Significance
Warsaw breakage syndrome, a developmental disorder caused by mutations in the conserved DDX11/ChlR1 DNA helicase, shows features of genome instability partly overlapping with those of Fanconi anemia (FA). Here, using avian cellular models of DDX11 deficiency, we find that DDX11 functions as backup to the FA pathway and facilitates, jointly with the checkpoint clamp 9-1-1, a homologous recombination pathway of DNA bulky-lesion repair that does not affect replication fork speed and stalled fork stability. DDX11 also promotes diversification of the immunoglobulin-variable gene locus by facilitating hypermutation and gene conversion at programmed abasic sites that constitute endogenous replication blocks. The results suggest commonality between postreplicative gap filling and replication through abasic sites and pinpoint DDX11 as a critical player in both these processes.
Keywords: homologous recombination, bulky lesions, abasic sites, mutagenesis, replication stress
Abstract
Warsaw breakage syndrome, a developmental disorder caused by mutations in the DDX11/ChlR1 helicase, shows cellular features of genome instability similar to Fanconi anemia (FA). Here we report that DDX11-deficient avian DT40 cells exhibit interstrand crosslink (ICL)-induced chromatid breakage, with DDX11 functioning as backup for the FA pathway in regard to ICL repair. Importantly, we establish that DDX11 acts jointly with the 9-1-1 checkpoint clamp and its loader, RAD17, primarily in a postreplicative fashion, to promote homologous recombination repair of bulky lesions, but is not required for intra-S checkpoint activation or efficient fork progression. Notably, we find that DDX11 also promotes diversification of the chicken Ig-variable gene, a process triggered by programmed abasic sites, by facilitating both hypermutation and homeologous recombination-mediated gene conversion. Altogether, our results uncover that DDX11 orchestrates jointly with 9-1-1 and its loader, RAD17, DNA damage tolerance at sites of bulky lesions, and endogenous abasic sites. These functions may explain the essential roles of DDX11 and its similarity with 9-1-1 during development.
Accurate DNA replication is critical for genome integrity and development. DNA lesions encountered during DNA replication challenge the stability of replication forks and are an important source of DNA replication stress (1). Cells are equipped with multiple DNA repair mechanisms, specific for different types of lesions, cell cycle phase, and in certain cases, cell/tissue type (2). For instance, abasic sites, estimated to be the most common spontaneously arising lesions in mammalian cells (3), can be repaired via accurate base excision repair. However, if encountered during replication, they are dealt with by specialized translesion synthesis (TLS) polymerases or by homologous/homeologous recombination-mediated bypass mechanisms. The processes by which abasic sites are tolerated or repaired have consequences for genome integrity and serve additional functions, such as generating Ig diversity in B cells (4). The diversification of Ig genes relies on activation-induced deaminase (AID)-mediated conversion of cytidine (dC) to uracil (dU) within regions of single-stranded DNA (ssDNA) (5), with dU being subsequently removed by uracil-DNA glycosylase and causing high local frequency of abasic sites. The arising abasic sites trigger somatic hypermutation, and in certain species including birds, homeologous recombination-mediated gene conversion with one of the 25 copies of upstream pseudogenes (6).
Complex lesions, such as DNA interstrand crosslinks (ICLs), also constitute replication blocks and require for repair the use of different types of repair enzymes implicated in ICL incision, TLS, and homologous recombination (HR) in subsequent fashion (7, 8). In vertebrate cells, the Fanconi anemia (FA) pathway is critical for ICL repair. Mutations in the so far 22 identified FA genes result in impaired ability of cells to deal with certain forms of DNA damage, such as endogenous formaldehyde (9), and lead to a hereditary disorder, FA, characterized by bone marrow failure, developmental abnormalities, and predisposition to cancer.
The central components of the FA pathway, FANCD2 and FANCI, interact with each other (10), and are monoubiquitylated by the FA core complex. FANCD2–FANCI ubiquitylation promotes lesion unhooking, causing the formation of a gapped DNA molecule, with the unhooked lesion in the gapped part, and a double strand break (DSB), repaired subsequently by TLS and HR (11). The FA pathway is strongly linked to the intra-S phase checkpoint function of ATR (12), with ATR-mediated phosphorylation of FANCD2–FANCI being required for the subsequent monoubiquitylation (10, 13). Accordingly, mutations in checkpoint factors often affect ICL repair, a phenotype used to identify new checkpoint components and for dissecting different functions of checkpoint mediators.
Warsaw breakage syndrome (WABS) is a disorder with mutations in the XPD family helicase DDX11/ChlR1 (14–17). WABS patient cells have features of FA (14, 17), but DDX11 has not been formally associated with FA or checkpoint signaling. DDX11 displays 5′–3′ DNA helicase activity in vitro, interacts with factors implicated in Okazaki fragment processing (18–20), and facilitates tolerance to cisplatin in human cells (21). However, the molecular functions of DDX11 remain incompletely understood.
Here, we found that DDX11-deficient avian cells show hallmarks of impaired ICL repair and genome instability. Mechanistically, DDX11 functions as backup to the FA pathway and facilitates an HR-related pathway of DNA repair that is not critical for replication fork speed and stability. Importantly, ddx11 is epistatic with mutations in the 9-1-1 checkpoint clamp loader, RAD17, in regard to DNA damage hypersensitivity, recombination defects, and chromosome aberrations. In similar trends to 9-1-1, DDX11 contributes to Ig variable gene (IgV) diversification via homeologous recombination-mediated gene conversion. Moreover, we find that DDX11 also contributes to hypermutation-mediated IgV gene diversification, and that the compensatory use of hypermutation in rad17 cells is dependent on DDX11. In conclusion, our findings identify a DDX11 helicase-dependent DNA repair pathway that supports and coordinates DNA damage tolerance of bulky replication lesions and of endogenous abasic sites in vertebrate cells. This function may explain the essential role of DDX11 during development (22, 23).
Results
DDX11 Helicase Facilitates Repair and Averts Genomic Instability Induced by ICLs.
