Abstract
The proto-oncogene β-catenin drives colorectal cancer (CRC) tumorigenesis. Casitas B-lineage lymphoma (c-Cbl) inhibits CRC tumor growth through targeting nuclear β-catenin by a poorly understood mechanism. In addition, the role of c-Cbl in human CRC remains largely underexplored. Using a novel quantitative histopathologic technique, we demonstrate that patients with high c-Cbl–expressing tumors had significantly better median survival (3.7 years) compared with low c-Cbl–expressing tumors (1.8 years; P = 0.0026) and were more than twice as likely to be alive at 3 years compared with low c-Cbl tumors (P = 0.0171). Our data further demonstrate that c-Cbl regulation of nuclear β-catenin requires phosphorylation of c-Cbl Tyr371 because its mutation compromises its ability to target β-catenin. The tyrosine 371 (Y371H) mutant interacted with but failed to ubiquitinate nuclear β-catenin. The nuclear localization of the c-Cbl–Y371H mutant contributed to its dominant negative effect on nuclear β-catenin. The biological importance of c-Cbl–Y371H was demonstrated in various systems, including a transgenic Wnt-8 zebrafish model. c-Cbl–Y371H mutant showed augmented Wnt/β-catenin signaling, increased Wnt target genes, angiogenesis, and CRC tumor growth. This study demonstrates a strong link between c-Cbl and overall survival of patients with CRC and provides new insights into a possible role of Tyr371 phosphorylation in Wnt/β-catenin regulation, which has important implications in tumor growth and angiogenesis in CRC.
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the third leading cause of cancer-related death in men and women in the United States.1, 2 The treatment of CRC is becoming increasingly personalized, guided by a combination of an individual's clinical, tumor, and molecular features.3, 4 Pathogenic mediators that regulate CRC tumorigenesis are desirable biomarkers to predict prognosis, recurrence of the disease, and response to therapy to improve the management of patients with CRC.
Aberrant hyperactivated Wnt/β-catenin signaling in the colonic epithelium plays an important role in the initiation and progression of CRC tumorigenesis, which is primarily mediated by nuclear β-catenin (transcriptionally active or mutant) in most patients with sporadic CRC.5, 6 Emerging data indicate casitas B-lineage lymphoma (c-Cbl) as a suppressor of Wnt signaling and CRC tumor growth.7 c-Cbl is a direct interactor and a ubiquitin E3 ligase for transcriptionally active and mutant β-catenin in the nucleus of CRC cells.8, 9 By targeting β-catenin, c-Cbl inhibits CRC tumor growth both in vitro and in animal xenograft models.7 Furthermore, c-Cbl levels in tumors negatively correlate with the levels of nuclear β-catenin in a cohort of patients with metastatic CRC (mCRC).7 Given its role in suppressing CRC tumor growth,10, 11 we posited that c-Cbl levels in CRC tumors could correlate with the overall survival in patients with CRC.
This study also mechanistically investigates the E3 ubiquitin ligase function of c-Cbl on nuclear β-catenin, an aspect of c-Cbl’s role that has remained poorly understood. The ubiquitin E3 ligase function of c-Cbl is tightly regulated by phosphorylation of Tyr371 residue on c-Cbl. Previous studies have found that the hypophosphorylated Tyr371 maintains c-Cbl in persistently autoinhibited conformation,10, 11 which hinders its ubiquitin ligase activity. Phosphorylation of this tyrosine residue is imperative for the conversion of the negative regulatory conformation to an active ubiquitin ligase. A homozygous mutation in c-Cbl c.1111T>C (p.371Y>H) is detected in 5% of patients with acute myeloid leukemia and myelodysplastic syndromes and is considered to be oncogenic in nature.10, 11, 12, 13 This mutation is suggested to lock c-Cbl in autoinhibited state to compromise its ubiquitin E3 ligase activity. This CBL mutation located in the helical linker was used to demonstrate the contribution of c-Cbl’s ubiquitin ligase activity to Wnt/β-catenin regulation. Although tumor growth and angiogenesis were used as biological models, they underscore the importance of ubiquitin E3 ligase activity of c-Cbl in events central to CRC tumorigenesis.14, 15
Materials and Methods
Study Design and Participants
In this retrospective, single-center, observational cohort study, 72 patients with stage IV CRC who had available tumor tissue samples for analysis and were treated from January 1, 2004, to December 31, 2014, at the Boston Medical Center were evaluated. These patients had presented with stage IV disease or developed stage IV disease after having been initially diagnosed with and treated for early-stage CRC. Demographic data, comorbidity scores, tumor clinical and molecular features, and treatment-related factors were obtained from the institutional cancer registry and the electronic medical record. Response to therapy was categorized based on the standard American Joint Committee on Cancer criteria.16 This group of patients represent a major proportion of a previously described patient cohort.7 The study was approved by the Boston Medical Center Institutional Review Board. Data were collected by medical record review; the extracted data were reconfirmed by two independent clinicians.
Immunohistochemistry and Image Quantification
Unstained, 6-μm, paraffin-embedded sections obtained from the Department of Pathology, Boston University School of Medicine, were processed and examined as described previously.7 A customized, color-based image segmentation pipeline was developed to estimate the amounts of c-Cbl expression in human CRC samples.7 Briefly, the pipeline consisted of basic image preprocessing, color-based segmentation, size-based morphologic filtering, and valid area computation of selected subregions within each image. The converted images were reduced in dimensionality and segmented into three clusters based on a distance-based clustering algorithm. A size-based filtering operation identified clusters to eliminate all connected components that have fewer than the threshold level of pixel area. The size of the resulting subregion was then estimated by computing the fraction of the nonzero pixels within the entire image, resulting in a measure of cytoplasmic c-Cbl content.
For this analysis, quantification of expression was normalized to the total tumor area in an image and reported as a value between 0 and 1.00. Patients with normalized c-Cbl expression values lower than the median were defined as having low c-Cbl expression, and patients with normalized c-Cbl values equal to or higher than the median were defined as having high c-Cbl expression.
Zebrafish Model
Adult male and female zebrafish (Danio rerio) fli–enhanced green fluorescent protein (eGFP)9 and heat shock–inducible Wnt-8–GFP (hs:Wnt-8–eGFP)17 were housed at 14:19-hour light-dark cycle at 26.5°C and 7.0 to 7.4 pH in a controlled multitank recirculating water system (Aquaneering, San Diego, CA). The Boston University School of Medicine animal welfare committee approved the experimental protocol. For the heat shock experiment, fish heterozygous for hs:Wnt-8–eGFP were out-crossed to wild-type (WT) fish. The capped RNA (100 pg) was injected to one- or two-cell-stage embryos. The embryos at 10 hours post fertilization (hpf) were treated at 38°C for 20 minutes and sorted at 24 hpf into nontransgenic and GFP-positive transgenic siblings, as described previously.17 The later embryos were harvested at 36 hpf for Western blotting.
For the angiogenesis assay, fli-eGFP fish were injected as above, and the images were obtained from 10 randomly selected fish per group at every experiment under the same light exposure setting and analyzed for the length of the tail vessels. The vessels were marked from the junction of the body and the tail going caudally using Image-Pro (Media Cybernetics, Rockville, MD) and averaged per group, as described previously.9, 18 The tail vessel plexus was marked and measured for fluorescent intensity using ImageJ version 1.43t (NIH, Bethesda, MD; http://imagej.nih.gov/ij).
Plasmids and Other Methods
Two hemagglutinin (HA)–tagged c-Cbl plasmids point mutation of Tyr371H to histidine (c-Cbl–Y371H) and deletion of Tyr371 (del371H) in pcDNA were obtained from Dr. Martin Settler (Dana Farber Cancer Institute, Boston, MA). Detailed methods of Western blotting, cell culture, antibodies, in vitro angiogenesis assay, ubiquitination assay, vascular endothelial growth factor (VEGF) enzyme-linked immunosorbent assay, and spheroid formation assay are described elsewhere.7, 8, 9
RT-PCR
The RNeasy minikit (Qiagen, Hilden, Germany) was used to extract total RNA from HT-29 cells that expressed various constructs, as described previously.7 A total of 0.5 μg of total RNA were converted to cDNA using a Sensiscript reverse transcription kit (Qiagen) followed by RT-PCR using prevalidated human AXIN2, MYC, and VEGF primers and SYBRgreen (Applied Biosystems, Foster City, CA). GAPDH served as a loading control. Levels of mRNA were determined using comparative Ct method. The primers used were as follows as described previously.19, 20, 21
AXIN2 forward primer, 5′-TTATGCTTTGCACTACGTCCCTCCA-3′; AXIN2 reverse primer, 5′-CGCAACATGGTCAACCCTCAGAC-3′; MYC forward primer, 5′-TCAAGAGGTGCCACGTCTCC-3′; Myc reverse primer, 5′-TCTTGGCAGCAGGATAGTCCTT-3′; VEGF forward primer, 5′-GCCTTGCCTTGCTGCTCTAC-3′; VEGF reverse primer, 5′-ATGATTCTGCCCTCCTCCTTC-3′; glyceraldehyde-3-phosphate dehydrogenase forward primer, 5′-CCATGTTCGTCATGGGTGTG-3′; glyceraldehyde-3-phosphate dehydrogenase reverse primer, 5′-GGTGCTAAGCAGTTGGTGGTG-3′.
