Significance
Plant pathogens, such as the infamous potato blight agent Phytophthora infestans, deliver effectors inside living plant cells to promote disease. Recent evidence suggests that some effectors will associate with endogenous negative regulators of immunity, or so-called susceptibility (S) factors, inside living plant cells. To date, little is known about how effectors exploit S factors to inactivate the immune system. We discovered that P. infestans effector Pi02860 uses the potato StNRL1 protein as an S factor by promoting its ability to target a positive regulator of immunity, StSWAP70, for proteasome-mediated degradation. Our results reveal that protecting StSWAP70 from degradation by reducing the activity or availability of NRL1 provides a means to attenuate late blight disease.
Keywords: potato blight, effector-triggered susceptibility, disease resistance, pathogenicity, virulence
Abstract
Plant pathogens deliver effectors into plant cells to suppress immunity. Whereas many effectors inactivate positive immune regulators, other effectors associate with negative regulators of immunity: so-called susceptibility (S) factors. Little is known about how pathogens exploit S factors to suppress immunity. Phytophthora infestans RXLR effector Pi02860 interacts with host protein NRL1, which is an S factor whose activity suppresses INF1-triggered cell death (ICD) and is required for late blight disease. We show that NRL1 interacts in yeast and in planta with a guanine nucleotide exchange factor called SWAP70. SWAP70 associates with endosomes and is a positive regulator of immunity. Virus-induced gene silencing of SWAP70 in Nicotiana benthamiana enhances P. infestans colonization and compromises ICD. In contrast, transient overexpression of SWAP70 reduces P. infestans infection and accelerates ICD. Expression of Pi02860 and NRL1, singly or in combination, results in proteasome-mediated degradation of SWAP70. Degradation of SWAP70 is prevented by silencing NRL1, or by mutation of Pi02860 to abolish its interaction with NRL1. NRL1 is a BTB-domain protein predicted to form the substrate adaptor component of a CULLIN3 ubiquitin E3 ligase. A dimerization-deficient mutant, NRL1NQ, fails to interact with SWAP70 but maintains its interaction with Pi02860. NRL1NQ acts as a dominant-negative mutant, preventing SWAP70 degradation in the presence of effector Pi02860, and reducing P. infestans infection. Critically, Pi02860 enhances the association between NRL1 and SWAP70 to promote proteasome-mediated degradation of the latter and, thus, suppress immunity. Preventing degradation of SWAP70 represents a strategy to combat late blight disease.
Plant innate immunity consists of two inducible layers that provide defense against microbial infections. The first of these, pathogen-associated molecular pattern (PAMP/Pattern)-triggered immunity (PTI), involves the detection of conserved microbial molecules, or the damage-associated products of microbial activity, by pattern-recognition receptors at the cell surface. Microbes successfully colonize plants by delivering effector proteins either to the inside (intracellular or cytoplasmic effectors) or outside (apoplastic effectors) of plant cells to attenuate PTI (1–3). The second inducible layer of immunity (effector-triggered immunity or ETI) involves the perception of effectors by cytoplasmic nucleotide-binding, leucine-rich repeat (NB-LRR) resistance (R) proteins (4). Effectors exploit a range of mechanisms to manipulate host immunity. Thus, identification of the targets of effectors in the host is essential to understand pathogen virulence and plant susceptibility. Many effector targets have been identified (1, 2, 5). The majority are positive regulators of immunity whose activity is suppressed as a consequence of effector interaction. In contrast, some targets either negatively regulate immunity or otherwise support a compatible interaction that benefits the pathogen. These targets in the host can be regarded as susceptibility (S) factors (6, 7).
Fungi and oomycetes are among the most damaging pathogens in agriculture. The oomycete Phytophthora infestans causes the devastating disease late blight on potato (Solanum tuberosum) (8, 9). P. infestans delivers so-called RXLR effectors into plant cells to modify host proteins to suppress immunity (5, 10). Some RXLR effectors target S factors. Examples include: the RXLR effector Pi04089, which interacts with a K-homology RNA-binding protein, KRBP1, to promote susceptibility (11); Pi04314/RD24, which interacts with three host isoforms of protein phosphatase 1 (PP1) catalytic subunits, forming unique holoenzymes that negatively regulate defense (12); and AVR2, which up-regulates a brassinosteroid-responsive basic helix-loop-helix transcription factor that suppresses immunity (13). Altering S-gene expression is a strategy particularly adopted and expanded by Xanthomonas species. For example, five Xanthomonas TAL effectors target the sugar transporter genes OsSWEET11 and OsSWEET14 that are S factors (14–16). Effectors from other bacteria (17–19) and from nematodes (20, 21) have also been reported to target S factors. However, little is understood about precisely how S factors are exploited by pathogen effectors to undermine immunity.
In a previous study (22) we identified a host target of RXLR effector Pi02860-potato NPH3/RPT2-LIKE1 protein (NRL1), a predicted substrate adaptor component of a CULLIN3-associated ubiquitin E3 ligase. NRL1 is an S factor, and is required for Pi02860 to enhance pathogen leaf colonization and suppress cell death triggered by perception of the P. infestans PAMP INF1 (22). However, the underlying basis for the susceptibility promoted by NRL1 is unknown. In this study, we performed a yeast two-hybrid (Y2H) screen that identified the guanine nucleotide exchange factor (GEF) SWAP70 as a candidate substrate of NRL1. SWAP70 acts as a positive regulator of immunity against P. infestans by promoting INF1-trigged cell death (ICD). We reveal that host protein NRL1 acts as an endogenous negative regulator of immunity by promoting proteasome-mediated turnover of SWAP70. Further analyses showed that Pi02860 promotes NRL1-mediated degradation of SWAP70. Loss of NRL1 by virus-induced gene silencing (VIGS) or expression of a dominant-negative dimerization-deficient NRL1 mutant in Nicotiana benthamiana prevented the degradation of SWAP70 by Pi02860. A mutation of Pi02860 that abolishs its interaction with NRL1 also prevent the degradation of SWAP70. Our results reveal how pathogens can exploit the endogenous negative regulators of immunity in the plant to promote disease.
Results
StNRL1 Forms Homodimers and Interacts with the Potato GEF StSWAP70.
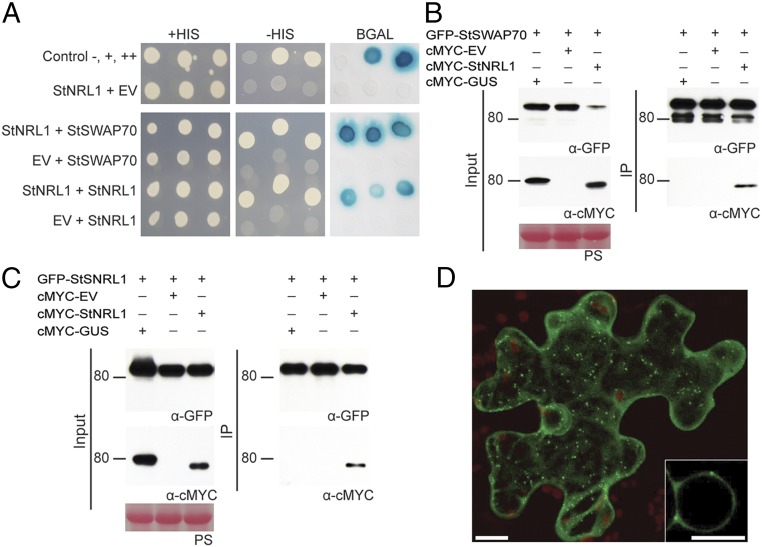
To explore the mechanism of Pi02860 action in plants, a Y2H screen was performed to identify candidate interactors of StNRL1. The Y2H library, made from cDNA of potato infected with P. infestans (23), was screened with a GAL4 DNA-binding domain–StNRL1 fusion (bait) construct to a depth of 4.6 × 106 yeast cotransformants. Thirty-four yeast colonies were recovered from selection plates that contained GAL4 activation domain (prey) fusions. These included seven clones containing 14-3-3 proteins, and one clone each encoding a protein phosphatase 2A regulatory subunit and a HAT4-like homeobox-leucine zipper protein (SI Appendix, Table S1). In addition, 11 clones yielded sequences corresponding to a potato plekstrin homology-diffuse β lymphoma homology (PH-DH) domain protein (SI Appendix, Table S1). The protein is highly similar to SWAP70 from rice and Arabidopsis (SI Appendix, Fig. S1), and is thus hereafter referred to as StSWAP70 (S. tuberosum SWAP70). SWAP70, a GEF, was reported to play a positive role in regulating both PTI and ETI (24, 25). It is thus a potential indirect target of the effector Pi02860 through its association with the S factor StNRL1, which is a predicted ubiquitin E3 ligase. We thus focused on SWAP70 in this study as a potential substrate of StNRL1. To confirm this interaction in yeast, a full-length StSWAP70 prey construct was tested pairwise with bait constructs for StNRL1 and the empty bait vector. While all transformants grew on the control plates, only yeast containing both StSWAP70 and StNRL1 were able to grow on the selection plates and activate the β-galactosidase (β-Gal) reporter (Fig. 1A).
