Significance
Snakes of the genus Thermophis are endemic to the Tibetan plateau and occur at elevations over 3,500 m and present an opportunity to study the genetics mechanisms of adaptation to high-elevation conditions in ectotherms. Here, we provide a de novo genome of the Tibetan hot-spring snake, Thermophis baileyi, and conduct a series of comparisons with other reptiles. We identify genes under positive selection and test properties of allelic variants of proteins that are involved in DNA damage repair and responses to hypoxia. Functional assays reveal convergent genetic mechanisms that underlie high-elevation adaptation in both endotherms and ectotherms.
Keywords: snakes, de novo genome, comparative genomics, positive selection, high-elevation adaptation
Abstract
Several previous genomic studies have focused on adaptation to high elevations, but these investigations have been largely limited to endotherms. Snakes of the genus Thermophis are endemic to the Tibetan plateau and therefore present an opportunity to study high-elevation adaptations in ectotherms. Here, we report the de novo assembly of the genome of a Tibetan hot-spring snake (Thermophis baileyi) and then compare its genome to the genomes of the other two species of Thermophis, as well as to the genomes of two related species of snakes that occur at lower elevations. We identify 308 putative genes that appear to be under positive selection in Thermophis. We also identified genes with shared amino acid replacements in the high-elevation hot-spring snakes compared with snakes and lizards that live at low elevations, including the genes for proteins involved in DNA damage repair (FEN1) and response to hypoxia (EPAS1). Functional assays of the FEN1 alleles reveal that the Thermophis allele is more stable under UV radiation than is the ancestral allele found in low-elevation lizards and snakes. Functional assays of EPAS1 alleles suggest that the Thermophis protein has lower transactivation activity than the low-elevation forms. Our analysis identifies some convergent genetic mechanisms in high-elevation adaptation between endotherms (based on studies of mammals) and ectotherms (based on our studies of Thermophis).
The Tibetan Plateau is the highest-elevation plateau on Earth, with an average elevation of more than 4,000 m. The inhospitality of its relatively extreme environment, including oxidative stress, UV radiation, and thermal extremes, has led to various adaptive responses in a variety of species. The biology of physiological responses to high-elevation stresses has been the subject of over a century of research (1). Recent advances in genomic technologies have opened up new opportunities to explore the genetic basis of adaptation to extreme habitats. Consequently, many recent studies have focused on genetic adaptations to high elevations, but most of these studies have been limited to endotherms (2–8), with only one genomic study of a high-elevation species of ectotherms, the frog Nanorana parkeri (9).
The genus Thermophis includes three closely related species (10): Thermophis baileyi, the Tibetan hot-spring snake; Thermophis zhaoermii, the Sichuan hot-spring snake; and Thermophis shangrila, the Shangri-La hot-spring snake. All three species are endemic to the Tibetan plateau and occur at elevations over 3,500 m (Fig. 1A). Although they are found around hot springs, these species still experience extreme environmental conditions, including low concentrations of molecular oxygen, high levels of UV radiation, and relatively dramatic fluctuations in temperature on a daily basis. Therefore, these species present an ideal opportunity to study the genetics of local adaptation to extreme conditions in ectotherms.
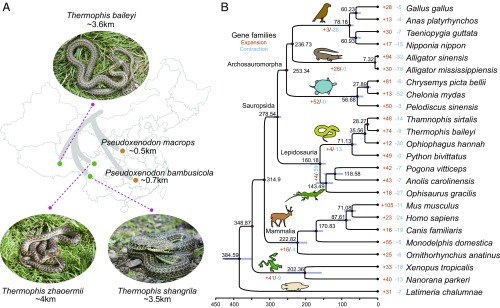
Fig. 1.
History of Thermophis snakes. (A) Geographical distribution and sampling locations of the snakes. (B) Phylogenetic position of Tibetan spring snake relative to other vertebrates. The branch lengths of the phylogenetic tree are scaled to estimated divergence time. Tree topology is supported by posterior probabilities of 1.0 for all nodes. The blue bars on the nodes indicate the 95% credibility intervals of the estimated posterior distributions of the divergence times. The red circles indicate the fossil calibration times used for setting the upper and lower bounds of the estimates. The number of significantly expanded (orange) and contracted (blue) gene families is designated on each branch.
Results and Discussion
Sequencing and Assembly of the Tibetan Hot-Spring Snake Genome.
A total of ∼325 Gb of clean Illumina HiSeq pair-ended reads was generated from a female T. baileyi, representing ∼185× coverage of the estimated 1.76-Gb genome (SI Appendix, Tables S1–S3). The resulting assembly is ∼1.74 Gb (98% of the estimated full genome) with a scaffold N50 value of 2.41 Mb (SI Appendix, Table S4) and is larger than those of the other snake species with sequenced genomes (SI Appendix, Table S10). Approximately 90% of the assembly was contained in the 889 longest scaffolds (>363 kb), with the largest spanning 18.66 Mb. This assembly captures more than 94% of the core eukaryotic genes (SI Appendix, Table S5). We identified ∼791 Mb of repetitive sequences, which are predominantly made up of LTRs and other unknown transposable elements (TEs) (SI Appendix, Table S12), comprising 45.28% of the T. baileyi genome assembly. This is the highest percentage of TE content among all five snakes with a sequenced genome (SI Appendix, Table S10). In total, 20,995 protein-coding genes were predicted using transcriptome sequencing data from five tissues, combined with de novo sequencing and homology-based strategies (detailed gene annotation methods are provided in SI Appendix).
