Significance
The circadian clock is involved in aging in animals, where mutations in core clock genes accelerate aging. However, little is known about the relationship between aging and the circadian clock in plants. Using the well-studied process of leaf senescence in Arabidopsis, a higher plant, as a model for aging, we show that the circadian clock has a critical role in regulating the aging process in plants. Specifically, we show that PSEUDO-RESPONSE REGULATOR 9 (PRR9), a core clock component, positively regulates leaf senescence. ORESARA 1 (ORE1), an aging regulator, is controlled by PRR9 via direct transcriptional activation and indirectly by suppressing miR164, a posttranscriptional repressor of ORE1, thus forming a coherent feed-forward regulatory loop.
Keywords: circadian clock, leaf senescence, PRR9, ORE1, miR164
Abstract
The circadian clock coordinates the daily cyclic rhythm of numerous biological processes by regulating a large portion of the transcriptome. In animals, the circadian clock is involved in aging and senescence, and circadian disruption by mutations in clock genes frequently accelerates aging. Conversely, aging alters circadian rhythmicity, which causes age-associated physiological alterations. However, interactions between the circadian clock and aging have been rarely studied in plants. Here, we investigated potential roles for the circadian clock in the regulation of leaf senescence in plants. Members of the evening complex in Arabidopsis circadian clock, EARLY FLOWERING 3 (ELF3), EARLY FLOWERING 4 (ELF4), and LUX ARRHYTHMO (LUX), as well as the morning component PSEUDO-RESPONSE REGULATOR 9 (PRR9), affect both age-dependent and dark-induced leaf senescence. The circadian clock regulates the expression of several senescence-related transcription factors. In particular, PRR9 binds directly to the promoter of the positive aging regulator ORESARA1 (ORE1) gene to promote its expression. PRR9 also represses miR164, a posttranscriptional repressor of ORE1. Consistently, genetic analysis revealed that delayed leaf senescence of a prr9 mutant was rescued by ORE1 overexpression. Thus, PRR9, a core circadian component, is a key regulator of leaf senescence via positive regulation of ORE1 through a feed-forward pathway involving posttranscriptional regulation by miR164 and direct transcriptional regulation. Our results indicate that, in plants, the circadian clock and leaf senescence are intimately interwoven as are the clock and aging in animals.
Aging and death are the inevitable fates of organisms. Aging is considered an evolved life history strategy specific to a given species, as each species exhibits its own characteristic life history of aging and death (1). Plants also undergo aging and death in a species-specific manner and show highly diverse life history strategies (2). Annual plants such as rice and Arabidopsis show a relatively short and well-defined seasonal lifespan. Trees can live thousands of years; however, their organs age and die, as observed in shedding of autumn leaves (3). The age-dependent senescence of leaves is critical for plants’ fitness and productivity. Leaf senescence mobilizes the nutrients accumulated through photosynthesis and nutrient uptake to newly developing leaves or seeds (4). Thus, when and how the aging process or age-dependent senescence of leaf organs proceeds critically affects fitness and productivity at the whole plant level. Plant-specific transcription factor (TF) families such as NAC (NAM, ATAF, and CUC) and WRKY control plant leaf senescence (5), implying that unique regulators of plant-specific TFs may underlie aging in plants.
Most organisms are influenced by the daily and annual cycles of light intensity and duration and temperature (6). Organisms accordingly coordinate their endogenous processes with these environmental cycles to enhance their fitness, leading to the evolution of circadian clocks in many organisms. The circadian clock is an endogenous biological clock that generates ∼24-h rhythms, regulating a large fraction of the transcriptome to cope with the daily environmental cycles (7). Plant circadian clocks also regulate many aspects of development and physiology throughout life, such as flowering (8). Plants are sessile and cannot avoid unfavorable conditions by mobility, so they likely experience greater pressure than animals to cope with daily environmental changes. Circadian clocks in plants have been suggested to function as a “mastermind” of plant life (9), coordinating internal physiological status with environmental cycles. In many systems, the 24-h cyclic rhythm is generated by interconnected feedback loops of the core circadian oscillator.
The physiological effect of aging encompasses the circadian clock system. In animals, aging is associated with the disruption of many circadian rhythms, such as a change of circadian period, delayed recovery from jet lag, and reduced amplitude of clock gene expression (10, 11). Conversely, the circadian clock also protects animals from aging. Disruption of circadian systems accelerates aging and reduces longevity. For example, deficiency of the animal core clock genes CLOCK or BMAL1 accelerated aging and several age-dependent phenotypes, such as cancers, in mice (12, 13). Mutations in Per1 and Per2, interconnected with CLOCK and BMAL1, also show premature aging (14). In plants, however, the interaction of circadian clock and aging has been rarely explored, despite the importance of both processes in plant physiology.
In this study, we reveal regulatory mechanisms that tightly link aging and the circadian clock in plants, using Arabidopsis leaf senescence as a model system. We report that several circadian clock genes are required for age-dependent and/or dark-induced leaf senescence. We find that the core clock component PRR9 positively regulates an aging regulator ORE1, directly via transcriptional activation and indirectly by suppressing miR164, a posttranscriptional repressor of ORE1. Importantly, prr9 mutant plants showed reduced ORE1 transcript levels and delayed age-induced leaf senescence. ORE1 overexpression restores age-associated senescence to prr9 mutant plants. These results suggest that the circadian clock may control leaf senescence by modulating ORE1 amplitude.