Fig. 1.
StNRL1 interacts with itself and StSWAP70 in yeast and in planta and localizes to vesicles. (A) Yeast coexpressing StNRL1 with itself or StSWAP70 grew on −HIS medium and yielded β-Gal activity, while those coexpressed with the control EV did not. The +HIS control shows all yeast were able to grow in the presence of histidine. (B and C) The interactions were further confirmed by co-IP assay using expression in N. benthamiana leaves. cMYC-StNRL1 associated with GFP-StSWAP70 and GFP-StNRL1, whereas cMYC-GUS did not. Expression of constructs in the leaves is indicated by a “+.” Protein size markers are indicated in kilodaltons, and protein loading is indicated by Ponceau stain (PS). (D) Confocal images showing that GFP-StSWAP70 is localized in the cytoplasm and in vesicles. (Scale bars, 10 μM.) The Inset is an enlarged image of the nucleus.
In addition, the Y2H screen with StNRL1 also generated 14 yeast cotransformants containing prey constructs expressing StNRL1 itself (SI Appendix, Table S1). Yeast expressing StNRL1 in both bait and prey vectors were able to grow on selection plates and activate the β-Gal reporter (Fig. 1A), suggesting that StNRL1 forms a homodimer. This is consistent with the role of BTB/POZ domain-containing proteins, which form homodimers as a core component of a CUL3-based ubiquitin protein ligase (E3) enzyme complex (26, 27).
To confirm that these interactions also occur in planta, coimmunoprecipitation (co-IP) assays were performed by expressing either GFP-StSWAP70 or GFP-StNRL1 with cMYC-GUS, cMYC-EV, or cMYC-StNRL1, pulling down with GFP-TRAP_M beads. Whereas all proteins were present in the relevant input samples, only cMYC-StNRL1 was coimmunoprecipitated in the presence of GFP-StSWAP70 (Fig. 1B and SI Appendix, Fig. S1) or GFP-StNRL1 (Fig. 1C and SI Appendix, Fig. S1). Previously, we showed that cMYC-StNRL1 did not bind nonspecifically to GFP beads (22). Thus, StNRL1 specifically associates with itself and with StSWAP70 in planta.
StSWAP70 Localizes to Endosomes.
GFP was fused to the N terminus of StSWAP70 (GFP-StSWAP70) and expressed in N. benthamiana. The fusion protein was observed to localize to vesicles, with cytoplasmic background (Fig. 1D). To further characterize the nature of the vesicles, a series of colocalization experiments were conducted with subcellular markers representing components of the endocytic cycle. Whereas GFP-StSWAP70 did not colocalize with a Golgi marker (ST-mRFP), it did strongly colocalize with Ara6 and Ara7 (SI Appendix, Fig. S2), which belong to the RAB5 GTPase family and are key regulators of endosomal trafficking, endocytosis, and vacuolar transport (28–30). Ara6-mRFP and mRFP-Ara7 have distinct but overlapping localization patterns (30) and label a variety of endosomal compartments, including early endosomes. We thus included the specific prevacuolar compartment (late endosome) marker PS1 cyan fluorescent protein (CFP) (31). We observed considerable colocalization of GFP-StSWAP70 with the prevacuolar compartment marker, demonstrating that late endosomes also are a site of GFP-StSWAP70 localization (SI Appendix, Fig. S2).
SWAP70 Positively Regulates Plant Immunity.
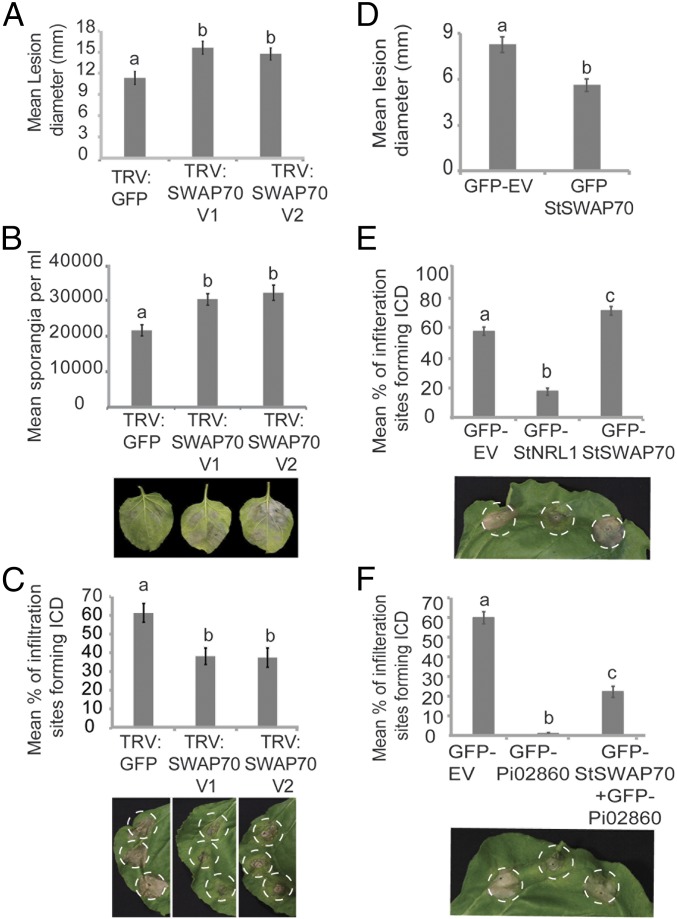
VIGS and transient overexpression were used to investigate the function of SWAP70 in immunity against P. infestans. VIGS was used to knock down the expression of the equivalent SWAP70 gene in N. benthamiana, which is a model host plant for late blight (5). A sequence, designated NbSWAP70, was identified in the N. benthamiana genome, encoding a protein 99% identical to StSWAP70 (SI Appendix, Fig. S1). Two independent VIGS constructs, TRV:SWAP70 V1 and TRV:SWAP70 V2, were generated to specifically silence NbSWAP70 by cloning two portions of the gene in the possible DH domain, which is downstream of the PH domain (SI Appendix, Fig. S3A). qRT-PCR was used to test silencing levels in each of three independent biological replicates and showed that transcript levels of NbSWAP70 were reduced by 50–80% in plants expressing each TRV:SWAP70 construct compared with plants expressing the TRV:GFP control (SI Appendix, Fig. S3B).
N. benthamiana plants expressing either TRV:SWAP70 V1 or TRV:SWAP70 V2 showed enhanced P. infestans leaf colonization, with significantly larger disease lesions (ANOVA, P < 0.001) developing compared with the TRV:GFP control plants (Fig. 2A). Moreover, significantly more P. infestans sporangia (ANOVA, P < 0.001) were produced (Fig. 2B). This indicates that StSWAP70 is detrimental to P. infestans infection.
Fig. 2.