Phylogenetic Relationships.
The available genomic datasets improve our understanding of the phylogeny and molecular evolution of these snakes, which have been difficult to place within the family Colubridae (10). Using 842 single-copy orthologous groups constructed from 12 reptilian genomes and 12 nonreptilian vertebrate genomes (SI Appendix, Table S19), we conducted Bayesian phylogenetic analyses (Fig. 1B), including time calibrations based on the fossil records (SI Appendix, Table S22). Our molecular clock analysis indicated that the split between Lepidosauria (snakes and lizards) and the Archosauromorpha (turtles, crocodilians, and birds) occurred at ∼278.5 Mya during the Permian period, which predates the Permian–Triassic extinction event (11) and is consistent with previous estimates (12). We also dated the divergence between the genus Thermophis and its colubrid relatives in the genus Thamnophis at ∼28 Mya and the divergence time of Thermophis-Python and Thermophis-Anolis at ∼71 Mya and ∼160 Mya, respectively.
Gene Family Evolution.
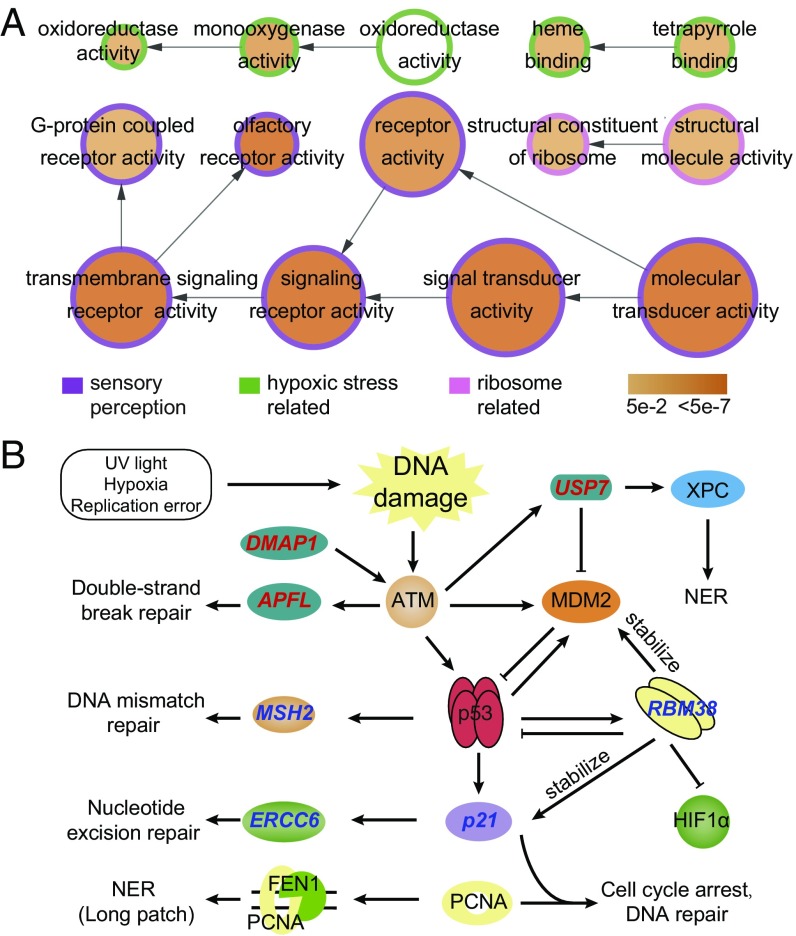
Frequent turnover of gene copy number has been proposed as a major mechanism underlying adaptive evolution (7, 13). Using OrthoMCL (14), we detected 92 gene clusters (including 354 genes) that were specific to the Tibetan hot-spring snake. These species-specific genes were significantly overrepresented in three major molecular functional categories (Fig. 2A and SI Appendix, Table S21). These categories include sensory perception (olfactory receptor activity, 32 genes, corrected P value = 4.00e-11; G protein-coupled receptor activity, 61 genes, corrected P value = 6.39e-03), reaction to hypoxic stress (such as heme binding, 27 genes, corrected P value = 5.34e-03; oxidoreductase activity, acting on paired donors, 11 genes, corrected P value = 6.96e-03), and ribosome-related genes (structural constituent of ribosome, 29 genes, corrected P value = 1.02e-02).
Fig. 2.
High-elevation adaptation in Tibetan hot-spring snake from comparative genomic analyses. (A) Overrepresented GO terms of Tibetan hot-spring snake-specific genes. Circle size is proportional to the number of genes in each category, whereas the color relates to the P value for the statistical significance of the enrichment. Links with low significance were removed manually to reduce complexity of the image. (B) Candidate targets of selection related to DNA damage repair in hot-spring snake. PSGs are labeled with red (upstream of p53 pathway) and blue (downstream).
We also identified 17 significantly expanded and two significantly contracted gene families in the Tibetan hot-spring snake compared with other vertebrates (Fig. 1B and SI Appendix, Table S23). The genes from the expanded gene families were mainly enriched in l-alanine-2-oxoglutarate aminotransferase activity (corrected P value = 2.74e-22), cholinesterase activity (corrected P value = 1.71e-16), heme binding (corrected P value = 7.07e-08), iron ion binding (corrected P value = 7.43e-07), oxidoreductase activity (corrected P value = 7.07e-08), monooxygenase activity (corrected P value = 0.04107), and carbon utilization (corrected P value = 5.03e-16) (SI Appendix, Table S24). The two contracted gene families were made up of olfactory receptor, family 5, subfamily U, member 1 and zinc finger protein 167 (SI Appendix, Table S25).