Statistical Analysis
Statistical analysis was performed using SPSS for Windows version 10.0 (SPSS Inc., Chicago, IL). Summary statistics are presented as means, medians, and SDs. Either a t-test or Mann-Whitney U-test was performed to compare the groups as appropriate. Differences in various clinicopathologic features were analyzed using the unpaired t-test and Fisher exact test. Kaplan-Meier analysis was performed to examine overall patient survival, which was calculated from the date of diagnosis to the date of death or date of last clinical visit. Median values of c-Cbl were used to divide the groups for survival analysis. Death was confirmed by review of the medical record. P < 0.05 was considered significant.
Results
c-Cbl Expression Correlates with the Overall Survival of Patients with mCRC
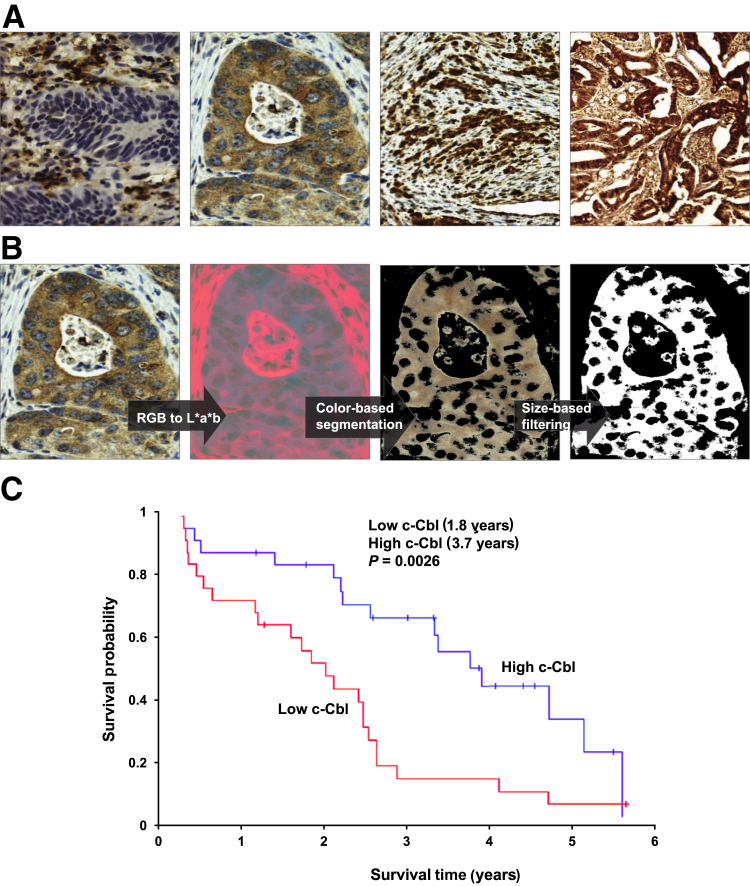
With the objective of analyzing the association between c-Cbl expression and CRC survival, the expression of c-Cbl was first examined by immunohistochemistry in a cohort of 72 patients with mCRC. The expression of c-Cbl widely varied in different patients (Figure 1A), which was quantitated using a color-based image segmentation pipeline,7 and this expression was normalized to the area of total tumor tissue on a slide (Figure 1B). Mean normalized c-Cbl expression in this set of tumor tissues was 0.740. With the thresholding based on the computed median value in 72 patients, 38 patients (52.78%) were found to have high c-Cbl expressing tumors, and 34 patients (47.22%) had low c-Cbl–expressing tumors. Of this cohort, Kaplan-Meier survival analysis was performed in 50 patients with survival data for the association between c-Cbl and overall survival in patients. Patients with high c-Cbl–expressing tumors had significantly better median overall survival compared with those with low c-Cbl–expressing tumors (3.7 versus 1.8 years; P = 0.003) (Figure 1C). Patients with high c-Cbl were more than twice as likely to be alive at 3 years compared with those with low c-Cbl; 3-year survival was 47.37% for high c-Cbl and 20.59% for low c-Cbl (P = 0.017). These differences were observed despite similar baseline, tumor-related characteristics (Table 1) between patients with low and high c-Cbl–expressing tumors. No significant differences were found in an overall response rate to therapy (51.61% in the high c-Cbl group compared with 54.17% in the low c-Cbl group, P = 0.85) (Table 2).
Figure 1.
c-Cbl correlates with the survival of metastatic colorectal cancer (mCRC) in humans. A: Four representative images of CRC tumor explants from 72 patients with stage IV CRC stained with c-Cbl antibody. The c-Cbl expression varies from low (left panel) to high (right panel). B: c-Cbl expression was estimated using color-based image segmentation pipeline. Two stages, including RGB color image conversion to L*a*b space followed by segmentation of the color image into three subregions using a clustering algorithm, are shown. C: The c-Cbl expression was divided into high and low expression based on the median normalized c-Cbl expression. The survival of patients with CRC was examined using Kaplan-Meier analysis based on c-Cbl expression. The survival curve for both these groups is shown. The plus sign refers to censored records. Original magnification, ×20 (A and B).
Table 1.
Baseline Characteristics of Patients with Metastatic Colorectal Cancer in the High c-Cbl and Low c-Cbl Groups
| Characteristic | High c-Cbl (n = 38) | Low c-Cbl (n = 34) | P value |
|---|---|---|---|
| Mean age, years | 61.03 | 57.74 | 0.30 |
| Sex, % | |||
| Male | 44.74 | 58.82 | 0.23 |
| Female | 55.26 | 41.18 | |
| Ethnicity, % | |||
| White | 39.47 | 32.35 | 0.53 |
| Black | 31.58 | 35.29 | 0.74 |
| Hispanic | 18.42 | 23.53 | 0.59 |
| Other | 10.53 | 8.82 | 0.81 |
| Mean Charlson Comorbidity Index | 7.00 | 6.90 | 0.86 |
| Primary site, % | |||
| Left-sided | 60.53 | 52.94 | 0.52 |
| Right-sided | 39.47 | 47.06 | 0.52 |
| Histologic subtype, % | |||
| Adenocarcinoma | 86.84 | 73.53 | 0.15 |
| Stage IV at presentation | 71.05 | 85.29 | 0.15 |
| Mean organs involved, n | 1.53 | 1.76 | 0.14 |
| Molecular features, % | |||
| KRAS mutant | 37.93 | 24.00 | 0.27 |
| BRAF mutant | 7.14 | 0.00 | 0.33 |
| Microsatellite unstable | 0.00 | 11.76 | 0.16 |
| Mean pretreatment carcinoembryonic antigen level | 317.10 | 139.20 | 0.55 |
c-Cbl, Casitas B-lineage lymphoma.
Table 2.
Treatment Received by Patients with Metastatic Colorectal Cancer in the High c-Cbl and Low c-Cbl Groups
| Treatment | High c-Cbl (n = 38) | Low c-Cbl (n = 34) | P value |
|---|---|---|---|
| Received surgery, % | 86.84 | 67.65 | 0.05 |
| Received chemotherapy, % | 81.58 | 70.59 | 0.27 |
| First-line chemotherapy, % | |||
| Chemotherapy regimen received | |||
| FOLFOX/CAPEOX, % | 80.65 | 83.33 | 0.80 |
| FOLFIRI/CAPEIRI, % | 3.23 | 12.50 | 0.19 |
| Biologics received | |||
| Bevacizumab | 54.84 | 70.83 | 0.23 |
| EGFR inhibitor | 12.90 | 0.00 | 0.07 |
| Mean cycles received, n | 7.39 | 7.58 | 0.87 |
| Response to first-line chemotherapy, AU | |||
| ORR | 51.61 | 54.17 | 0.85 |
| PR | 45.16 | 33.33 | 0.37 |
| CR | 6.45 | 20.83 | 0.11 |
AU, arbitrary units; CAPEIRI, capecitabine, irinotecan; CAPEOX, capecitabine, oxaliplatin; c-Cbl, Casitas B-lineage lymphoma; CR, complete response; FOLFIRI, 5-fluorouracil, leucovorin, irinotecan; FOLFOX, 5-fluorouracil, leucovorin, oxaliplatin; ORR, overall response rate; PR, partial response.