StSWAP70 is a positive regulator of immunity. (A) VIGS of NbSWAP70 enhances P. infestans leaf colonization. Graph shows increased P. infestans lesion sizes on plants expressing TRV:SWAP70 V1 or TRV:SWAP70 V2, compared with a TRV:GFP control (ANOVA, P < 0.002; n = 228). (B) Graph showing an increase in the average numbers of sporangia per milliliter recovered from infected leaves of plants expressing TRV:SWAP70 V1 or TRV:SWAP70 V2, compared with the TRV:GFP control plants (ANOVA, P < 0.001; n = 33). Example leaves below the graph show P. infestans lesion development on control, TRV:NRL1 V1, or TRV:NRL1 V2 plants. (C) SWAP70 VIGS results in a significant decrease in ICD in TRV:SWAP70 V1 and TRV:SWAP70 V2 plants compared with the TRV:GFP control at 6 dpi (ANOVA, P < 0.001; n = 55). Example leaves below the graph show INF1 cell death development on TRV:GFP control, TRV:NRL1 V1, or TRV:NRL1 V2 plants (left to right). (D) Overexpression of GFP-StSWAP70 significantly decreased P. infestans lesion size compared with the GFP control empty (GFP-EV) (ANOVA, P < 0.001; n = 155). (E) Transient overexpression of GFP-StSWAP70 in N. benthamiana accelerates ICD (ANOVA, P < 0.001; n = 56) compared with free GFP or GFP-StNRL1. Representative leaf image below the graph shows ICD following overexpression of each construct, as indicated, in N. benthamiana. (F) The transient overexpression of StSWAP70 reduces Pi02860 suppression of ICD (ANOVA, P < 0.001; n = 56). Below the graph is a representative leaf image showing ICD following overexpression of each construct, as indicated, in N. benthamiana. The results shown in A to F are combinations of at least five individual biological replicates and error bars show ±SE. Letters on the graphs denote statistically significant differences (ANOVA).
Because both Pi02860 and StNRL1 were observed to suppress ICD when transiently expressed in N. benthamiana (22), this prompted us to investigate whether StSWAP70 is required for ICD. Constructs expressing INF1 were agroinfiltrated into plants expressing each of the VIGS constructs and cell death was scored at 6 d postinoculation (dpi). At 6 dpi, a significant decrease (ANOVA, P < 0.001) in ICD was observed on TRV:SWAP70 VIGS plants compared with TRV:GFP control plants (Fig. 2C). The same assay was carried out to examine the cell death triggered by coexpression of the tomato Cf4 resistance gene with Cladosporium fulvum Avr4, which was shown not to be suppressed by either Pi02860 or StNRL1 (22). As anticipated, there were no significant differences in Cf4-Avr4 cell death in TRV:SWAP70 plants compared with TRV:GFP plants at 6 dpi (SI Appendix, Fig. S3 C and D). These results confirm observations in the literature that SWAP70 is a positive regulator of immunity (24, 25) and reveal that it is required for ICD.
We investigated whether StSWAP70 overexpression could alter levels of P. infestans colonization or ICD. Transient expression in N. benthamiana of GFP-StSWAP70, followed by pathogen challenge, was found to result in significantly smaller P. infestans lesions (ANOVA, P < 0.05) compared with free GFP expression (Fig. 2D), which is consistent with it acting as a positive regulator of immunity. Moreover, GFP-StSWAP70 coexpression with INF1 significantly (ANOVA, P < 0.001) accelerated ICD compared with free GFP (Fig. 2E), whereas GFP-StNRL1 expression had a significantly suppressive effect on ICD (ANOVA, P < 0.001), as shown recently (22).
Previously, we demonstrated that Pi02860 targets the S factor StNRL1 to suppress ICD (22). We thus investigated whether StSWAP70 overexpression had any effect on the ability of Pi02860 to suppress ICD. Indeed, we observed that the strong suppression of ICD by GFP-Pi02860 was reduced significantly (ANOVA, P < 0.001) by coexpression with GFP-StSWAP70 (Fig. 2F). Taken together, VIGS and transient overexpression of StSWAP70 reveal that it is a positive regulator of the ICD immune response and thus contributes to impeding P. infestans infection. This raises the question of whether StSWAP70 is the true virulence target of P. infestans, and whether the S factor StNRL1 is an endogenous negative regulator of StSWAP70 that is exploited by the effector Pi02860.
Pi02860 and StNRL1 Promote StSWAP70 Turnover.
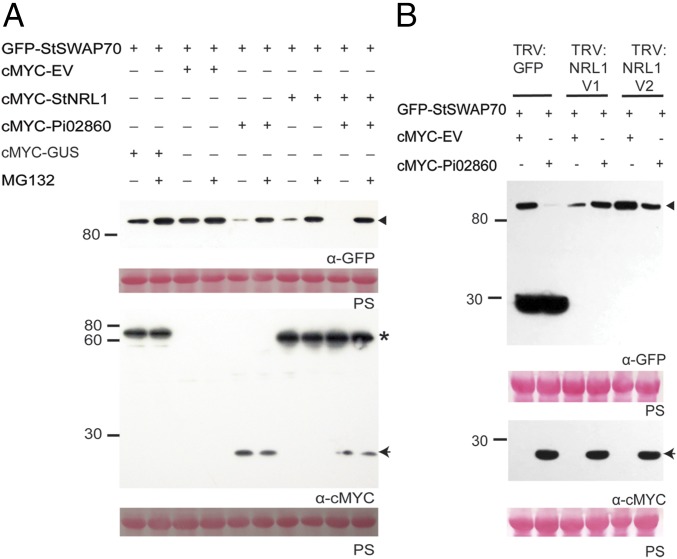
To ascertain the biological significance in defense suppression of the interaction between putative ubiquitin E3 ligase StNRL1 and StSWAP70, we investigated the protein levels of SWAP70 in N. benthamiana when coexpressed with StNRL1 in the presence or absence of Pi02860, and also with or without treatment with the 26S proteasome inhibitor MG132. Immunoblot analysis revealed that the abundance of GFP-StSWAP70 was reproducibly decreased when coexpressed with cMYC-Pi02860 or cMYC-StNRL1, but this effect was prevented by MG132. No such reduction in SWAP70 level was seen when GFP-StSWAP70 was coexpressed with cMYC-GUS, or coexpressed with cMYC-empty vector (EV) (Fig. 3A and SI Appendix, Fig. S4). Interestingly, the protein level of GFP-StSWAP70 was consistently barely detectable when coexpressed with StNRL1 and Pi02860 together, and this degradation was again attenuated by MG132 treatment (Fig. 3A and SI Appendix, Fig. S4). These observations suggest that the GEF, StSWAP70, is destabilized by Pi02860 and StNRL1 through proteasome-dependent degradation.
Fig. 3.
Pi02860 promotes proteasome-mediated degradation of StSWAP70 in an NRL1-dependent manner. (A) Coexpression of GFP-StSWAP70 with cMYC-StNRL1 and/or cMYC-Pi02860 causes reduced StSWAP70 abundance, which was prevented by MG132 treatment. (B) GFP-StSWAP70 was transiently expressed alone or with cMYC-Pi02860 in TRV:GFP and silenced TRV:NRL1 V1 and TRV:NRL V2 N. benthamiana leaves. The silencing of NRL1 shows attenuation of Pi02860-mediated degradation of StSWAP70 compared with TRV:GFP. Free GFP specific to TRV: GFP-expressing plants are detected below 30 kDa, indicting GFP is not silenced. Expression of constructs in the leaves is indicated by a “+.” Protein size markers are indicated in kilodaltons, and protein loading is indicated by Ponceau stain. Immunoblots with anti-GFP show protein fusion of GFP-StSWAP70 (triangle) and the anti-cMYC shows protein fusion of cMYC-StNRL1 (star) and cMYC-Pi02860 (arrow) of the expected size.
Pi02860-Mediated Degradation of StSWAP70 Is NRL1-Dependent.
