Significance
The molecular mechanisms leading to the creation and maintenance of the latent HIV reservoir remain incompletely understood. Unbiased shRNA screens showed that the estrogen receptor acts as a potent repressor of proviral reactivation in T cells. Antagonists of ESR-1 activate latent HIV-1 proviruses while agonists, including β-estradiol, potently block HIV reactivation. Using a well-matched set of male and female donors, we found that ESR-1 plays an important role in regulating HIV transcription in both sexes. However, women are much more responsive to estrogen and appear to harbor smaller inducible RNA reservoirs. Accounting for the impact of estrogen on HIV viral reservoirs will therefore be critical for devising curative therapies for women, a group representing 51% of global HIV infections.
Keywords: HIV-1 latency, HIV-1 reservoir, estrogen receptor, latency-reversing agents, selective estrogen receptor modulators
Abstract
Unbiased shRNA library screens revealed that the estrogen receptor-1 (ESR-1) is a key factor regulating HIV-1 latency. In both Jurkat T cells and a Th17 primary cell model for HIV-1 latency, selective estrogen receptor modulators (SERMs, i.e., fulvestrant, raloxifene, and tamoxifen) are weak proviral activators and sensitize cells to latency-reversing agents (LRAs) including low doses of TNF-α (an NF-κB inducer), the histone deacetylase inhibitor vorinostat (soruberoylanilide hydroxamic acid, SAHA), and IL-15. To probe the physiologic relevance of these observations, leukapheresis samples from a cohort of 12 well-matched reproductive-age women and men on fully suppressive antiretroviral therapy were evaluated by an assay measuring the production of spliced envelope (env) mRNA (the EDITS assay) by next-generation sequencing. The cells were activated by T cell receptor (TCR) stimulation, IL-15, or SAHA in the presence of either β-estradiol or an SERM. β-Estradiol potently inhibited TCR activation of HIV-1 transcription, while SERMs enhanced the activity of most LRAs. Although both sexes responded to SERMs and β-estradiol, females showed much higher levels of inhibition in response to the hormone and higher reactivity in response to ESR-1 modulators than males. Importantly, the total inducible RNA reservoir, as measured by the EDITS assay, was significantly smaller in the women than in the men. We conclude that concurrent exposure to estrogen is likely to limit the efficacy of viral emergence from latency and that ESR-1 is a pharmacologically attractive target that can be exploited in the design of therapeutic strategies for latency reversal.
Patients adhering to combination antiretroviral therapy (ART) regimens have minimal plasma viremia, but unfortunately HIV-1 still persists due to the existence of a tiny population of transcriptionally silent viruses (1–3) that reside primarily in memory CD4+ T cells in the peripheral blood (4) and tissues (5). Latently infected T cells are already established during the acute phase of infection when viremia is highest, and early initiation of ART cannot prevent the seeding of this pool of latently infected cells (6). The latent reservoir is very stable, with an apparent half-life of 44 mo in the presence of ART (7, 8). Recent studies of proviral integration sites have provided strong evidence for clonal expansion of specific proviruses, suggesting that the reservoirs can be replenished through homeostatic proliferation (9, 10).
Devising strategies that safely and effectively eradicate the latent reservoir is the challenging goal of current HIV-1 “cure” efforts (11, 12). In quiescent T cells, latency is maintained through restrictions imposed on both the cellular transcription machinery, due to sequestration of the transcriptional elongation factor P-TEFb and the transcriptional initiation factors including NF-κB and NF-AT and the imposition of epigenetic silencing by histone methyltransferases (13). These complementary restrictions make it difficult to reverse proviral latency with single agents (14). Clinical studies examining candidate latency-reversal agents (LRAs) have demonstrated transient increases in cell-associated HIV-1 RNA, but none of these agents has led to a significant change in integrated HIV-1 DNA or the inducible reservoir (15).
Using an unbiased, genome-wide shRNA screen to identify pathways relevant to latency, we identified estrogen receptor-1 (ESR-1) as a cellular factor critical for the maintenance of HIV-1 latency. Although earlier reports suggested that estrogen can inhibit HIV-1 transcription (16, 17) in activated cells, the association of ESR-1 with latency control was unexpected and may underlie the observation that plasma HIV RNA levels tend to be lower in women than men (18).
Our group established a cohort of 52 well-matched reproductive-age women and men with comparable clinical and demographic features. Using a subset of women and men from this cohort who underwent leukapheresis, we showed that latent proviruses can be partially reactivated in both sexes in response to selective estrogen receptor modulators (SERMs) and potently inhibited by hormone exposure. Females showed significantly higher levels of inhibition in response to β-estradiol and higher reactivity in response to SERMs than males and had larger inducible RNA reservoirs. These data demonstrate physiologically determined differences in the behavior of the proviral reservoirs in women and men, highlighting the need to design cure-related clinical studies and therapeutic regimens that effectively target both sexes (19).
Results
ESR-1 Regulates HIV-1 Latency.
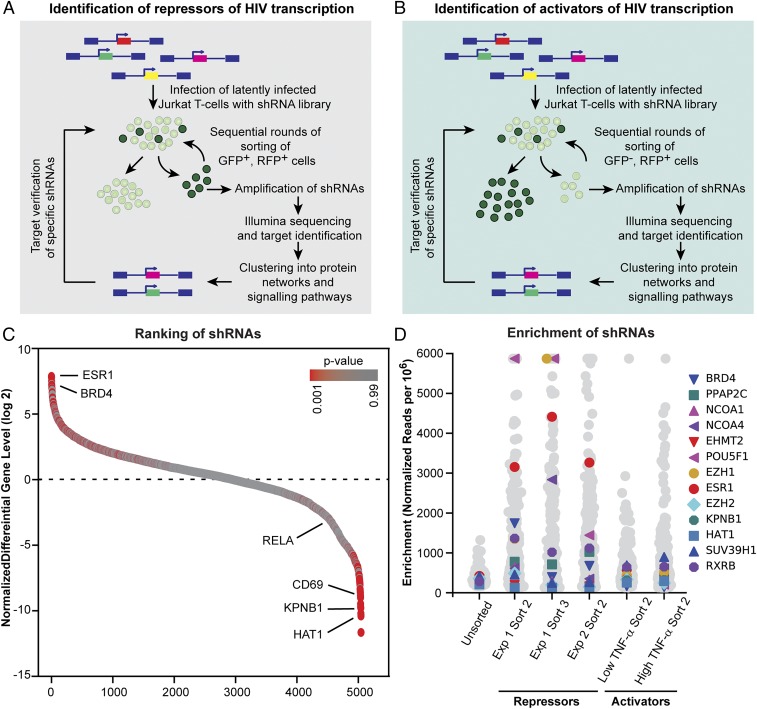
An unbiased genome-wide shRNA screen for cellular factors required for the maintenance of HIV-1 latency demonstrated that ESR-1 is one of the key factors regulating latency. As shown in Fig. 1 A and B, a synthetic lentiviral shRNA library from Cellecta, Inc. containing 82,500 shRNAs targeting 15,439 mRNA sequences in three modules was used to infect 2D10 cells, which are a latently infected Jurkat T cell line (20–22). Cells carrying reactivated proviruses (repressor screen) or cells that failed to reactivate after stimulation by TNF-α (activator screen) were then purified by sequential rounds of sorting. This resulted in a progressive enrichment for clones expressing shRNAs that either led to constitutive reactivation of HIV-1 or blocked HIV-1 reactivation by TNF-α (SI Appendix, Fig. S1A).
Fig. 1.
ESR-1 is required to maintain HIV-1 latency. (A) Repressor screen. Latently infected Jurkat 2D10 cells (d2EGFP reporter) were infected with a shRNA library (RFP reporter). GFP+, RFP+ cells were purified by three sequential rounds of sorting. (B) Activator screen. Cells were stimulated with TNF-α before sorting for d2EGFP−, RFP+ cells. After each round of sorting, genomic DNA was isolated, and shRNAs were identified by nested PCR amplification with barcode-specific primers followed by Illumina sequencing. (C) Ratio between normalized abundance values of shRNAs from the repressor screen (d2EGFP+) and activator screen (d2EGFP−) candidates. Data are from the module 1 screen (14,849 shRNAs identified). (D) The level of enrichment of ESR-1 and a number of additional genes with known roles in HIV latency is shown for three repressor screen datasets from two different rounds of selection (sorts 2 and 3) and two independent experiments. The initial level of representation of each gene in the transfected cell population (sort 0) and the results of two activator screen datasets performed after stimulation with either 1 ng or 6 ng/mL of TNF-α are shown as controls. The level of enrichment of each gene is the number of RNA-seq reads mapped to the gene per million total mapped reads, flattened to a maximum enrichment value of 6,000 reads per million.