Several studies found an association of race on CRC survival.22, 23 Our recent work showed that black patients with mCRC at Boston Medical Center had a significantly lower survival compared with white patients.23 Therefore, we examined whether the association of c-Cbl expression and survival differed according to race. Among black patients with mCRC, those with low and high c-Cbl expression (divided based on median normalized c-Cbl expression) had a median overall survival of 2.1 and 3.8 years, respectively (P = 0.0284). Although a similar trend was noted in white patients, this difference fell short of reaching statistical significance (P = 0.0558). These data indicate that c-Cbl expression exerts its effect on CRC survival in different races but statistically more in black patients with to white patients with mCRC.
c-Cbl Tyr371 Mutation Up-Regulates Nuclear β-Catenin
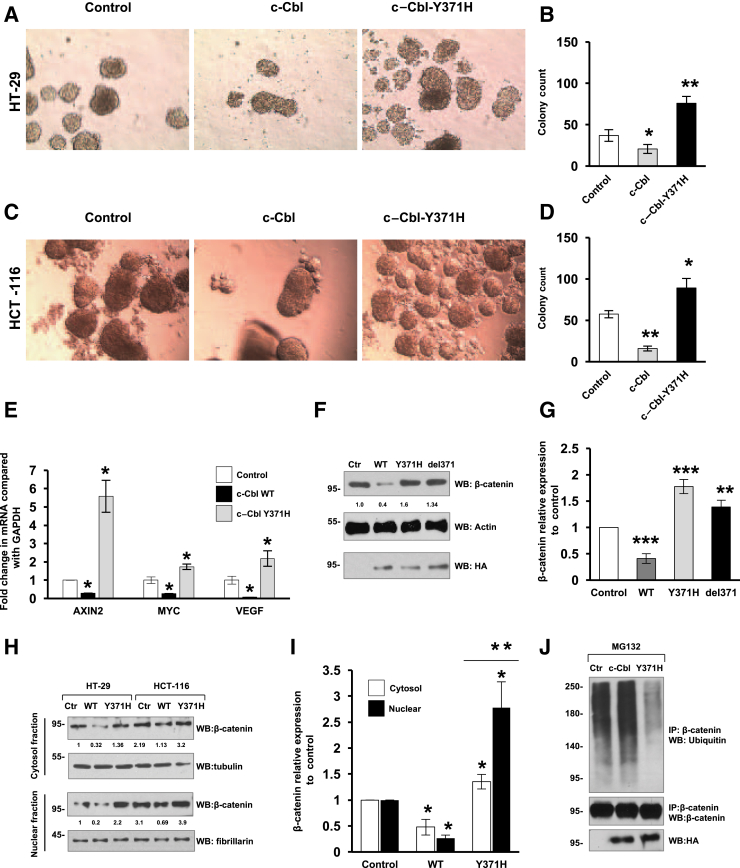
Having observed a correlation of c-Cbl levels with human CRC survival, the role of c-Cbl in CRC pathogenesis was further investigated. Wnt/β-catenin is an important mediator of CRC tumorigenesis,6 and c-Cbl is a ubiquitin E3 ligase of β-catenin8, 9 with a poorly understood mechanism. A naturally occurring ubiquitin ligase–deficient oncogenic c-Cbl–Y371H mutant, which is known to drive other cancers, was used.10, 12, 24 The effect of c-Cbl–Y371H was first examined in an in vitro tumor formation (spheroids) assay in CRC cell lines. Two different CRC cell lines, HT-29 and HCT-116, that expressed c-Cbl WT or c-Cbl–Y371H were grown in low adhesion plates. Compared with c-Cbl WT, significantly more spheroids were observed in c-Cbl–Y371H that expressed HT-29 (P = 0.001) and HCT-116 cell lines (P = 0.04), whereas WT c-Cbl significantly reduced spheroids in these cell lines (P = 0.02 for HT-29 and P = 0.003 for HCT-116) (Figure 2, A–D). Next, the effect of c-Cbl WT and Y371H mutants was examined on three prevalidated Wnt target genes in CRC: AXIN2, MYC, and VEGF.19, 20, 21 Although a significant suppression of Wnt target genes was observed with c-Cbl WT, c-Cbl–Y371H up-regulated all of them, with the most pronounced effect on AXIN2 (Figure 2E). These data suggest that unlike WT c-Cbl, c-Cbl–Y371H induces Wnt target genes and augments in vitro growth of CRC cells.
Figure 2.
Casitas B-lineage lymphoma (c-Cbl)–Y371H serves as a dominant negative for β-catenin. A: HT-29 colorectal cancer cell lines expressing control (Ctr), c-Cbl, or c-Cbl–Y371H constructs were suspended in ultralow adhesion plates and imaged after 72 hours. B: Mean number of spheroids in each group. C: Random images taken per well after 72 hours of HCT-116 colorectal cancer cell lines that express different constructs. D: Mean number of spheroids in each group. E: HT-29 cells were investigated for the Wnt target genes using quantitative RT-PCR reactions for AXIN2, MYC, and vascular endothelial growth factor (VEGF). The CT values were generated. F: HT-29 cells expressing different c-Cbl constructs, including c-Cbl wild-type (WT) and c-Cbl–Y371H or c-Cbl deletion 371 (del371), were lyzed and investigated as shown. Equal amounts of whole cell lysate were investigated with antihemagglutinin antibody. The numbers below the Western blot images represent the amount of β-catenin protein normalized to the loading control, which was measured using ImageJ version 1.43t. G: Mean normalized β-catenin protein levels. H: HT-29 and HCT-116 cells that expressed c-Cbl WT and c-Cbl–Y371H (Y371H) mutants were subjected to fractionation. Tubulin and fibrillarin served as markers of cytosol and nuclear fractions, respectively. I: Mean normalized β-catenin to the loading control of the respective subcellular fraction. J: HT-29 cells that expressed c-Cbl constructs were treated with 10 μmol/L of MG132 for 16 hours before fractionation. The nuclear fractions were immunoprecipitated using anti–β-catenin antibody and probed using antiubiquitin antibody. The stripped blot was reexamined for β-catenin and 5% of cell lysate was examined separately using antihemagglutininantibody. Data are expressed as means ± SEM (B, D, and F) and means ± SD (G and I). n = 3 (A, 3 experiments performed in triplicate); n = 3 (E, 3 independent experiments performed in duplicate); n = 3 (F–I); n = 4 (J). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus control (t-test). Original magnification, ×40 (A and C). IP, immunoprecipitation; WB, Western blot.
Because c-Cbl suppresses CRC tumor growth by down-regulating β-catenin,7 we posited that c-Cbl–Y371H mutation may regulate nuclear β-catenin in a manner different compared with WT c-Cbl. The levels of β-catenin were examined in HT-29–expressing WT c-Cbl and c-Cbl–Y371H or c-Cbl–delY371. Although a significant down-regulation of β-catenin was observed in the whole cell lysate with the WT c-Cbl (60% to 70%, P < 0.001), both the c-Cbl–Y371H (70% increase, P < 0.0001) and delY371 (34% increase, P < 0.006) mutants had a significant increase in β-catenin (Figure 2, F and G). Cell fractionation experiments further showed a more pronounced effect of c-Cbl–Y371H in the nuclear fraction of β-catenin. WT c-Cbl reduced β-catenin by 70% to 80% in both the fractions in the HT-29 and HCT-116 cell lines (Figure 2, H and I). On the other hand, c-Cbl–Y371H increased nuclear β-catenin threefold higher compared with cytosolic β-catenin (P = 0.008). c-Cbl targets its substrate to ubiquitination followed by proteasomal degradation. To examine the ubiquitination of nuclear β-catenin, we treated the cells with MG132, a proteasomal inhibitor, and the immunoprecipitated nuclear β-catenin was searched for ubiquitin.8, 25 The results showed that WT c-Cbl increased the nuclear β-catenin ubiquitination by 35% to 40% (P = 0.04), whereas c-Cbl–Y371H reduced β-catenin ubiquitination by 63% to 75% (P < 0.001) compared with the control (Figure 2J).
