Significance
In humans, SEC23B deficiency results in congenital dyserythropoietic anemia type II, a disease of abnormal red blood cell development, while SEC23A deficiency results in cranio-lenticulo-sutural-dysplasia, a disease characterized by bone abnormalities due to defective collagen secretion (but no red blood cell defect). In this study, we show that SEC23A and SEC23B overlap in function, and that the disparate phenotypes of SEC23A/SEC23B deficiency within and across species are likely due to evolutionary shifts in gene-expression programs, rather than distinct functions of the SEC23 paralogs. Our studies provide a rationale for increased SEC23A or SEC23B expression as a therapeutic strategy for congenital dyserythropoietic anemia type II or cranio-lenticulo-sutural-dysplasia, respectively.
Keywords: endoplasmic reticulum, vesicular transport proteins, mice, transgenic, SEC23
Abstract
Approximately one-third of the mammalian proteome is transported from the endoplasmic reticulum-to-Golgi via COPII-coated vesicles. SEC23, a core component of coat protein-complex II (COPII), is encoded by two paralogous genes in vertebrates (Sec23a and Sec23b). In humans, SEC23B deficiency results in congenital dyserythropoietic anemia type-II (CDAII), while SEC23A deficiency results in a skeletal phenotype (with normal red blood cells). These distinct clinical disorders, together with previous biochemical studies, suggest unique functions for SEC23A and SEC23B. Here we show indistinguishable intracellular protein interactomes for human SEC23A and SEC23B, complementation of yeast Sec23 by both human and murine SEC23A/B, and rescue of the lethality of sec23b deficiency in zebrafish by a sec23a-expressing transgene. We next demonstrate that a Sec23a coding sequence inserted into the murine Sec23b locus completely rescues the lethal SEC23B-deficient pancreatic phenotype. We show that SEC23B is the predominantly expressed paralog in human bone marrow, but not in the mouse, with the reciprocal pattern observed in the pancreas. Taken together, these data demonstrate an equivalent function for SEC23A/B, with evolutionary shifts in the transcription program likely accounting for the distinct phenotypes of SEC23A/B deficiency within and across species, a paradigm potentially applicable to other sets of paralogous genes. These findings also suggest that enhanced erythroid expression of the normal SEC23A gene could offer an effective therapeutic approach for CDAII patients.
In vertebrates, ∼8,000 secretory proteins (cargos) are cotranslationally inserted into the endoplasmic reticulum (ER) and subsequently transported to the Golgi apparatus via coat protein-complex II (COPII)-coated vesicles before reaching their final destination (endosomes, lysosomes, plasma membrane, or the extracellular space) (1–6). SAR1 and SEC23-SEC24 heterodimers form the inner COPII coat, with the outer coat composed of SEC13-SEC31 heterotetramers. COPII coat assembly begins with GTP-bound SAR1 recruiting cytoplasmic SEC23-SEC24 heterodimers to the ER membrane (7, 8), through direct interaction between SAR1 and SEC23. SAR1-SEC23-SEC24 forms the prebudding complex, which recruits cargo proteins (9–15) either via direct interaction (3, 10, 16–18) or via interaction with intermediary proteins (termed receptors or adaptors) (3, 18–22). Following cargo selection, SEC13-SEC31 heterotetramers are recruited, forming the outer COPII coat. SEC23 functions as a GTPase activating protein stimulating SAR1-GTP hydrolysis, a process required for vesicle fission in vivo (4, 7). The SEC23 GTPase activity is enhanced by SEC31 (23).
The mammalian genome encodes multiple paralogs for SEC23, SEC24, SAR1, and SEC31. These paralogs have been proposed to contribute to COPII coat diversity, facilitating distinct cargo selectivity and tissue-specific secretory repertoires. Alternative splicing has been described for SEC31 and SEC24C (24, 25) and might additionally contribute to COPII diversity and cargo selectivity. Alhough the SEC23 gene duplication is estimated to have occurred ∼615 Mya, the two mammalian paralogs, SEC23A and SEC23B, encode proteins that are ∼85% identical at the amino acid sequence level. The sequence identity between the mouse and human orthologs for SEC23A and SEC23B are ∼98% and ∼97%, respectively.
Mutations in human SEC23A result in cranio-lenticulo-sutural dysplasia (CLSD), an autosomal recessive disease characterized by abnormal closure of cranial fontanelles, dysmorphic facial features, skeletal abnormalities, and sutural cataracts (26, 27), features thought to result from defects in collagen secretion. In contrast, loss of SEC23B function results in congenital dyserythropoietic anemia type II (CDAII), an autosomal recessive disease characterized by anemia and increased bi/multinucleated erythroid precursors in the bone marrow (BM) (28).
In mice, germline Sec23a deletion results in an open neural tube defect and compromised extraembryonic membranes, likely due to impaired collagen secretion, a phenotype reminiscent of human CLSD (29). In contrast, mice with germline SEC23B deficiency die perinatally, exhibiting massive pancreatic degeneration (30–32) with no anemia or other features of CDAII (30, 33). In both instances, partially compensatory increases in the remaining paralog were shown to be posttranslational, likely due to enhanced stability of SEC23/SEC24 heterodimers (29).
Previous reports suggest unique functions for SEC23A and SEC23B (29, 34–37), potentially due to distinct sets of cargo proteins. Additionally, human SEC23A but not SEC23B was reported to complement yeast SEC23 (35), consistent with distinct function of the two mammalian paralogs.
In surprising contrast to these earlier findings, we now report indistinguishable intracellular interactomes for human SEC23A and SEC23B and also show complementation of yeast SEC23 by both human and murine SEC23A/B paralogs. We further show that the lethality resulting from sec23b disruption in zebrafish can be rescued by a sec23a-expressing transgene. Finally, we demonstrate that the Sec23a coding sequence inserted into the endogenous murine Sec23b locus fully rescues the mortality and severe pancreatic phenotype of SEC23B-deficiency in the mouse.
Results
The SEC23A and SEC23B Interactomes Are Indistinguishable.
Recruitment of SAR1-SEC23-SEC24 to the cytoplasmic face of the ER membrane forms the inner coat of the COPII vesicle. Interaction of SEC23 with each of the four SEC24 mammalian paralogs (SEC24A–D) provides a potential for functional differentiation between SEC23A and SEC23B. However, we detected no differences in paralog specificity by coimmunoprecipitation (SI Appendix, Fig. S1).
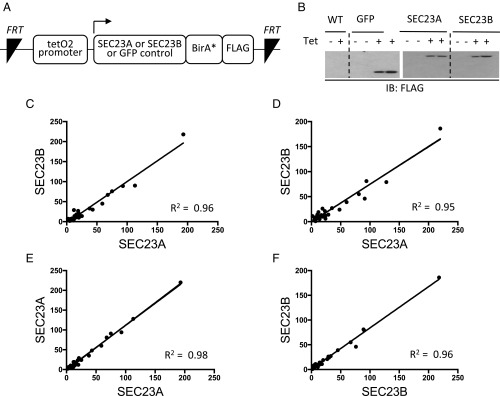
To comprehensively characterize the full set of interacting proteins for each SEC23 paralog, we performed two independent “BioID” (proximity-dependent biotinylation) experiments in HEK293 cells equivalently expressing either BirA*-tagged SEC23A, SEC23B, or a GFP control (Fig. 1 A and B). As expected, known SEC23-interacting proteins were identified, including the COPII structural proteins SEC24, SAR1, SEC13, and SEC31. Comparison of the SEC23A to SEC23B interactomes revealed a remarkably high correlation in two independent experiments (Fig. 1 C and D and SI Appendix, Fig. S2), equivalent to the correlation between biological replicates for SEC23A (Fig. 1E) or SEC23B (Fig. 1F). Thus, the comprehensive interactomes of SEC23A and SEC23B in HEK293 cells are indistinguishable by BioID.