Results
Age-Dependent Leaf Senescence and Flowering Time Are Coordinately Controlled by the Circadian Clock.
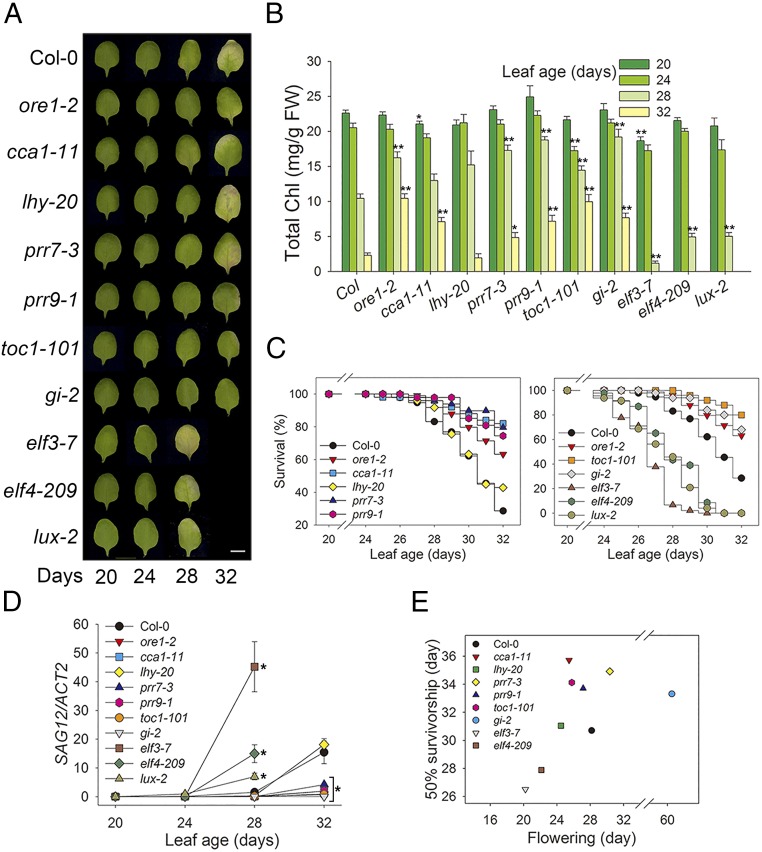
In Arabidopsis, the circadian clock that generates the daily cycles includes many core clock components in the “morning” (CCA1, LHY, PRR7, and PRR9) and “evening” (TOC1, GI, ELF3, ELF4, and LUX) loops. To test the interaction between the circadian system and leaf senescence in Arabidopsis, we examined leaf senescence, which is physiologically and genetically well defined in Arabidopsis leaves (4), in mutants deficient in these nine core circadian clock components. We first examined leaf yellowing, a visible indicator which reflects chloroplast senescence of mesophyll cells (Fig. 1A). At 28 d of leaf age, elf3-7, elf4-209, and lux-2 showed greater yellowing compared with wild-type Columbia (Col-0). In contrast, other clock mutants (cca1-1, prr7-3, prr9-1, toc1-101, and gi-2) remained green until 32 d of leaf age, showing delayed senescence compared with wild type. Of the clock mutants tested, only lhy-20 showed no change in leaf yellowing. Chlorophyll content and survival were consistent with the leaf yellowing phenotypes (Fig. 1 B and C). Expression of the cysteine protease-encoding SENESCENCE-ASSOCIATED GENE 12 (SAG12), a representative leaf senescence marker in plants, also was positively correlated with the leaf yellowing phenotypes of clock mutants (Fig. 1D). The observation that eight of nine circadian clock mutants affected leaf senescence indicates a strong interplay between the circadian clock and aging in plant leaves.
Fig. 1.
Circadian clock genes are associated with age-dependent senescence. (A) Chlorophyll (Chl) loss in Col-0, ore1-2 (positive control), and clock mutants. The photographs show representative of the third and fourth rosette leaves at the indicated days after emergence. (Scale bar: 0.5 cm.) (B–D) Analysis of chlorophyll content (B), survivorship (C), and expression of age-associated gene (SAG12) (D) in leaves of the indicated genotypes at the indicated days. (E) Association between flowering and senescence in Col-0 and clock mutants. Data are presented as the mean ± SEM of biological triplicates. *P < 0.05, **P < 0.01; t test.
The circadian system controls photoperiodic flowering (8, 15), which can in turn influence age-dependent leaf senescence. Given that age-dependent senescence is controlled by the circadian clock, we examined whether there is a correlated change in flowering and senescence timing in the clock mutants. For this test, we generated survival and flowering curves (SI Appendix, Fig. S1) and measured 50% survival and flowering by regression analyses of the curves. We observed a strong correlation between flowering and leaf senescence timing (correlation coefficient r2 = 0.554, P = 0.021) except for the gi-2 mutant which has an extremely late flowering phenotype (Fig. 1E). Thus, these two developmental processes may be coupled by the circadian clock in Arabidopsis.
The Circadian Clock Regulates Dark-Induced Leaf Senescence.