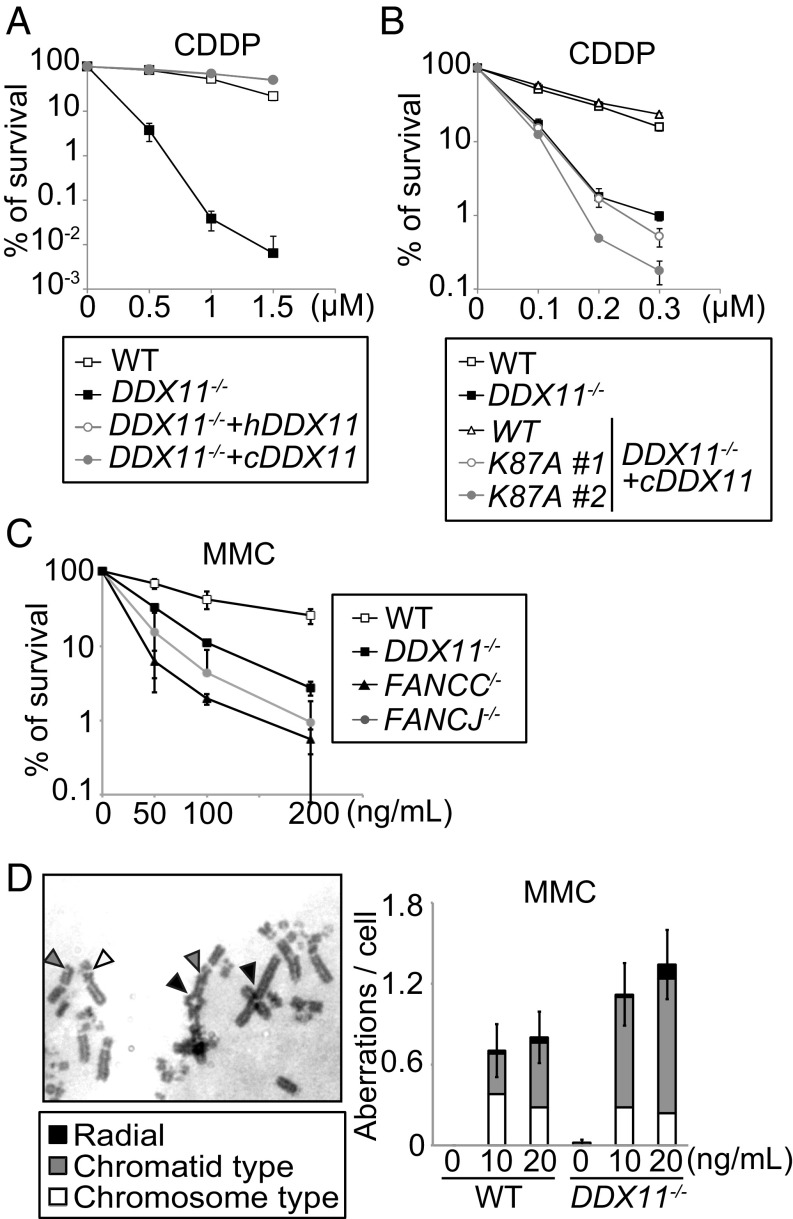
To examine the effect of DDX11 deficiency in vertebrate cells, we established DT40 knockout cell lines (24). DDX11−/− cells (hereafter ddx11) proliferate normally (24), but are hypersensitive toward cisplatin (CDDP), which causes intra- and interstrand crosslinks (ICLs), and methylmethane sulfonate (MMS), which causes bulky lesions (Fig. 1A and SI Appendix, Fig. S1A). The sensitivity of ddx11 mutants is complemented by expressing chicken and human DDX11 (Fig. 1A and SI Appendix, Fig. S1A), but not by expressing a helicase-dead, K87A variant of chicken DDX11 (cDDX11), carrying inactivating mutations in a conserved lysine residue located in the Walker A motif (Fig. 1B and SI Appendix, Fig. S1B), despite similar levels of expression of these variants (SI Appendix, Fig. S1C). These effects are observed in both colony formation (Fig. 1A and SI Appendix, Fig. S1A) and cellular viability assays (Fig. 1B and SI Appendix, Fig. S1B).
Fig. 1.
DDX11 helicase facilitates ICL repair. (A and B) CDDP sensitivity of DDX11 mutants complemented or not with DDX11 variants. Colony/cell survival percentage is displayed as the ratio of the number of surviving colonies (A) or cells (B) following CDDP treatment relative to the untreated control. Each line and error bar represents the mean value and SD from two independent experiments, respectively. In A please note overlap between ddx11 clones expressing human or chicken DDX11. (C) Sensitivity of cells as in B. (D) MMC-induced chromosomal aberrations. Each bar and error bar represents the mean value and SD from 50 cells.
In similar trends with FA mutants, such as fancc and fancj, ddx11 cells are defective in recovery from a transient exposure to mitomycin C (MMC) (Fig. 1C). Moreover, after exposure to MMC, the frequency of chromosome abnormalities increased, especially in regard to chromatid breaks (Fig. 1D). Thus, DDX11 is important in averting genome instability induced by ICLs.
DDX11 Functions as Backup to the FA Pathway in ICL Repair.
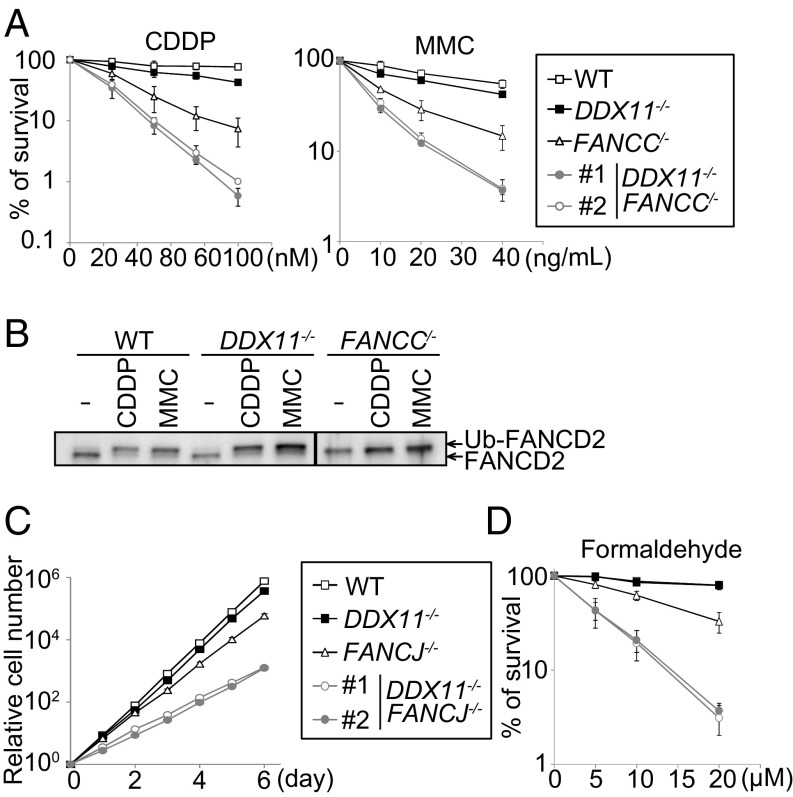
To examine the phenotypic relationship between DDX11 and the FA pathway, we established double knockout mutants between DDX11 and FANCC, an FA core component required for FANCD2–FANCI ubiquitylation (25). Double mutant fancc ddx11 cells exhibited much greater sensitivity to cisplatin and MMC than either single mutant (Fig. 2A). Differently from fancc mutants, ddx11 cells were proficient in inducing FANCD2 ubiquitylation in response to cisplatin and MMC (Fig. 2B). Thus, DDX11 is important for ICL repair, acting in parallel with FA or downstream of FANCD2 ubiquitylation in the FA pathway.