Analysis of the shRNA repressor screen data obtained from next-generation sequencing (NGS) of samples from three sequential rounds of enrichment was performed using edgeR (23). To help identify and eliminate genes that were enriched due to their impact on cell growth and other confounding factors in cell-based screens, the ratio of the normalized abundance values of shRNAs identified by the two opposing selection conditions was calculated (Fig. 1C). The results from all shRNAs targeting the same gene (three to six per gene) were summarized to obtain gene-level enrichment data per replicate group. False-discovery rates (FDRs) and P values were calculated from the enrichment values obtained from the repressor–activator screen comparison. ESR-1 was the second most enriched gene in the repressor screen, with enrichment levels ranging between eightfold after the first round of sorting (FDR = 0.008) to 40-fold after the second round of sorting and 550-fold after the third round of sorting (FDR = 9E-05–2E-05) (Fig. 1 C and D).
Bee swarm plots were also used to illustrate the high level of enrichment of ESR-1 achieved for three independent repressor screen datasets from two different rounds of selection (dataset 1, sorts 2 and 3) and two independent experiments (dataset 2, sort 2) (Fig. 1D). By contrast, there was no significant enrichment of ESR-1 in the unsorted cell population or in two independent activator screens performed with TNF-α.
A number of previously identified HIV-1 latency factors served to validate the procedure (Fig. 1D). In particular, BRD4, which competes for Tat and is the key target of the LRA JQ1 (24), was reproducibly highly ranked in the repressor screen and is known to induce latent HIV after shRNA knockdown (25).
To confirm ESR-1 as a hit, knockdown of the ESR-1 transcript by the specific shRNA sequence from the screening library was performed. Successive rounds of sorting led to the isolation of a constitutively reactivated proviral population (93.08%) compared with 2.84% reactivation using a scrambled shRNA control (SI Appendix, Fig. S1B). Since HIV-1 induction in Jurkat cells is exquisitely sensitive to the activation of NF-κB, an important control to demonstrate the specificity of the ESR-1 shRNA knockdowns was to rule out induction of NF-κB–p65 in the shRNA-treated cells. Flow cytometry-based image-stream analysis (SI Appendix, Fig. S1C) and immunoblot analysis of nuclear extracts (SI Appendix, Fig. S1D) showed no nuclear localization of NF-κB–p65 in either scrambled shRNA or ESR-1 shRNA-knockdown cells.
In addition to ESR-1 itself, the shRNA screen identified a number of interacting proteins that also ranked highly in the repressor screen. A GeneMANIA-generated network (SI Appendix, Fig. S2A) identified several molecules that ranked at the top of the screening list which also are known to directly modulate or influence the biological activity of ESR-1, including the nuclear receptor coactivators 1 and 3 (NCOA1 and 3). Heat maps from an independent analysis of the ESR-1 interactome using the list of ESR-1–containing complexes in the CORUM (26) and PCDq databases (27) and Gene Ontology (GO) terms related to estrogen signaling similarly indicated the enrichment of ESR-1–interacting proteins in the shRNA screen (SI Appendix, Fig. S3).
Pharmacological Control of HIV Latency in Primary Cells Using ESR-1 Antagonists and Agonists.
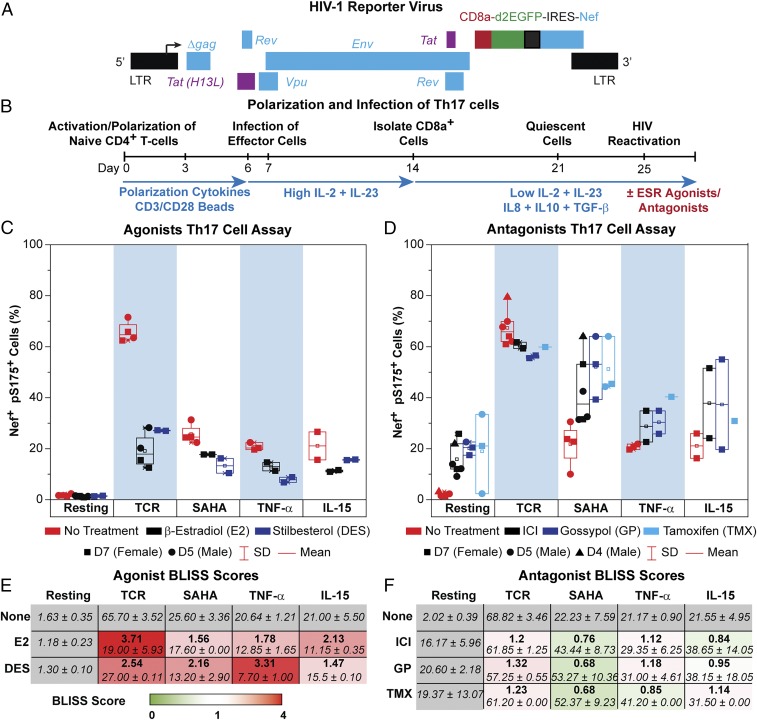
The quiescent effector cell latency (QUECEL) model (Fig. 2) is a robust primary cell model for HIV latency (20, 28) using polarized effector memory T cell populations (29, 30). Briefly, naive helper T cells were polarized to a Th17 polarization phenotype, which represents the most abundant effector T cell population in the lamina propria of the gastrointestinal tract and a major reservoir for HIV (31, 32). Peripheral blood mononuclear cell (PBMC) populations with the highest frequency of HIV-1 proviruses are detected in Th1 cells, followed closely by Th17 cells (33).
Fig. 2.
Synergism and antagonism of SERMs and LRAs in the QUECEL (Th17) primary cell model. (A) HIV-1 reporter virus. (B) Experimental design. (C) Assay of ESR-1 agonists. Cells were stimulated overnight through the TCR or by 500 nM SAHA, 10 ng/mL TNF-α, or 10 ng/mL IL-15. The addition of 300 pg/mL β-estradiol or 1 nM stilbesterol greatly reduced the reactivation of latent HIV-1 by each of these LRAs. Symbols indicate separate experiments using various donors: D7 (female; squares), D5 (male; circles), and D4 (male; triangles). Reactivation was measured by flow cytometry scoring for Nef+, pSer175+ cells. (D) Antagonists of ESR-1 are either additive or synergistic when combined with LRAs. Cells were activated as described above in the absence or presence of SERMs (ICI and TMX) and the NCOA1/3 inhibitor gossypol. (E) Table showing the average fraction of Nef+, pSer175+ cells following treatment with LRAs and ESR-1 agonists using the data in A and BLISS scores (bold). Table cells are color-coded based on the degree of antagonism (red, BLISS >1). (F) Table showing the average fraction of Nef+, pSer175+ cells following treatment with LRAs and ESR-1 antagonists, using the data in B. BLISS scores are indicated in bold. Table cells are color-coded based on the degree of synergism (green, BLISS <1), additivity (white, BLISS = 1), or antagonism (red, BLISS >1).
The expanding polarized T cells were infected using a single-round reporter virus carrying the tat, rev, env, and nef genes (21, 22) and a CD8a–EGFP fusion protein that permitted the purification of infected cells with magnetic beads (Fig. 2A). The HIV-1–infected cells are forced into quiescence by culturing the cells in restrictive cytokine medium (low IL-2 and IL-23) supplemented with the antiinflammatory cytokines IL8, IL-10, and TGF-β (Fig. 2B). The quiescent cells were in the Go phase of the cell cycle, showed negligible 5-ethynyl-2′-deoxyuridine (EDU) incorporation, and were negative for Ki67 and cyclin D3 (SI Appendix, Fig. S4A).
Once the cells become quiescent, the level of HIV-1 Nef protein is reduced to almost undetectable levels (<1.5%), indicative of HIV-1 latency (Fig. 2C and SI Appendix, Fig. S4B), but a moderate level of GFP persists in the quiescent cells because of the high stability of the CD8a–GFP fusion protein, which serves as a marker of infection (SI Appendix, Fig. S4B). Stimulation through the T cell receptor (TCR) by α-CD3/α-CD28 Dynal magnetic beads, induced Nef and cycling markers dramatically (>79%) (SI Appendix, Fig. S4). We therefore routinely use Nef, rather than CD8a–GFP, as a marker of proviral transcription. Since LRAs do not always force the cells into proliferation, we use a P-TEFb–specific marker, pSer175 CDK9, to identify cells that have become activated (SI Appendix, Fig. S4B) (25, 34). Accordingly, in the QUECEL assays presented here, we have scored Nef and pSer175 doubly positive cells (Fig. 2).