Fig. 1.
SEC23A and SEC23B exhibit indistinguishable interactomes in HEK293 cells. (A) The constructs shown here, directing expression of SEC23A or SEC23B fused to BirA* and a FLAG tag under the control of a tetracycline-inducible promoter, were stably inserted into Flp-In T-REX HEK293 cells. (B) FLAG immunoblotting following tetracycline induction. (C–F) Following tetracycline and biotin administration, the full sets of SEC23A and SEC23B interactomes were each determined with BioID, in two independent experiments. Each “interacting protein” is represented by a dot and the spectral counts are indicated on the x and y axes. Pairwise comparisons of spectral counts for a given protein are shown across paralogs (C and D) or between biologic replicas for the same paralog (E and F).
Mouse and Human SEC23 Paralogs Complement the Yeast Sec23 Protein.
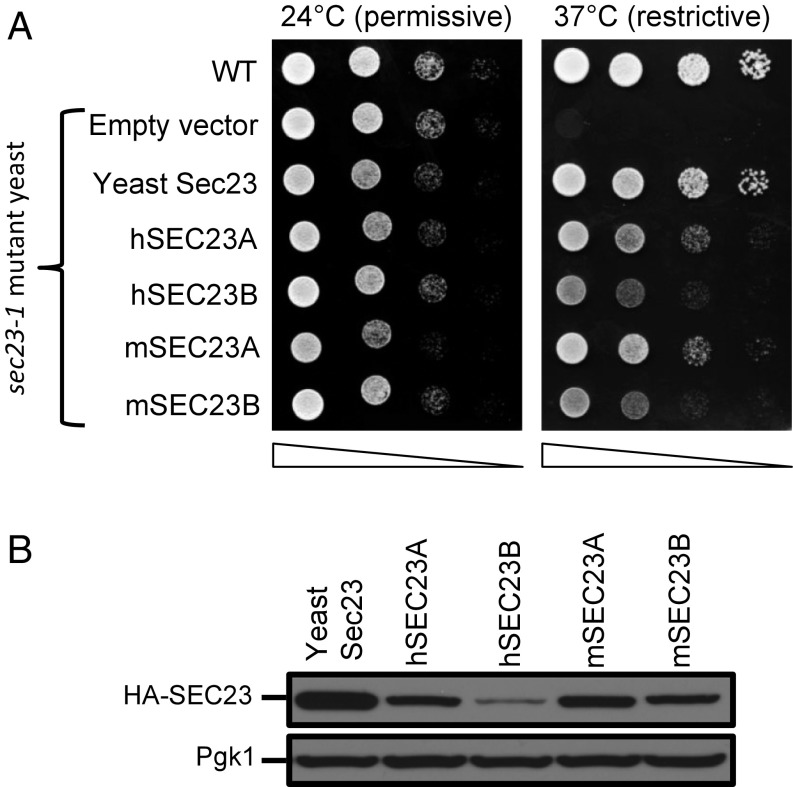
Human SEC23A but not human SEC23B was previously reported to complement the yeast Sec23 protein (35), a puzzling finding in light of the above evidence for indistinguishable SEC23A/B interactomes in mammalian cells. We thus retested complementation in a temperature sensitive sec23-1 mutant yeast strain using both human (h) and mouse (m) SEC23A and -B sequences. Indeed, all four mammalian sequences (hSEC23A, hSEC23B, mSEC23A, and mSEC23B) rescued growth of the temperature-sensitive sec23-1 mutant yeast at the restrictive temperature (Fig. 2A). The discrepancy between these findings and the previous report (35) is likely explained by poorer expression of hSEC23B in yeast compared with hSEC23A (Fig. 2B and SI Appendix, Fig. S3), potentially due to differences in codon usage and protein stability.
Fig. 2.
Human and mouse SEC23A and SEC23B complement the yeast Sec23 protein. (A) The coding sequences of the human (h) or mouse (m) SEC23 paralogs (as well as yeast Sec23) were cloned with a 3′ sequence encoding an HA-tag into the pRS416 vector downstream of a ZEO1 promoter. In contrast to WT yeast, temperature-sensitive sec23-1 mutant yeast transfected with a control “empty vector” grow at the permissive (24 °C) but not at the restrictive (37 °C) temperature. Growth of temperature-sensitive sec23-1 mutant yeast expressing h or m SEC23A/SEC23B (or yeast Sec23) was determined at the permissive and restrictive temperatures. Yeast cultures were plated as 10-fold serial dilutions. (B) Immunoblotting for HA (with Pgk1 as loading control) detects relative expression for each construct.
Sec23b Deficiency Does Not Produce an Erythroid Phenotype in Zebrafish.
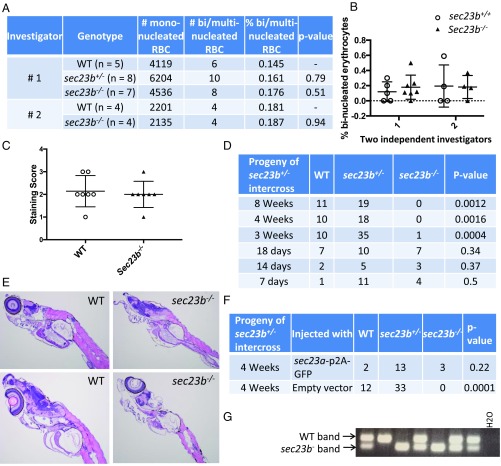
The zebrafish (ZF) genome also contains two sec23 paralogs (sec23a and sec23b). SEC23B deficiency in humans results in CDAII, a disease characterized by anemia and increased binucleated erythroid precursors (6), with sec23b knockdown in ZF previously reported to similarly result in an increased percentage of circulating binucleated erythrocytes (28). In contrast, SEC23B deficiency in mice results in perinatal mortality and massive pancreatic degeneration but no findings comparable to CDAII (30–33). To address these discordant phenotypes across species, we generated ZF heterozygous for a 53-bp deletion in exon 5 of sec23b (sec23b+/−) by CRISPR/Cas9 genome editing. No difference in the percentage of circulating binucleated erythrocytes was noted at day 16 between sec23b−/− ZF compared with WT and sec23b+/− ZF clutchmates (Fig. 3 A and B). Similarly, day 7 sec23b−/− ZF exhibited no difference in hemoglobin content (as assessed by o-dianisidine staining) compared with WT controls (Fig. 3C and SI Appendix, Fig. S4B). However, no sec23b−/− ZF survived beyond 3 wk of age (Fig. 3D), although no gross or histologic abnormalities were detected at these time points (including in the pancreas), with the exception of reduced overall ZF size (Fig. 3E and SI Appendix, Fig. S4A).
Fig. 3.