Fig. 2.
DDX11 functions as backup to the FA pathway in ICL repair. (A and D) Sensitivity of cells as in Fig. 1B. (B) Assessment of FANCD2 monoubiquitination after treatment with MMC or CDDP. (C) Growth curves.
The FA pathway is required to act on metabolic formaldehyde (9), which can cause formation of DNA adducts. ddx11 cells showed mild sensitivity to formaldehyde, but strongly aggravated the sensitivity of fancc mutants (SI Appendix, Fig. S2A). We also established double mutants between DDX11 and FANCJ, as FANCJ is the main helicase associated with the FA pathway, and both DDX11 and FANCJ are orthologs of budding yeast Chl1 (26). ddx11 fancj cells showed slow proliferation (Fig. 2C), with accumulation of cells in sub-G1 (SI Appendix, Fig. S2B), and exhibited synergistic sensitivity to formaldehyde (Fig. 2D), suggesting compensatory functions of DDX11 and FANCJ in proliferation and DNA repair. Thus, DDX11 is dispensable for FANCD2–FANCI ubiquitylation and acts as backup to the FA pathway in ICL repair.
DDX11 Facilitates DNA Repair by Homologous Recombination.
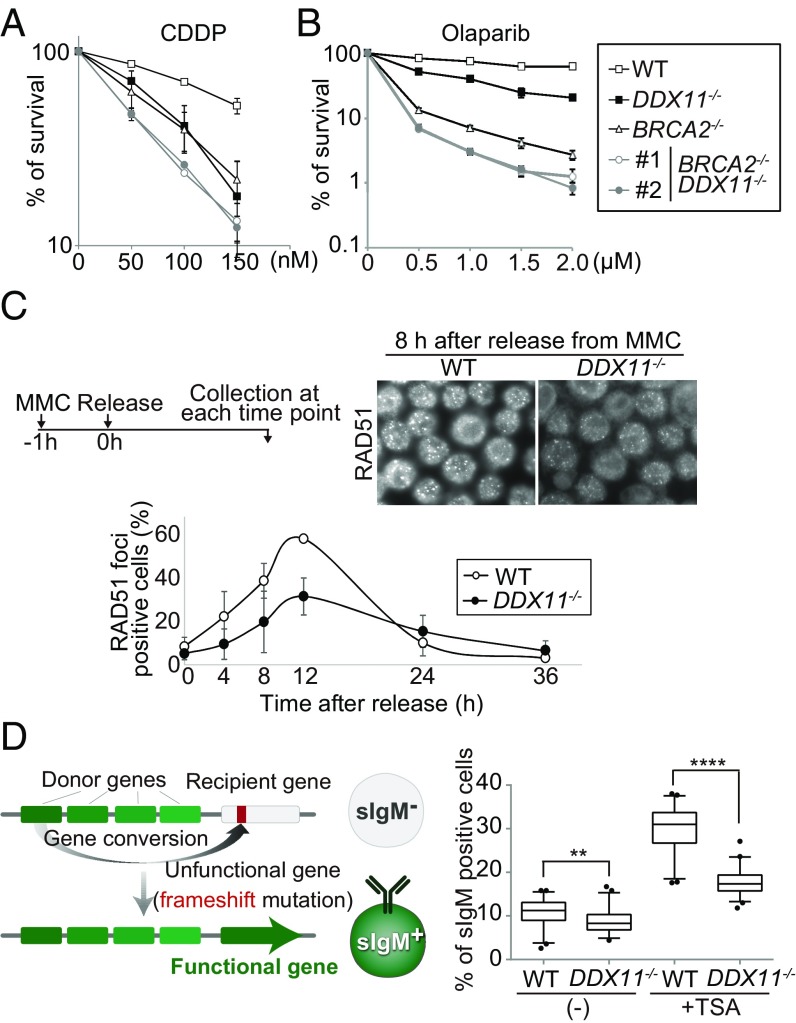
In line with previous reports (27), we found that DDX11 helicase is required to provide resistance against the PARP inhibitor, olaparib (SI Appendix, Fig. S3A), which causes lesions that are repaired primarily by HR. To further investigate connections with HR, we established double mutants between DDX11 and BRCA2, which is critical for HR and implicated in FA (28). ddx11 brca2 cells showed additive sensitivity in regard to both cisplatin and olaparib (Fig. 3 A and B). These results suggest a role for DDX11 in a repair mechanism that responds to olaparib, but which presents distinct features from the BRCA2- and FA core-mediated pathways. Of interest, ddx11 cells are sensitive to the UV mimetic 4NQO (SI Appendix, Fig. S3B), suggesting general roles of DDX11 in the repair of bulky lesions, rather than specificity toward ICLs.
Fig. 3.