The extensive armamentarium of therapeutically developed ESR-1 modulators provides informative tools to study the role of ESR-1 in the repression of HIV-1 transcription in the QUECEL (Th17) model (Fig. 2 and SI Appendix, Fig. S5) and Jurkat T cells (SI Appendix, Fig. S2). In both systems, ESR-1 agonists are potent repressors of HIV proviral reactivation, while ESR-1 antagonists (SERMS) are weak activators by themselves and potentiate activation by other LRAs, such as the histone deacetylase inhibitor (HDACi) vorinostat (soruberoylanilide hydroxamic acid, SAHA), TNF-α, and IL-15.
Ordinary tissue-culture media containing FBS can be a source of β-estradiol, and phenol red is an estrogen analog. To control for this in our ex vivo experiments, we used medium without phenol red and charcoal-stripped FBS. The levels of β-estradiol were measured in all media using a sensitive ELISA (SI Appendix, Fig. S6). All media used in these studies contained less than 10 pg/mL β-estradiol, which is at the level of detection for the assay. We also used the ELISA to confirm the accuracy of the added β-estradiol levels.
The impact of a wide range of ESR-1 agonists and SERMs (antagonists) were tested in the Th17 model using cells from three separate donors, two male and one female (Fig. 2 and SI Appendix, Fig. S5). β-Estradiol and the ESR-1 synthetic agonist diethylstilbestrol (DES) were extremely potent blockers of TCR-mediated proviral reactivation, and both compounds partially blocked viral induction by SAHA, TNF-α, and IL-15 (Fig. 2C). The ESR-1 antagonists tamoxifen (TMX) and fulvestrant (ICI) had the opposite effect, partially reactivating latent HIV-1, with additive effects on proviral reactivation when combined with either SAHA or IL-15 (Fig. 2 and SI Appendix, Fig. S5). For example, treatment of the quiescent Th17 cells with 1 μM TMX or 500 nM SAHA activated 21.29% and 22.56% of the cells, respectively, whereas the combination of TMX and SAHA activated 51.25% of the cells (Fig. 2D).
The NCOA1 and NCOA3 inhibitor gossypol, an agent under investigation as a male oral contraceptive (35), partially reactivated the latent proviruses as a solo agent and sensitized cells to reactivation by SAHA, TNF-α, and IL-15 (Fig. 2D and SI Appendix, Fig. S5C).
Calculation of Bliss scores showed potent repression of HIV transcription by the ESR-1 agonists (Fig. 2E) and additive or occasionally (depending on the donor) synergistic effects of ESR-1 antagonists on proviral reactivation with SAHA, TNF-α, and IL-15 (Fig. 2F). Titrations of the SERMs ICI and TMX in the presence and absence of SAHA confirmed the additive effects of these agents (SI Appendix, Fig. S5D).
ESR-1 Accumulates on the LTRs of Latent Proviruses in Primary Cells.
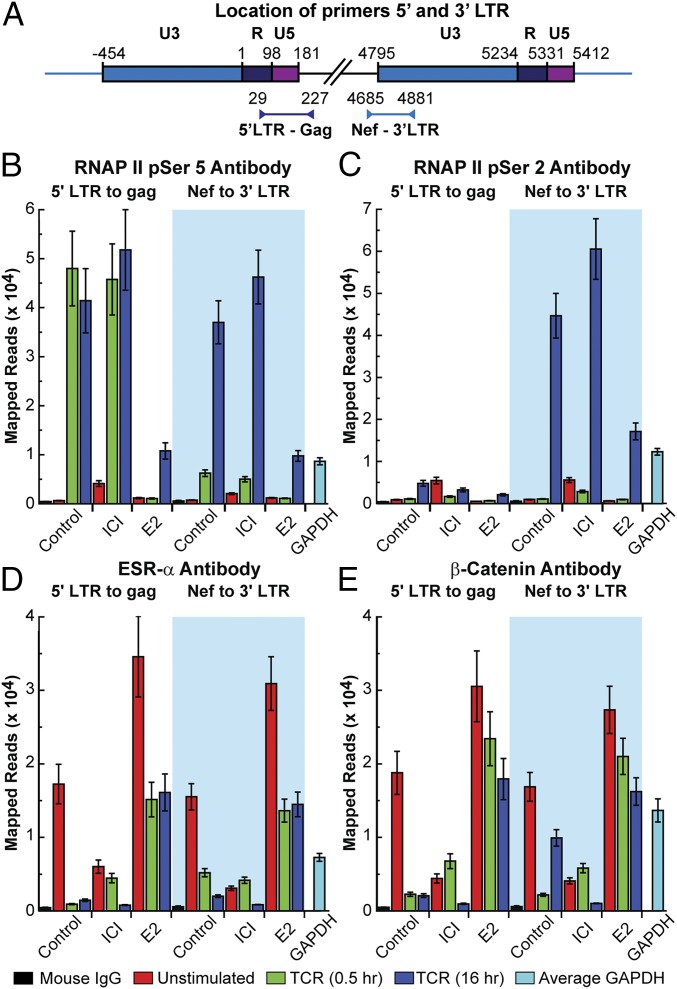
We were able to obtain sufficient numbers of Th17 cells from a healthy male donor (donor 7) to measure the occupancy of ESR-1 on the HIV-1 LTR by sensitive ChIP assays based on ion torrent sequencing (Fig. 3) (22). LTR-specific primer sets each carrying one primer containing unique viral sequences in Gag or Nef that are adjacent to the 5′ or 3′ LTR and a second primer within the LTR were used to distinguish between the two LTRs (Fig. 3A) (22). ChIP assays were performed on quiescent cells, and cells were stimulated for 0.5 or 16 h via the TCR in the presence or absence of ICI or β-estradiol.
Fig. 3.
ChIP assay demonstrating repression of HIV-1 transcription by ESR-1. Latently infected Th17 cells were activated through the TCR by CD3/CD28-coated magnetic beads for either 0.5 or 16 h in the presence or absence of 1 μM ICI or 300 pg/mL β-estradiol. (A) Location of primers used to detect protein interactions at the 5′ and 3′ LTRs of the HIV-1 provirus. (B) RNAP II pSer5. (C) RNAP II pSer 2. (D) ESR-1. (E) β-catenin.
The distribution of RNA polymerase II (RNAP II) on the HIV-1 provirus was determined using antibodies that detect the pSer5 and pSer2 phosphorylated forms of RNAP II (Fig. 3 B and C) (22). The combination of these antibodies allow us to distinguish between RNAP II paused proximally to the HIV-1 promoter (pSer-5) and elongating RNAP II transcription complexes that have been activated by Tat/P-TEFb (pSer-2 plus pSer-5) (22). In the quiescent cells, minimal RNAP II was detected by the pSer5 antibody at the 5′ LTR and 3′ LTRs due to the highly restricted transcription in latently infected cells (Fig. 3B). This is also consistent with our previous observation that only hypophosphorylated RNAP II accumulates at the 5′ LTR of latently infected cells (36). Stimulation of the cells through the TCR resulted in a large accumulation of promoter-proximal RNAP II (pSer5) at both 30 min and 16 h. Essentially all the RNAP II (pSer 5) is associated with the 5′ LTR after 30-min induction by TCR. However, after 16 h of induction, there is significant accumulation of RNAP II at the downstream LTR, in agreement with our previous observation that there is a 4-h lag before the induction of Tat from latent proviruses (24). Consistent with these results, RNAP II (pSer2) accumulates at high levels on the 3′ LTR only after 16 h of TCR induction (Fig. 3C).
ICI is able to increase RNAP II (pSer5) accumulation in latently infected cells, which is indicative of its impact in reversing blocks imposed by ESR-1 (Fig. 3B). ICI also significantly increases the levels of RNAP II (pSer5 and pSer2) at the 3′ LTR, which is a measure of enhanced transcription (Fig. 3 B and C), at 16 h after the initial TCR induction. The ChIP assay also confirmed direct transcriptional repression imposed by β-estradiol, which leads to a dramatic reduction of RNAP II (pSer5) at the 5′ and 3′ LTRs and reduction of RNAP II (pSer2) at the 3′ LTR after stimulation by TCR (Fig. 3 B and C).
Using the same samples, the distribution of ESR-1 (Fig. 3D) and β-catenin (Fig. 3E), which has been reported to be part of a corepressor complex recruited by ESR-1 in activated T cells, (17) was measured. Importantly, the quiescent cells accumulate ESR-1 at both LTRs, and ESR-1 levels rise in the presence of added β-estradiol, consistent with the role of ESR-1 as a repressor of HIV-1 transcription. ESR-1 levels fall dramatically in the presence of ICI or after activation via the TCR. β-Catenin levels fluctuate in parallel with ESR-1 levels, and both ESR-1 and β-catenin levels are inversely correlated with RNAP II levels (Fig. 3 D and E).