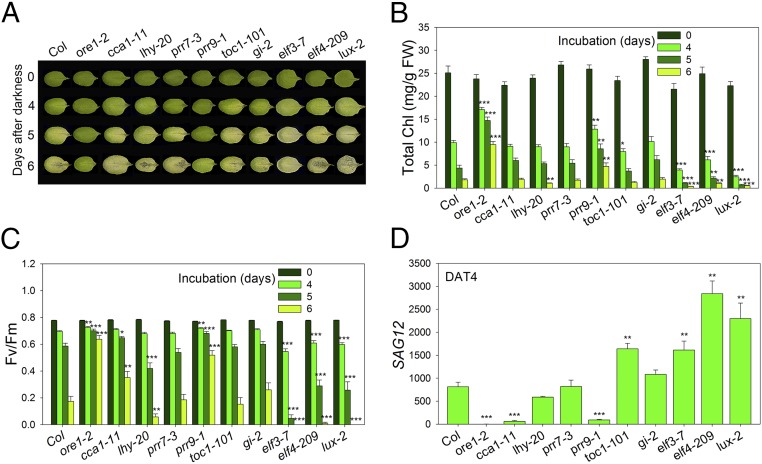
Because age-dependent leaf senescence can be influenced by other developmental processes such as flowering, we aimed to identify clock components that regulate other forms of leaf senescence. Leaf senescence is regulated not only by the internal factor of aging, but also by many external factors, including hormones, pathogen attacks, drought, high salinity, extreme temperature, and darkness (4). Dark-induced senescence displays many similarities to age-dependent senescence, such as chlorophyll and protein degradation (16). However, transcriptomic analysis revealed that age-dependent versus dark-induced senescence triggers significantly different gene expression profiles and signaling pathways (5, 16). To investigate whether the circadian clock regulates dark-induced senescence, we exposed the third and fourth rosette leaves, detached from 3-wk-old wild-type and nine core clock mutant plants, to darkness. We found that elf3-7, elf4-209, and lux-2 showed significantly faster dark-induced senescence than wild-type (Col-0), consistent with their effects on age-dependent senescence and with previous findings that ELF3 suppresses senescence in darkness (17). However, most of clock mutants (cca1-1, lhy-20, prr7-3, toc1-101, and gi-2) showed similar rates of senescence in darkness. Of the clock mutants tested, only prr9-1 displayed significantly delayed dark-induced senescence compared with wild type (Fig. 2A). Quantitative assays of chlorophyll content, photochemical efficiency, and SAG12 expression were consistent with the leaf yellowing phenotypes of prr9-1 (Fig. 2 B–D). Thus, PRR9 promotes both dark-induced and age-dependent leaf senescence.
Fig. 2.
Dark-induced senescence phenotype of clock mutants in Arabidopsis. (A) Representative leaves of Col-0, ore1-2 (positive control), and clock mutant plants after incubation in darkness at the indicated days. (B and C) Analysis of chlorophyll content (B) and photochemical efficiency of PSII (C) of detached leaves of the indicated genotypes at the indicated days after dark incubation. (D) Expression of the senescence-induced SAG12 gene at day 4 after dark treatment (DAT4) relative to DAT0 in leaves of the indicated genotypes. Data are presented as the mean ± SEM of biological triplicates. *P < 0.05, **P < 0.01, ***P < 0.001; t test.
The Circadian Clock Regulates Several Senescence-Related Transcription Factors.
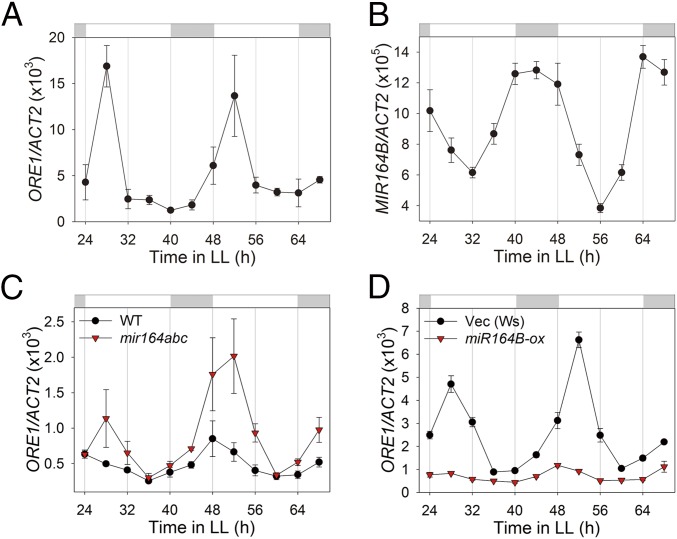
The aging process in Arabidopsis is regulated by senescence-related NAC and WRKY transcription factor families, among others (18). Therefore, we tested whether genes in these two families are regulated by the circadian clock to mediate clock-controlled aging. We selected 8 NAC and 4 WRKY genes associated with the Gene Ontology (GO) term “aging” (19), screened them for potential circadian regulation in the Diurnal database (20), and found that six of them oscillated in a circadian clock-dependent manner (SI Appendix, Fig. S2). We also evaluated the expression of all 12 genes by qRT-PCR and confirmed that at least three NAC (ANAC029, 083, 092, also called ORE1) and two WRKY (WRKY22, 54) genes showed clear circadian expression patterns with distinct amplitudes, phases of peak expression, and waveforms (Fig. 3A and SI Appendix, Fig. S3). The circadian regulation of 5 of 12 senescence-related transcription factors further supports the circadian clock influence on the aging process.