Genes Under Positive Selection.
Our first step in identifying potential genomic adaptations was to identify genes that are under positive selection in the Tibetan hot-spring snake lineages (15). We identified 308 positively selected genes (PSGs) in Tibetan hot-spring snake lineages (Materials and Methods and SI Appendix, Table S26). Of 517 genes annotated in the whole genome and involved in the “response to DNA damage stimulus” functional category, 12 showed evidence of positive selection in the Tibetan hot-spring snake [P = 0.0239, Fisher test in topGO (16)], including ERCC6, SMARCAL1, MEIOB, ING4, RBBP5, DMAP1, MSH2, p21, CCNT2, USP7, RBM38, and APLF (SI Appendix, Table S28). Interestingly, most of these genes are located upstream in the p53 pathway or are the targets of the p53 protein (Fig. 2B).
Three PSGs were identified on the upstream of p53 activating pathway (Fig. 2B). DMAP1 is required for ataxia telangiectasia mutated (ATM) activation (17) and is recruited to damaged sites to form complexes with gamma-H2AX and replication factors, including proliferating cell nuclear antigen (PCNA) (18). APLF promotes the assembly and activity of nonhomologous end-joining protein complexes (19, 20), which are critical for the maintenance of genetic integrity and repairing DNA double-strand breaks. USP7 is a direct antagonist of MDM2 and can deubiquitinate p53 and protect p53 from MDM2-mediated degradation. USP7 also regulates nucleotide excision repair (NER) by deubiquitinating the xeroderma pigmentosum complementation group C protein (XPC), which is a crucial damage-recognition factor that binds to helix-distorting DNA lesions and initiates NER and is ubiquitinated during the early stage of NER for UV-induced DNA lesions.
When DNA has sustained damage, p53 expression can be stimulated and then can activate DNA repair proteins, including MSH2 (21), ERCC6 (22), RBM38 (23), and p21 (24) (Fig. 2B). ERCC6 is one of the main enzymes involved in repairing the genome when specific genes undergoing transcription are damaged and serves as a transcription-coupled excision repair gene. In addition to the NER pathway, ERCC6 is involved in the base excision repair (BER) pathway by stimulating the apurinic/apyrimidinic (AP) site incision activity of AP endonuclease independently of adenosine triphosphate (25). RBM38 is a target of p53 and a negative regulator of p53 and MDM2 (26). Interestingly, RBM38 is required for maintaining the stability of the basal and stress-induced p21 transcript (27) and also regulates HIF1α expression via mRNA translation (23). MSH2 is a caretaker gene that is responsible for DNA mismatch repair and is involved in many different forms of DNA repair, including transcription-coupled repair (28), homologous recombination (29), and BER (30). p21 interacts with proliferating cell nuclear antigen (PCNA) and plays a regulatory role in S-phase DNA replication and DNA damage repair (31).
The exposure of Tibetan hot-spring snakes to UV radiation and hypoxia in high-altitude environments may cause DNA damage. The finding of a group of genes involved in DNA repair under positive selection is consistent with the high levels of exposure to UV radiation and hypoxia that Tibetan hot-spring snakes are subject to in high-altitude environments and suggests that these genes have functionally diverged between lowland and high-altitude snake lineages.
Shared Amino Acid Replacements in Thermophis Genomes.
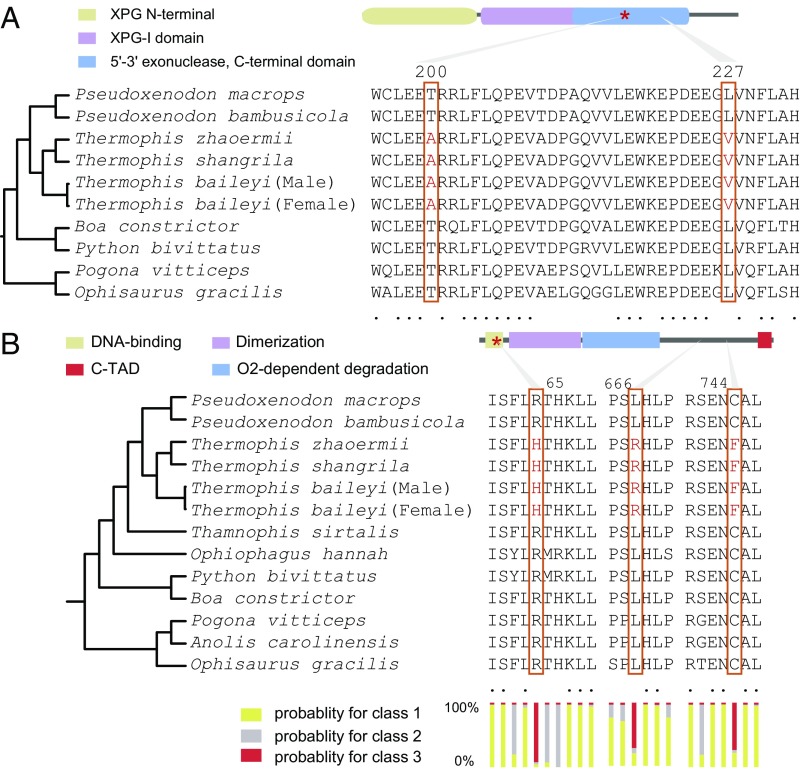
We next compared sequences of proteins identified as being under positive selection in high-elevation Thermophis to the sequences of the homologous proteins in nine low-elevation species of lizards and snakes (SI Appendix, Tables S2 and S10). We used the algorithms SIFT (32) and PolyPhen-2 (33) to examine the potential functional effects of amino acid replacements that were unique to Thermophis compared with the other species. We identified 27 unique amino acid replacements in 27 different proteins of Thermophis that were predicted to impact function by both the SIFT and PolyPhen-2 algorithms (SI Appendix, Fig. S10). Two of these proteins, NT5C2 and NT5DC3, are involved in 5′-nucleotidase activity; three proteins, RNF41, CARNS1, and FEN1 (Figs. 2B and 3A), have functional associations with reactive oxygen species and DNA damage; and five proteins, FKB1A, ITSN2, CHST1, WDFY4, and IL4RA, are related to immunity. Five of six members of the hypoxia-inducible factors (HIFs) superfamily, which are transcription factors that respond to decreases in available oxygen in the cellular environment, were found to have 11 shared amino acid replacements (Fig. 3B and SI Appendix, Fig. S11), and an amino acid replacement at position 65 of the endothelial PAS domain protein 1 (EPAS1) was predicted as possibly damaging by PolyPhen-2 (Fig. 3B).