As a control for the impact of these drugs on total cellular ESR-1 levels, CD4 T cells from one male and one female donor were treated similarly as in the ChIP assay. Nuclear extracts were collected, and ESR-1 expression levels were detected via Western blot (SI Appendix, Fig. S6C). Both donors had similar levels of ESR-1 protein, which remained relatively constant under all conditions, although there was a general increase in ESR-1 levels at 16 h (SI Appendix, Fig. S6D).
Women Have Reduced Inducible RNA Reservoirs Compared with Men.
The preceding molecular experiments demonstrate a direct role for ESR-1 in the silencing of HIV-1. These observations led us to hypothesize that there would be sex-specific differences in both the size of the inducible HIV-1 reservoir and the response to LRAs in the presence and absence of hormones and SERMs. We obtained leukapheresis samples from six women and six men from a cohort of well-matched reproductive-aged females and males that has been extensively characterized as part of a study of gender differences. As shown in Table 1, the men and women in this group, as in the larger cohort, were matched by age, CD4 nadir, pretreatment viral load, time of suppression, and duration of infection. None of the women used hormonal contraception.
Table 1.
Demographics and clinical parameters of study subjects
| Parameter | Women | Men | P value |
| Age, y (range) | 44 (30–48) | 36.5 (32–54) | 0.6 |
| CD4 nadir in cells/µL (range) | 213.5 (76–661) | 210 (129–480) | 0.9 |
| Log maximum pretreatment viral load in copies/mL (range) | 4.63 (4.05–5.41) | 5.19 (2.85–5.70) | 0.5 |
| Duration of viral suppression, mo (range) | 37 (19–131) | 37 (24–98) | 0.9 |
| Duration of HIV-1 infection, y (range) | 11.5 (5–16) | 8 (5–16) | 0.5 |
For each variable the median and range of values are shown. P values are shown for the comparison of men and women using Mann–Whitney U nonparametric statistics. The use of hormonal contraception was an exclusion criterion for the women.
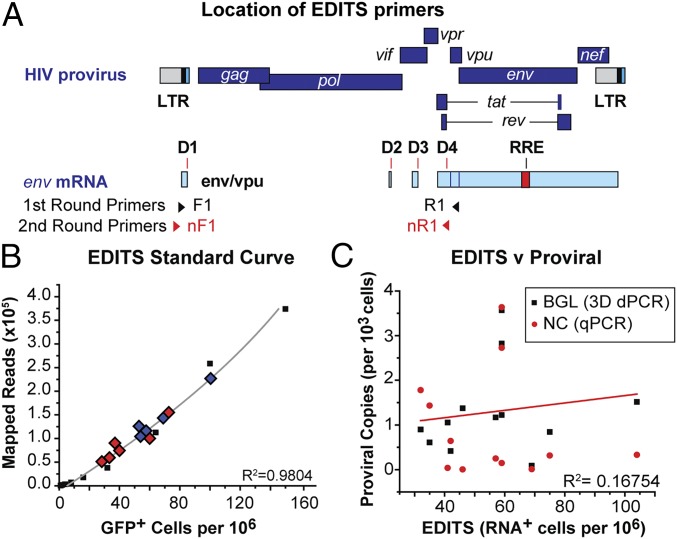
To address difficulties in quantitation of the inducible HIV-1 reservoir, we developed an NGS-based protocol called “EDITS” (envelope detection by induced transcription-based sequencing) to measure inducible cell-associated HIV-1 RNA (Fig. 4). Briefly, 1.25 × 106 resting memory cells from the virally suppressed (<20 copies/mL) HIV-1–infected donors were induced by a range of LRAs and by engagement of the TCR. mRNA was isolated from the cells, and cDNA corresponding to the multiply spliced env mRNA was amplified by nested PCR using a forward primer upstream of the major splice donor site combined with a reverse primer permitting identification of splice junctions spanning diverse regions of the genome (Fig. 4A) (37, 38). The samples treated under the various conditions and from different patients were bar-coded, pooled, and sequenced simultaneously. This approach is efficient with respect to time and sequencing costs and allows accurate comparisons since input cDNA levels are effectively normalized.
Fig. 4.
EDITS assay for measuring RNA induction in patient cells. (A) Location of primers used to detect HIV-1 Env mRNA. (B) Quantitative measurement of inducible cells using EDITS. The calibration curve was produced by sorting specific numbers of activated primary memory cells infected with a replication-competent HIV-1NL4-3 GFP-reporter virus. TCR-induced cells from 11 patients are plotted, showing the range of values obtained by EDITS. Blue diamonds are males, and red diamonds are females. (C) Proviral DNA viral loads do not correlate with EDITS reservoir measurements. Proviral DNA levels were determined using either a qPCR assay (red circles) or a digital droplet DNA assay (black squares), which was more sensitive and able to detect proviral DNA in all of the samples.
Because latently infected cells carry only one provirus on average (39), the frequency of RNA reads obtained by sequencing is proportional to the number of inducible cells in each of the wells. The readouts from the EDITS assay therefore can be used to estimate the total number of inducible cells in a sample by comparing sequencing reads to a calibration curve produced by sorting specific numbers of TCR-activated primary memory cells infected with a replication-competent HIV-1 virus carrying a GFP reporter (Fig. 4B). The accuracy of the EDITS measurements was verified for three donors by using limiting dilution assays (SI Appendix, Fig. S7 A and B) (40). There was reasonable agreement between the estimates obtained by maximum likelihood estimates of the diluted samples and quantitation by the EDITS method; however, the maximum likelihood estimates had very large confidence limits that obscured the differences between samples measured by the EDITS results, which had SDs of <5%.
As expected there was no correlation between estimates of HIV proviral DNA loads obtained by qPCR and droplet digital PCR (ddPCR) and the EDITS assay, due to the presence of defective genomes within the peripheral HIV reservoir which typically resulted in a 10-fold excess of genomes (Fig. 4C).
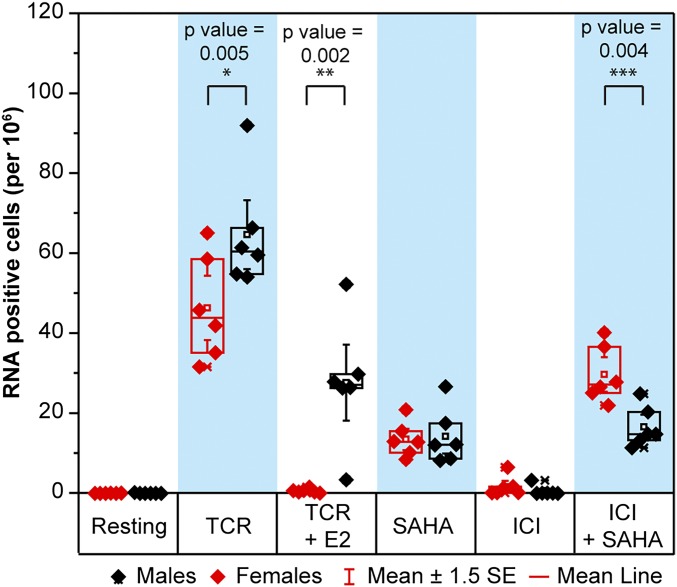
When the inducible RNA reservoir size as measured by the EDITS assay was compared between the women and men, lower levels of inducible RNA were reproducibly found in cells derived from women, and even within this comparatively small sample the accuracy of the EDITS analysis resulted in a highly statistically significant value (P = 0.005) (Fig. 5). The data shown in Fig. 5 include data from six male and six female donors obtained using two sequencing platforms with three technical replicates per condition (i.e., biological and technical replicates). The data were analyzed using linear mixed effects models suitable for analysis of RNA-sequencing (RNA-seq)–based data, with donors as random variables and sequencing platforms as blocking variables. In SI Appendix, Fig. S8, the TCR induction data used to generate Fig. 5 are presented for each individual patient. This illustrates the high reproducibility of the EDITS assay for the biological and technical replicates as well as the minimal background signals seen before induction by LRAs.
Fig. 5.
EDITS estimate of the inducible RNA reservoir size in females and males. EDITS assays were performed on unstimulated cells or cells activated by TCR stimulation (maximal activation) in the presence and absence of 300 pg/mL β-estradiol, 1 μM ICI, 500 nM SAHA, or a combination of ICI and SAHA. For each donor, the EDITS assays were performed on three separate occasions using separate aliquots (biological replicates). Libraries were prepared from the isolated RNAs and were sequenced in two separate runs (technical replicates). Solid circles: PGM sequencer. Open symbols: S5 sequencer. Males respond statistically better than females to TCR stimulation (P value = 0.005), indicating a larger inducible reservoir size.
Impact of SERMs and β-Estradiol on the Reversal of HIV-1 Latency ex Vivo.