DDX11 facilitates DNA repair by homologous recombination. (A and B) Sensitivity of cells as in Fig. 1B. (C) RAD51 focus formation after MMC treatment. Nuclei containing more than four bright foci were defined as foci positive and at least 200 cells were scored for each preparation. The error bars indicate SD from three independent experiments. (D) The scheme of sIgM assay of gene conversion (Left) and the rate of sIgM-positive cells (Right). In the box plots, the middle line indicates the median value; the box shows the 25th and 75th percentiles; the bars, the 5th and 95th percentiles. P values were calculated by Student’s t test. **P ≤ 0.01, ****P ≤ 0.0001.
Many HR-related FA factors contribute to DSB repair. ddx11 cells display sensitivity toward drugs that cause topological stress and ultimately induce DSBs, such as camptothecin, bleomycin, and etoposide (SI Appendix, Fig. S3 C–E), but are proficient in DSB-induced recombination, as measured with a GFP reporter assay (SI Appendix, Fig. S3F). In this assay, the negative effects on HR caused by overexpression of the chicken BRC4 repeat of BRCA2 was used as control (29).
To examine whether DDX11 may contribute to HR repair by affecting RAD51 focus formation, we measured RAD51 foci after a short treatment of cells with MMC, followed by recovery. Notably, RAD51 foci were decreased in ddx11 mutants compared with WT control cells (Fig. 3C). To further address a possible role for DDX11 in endogenous recombination-mediated processes, we examined its role in Ig gene conversion, measured as the gain of surface IgM (sIgM) expression. DT40 cells carry a frameshift mutation in the Ig light-chain variable (IgVλ) segment, but gene conversion from pseudo-V segments removes the frameshift mutation, causing sIgM expression (Fig. 3D, Left). WT and ddx11 clones were expanded for 14 d, in the absence or presence of tricostatin A (TSA), a histone deacetylase inhibitor that increases the frequency of Ig gene conversion in DT40 cells (30), thus allowing detection of rare gene-conversion events. In both spontaneous and TSA conditions, ddx11 mutants showed significant reduction in the frequency of Ig gene conversion events compared with WT cells (Fig. 3D). In all, the results suggest that DDX11 facilitates HR repair of bulky lesions and gene conversion at the IgV region, but is dispensable for initiating HR at DSBs.
DDX11 Acts Jointly with 9-1-1 to Facilitate Postreplicative DNA Repair.
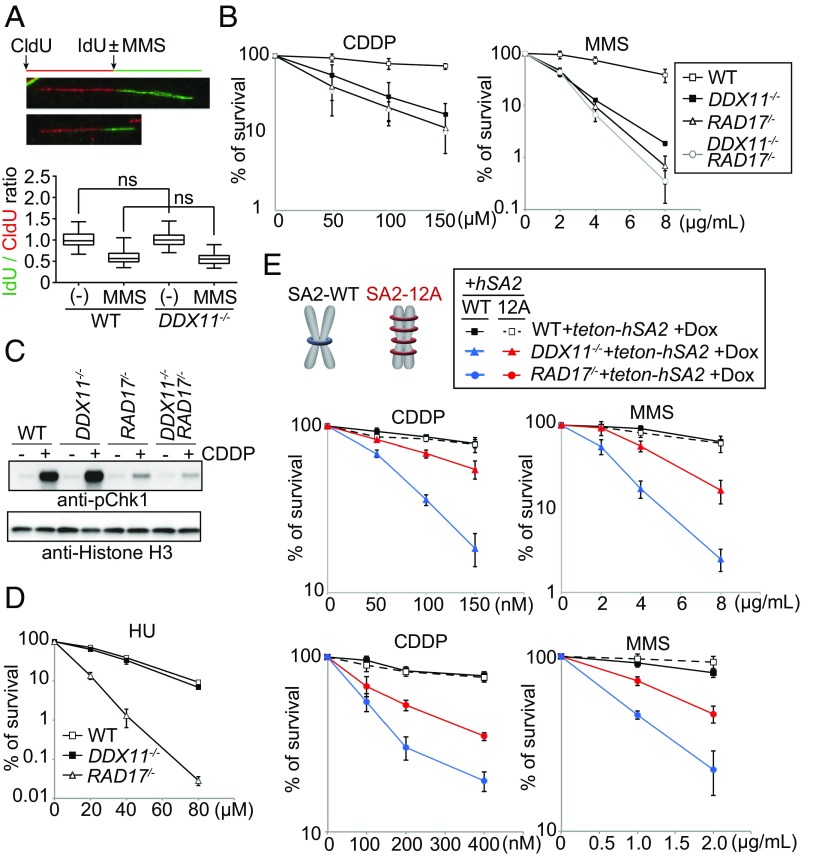
To address whether DDX11 repair functions are manifested at the fork, in which case their absence will delay fork speed, we used CldU/IdU double labeling, with each labeling for the same amount of time. In unperturbed conditions, the ratio is close to 1, whereas in DNA damaging conditions, the ratio is smaller than 1. We did not observe significant differences between WT and ddx11 cells in either condition (Fig. 4A). Moreover, DDX11 is not essential for replication fork stability, as deduced from the lack of sensitivity of ddx11 cells to aphidicolin, a DNA polymerase inhibitor (SI Appendix, Fig. S4A and see below). Taken together, these results suggest that DDX11 may manifest its DNA repair functions primarily postreplicatively.
Fig. 4.
DDX11 acts jointly with 9-1-1 in postreplicative DNA repair. (A) DNA replication speed in the presence of DNA damage. The ratio of IdU/CldU track was plotted. For each sample, more than 70 fibers were measured (WT untreated, n = 102; WT MMS n = 78; ddx11 untreated, n = 90; ddx11 MMS, n = 85). In the box plots, the middle line indicates the median value; the box shows the 25th and 75th percentiles; the bars, the 5th and 95th percentiles. (B) Sensitivity of cells as in Fig. 1B. Note overlap between rad17 and ddx11 rad17 curves upon CDDP treatment. (C) Chk1 phosphorylation after CDDP treatment. Histone H3 was used as a loading control. (D and E) Sensitivity of cells of the indicated genotypes as in Fig. 1B.
Because PrimPol has been implicated in reinitiating replication upon different types of replication lesions in eukaryotic cells (31), we also generated ddx11 primpol double mutants. These mutants showed additive sensitivity toward cisplatin and MMS (SI Appendix, Fig. S4B), suggesting that DDX11 affects a step in DNA damage tolerance that is distinct from the replication restart step and TLS functions mediated by PrimPol. This step is likely independent of proliferating cell nuclear antigen ubiquitylation, as we found this modification to be proficient in ddx11 cells (SI Appendix, Fig. S4C).