NRL1 is a susceptibility factor, the activity of which is required for Pi02860 to promote P. infestans colonization and suppress ICD (22). To determine whether Pi02860-mediated degradation of StSWAP70 is NRL1-dependent, we thus investigated StSWAP70 protein level on NRL1 silenced N. benthamiana. We coinfiltrated Agrobacteria expressing GFP-StSWAP70 with either cMYC-EV or cMYC-Pi02860 into plants expressing TRV:GFP (control), or VIGS constructs TRV:NRL1 V1 or TRV:NRL1 V2 (22) to silence NbNRL1. Immunoblot analysis showed that GFP-StSWAP70 is stable when coexpressed with cMYC-EV either on the TRV:GFP plants or NbNRL1-silenced plants (Fig. 3B and SI Appendix, Fig. S5). As anticipated, GFP-StSWAP70 abundance was reduced by cMYC-Pi02860 but not cMYC-EV on the TRV:GFP control plants. In contrast, GFP-StSWAP70 abundance was reproducibly unaltered when coexpressed with cMYC-Pi02860 on the VIGS plants in which NbNRL1 was silenced (expressing TRV:NRL1 V1 and TRV:NRL1 V2) (Fig. 3B and SI Appendix, Fig. S5), demonstrating that silencing of NRL1 prevented Pi02860-stimulated turnover of StSWAP70. This indicates that proteasome-mediated degradation of StSWAP70 is NRL1-dependent.
Proteasome-Mediated Degradation of StSWAP70 Requires Dimerization of StNRL1.
NRL1 belongs to a large protein family that includes the functionally characterized members NPH3 and RPT2 in Arabidopsis, which are required for ubiquitination, internalization, and proteasome-mediated turnover of phototropins to regulate blue-light signaling (32). The BTB/POZ domain in NPH3 promotes an association with CULLIN3 (CUL3), forming a substrate adaptor in a CRL3 (for Cullin-RING ubiquitin ligase 3)-NPH3 complex (CRL3NPH3) that targets phototropin 1 (phot1) for ubiquitination (32, 33). As a negative regulator of immunity, NRL1 is thus predicted to act as a substrate adaptor in a CRL3NRL1 complex that targets proteins associated with immunity for degradation (22).
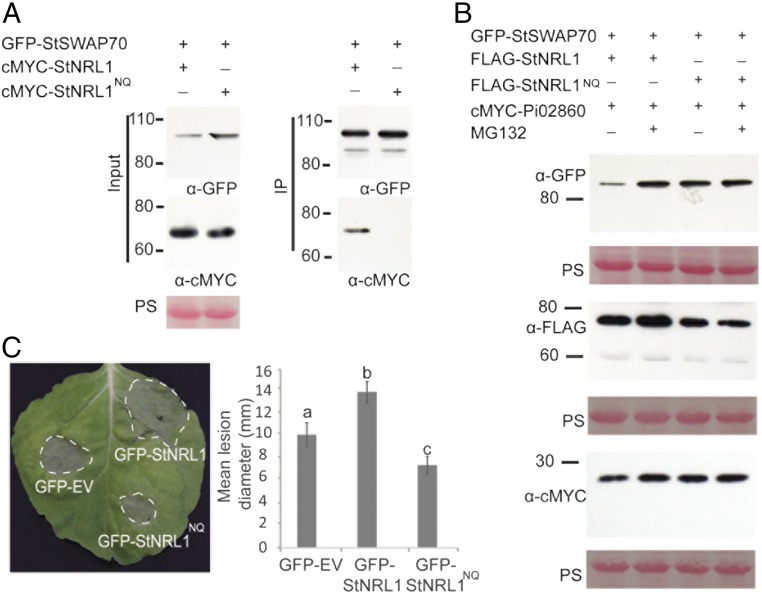
Dimerization of BTB proteins is required for selection of substrates for ubiquitination. Thus, mutations in conserved pocket residues in the BTB domain that prevent or reduce dimerization result in loss of substrate interaction and ubiquitination (34, 35). Indeed, we have recently shown that mutation to prevent dimerization of the BTB/POZ domain in E3 ligase POB1 prevents interaction with, and proteasome-mediated turnover of, its substrate PUB17 (36). We identified the conserved residues in StNRL1 (Asp28 and Lys42) (SI Appendix, Fig. S6A) and mutated Asp28 (D28) to Asn (N) and Lys42 (K42) to Gln (Q), generating StNRL1NQ mutant. The effect of this mutation on StNRL1 dimerization was tested using co-IP. When the fusion constructs GFP–StNRL1 and cMYC-StNRL1 were coexpressed, the latter was strongly coimmunoprecipitated. In contrast, GFP-StNRL1 or GFP-StNRL1NQ showed considerably weakened associations with cMYC-StNRL1NQ (SI Appendix, Fig. S6B). This confirms the effect of the D28N/K42Q mutation in attenuating NRL1-mediated dimerization in planta. As anticipated, whereas GFP-StSWAP70 associated with cMYC-StNRL1 wild-type, which was accompanied by reduced GFP-StSWAP70 abundance, no such association was observed between GFP-StSWAP70 and cMYC-StNRL1NQ mutant (Fig. 4A), suggesting that dimerization is required for StNRL1 to associate with StSWAP70. However, the interaction of the GFP-Pi02860 effector fusion was maintained in planta with the FLAG-StNRL1NQ mutant (SI Appendix, Fig. S6C).
Fig. 4.
Pi02860-mediated degradation of StSWAP70 requires dimerization of StNRL1. (A) IP of protein extracts from N. benthamiana leaves transiently expressing GFP-StSWAP70 with either cMYC-StNRL1 or cMYC-StNRL1NQ using GFP-Trap confirmed that cMYC-StNRL1 associates with GFP-StSWAP70 but the dimerization mutant cMYC-StNRL1NQ does not. IP was analyzed by immunoblotting using an anti-GFP antibody showing protein fusion of GFP-StSWAP70 and the anti-cMYC antibody showing protein fusions of cMYC-StNRL1 and cMYC-StNRL1NQ of the expected size. (B) The dimerization mutant StNRL1NQ prevents degradation of StSWAP70. Transient coexpression of GFP-StSWAP70 and cMYC-Pi02860 in the presence of either wild-type FLAG-StNRL1 or mutant FLAG-StNRL1NQ in N. benthamiana leaves, with or without MG132 treatment. Immunoblots with anti-GFP show stable protein fusion of GFP-StSWAP70, the anti-cMYC antibody shows protein fusion of cMYC-Pi02860 and anti-FLAG antibody shows stable protein fusions of FLAG-NRL1 and FLAG-StNRL1 of the expected size. (C) Expression of wild-type GFP-StNRL1 significantly enhances P. infestans infection compared with the control GFP-EV, whereas the dimerization mutant GFP-StNRL1NQ reduces P. infestans colonization. The results shown are combinations of at least five individual biological replicates (ANOVA, P < 0.05; n = 82 per construct) and error bars show SE. Letters on the graph denote statistically significant differences. Representative leaf image showing P. infestans lesions following overexpression of each construct, as indicated, in N. benthamiana.
To examine the effect of StNRL1NQ mutation on the ability of Pi02860 to mediate SWAP70 degradation, FLAG-StNRL1NQ and FLAG-StNRL1 were coexpressed in N. benthamiana with GFP-StSWAP70 and cMYC-Pi02860, with or without MG132 treatment (Fig. 4B and SI Appendix, Fig. S7). As expected, GFP-StSWAP70 protein abundance was reproducibly reduced by coexpression with cMYC-Pi02860 and FLAG-StNRL1, and this was inhibited by MG132 (Fig. 4B and SI Appendix, Fig. S7). In contrast, GFP-StSWAP70 protein abundance was unaffected by coexpression with FLAG-NRL1NQ mutant and cMYC-Pi02860 (Fig. 4B and SI Appendix, Fig. S7). This indicates that StNRL1NQ acts as a dominant-negative mutation; although its interaction is maintained with the effector Pi02860, its homodimerization is attenuated and thus StNRL1NQ fails to interact with StSWAP70 to mediate its degradation.