Fig. 3.
ORE1 is posttranscriptionally regulated by clock-controlled miR164. (A and B) mRNA abundance of ORE1 (ANAC092) (A) and MIR164B (B) in young (3 wk old) Col-0 leaves under LL (n = 3). (C) mRNA abundance of ORE1 in wild-type and mir164abc mutant plants under LL (n = 3). (D) mRNA abundance of ORE1 in vector control (Vec) and a miR164B-overexpressing line (miR164B-ox) in young (3 wk old) leaves under LL (n = 3). ACT2, internal control. Data are presented as the mean ± SEM of biological triplicates.
ORE1 Is Negatively Regulated by the Clock-Controlled miR164.
ORE1/ANAC092 is a well-established positive regulator of the aging process that showed a robust circadian expression pattern (Fig. 3A). ORE1 is down-regulated posttranscriptionally by miR164 and up-regulated transcriptionally by EIN3 during aging through a trifurcate feed-forward pathway, which affects age-dependent cell death (21, 22). We tested whether the circadian clock controls expression of miR164, which would in turn negatively regulate expression of ORE1. We found that MIR164B, the major among three miR164 isoforms (miR164a, b, and c) showed a robust circadian expression pattern, peaking antiphasic to ORE1 (Fig. 3B); MIR164B and ORE1 showed peak expression at early night and early morning time points, respectively. This observation suggests that antiphasic expression of miR164 contributes to the circadian expression of ORE1. We confirmed that miR164 negatively controls the amplitude of circadian expression of ORE1 by using a mir164abc triple knockout mutant line and a line overexpressing miR164B (miR164B-ox). In the mir164abc null mutant, the circadian amplitude of ORE1 mRNA was over twofold higher than that of wild type (Fig. 3C). In contrast, the circadian amplitude of ORE1 mRNA was greatly (over fivefold) attenuated in the miR164B-ox line (Fig. 3D). These results demonstrate that the circadian clock negatively regulates the amplitude of ORE1 mRNA levels via antiphasic expression of miR164.
PRR9 Directly Promotes Cyclic Transcription of ORE1.
miR164 suppresses the amplitude of ORE1 mRNA levels, indicating that other mechanisms must promote circadian expression of ORE1. To investigate whether ORE1 promoter activity is under circadian control, we used transgenic reporter plants in which the ORE1 promoter drives luciferase (LUC) expression. We observed robustly rhythmic luciferase activity in young (3 wk old) and aged (5 wk old) transgenic plants under continuous light (LL) (SI Appendix, Fig. S4), revealing that the circadian clock regulates ORE1 promoter activity.
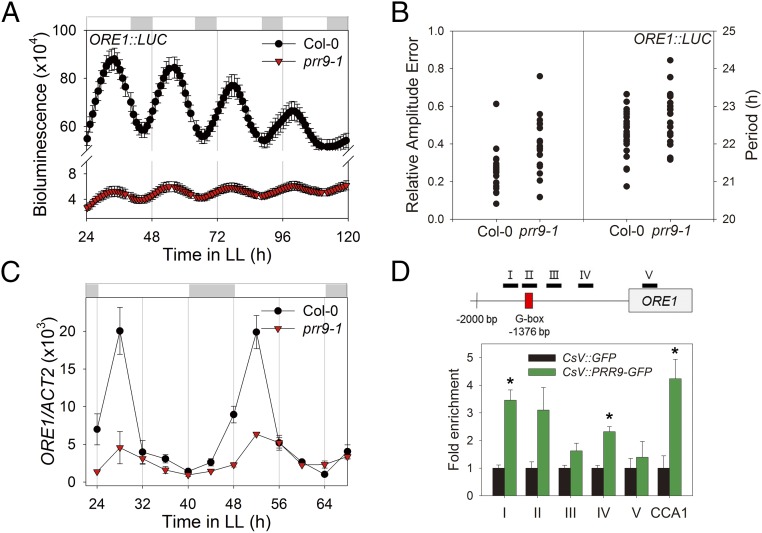
Most of the circadian core clock components are transcription factors (23). To investigate whether these circadian components regulate the ORE1 promoter through direct binding, we performed yeast one-hybrid (Y1H) assays. We tested six core clock components, CCA1, LHY, PRR7, PRR9, TOC1, and GI, and found that PRR9, CCA1, and GI bind to the ORE1 promoter in yeast (SI Appendix, Fig. S5A). We then examined the ORE1 transactivation capacity of these three clock components using an Arabidopsis protoplast reporter assay. Among these three, PRR9 activated the ORE1 promoter in planta twofold or more compared with CCA1, GI, and vector alone (SI Appendix, Fig. S5B), suggesting that PRR9 is a direct activator of the ORE1 promoter. To investigate whether these transcription factors regulate diurnal ORE1 transcript levels, we monitored the expression of ORE1 in leaves from cca1-11, prr9-1, and gi-2 mutant plants versus wild type. We found that ORE1 mRNA levels were affected in prr9-1 and in gi-2 mutant plants, with the peak expression of ORE1 mRNA being reduced more than twofold in the mutants compared with that in wild type (SI Appendix, Fig. S6). Further, cyclic ORE1:LUC activity (Fig. 4 A and B) and overall ORE1 transcript levels (Fig. 4C) were greatly reduced in the prr9-1 mutant in the free-running condition of constant light. Together, these data suggest that PRR9 is a direct positive regulator of circadian ORE1 transcription.