The 9-1-1 checkpoint clamp promotes HR and facilitates repair of various types of replication lesions, including those triggered by ICLs, bulky adducts, and ssDNA gaps (32, 33). Because these features are overlapping to what we uncovered here for ddx11, we next analyzed the functional interaction between DDX11 and 9-1-1. Strikingly, we found epistasis between DDX11 and the 9-1-1 checkpoint clamp loader, RAD17, in regard to cisplatin and MMS sensitivity (Fig. 4B).
Next, we analyzed whether DDX11 would act similarly with 9-1-1, or redundantly with it, to facilitate intra-S checkpoint activation. Differently from RAD17, DDX11 was not required for efficient checkpoint activation upon DNA damage as measured by Chk1 phosphorylation (Fig. 4C), and did not affect the ability of cells to slow down S phase progression in the presence of DNA damage (SI Appendix, Fig. S4D). Importantly, DDX11 was also largely dispensable for survival following hydroxyurea (HU)-induced dNTP pool depletion (Fig. 4D), in contrast to the strong effects caused by the rad17 mutation (Fig. 4D) or inhibition of Chk1 (SI Appendix, Fig. S4E), which functions downstream of 9-1-1 and ATR in the checkpoint response. DDX11 was proposed to function together with Tim–Tipin to mediate fork restart upon HU treatment (34). While the checkpoint proficiency and the modest sensitivity of ddx11 cells to HU (Fig. 4 C and D) do not exclude roles in fork protection and fork restart upon HU treatment, here we focused on the joint role of 9-1-1 and DDX11 in DNA repair.
Next, we tested whether the lesions remaining in ddx11 and rad17 cells are amenable to postreplicative HR repair, a pathway induced by improving sister chromatid cohesion (SCC) in G2/M, after the bulk of DNA replication is complete. We improved postreplicative SCC by inhibiting a prophase pathway of cohesin release relying on Plk1-mediated phosphorylation of the cohesin subunit, SA2 (35) (Fig. 4E). We found that expression of SA2-12A can partly rescue the sensitivity of ddx11 and rad17 cells to cisplatin and MMS (Fig. 4E). We note that SA2 and SA2-12A were expressed at similar levels and strictly induced by doxycycline (SI Appendix, Fig. S5A). Similar effects were observed when the consequences of SA2-12A overexpression were compared with cells that do not induce its expression (SI Appendix, Fig. S5B). Thus, the lesions accumulating in ddx11 and rad17 cells can be repaired postreplicatively by alternate HR pathways.
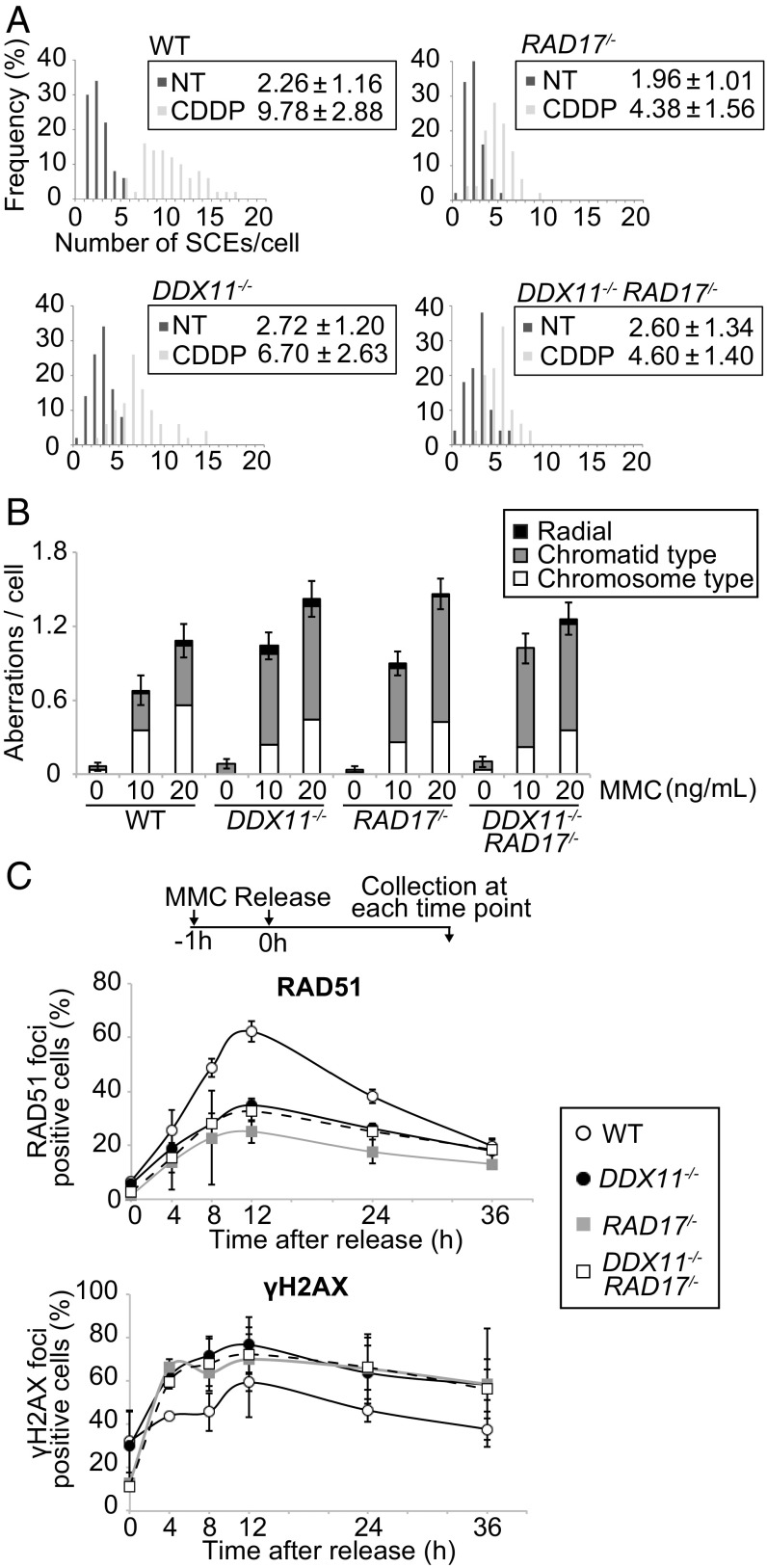
Because HR repair of DNA damage leads to increased sister chromatid exchanges (SCEs) (36), we measured whether DDX11 and RAD17 were required for this process. Both ddx11 and rad17 mutations did not affect the frequency of SCEs in unperturbed conditions, but impaired the ability of cells to efficiently induce SCEs following treatment with cisplatin, in an epistatic manner (Fig. 5A).