The EDITS assay was also used to study the impact of β-estradiol on proviral reactivation and possible synergistic or additive effects of a SERM and an LRA, in this case, ICI and SAHA (Fig. 5). These compounds also were tested in the QUECEL (Th17) assay and Jurkat (2D10) models. Data from both models show a high degree of correlation with each other (R2 = 0.923) and the EDITS assay (R2 = 0.935) (SI Appendix, Fig. S7 C and D), indicating that they share a common mechanism in each system.
Gender-Specific Differences.
These experiments revealed striking differences in the responsiveness of females compared with males to ESR-1 agonists and antagonists (Fig. 5 and SI Appendix, Fig. S8). In women, the addition of 300 pg/mL β-estradiol, which is a level comparable to peak levels in healthy cycling women (the peak level noted in plasma from the leukapheresis samples was 395 pg/mL), completely abolished responses to TCR stimulation (mean 97.7%). By contrast, in men, β-estradiol was still a potent inhibitor of reactivation but reduced TCR stimulation levels by only 47.5% (P = 0.0013 vs. women). ICI was a poor inducer in women and was inactive in men. When combined with SAHA, ICI showed synergistic induction in women (mean increase 17.9%) but did not substantially increase proviral reactivation in men (mean increase 2.9%, P < 0.0001). This pattern of enhanced responses to β-estradiol and SERMs is evident in each of the women compared with each of the men.
To confirm the synergistic effects of SERMs and SAHA in women, but not in men, reciprocal titrations of TMX and SAHA were performed using three representative donors (SI Appendix, Fig. S9 A and B). The data show the expected EC50 values for each drug and confirm the synergistic interactions between SAHA and TMX in women. Three distinct SERMs (TMX, raloxifene, and ICI), showed similar synergistic behavior when combined with SAHA in women, but not in men (SI Appendix, Fig. S9C).
To further verify the role of the estrogen receptor pathway in postmenopausal women, we examined cryopreserved PBMCs from four women on fully suppressive ART followed as part of a longitudinal observational cohort (A5321). These samples were stimulated ex vivo and analyzed by the EDITS assay, as described above (SI Appendix, Fig. S10), except that concanavalin A was used in place of TCR stimulation as the maximal stimulation condition. These results demonstrated that postmenopausal women retained the strong susceptibility to estrogen-mediated repression of transcription. Although these participants also showed an increase in HIV-1 RNA production after ex vivo treatment with TMX, the levels of HIV-1 RNA production induced by SAHA were similar to the levels induced by the combination of TMX and SAHA, as was previously observed in men.
Discussion
ESR-1 Is a Key Regulator of HIV-1 Latency.
The need to develop novel therapeutics to attack the latently infected population during HIV infection and prevent a rebound of virus after termination of suppressive ART is a widely recognized goal of efforts at HIV-1 cure (11, 12). Clinical trials testing candidate LRAs have demonstrated transient increases in intracellular HIV-1 RNA after treatment of study participants with a variety of HDACis [SAHA (41, 42), panobinostat (43), romidepsin (44, 45)] and the phosphatase and tensin homolog (PTEN) inhibitor, disulfiram (46). However, despite this proof of concept, all these agents are highly inefficient ex vivo compared with the maximal induction observed with TCR stimulation. Furthermore, even after maximal activation of T cells ex vivo, a significant proportion of proviruses remain uninduced (14), highlighting the inadequacy of current strategies.
To identify host factors controlling HIV-1 latency in an unbiased manner, we performed shRNA library screens for repressors of HIV-1 transcription using a sequencing strategy as a readout (47, 48). ESR-1 was reproducibly found among highest the hits in multiple screens. Subsequent bioinformatic analyses demonstrated that ESR-1 is a key cellular factor for regulating HIV-1 latency.
The importance of ESR-1 in regulating HIV-1 latency has been largely overlooked. Female sex hormones and their receptors are likely to have direct effects on HIV-1 replication at the molecular level (17). However, previous mechanistic studies have led to seemingly contradictory results, with low levels of estrogen reported to activate the HIV-1 LTR (49) while higher levels are inhibitory (17, 50). None of these studies addressed the role of estrogen and ESR-1 in maintaining latency in quiescent T cells, which have a very different physiology from activated and transformed cells.
Using two complementary models of HIV-1 latency, Jurkat T cells and QUECEL (Th17 cells), and subsequently by the direct assessment of effects in primary samples from a unique cohort of chronically infected patients on treatment using the EDITS assay, we showed that agonists of ESR-1, including β-estradiol, are potent repressors of HIV-1 reactivation. By contrast, the SERMs TMX, raloxifene, and ICI partially reverse latency and sensitize the provirus for reactivation. The impacts of the SERMs and hormones in each of these systems are highly correlated, and this strongly suggests that ESR-1 acts directly on HIV transcription, independently of the cellular background.
Mechanistically, we showed in primary T cells that latency reversal by down-regulation of ESR-1 is a result of the recruitment of repressor complexes to the HIV-1 LTR by ESR-1. ChIP assays showed that latent proviruses rapidly accumulate RNAP II at the 5′ LTR after TCR induction, but little RNAP II reaches the 3′ LTR until Tat accumulates in the cells (22, 24). The SERM ICI enhances RNAP II recruitment at the HIV 5′ LTR and elongates RNAP reaching the 3′ LTR, while β-estradiol strongly inhibits TCR-mediated reactivation. Consistent with this transcriptional pattern, ESR-1 levels at the LTR fluctuate as expected upon pharmacologic modulation (i.e., increase with SERMs and decrease with hormone). Interestingly, β-catenin, which has been associated with a corepressor complex recruited by ESR-1 (17), fluctuates in parallel with ESR-1.
There are no obvious ESR-1–binding sites on the HIV-1 LTR, suggesting that the recruitment may be indirect (51). A potential mediator of ESR-1 recruitment is Sp1, which binds constitutively to three sites in the HIV-1 core promoter (52). Interactions between Sp1 and ESR-1 and ESR-2 have been reported to recruit corepressor complexes at a number of genetic loci (53, 54). Alternatively, TCF-4 may mediate ESR-1 recruitment to the HIV-1 LTR (17, 55). However, neither TCF-4 nor β-catenin appeared as reproducible hits in the shRNA screens. Although the recruitment mechanism remains to be clarified, our work clearly demonstrates that the recruitment of ESR-1 directly suppresses HIV-1 transcription in quiescent cells and is required to maintain HIV-1 latency.
Women Have Lower Inducible HIV-1 RNA Reservoirs than Men.
Women represent half of the 36.7 million people currently living with HIV-1 [Joint United Nations Programme on HIV/AIDS (UNAIDS), 2017, www.unaids.org/en/resources/documents/2017/2017_data_bank], and young women 15–24 y old represent 30% of new HIV-1 infections in key regions, a rate almost twice as high as young men. Numerous studies have shown considerable sex differences in the manifestations of HIV-1 (18), although it is unclear whether these are due to intrinsic differences in immunological responses, differential susceptibility to the virus, behavioral modulators, effects due to hormonal differences, or a combination of all these factors (56–58).
The EDITS assay was used to measure both the total inducible RNA reservoir size and the impact of SERMs and hormone on HIV-1 latency reversal using a well-matched cohort of men and women composed of participants in the University of California, San Francisco (UCSF) SCOPE cohort and patients enrolled in the San Francisco General Hospital Ward 86 HIV clinic. Females were excluded from this study if they were on hormonal contraceptive therapy to eliminate the possible in vivo effects of contraception on the HIV reservoir.
The reservoir size was estimated by comparing the levels of induced RNA following TCR stimulation with a standard curve created using individually sorted productively infected cells. Treating the same cells with LRAs leads to partial reactivation of the population, and this provides a measure of the potency of these drugs. Consistent with the EDITS results, using the related Tat/rev-induced limiting-dilution assay (TILDA) (59) from a partially overlapping subset of 22 men and women from the same cohort, women showed an approximately twofold lower level of inducible HIV-1 RNA when controlled for HIV-1 proviral reservoir size. However, the confidence limits of the TILDA, which is a limiting-dilution assay, are considerably higher than those of the EDITS assay.
Sequencing-based assays, such as EDITS, provide an accurate and quantitative alternative for measuring levels of inducible viral RNA (60, 61). Although these assays do not directly measure the number of cells capable of producing infectious virions (because some of the signals may arise from defective proviruses in the samples), they can be regarded a providing an upper limit for the inducible reservoir size. It was originally suggested that less than 10% of cells producing viral RNA gave rise to infectious virus (62), but a recent highly sensitive quantitative viral outgrowth assay (Q-VOA) suggests that between 20–70% of the cells able to produce HIV-1 RNA produce infectious virus particles (63).