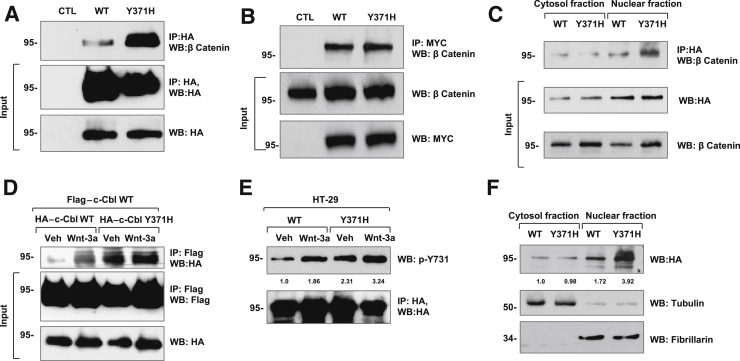
c-Cbl–Y371H Shows Higher Homodimerization and Binding to β-Catenin
Because HT-29 cells express endogenous c-Cbl,7 an increase in nuclear β-catenin by c-Cbl–Y371H suggests its dominant negative effect. The nuclear regulation of β-catenin was further examined to determine whether this dominant negative effect is mediated through binding of c-Cbl–Y371H to β-catenin. Indeed, c-Cbl–Y371H interacted with endogenous β-catenin (Figure 3A). Although the interaction of c-Cbl–Y371H with β-catenin seemed higher compared with the WT c-Cbl, this comparison is confounded by the reduced nuclear β-catenin levels by WT c-Cbl7 (Figure 3A). To avoid this confounding effect, we compared the interaction of nuclear β-catenin and Y371H with another E3 ligase–deficient c-Cbl–70Z mutant (Figure 3B). The interaction of nuclear β-catenin with these mutants of c-Cbl was comparable. In line with a greater increase in nuclear β-catenin (Figure 2, H and I), the binding of c-Cbl–Y371H with nuclear β-catenin was twofold higher compared with cytosolic β-catenin (P = 0.008) (Figure 3C). Collectively, all the above findings supported a dominant negative role of c-Cbl–Y371H because it interacted with β-catenin but failed to degrade it.
Figure 3.
Casitas B-lineage lymphoma (c-Cbl)–Y371H mutant exhibits increased β-catenin interaction, dimerization, phosphorylation, and nuclear localization. A: HT-29 cells expressing c-Cbl wild-type (WT) and c-Cbl–Y371H (Y371H) were lyzed and immunoprecipitated (IP) using anti–β-catenin antibody and investigated with hemagglutinin (HA)–tagged antibody. The stripped blot was reinvestigated using anti-HA–tagged antibody. Five percentage of the lysates were probed for HA-tagged c-Cbl as input. B: HT-29 cells transfected with Myc-tagged c-Cbl–70Z and Y371H underwent IP using Myc-tagged antibody, and the eluents were examined for β-catenin. Five percentage of lysates were examined as inputs. C: HT-29 cells that expressed c-Cbl or c-Cbl–Y371H were fractionated, each fraction underwent IP using anti–HA-tagged antibody, and eluents were investigated using anti–β-catenin antibody and vice versa. Five percentage of the lysates were probed as inputs. D: HT-29 cells co-expressing the FLAG-tagged WT c-Cbl and HA-tagged c-Cbl WT or HA-tagged c-Cbl–Y371H were serum starved and treated with 50 ng/mL of Wnt-3a ligand for 3 hours. The whole cell lysates that underwent IP using anti–FLAG antibody were investigated using anti-HA antibody. The blot was stripped and reinvestigated with anti-FLAG antibody. Five percentage of the lysate was probed as an input. E: HT-29 cells that expressed c-Cbl WT and c-Cbl–Y371H were serum starved overnight and then stimulated with 50 ng/mL of Wnt-3a. The lysates underwent IP with anti-HA antibody and were investigated with Tyr731 antibody. The blot was stripped and reinvestigated with anti-HA antibody. F: HT-29 cells that expressed different c-Cbl constructs were fractionated. The fractions were probed using HA-tagged and tubulin antibodies, where tubulin served as a marker of cytosolic fraction. Equal amounts of lysates were investigated for fibrillarin, which served as a marker of nuclear fraction. β-catenin levels in both the fractions were normalized to their respective markers. n = 3 (A, C, and D–F); n = 2 (B). Veh, vehicle; WB, Western blot.
c-Cbl dimerization enhances binding to β-catenin, both of which are further increased with Wnt activation.9 Because the above data supported a dominant negative effect of c-Cbl–Y371H in CRC cells in the presence of WT c-Cbl,7 the dimerization of c-Cbl–Y371H with WT c-Cbl was examined using various tagged constructs of c-Cbl under different states of Wnt activation. In vehicle-treated condition, HA-tagged c-Cbl–Y371H exhibited close to twofold higher dimerization with FLAG-tagged WT c-Cbl compared with HA-tagged WT c-Cbl (P = 0.001). The dimerization of WT c-Cbl and c-Cbl–Y371H was further augmented by 40% with Wnt signaling (Figure 3D).
Our previous work showed that Wnt activation enhances c-Cbl dimerization through phosphorylation of Tyr731 on c-Cbl.9 Therefore, the status of Tyr731 was compared on WT c-Cbl and c-Cbl–Y371H in different states of Wnt activity. In vehicle-treated cells, close to twofold higher levels of phosphorylation of Tyr731 was observed in c-Cbl–Y371H compared with WT c-Cbl, which was further augmented with Wnt activation (Figure 3E). These data suggest a higher phosphorylation of Tyr731 in the c-Cbl–Y371H mutant. Because both dimerization and Tyr731 phosphorylation regulate c-Cbl nuclear translocation,9 a higher amount of c-Cbl–Y371H was anticipated in the nuclear fraction compared with WT c-Cbl. Indeed, compared with WT c-Cbl, an almost 2.5-fold higher amount of c-Cbl–Y371H was found in the nuclei of CRC cells (P = 0.032) (Figure 3F). These data suggest that c-Cbl–Y371H exhibits higher dimerization and nuclear localization, which explains its higher binding and dominant negative effect on nuclear β-catenin.
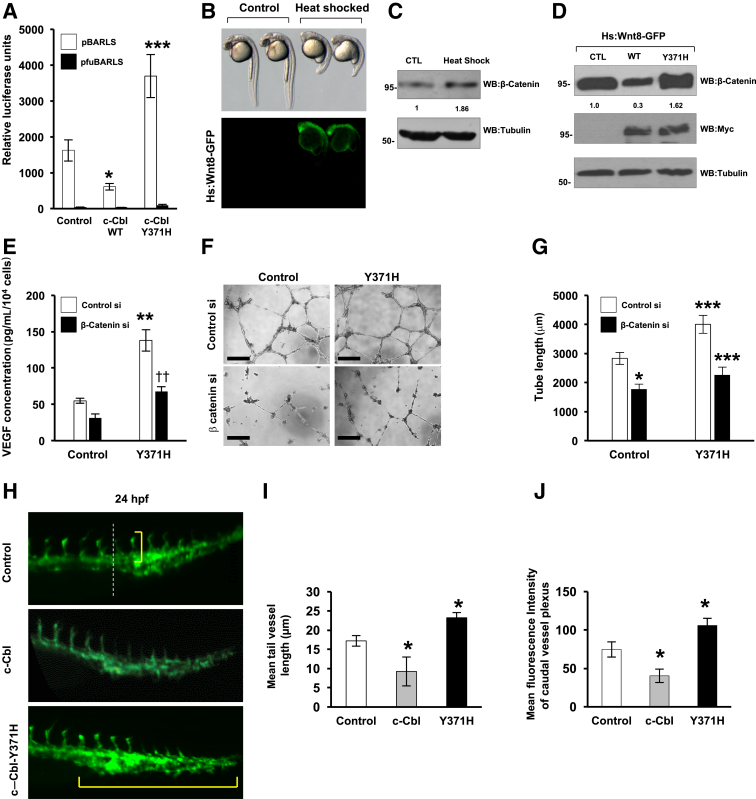
c-Cbl–Y371H Enhances Wnt Activity
Because c-Cbl–Y371H increases nuclear β-catenin, it is likely to also induce Wnt activity. Indeed, in cells that express a β-catenin–responsive promoter tethered to the luciferase reporter, a significant increase in Wnt activity was noted in the presence of c-Cbl–Y371H (P < 0.001), whereas c-Cbl WT suppressed Wnt activity (P = 0.02) (Figure 4A). The role of c-Cbl–Y371H in the Wnt active state was further confirmed using a transgenic zebrafish model (hs:Wnt8-GFP), which harbors WNT8 under a heat shock promoter. Induction of the Wnt-8 ligand with heat shock is a validated model of Wnt signaling activation.26, 27 As previously described, the heat shock of these embryos resulted in a distinct GFP signal in the neuroectoderm and a range of phenotypes, including small eyes, loss of eyes, and reduction or curling up of the trunk and tail, which were all suggestive of dorsalized embryos that had hallmarks of Wnt activation (Figure 4B).17 This finding was accompanied by close to a 80% to 90% increase in β-catenin expression (Figure 4C). These heat shocked embryos injected with different c-Cbl constructs were analyzed for levels of β-catenin. WT c-Cbl reduced zebrafish β-catenin by 70% to 80%, whereas c-Cbl–Y371H up-regulated zebrafish β-catenin by 60% to 73% (P = 0.04) (Figure 4D). These results corroborate with the dominant negative role of c-Cbl–Y371H observed in an in vitro cell culture model.