Fig. 5.
DDX11 and 9-1-1 jointly promote homologous recombination and avert genomic instability. (A) Spontaneous (nontreated, NT) and damage (CDDP)-induced SCE. The scores represent the mean value and SD from 50 samples, respectively. (B) Chromosomal aberrations following MMC treatment as in Fig. 1D. (C) RAD51 and γH2AX focus formation in cells of the indicated genotype following MMC treatment as in Fig. 3C. The average values were plotted, and the bars indicate the values from two independent experiments.
Next, we asked whether DDX11 and 9-1-1 play similar or distinct roles in averting genome instability induced by MMC (Fig. 1D). We observed similar and nonadditive effects in single and double mutants, with increased incidence in chromatid breaks upon DNA damage (Fig. 5B). Moreover, defects in RAD51 focus formation and accumulation of γH2AX foci following MMC treatment appeared similar in rad17, ddx11, and ddx11 rad17 double mutants (Fig. 5C). In all, these results indicate that DDX11 and 9-1-1 act jointly to mediate efficient HR-mediated repair in response to replication lesions.
DDX11 Facilitates Ig Hypermutation and Gene Conversion.
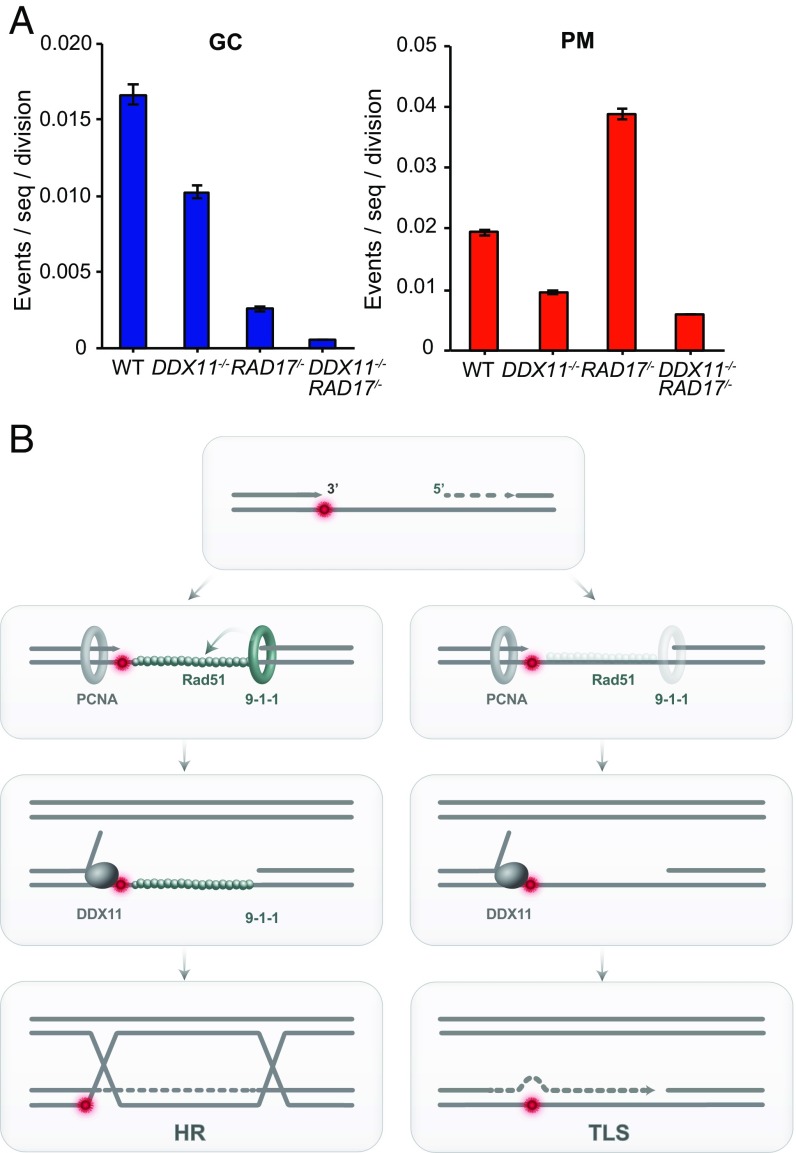
To better analyze the in vivo role played by DDX11 at endogenous abasic sites (Fig. 3D), and its relationship with RAD17, we examined the diversification of the IgVλ region in WT, ddx11, rad17, and ddx11 rad17 mutants overexpressing AID, required for programmed induction of abasic sites at this locus (37, 38). We sequenced IgVλ gene libraries from expanded clones and analyzed the contributions of TLS-mediated hypermutation and gene conversion in this process (SI Appendix, Fig. S6). In a similar trend with rad17, and in line with the sIgM assay of gene conversion (Fig. 3D), we found a reduction in gene conversion events in ddx11 cells compared with WT (Fig. 6A). The reduction in gene conversion events was stronger in rad17 cells, and in line with previous reports (32, 39), and ddx11 rad17 showed similar trends with rad17 mutants (Fig. 6A). rad17 mutants compensate for the reduction in gene conversion by increasing hypermutation (Fig. 6A and ref. 32), similarly with mutations in RAD51 paralogues (39). Interestingly, ddx11 mutants showed reduction in both hypermutation and gene conversion, and the strongly increased hypermutation events in rad17 cells depended in large part on DDX11 (Fig. 6A). These results indicate that DDX11 promotes both gene conversion and TLS-mediated repair of abasic sites.
Fig. 6.
DDX11 facilitates replication through abasic sites and repair of bulky lesions. (A) Rate of gene conversion (GC) and hypermutation (PM). (B) Model for the DDX11 role in promoting homologous recombination jointly with 9-1-1 and translesion synthesis-mediated bypass in the absence of 9-1-1 (see text for details).