Fig. 3.
Shared amino acid replacements in Thermophis genomes. (A) Variation in the FEN1 protein among two species of lizards and seven species of snakes. Two amino acid replacements in Thermophis compared with all low-elevation species of lizards and snakes are shown in red and marked with boxes. The red star marks the amino acid replacement that affects protein function as predicted by the SIFT and PolyPhen-2 algorithms. (B) Variation in the EPAS1 protein among three species of lizards and nine species of snakes. Three amino acid replacements in Thermophis compared with all lowland species of lizards and snakes are shown in red. The red star marks the amino acid replacement that affects protein function as predicted by the PolyPhen-2 algorithm. The lower bars indicate posterior probabilities for three different site classes as calculated using codeml of PAML. Class 1 sites are evolving under purifying selection (very low rate of nonsynonymous compared with synonymous substitutions), class 2 sites are evolving under neutral selection (similar rate of nonsynonymous and synonymous substitutions), and class 3 sites are under positive selection (high rate of nonsynonymous compared with synonymous substitutions).
Adaptation to UV Radiation.
Increased exposure to UV radiation in high-elevation environments may cause DNA damage, and DNA damage response and repair pathways may show functional adaptations. Indeed, we found changes in several genes involved in such pathways. Specifically, we identified two amino acid replacements in flap structure-specific endonuclease 1 (FEN1, Fig. 3A) of Thermophis compared with all of the lowland species of reptiles. This protein is involved in processing intermediates of Okazaki fragment maturation, long-patch BER, telomere maintenance, and stalled replication fork rescue (34). The FEN1–XPG complex also displays substantial NER activity in vivo (35). NER is a particularly important excision mechanism that removes DNA damage induced by UV.
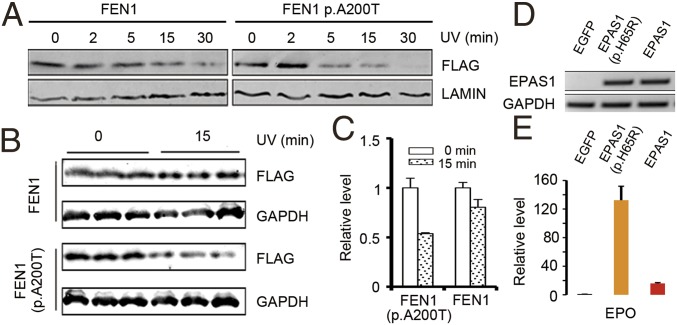
To test the potential functional differences of the FEN1 protein in Thermophis compared with all of the lowland species (Fig. 3A), we compared the in vitro stability of these two protein variants under UV radiation. The Thermophis protein has two amino acid replacements compared with the lowland species, one of which (an alanine-to-threonine replacement at position 200) is predicted by the SIFT (32) and PolyPhen-2 (33) algorithms to impact the protein’s function. We expressed the Tibetan Thermophis-specific allele of FEN1, as well as the allele found in all of the lowland species, in human embryonic kidney cells (HEK293) using plasmids. We then irradiated each cell type with UV light and measured each protein’s stability using immunoblots to detect protein concentration over exposure times. Our UV irradiation assays showed that the lowland protein variant of FEN1 is less stable than the Thermophis variant after irradiation with UV light, suggesting that the amino acid replacement in Thermophis at position 200 represents adaptation to high UV exposure (Fig. 4 A–C).
Fig. 4.
Functional assays of the FEN1 (A–C) and EAPS1 (D and E) variants shared by the high-elevation Thermophis snakes. (A) Immunoblots of N-terminally FLAG-tagged FEN1 (Thermophis allele) and FEN1 p.A200T (low-elevation allele) in a time course of HEK293 cells irradiated with UV of 40 J⋅m−2⋅min−1. LAMIN is shown as a loading control. The lysis solutions of the cells were immunoblotted with anti-FLAG or anti-LAMIN antibody. (B) Immunoblot of N-terminally FLAG-tagged FEN1 (Thermophis allele) and FEN1 p.A200T (low-elevation allele) of HEK293 cells irradiated with UV of 40 J⋅m−2⋅min−1 for 0 and 15 min. GAPDH is shown as a loading control. The lysis solutions of the cells were chosen to immunoblot with anti-FLAG or anti-GAPDH antibody, respectively. (C) Quantification of FEN1 (Thermophis allele) and FEN1 p.A200T (low-elevation allele) expression levels shown in B. Data are presented as the mean ± SEM. (D) Semiquantitative RT-PCR analysis confirmed that the plasmids were successfully transfected and the cDNA were successfully transcribed. The GAPDH is reference gene for semiquantitative RT-PCR analysis. EGFP is the negative control, EPAS1 (p.H65R) is the low-elevation allele, and EPAS1 is the Thermophis allele. (E) Real-time qPCR analysis of EPO expression in 293T cell line transfected with different plasmids. The cells transfected with the low-elevation allele plasmid had higher EPO expression level. EGFP is the negative control, EPAS1 (p.H65R) is the low-elevation allele, and EPAS1 is the Thermophis allele.