Figure 4.
Casitas B-lineage lymphoma (c-Cbl)–Y371H is a Wnt inducer that up-regulates β-catenin. A: Cell lysates of HT-29 cells co-expressing hemagglutinin (HA)–tagged c-Cbl wild-type (WT) or c-Cbl–Y371H and the TCF-responsive promoter reporter pBARLS and nonresponsive control reporter pfuBARLS tethered to luciferase reporter were analyzed for luciferase assay.26 Activity of the Wnt signaling pathway was quantified by measuring relative firefly luciferase units normalized to protein concentration. Luciferase activity was noted in the presence of c-Cbl-Y371H, whereas c-Cbl WT suppresses Wnt activity. B: Hs:Wnt-8–enhanced green fluorescent protein (eGFP) transgenic zebrafish embryos were heat shocked at 38°C for 20 minutes at 10 hours post fertilization (hpf) and imaged for 24 hpf. C: Ten deyolked control and heat shocked embryos were lyzed at 36 hpf and investigated for β-catenin. Tubulin served as a loading control. D: Hs:Wnt-8–eGFP transgenic zebrafish embryos were injected with capped mRNA of Myc-tagged c-Cbl and Myc-tagged c-Cbl–Y371H at one cell stage and then heat shocked. Ten deyolked eGFP-positive embryos harvested at 36 hpf were investigated for β-catenin and tubulin. Equal amounts of lysates were investigated separately with anti-Myc–tagged antibody. E: Primary human umbilical venin endothelial cells (HUVECs) co-transfected with c-Cbl–Y371H construct and control and β-catenin siRNA (si) oligos were serum starved overnight followed by stimulation with 50 ng/mL of Wnt-3a ligand. The media of endothelial cells collected after 24 hours of Wnt treatment were analyzed for vascular endothelial growth factor (VEGF) levels. F: HUVECs co-expressing control (CTL) or HA-tagged c-Cbl– Y371H and control and β-catenin si oligos were serum starved and treated with 50 ng/mL of Wnt-3a. Cells were then seeded in a 96-well plate coated with Matrigel and analyzed for tube formation within 24 hours. G: The tube lengths were measured in the Matrigel with ImageJ version 1.43t. H: Fli-eGFP transgenic zebrafish two-cell-stage embryos injected with different mRNA. LacZ-injected embryos served as controls. Representative images from randomly selected fish from total injected at 24 hpf are shown (Table 3). The dashed white line represents the junction of main body and tail, after which the tail vessels are measured (white dotted line). The yellow brackets indicate the caudal vessel plexus. I: Mean length of tail vessels of 10 randomly selected fish from total injected. The images under same light exposure settings were obtained from 10 randomly selected fish per group from total live fish and analyzed for the length of the tail vessels. The tail vessels were marked from the junction of the body and the tail (white dotted line in H) going caudally using Image-Pro and averaged per group.9, 18J: The images of fish under same light exposure settings were obtained for 10 randomly selected fish per group from total live fish. The region of interest was marked and analyzed for the intensity of caudal vessel plexus using ImageJ. Mean intensity is shown. Data are expressed as means ± SEM (A, E, G, I, and J). n = 3 (A, C, D, F, H–J); n = 62 heat shocked and 80 control embryos (B); n = 2 (G, six images from two separate experiments performed in triplicate). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus control (t-test with Bonferroni correction). Scale bars = 100 μm. Original magnification, ×5 (B and H).
c-Cbl–Y371H Enhances Angiogenesis in a Wnt/β-Catenin Manner
Angiogenesis is important in CRC growth, progression, and metastasis, and antiangiogenic agents constitute the standard of care for the management of patients with CRC.14, 15 Wnt/β-catenin signaling regulates angiogenesis through different mechanisms, including target genes such as VEGF.8, 9, 28, 29 Therefore, the effect of c-Cbl–Y371H was further examined in angiogenesis models and β-catenin mediation explored. To this end, we examined the levels of VEGF in media of human umbilical vein endothelial cells expressing c-Cbl–Y371H mutant, and its dependence on β-catenin was evaluated using β-catenin siRNAoligos, as previously described (Figure 4E).8 The data indicate that the c-Cbl–Y371H mutant increased VEGF levels in media by threefold (P = 0.003), which was substantially suppressed close to baseline with β-catenin silencing (P = 0.01). In line with the VEGF levels, an in vitro angiogenesis assay showed that the c-Cbl–Y371H increased the tube length by 80% (P = 0.003), which was reduced to baseline with β-catenin silencing (Figure 4, F and G). This effect was further confirmed in a transgenic zebrafish, Fli-eGFP, which is a genetically engineered zebrafish expressing eGFP in endothelial cells and has been used as a tool to examine the function of c-Cbl in vivo.9, 18 In this model, the number and the length of tail vessels and caudal vessel plexus serve as biological read-outs. Zebrafish injected with c-Cbl–Y371H had a significant (60%) increase in the length (P = 0.001) and 40% increase in caudal vessel plexus (P = 0.03) compared with the control (Figure 4, H–J and Table 3). All these findings indicate that c-Cbl–Y371H mutant enhances the angiogenesis in a β-catenin–dependent manner and support the Wnt inducer activity of c-Cbl–Y371H.
Table 3.
Number of Injected Fli-eGFP Zebrafish
| Capped RNA | Zebrafish exhibiting angiogenesis phenotype in total number of live zebrafish, n (%) |
|---|---|
| LacZ | 54 (3.6) |
| c-Cbl | 69 (73) |
| c-Cbl–Y371H | 73 (82) |
c-Cbl, Casitas B-lineage lymphoma.
Discussion
Consistent with the role of c-Cbl as a negative regulator of Wnt/β-catenin signaling and CRC tumor growth, this study demonstrates that higher c-Cbl expression in CRC tumors correlates with a better overall survival of patients with mCRC. Using different model systems, we also found the importance of the ubiquitin E3 ligase function of c-Cbl in regulating several crucial aspects of CRC, such as Wnt/β-catenin activity and CRC tumor growth and angiogenesis using a naturally occurring ubiquitin ligase–deficient and an oncogenic mutant of c-Cbl (c-Cbl–Y371H).12, 13, 24, 30 Although the c-Cbl–Y371H mutant interacted with β-catenin and exhibited the molecular features that enhanced the mutant's nuclear localization, it did not result in an increased degradation of nuclear β-catenin, a finding with implications in CRC tumorigenesis.
c-Cbl is a ubiquitin E3 ligase for several receptor tyrosine kinases, such as c-Met and epidermal growth factor receptor (EGFR), and nonreceptor protein tyrosine kinases, including Src family kinases,31, 32 which are known contributors to CRC pathogenesis.33, 34, 35 The significant correlation of c-Cbl levels in tumors with overall survival of the patients with mCRC is likely to be contributed to the key role of c-Cbl in down-regulating mutant and active nuclear β-catenin as well as several other oncoproteins (such as tyrosine kinases) important in CRC.33, 34, 35 The current study demonstrates an intriguing difference in the association of c-Cbl expression and mCRC survival in various racial groups. Black patients had a significantly different overall survival between low and high c-Cbl–expressing tumors; however, this difference did not reach statistical significance in white patients. Although this study was not powered to address the differences in such mediators among different races, this hypothesis-generating observation supports future studies on sufficiently powered racially diverse larger cohorts to investigate various pathogenic regulators to explain racial disparity in CRC survival.
Although both low and high c-Cbl groups were fairly balanced in regard to baseline characteristics and treatment received, a larger proportion of patients with high c-Cbl expression had undergone surgery as part of their treatment compared with those with low c-Cbl expression. Although this difference barely approached significance (P = 0.0504) (Table 1), it would seem unlikely to confound the prominent survival disparity between the high and low c-Cbl groups (Figure 1C). Chemotherapy, especially an EGFR inhibitor, can also potentially confound the results. For example, EGFR inhibitors can influence the MET receptor activity.36, 37 In gastric cancer, perturbation of MET activity regulated c-Cbl,38 which might also be the case in CRC. Thus, treatment with EGFR inhibitors may change the c-Cbl expression in CRC. However, this confounding effect is unlikely because there was no statistical difference in the number of patients treated with EGFR inhibitors between the two groups (Table 2). A trend toward lower metastatic disease burden was also noted in patients with high c-Cbl (P = 0.15) (Table 1), which may explain why the group of high c-Cbl–expressing tumors underwent more potentially curative surgery. Pending validation in larger cohorts, these data also suggest that the patients with high c-Cbl–expressing tumors may exhibit less aggressive disease.