Mortality in Sec23b-deficient ZF between 18 and 21 d is rescued by Sec23a. (A and B) sec23b heterozygous (sec23b+/−) ZF with a 53-bp deletion were generated with CRISPR/Cas9. Binucleated erythrocyte counts from peripheral blood of ZF offspring from a sec23b+/− × sec23b+/− intercross were evaluated by two independent investigators blinded to the ZF genotype. (C) sec23b−/− ZF exhibit indistinguishable o-dianisidine staining compared with WT clutchmate controls. Scores 1–3 correspond to staining intensities (3 = highest staining intensity). (D) sec23b−/− ZF (generated from sec23b+/− intercrosses) die between 18 and 21 d. (E) H&E stains of sec23b−/− and WT ZF at day 16. sec23b−/− ZF appear histologically normal. (Magnification: 10×.) (F) One-cell–stage embryos generated from sec23b+/− ZF intercrosses were injected with a vector (designed to integrate in the ZF genome) that expresses Sec23a and GFP or GFP alone as control. Genotyping at 4 wk of age demonstrates rescue of sec23b−/− mortality by Sec23a. (G) PCR genotyping assay demonstrating 2 sec23b−/− ZF, expressing Sec23a from a ubiquitin promoter, alive at 4 wk of age.
Sec23a Expression Rescues the Lethality of Sec23b Deficiency in ZF.
To determine if Sec23a could rescue the early mortality of Sec23b-deficient ZF, one-cell–stage embryos generated from sec23b+/− intercrosses were injected with a vector (designed to integrate into the ZF genome) expressing Sec23a and GFP or GFP alone under the control of a ubiquitin promoter (SI Appendix, Fig. S4C). Genotyping of transgene-expressing ZF (identified by GFP positivity) (SI Appendix, Fig. S4D) demonstrated that Sec23a rescues the mortality of Sec23b-deficiency in ZF (Fig. 3 F and G).
Generation of Mice Expressing Sec23a from the Sec23b Locus.
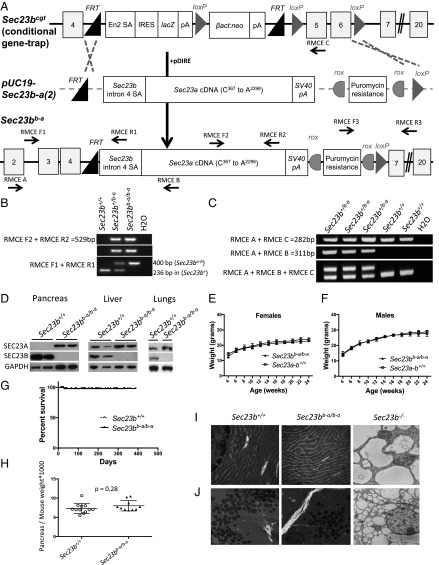
The ZF results suggest at least partial overlap in function between SEC23A and SEC23B. To characterize the capacity of the SEC23A protein to replace SEC23B in mammals in vivo, we substituted Sec23a coding sequences for Sec23b (at the endogenous Sec23b genomic locus). Although efforts to engineer this allele by direct oocyte injection were unsuccessful (SI Appendix, Fig. S5A), embryonic stem (ES) cells carrying the Sec23bb-a allele were generated as shown in Fig. 4A, and used to obtain mice carrying the Sec23bb-a allele. Sec23b+/b-a mice (Fig. 4 B and C) were observed at the expected Mendelian ratio in N2 progeny (Table 1, Sec23b+/b-a × Sec23b+/+). Sec23b+/b-a mice were indistinguishable from their WT control littermates, exhibiting normal growth, fertility, and gross appearance, and no abnormalities on standard pathology evaluation.
Fig. 4.
Mice expressing Sec23a from the Sec23b locus (Sec23bb-a/b-a) exhibit normal growth and survival. (A) Design of the replacement vector pUC19-Sec23b-a(2), which was coelectroporated with pDIRE into Sec23b+/cgt embryonic stem cells. Arrows indicate locations of the genotyping primers. (B) PCR demonstrates the expected band sizes for correct Sec23bb-a 5′ and 3′ insertions. (C) RT-PCR identifies the expected splicing of Sec23b exon4 into the Sec23a cDNA. (D) SEC23B is the predominantly expressed SEC23 paralog in the WT mouse pancreas. In contrast, mice with insertion of the Sec23a coding sequence into the Sec23b locus have no detectable SEC23B in their pancreas tissues by Western blot, with a high level of SEC23A detected. Similarly, SEC23A is the exclusively expressed SEC23 paralog in other tissues from Sec23bb-a/b-a mice, including liver and lung. (E and F) Sec23bb-a/b-a female and male mice exhibit normal growth compared with WT littermate controls. (G) Sec23bb-a/b-a mice exhibit normal survival compared with WT littermate controls. (H) Pancreas tissues from Sec23bb-a/b-a mice exhibit normal weights compared with WT littermate controls. (I and J) Normal appearance of ER (I) and zymogen granules (J) in Sec23bb-a/b-a (and WT Sec23b+/+) compared with Sec23b−/− pancreas tissues by electron microscopy. [Magnification: 25,000× (I); 7,900× (J).]
Table 1.
Mouse crosses with the Sec23bb-a allele
| Mouse crosses and genotypes | Expected, % | Observed at weaning, % | Observed at E14.5, % | Observed at E13.5, % |
| Sec23b+/b-a × Sec23b+/+* | ||||
| Sec23b+/+ | 50 | 47.8 (155) | ||
| Sec23b+/b-a | 50 | 52.2 (169) | ||
| Sec23b+/b-a × Sec23b+/b-a† | ||||
| Sec23b+/+ | 25 | 23.4 (67) | ||
| Sec23b+/b-a | 50 | 50.5 (145) | ||
| Sec23bb-a/b-a | 25 | 26.1 (75) | ||
| Sec23a+/− × Sec23b+/b-a‡ | ||||
| Sec23a+/+ Sec23b+/+ | 25 | 29.1 (23) | ||
| Sec23a+/+ Sec23b+/b-a | 25 | 21.5 (17) | ||
| Sec23a+/− Sec23b+/+ | 25 | 29.1 (23) | ||
| Sec23a+/− Sec23b+/b-a | 25 | 20.3 (16) | ||
| Sec23a+/− Sec23b+/b-a × Sec23a+/−§ | ||||
| Sec23a+/+ Sec23b+/+ | 16.7 | 16.1 (14) | 11.1 (2) | 12.2 (5) |
| Sec23a+/+ Sec23b+/b-a | 16.7 | 19.5 (17) | 11.1 (2) | 9.8 (4) |
| Sec23a+/− Sec23b+/+ | 16.7 | 28.7 (25) | 22.2 (4) | 53.6 (22) |
| Sec23a+/− Sec23b+/b-a | 16.7 | 35.6 (31) | 55.6 (10) | 24.4 (10) |
| Sec23a−/− Sec23b+/+ | 16.7 | 0 (0) | 0 (0) | 0 (0) |
| Sec23a−/− Sec23b+/b-a | 16.7 | 0 (0) | 0 (0) | 0 (0) |
No. of mice is expressed in parentheses.
Observed at weaning, n = 324; P = 0.73 (P value calculated for Sec23b+/b-a vs. WT mice).
Observed at weaning, n = 287; P = 0.89 (P value calculated for Sec23bb-a/b-a vs. all other genotypes).
Observed at weaning, n = 79; P = 0.48 (P value calculated for Sec23a+/− Sec23b+/b-a vs. all other genotypes).
Observed at weaning, n = 87 (P < 0.0001); observed at E14.5, n = 18 (P = 0.037); observed at E1.5, n = 41 (P = 0.0007). P values calculated for Sec23a−/− Sec23b+/b-a mice vs. all other genotypes.
The Sec23bb-a Allele Rescues the Mortality of Sec23b−/− Mice.
