small J. 1945a. Tables to illustrate the geological history of species‐number in diatoms. Proceedings of the Royal Irish Academy. Section B: Biological, Geological, and Chemical Science 50: 295–309. [Google Scholar]
- Small J. 1945b. Quantitative evolution: VIII. Numerical analysis of tables to illustrate the geological history of species number in diatoms; an introductory summary. Proceedings of the Royal Irish Academy. Section B: Biological, Geological, and Chemical Science 51: 53–80. [Google Scholar]
- Small J. 1950. Quantitative evolution: XVI. Increase of species‐number in diatoms. Annals of Botany 14: 91–113. [Google Scholar]
- Smith SA, O'Meara BC. 2012. treePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28: 2689–2690. [DOI] [PubMed] [Google Scholar]
- Sorhannus U, Fox MG. 2012. Phylogenetic analyses of a combined data set suggest that the Attheya lineage is the closest living relative of the pennate diatoms (Bacillariophyceae). Protist 163: 252–262. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank DC, Eastman JM, Pennell MW, Soltis PS, Soltis DE, Hinchliff CE, Brown JW, Sessa EB, Harmon LJ. 2015. Nested radiations and the pulse of angiosperm diversification: increased diversification rates often follow whole genome duplications. New Phytologist 207: 454–467. [DOI] [PubMed] [Google Scholar]
- Theriot EC, Ashworth MP, Nakov T, Ruck E, Jansen RK. 2015. Dissecting signal and noise in diatom chloroplast protein encoding genes with phylogenetic information profiling. Molecular Phylogenetics and Evolution 89: 28–36. [DOI] [PubMed] [Google Scholar]
- Togashi T, Bartelt JL, Yoshimura J, Tainaka K‐I, Cox PA. 2012. Evolutionary trajectories explain the diversified evolution of isogamy and anisogamy in marine green algae. Proceedings of the National Academy of Sciences 109: 13692–13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen L. 1973. A new evolutionary law. Evolutionary Theory 1: 1–30. [Google Scholar]
- de Vos JM, Hughes CE, Schneeweiss GM, Moore BR, Conti E. 2014. Heterostyly accelerates diversification via reduced extinction in primroses. Proceedings of the Royal Society of London B: Biological Sciences 281: 20140075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Phylogeny of 1151 diatoms and 20 outgroups reconstructed using an 11‐gene dataset.
Fig. S2 Diatom diversification estimated from first–last occurrence data for Cenozoic fossils.
Fig. S3 Granularity of fossil data and its effect on the phylogenetic estimates of diversification.
Fig. S4 Diatom diversification at predefined ranks with diversity data from AlgaeBase.
Fig. S5 Discrete shifts and temporal trends of diversification across diatoms with diversity data from AlgaeBase.
Table S1 Sequence data for diatoms and Parmales
Table S2 Properties of the phylogenetic dataset, partitioning and model selection
Table S3 ;Minimum and maximum bounds for the calibration of internal nodes