Although the c-Cbl–Y371H mutant was used in this work to demonstrate the importance of the ubiquitin E3 ligase activity of c-Cbl in regulating nuclear β-catenin, the results of this study have implications in CRC tumorigenesis even though the presence of a c-Cbl–Y371H mutation in CRC remains unknown. It is likely that c-Cbl Tyr371 may be phosphorylated in CRC, considering that several important tyrosine kinases, such as Src, MET, and EGFR, are constitutively activated in CRC33, 39, 40, 41, 42, 43 and can potentially contribute to Tyr371 phosphorylation. The phosphorylation of Tyr371 is anticipated to relieve the autoinhibitory conformation of c-Cbl, leading to its activation.10, 11 c-Cbl Tyr371 phosphorylation and the ensuing increased ubiquitin E3 ligase activity are anticipated to down-regulate β-catenin. On the contrary, CRC is characterized by an increase in the expression of β-catenin.5, 6 This raises the possibility of a hypophosphorylated state of c-Cbl Tyr371 residue, which will maintain c-Cbl in an autoinhibited form and suppress its ubiquitin ligase activity. It is conceivable that this effect might be mediated through tyrosine phosphatases because several of them are known to be up-regulated in CRC.44, 45, 46
Higher dimerization propensity of c-Cbl–Y371H has potential implications in CRC. It is likely that in a CRC cell c-Cbl molecules may exhibit different phosphorylation states of Tyr371 residue. c-Cbl molecules phosphorylated at Tyr371 are poised to ubiquitinate β-catenin, whereas those hypophosphorylated at Tyr371 will continue to assume an autoinhibited structure and will fail to ubiquitinate β-catenin. On dimerization with WT c-Cbl, hypophosphorylated Tyr371 c-Cbl will serve as a dominant negative for WT c-Cbl for degrading β-catenin and will augment Wnt activity in CRC cells.
Although the hs:Wnt8-GFP zebrafish model supports the physiologic role of the ubiquitin E3 ligase function of c-Cbl in the Wnt-on phase,8 it provides potentially interesting insight into the role of c-Cbl in the pathogenesis of CRC. Wnt activity in CRC cells is maintained from within (from APC and CTNNB1 mutations) and outside cells. The agonistic Wnt-2 and Wnt-5a ligands are elevated in CRC tissue and are increased during the progression from adenoma to carcinoma47, 48, 49 to maintain higher levels of Wnt activity in colon cancer cells through autocrine and paracrine signaling.50 In addition, Wnt-2–mediated signaling in fibroblasts also occurs to promote CRC progression.51 All these observations implicate secreted Wnt ligands by different cell types in CRC tissue to create feedforward loops to augment CRC progression. The hs:Wnt8-GFP zebrafish model indicates that mutant c-Cbl at Tyr371 failed to down-regulate β-catenin in cells with active Wnt status induced by a Wnt ligand. These data suggest that in CRC cells with active Wnt status from Wnt ligands, hypophosphorylated c-Cbl protein at Tyr371 will impair the degradation of β-catenin. Collectively, the models of Wnt activation from outside (hs:Wnt8-GFP zebrafish) and within cells (HT-29 with inactivating mutation in APC) underscore the phosphorylation status of c-Cbl Tyr371 as an important determinant of regulation of β-catenin irrespective of the mode of Wnt activation.
A homozygous CBL mutation at c.1111T>C (p.371Y>H) was identified as a germline missense mutation in a cohort of patients with impaired growth, developmental delay, cryptorchidism, and a predisposition to juvenile myelomonocytic leukemia.24 In that study, c-Cbl p.371Y>H induced cytokine-independent growth of hematopoietic cells and enhanced phosphorylation of signaling molecules ERK, AKT, and S6.24, 30 However, this effect was observed in hematopoietic cells silenced for CBL expression or in the presence of the deletion of the normal copy of CBL-B, supporting c-Cbl p.371Y>H as a gain-of-function mutation.24, 30 Recent work from Nadeau et al12 supported the dominant negative mechanism of c-Cbl–Y371H. They proposed a model wherein c-Cbl–Y371H can potentially compete with the remaining copy of WT CBL or CBL-B for protein tyrosine kinases to hyperactivate signaling downstream of oncoproteins, including EGFR.12 Although the Wnt/β-catenin pathway was not specifically examined in those studies,12, 24, 30 the current body of work suggests its possible contribution in the pathogenesis of c-Cbl–Y371H–mediated diseases. Wnt activation may also explain c-Cbl–mediated phosphorylation of ERK, AKT, and S6 kinase.52, 53 The data from the study by Nadeau et al12 further support the involvement of β-catenin.12 A substantial reduction in the ability of the c-Cbl–Y371H mutant to promote hyperproliferation of hematopoietic cells was noted with a deletion of a part of its C-terminus. Because the C-terminus of c-Cbl mediates its interaction with β-catenin,8, 9 it is conceivable that the loss of regulation of β-catenin may contribute in part to the promoted hyperproliferation of hematopoietic cells and other manifestations observed with CBL mutation at c.1111T>C (p.371Y>H). However, this hypothesis needs further investigation.
Although this study examined the expression of c-Cbl in CRC tumor tissue, its posttranslational modifications, including Tyr371 phosphorylation, can be examined in a similar manner. However, lack of a phospho-Tyr371–specific antibody at this time precluded direct testing of this phosphorylation site in tumor tissues. Although the current study focused on the primary tumor, it would be interesting to examine alterations of c-Cbl expression or its posttranslational modifications in metastatic tissue and after therapeutic interventions. A similar analysis of other known E3 ligases of β-catenin, such as β-TrCP and Jade-1, will also provide a greater insight into the regulation of Wnt/β-catenin in patients with CRC.25, 54, 55 These future studies will use a large cohort of patients with CRC to detect c-Cbl mutations, as observed in another solid organ cancer.56
In conclusion, an association of tumor levels of c-Cbl and overall survival of patients with mCRC strengthens the role of c-Cbl as a negative regulator of CRC tumor growth.9 These findings demonstrate Wnt/β-catenin signaling as a potential downstream mediator of the effects of CBL mutation at c.1111T>C (p.371Y>H) and validate the molecular features of c-Cbl, which are critical to its effect on nuclear β-catenin. c-Cbl’s ubiquitin ligase activity regulates several aspects of CRC, such as tumor growth and angiogenesis and overall patient survival, and is worthy of further exploration in CRC.
Acknowledgments
We thank Dr. Martin Settler (Dana Farber Cancer Institute) for sharing pcDNA c-Cbl–Y371H and Y371H deletion constructs, Dr. Hui Feng (Boston University School of Medicine) for hs:Wnt8-GFP transgenic zebrafish, and the core facilities of the Department of Medicine of Boston Medical Center and Boston University School of Medicine and the Department of Pathology of Boston University School of Medicine for their assistance.
Footnotes
Supported by National Cancer Institute grant RO1CA175382 (V.C.C.), NIH grants R01 HL132325 (V.C.C.), R21CA191970 (N.R.), R21CA193958 (N.R.), and R00HD071968-03 (D.C.), the Boston University Evans Faculty Merit award (V.C.C.), Hariri Research Award 2016-10-009 from the Hariri Institute of Computing, Boston University (V.B.K.), American Heart Association's Scientist Development Grant 17SDG33670323 (V.B.K.), T32 training in renal biology grant T32 DK007053-44 (C.L.), T32 training in cardiovascular biology grant T32 HL007224-40 (J.W.), T32 training in immunobiology of trauma grant T32 GM086308-06A1 (N.A.), and Undergraduate Research Opportunities Program award from Boston University (S.G. and Y.J.).
S.K. and S.Y.L. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2018.05.007.