To investigate whether the StNRL1NQ mutant could suppress ICD, GFP-EV (control), GFP-StNRL1NQ, and GFP-StNRL1NQ were each transiently coexpressed with INF1 on N. benthamiana leaves. The ability of GFP-StNRL1NQ to suppress ICD was significantly reduced (ANOVA, P < 0.001) compared with the GFP-StNRL1 wild-type (SI Appendix, Fig. S8A). Previously, we have shown that silencing of NRL1 in N. benthamiana prevented effector Pi02860 from enhancing colonization (22). We thus investigated the impact of the NRL1NQ mutant on late blight disease. Critically, using a high inoculum of P. infestans zoospores, whereas transient expression of GFP-StNRL1 led to enhanced P. infestans colonization, expression of GFP-StNRL1NQ mutant attenuated P. infestans leaf colonization compared with the GFP control (Fig. 4C). These results indicate that both the dimerization of StNRL1 and its ability to interact with StSWAP70 are essential for its function in suppressing the ICD immune response and thus in enhancing P. infestans infection.
To examine whether the StNRL1NQ mutant had an effect on the ability of Pi02860 to suppress ICD, INF1 was coexpressed in N. benthamiana leaves with GFP-EV control, GFP-Pi02860 alone, GFP-Pi02860 with wild-type GFP-StNRL1, or GFP-Pi02860 with GFP-StNRL1NQ. At 6 dpi, GFP-Pi02860 alone suppressed ICD to a significant level (P < 0.001) compared with free GFP (SI Appendix, Fig. S8B), as anticipated. However, the suppression of ICD was significantly enhanced (ANOVA, P < 0.5) by coexpression of GFP-Pi02860 with wild-type GFP-NRL1, whereas the suppression of ICD was significantly reduced (ANOVA, P < 0.5) when GFP-Pi02860 was coexpressed with GFP-StNRL1NQ (SI Appendix, Fig. S8C). This confirms that the dimerization-deficient NRL1NQ acts as a dominant-negative to reduce suppression of ICD by Pi02860.
A Pi02860 Mutant That Fails to Interact with StNRL1 Does Not Promote StSWAP70 Degradation.
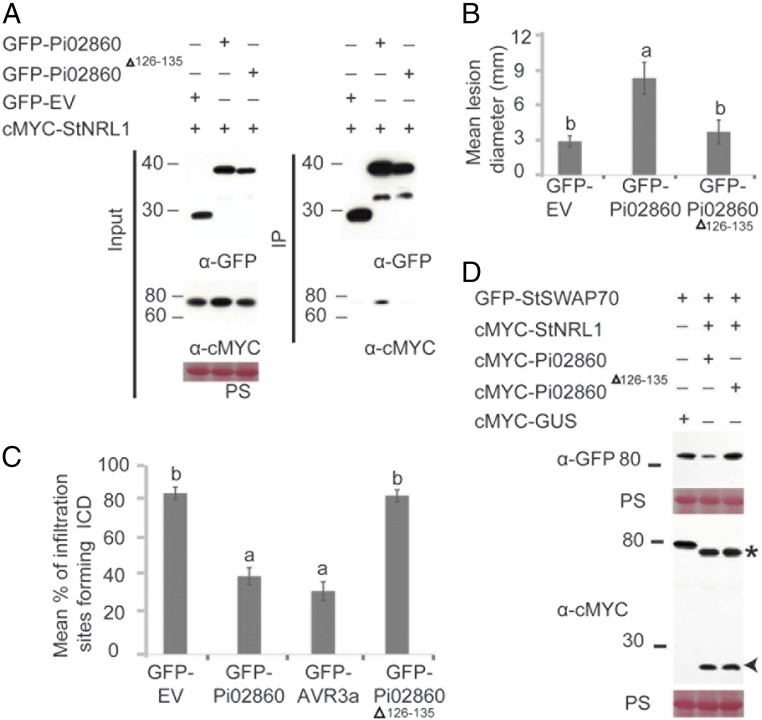
To provide additional genetic evidence linking Pi02860 activity to StNRL1 interaction and StSWAP70 turnover, we attempted to make an effector mutant that was unable to interact with StNRL1. Because the C terminus of RXLR effectors is regarded as the effector domain, we made sequential 5- and 10-amino acid deletions from the C terminus (SI Appendix, Fig. S9A) and tested whether interaction with StNRL1 was maintained using the Y2H assay. We found that, whereas a 5-amino acid deletion (Pi02860Δ131–135) retained weak interaction indicated by activation of the β-Gal assay, a 10-amino acid deletion (Pi02860Δ126–135) no longer interacted (SI Appendix, Fig. S9B). We confirmed that the mutant Pi02860Δ126–135 form also failed to interact with StNRL1 in planta, although it was as stable as the wild-type Pi02860 form (Fig. 5A). Unlike the wild-type effector, Pi02860Δ126–135 also failed to enhance P. infestans colonization of N. benthamiana (Fig. 5B), suppress ICD (Fig. 5C), or promote turnover of StSWAP70 (Fig. 5D).
Fig. 5.
A Pi02860 mutant fails to interact with NRL1 or promote StSWAP70 degradation. (A) IP of protein extracts from N. benthamiana leaves transiently expressing cMYC-StNRL1 with either GFP-EV, GFP-Pi02860, or GFP-Pi02860Δ126–135 using GFP-Trap confirmed that cMYC-StNRL1 associates with GFP-Pi02860 but not with GFP-Pi02860Δ126–135. IP was analyzed by immunoblotting using an anti-GFP antibody showing protein fusions GFP-Pi02860, GFP-Pi02860Δ126–135, GFP-EV, and the anti-cMYC antibody showing protein fusions of cMYC-StNRL1 of the expected size. (B) Expression of wild-type GFP-Pi02860 significantly enhances P. infestans infection compared with the control GFP-EV. However, the Pi02860Δ126–135 fails to enhance P. infestans colonization. The results shown are combinations of three individual biological replicates (ANOVA, P < 0.001; n = 75 per construct) and error bars show SE. Letters on the graph denote statistically significant differences. (C) Suppression of ICD by Pi02860Δ126–135 is significantly reduced (P < 0.001, n = 45). The graph shows the mean percentage of infiltration sites forming ICD by transient expression of INF1 with GFP-EV, GFP-Pi02860, GFP-PiAvr3a, or GFP-Pi02860Δ126–135. The graph represents the combined data from three biological replicates. Letters on the graph denote statistically significant differences and error bars show SE. (D) The Pi02860Δ126–135 mutant fails to degrade StSWAP70 by StNRL1. GFP-StSWAP70 and cMYC-StNRL1 were transiently coexpressed with either wild-type cMYC-Pi02860 or mutant cMYC-Pi02860Δ126–135 in N. benthamiana leaves. Immunoblots with anti-GFP show the protein fusion of GFP-StSWAP70; the anti-cMYC antibody shows protein fusion of cMYC-Pi02860 and cMYC-Pi02860Δ126–135 of the expected size.
Pi02860 Enhances the Association Between StNRL1 and StSWAP70.