Hypoxia Adaption.
In their high-elevation environments, Thermophis species are exposed not only to higher UV radiation intensity but also to hypoxia. EPAS1, one of the six members of the HIF superfamily, has been demonstrated to be under selection for adaptation to hypoxia at high elevations in Tibetan mammalian species, including humans (6, 36) and the Tibetan Mastiff (37). We identified three shared amino acid replacements in EPAS1 of Thermophis, and the branch-site model implemented in PAML (38) revealed significant positive selection of the EPAS1 gene in these snakes (likelihood ratio test, P = 0.025; Fig. 3B). Among the three amino acid replacements, the histidine-to-arginine replacement at position 65 was predicted to be “damaging” to function by PolyPhen-2 (note that “damaging” in this context means “change in function”), and the codeml function of PAML found that this position was likely to be under positive selection for change (P = 0.922; Fig. 3B).
The amino acid replacement at position 65 of EPAS1 is located in the DNA-binding domain, where it is expected to affect the expression of the hypoxia-related protein erythropoietin (EPO) (39). To test the potential functional effects of this replacement in EPAS1, we compared EPAS1-targeted EPO expression in Thermophis compared with the lowland reptile species. Using three plasmids including pIRES2-EPAS1-EGFP (Thermophis allele), pIRES2-EPAS1p.His65Arg-EGFP (low-elevation allele), and pIRES2-EGFP (negative control) (Fig. 4D), our qPCR results showed that the endogenous expression of EPO in a 293T cell line (a variant cell line derived from human embryonic kidney cells 293) was much higher in cells transfected by the “low-elevation allele” plasmid than the “Thermophis allele” plasmid (Fig. 4E), reflecting much lower transactivation activity in the Thermophis protein variant.
Notably, EPAS1 mutations in patients presenting with polycythemia are associated with increased EPAS1 activity, increased protein half-life (40), and increased expression of hypoxia-related genes (41). Mutations in this gene are also associated with erythrocytosis (42), pulmonary hypertension, and chronic mountain sickness (43). However, there is also evidence that some variants of this gene provide protection for people living at high elevation (2, 6, 36). In Tibetan humans, the Tibetan EPAS1 allele is associated with lower erythrocyte quantities, and correspondingly lower hemoglobin levels. As elevated erythrocyte production is a common response to hypoxic stress, the Tibetan EPAS1 allele may have lower transactivation activity than the lowland allele and may be able to maintain sufficient oxygenation of tissues at high elevation without the need for increased erythrocyte levels (6). Therefore, the lower transactivation activity of EPAS1 in Thermophis suggests a similar adaption to high-elevation environments.
Our comparative genomics analyses have identified several convergent genetic mechanisms in high-elevation adaptation between endotherms and ectotherms. Moreover, the assembled genome of Thermophis, the first genome of a high-elevation squamate, provides useful genomic resources for further investigating adaptation to high elevations in ectotherms.
Materials and Methods
SI Appendix has additional information relating to the methodologies described below.
Materials and Genome Sequencing.
Blood samples acquired from a female Tibetan hot-spring snake (T. baileyi, sample name 1-13) captured in Yangbajing, Xizang, China were used for de novo genome sequencing. Whole-genome shotgun sequencing was employed and short paired-end inserts (280 bp and 450 bp) and long mate-paired inserts (2 kb, 5 kb, and 10 kb) were subsequently constructed using a standard protocol provided by Illumina. Six tissues (liver, brain, heart, lung, muscle, and ovary) were collected from the same Tibetan hot-spring snake individual (individual name 1-13), and total RNA was extracted from pooled tissues and a single cDNA library was constructed.
Samples used for whole-genome resequencing were acquired from three Thermophis and two false cobra (Pseudoxenodon) individuals. A male Tibetan hot-spring snake (T. baileyi) sample, a female Sichuan hot-spring snake (T. zhaoermii) sample, and a female Shangri-La hot-spring snake (T. shangrila) sample were obtained from Yangbajing, Xizang, China, Quhe village, Litang, Sichuan, China, and Tianshengqiao, Shangri-La, Yunnan, China, respectively. A female large-eyed false cobra (Pseudoxenodon macrops, www.iucnredlist.org/details/191926/0) and male bamboo false cobra (Pseudoxenodon bambusicola, www.iucnredlist.org/details/192230/0) were collected from Honghe, Yunnan and Quanzhou, Fujian, China, respectively. For each of the five resequenced snakes, two DNA libraries with an average insert size of 450 bp were constructed. Each library was prepared according to the appropriate Illumina’s protocols and was sequenced using the HiSEq 2000 instrument.
Genomes were assembled and then annotated using various bioinformatic tools. Detailed information is provided in SI Appendix.
Identification of Gene Families.