Supplemental Data
References
- 1.Siegel R., Desantis C., Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Stein A., Hiemer S., Schmoll H.J. Adjuvant therapy for early colon cancer: current status. Drugs. 2011;71:2257–2275. doi: 10.2165/11594490-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 3.Catenacci D.V., Kozloff M., Kindler H.L., Polite B. Personalized colon cancer care in 2010. Semin Oncol. 2011;38:284–308. doi: 10.1053/j.seminoncol.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yothers G., Song N., George T.J., Jr. Cancer hallmark-based gene sets and personalized medicine for patients with stage II colon cancer. JAMA Oncol. 2016;2:23–24. doi: 10.1001/jamaoncol.2015.3614. [DOI] [PubMed] [Google Scholar]
- 5.Markowitz S.D., Bertagnolli M.M. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogelstein B., Fearon E.R., Hamilton S.R., Kern S.E., Preisinger A.C., Leppert M., Nakamura Y., White R., Smits A.M., Bos J.L. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 7.Shashar M., Siwak J., Tapan U., Lee S.Y., Meyer R.D., Parrack P., Tan J., Khatami F., Francis J., Zhao Q., Hartshorn K., Kolachalama V.B., Rahimi N., Chitalia V. c-Cbl mediates the degradation of tumorigenic nuclear beta-catenin contributing to the heterogeneity in Wnt activity in colorectal tumors. Oncotarget. 2016;7:71136–71150. doi: 10.18632/oncotarget.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chitalia V., Shivanna S., Martorell J., Meyer R., Edelman E., Rahimi N. c-Cbl, a ubiquitin E3 ligase that targets active beta-catenin: a novel layer of Wnt signaling regulation. J Biol Chem. 2013;288:23505–23517. doi: 10.1074/jbc.M113.473801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shivanna S., Harrold I., Shashar M., Meyer R., Kiang C., Francis J., Zhao Q., Feng H., Edelman E.R., Rahimi N., Chitalia V.C. The c-Cbl ubiquitin ligase regulates nuclear beta-catenin and angiogenesis by its tyrosine phosphorylation mediated through the Wnt signaling pathway. J Biol Chem. 2015;290:12537–12546. doi: 10.1074/jbc.M114.616623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dou H., Buetow L., Hock A., Sibbet G.J., Vousden K.H., Huang D.T. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat Struct Mol Biol. 2012;19:184–192. doi: 10.1038/nsmb.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kassenbrock C.K., Anderson S.M. Regulation of ubiquitin protein ligase activity in c-Cbl by phosphorylation-induced conformational change and constitutive activation by tyrosine to glutamate point mutations. J Biol Chem. 2004;279:28017–28027. doi: 10.1074/jbc.M404114200. [DOI] [PubMed] [Google Scholar]
- 12.Nadeau S.A., An W., Mohapatra B.C., Mushtaq I., Bielecki T.A., Luan H., Zutshi N., Ahmad G., Storck M.D., Sanada M., Ogawa S., Band V., Band H. Structural determinants of the gain-of-function phenotype of human leukemia-associated mutant CBL oncogene. J Biol Chem. 2017;292:3666–3682. doi: 10.1074/jbc.M116.772723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiba N., Hasegawa D., Park M.J., Murata C., Sato-Otsubo A., Ogawa C., Manabe A., Arakawa H., Ogawa S., Hayashi Y. CBL mutation in chronic myelomonocytic leukemia secondary to familial platelet disorder with propensity to develop acute myeloid leukemia (FPD/AML) Blood. 2012;119:2612–2614. doi: 10.1182/blood-2011-02-333435. [DOI] [PubMed] [Google Scholar]
- 14.Kos M., Dabrowski A. Tumour's angiogenesis–the function of VEGF and bFGF in colorectal cancer. Ann Univ Mariae Curie Sklodowska Med. 2002;57:556–561. [PubMed] [Google Scholar]
- 15.Kircher S.M., Nimeiri H.S., Benson A.B., 3rd Targeting angiogenesis in colorectal cancer: tyrosine kinase inhibitors. Cancer J. 2016;22:182–189. doi: 10.1097/PPO.0000000000000192. [DOI] [PubMed] [Google Scholar]
- 16.Eisenhauer E.A., Therasse P., Bogaerts J., Schwartz L.H., Sargent D., Ford R., Dancey J., Arbuck S., Gwyther S., Mooney M., Rubinstein L., Shankar L., Dodd L., Kaplan R., Lacombe D., Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 17.Weidinger G., Thorpe C.J., Wuennenberg-Stapleton K., Ngai J., Moon R.T. The Sp1-related transcription factors sp5 and sp5-like act downstream of Wnt/beta-catenin signaling in mesoderm and neuroectoderm patterning. Curr Biol. 2005;15:489–500. doi: 10.1016/j.cub.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 18.Hartsough E.J., Meyer R.D., Chitalia V., Jiang Y., Marquez V.E., Zhdanova I.V., Weinberg J., Costello C.E., Rahimi N. Lysine methylation promotes VEGFR-2 activation and angiogenesis. Sci Signal. 2013;6:ra104. doi: 10.1126/scisignal.2004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan D., Wiesmann M., Rohan M., Chan V., Jefferson A.B., Guo L., Sakamoto D., Caothien R.H., Fuller J.H., Reinhard C., Garcia P.D., Randazzo F.M., Escobedo J., Fantl W.J., Williams L.T. Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/beta -catenin signaling is activated in human colon tumors. Proc Natl Acad Sci U S A. 2001;98:14973–14978. doi: 10.1073/pnas.261574498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueno K., Hiura M., Suehiro Y., Hazama S., Hirata H., Oka M., Imai K., Dahiya R., Hinoda Y. Frizzled-7 as a potential therapeutic target in colorectal cancer. Neoplasia. 2008;10:697–705. doi: 10.1593/neo.08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herbst A., Jurinovic V., Krebs S., Thieme S.E., Blum H., Goke B., Kolligs F.T. Comprehensive analysis of beta-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/beta-catenin signaling. BMC Genomics. 2014;15:74. doi: 10.1186/1471-2164-15-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robbins A.S., Siegel R.L., Jemal A. Racial disparities in stage-specific colorectal cancer mortality rates from 1985 to 2008. J Clin Oncol. 2012;30:401–405. doi: 10.1200/JCO.2011.37.5527. [DOI] [PubMed] [Google Scholar]
- 23.Tapan U., Lee S.Y., Weinberg J., Kolachalama V.B., Francis J., Charlot M., Hartshorn K., Chitalia V. Racial differences in colorectal cancer survival at a safety net hospital. Cancer Epidemiol. 2017;49:30–37. doi: 10.1016/j.canep.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niemeyer C.M., Kang M.W., Shin D.H., Furlan I., Erlacher M., Bunin N.J., Bunda S., Finklestein J.Z., Sakamoto K.M., Gorr T.A., Mehta P., Schmid I., Kropshofer G., Corbacioglu S., Lang P.J., Klein C., Schlegel P.G., Heinzmann A., Schneider M., Stary J., van den Heuvel-Eibrink M.M., Hasle H., Locatelli F., Sakai D., Archambeault S., Chen L., Russell R.C., Sybingco S.S., Ohh M., Braun B.S., Flotho C., Loh M.L. Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat Genet. 2010;42:794–800. doi: 10.1038/ng.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chitalia V.C., Foy R.L., Bachschmid M.M., Zeng L., Panchenko M.V., Zhou M.I., Bharti A., Seldin D.C., Lecker S.H., Dominguez I., Cohen H.T. Jade-1 inhibits Wnt signalling by ubiquitylating beta-catenin and mediates Wnt pathway inhibition by pVHL. Nat Cell Biol. 2008;10:1208–1216. doi: 10.1038/ncb1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biechele T.L., Moon R.T. Assaying beta-catenin/TCF transcription with beta-catenin/TCF transcription-based reporter constructs. Methods Mol Biol. 2008;468:99–110. doi: 10.1007/978-1-59745-249-6_8. [DOI] [PubMed] [Google Scholar]
- 27.Goessling W., North T.E., Lord A.M., Ceol C., Lee S., Weidinger G., Bourque C., Strijbosch R., Haramis A.P., Puder M., Clevers H., Moon R.T., Zon L.I. APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Dev Biol. 2008;320:161–174. doi: 10.1016/j.ydbio.2008.05.526. [DOI] [PubMed] [Google Scholar]
- 28.Goessling W., North T.E., Loewer S., Lord A.M., Lee S., Stoick-Cooper C.L., Weidinger G., Puder M., Daley G.Q., Moon R.T., Zon L.I. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136:1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dejana E. The role of wnt signaling in physiological and pathological angiogenesis. Circ Res. 2010;107:943–952. doi: 10.1161/CIRCRESAHA.110.223750. [DOI] [PubMed] [Google Scholar]