Fig. 4.
ORE1 is directly regulated by PRR9, a core clock component. (A) ORE1 promoter activity under LL in Col-0 and the prr9-1 mutant. Luminescence intensity from ORE1:LUC in excised leaves (mean ± SEM, n = 24). (B) Relative amplitude error (RAE; a measure of the strength of rhythmicity, where RAE = 0 for a perfect sine wave and RAE = 1.0 defines the limit of a statistically significant oscillation) of ORE1:LUC in Col-0 and prr9-1 mutant was analyzed by fast Fourier transform-nonlinear least-squares (FTT-NLLS) analysis (Left, n = 24). Period lengths of ORE1:LUC were computed from an experiment in Col-0 and prr9-1 (Right). (C) mRNA abundance of ORE1 in Col-0 and prr9-1 (mean ± SEM, n = 3). ACT2, internal control. (D) ORE1 promoter binding affinity of PRR9 in a CsV:PRR9-GFP transgenic line. Diagram for the ORE1 amplicons (I–V) in ChIP assay are indicated. Red box indicates G box (CACGTG). Two-week-old seedlings were harvested 4 h after lights on. The fold enrichment is a ratio of CsV:PRR9-GFP normalized to CsV:GFP plants. The binding of PRR9-GFP to the CCA1 promoter is used as a positive control (mean ± SEM, n = 3). Asterisk indicate statistically significant difference from CsV:GFP (t test, *P < 0.05).
To identify the PRR9-binding domain(s) within the ORE1 promoter, we generated overexpressed PRR9 transgenic plants driven by the CsV promoter in the prr9-1 mutant to perform chromatin immunoprecipitation (ChIP)-PCR. CsV:PRR9 (PRR9-ox) rescued the delayed age-dependent leaf senescence phenotype of prr9-1 (SI Appendix, Fig. S7). In addition, ChIP-PCR experiments using prr9; PRR9-ox plants showed enrichment of PRR9 binding to the CCA1 promoter (Fig. 4D), consistent with earlier study (24). PRR9 associated with two regions within the ORE1 promoter (Fig. 4D). PRR9 transcriptionally represses several clock genes, including CCA1, by binding to G-box elements. The ORE1 promoter contains a single G-box element at −1376 bp (Fig. 4D). To test whether the G box in the ORE1 promoter is associated with transcriptional regulation of ORE1 by PRR9, we performed yeast one-hybrid assays with mutated G-box element and found that PRR9 binds to the mutated ORE1 promoter lacking the G box (SI Appendix, Fig. S5C). In a protoplast reporter assay, we found that PRR9-mediated transcriptional induction was about 2.5-fold for both the wild-type and mutated ORE1 promoters. However, the basal expression level of the mutated ORE1 promoter was elevated relative to the wild-type promoter (SI Appendix, Fig. S5D). Thus, we conclude that PRR9 positively regulates the ORE1 promoter through a novel mechanism distinct from binding the G-box element.
To further confirm the role of PRR9 on the expression of ORE1 as a transcriptional activator, we generated PRR9-LUC transgenic plants in which this translational fusion of LUC to the C terminus of PRR9 was under the control of an ecdysone agonist-inducible promoter, which allows inducible expression of PRR9-LUC with the chemical inducer, methoxyphenozide (MOF) (25). Compared with mock treatment (DMSO), MOF treatment not only decreased CCA1 transcript level, consistent with the direct repression of CCA1 expression by PRR9 as shown in a previous study (24), but also increased the ORE1 transcript level (SI Appendix, Fig. S5E). The increase in ORE1 expression in response to transient induction of PRR9 supports the notion that PRR9 acts as a direct transcriptional activator of ORE1.
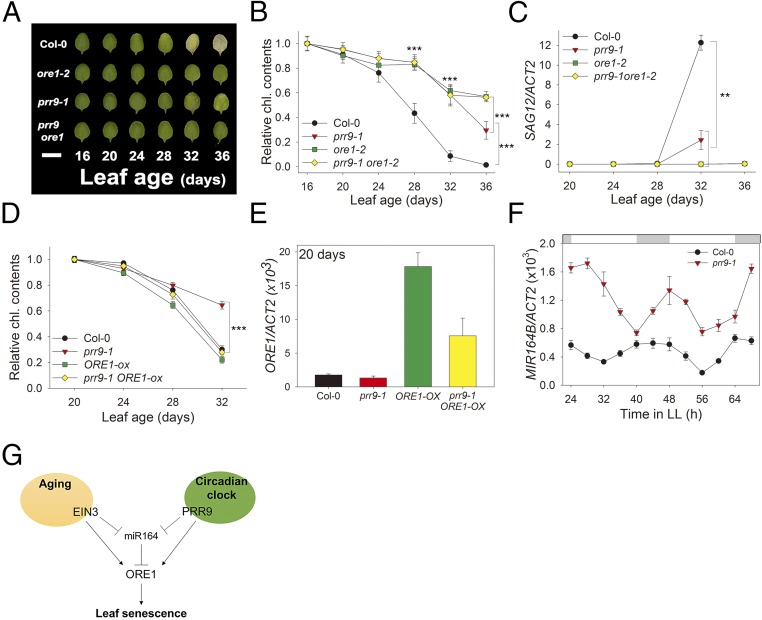
ORE1 Is Epistatic to PRR9 for Regulating Leaf Senescence.