Protein sequences of 23 species of vertebrate were downloaded from Ensembl (release version 78) and NCBI (SI Appendix, Table S19). Only the longest transcript was selected for each gene locus with alternative splicing variants. Orthologous groups were constructed by ORTHOMCL (14) v2.0.9 using the default settings based on the filtered BLASTP results (SI Appendix, Table S20). Genes that could not be clustered into any gene family and for which only one species sample was available were species-specific within our sample. Gene Ontology (GO) terms that were statistically significantly overrepresented among the Tibetan hot-spring snake-specific genes were identified using BiNGO (44) in Cytoscape (45) by conducting a hypergeometric test. The entire GO annotations of the Tibetan hot-spring snake genes were assigned as a reference set, and the Benjamini and Hochberg false discovery rate (FDR) correction was applied (SI Appendix, Table S21).
Phylogenetic Tree Construction and Divergence Time Estimation.
Single-copy gene families were retrieved from the ORTHOMCL (14) results and used for phylogenetic tree construction. Conserved coding sequences (CDS) alignments of each single family were concatenated to generate a matrix of unambiguously aligned nucleotides. Fourfold degenerate nucleotide sites (4DTV) were extracted from these super genes, and MrBayes3.22 (46) was used to generate a Bayesian tree with the GTR+I+Γ model using 4DTV (SI Appendix, Fig. S9). The concatenated supergenes were separated into three categories that corresponded with the first, second, and third codon sites in the CDS. Divergence times were estimated under a relaxed clock model using the MCMCTREE program in the PAML4.7 package (38). We ran the program twice for each data type to confirm that the results were similar between runs. The constraints are listed in SI Appendix, Table S22.
Identification of Expanded and Contracted Gene Families.
Gene family expansion and contraction analyses were performed using CAFÉ 3.1 (47). For each branch and node, an “expanded and contracted gene family” with both an overall P value (family-wide P value) and an exact P value (Viterbi method) ≤0.01 was defined as a “significantly expanded and contracted gene family.” Significantly overrepresented GO terms among these significantly expanded gene families were identified using the topGO (16) package in R programming language (https://www.r-project.org/), and the Benjamini and Hochberg FDR correction was applied. Significantly overrepresented GO terms were identified with corrected P values of ≤0.05.
Identification of PSGs.
To identify potential PSGs in the Tibetan hot-spring snake lineage, gene families of the Tibetan hot-spring snake and seven other species [Gallus gallus, Alligator sinensis (48), Chelonia mydas (12), Thamnophis sirtalis (49), Python bivittatus (50), Ophiophagus hannah (51), and Anolis carolinensis (52)] were retrieved from the ORTHOMCL (14) results. Conserved CDS alignments of each single-copy gene family were extracted by Gblocks (53) and used for further identification of PSGs. The branch-site model of CODEML in PAML 4.7 (38) was used to test for potentially PSGs, with the Tibetan hot-spring snake set as the foreground branch and the others as background branches. A likelihood ratio test was then performed, and the P values were further corrected for multiple testing by conducting FDR test with a Bonferroni correction. The genes with corrected P value <0.01 and containing at least one positively selected site (posterior probability ≥0.99, Bayes empirical Bayes analysis) were defined as PSGs. Significantly overrepresented GO terms among the PSGs were identified using topGO (16), and significantly overrepresented GO terms were identified with corrected P values of ≤0.05.
Shared Amino Acid Substitutions in Hot-Spring Snakes.
Tibetan hot-spring snake proteomes were independently aligned with those of Ophisaurus gracilis (54), A. carolinensis (52), Pogona vitticeps (55), Ophiophagus hannah (51), P. bivittatus (50), Boa constrictor (platanus.bio.titech.ac.jp/platanus-assembler/platanus-1-2-1), and Thamnophis sirtalis (49) using BLASTP. Reciprocal best hits (RBHs) were extracted from each pair, and RBHs from each pair were merged into groups according to their homology with the Tibetan hot-spring snake. Groups with fewer than two lizard and two snake species were removed. Tibetan hot-spring snake-specific amino acid substitutions were extracted from the conserved amino acid alignments, and the amino acids at these substitution sites were checked in the five resequenced snakes using multiple genome alignments of the Tibetan hot-spring snake and the five resequenced genome assemblies. If the amino acids in one site were the same in all four hot-spring snake individuals and differed to those in the lizard and lowland snake species, they were defined as Thermophis-specific amino acid substitutions. The functional effects of these Thermophis-specific amino acid substitutions were further evaluated by SIFT (32) and PolyPhen-2 (33). In total, we identified 27 sites (from 27 genes) that were predicted as “DELETERIOUS” by SIFT and predicted as “damaging” by PolyPhen-2 (Fig. 3A and SI Appendix, Fig. S10 and Table S29). Sites of 12 PSGs that may play a role in DNA damage repair in the Tibetan hot-spring snake (SI Appendix, Table S28) were also checked in the five resequenced snakes (SI Appendix, Table S2).
Functional Assay of Flap Structure-Specific Endonuclease 1 (FEN1).
HEK293 cells were cultivated in DMEM (C11995500BT; Gibco) with 10% FBS (10099141; Gibco) and were then transiently transfected with pCMV-3×FLAG-FEN1 (Thermophis allele) or pCMV-3×FLAG-FEN1 p.Ala200Thr (low-elevation allele). Twenty-four hours after transfection (Lipofectamine 3000 Transfection Reagent, L3000015; Gibco), the cells were passage-cultured into five plates equally. After 48 h, the cells were irradiated with UV of 40 J⋅m−2⋅min−1 for 0, 2, 5, 15, and 30 min, harvested, and immediately lysed (RIPA Lysis Buffer, P0013B; Beyotime and PMSF, ST506; Beyotime), and subjected to immunoblot with anti-FLAG (1804; Sigma) or anti-LAMIN (ab83472; Abcam) or anti-GAPDH (KM9002T; SUNGENE BIOTECH) antibodies. Three biological replicates were used to produce grayscale images by Quantity One.