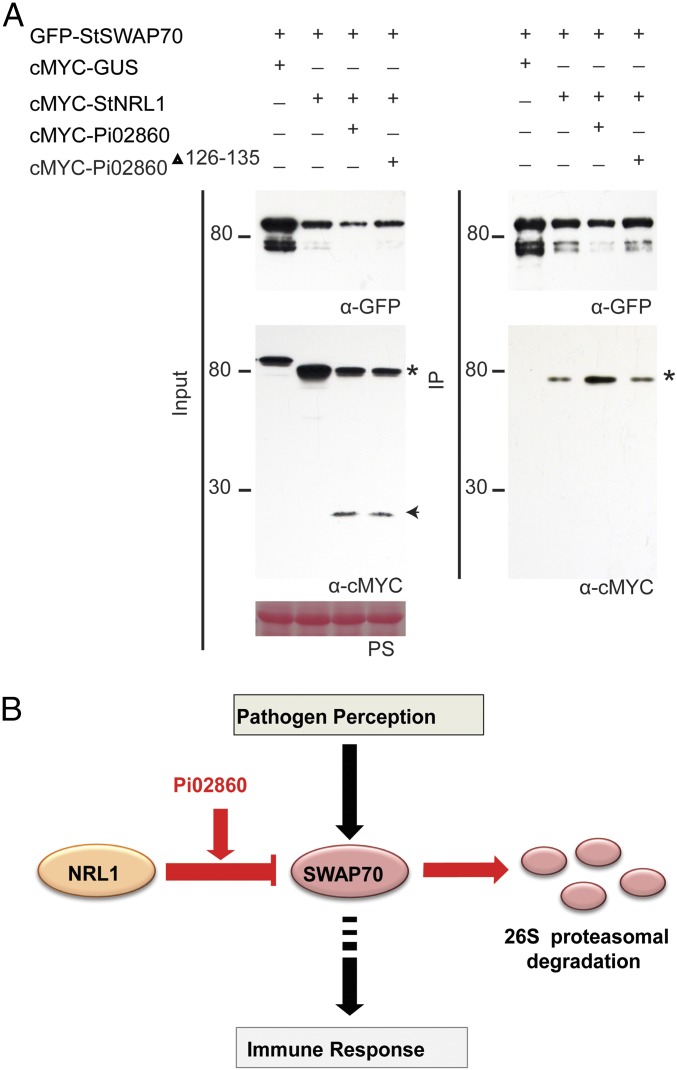
To examine the effect of Pi02860 on the interaction between StNRL1 and StSWAP70, we conducted a co-IP assay of GFP-StSWAP70 and cMYC-StNRL1 in the presence or absence of cMYC-Pi02860. GFP-StSWAP70 was coexpressed with cMYC-EV, with cMYC-GUS, with cMYC-StNRL1 alone, or with cMYC-StNRL1 and cMYC-Pi02860 together and pulled down using GFP-TRAP_M beads. As observed previously in Fig. 3, while all proteins were present in the relevant input fractions, the protein level of GFP-StSWAP70 was decreased by coexpression with cMYC-StNRL1, and was further reduced when coexpressed with both cMYC-StNRL1 and cMYC-Pi02860 (Fig. 6 and SI Appendix, Fig. S10). As observed in Fig. 1C, cMYC-StNRL1 was coimmunoprecipitated with GFP-StSWAP70. Critically, however, reproducibly more cMYC-StNRL1 protein was immunoprecipitated by GFP-StSWAP70 in the presence of Pi02860 even though there is a corresponding decrease in the abundance of GFP-StSWAP70 in the input samples (Fig. 6 and SI Appendix, Fig. S10). However, the mutated effector Pi02860Δ126–135 was unable to enhance the association of StNRL1 with StSWAP70 (Fig. 6).
Fig. 6.
Pi02860 enhances StNRL1 and StSWAP70 association in planta. (A) IP of protein extracts from N. benthamiana leaves transiently expressing GFP-StSWAP70 with either cMYC-GUS, cMYC-StNRL1, cMYC-StNRL1 + cMYC-Pi02860, or cMYC-StNRL1 + cMYC-Pi02860Δ126–135 using GFP-Trap shows reduced protein levels of GFP-StSWAP70 in the presence of cMYC-StNRL1, which are further decreased when coexpressed with both cMYC-StNRL1 and cMYC-Pi02860 but not with cMYC-StNRL1 + cMYC-Pi02860Δ126–135. Increased cMYC-StNRL1 protein pull downs with GFP-StSWAP70 in the presence of cMYC-Pi02860 indicates that Pi02860 enhances the association between StSWAP70 and StNRL1. Expression of constructs in the leaves is indicated by a “+.” Protein size markers are indicated in kilodaltons, and protein loading is indicated by Ponceau stain. Immunoblots with anti-GFP shows protein fusion of GFP-StSWAP70 and the anti-cMYC shows protein fusion of cMYC-StNRL1 (star) and cMYC-Pi02860 (arrow) of the expected size. (B) Model for the action of Pi02860 and NRL1 (red) on immunity regulated by SWAP70 (black). Perception of pathogen molecules (such as the PAMP INF1) leads to the activation of SWAP70 which, in turn, positively regulates immunity (24, 25). NRL1, an S factor and predicted CRL ubiquitin E3 ligase component (22), negatively regulates immunity by mediating proteasome-dependent degradation of SWAP70. Effector Pi02860 enhances interaction between NRL1 and SWAP70, promoting increased turnover of SWAP70 to suppress immunity.
Discussion
We have recently shown that the RXLR effector Pi02860 promotes virulence by interacting with the CUL3 E3 ligase substrate adaptor protein StNRL1, exploiting its ability to negatively regulate immunity (22). Here we show that the Rho-GEF protein StSWAP70, a positive regulator of immunity, interacts with StNRL1 and is a substrate for proteasome-mediated degradation that is dependent on StNRL1.
The Y2H screen with StNRL1 revealed not only StSWAP70 as an interactor but also NRL1 itself, 14-3-3 proteins, PP2A, and a HAT4-like homeobox protein (SI Appendix, Table S1). Interestingly, 14-3-3 proteins (37, 38), PP2A (39), and NRLs (37), have all been found to directly interact with blue-light phototropin receptors Phot1 and Phot2 in yeast and in planta. Interaction between PP2A and Phot2 has been implicated in blue-light–induced chloroplast movement (40). Critically, the Arabidopsis ortholog of StNRL1, NCH1, has recently been shown to be required for light-stimulated chloroplast accumulation (41). Future work will reveal whether StNRL1 plays a role in chloroplast movement responses to light, or indeed to pathogen challenge, and whether any of the interacting proteins in addition to SWAP70 are potential substrates for proteasome-mediated degradation. However, SWAP70 dominated the Y2H screen along with NRL1 itself, and became our focus here, as SWAP70 has been reported to play a positive role in regulating immunity (24, 25).
In our work we show that the effector Pi02860 enhances the interaction between the CUL3 E3 ligase adaptor protein StNRL1 and StSWAP70, promoting a rapid turnover of StSWAP70, leading to susceptibility (Fig. 6B). The pathogen effector Avr-Piz-t from Magnaporthe oryzae also directly interacts with ubiquitin E3 ligases APIP6 and APIP10 in rice (42, 43). However, in contrast to NRL1, APIP6 and APIP10 are positive regulators of PTI and interaction with Avr-Piz-t results in their degradation, and consequent PTI suppression. Interestingly, the resistance protein Piz-t is a substrate for ubiquitination and degradation by APIP10. Therefore, APIP10 degradation promoted by Avr-Piz-t results in increased stability of Piz-t, leading to ETI (43).
The phytoplasma effector Sap54 also mediates degradation of a host protein. Sap54 interacts with MADS-domain transcription factors (MTFs), which regulate floral development, and also interacts with members of the RADIATION SENSITIVE23 (RAD23) family, which shuttle substrates to the proteasome for degradation (44). As a consequence, the MTFs are degraded. No known association between RAD23 and MTFs exist, and RAD23 proteins are not known to regulate flowering and Maclean et al. (44) concluded that Sap54 provided a “short-circuit,” bringing together two plant proteins that do not normally associate. In contrast, StNRL1 interacts directly in Y2H with StSWAP70, an interaction that is maintained in planta. Moreover, in the absence of the effector Pi02860, StNRL1 is an endogenous negative regulator of immunity that promotes StSWAP70 turnover. The effector Pi02860 was observed to enhance the StNRL1–StSWAP70 interaction. It is possible that, during an immune response, negative regulators such as StNRL1 are simultaneously inactivated, or are prevented from targeting substrates that are required for immunity. Future work will investigate how StNRL1 is regulated and whether Pi02860 interferes with that, or whether the observed strengthening of StNRL1–StSWAP70 interaction is its primary mode-of-action.