The EDITS data also demonstrate significant sex differences in the response to ex vivo exposure to hormones and SERMs. Repression of HIV-1 transcription by β-estradiol is much more pronounced in women than in men. Similarly, women are more responsive than men to HIV-1 induction by SERMs. Intrinsic differences in the levels of estrogen receptors and their cofactors in T cells conditioned by distinct in vivo hormonal environments in men and women may account for these differential responses to hormones and SERMs. Further work is needed to determine whether these pathways are similarly operative in vivo where pharmacologic exposure to antagonists or hormones could be sustained.
Although the impact of drugs on HIV latency generally correlated extremely well between the patient samples, these sex differences were not obvious in our model systems. Jurkat T cells and most of our healthy donors are male, so we may not have had sufficient statistical power to distinguish between the genders. However, it may also be possible that some subtle features of the regulation are lost in the transformed cells and after the extensive manipulations required for the QUECEL model, which has higher basal activation and is more responsive to LRAs than are patient cells.
It remains to be determined whether the reduced reservoir seen in women is a direct consequence of the profound inhibition of HIV-1 reactivation and replication by β-estradiol, whether better immunological control by women leads to a reduction in the reservoir size, or whether a combination of these factors is at play.
Impact of Sex on HIV-1 Eradication Strategies.
Our results suggest that concurrent exposure to estrogen may limit the efficacy of LRAs. There has been a consistent underrepresentation of women in trials relevant to cure (64). Given the small size and exploratory nature of clinical trials of cure to date, the inclusion of women in small clinical trials may lead to skewed results if hormone exposure is not considered. Furthermore, since reactivation of the HIV-1 reservoir is potently inhibited by estrogen, studies of the impact of hormonal contraception on reservoir establishment and its long-term maintenance should be undertaken. Similarly, it will be important to investigate whether hormonal fluctuations during the menstrual cycle have an impact on residual viremia in well-suppressed patients.
Hormone-modulating therapy with various SERMs, including ICI, TMX, and raloxifene, is routinely used in the context of breast cancer treatment (65–72). Repurposing these extensively used drugs suggests a practical route to exploit the potentially synergistic activities of SERMs and other latency-reversal classes, such as HDACi, for HIV eradication efforts. While in vitro and ex vivo studies demonstrate an important role of hormones in latency reversal, the complexity of the system obligates further clinical investigation. The work detailed here has provided the scientific underpinning for a preliminary trial of the effects of TMX and vorinostat on the HIV reservoir among postmenopausal HIV-infected women, AIDS Clinical Trials Group (ACTG) study A5366, Selective Estrogen Receptor Modulators to Enhance the Efficacy of Viral Reactivation with Histone Deacetylase Inhibitors (ClinicalTrials.gov Identifier: NCT03382834). Further investigations to elucidate sex differences in latency reversal and HIV persistence are mandatory to extend the benefits of the HIV cure effort to all men and women impacted by the global epidemic.
Materials and Methods
Cell Lines and Tissue-Culture Conditions.
Latently infected Jurkat 2D10 cells were used for the shRNA screens and initial characterization of SERMs (21). To minimize exposure to estrogen and estrogen mimics, all cells used in these experiments were maintained in phenol red-free HyClone RPMI medium with l-glutamine, 10% charcoal-stripped FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin in 5% CO2 at 37 °C. Levels of β-estradiol in all media were confirmed by ELISAs (SI Appendix, Fig. S6).
Cells were pretreated with estrogen receptor agonists or antagonists for 2 h before additional stimuli. Estrogen receptor agonists were 300 pg/mL β-estradiol (E2758-1G; Sigma) and 1 nM stilbesterol (ab142421; Abcam). The estrogen receptor antagonists ICI (10-471; R&D Systems), tamoxifen citrate (09-991-00; R&D Systems), and raloxifene hydrochloride (22-805-0; R&D Systems) were used at 1 µM for QUECEL experiments and at 2.5 µM for the Jurkat T cell experiments.
QUECEL (Th17) Cell Model for HIV-1 Latency.
Polarized Th17 cells were generated from naive CD4+ T cells that were negatively isolated from PBMCs using the Human Naive CD4+ T Cell Enrichment Kit (19155RF; STEMCELL Technologies) as described (20). Afterward, cells were infected with VSV-G pseudotyped HIV-1 Nef+ virus, which expresses CD8a-d2EGFP. HIV-1–infected cells were purified using the anti-CD8a selection kit (18953; STEMCELL Technologies). Cells were then forced into quiescence with culture in medium supplemented with 15 IU/mL IL-2, 12.5 ng/mL IL-23, 10 ng/mL TGF-β, 50 ng/mL IL-10, and 50 ng/mL IL-8. Entry of the cells into quiescence was confirmed using flow assays showing the loss of pS175 CDK9 (25) and the reduction of cyclin D3 levels. Maximal reactivation of proviral gene expression was achieved by incubating cells overnight with Dynabeads Human T-Activator CD3/CD28 (25 µL per 106 cells). The high reproducibility of the assay permitted us to combine data from multiple experiments without normalization.
Flow Cytometry Analysis.
Cells were analyzed using a BD LSRFortessa flow cytometry instrument. For intracellular staining, samples were fixed in 4% formaldehyde (Electron Microscope Sciences) for 15 min at room temperature. Cells were permeabilized and stained using an antibody mixture containing AF647-HIV-1 EH1 Nef [the gift of James A. Hoxie, Perelman School of Medicine, University of Pennsylvania, Philadelphia (73)], AF750-pSer175 CDK9 (generated by Covance for the J.K. laboratory), and TRITC-Cyclin D3 (Santa Cruz Biotechnologies). Data were analyzed using WinList 3D version 9.
shRNA Library Screening.
The human module shRNA library in a lentiviral vector was obtained from Cellecta, Inc. For the repressor screen, latent HIV 2D10 cells were infected in parallel with each of the shRNA library modules and were grown in puromycin selection medium for 14 d with a regular change of fresh medium. Double-positive [d2EGFP+ (HIV-1 expression), RFP+ (shRNA vector)] cells representing the constitutively reactivated population were grown in puromycin selection medium for 7 d before the next round of cell sorting. Three consecutive cycles of sorting were performed. After each step of purification, 2–5 × 106 cells were harvested, and genomic DNA was isolated using a Qiagen Blood and DNA mini Kit. For the activator screen, latently infected 2D10 cells were infected with the shRNA library, as above. The day before each stage of cell sorting, the cells were stimulated with TNF-α, and the d2GFP−, RFP+ cells were purified and genomic DNA was isolated, as above.
Isolated genomic DNA was used as a template to amplify the barcode sequences of shRNAs by nested PCR. Agarose gel (1%)-purified amplified barcode sequences were then sequenced by Illumina high-throughput sequencers, and the sequences were deconvoluted to their target mRNA sequences using software tools provided by Cellecta, Inc.
Robo ChIP.
ChIP was performed on 106 cells per condition using our previously described Robo Chip protocol (22). Antibodies used for immunoprecipitations included anti-RNAP II pSer5 (ab5131; Abcam), anti-RNAP II pSer2 (ab5095; Abcam), anti–ESR-1 (SAB4300344; Sigma), and anti–β-catenin (610154; BD Biosciences).
Study Cohort.
Study participants were enrolled through the UCSF SCOPE cohort between March 2014 and April 2015. Clinical and demographic information was collected, and reproductive health questionnaires were completed with study staff. A subset of participants (six women and six men) returned for leukapheresis. All subjects provided informed consent, and the study was reviewed and approved by the UCSF institutional review board. Additional samples from postmenopausal women were obtained through the ACTG longitudinal study A5321 for testing ex vivo HIV-1 RNA induction in the presence of hormones and antagonists.
RNA Induction Assay (EDITS Assay).
Well-suppressed PBMCs from the patients in the cohort were obtained by leukapheresis. CD4+ memory cells were negatively isolated using robotic magnetic bead isolation using the EasySep Human Memory CD4 T Cell Enrichment Kit (19157; STEMCELL Technologies). For each determination, 1.25 × 106 cells were used. Cells were induced with Dynabeads Human T-Activator CD3/CD28 (25 µL per 106 cells), IL-15 (50 ng/mL), or SAHA (500 nM) overnight in the presence or absence of the indicated SERMs and ESR-1 agonists. Total RNA was isolated using the RNeasy purification system (74134; Qiagen) following the manufacturer’s protocol. The total sample was eluted using 20 μL of RNase-free water. The entire sample was used as template in a one-step RT-PCR (AB-4104A; Thermo Scientific) using the following cycling conditions: 1 cycle at 50 °C for 15 min, 1 cycle at 95 °C for 15 min, 35 cycles of 95 °C for 15 s and 60 °C for 30 s, and 72 °C for 30 s.