An intercross between mice heterozygous for the Sec23bb-a allele generated the expected number of Sec23bb-a/b-a offspring at weaning (Table 1, Sec23b+/b-a × Sec23b+/b-a). In WT mice, SEC23B is the predominantly expressed SEC23 paralog in the pancreas (Fig. 4D). Consistent with substitution of the SEC23A coding sequence for SEC23B, no SEC23B immunoreactive material was detected in pancreas tissues of Sec23bb-a/b-a mice, with high levels of SEC23A protein observed (Fig. 4D), in contrast to WT mice, which exhibit the opposite pattern. SEC23B was also undetectable in SEC23bb-a/b-a liver and lung tissues (Fig. 4D).
The Sec23bb-a Allele Rescues the Pancreatic Phenotype of Sec23b−/− Mice.
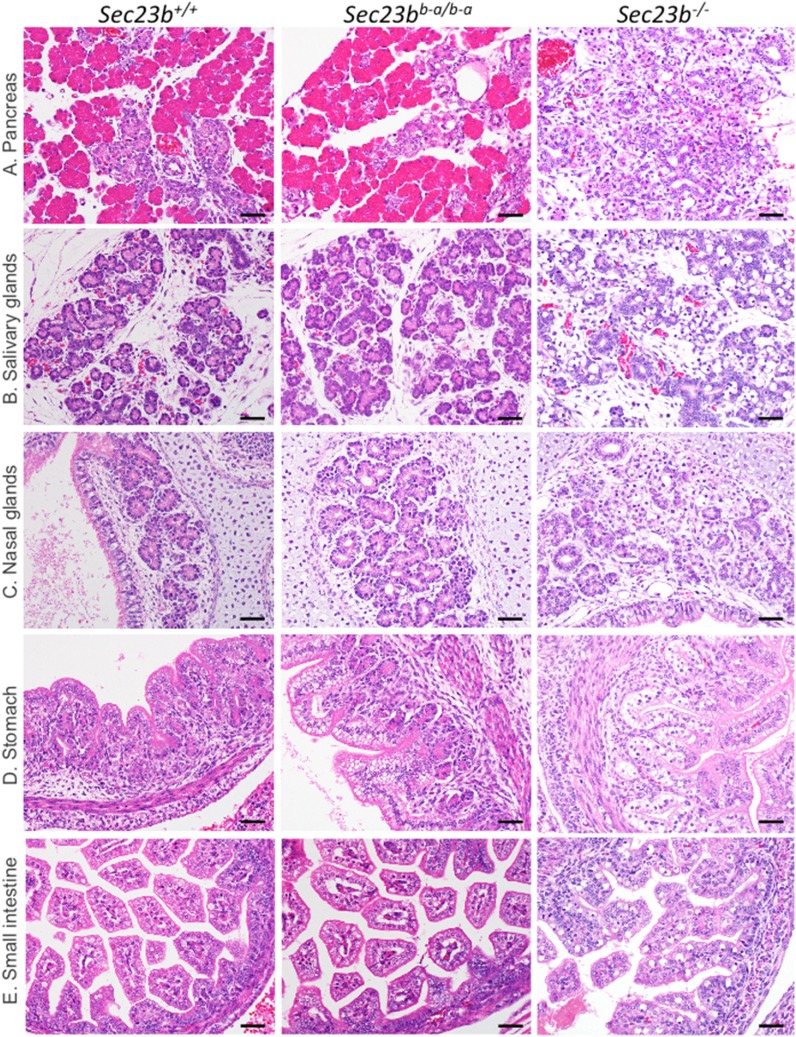
Sec23bb-a/b-a mice exhibited normal growth (Fig. 4 E and F) and fertility, normal gross appearance, no abnormalities on histopathologic evaluation (Fig. 5 and SI Appendix, Fig. S6), and normal overall survival (up to 1.5 y of follow-up) (Fig. 4G). In contrast to the pancreatic degeneration and distended ER observed in Sec23b−/− mice (32, 33), pancreas tissue from Sec23bb-a/b-a mice exhibited normal weight (Fig. 4H) and histology (Fig. 5A), indistinguishable from pancreas of WT littermate controls. The Sec23bb-a/b-a pancreatic ER also appeared normal by transmission electron microscopy (Fig. 4I), with intact acinar cell zymogen granules (Fig. 4J), in contrast to the appearance in Sec23b−/− mice (Fig. 4 I and J). Other secretory tissues that were perturbed in SEC23B-deficient mice (including salivary glands, nasal glands, gastric epithelium, and intestinal cells) (32) also appeared entirely normal histologically in Sec23bb-a/b-a mice (Fig. 5 B–E).
Fig. 5.
Mice expressing Sec23a from the Sec23b locus exhibit normal pancreas histology. (A–E) In contrast to Sec23b−/− mice, Sec23bb-a/b-a mice exhibit normal (A) pancreas, (B) salivary glands, (C) nasal glands, (D) stomach, and (E) small intestine histology, similar to WT (Sec23b+/+) littermate controls. (Scale bars, 40 μm.)
Taken together, these findings demonstrate that the SEC23A protein fully rescues the SEC23B-deficient phenotype in mice, when expressed under the endogenous regulatory elements of the Sec23b gene. Although mice homozygous for the latter genetically engineered Sec23b locus (expressing only SEC23A and no intact SEC23B protein) appear entirely normal and indistinguishable from WT controls, a subtle phenotypic difference or key contribution from the residual SEC23B N-terminal tail retained in the SEC23B-A protein (Fig. 4A and SI Appendix, Fig. S5B) cannot be excluded. Nevertheless, these data demonstrate complete (or near complete) overlap in function between the two SEC23 paralogs.
The Sec23bb-a Allele Fails to Complement Disruption of Sec23a in Sec23a−/− Mice.
Mice with germline SEC23A deficiency die at midembryogenesis (29). To test the capacity of the Sec23bb-a allele to rescue the lethality of mice homozygous for an inactivated endogenous Sec23a allele (Sec23a−/− mice), Sec23b+/b-a mice were crossed with Sec23a+/− mice. Although the expected number of Sec23b+/b-a Sec23a+/− pups from a Sec23b+/b-a × Sec23a+/− cross were present at weaning (Table 1, Sec23a+/− × Sec23b+/b-a), no Sec23b+/b-a Sec23a−/− progeny were observed at weaning or at embryonic day (E) 13.5 (Table 1, Sec23a+/− Sec23b+/b-a × Sec23a+/−). Thus, SEC23A expression under control of Sec23b transcriptional regulatory sequences (Sec23bb-a) fails to complement loss of expression from the Sec23a locus.
The SEC23B/SEC23A Expression Programs Have Shifted During Evolution.
Although the above data suggest functional equivalence for SEC23A and SEC23B in yeast, ZF, and mice, the disparate phenotypes of SEC23B deficiency in humans and mice remain unexplained. To address this question, the relative expression of SEC23B:SEC23A in WT tissues from both humans and mice was determined by qRT-PCR. This ratio was higher in human BM (9.7) compared with human pancreas (1.8), although higher in mouse pancreas (12.7) compared with mouse BM (2.6), consistent with the pathology of SEC23B deficiency in these two species. The differential expression of SEC23A/B in human and murine erythroid cells is consistent with previously published results (38, 39). In addition, a previous analysis of chromatin occupancy in erythroid cells demonstrated a state of transcriptional repression at the human SEC23A locus relative to the mouse (40). SEC23B deficiency also results in a salivary gland defect in mice (Fig. 5B), although this has not been observed in humans. Consistent with this pattern, SEC23B is the predominantly expressed paralog in the murine salivary gland (32), while in humans the SEC23B:SEC23A mRNA expression ratio is ∼0.27 (www.gtexportal.org/).