Discussion
The importance of the FA pathway in repairing complex lesions such as ICLs is now well established. The existence of genetic disorders that have clinical and cellular overlaps with FA indicates the presence of pathways that can compensate in certain contexts for FA and/or act on other types of endogenous lesions. Our study highlighted that one such pathway is defined by the highly conserved DDX11/ChlR1 helicase mutated in the WABS genetic disorder. The genetic and cell-based analysis provided in this study, with cellular models of WABS, FA subtypes (FANCC, FANCJ, and BRCA2), and combinations, suggests that DDX11 has roles in averting genome instability and in promoting DNA repair of ICLs, but this function is genetically distinct and secondary in importance to the one of canonical FA components. Importantly, our study suggests that DDX11 also facilitates DNA damage tolerance of bulky lesions and replication through endogenous abasic sites, in a largely postreplicative manner and in collaboration with 9-1-1.
Although previous studies reported accumulation of postreplicative gaps in human and mouse cells in response to UV damage (40–43) or upon RAD51 depletion in Xenopus egg extracts (44), little is known about how postreplicative repair is carried out in vertebrate cells. Our work indicates that DDX11 collaborates with the checkpoint clamp 9-1-1 and its loader, RAD17, in the postreplicative HR repair of bulky lesions, and in facilitating gene conversion induced by abasic sites, but without affecting checkpoint activation. Based on the current findings, we propose that 9-1-1, in addition to its role as lesion sensor and activator of ATR, participates together with DDX11 in the postreplicative bypass of replication lesions that are not preferential substrates for the canonical FA pathway or other fork stabilization mechanisms. We note that a noncanonical role for 9-1-1 in the postreplicative HR-mediated repair of replication lesions was observed in budding yeast (45), and that 9-1-1 repair defects can be overcome by overexpressing RAD51 (46). We propose a model in which the action of 9-1-1 favors the formation of the RAD51 filament on postreplicative gaps, while DDX11 promotes the unwinding of the stalled 3′ end (Fig. 6B). The intermediate formed by the joint actions of 9-1-1 and DDX11 may be efficiently matured into an HR intermediate, while in the absence of 9-1-1, the 3′ end exposed by the action of the DDX11 helicase may be more readily engaged by TLS polymerases (Fig. 6B).
A joint role for DDX11 and 9-1-1 in the HR repair of replication lesions and abasic sites may explain why their individual depletion/knockout leads to very similar developmental defects in mouse, with the somitic mesoderm being especially sensitive to the genomic instability produced by their individual mutations (22, 23) and resembling the ones caused by mutations in Brca2 and Palb2 (47, 48). These repair functions are likely critical in situations of fast proliferation, such as during early stages of development. We also found that DDX11 mitigates replication stress in FANCJ-defective cells and is critical for the repair of olaparib- and cisplatin-induced DNA damage in BRCA2 and FA-defective cells. Thus, another implication of our findings is that DDX11 is important for limiting replication stress in FA and BRCA mutated cells, and therefore it may be vital for the survival of these and other cancers, which often suffer from replication stress (1). Based on our findings, we propose that the DNA repair pathway mediated by DDX11 has important functions related to replication of endogenous abasic sites and DNA damage tolerance of bulky lesions. This pathway is critical for human disease and may offer new opportunities for cancer treatment.
Materials and Methods
The methodology employed was described in refs. 24 and 29 except for the DNA fiber analysis, which was performed largely as described in ref. 49, and Ig gene diversification experiments, which were performed as in ref. 38. A complete list of reagents and detailed methodology are available in SI Appendix.
Supplementary Material
Acknowledgments
We thank S. Takeda, H. Arakawa, J. M. Peters, M. Kobayashi, K. Yamamoto, and H. Kitao for reagents; B. Szakal for help with the art work; J. Sale and all laboratory members for discussion; and the tissue culture and imaging facility at the FIRC Institute of Molecular Oncology. This work was supported by Fondazione Telethon Grant (GGP12160), the Italian Association for Cancer Research Grants (IG 14171 and IG 18976), and the European Research Council (starting Grant 242928 and consolidator Grant 682190) (to D.B.), and Japanese Society for Promotion of Science Grant KAKENHI (JP 16H02957) (to K.H.). M.O. was supported by an internal grant from Tokyo Metropolitan University for visiting graduate students.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803110115/-/DCSupplemental.
References
- 1.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 2.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 4.Sale JE. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012708. doi: 10.1101/cshperspect.a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romanello M, Schiavone D, Frey A, Sale JE. Histone H3.3 promotes IgV gene diversification by enhancing formation of AID-accessible single-stranded DNA. EMBO J. 2016;35:1452–1464. doi: 10.15252/embj.201693958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arakawa H, Buerstedde JM. Activation-induced cytidine deaminase-mediated hypermutation in the DT40 cell line. Philos Trans R Soc Lond B Biol Sci. 2009;364:639–644. doi: 10.1098/rstb.2008.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: New players and new functions. Nat Rev Mol Cell Biol. 2016;17:337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 8.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pontel LB, et al. Endogenous formaldehyde is a hematopoietic stem cell genotoxin and metabolic carcinogen. Mol Cell. 2015;60:177–188. doi: 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knipscheer P, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomida J, et al. A novel interplay between the Fanconi anemia core complex and ATR-ATRIP kinase during DNA cross-link repair. Nucleic Acids Res. 2013;41:6930–6941. doi: 10.1093/nar/gkt467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishiai M, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15:1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Lelij P, et al. Warsaw breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am J Hum Genet. 2010;86:262–266. doi: 10.1016/j.ajhg.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capo-Chichi JM, et al. Identification and biochemical characterization of a novel mutation in DDX11 causing Warsaw breakage syndrome. Hum Mutat. 2013;34:103–107. doi: 10.1002/humu.22226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey C, Fryer AE, Greenslade M. Warsaw breakage syndrome–A further report, emphasising cutaneous findings. Eur J Med Genet. 2015;58:235–237. doi: 10.1016/j.ejmg.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Eppley S, Hopkin RJ, Mendelsohn B, Slavotinek AM. Clinical report: Warsaw breakage syndrome with small radii and fibulae. Am J Med Genet A. 2017;173:3075–3081. doi: 10.1002/ajmg.a.38382. [DOI] [PubMed] [Google Scholar]