Primers were designed to bind to either side of the HIV-1 Env RNA splice junction at positions 648 and 666 (forward primer AGGGACCTGAAAGCGAAAG) and positions c6340 and c6359 (reverse primer CCACACAGGTACCCCATAAT), yielding a product of 168 bp from the spliced env mRNA. Using these primers allowed us to detect only late spliced viral transcripts and eliminated any potential proviral DNA amplification because of the wide spacing of the primers on the viral genome. The primer-binding regions are located in highly conserved regions of HIV-1 based on the Los Alamos National Laboratory HIV-1 Database.
In addition to the priming sequence, the reverse primer has a synthetic GEX R-AATGATACGGCGACCACC sequence placed directly after the priming region to allow further amplification using nested PCR. After cDNA synthesis and PCR, 2 μL of the reaction was used as template for a subsequent round of nested PCR using a high-fidelity Phusion Flash polymerase (F548; Thermo Scientific). The amplification protocol was as follows: 1 cycle at 95 °C for 30 s, 35 cycles of 95 °C for 1 s, 62 °C for 5 s, and 72 °C for 5 s, and 1 cycle of 72 °C for 1 min. The nested primers were designed to bind to positions 757 and 776 of HIV-1 (forward primer AAATTTTGACTAGCGGAGGC and nested GEX reverse primer AATGATACGGCGACCACC) and were added during the first round of PCR.
To allow for NGS, ion torrent A forward and Trp reverse adapters were added to the nested primer sets, and a unique barcode was added in the forward primer to allow multiplexing of samples. Samples were then pooled and purified up using the GeneJET NGS cleanup kit (FERK0852; Thermo Scientific) to remove residual free primers. Three hundred picograms of the pooled sample were then sequenced using an ion torrent sequencing system following the manufacturer’s protocol. Sequencing was performed on either an S5 sequencer (Thermo Fisher) using a 540 chip (80 million reads) or a PGM sequencer (Thermo Fisher) using the 318 chip (8 million reads).
Barcodes were separated by sample using the Ion Torrent Browser, and all reads were filtered to remove short products (<80 bp); only reads that contained the GEX reverse sequence were retained. The filtered reads were then mapped to a synthetically spliced HXB2 sequence, and total mapped reads were scored. The number of mapped reads was then converted into the equivalent number of cells harboring HIV-1 per 106 cells using a standard curve prepared from activated memory cells infected with replication-competent HIV-1–GFP virus and then were sorted by flow cytometry into single wells of a 96-well plate. Samples for the standard curve contained between 1 and 300 infected cells per well and 1.25 × 106 uninfected cells. The samples were processed, barcoded, pooled with the experimental EDITS samples, and sequenced.
3D Digital PCR Proviral DNA Assay.
DNA was isolated from memory T cells of HIV+ donors (DNeasy Blood and Tissue Kit; Qiagen) and was quantified by the Qubit HS dsDNA assay (Fisher Scientific). Up to 2 µg of cellular DNA was mixed with QuantStudio 3D Digital PCR Master Mix v2 and 1× HIV-1 5′ LTR primer/probe mix (TaqMan Expression Assay; Life Technologies) and spread evenly onto two separate 20-K v2 chips with a QuantStudio 3D Digital PCR chip loader. Chips were then filled with immersion fluid and sealed. HIV 5′ LTR primers in this assay amplify a 128-bp product that maps to nucleotide positions 588–716 of the HXB2 genome. Cellular DNA from uninfected donors served as the negative control. Loaded chips were thermally cycled on a ProFlex 2× flat PCR system (Thermo Fisher Scientific) using the manufacturer’s suggested conditions (96 °C for 10 min, 39 cycles of 60 °C for 2 min and 98 °C for 30 s, and 60 °C for 2 min) and were held at 10 °C. Cycled chips were read with a QuantStudio 3D Digital PCR Instrument and were analyzed using QuantStudio 3D AnalysisSuite Software. Thresholds for positive wells on all chips were based on nonspecific fluorescence from a negative control DNA chip. Results given in HIV copies per microliter were then translated to HIV copies per 106 cells. The ddPCR determinations correlated nicely with qPCR performed on parallel samples using standard methods but were more sensitive and therefore were able to quantify proviral DNA in various samples where it was undetectable by qPCR.
Supplementary Material
Acknowledgments
We thank Dr. Bryan Nikolai (Baylor University) for discussions and advice, Dr. James W. Jacobberger, Mike Sramkoski, Jonida Toska, and Amy Graham for assistance with cell sorting and image stream analysis, and Cellecta, Inc., for providing the human modules of the shRNA library.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. SRP149947).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803468115/-/DCSupplemental.
References
- 1.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 2.Chun TW, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong JK, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 4.Finzi D, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 5.Yukl SA, et al. Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J Infect Dis. 2010;202:1553–1561. doi: 10.1086/656722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chun T-W, et al. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci USA. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strain MC, et al. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: Intrinsic stability predicts lifelong persistence. Proc Natl Acad Sci USA. 2003;100:4819–4824. doi: 10.1073/pnas.0736332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siliciano JD, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 9.Maldarelli F, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner TA, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richman DD, et al. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 12.Trono D, et al. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science. 2010;329:174–180. doi: 10.1126/science.1191047. [DOI] [PubMed] [Google Scholar]
- 13.Mbonye U, Karn J. Transcriptional control of HIV latency: Cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology. 2014;454–455:328–339. doi: 10.1016/j.virol.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho YC, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasmussen TA, Lewin SR. Shocking HIV out of hiding: Where are we with clinical trials of latency reversing agents? Curr Opin HIV AIDS. 2016;11:394–401. doi: 10.1097/COH.0000000000000279. [DOI] [PubMed] [Google Scholar]
- 16.Katagiri D, Hayashi H, Victoriano AF, Okamoto T, Onozaki K. Estrogen stimulates transcription of human immunodeficiency virus type 1 (HIV-1) Int Immunopharmacol. 2006;6:170–181. doi: 10.1016/j.intimp.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 17.Szotek EL, Narasipura SD, Al-Harthi L. 17β-estradiol inhibits HIV-1 by inducing a complex formation between β-catenin and estrogen receptor α on the HIV promoter to suppress HIV transcription. Virology. 2013;443:375–383. doi: 10.1016/j.virol.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meditz AL, et al. Sex, race, and geographic region influence clinical outcomes following primary HIV-1 infection. J Infect Dis. 2011;203:442–451. doi: 10.1093/infdis/jiq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curno MJ, et al. A systematic review of the inclusion (or exclusion) of women in HIV research: From clinical studies of antiretrovirals and vaccines to cure strategies. J Acquir Immune Defic Syndr. 2016;71:181–188. doi: 10.1097/QAI.0000000000000842. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen K, Das B, Dobrowolski C, Karn J. Multiple histone lysine methyltransferases are required for the establishment and maintenance of HIV-1 latency. MBio. 2017;8:e00133-17. doi: 10.1128/mBio.00133-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson R, et al. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J Virol. 2008;82:12291–12303. doi: 10.1128/JVI.01383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jadlowsky JK, et al. Negative elongation factor is required for the maintenance of proviral latency but does not induce promoter-proximal pausing of RNA polymerase II on the HIV long terminal repeat. Mol Cell Biol. 2014;34:1911–1928. doi: 10.1128/MCB.01013-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai Z, et al. edgeR: A versatile tool for the analysis of shRNA-seq and CRISPR-Cas9 genetic screens. F1000 Res. 2014;3:95. doi: 10.12688/f1000research.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim YK, Mbonye U, Hokello J, Karn J. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J Mol Biol. 2011;410:896–916. doi: 10.1016/j.jmb.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mbonye UR, et al. Phosphorylation of CDK9 at Ser175 enhances HIV transcription and is a marker of activated P-TEFb in CD4(+) T lymphocytes. PLoS Pathog. 2013;9:e1003338. doi: 10.1371/journal.ppat.1003338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruepp A, et al. CORUM: The comprehensive resource of mammalian protein complexes–2009. Nucleic Acids Res. 2010;38:D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kikugawa S, et al. PCDq: Human protein complex database with quality index which summarizes different levels of evidences of protein complexes predicted from h-invitational protein-protein interactions integrative dataset. BMC Syst Biol. 2012;6(Suppl 2):S7. doi: 10.1186/1752-0509-6-S2-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das B, et al. Short chain fatty acids potently induce latent HIV-1 in T-cells by activating P-TEFb and multiple histone modifications. Virology. 2015;474:65–81. doi: 10.1016/j.virol.2014.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosque A, Famiglietti M, Weyrich AS, Goulston C, Planelles V. Homeostatic proliferation fails to efficiently reactivate HIV-1 latently infected central memory CD4+ T cells. PLoS Pathog. 