The observation that PRR9 activates ORE1 transcription is consistent with the delayed leaf senescence phenotype of prr9-1 mutants (Figs. 1 and 2), as reduced expression of ORE1 also delayed senescence (Fig. 4C and SI Appendix, Fig. S6) (21). To elucidate the genetic relationship between PRR9 and ORE1 in controlling leaf senescence, we performed an age-dependent leaf senescence assay. Wild-type leaves displayed yellowing from 28 d of age, whereas prr9-1 mutant leaves remained green until 32 d of age. Importantly, ore1-2 and prr9; ore1 mutant leaves remained green until 36 d of age (Fig. 5A). These distinct leaf yellowing phenotypes were consistent with a quantitative assay of chlorophyll content (Fig. 5B), and with expression of SAG12 (Fig. 5C). These mutants incubated in the dark behaved the same as the mutants analyzed during age-dependent senescence (SI Appendix, Fig. S8), suggesting that PRR9 mediates age-dependent and dark-induced leaf senescence primarily through ORE1. To investigate whether PRR9 activates ORE1 to mediate senescence phenotypes, we tested whether ORE1 overexpression could rescue prr9 mutant phenotypes. Indeed, prr9; ORE1-ox plant leaves showed age-dependent senescence similar to wild-type and ORE1-ox plant leaves, revealing that ORE1 is epistatic to PRR9 for controlling leaf senescence (Fig. 5D).
Fig. 5.
PRR9 positively regulates leaf senescence via ORE1. (A) Chlorophyll loss in Col-0, ore1-2, prr9-1, and the prr9-1ore1-2 double mutant. The photographs show representative third and fourth rosette leaves at the indicated days after leaf emergence. (Scale bar: 1 cm.) (B) The chlorophyll contents of the indicated genotypes were measured from the third and fourth leaves at indicated days (mean ± SE, n = 10). Asterisks indicate statistically significant difference from Col-0 (t test, ***P < 0.001). (C) The expression of SAG12 gene in plants of the indicated genotypes at the indicated days (mean ± SEM, n = 3). Asterisks indicate statistically significant difference from Col-0 (t test, **P < 0.01). (D) The chlorophyll content of the third and fourth leaves of the indicated genotypes was measured at indicated days (mean ± SEM, n = 8). Asterisks indicate statistically significant difference from Col-0 (t test, ***P < 0.001). (E) The expression of ORE1 gene in plants of the indicated genotypes at the indicated days (mean ± SEM, n = 3). (F) Level of MIR164B transcript in Col-0 and prr9-1 in 20-d-old leaves under LL (mean ± SEM, n = 3). ACT2, internal control. (G) A trifurcate feed-forward pathway model for regulating leaf senescence by aging and circadian clock. PRR9 positively regulates ORE1 with a circadian rhythm at both the transcriptional and posttranscriptional levels. Posttranscriptional repression by clock-controlled miR164 negatively regulates ORE1 mRNA level. Aging activates ORE1 expression by a similar trifurcate feed-forward pathway; direct binding of EIN3 to the ORE1 and miR164 promoters activates ORE1 expression directly and indirectly, via repression of miR164, a repressor of ORE1, to promote age-dependent senescence.
PRR9 Indirectly Regulates ORE1 Expression via miR164.
Interestingly, we found that ORE1 transcript levels were reduced in 20-d-old prr9; ORE1-ox leaves compared with ORE1-ox leaves (Fig. 5E), suggesting that PRR9 also regulates ORE1 transcript abundance independently of direct transcriptional activation, possibly at the posttranscriptional level. Since expression of miR164 is also controlled by the circadian clock, we measured MIR164B transcript levels in young (3 wk old) wild-type and prr9 mutant leaves in LL. The amplitude of MIR164B was dramatically increased in the prr9 mutant compared with wild type (Fig. 5F), consistent with PRR9 functioning as a negative regulator of miR164 expression.
To test whether PRR9 regulates miR164 expression directly, we performed a Y1H assay and found that PRR9 binds to the MIR164B promoter region directly (SI Appendix, Fig. S9A). To evaluate the effect of PRR9 on the expression of miR164, we used inducible PRR9-LUC plants. In the presence of MOF, MIR164B transcript level was decreased (SI Appendix, Fig. S9B), indicating that PRR9 functions as a direct transcriptional repressor of miR164B. Collectively, these data suggest that PRR9 tightly regulates leaf senescence by dual control of ORE1, directly at the level of transcriptional activation of ORE1 and indirectly at the posttranscriptional level, via direct suppression of miR164, a repressor of ORE1. This coherent feed-forward loop between a circadian clock component and an aging regulator coordinates leaf senescence.