Functional Assay of Endothelial PAS Domain Protein 1 (EPAS1).
To investigate whether endogenous EPO transcriptional up-regulation differs between EPAS1 (Thermophis allele) and EPAS1 p.His65Arg (low-elevation allele), we constructed three plasmids including pIRES2-EPAS1-EGFP (Thermophis allele), pIRES2-EPAS1p.His65Arg-EGFP (low-elevation allele), and pIRES2-EGFP (negative control). These plasmids were overexpressed in 293T cells. For mRNA extraction, 5 × 104 293T cells were plated on six-well plates in triplicate, and when the cells reached 50% confluence the plasmids (1 μg per well) were transfected to the cells after 48 h using Lipofectamine 3000 (L3000015; Thermo Fisher).
A semiquantitative RT-PCR was performed using first-strand cDNA (RevertAid H Minus First Strand cDNA Synthesis Kit, K1631; Thermo) that was synthesized from total RNA samples (Total RNA Kit II, R6934-01; Omega) and GoTaq Colorless Master Mix (M7133; Promega) to ascertain whether the plasmids were successfully transfected. The amplified products were separated on 1% agarose gels, stained with Goldview (090804; SBS), and photographed.
Real-time qPCR was performed using SYBR Premix Ex Taq II (RR820A ; Takara) with first-strand cDNA to evaluate EPO expression. The primers used, annealing temperatures, and expected product sizes are described in SI Appendix, Table S30.
Supplementary Material
Acknowledgments
This study was supported by Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) Grant XDB31000000; National Natural Science Foundation of China Grants 31722049 and 31772434; Key Research Program of Frontier Sciences, CAS Grant QYZDB-SSW-SMC058; the Youth Innovation Promotion Association of CAS; Southeast Asia Biodiversity Research Institute, CAS Grant Y4ZK111B01; Fund of the State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, CAS Grant GREKF16-15; CAS Key Technology Talent Program (Y.-D.G.); and CAS President’s International Fellowship Initiative 2017DB0016 (D.M.H.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The hot-spring snakes raw reads and genome assembly reported in this paper have been deposited in the NCBI database (Bioproject no. PRJNA473624). The transcriptome and the whole-genome resequencing raw reads reported in this paper have been deposited in the NCBI Sequence Read Archive database (SRA accession no. SRP150039). This whole-genome shotgun project has been deposited in the GenBank database (accession no. QLTV00000000). The version described in this paper is version QLTV01000000.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805348115/-/DCSupplemental.
References
- 1.Cheviron ZA, Brumfield RT. Genomic insights into adaptation to high-altitude environments. Heredity (Edinb) 2012;108:354–361. doi: 10.1038/hdy.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall CM, et al. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci USA. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bigham A, et al. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6:e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng Y, et al. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol. 2011;28:1075–1081. doi: 10.1093/molbev/msq290. [DOI] [PubMed] [Google Scholar]
- 5.Simonson TS, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- 6.Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu Q, et al. The yak genome and adaptation to life at high altitude. Nat Genet. 2012;44:946–949. doi: 10.1038/ng.2343. [DOI] [PubMed] [Google Scholar]
- 8.Ge R-L, et al. Draft genome sequence of the Tibetan antelope. Nat Commun. 2013;4:1858. doi: 10.1038/ncomms2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y-B, et al. Whole-genome sequence of the Tibetan frog Nanorana parkeri and the comparative evolution of tetrapod genomes. Proc Natl Acad Sci USA. 2015;112:E1257–E1262. doi: 10.1073/pnas.1501764112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng L, Lu C, Huang S, Guo P, Zhang Y. A new species of the genus Thermophis (Serpentes: Colubridae) from Shangri-La, Northern Yunnan, China, with a proposal for an eclectic rule for species delimitation. Asian Herpetol Res. 2014;5:228–239. [Google Scholar]
- 11.Chen Z, Benton M. The timing and pattern of biotic recovery following the end-Permian mass extinction. Nat Geosci. 2012;10:375–383. [Google Scholar]
- 12.Wang Z, et al. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat Genet. 2013;45:701–706. doi: 10.1038/ng.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 14.Fischer S, et al. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics. 2011;Chap 6:Unit 6.12.1-19. doi: 10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y, et al. Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc Natl Acad Sci USA. 2017;114:1081–1086. doi: 10.1073/pnas.1613870114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexa A, Rahnenfuhrer J. 2010. topGO: Enrichment Analysis for Gene Ontology, R Package Version 2(0) (Bioconductor)
- 17.Penicud K, Behrens A. DMAP1 is an essential regulator of ATM activity and function. Oncogene. 2014;33:525–531. doi: 10.1038/onc.2012.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Negishi M, Chiba T, Saraya A, Miyagi S, Iwama A. Dmap1 plays an essential role in the maintenance of genome integrity through the DNA repair process. Genes Cells. 2009;14:1347–1357. doi: 10.1111/j.1365-2443.2009.01352.x. [DOI] [PubMed] [Google Scholar]
- 19.Fenton AL, Shirodkar P, Macrae CJ, Meng L, Koch CA. The PARP3- and ATM-dependent phosphorylation of APLF facilitates DNA double-strand break repair. Nucleic Acids Res. 2013;41:4080–4092. doi: 10.1093/nar/gkt134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grundy GJ, et al. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 2013;32:112–125. doi: 10.1038/emboj.2012.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scherer SJ, Welter C, Zang K-D, Dooley S. Specific in vitro binding of p53 to the promoter region of the human mismatch repair gene hMSH2. Biochem Biophys Res Commun. 1996;221:722–728. doi: 10.1006/bbrc.1996.0663. [DOI] [PubMed] [Google Scholar]