2011;7:e1002288. doi: 10.1371/journal.ppat.1002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bosque A, Planelles V. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood. 2009;113:58–65. doi: 10.1182/blood-2008-07-168393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 32.Gosselin A, et al. HIV persists in CCR6+CD4+ T cells from colon and blood during antiretroviral therapy. AIDS. 2017;31:35–48. doi: 10.1097/QAD.0000000000001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee GQ, et al. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J Clin Invest. 2017;127:2689–2696. doi: 10.1172/JCI93289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mbonye U, et al. Cyclin-dependent kinase 7 (CDK7)-mediated phosphorylation of the CDK9 activation loop promotes P-TEFb assembly with Tat and proviral HIV reactivation. J Biol Chem. 2018;293:10009–10025. doi: 10.1074/jbc.RA117.001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, et al. Small molecule inhibition of the steroid receptor coactivators, SRC-3 and SRC-1. Mol Endocrinol. 2011;25:2041–2053. doi: 10.1210/me.2011-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim YK, et al. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J. 2006;25:3596–3604. doi: 10.1038/sj.emboj.7601248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carrera C, Pinilla M, Pérez-Alvarez L, Thomson MM. Identification of unusual and novel HIV type 1 spliced transcripts generated in vivo. AIDS Res Hum Retroviruses. 2010;26:815–820. doi: 10.1089/aid.2010.0011. [DOI] [PubMed] [Google Scholar]
- 38.Pasternak AO, et al. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J Clin Microbiol. 2008;46:2206–2211. doi: 10.1128/JCM.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Josefsson L, et al. Majority of CD4+ T cells from peripheral blood of HIV-1-infected individuals contain only one HIV DNA molecule. Proc Natl Acad Sci USA. 2011;108:11199–11204. doi: 10.1073/pnas.1107729108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenbloom DI, et al. Designing and interpreting limiting dilution assays: General principles and applications to the latent reservoir for human immunodeficiency virus-1. Open Forum Infect Dis. 2015;2:ofv123. doi: 10.1093/ofid/ofv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Archin NM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elliott JH, et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog. 2014;10:e1004473. doi: 10.1371/journal.ppat.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rasmussen TA, et al. Activation of latent human immunodeficiency virus by the histone deacetylase inhibitor panobinostat: A pilot study to assess effects on the central nervous system. Open Forum Infect Dis. 2015;2:ofv037. doi: 10.1093/ofid/ofv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei DG, et al. Histone deacetylase inhibitor romidepsin induces HIV expression in CD4 T cells from patients on suppressive antiretroviral therapy at concentrations achieved by clinical dosing. PLoS Pathog. 2014;10:e1004071. doi: 10.1371/journal.ppat.1004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Søgaard OS, et al. The depsipeptide romidepsin reverses HIV-1 latency in vivo. PLoS Pathog. 2015;11:e1005142. doi: 10.1371/journal.ppat.1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spivak AM, et al. A pilot study assessing the safety and latency-reversing activity of disulfiram in HIV-1-infected adults on antiretroviral therapy. Clin Infect Dis. 2014;58:883–890. doi: 10.1093/cid/cit813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shtutman M, et al. Function-based gene identification using enzymatically generated normalized shRNA library and massive parallel sequencing. Proc Natl Acad Sci USA. 2010;107:7377–7382. doi: 10.1073/pnas.1003055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Q, et al. Novel high throughput pooled shRNA screening identifies NQO1 as a potential drug target for host directed therapy for tuberculosis. Sci Rep. 2016;6:27566. doi: 10.1038/srep27566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mavoungou D, Poaty-Mavoungou V, Akoume MY, Ongali B, Mavoungou E. Inhibition of human immunodeficiency virus type-1 (HIV-1) glycoprotein-mediated cell-cell fusion by immunor (IM28) Virol J. 2005;2:9. doi: 10.1186/1743-422X-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bourinbaiar AS, Nagorny R, Tan X. Pregnancy hormones, estrogen and progesterone, prevent HIV-1 synthesis in monocytes but not in lymphocytes. FEBS Lett. 1992;302:206–208. doi: 10.1016/0014-5793(92)80441-i. [DOI] [PubMed] [Google Scholar]
- 51.Pereira LA, Bentley K, Peeters A, Churchill MJ, Deacon NJ. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28:663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mbonye U, Karn J. Control of HIV latency by epigenetic and non-epigenetic mechanisms. Curr HIV Res. 2011;9:554–567. doi: 10.2174/157016211798998736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartella V, et al. Estrogen receptor beta binds Sp1 and recruits a corepressor complex to the estrogen receptor alpha gene promoter. Breast Cancer Res Treat. 2012;134:569–581. doi: 10.1007/s10549-012-2090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hao H, et al. Estrogen-induced and TAFII30-mediated gene repression by direct recruitment of the estrogen receptor and co-repressors to the core promoter and its reversal by tamoxifen. Oncogene. 2007;26:7872–7884. doi: 10.1038/sj.onc.1210592. [DOI] [PubMed] [Google Scholar]
- 55.Henderson LJ, Narasipura SD, Adarichev V, Kashanchi F, Al-Harthi L. Identification of novel T cell factor 4 (TCF-4) binding sites on the HIV long terminal repeat which associate with TCF-4, β-catenin, and SMAR1 to repress HIV transcription. J Virol. 2012;86:9495–9503. doi: 10.1128/JVI.00486-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meier A, et al. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat Med. 2009;15:955–959. doi: 10.1038/nm.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wira CR, Fahey JV. A new strategy to understand how HIV infects women: Identification of a window of vulnerability during the menstrual cycle. AIDS. 2008;22:1909–1917. doi: 10.1097/QAD.0b013e3283060ea4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodriguez-Garcia M, et al. Estradiol reduces susceptibility of CD4+ T cells and macrophages to HIV-infection. PLoS One. 2013;8:e62069. doi: 10.1371/journal.pone.0062069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Procopio FA, et al. A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine. 2015;2:874–883. doi: 10.1016/j.ebiom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med. 2014;20:425–429. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vandergeeten C, et al. Interleukin-7 promotes HIV persistence during antiretroviral therapy. Blood. 2013;121:4321–4329. doi: 10.1182/blood-2012-11-465625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ho YC, Laird GM, Siliciano RF. Measuring reversal of HIV-1 latency ex vivo using cells from infected individuals. Proc Natl Acad Sci USA. 2014;111:6860–6861. doi: 10.1073/pnas.1405194111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanyal A, et al. Novel assay reveals a large, inducible, replication-competent HIV-1 reservoir in resting CD4+ T cells. Nat Med. 2017;23:885–889. doi: 10.1038/nm.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnston RE, Heitzeg MM. Sex, age, race and intervention type in clinical studies of HIV cure: A systematic review. AIDS Res Hum Retroviruses. 2015;31:85–97. doi: 10.1089/aid.2014.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stossi F, Likhite VS, Katzenellenbogen JA, Katzenellenbogen BS. Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. J Biol Chem. 2006;281:16272–16278. doi: 10.1074/jbc.M513405200. [DOI] [PubMed] [Google Scholar]
- 66.Yeh WL, Lin HY, Wu HM, Chen DR. Combination treatment of tamoxifen with risperidone in breast cancer. PLoS One. 2014;9:e98805. doi: 10.1371/journal.pone.0098805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Butt AJ, Sutherland RL, Musgrove EA. Live or let die: Oestrogen regulation of survival signalling in endocrine response. Breast Cancer Res. 2007;9:306. doi: 10.1186/bcr1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Doisneau-Sixou SF, et al. Estrogen and antiestrogen regulation of cell cycle progression in breast cancer cells. Endocr Relat Cancer. 2003;10:179–186. doi: 10.1677/erc.0.0100179. [DOI] [PubMed] [Google Scholar]
- 69.Mooney LM, Al-Sakkaf KA, Brown BL, Dobson PR. Apoptotic mechanisms in T47D and MCF-7 human breast cancer cells. Br J Cancer. 2002;87:909–917. doi: 10.1038/sj.bjc.6600541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dolfi SC, et al. Fulvestrant treatment alters MDM2 protein turnover and sensitivity of human breast carcinoma cells to chemotherapeutic drugs. Cancer Lett. 2014;350:52–60. doi: 10.1016/j.canlet.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suddek GM. Protective role of thymoquinone against liver damage induced by tamoxifen in female rats. Can J Physiol Pharmacol. 2014;92:640–644. doi: 10.1139/cjpp-2014-0148. [DOI] [PubMed] [Google Scholar]
- 72.Fan P, Craig Jordan V. Acquired resistance to selective estrogen receptor modulators (SERMs) in clinical practice (tamoxifen & raloxifene) by selection pressure in breast cancer cell populations. Steroids. 2014;90:44–52. doi: 10.1016/j.steroids.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang AH, Hoxie JA, Cassol S, O’Shaughnessy M, Jirik F. Construction of single-chain antibodies that bind an overlapping epitope of HIV-1 Nef. FEBS Lett. 1998;441:307–312. doi: 10.1016/s0014-5793(98)01569-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.