- 18.Hirota Y, Lahti JM. Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase. Nucleic Acids Res. 2000;28:917–924. doi: 10.1093/nar/28.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farina A, et al. Studies with the human cohesin establishment factor, ChlR1. Association of ChlR1 with Ctf18-RFC and Fen1. J Biol Chem. 2008;283:20925–20936. doi: 10.1074/jbc.M802696200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y, Sommers JA, Khan I, de Winter JP, Brosh RM., Jr Biochemical characterization of Warsaw breakage syndrome helicase. J Biol Chem. 2012;287:1007–1021. doi: 10.1074/jbc.M111.276022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah N, et al. Roles of ChlR1 DNA helicase in replication recovery from DNA damage. Exp Cell Res. 2013;319:2244–2253. doi: 10.1016/j.yexcr.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cota CD, García-García MJ. The ENU-induced cetus mutation reveals an essential role of the DNA helicase DDX11 for mesoderm development during early mouse embryogenesis. Dev Dyn. 2012;241:1249–1259. doi: 10.1002/dvdy.23810. [DOI] [PubMed] [Google Scholar]
- 23.Weiss RS, Enoch T, Leder P. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 2000;14:1886–1898. [PMC free article] [PubMed] [Google Scholar]
- 24.Abe T, et al. Chromatin determinants of the inner-centromere rely on replication factors with functions that impart cohesion. Oncotarget. 2016;7:67934–67947. doi: 10.18632/oncotarget.11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meetei AR, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 26.Bharti SK, et al. Molecular functions and cellular roles of the ChlR1 (DDX11) helicase defective in the rare cohesinopathy Warsaw breakage syndrome. Cell Mol Life Sci. 2014;71:2625–2639. doi: 10.1007/s00018-014-1569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoepker C, et al. DNA helicases FANCM and DDX11 are determinants of PARP inhibitor sensitivity. DNA Repair (Amst) 2015;26:54–64. doi: 10.1016/j.dnarep.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: The roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7:a016600. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe T, Branzei D. High levels of BRC4 induced by a Tet-On 3G system suppress DNA repair and impair cell proliferation in vertebrate cells. DNA Repair (Amst) 2014;22:153–164. doi: 10.1016/j.dnarep.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seo H, et al. Rapid generation of specific antibodies by enhanced homologous recombination. Nat Biotechnol. 2005;23:731–735. doi: 10.1038/nbt1092. [DOI] [PubMed] [Google Scholar]
- 31.Bianchi J, et al. PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol Cell. 2013;52:566–573. doi: 10.1016/j.molcel.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saberi A, et al. The 9-1-1 DNA clamp is required for immunoglobulin gene conversion. Mol Cell Biol. 2008;28:6113–6122. doi: 10.1128/MCB.00156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim PX, et al. Genome protection by the 9-1-1 complex subunit HUS1 requires clamp formation, DNA contacts, and ATR signaling-independent effector functions. J Biol Chem. 2015;290:14826–14840. doi: 10.1074/jbc.M114.630640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calì F, Bharti SK, Di Perna R, Brosh RM, Jr, Pisani FM. Tim/Timeless, a member of the replication fork protection complex, operates with the Warsaw breakage syndrome DNA helicase DDX11 in the same fork recovery pathway. Nucleic Acids Res. 2016;44:705–717. doi: 10.1093/nar/gkv1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hauf S, et al. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonoda E, et al. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol Cell Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohzaki M, et al. DNA polymerases nu and theta are required for efficient immunoglobulin V gene diversification in chicken. J Cell Biol. 2010;189:1117–1127. doi: 10.1083/jcb.200912012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirota K, et al. Simultaneous disruption of two DNA polymerases, Polη and Polζ, in Avian DT40 cells unmasks the role of Polη in cellular response to various DNA lesions. PLoS Genet. 2010;6:e1001151. doi: 10.1371/journal.pgen.1001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sale JE, Calandrini DM, Takata M, Takeda S, Neuberger MS. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 2001;412:921–926. doi: 10.1038/35091100. [DOI] [PubMed] [Google Scholar]
- 40.Lehmann AR. Postreplication repair of DNA in ultraviolet-irradiated mammalian cells. J Mol Biol. 1972;66:319–337. doi: 10.1016/0022-2836(72)90418-4. [DOI] [PubMed] [Google Scholar]
- 41.Lehmann AR, et al. Repair of ultraviolet light damage in a variety of human fibroblast cell strains. Cancer Res. 1977;37:904–910. [PubMed] [Google Scholar]
- 42.Buhl SN, Stillman RM, Setlow RB, Regan JD. DNA chain elongation and joining in normal human and xeroderma pigmentosum cells after ultraviolet irradiation. Biophys J. 1972;12:1183–1191. doi: 10.1016/S0006-3495(72)86154-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buhl SN, Setlow RB, Regan JD. Recovery of the ability to synthesize DNA in segments of normal size at long times after ultraviolet irradiation of human cells. Biophys J. 1973;13:1265–1275. doi: 10.1016/S0006-3495(73)86061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karras GI, et al. Noncanonical role of the 9-1-1 clamp in the error-free DNA damage tolerance pathway. Mol Cell. 2013;49:536–546. doi: 10.1016/j.molcel.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 46.Shinohara M, Sakai K, Ogawa T, Shinohara A. The mitotic DNA damage checkpoint proteins Rad17 and Rad24 are required for repair of double-strand breaks during meiosis in yeast. Genetics. 2003;164:855–865. doi: 10.1093/genetics/164.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki A, et al. Brca2 is required for embryonic cellular proliferation in the mouse. Genes Dev. 1997;11:1242–1252. doi: 10.1101/gad.11.10.1242. [DOI] [PubMed] [Google Scholar]
- 48.Rantakari P, et al. Inactivation of Palb2 gene leads to mesoderm differentiation defect and early embryonic lethality in mice. Hum Mol Genet. 2010;19:3021–3029. doi: 10.1093/hmg/ddq207. [DOI] [PubMed] [Google Scholar]
- 49.Hosono Y, et al. Tipin functions in the protection against topoisomerase I inhibitor. J Biol Chem. 2014;289:11374–11384. doi: 10.1074/jbc.M113.531707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.