- 30.Rahimi N. A role for protein ubiquitination in VEGFR-2 signalling and angiogenesis. Biochem Soc Trans. 2009;37:1189–1192. doi: 10.1042/BST0371189. [DOI] [PubMed] [Google Scholar]
- 31.Ogawa S., Sanada M., Shih L.Y., Suzuki T., Otsu M., Nakauchi H., Koeffler H.P. Gain-of-function c-CBL mutations associated with uniparental disomy of 11q in myeloid neoplasms. Cell Cycle. 2010;9:1051–1056. doi: 10.4161/cc.9.6.11034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohapatra B., Ahmad G., Nadeau S., Zutshi N., An W., Scheffe S., Dong L., Feng D., Goetz B., Arya P., Bailey T.A., Palermo N., Borgstahl G.E., Natarajan A., Raja S.M., Naramura M., Band V., Band H. Protein tyrosine kinase regulation by ubiquitination: critical roles of Cbl-family ubiquitin ligases. Biochim Biophys Acta. 2013;1833:122–139. doi: 10.1016/j.bbamcr.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh A.J., Meyer R.D., Navruzbekov G., Shelke R., Duan L., Band H., Leeman S.E., Rahimi N. A critical role for the E3-ligase activity of c-Cbl in VEGFR-2-mediated PLCgamma1 activation and angiogenesis. Proc Natl Acad Sci U S A. 2007;104:5413–5418. doi: 10.1073/pnas.0700809104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J., Elfiky A., Han M., Chen C., Saif M.W. The role of Src in colon cancer and its therapeutic implications. Clin Colorectal Cancer. 2014;13:5–13. doi: 10.1016/j.clcc.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi H., Bilchik A., Saha S., Turner R., Wiese D., Tanaka M., Kuo C., Wang H.J., Hoon D.S. c-MET expression level in primary colon cancer: a predictor of tumor invasion and lymph node metastases. Clin Cancer Res. 2003;9:1480–1488. [PubMed] [Google Scholar]
- 36.Spano J.P., Lagorce C., Atlan D., Milano G., Domont J., Benamouzig R., Attar A., Benichou J., Martin A., Morere J.F., Raphael M., Penault-Llorca F., Breau J.L., Fagard R., Khayat D., Wind P. Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann Oncol. 2005;16:102–108. doi: 10.1093/annonc/mdi006. [DOI] [PubMed] [Google Scholar]
- 37.Song N., Liu S., Zhang J., Liu J., Xu L., Liu Y., Qu X. Cetuximab-induced MET activation acts as a novel resistance mechanism in colon cancer cells. Int J Mol Sci. 2014;15:5838–5851. doi: 10.3390/ijms15045838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Safaie Qamsari E., Safaei Ghaderi S., Zarei B., Dorostkar R., Bagheri S., Jadidi-Niaragh F., Somi M.H., Yousefi M. The c-Met receptor: implication for targeted therapies in colorectal cancer. Tumour Biol. 2017;39 doi: 10.1177/1010428317699118. 1010428317699118. [DOI] [PubMed] [Google Scholar]
- 39.Lai A.Z., Durrant M., Zuo D., Ratcliffe C.D., Park M. Met kinase-dependent loss of the E3 ligase Cbl in gastric cancer. J Biol Chem. 2012;287:8048–8059. doi: 10.1074/jbc.M112.339820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vlahovic G., Crawford J. Activation of tyrosine kinases in cancer. Oncologist. 2003;8:531–538. doi: 10.1634/theoncologist.8-6-531. [DOI] [PubMed] [Google Scholar]
- 41.Craven R.J., Xu L.H., Weiner T.M., Fridell Y.W., Dent G.A., Srivastava S., Varnum B., Liu E.T., Cance W.G. Receptor tyrosine kinases expressed in metastatic colon cancer. Int J Cancer. 1995;60:791–797. doi: 10.1002/ijc.2910600611. [DOI] [PubMed] [Google Scholar]
- 42.Spano J.P., Fagard R., Soria J.C., Rixe O., Khayat D., Milano G. Epidermal growth factor receptor signaling in colorectal cancer: preclinical data and therapeutic perspectives. Ann Oncol. 2005;16:189–194. doi: 10.1093/annonc/mdi057. [DOI] [PubMed] [Google Scholar]
- 43.Resnick M.B., Routhier J., Konkin T., Sabo E., Pricolo V.E. Epidermal growth factor receptor, c-MET, beta-catenin, and p53 expression as prognostic indicators in stage II colon cancer: a tissue microarray study. Clin Cancer Res. 2004;10:3069–3075. doi: 10.1158/1078-0432.ccr-03-0462. [DOI] [PubMed] [Google Scholar]
- 44.Sekharam M., Zhao H., Sun M., Fang Q., Zhang Q., Yuan Z., Dan H.C., Boulware D., Cheng J.Q., Coppola D. Insulin-like growth factor 1 receptor enhances invasion and induces resistance to apoptosis of colon cancer cells through the Akt/Bcl-x(L) pathway. Cancer Res. 2003;63:7708–7716. [PubMed] [Google Scholar]
- 45.Asbagh L.A., Vazquez I., Vecchione L., Budinska E., De Vriendt V., Baietti M.F., Steklov M., Jacobs B., Hoe N., Singh S., Imjeti N.S., Zimmermann P., Sablina A., Tejpar S. The tyrosine phosphatase PTPRO sensitizes colon cancer cells to anti-EGFR therapy through activation of SRC-mediated EGFR signaling. Oncotarget. 2014;5:10070–10083. doi: 10.18632/oncotarget.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai P., Guo W., Yuan H., Li Q., Wang W., Sun Y., Li X., Gu Y. Expression and clinical significance of tyrosine phosphatase SHP-2 in colon cancer. Biomed Pharmacother. 2014;68:285–290. doi: 10.1016/j.biopha.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 47.Hou P.C., Li Y.H., Lin S.C., Lin S.C., Lee J.C., Lin B.W., Liou J.P., Chang J.Y., Kuo C.C., Liu Y.M., Sun H.S., Tsai S.J. Hypoxia-induced downregulation of DUSP-2 phosphatase drives colon cancer stemness. Cancer Res. 2017;77:4305–4316. doi: 10.1158/0008-5472.CAN-16-2990. [DOI] [PubMed] [Google Scholar]
- 48.Holcombe R.F., Marsh J.L., Waterman M.L., Lin F., Milovanovic T., Truong T. Expression of Wnt ligands and Frizzled receptors in colonic mucosa and in colon carcinoma. Mol Pathol. 2002;55:220–226. doi: 10.1136/mp.55.4.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park J.K., Song J.H., He T.C., Nam S.W., Lee J.Y., Park W.S. Overexpression of Wnt-2 in colorectal cancers. Neoplasma. 2009;56:119–123. doi: 10.4149/neo_2009_02_119. [DOI] [PubMed] [Google Scholar]
- 50.Smith K., Bui T.D., Poulsom R., Kaklamanis L., Williams G., Harris A.L. Up-regulation of macrophage wnt gene expression in adenoma-carcinoma progression of human colorectal cancer. Br J Cancer. 1999;81:496–502. doi: 10.1038/sj.bjc.6690721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voloshanenko O., Erdmann G., Dubash T.D., Augustin I., Metzig M., Moffa G., Hundsrucker C., Kerr G., Sandmann T., Anchang B., Demir K., Boehm C., Leible S., Ball C.R., Glimm H., Spang R., Boutros M. Wnt secretion is required to maintain high levels of Wnt activity in colon cancer cells. Nat Commun. 2013;4:2610. doi: 10.1038/ncomms3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kramer N., Schmollerl J., Unger C., Nivarthi H., Rudisch A., Unterleuthner D., Scherzer M., Riedl A., Artaker M., Crncec I., Lenhardt D., Schwarz T., Prieler B., Han X., Hengstschlager M., Schuler J., Eferl R., Moriggl R., Sommergruber W., Dolznig H. Autocrine WNT2 signaling in fibroblasts promotes colorectal cancer progression. Oncogene. 2017;36:5460–5472. doi: 10.1038/onc.2017.144. [DOI] [PubMed] [Google Scholar]
- 53.Morris S.L., Huang S. Crosstalk of the Wnt/beta-catenin pathway with other pathways in cancer cells. Genes Dis. 2016;3:41–47. doi: 10.1016/j.gendis.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu D., Pan W. GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem Sci. 2010;35:161–168. doi: 10.1016/j.tibs.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winston J.T., Strack P., Beer-Romero P., Chu C.Y., Elledge S.J., Harper J.W. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan Y.H., Krishnaswamy S., Nandi S., Kanteti R., Vora S., Onel K., Hasina R., Lo F.Y., El-Hashani E., Cervantes G., Robinson M., Hsu H.S., Kales S.C., Lipkowitz S., Karrison T., Sattler M., Vokes E.E., Wang Y.C., Salgia R. CBL is frequently altered in lung cancers: its relationship to mutations in MET and EGFR tyrosine kinases. PLoS One. 2010;5:e8972. doi: 10.1371/journal.pone.0008972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.