Discussion
In this report, we investigated the molecular interactions between the circadian clock and aging in Arabidopsis leaves. Several of the core circadian components affect age-dependent leaf senescence (Fig. 1). Age-dependent leaf senescence can be influenced by developmental processes such as flowering. Indeed, we found that the age-dependent leaf senescence phenotypes of clock mutants showed significant correlation with flowering time, as expected since the circadian clock controls photoperiodic flowering (15, 26). To identify clock components directly involved in senescence, independent of flowering, we evaluated the effect of clock mutants on dark-induced senescence. Three evening complex (EC) mutants (elf3, elf4, and lux) showed significantly early senescence in darkness, whereas prr9 mutants showed significantly delayed senescence under these conditions (Fig. 2). Because the evening complex is a known repressor of PRR9 expression (27, 28), we attribute the early senescence of EC mutants at least in part to a relief of inhibition of PRR9 and consequent induction of ORE1. As plant aging and senescence are highly programmed processes that recycle nutrients to sink sources such as developing seeds, positive regulation of senescence by the circadian clock may be essential for fitness in plants.
Circadian regulation of several plant-specific senescence-related transcription factors (Fig. 3 and SI Appendix, Fig. S2) raised the possibility that clock components directly regulate their expression. We found that ORE1 promoter activity and transcript levels show circadian rhythmic patterns (Figs. 3 and 4). ORE1 is posttranscriptionally regulated by miR164 (21). miRNAs also regulate the circadian system in animals, where expression of various miRNAs is clock regulated, mediating the circadian clock system by regulating core clocks, inputs, and outputs (29, 30). In Arabidopsis, several miRNAs (miR167, miR168, miR171, and miR398) show rhythmic expression patterns in diurnal conditions, but do not appear to be directly regulated by the circadian clock (31). Here, we showed that miR164 is under circadian regulation, is expressed in an antiphasic manner relative to its target, ORE1, and negatively regulates ORE1 (Fig. 3). Posttranscriptional regulation via miRNA in the circadian clock system thus represents a general regulatory mechanism in both plants and animals. Circadian expression of ORE1 is negatively regulated by clock-controlled miR164 at the posttranscriptional level, but positively regulated by PRR9, a core clock component, at the transcriptional level. Moreover, MIR164B levels were negatively regulated by PRR9 (Fig. 5F and SI Appendix, Fig. S9B). This regulatory mechanism has identical characteristics to the trifurcate feed-forward pathway for regulation of age-dependent senescence in Arabidopsis (Fig. 5G). EIN3 induces ORE1 in an age-dependent manner and simultaneously suppresses the expression of miR164, which negatively regulates ORE1 (21, 22). In this model, the circadian system activates expression of a key positive regulator of aging, ORE1, through a core circadian transcription factor, PRR9, thereby affecting the aging process (Fig. 5). ORE1 is under circadian control but its overall expression level increases with leaf age. On the other hand, ORE1 still shows a circadian expression pattern in the absence of PRR9 (Fig. 4 A and C), suggesting that other clock components contribute to ORE1 regulation. Recent studies have proposed other trifurcate feed-forward pathways for regulating leaf senescence in Arabidopsis (32, 33), suggesting that this regulation may be required to fine tune leaf senescence.
Materials and Methods
Plant Materials and Growth Condition.
All clock mutants used in this study: cca1-11, lhy-20, prr7-3, prr9-1, and toc1-101 (34), gi-2 and elf4-209 (35), and elf3-7 (36) are in the Columbia (Col-0) background. The ore1-2, mir164abc, and miR164B-ox line were described previously (21). To generate the ORE1:LUC transgenic line, a 2.4-kb ORE1 promoter was cloned into the gZPXomegaLUC vector (37) to fuse with the firefly luciferase gene and it was introduced into Col-0 plants by Agrobacterium tumefaciens-mediated transformation. All plants expressing luciferase were generated by genetic crossing. See SI Appendix, SI Materials and Methods for complete details.
Assay of Age-Dependent Senescence.
Leaf senescence was assayed as described previously with minor modifications (21). Chlorophyll was extracted from individual leaves by heating in 95% ethanol at 80 °C. The chlorophyll concentration per fresh weight of leaf tissue was calculated as described previously (38). Survival of the leaves in clock mutants was performed with leaf age; leaves with 50% of leaf area yellowed were counted as senesced. See SI Appendix, SI Materials and Methods for complete details.
Details of the experimental programs for analysis of gene expression, luminescence assay, yeast one-hybrid assay, transient expression assay, ChIP-qPCR assay, and chemical induction of PRR9 in inducible PRR9-LUC transgenic plants are provided in SI Appendix, SI Materials and Methods. The primers used in this study are listed in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
We thank K. H. Suh, M. J. Chae, J. Y. Kim, and S. Seo for their technical assistance; M. Yeom for generation of the inducible PRR9-LUC transgenic plants; and Life Science Editors for editorial assistance. This work was supported by Project Code (IBS-R013-D1) (to H.K., H.J.K., Q.T.V., S.J., S.H., and H.G.N.) and by the National Science Foundation (IOS-1257722) (to C.R.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722407115/-/DCSupplemental.