We show that StSWAP70 is a positive regulator of immunity, in the form of ICD, and that silencing it leads to enhanced P. infestans infection. Previously it has been shown in rice that OsSWAP70 interacts with Rho GTPase OsRac1 and its DH domain activates OsRac1 by promoting dissociation of GDP to facilitate the association of GTP (24). The interconversion of GDP-bound inactive and GTP-bound active forms of Rac1 acts as a molecular switch in the regulation of many cellular processes, including PTI and ETI (45). Rac1 can be activated by different GEFs, including SWAP70, that are themselves activated via phosphorylation by distinct immune receptors (45, 46). In rice, for example, OsRacGEF1 has been shown to act downstream of the chitin receptor complex OsCEBiP/OsCERCK1 to activate OsRac1 (46). Here we show that StSWAP70 is required for ICD but not for cell death triggered by perception of C. fulvum Avr4 by Cf4. It is thus possible that StSWAP70 is activated via phosphorylation by receptors that perceive elicitins such as INF1.
As a GEF, SWAP70 is implicated in activating small GTPases, such as Rac1, but precisely how it regulates immunity is poorly understood. It is important to note that the interaction between NRL1 and Pi02860 was shown to occur primarily at the host plasma membrane (22). This is consistent with other NRLs, such as NPH3, which interact with ARF-GEF proteins to regulate endocytosis of the auxin PIN receptors (32). It was thus perhaps unsurprising to find that StSWAP70 localized to endosomes (Fig. 1), and future work should investigate the impact of Pi02860 and StNRL1 upon the endocytosis of the elicitin receptor ELR. Receptor internalization is required for immune signaling (47). The endocytic cycle has been shown to be a target for pathogens. For example, the type III effector HopM1 from Pseudomonas syringae promotes the proteasome-dependent degradation of AtMIN7, a host ADP ribosylation factor and GEF that is required to regulate vesicle trafficking during PTI and ETI (48, 49). The exact mode-of-action of HopM1 remains unknown, and it will be interesting to see whether an E3 ligase such as NRL1 is coopted for its activity in removing AtMIN7.
Suppression of ICD has been observed in the presence of several P. infestans RXLR effectors. AVR3a suppresses it by targeting the ubiquitin E3 ligase CMPG1 and preventing its normal activity (23). More recently, we have shown that AVR2 activates a transcriptional regulator, CHL1, which is an endogenous negative regulator of ICD (13), and Pi17316 interacts directly with the potato VASCULAR HIGHWAY 1 (VH1)-interacting kinase (StVIK) as an S factor to suppress ICD (50). Now we show that the effector Pi02860 suppresses ICD by targeting the GEF protein SWAP70 for degradation by NRL1, an E3 ligase related to the blue-light regulators NPH3 and RPT2. This emphasizes the importance of the ICD immune pathway to late blight resistance, and highlights the diverse range of mechanisms employed to suppress it.
Recently, P. infestans effectors have been shown to target and exploit S factors to negatively regulate immunity (reviewed in ref. 5). Effector Pi04314 forms unique holoenzymes with PP1c, presumably to dephosphorylate host proteins to suppress immunity (12). To date, the targets of Pi04314-PP1c holoenzymes have not been identified. Pi04089 effector targets the K-homology RNA binding protein KRBP1, increasing its stability and potentially promoting its activity (11). However, the precise RNAs that interact with KRBP1 have yet to be identified. Here, we detail the mechanism underlying the activity of Pi02860, which interacts with the ubiquitin E3 ligase NRL1, and reveal that SWAP70 is then targeted for removal to suppress immunity (Fig. 6B). Critically, preventing the degradation of SWAP70 either by silencing NRL1 or by expression of the dimerization-deficient mutant NRL1NQ, which interacts with the effector Pi02860 but not the substrate SWAP70, provides a strategy to combat potato late blight.
Materials and Methods
Plant Materials and Growth Conditions.
N. benthamiana plants were grown under a 16-h day at 22 °C and an 8-h night at 18 °C; when the ambient light dropped below 200 Wm−2, supplementary lighting was automatically provided, and above 450 Wm−2, shading was automatically provided. Three-week-old plantlets were transplanted and grown in individual pots in a greenhouse at 20–26 °C, as described in ref. 23.
Plasmid Constructs.
Full-length StSWAP70 was cloned from potato cDNA with gene-specific primers modified to contain the Gateway (Invitrogen) attB recombination sites. PCR products were purified and recombined into pDONR201 (Invitrogen) to generate entry clones via BP reactions using Gateway technology (Invitrogen). Primer sequences are shown in SI Appendix, Table S2. Protein fusions were made by recombining the entry clones with the following plant expression vectors using LR clonase (Invitrogen). N-terminal GFP, mRFP, cMYC, FLAG fusions were made by recombining the entry clones with pB7WGF2, pK7WGR2 and pGWB18, PGWB412 respectively.
Mutagenesis.
The StNRLNQ mutant was generated according to the manufacturer’s protocol QuickChangeh Site-Directed Mutagenesis Kit (Stratagene) using pDonr201-StNRL1 as a template. The primers sequences used for mutation are shown in SI Appendix, Table S2. Two conserved amino acids of StNRL1, Asp at 28 position (D28) to Asn (N) and Lys at 42 position (K42) to Gln (Q), generating StNRL1NQ mutant, resulting in the mutant StNRL1D28N/K42Q. The mutant StNRL1 was recombined, using LR clonase, into pB7WGF2, PGWB412, or PGWB18 for in planta assays.
The Pi02860 in which we removed the 10 or 5 amino acids in the C terminus were cloned using PUC57-Pi02860 without Signal peptide as a template, generating pDonr201-Pi02860∆126–135 or pDonr201-Pi02860∆131–135. The primers sequences used for mutation are shown in SI Appendix, Table S2. The pDonr201-Pi02860∆126–135 or pDonr201-Pi02860∆131–135 mutants were recombined, using LR clonase, into pB7WGF2 or PGWB18 for in planta assays.
P. infestans Infection Assay.
P. infestans 88069 was grown in Petri dishes (90-mm diameter) of rye agar medium at 19 °C. The P. infestans strains grown for 13 d are used for pathology tests. The plates were flooded with 3 mL H2O, and scraped to release sporangia. The sporangial suspension was poured into a Falcon tube, and spun at 2,750 rpm for 10 min at 4 °C. Sporangia numbers were counted using a hemocytometer then adjusted to 50,000 sporangia per milliliter for VIGS plant infection and was elevated to 80,000 sporangia per milliliter for agroinfiltrated N. benthamiana leaves in Fig. 4C. Ten-microliter droplets were inoculated onto the abaxial side of detached N. benthamiana leaves (four sites per leaf) on moist tissue in sealed boxes. The infection assays were carried out as described in refs. 22 and 51. The lesions were measured at 7 dpi and expressed as mean lesion diameter compared with the GFP control plants. For VIGS plants, Sporangia were recovered from the lesions by immersing the silenced leaves in 3 mL H2O. The number of sporangia recovered from each leaf was counted using a hemocytometer and was expressed as sporangia per milliliter.
TRV-Based VIGS in N. benthamiana.
VIGS constructs were made by cloning 265-bp and 307-bp PCR fragments of NbSWAP70 into pBinary tobacco rattle virus (TRV) vectors (52) between HpaI and EcoRI sites in the antisense orientation (SI Appendix, Fig. S3A). Primer sequences are shown in SI Appendix, Table S2. BLAST analysis of this sequence against the P. infestans genome (https://www.broadinstitute.org/scientific-community/science/programs/genome-sequencing-and-analysis/update-our-microbial-eukaryotes) did not reveal any matches that could initiate silencing in the pathogen. A TRV construct expressing GFP described previously was used as a control (52–54). The two largest leaves of four-leaf–stage N. benthamiana plants were pressure-infiltrated with LBA4404 Agrobacterium tumefaciens strains containing a mixture of RNA1 (OD600 = 0.4) and each SWAP70 VIGS construct or the GFP control at OD600 = 0.5. Plants were used for assays or to check gene-silencing levels by qRT-PCR 3 wk later. A. tumefaciens transient expression in combination with P. infestans infection were carried out as described previously (22, 51, 53).
Agrobacterium-Mediated Transient Expression.