Taken together, these results suggest that the disparate human/mouse phenotypes are due to an evolutionary shift in the tissue-specific expression programs between mouse and human for these two functionally equivalent proteins.
Discussion
The genes encoding a number of components of the COPII machinery have been duplicated during vertebrate evolution, with evidence for unique protein function for the corresponding paralogous proteins. In the case of SEC23, the distinct and highly specific phenotypes of SEC23A and SEC23B deficiency in humans and mice, together with evidence for paralog-specific contributions to collagen (29) and EGFR (36) transport, as well as a potential role for SEC23B in oncogenesis (41), suggest unique functions for each of these paralogous proteins.
However, our data demonstrate indistinguishable interactomes for SEC23A and SEC23B in HEK293 cells and complementation of yeast Sec23 function by both human and murine SEC23A and SEC23B paralogs. We also show that transgenic germline sec23a expression rescues the mortality of sec23b disruption in ZF and that SEC23A rescues the mortality of SEC23B-deficient mice when expressed under the control of the endogenous Sec23b transcription machinery. These data strongly suggest complete overlap in function between the SEC23A and SEC23B proteins, despite their origin via an ancient gene duplication preceding the origin of vertebrates. The high degree of sequence conservation between SEC23A/B (∼85% amino acid identity) is also consistent with our findings, suggesting strong selection at the level of protein function.
Gene duplications are frequent evolutionary events, with the duplicated copies most commonly accumulating loss-of-function mutations with subsequent disappearance from the genome (42–44). Less frequently, paralogous gene copies become fixed either through neofunctionalization (acquisition of a new function by one of the paralogous gene copies), or subfunctionalization (the division of the functions of the ancestral gene between the paralogous copies). The latter process can occur either at the level of protein function or gene expression. A recent analysis suggests that subfunctionalization of expression evolves slowly and is rare among recently duplicated genes pairs, although is a more common mechanism among duplicated transcription factors (45). Consistent with this model, functional equivalence for a number of transcription factor paralogs has been demonstrated in mice by gene replacement (46–49), although overlap is often incomplete (46, 49).
Our evidence for functional equivalence between the SEC23A and SEC23B proteins suggests that the evolutionary subfunctionalization of these two genes occurred at the level of their gene-expression programs rather than protein function, with select cell types manifesting greater dependence on expression of one or the other paralog. Further shifts in the SEC23A and SEC23B expression programs during mammalian evolution could account for the disparate phenotypes of SEC23A and SEC23B deficiency in mice and humans. Consistent with this hypothesis, human SEC23B is the predominantly expressed paralog in the BM, consistent with the phenotype of CDAII, while SEC23A is the more strongly expressed paralog in calvarial osteoblasts (50), consistent with the CLSD phenotype. In contrast, SEC23B is the predominant paralog in the mouse pancreas, consistent with the pancreatic phenotype observed in murine SEC23B deficiency (30–32).
Similar subfunctionalization at the level of gene expression might also explain highly tissue-restricted phenotypes for other genetic diseases due to loss-of-function in paralogous genes encoding other COPII subunits or other core components of the cellular machinery. This paradigm might also explain other examples of highly discordant phenotypes observed between animal models and humans for various other diseases (51).
Finally, the apparent complete rescue of the SEC23B-deficient phenotype in mice by activating SEC23A expression in SEC23B-dependent tissues suggests that therapeutic strategies to increase the expression of SEC23A in human hematopoietic precursor cells could ameliorate the CDAII phenotype. Such approaches could include delivery of modified CRISPR/Cas9 or transcription factor constructs designed to specifically activate the SEC23A gene (52–55). This paradigm could also be applicable to a number of other human disorders due to mutations in functionally overlapping paralogous genes.
Methods
Evolutionary Sequence Analysis of Sec23 Paralogs.
The gene tree for SEC23A/SEC23B was examined on Ensembl (https://useast.ensembl.org/Homo_sapiens/Gene/Compara_Tree?collapse=12002157%2C12002179%2C12001773%2C12001917%2C12001992%2C12002187%2C12001885%2C12001925%2C12002033%2C12002044%2C12002005%2C12001805%2C12001978%2C12002051%2C12002189%2C12002084%2C12002151%2C12001996%2C12002037%2C12002091%2C12001765%2C12002146%2C12002175%2C12001947%2C12001985%2C12001831;db=core;g=ENSG00000100934;r=14:39031919-39109646). This analysis estimates the Sec23 duplication as having occurred ∼615 Mya.
Coimmunoprecipitation.
FLAG-tagged SEC23A and SEC23B, and RFP-tagged SEC24A, SEC24B, SEC24C, and SEC24D were cloned into the PED plasmid. Expression vectors were transfected into COS cells using FUGENE HD (Promega) per the manufacturer’s instructions. FLAG coimmunoprecipitation with anti-FLAG M2 beads (Sigma) was performed as per the manufacturer’s instructions.
Generation of Stable Cell Lines.
The coding sequences of human SEC23A and SEC23B were cloned into the pENTR vector (K2400-20; Invitrogen) per the manufacturer’s instructions and then into the pcDNA5-pDEST-BirA-FLAG-C-term vector by Gateway cloning. Bait proteins (SEC23A or SEC23B) were stably expressed in Flp-In T-REx HEK293 cells as previously described (56). Stable pools of integrated cells were selected with 200 µg/mL hygromycin. For BioID experiments, cells were treated with tetracycline (1 µg/mL) to induce gene expression and with biotin (50 µM) for 24 h and were subsequently harvested for analysis, as previously described (57).
Affinity Purification and BioID Mass Spectrometry.
Cell lysates were prepared and biotinylated proteins were captured on streptavidin beads and subjected to trypsin digestion, as previously described (57). Tryptic peptides were suspended in 5% formic acid and stored at −80 °C until analysis by MS. Tryptic peptides were analyzed on a TripleTOF 5600 (Sciex) instrument, as previously described (56). Two biological replicates were analyzed for each sample and for controls, and data analysis was performed as follows. Raw MS files were converted to mzXML and searched using the Mascot and Comet search engines. A protein database consisting of the human and adenovirus complements of RefSeq v57 supplemented by common contaminants and epitope-tagged sequences was appended with an equal number of decoy sequences. For peptide identification, a 30-ppm precursor ion and 0.15-Da fragment ion tolerance were specified, and Asn and Gln deamidation and Met oxidation were allowed as variable modifications for searches with Comet and Mascot, essentially as described. Search results were further processed using PeptideProphet to generate a list of peptides, which were then assembled to protein lists using ProteinProphet. The resulting protein lists were then processed using Prohits (default parameters) to generate a spectral count matrix. The spectral count matrix was formatted according to CRAPome/REPRINT specifications (manually) and uploaded onto reprint-apms.org. Protein interactions were scored using two enrichment scoring approaches, empirical fold-change score (FC) and SAINT (SP). A list of high-confidence interactions was generated by applying the following filters: FC ≥ 10; SP ≥ 0.96; abundance ≥ 2. Finally, high-scoring interactions were visualized as a network (using the network generation utility of REPRINT). Raw abundances of filtered interactions in each replicate were plotted against others to evaluate correlation of prey abundances.
The MS dataset has been assigned the MassIVE ID MSV000081703 and is available for FTP download at ftp://MSV000081703@massive.ucsd.edu. The dataset was assigned the ProteomeXchange Consortium (proteomecentral.proteomexchange.org/cgi/GetDataset) identifier PXD008180.