References
- 1.Jones OR, et al. Diversity of ageing across the tree of life. Nature. 2014;505:169–173. doi: 10.1038/nature12789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler PB, et al. Functional traits explain variation in plant life history strategies. Proc Natl Acad Sci USA. 2014;111:740–745. doi: 10.1073/pnas.1315179111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guarente L, Ruvkun G, Amasino R. Aging, life span, and senescence. Proc Natl Acad Sci USA. 1998;95:11034–11036. doi: 10.1073/pnas.95.19.11034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim PO, Kim HJ, Nam HG. Leaf senescence. Annu Rev Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- 5.Breeze E, et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell. 2011;23:873–894. doi: 10.1105/tpc.111.083345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merrow M, Spoelstra K, Roenneberg T. The circadian cycle: Daily rhythms from behaviour to genes. EMBO Rep. 2005;6:930–935. doi: 10.1038/sj.embor.7400541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 8.de Montaigu A, Tóth R, Coupland G. Plant development goes like clockwork. Trends Genet. 2010;26:296–306. doi: 10.1016/j.tig.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez SE, Kay SA. The plant circadian clock: From a simple timekeeper to a complex developmental manager. Cold Spring Harb Perspect Biol. 2016;8:a027748. doi: 10.1101/cshperspect.a027748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monk TH. Aging human circadian rhythms: Conventional wisdom may not always be right. J Biol Rhythms. 2005;20:366–374. doi: 10.1177/0748730405277378. [DOI] [PubMed] [Google Scholar]
- 11.Hofman MA, Swaab DF. Living by the clock: The circadian pacemaker in older people. Ageing Res Rev. 2006;5:33–51. doi: 10.1016/j.arr.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Dubrovsky YV, Samsa WE, Kondratov RV. Deficiency of circadian protein CLOCK reduces lifespan and increases age-related cataract development in mice. Aging (Albany NY) 2010;2:936–944. doi: 10.18632/aging.100241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee CC. The circadian clock and tumor suppression by mammalian period genes. Methods Enzymol. 2005;393:852–861. doi: 10.1016/S0076-6879(05)93045-0. [DOI] [PubMed] [Google Scholar]
- 15.Putterill J. Flowering in time: Genes controlling photoperiodic flowering in Arabidopsis. Philos Trans R Soc Lond B Biol Sci. 2001;356:1761–1767. doi: 10.1098/rstb.2001.0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchanan-Wollaston V, et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005;42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- 17.Sakuraba Y, et al. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun. 2014;5:4636. doi: 10.1038/ncomms5636. [DOI] [PubMed] [Google Scholar]
- 18.Woo HR, Kim HJ, Nam HG, Lim PO. Plant leaf senescence and death–Regulation by multiple layers of control and implications for aging in general. J Cell Sci. 2013;126:4823–4833. doi: 10.1242/jcs.109116. [DOI] [PubMed] [Google Scholar]
- 19.Carbon S, et al. AmiGO Hub; Web Presence Working Group AmiGO: Online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mockler TC, et al. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 21.Kim JH, et al. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science. 2009;323:1053–1057. doi: 10.1126/science.1166386. [DOI] [PubMed] [Google Scholar]
- 22.Kim HJ, et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J Exp Bot. 2014;65:4023–4036. doi: 10.1093/jxb/eru112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nohales MA, Kay SA. Molecular mechanisms at the core of the plant circadian oscillator. Nat Struct Mol Biol. 2016;23:1061–1069. doi: 10.1038/nsmb.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamichi N, et al. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koo JC, Asurmendi S, Bick J, Woodford-Thomas T, Beachy RN. Ecdysone agonist-inducible expression of a coat protein gene from tobacco mosaic virus confers viral resistance in transgenic Arabidopsis. Plant J. 2004;37:439–448. doi: 10.1046/j.1365-313x.2003.01869.x. [DOI] [PubMed] [Google Scholar]
- 26.Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T. Photoperiodic flowering: Time measurement mechanisms in leaves. Annu Rev Plant Biol. 2015;66:441–464. doi: 10.1146/annurev-arplant-043014-115555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helfer A, et al. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol. 2011;21:126–133. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixon LE, et al. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol. 2011;21:120–125. doi: 10.1016/j.cub.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Tian G, Li Z, Zheng L. The crosstalk between miRNA and mammalian circadian clock. Curr Med Chem. 2015;22:1582–1588. doi: 10.2174/0929867322666150227155009. [DOI] [PubMed] [Google Scholar]
- 30.Mehta N, Cheng HY. Micro-managing the circadian clock: The role of microRNAs in biological timekeeping. J Mol Biol. 2013;425:3609–3624. doi: 10.1016/j.jmb.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 31.Siré C, Moreno AB, Garcia-Chapa M, López-Moya JJ, San Segundo B. Diurnal oscillation in the accumulation of Arabidopsis microRNAs, miR167, miR168, miR171 and miR398. FEBS Lett. 2009;583:1039–1044. doi: 10.1016/j.febslet.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 32.Sakuraba Y, Kim YS, Han SH, Lee BD, Paek NC. The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell. 2015;27:1771–1787. doi: 10.1105/tpc.15.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song Y, et al. Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol Plant. 2014;7:1776–1787. doi: 10.1093/mp/ssu109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H, Kim Y, Yeom M, Lim J, Nam HG. Age-associated circadian period changes in Arabidopsis leaves. J Exp Bot. 2016;67:2665–2673. doi: 10.1093/jxb/erw097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim Y, et al. GIGANTEA and EARLY FLOWERING 4 in Arabidopsis exhibit differential phase-specific genetic influences over a diurnal cycle. Mol Plant. 2012;5:678–687. doi: 10.1093/mp/sss005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McWatters HG, Bastow RM, Hall A, Millar AJ. The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature. 2000;408:716–720. doi: 10.1038/35047079. [DOI] [PubMed] [Google Scholar]
- 37.Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA. A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell. 2001;13:2659–2670. doi: 10.1105/tpc.010332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lichtenthaler HK. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:350–382. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.