- 22.Yu A, Fan H-Y, Liao D, Bailey AD, Weiner AM. Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2, and 5S genes. Mol Cell. 2000;5:801–810. doi: 10.1016/s1097-2765(00)80320-2. [DOI] [PubMed] [Google Scholar]
- 23.Cho S-J, et al. Hypoxia-inducible factor 1 alpha is regulated by RBM38, a RNA-binding protein and a p53 family target, via mRNA translation. Oncotarget. 2015;6:305–316. doi: 10.18632/oncotarget.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouvard V, et al. Tissue and cell-specific expression of the p53-target genes: Bax, fas, mdm2 and waf1/p21, before and following ionising irradiation in mice. Oncogene. 2000;19:649–660. doi: 10.1038/sj.onc.1203366. [DOI] [PubMed] [Google Scholar]
- 25.Wong H-K, et al. Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007;35:4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Xu E, Chen X. Regulation of Mdm2 mRNA stability by RNA-binding protein RNPC1. Oncotarget. 2013;4:1121–1122. doi: 10.18632/oncotarget.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shu L, Yan W, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev. 2006;20:2961–2972. doi: 10.1101/gad.1463306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellon I, Rajpal DK, Koi M, Boland CR, Champe GN. Transcription-coupled repair deficiency and mutations in human mismatch repair genes. Science. 1996;272:557–560. doi: 10.1126/science.272.5261.557. [DOI] [PubMed] [Google Scholar]
- 29.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 30.Pitsikas P, Lee D, Rainbow AJ. Reduced host cell reactivation of oxidative DNA damage in human cells deficient in the mismatch repair gene hMSH2. Mutagenesis. 2007;22:235–243. doi: 10.1093/mutage/gem008. [DOI] [PubMed] [Google Scholar]
- 31.Karimian A, Ahmadi Y, Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 33.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chap 7:Unit7.20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balakrishnan L, Bambara RA. Flap endonuclease 1. Annu Rev Biochem. 2013;82:119–138. doi: 10.1146/annurev-biochem-072511-122603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hohl M, et al. Domain swapping between FEN-1 and XPG defines regions in XPG that mediate nucleotide excision repair activity and substrate specificity. Nucleic Acids Res. 2007;35:3053–3063. doi: 10.1093/nar/gkm092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanaoka M, et al. Genetic variants in EPAS1 contribute to adaptation to high-altitude hypoxia in Sherpas. PLoS One. 2012;7:e50566. doi: 10.1371/journal.pone.0050566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miao B, Wang Z, Li Y. Genomic analysis reveals hypoxia adaptation in the Tibetan mastiff by introgression of the gray wolf from the Tibetan plateau. Mol Biol Evol. 2017;34:734–743. doi: 10.1093/molbev/msw274. [DOI] [PubMed] [Google Scholar]
- 38.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 39.Patel SA, Simon MC. Biology of hypoxia-inducible factor-2α in development and disease. Cell Death Differ. 2008;15:628–634. doi: 10.1038/cdd.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhuang Z, et al. Somatic HIF2A gain-of-function mutations in paraganglioma with polycythemia. N Engl J Med. 2012;367:922–930. doi: 10.1056/NEJMoa1205119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang C, et al. Novel HIF2A mutations disrupt oxygen sensing, leading to polycythemia, paragangliomas, and somatostatinomas. Blood. 2013;121:2563–2566. doi: 10.1182/blood-2012-10-460972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Percy MJ, et al. Novel exon 12 mutations in the HIF2A gene associated with erythrocytosis. Blood. 2008;111:5400–5402. doi: 10.1182/blood-2008-02-137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gale DP, Harten SK, Reid CD, Tuddenham EG, Maxwell PH. Autosomal dominant erythrocytosis and pulmonary arterial hypertension associated with an activating HIF2 α mutation. Blood. 2008;112:919–921. doi: 10.1182/blood-2008-04-153718. [DOI] [PubMed] [Google Scholar]
- 44.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 45.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 47.De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: A computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 48.Wan Q-H, et al. Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res. 2013;23:1091–1105. doi: 10.1038/cr.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castoe TA, et al. A proposal to sequence the genome of a garter snake (Thamnophis sirtalis) Stand Genomic Sci. 2011;4:257–270. doi: 10.4056/sigs.1664145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castoe TA, et al. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci USA. 2013;110:20645–20650, and erratum (2014) 111:3194. doi: 10.1073/pnas.1314475110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vonk FJ, et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci USA. 2013;110:20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alföldi J, et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477:587–591. doi: 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 54.Song B, et al. A genome draft of the legless anguid lizard, Ophisaurus gracilis. Gigascience. 2015;4:17. doi: 10.1186/s13742-015-0056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Georges A, et al. High-coverage sequencing and annotated assembly of the genome of the Australian dragon lizard Pogona vitticeps. Gigascience. 2015;4:45. doi: 10.1186/s13742-015-0085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.