Sec23 Complementation in Yeast.
The coding sequences of the mouse and human SEC23 paralogs, (as well as control yeast sec23) were cloned into the yeast expression vector pRS416, with a 3′ sequence encoding an HA tag. The sequence integrity of all constructs was confirmed by Sanger sequencing. Vectors with the following promoters were used: the ZEO1 constitutive promoter and the CUP1 copper-inducible promoter, resulting in a range of expressions of the SEC23 proteins. The latter vectors were transduced into a temperature-sensitive sec23-1 yeast mutant in the BY4741 background (58), which expresses the functional endogenous yeast Sec23 protein at the permissive temperature (24 °C) but not at the restrictive temperature (37 °C).
Generation of Sec23b-Deficient ZF Using CRISPR/Cas9.
The sec23b locus was identified in the ZF genomic sequence assembly (59). A guide RNA (gRNA) targeting the ZF sec23b exon 5 sequence (ggtggacacatgtctggagg) was cloned into the pDR274 vector, as previously described (60). gRNA was then in vitro transcribed using the T7 Quick High Yield RNA Synthesis kit (New England Biolabs) and purified using the RNA Clean and Concentrator kit (Zymo Research). gRNA was coinjected with Cas9 RNA, generated as previously described (60) into single-cell ZF embryos. ZF were genotyped for indels with primers ZF sec23b CRISPR F and ZF sec23b CRISPR R (SI Appendix, Table S1) spanning the expected CRISPR cut site. ZF heterozygous for a 53-bp deletion in exon 5 of sec23b (sec23b+/−) were generated. sec23b+/− ZF were intercrossed to generate Sec23b-deficient ZF (sec23b−/−). Blood collected from WT, sec23b+/−, and sec23b−/− ZF was stained with the HEMA 3 kit (Fisher) and evaluated under light microscopy by two independent investigators blinded to the ZF genotype.
Generation of Transgenic ZF Expressing sec23a Under the Control of a Ubiquitin Promoter.
The ZF sec23a cDNA was cloned downstream of the ubiquitin promoter into the JS218 vector (61) (SEC23A-GFP). The correct sec23a sequence was confirmed by Sanger sequencing. A p2A sequence separates the sec23a cDNA and the GFP coding sequence, resulting in ubiquitous sec23a and GFP expression from the same mRNA molecule (SI Appendix, Fig. S4C).
SEC23A-GFP or JS218 (in which GFP alone is expressed ubiquitously) were coinjected with transposase mRNA, as previously described (61), into one-cell–stage embryos generated from sec23b+/− ZF intercrosses to generate transgenic ZF ubiquitously expressing SEC23A and GFP or GFP alone. The latter ZF were genotyped with primers ZF sec23b CRISPR F and ZF sec23b CRISPR R (listed in SI Appendix, Table S1).
Cloning the Sec23b-a dRMCE Cassette into pUC19.
The Sec23b-a cassette was generated by assembling the following sequences from the 5′ to 3′ direction: an FRT sequence, the endogenous Sec23b intron 4 splice acceptor sequence, Sec23a from C367 to A2298 of the cDNA coding sequence (encoding murine SEC23A starting at Arg123), and the SV40 polyA sequence present in the Sec23bcgt allele (30, 62). The above Sec23b-a cassette was inserted into pUC19 at the HindIII and SalI restriction sites, producing pUC19-Sec23b-a(1) (SI Appendix, Fig. S5A) and a puromycin resistance cassette flanked by rox sites was inserted 3′ of the SV40 polyA sequence using PmeI and AscI restriction sites added to the vector by PCR, resulting in pUC19-Sec23b-a(2) (Fig. 4A), with sequence integrity confirmed by Sanger sequencing.
Plasmid Purification and Microinjections.
The pDIRE plasmid driving expression of both FLPo and iCRE was obtained from Addgene (#26745). The pCAGGS-FLPo and pCAGGS-iCre plasmids, which contain the CAG promoter/enhancer driving the expression of FLPo and iCre recombinase, respectively, were generated as previously described (63). Plasmids were purified using the Qiagen EndoFree Plasmid Maxi Kit (#12362; Qiagen) or the Machery-Nagel NucleoBond Xtra Maxi EF kit (#740424.10; Machery-Nagel), per the manufacturers’ instructions.
Zygotes generated from the in vitro fertilization of C57BL/6J oocytes with sperm from Sec23b+/− male mice (33), were coinjected with pUC19-Sec23b-a(1) and pDIRE plasmids and then implanted in pseudopregnant foster mothers. Pups were genotyped at 2 wk of age. All microinjections were performed at the University of Michigan Transgenic Animal Model Core.
Transient Electroporation of ES Cells.
The ES cell clone EPD0237_3_G06 heterozygous for the Sec23bcgt allele (33) was obtained from the European Conditional Mouse Mutagenesis Program (62) and coelectroporated with pUC19-Sec23b-a(2) and either pDIRE or a combination of pCAGGS-FLPo and pCAGGS-iCre, as previously described (63). One week postelectroporation, individual ES cells were plated in 96-well plates. Cells were expanded and plated in triplicates for DNA analysis, frozen stocks, and puromycin selection. Following puromycin selection, 288 single cell ES clones were evaluated demonstrating correct targeted insertion of the dRMCE replacement vector in 13 of 288 independent clones.
Generation of Sec23b+/b-a Mice.
Three independent EPD0237_3_G06 ES cell clones correctly targeted with pUC19-Sec23b-a(2) were cultured, expanded, and then microinjected into albino C57BL/6 blastocysts to generate Sec23b+/b-a mice (Fig. 4A). The chimeric progeny were crossed to B6(Cg)-Tyrc-2J/J mice (Jackson Laboratory stock #000058) and germline transmission was confirmed by genotyping the progeny for the Sec23b+/b-a allele. The Sec23b+/b-a allele was maintained on a C57BL/6J background.
Mouse Genotyping.
Genotyping of the Sec23bb-a allele was done on genomic DNA extracted from tail biopsies using primers RMCE F2 and RMCE R2, which detect the dRMCE replacement construct. These primers are located in Sec23a exons 10 and 14, respectively, and therefore should not yield a detectable product from the WT Sec23a allele. To identify potential random insertions of the dRMCE allele, PCR genotyping was also performed with primers RMCE F1 and RMCE R1 to confirm correct insertion of the Sec23b-a(1) and Sec23b-a(2) alleles at the 5′ end, and with primers RMCE F3 and RMCE R3 to confirm correct placement of the Sec23b-a(2) allele at the 3′ end. All mice with a visible PCR product with primers RMCE F2 and RMCE R2 also had PCR products with primers RMCE F1-RMCE R1 and primers RMCE F3-RMCE R3, effectively excluding random insertion.
Genotyping for the Sec23bb-a allele was also performed by RT-PCR on RNA isolated from mouse tail biopsies, using a three-primer competitive PCR assay, with a forward primer (RMCE A) located in exon 2 of Sec23b and two reverse primers, one located in the Sec23a cDNA (RMCE B) and one located in exon 5 of Sec23b (RMCE C). This competitive PCR assay should generate a 282-bp band from the WT allele and a 311-bp band from the Sec23bb-a allele (Fig. 4C), which were resolved on a 2% (wt/vol) agarose gel. The location of the primers is indicated in Fig. 4A and SI Appendix, Fig. S5A and the primer sequences are listed in SI Appendix, Table S1.
RT-PCR.
RNA was isolated from tissues using the RNeasy Mini kit (Qiagen), and reverse transcription was performed using the SuperScript first strand synthesis system for RT-PCR (Invitrogen) with random primers.
Western Blot and Antibodies.
Total cell lysates were prepared and Western blots were performed as previously described (33). Rabbit anti-SEC23A and anti-SEC23B antibodies were generated as previously described (33), and mouse anti-GAPDH and anti–RAL-A antibodies were obtained from Millipore and BD Biosciences, respectively. Anti-FLAG, anti-RFP, and anti-HA antibodies were obtained from Cell Signaling Technology, Abcam, and Sigma, respectively.
Complete Blood Counts.
Peripheral blood was collected from the retro-orbital venous sinuses of anesthetized mice as previously described (30), and complete blood counts were performed using the Advia120 whole-blood analyzer (Siemens), as previously described (33).
Animal Care.
Mice and zebrafish were housed at the University of Michigan and all animal care and procedures were in accordance with the Principles of Laboratory and Animal Care, established by the National Society of Medical Research. Mice had free access to water and food and were kept in cages in a 12-h light/dark cycle. Zebrafish were maintained in a recirculating system in a 14-h light/10-h dark cycle. All animal protocols used in this study were approved by the University of Michigan Committee on the Use and Care of Animals.
Histology, Cytology, o-dianisidine Staining, and Electron Microscopy.
For histological analysis, mouse tissues or ZF embryos were harvested and immediately fixed in 4% paraformaldehyde or aqueous buffered zinc formalin (Z-fix, Anatech). Tissues were then routinely processed, paraffin-embedded, sectioned, and stained with H&E. BM smears were prepared using a brush technique, air dried, and stained with a modified Romanowsky staining (Diff-Quik) technique. ZF embryos were stained with o-dianisidine, as previously described (64). For electron microscopy, mouse pancreas tissues were prepared as previously described (30, 31) and sections were examined on a Philips CM100 electron microscope. Images were recorded as previously described (30, 31).
Statistical Analysis.
The statistical significance of deviation from the expected Mendelian ratio of mouse crosses was assessed using the χ2 test. The statistical significance of the differences between parameters among different genotype groups was assessed using the Student’s t test. The difference in overall survival between mice of distinct genotypes was assessed using the Kaplan–Meier method.
Supplementary Material
Acknowledgments
The authors thank Elizabeth Hughes, Corey Ziebell, Galina Gavrilina, Wanda Filipiak, Keith Childs, and Debora VanHeyningen for microinjections and preparation of embryonic stem cell–mouse chimeras; and Wendy Rosebury-Smith and Kathy Toy of the In Vivo Animal Core for their necropsy and histology expertise. This work was supported by National Institute of Health Grants R01 HL039693, R35-HL135793, and P01-HL057346 (to D.G.), K08 HL128794 (to R.K.), R01 GM053396 (to D.J.K.), R01 HL 124232 (to J.A.S.), R01 GM094231 (to A.I.N.), R01 HL094505 (to B.Z.), and T32 HL007622 (to A.C.W.); Canadian Institutes of Health Research Foundation Grant FDN 143301 (to A.-C.G.); and by MCubed, a research seed-funding program for faculty at the University of Michigan (to R.K., J.A.S., and D.J.K.). Support for the Transgenic Animal Model Core of the University of Michigan’s Biomedical Research Core Facilities was provided by the University of Michigan Cancer Center (NIH Grant P30 CA046592). R.K. and J.A.S. are recipients of American Society of Hematology Scholar Awards. A.C.W. was supported by a National Hemophilia Foundation-Shire Clinical Fellowship Award. G.G.H. was supported by a Parkinson Canada Basic Research Fellowship. A.-C.G. is the Canada Research Chair in Functional Proteomics. D.G. is a Howard Hughes Medical Institute investigator.
Footnotes
The authors declare no conflict of interest.
Data deposition: The mass spectrometry dataset has been assigned the MassIVE ID MSV000081703 and is available for FTP download at: ftp://MSV000081703@massive.ucsd.edu. The dataset has been deposited in the ProteomeXchange Consortium, proteomecentral.proteomexchange.org/cgi/GetDataset) (identifier PXD008180).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805784115/-/DCSupplemental.
References
- 1.Kanapin A, et al. RIKEN GER Group; GSL Members Mouse proteome analysis. Genome Res. 2003;13:1335–1344. doi: 10.1101/gr.978703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 2011;14:20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]
- 3.Barlowe C, Helenius A. Cargo capture and bulk flow in the early secretory pathway. Annu Rev Cell Dev Biol. 2016;32:197–222. doi: 10.1146/annurev-cellbio-111315-125016. [DOI] [PubMed] [Google Scholar]
- 4.Bannykh SI, Rowe T, Balch WE. The organization of endoplasmic reticulum export complexes. J Cell Biol. 1996;135:19–35. doi: 10.1083/jcb.135.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orci L, et al. Mammalian Sec23p homologue is restricted to the endoplasmic reticulum transitional cytoplasm. Proc Natl Acad Sci USA. 1991;88:8611–8615. doi: 10.1073/pnas.88.19.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khoriaty R, Vasievich MP, Ginsburg D. The COPII pathway and hematologic disease. Blood. 2012;120:31–38. doi: 10.1182/blood-2012-01-292086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee MC, et al. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 8.Matsuoka K, et al. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 9.Aridor M, et al. The Sar1 GTPase coordinates biosynthetic cargo selection with endoplasmic reticulum export site assembly. J Cell Biol. 2001;152:213–229. doi: 10.1083/jcb.152.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aridor M, Weissman J, Bannykh S, Nuoffer C, Balch WE. Cargo selection by the COPII budding machinery during export from the ER. J Cell Biol. 1998;141:61–70. doi: 10.1083/jcb.141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mancias JD, Goldberg J. The transport signal on Sec22 for packaging into COPII-coated vesicles is a conformational epitope. Mol Cell. 2007;26:403–414. doi: 10.1016/j.molcel.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Miller E, Antonny B, Hamamoto S, Schekman R. Cargo selection into COPII vesicles is driven by the Sec24p subunit. EMBO J. 2002;21:6105–6113. doi: 10.1093/emboj/cdf605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller EA, et al. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- 14.Mossessova E, Bickford LC, Goldberg J. SNARE selectivity of the COPII coat. Cell. 2003;114:483–495. doi: 10.1016/s0092-8674(03)00608-1. [DOI] [PubMed] [Google Scholar]
- 15.Cox NJ, et al. Dynamic glycosylation governs the vertebrate COPII protein trafficking pathway. Biochemistry. 2018;57:91–107. doi: 10.1021/acs.biochem.7b00870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuehn MJ, Herrmann JM, Schekman R. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- 17.Votsmeier C, Gallwitz D. An acidic sequence of a putative yeast Golgi membrane protein binds COPII and facilitates ER export. EMBO J. 2001;20:6742–6750. doi: 10.1093/emboj/20.23.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fromme JC, Orci L, Schekman R. Coordination of COPII vesicle trafficking by Sec23. Trends Cell Biol. 2008;18:330–336. doi: 10.1016/j.tcb.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Appenzeller C, Andersson H, Kappeler F, Hauri HP. The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat Cell Biol. 1999;1:330–334. doi: 10.1038/14020. [DOI] [PubMed] [Google Scholar]
- 20.Muñiz M, Nuoffer C, Hauri HP, Riezman H. The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J Cell Biol. 2000;148:925–930. doi: 10.1083/jcb.148.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powers J, Barlowe C. Erv14p directs a transmembrane secretory protein into COPII-coated transport vesicles. Mol Biol Cell. 2002;13:880–891. doi: 10.1091/mbc.01-10-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang B, et al. Bleeding due to disruption of a cargo-specific ER-to-Golgi transport complex. Nat Genet. 2003;34:220–225. doi: 10.1038/ng1153. [DOI] [PubMed] [Google Scholar]
- 23.Antonny B, Madden D, Hamamoto S, Orci L, Schekman R. Dynamics of the COPII coat with GTP and stable analogues. Nat Cell Biol. 2001;3:531–537. doi: 10.1038/35078500. [DOI] [PubMed] [Google Scholar]
- 24.Adams EJ, Chen XW, O’Shea KS, Ginsburg D. Mammalian COPII coat component SEC24C is required for embryonic development in mice. J Biol Chem. 2014;289:20858–20870. doi: 10.1074/jbc.M114.566687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stankewich MC, Stabach PR, Morrow JS. Human Sec31B: A family of new mammalian orthologues of yeast Sec31p that associate with the COPII coat. J Cell Sci. 2006;119:958–969. doi: 10.1242/jcs.02751. [DOI] [PubMed] [Google Scholar]
- 26.Boyadjiev SA, et al. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat Genet. 2006;38:1192–1197. doi: 10.1038/ng1876. [DOI] [PubMed] [Google Scholar]
- 27.Boyadjiev SA, et al. Cranio-lenticulo-sutural dysplasia associated with defects in collagen secretion. Clin Genet. 2011;80:169–176. doi: 10.1111/j.1399-0004.2010.01550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarz K, et al. Mutations affecting the secretory COPII coat component SEC23B cause congenital dyserythropoietic anemia type II. Nat Genet. 2009;41:936–940. doi: 10.1038/ng.405. [DOI] [PubMed] [Google Scholar]
- 29.Zhu M, et al. Neural tube opening and abnormal extraembryonic membrane development in SEC23A deficient mice. Sci Rep. 2015;5:15471. doi: 10.1038/srep15471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khoriaty R, et al. Pancreatic SEC23B deficiency is sufficient to explain the perinatal lethality of germline SEC23B deficiency in mice. Sci Rep. 2016;6:27802. doi: 10.1038/srep27802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khoriaty R, et al. SEC23B is required for pancreatic acinar cell function in adult mice. Mol Biol Cell. 2017;28:2146–2154. doi: 10.1091/mbc.E17-01-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tao J, et al. SEC23B is required for the maintenance of murine professional secretory tissues. Proc Natl Acad Sci USA. 2012;109:E2001–E2009. doi: 10.1073/pnas.1209207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khoriaty R, et al. Absence of a red blood cell phenotype in mice with hematopoietic deficiency of SEC23B. Mol Cell Biol. 2014;34:3721–3734. doi: 10.1128/MCB.00287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marti L, Fornaciari S, Renna L, Stefano G, Brandizzi F. COPII-mediated traffic in plants. Trends Plant Sci. 2010;15:522–528. doi: 10.1016/j.tplants.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 35.Paccaud JP, et al. Cloning and functional characterization of mammalian homologues of the COPII component Sec23. Mol Biol Cell. 1996;7:1535–1546. doi: 10.1091/mbc.7.10.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scharaw S, et al. The endosomal transcriptional regulator RNF11 integrates degradation and transport of EGFR. J Cell Biol. 2016;215:543–558. doi: 10.1083/jcb.201601090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng Y, et al. Unique COPII component AtSar1a/AtSec23a pair is required for the distinct function of protein ER export in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2015;112:14360–14365. doi: 10.1073/pnas.1519333112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An X, et al. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood. 2014;123:3466–3477. doi: 10.1182/blood-2014-01-548305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pishesha N, et al. Transcriptional divergence and conservation of human and mouse erythropoiesis. Proc Natl Acad Sci USA. 2014;111:4103–4108. doi: 10.1073/pnas.1401598111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulirsch JC, et al. Altered chromatin occupancy of master regulators underlies evolutionary divergence in the transcriptional landscape of erythroid differentiation. PLoS Genet. 2014;10:e1004890. doi: 10.1371/journal.pgen.1004890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yehia L, et al. Germline heterozygous variants in SEC23B are associated with Cowden syndrome and enriched in apparently sporadic thyroid cancer. Am J Hum Genet. 2015;97:661–676. doi: 10.1016/j.ajhg.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conant GC, Wolfe KH. Turning a hobby into a job: How duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 43.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 44.Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- 45.Lan X, Pritchard JK. Coregulation of tandem duplicate genes slows evolution of subfunctionalization in mammals. Science. 2016;352:1009–1013. doi: 10.1126/science.aad8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouchard M, Pfeffer P, Busslinger M. Functional equivalence of the transcription factors Pax2 and Pax5 in mouse development. Development. 2000;127:3703–3713. doi: 10.1242/dev.127.17.3703. [DOI] [PubMed] [Google Scholar]
- 47.Greer JM, Puetz J, Thomas KR, Capecchi MR. Maintenance of functional equivalence during paralogous Hox gene evolution. Nature. 2000;403:661–665. doi: 10.1038/35001077. [DOI] [PubMed] [Google Scholar]
- 48.Hanks M, Wurst W, Anson-Cartwright L, Auerbach AB, Joyner AL. Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science. 1995;269:679–682. doi: 10.1126/science.7624797. [DOI] [PubMed] [Google Scholar]
- 49.Malynn BA, et al. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000;14:1390–1399. [PMC free article] [PubMed] [Google Scholar]
- 50.Fromme JC, et al. The genetic basis of a craniofacial disease provides insight into COPII coat assembly. Dev Cell. 2007;13:623–634. doi: 10.1016/j.devcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kahn ML, et al. A dual thrombin receptor system for platelet activation. Nature. 1998;394:690–694. doi: 10.1038/29325. [DOI] [PubMed] [Google Scholar]
- 52.Horlbeck MA, et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 2016;5:e19760. doi: 10.7554/eLife.19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao HK, et al. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell. 2017;171:1495–1507.e15. doi: 10.1016/j.cell.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Traxler EA, et al. A genome-editing strategy to treat β-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat Med. 2016;22:987–990. doi: 10.1038/nm.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lambert JP, Tucholska M, Go C, Knight JD, Gingras AC. Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J Proteomics. 2015;118:81–94. doi: 10.1016/j.jprot.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hesketh GG, Youn JY, Samavarchi-Tehrani P, Raught B, Gingras AC. Parallel exploration of interaction space by BioID and affinity purification coupled to mass spectrometry. Methods Mol Biol. 2017;1550:115–136. doi: 10.1007/978-1-4939-6747-6_10. [DOI] [PubMed] [Google Scholar]
- 58.Li Z, et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol. 2011;29:361–367. doi: 10.1038/nbt.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howe K, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503, and erratum (2014) 505:248. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu Z, et al. Genome editing of factor X in zebrafish reveals unexpected tolerance of severe defects in the common pathway. Blood. 2017;130:666–676. doi: 10.1182/blood-2017-02-765206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Osterwalder M, et al. Dual RMCE for efficient re-engineering of mouse mutant alleles. Nat Methods. 2010;7:893–895. doi: 10.1038/nmeth.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kafina MD, Paw BH. Using the zebrafish as an approach to examine the mechanisms of vertebrate erythropoiesis. Methods Mol Biol. 2018;1698:11–36. doi: 10.1007/978-1-4939-7428-3_2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.