Agrobacterium-mediated transient expression by agroinfiltration was performed as described previously (12, 22, 55, 56). A. tumefaciens strain AGL1 or GV3101 containing constructs were grown overnight in yeast-extract and beef (YEB) medium with appropriate antibiotics at 28 °C. The bacteria were pelleted, resuspended in infiltration buffer (10 mM MES, 10 mM MgCl2, and 200 mM acetosyringone), and adjusted to the required OD600 before infiltration into N. benthamiana leaves [the OD600 was generally 0.005–0.01 for confocal-imaging purposes, and 0.5 for immunoblots, IP, and homologous recombination (HR) activity assays]. For coexpression of multiple constructs, agrobacterial suspensions carrying the different constructs were thoroughly mixed before infiltration. For agroinfiltration and infection assays, Agrobacterium suspensions at concentrations of OD600 = 0.15 were infiltrated into leaves and each infiltration site was inoculated with 10 µL of P. infestans inoculum at 80,000 sporangia per milliliter after 1-d infiltration. Lesion sizes were measured at 7 dpi. The number of positive HRs (i.e., more than 50% of the inoculated sites forming clear cell death) were counted as described previously (56) and expressed as the mean percentage of total inoculations per plant. One-way ANOVA was performed to determine statistically significant differences.
Immunoprecipitation.
N-terminal GFP-StSWAP70/cMYC-StNRL1 with or without cMYC-Pi02860, GFP-StNRL1/cMYC-StNRL1, mRFP-StNRL1/FLAG-StNRL1, mRFP-StNRL1NQ/FLAG-StNRL1NQ, GFP-StSWAP70/cMYC-StNRL1, GFP-StSWAP70/cMYC-StNRL1NQ, and all of the control combinations (Figs. 1C, 4B, and 5C, and SI Appendix, Figs. S5 and S7) were overexpressed in N. benthamiana using Agrobacterium-mediated expression. Leaf samples were collected at 48-h postinfiltration and proteins were extracted using GTEN [10% (vol/vol) glycerol, 25 mM Tris⋅HCl (pH 7.5), 1 mM EDTA, 150 mM NaCl] buffer with 10 mM DTT, protease inhibitor mixture, 1 mM phenylmethyl sulphonyl fluoride, and 0.2% Nonidet P-40. The fusions of GFP-tagged SWAP70/cMYC-StNRL1 with or without cMYC-Pi02860, GFP-StSWAP70/cMYC-StNRL1, and GFP-StSWAP70/cMYC-StNRL1NQ were immunoprecipitated using GFP-Trap-M magnetic beads (Chromotek). The mRFP-StNRL1/FLAG-StNRL1 or mRFP-StNRL1NQ/FLAG-StNRL1NQ were immunoprecipitated using mRFP-Trap-M magnetic beads (Chromotek). The resulting samples were separated by PAGE and Western-blotted. Immunoprecipitated GFP fusions, c-Myc, FLAG, or mRFP fusions were detected using appropriate antisera (Santa Cruz Biotechnology). The IP protocol is described in Boevink et al. (12).
Plant Treatments and Western Blotting.
Protein fusions were overexpressed in N. benthamiana using Agrobacterium-mediated expression. Leaf samples were collected at 48-h postinfiltration and MG132 (10 mM stock dissolved in DMSO, diluted to 100 μM for working solution in buffer) solutions were infiltrated 8 h before collecting samples, using DMSO + buffer as a control. The leaf samples treated with MG132 were floated on MG132 solution for the 8 h after infiltration. Proteins were extracted using GTEN buffer and then were mixed with 2× SDS/PAGE sample buffer and analyzed by immunoblotting. Samples were loaded onto a 4–12% Bis⋅Tris NuPAGE Novex gel run with 1× MES SDS running buffer for 30 min at 80 V and 2 h at 120 V (Invitrogen). Gels were blotted onto a nitrocellulose membrane for 1.5 h at 30 V and stained with Ponceau solution to show loading and transfer. Membranes were blocked in 5% milk in 1× PBST [PBS (137 mM NaCl, 12 mM Phosphate, 2.7 mM KCl, pH 7.4) with Tween-20 0.2% (vol/vol)] for 1–2 h before addition of the primary antibodies overnight: either a monoclonal GFP antibody raised in mouse at 1:1,000 dilution (cat. no. G1546; Sigma-Aldrich), a monoclonal anti-cMYC antibody raised in mouse at 1:1,000 (cat. no. SC-40; Santa Cruz), a polyclonal anti-mRFP antibody raised in rabbit (cat. no. 5F8; Chromotek) at 1:4,000, or a Monoclonal ANTI-FLAG M2 antibody produced in mouse at 1:8,000 (F3165-0.2MG; Sigma). The membrane was washed with 1× PBST (0.2% Tween 20) five times for 10-min each before addition of the secondary antibody at 1:8,000 dilution with either anti-mouse Ig-HRP antibody (cat. no. A9044; Sigma-Aldrich) or anti-rat Ig-HRP antibody (cat. no. ab6836; Abcam) for 1 h. ECL (Amersham) detection was used according to the manufacturer’s instructions.
Y2H.
A Y2H screen was performed in Saccharomyces cerevisiae strain MaV203 as described in ref. 53 using the Invitrogen ProQuest system. DNA-binding domain “bait” fusions were generated by recombination between pDonr201-StNRL1 and pDEST32, resulting in pDest32-StNRL1. This construct was transformed into yeast strain MaV203, and nutritional selection used to recover transformants. A single transformant was grown up and used to prepare competent yeast cells, which were transformed with a potato Y2H “prey” library, as described in ref. 12. Interactions were confirmed using reporter gene assays, namely the ability to grow on media missing histidine (−HIS), and screening for gain of β-Gal. The full-length coding sequence of the candidate interacting prey sequence, StSWAP70 [XM_006364745.1 (T398A, T917C); XP_006364807.1 (L133Q, L306S)], was cloned and retested with pDEST32-StNRL1 and pDEST32-EV as a control to rule out the possibility that the observed reporter gene activation had resulted from interactions between the prey and the DNA-binding domain of the bait construct or the DNA-binding activity of the prey itself. StNRL1 was also cloned into the prey vector to obtain pDEST22-StNRL1, which was tested pairwise for an interaction with pDEST32-StNRL1 using pDEST22- EV as a control.
Confocal Imaging.
A. tumefaciens containing GFP-StSWAP70 was coinfiltrated at a low OD600 (0.01–0.05) with different subcellular markers (ST-mRFP, mRFP-Ara7, Ara6-mRFP, CFP-PS1) (57) into leaves of 4-wk-old N. benthamiana plants. Cells expressing fluorescent protein fusions were observed using a Zeiss 710 confocal microscope with a PL APO 40×/1.0 water dipping objective at no more than 2 d postinfiltration. GFP was excited by 488-nm light and its emissions were detected between 500 and 530 nm. mRFP was excited with 561 nm and its emissions were detected between 600 and 630 nm. CFP was excited with 405 nm and its emission was detected between 440 and 480 nm.
Gene-Expression Assay.
Total RNA was extracted from the leaves of N. benthamiana VIGS plants with an RNeasy plant kit (Qiagen) according to the manufacturer’s recommendations. Two milligrams of RNA was used for first-strand cDNA synthesizing using SuperScript II RNase HReverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. Real-time qRT-PCR was performed using Power SYBR Green and run on a Chromo4 thermal cycler (MJ Research) using Opticon Monitor 3 software. Primer sequences are given in SI Appendix, Table S2. Primers were designed outside the region of cDNA targeted for silencing. Gene expression levels were calculated by a comparative Ct method, as described by ref. 58.
Supplementary Material
Acknowledgments
The authors received financial support from the Biotechnology and Biological Sciences Research Council (Grants BB/G015244/1, BB/K018183/1, BB/L026880/1), and the Scottish Government Rural and Environment Science and Analytical Services Division.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808585115/-/DCSupplemental.
References
- 1.Büttner D. Behind the lines-actions of bacterial type III effector proteins in plant cells. FEMS Microbiol Rev. 2016;40:894–937. doi: 10.1093/femsre/fuw026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanver D, et al. Ustilago maydis effectors and their impact on virulence. Nat Rev Microbiol. 2017;15:409–421. doi: 10.1038/nrmicro.2017.33. [DOI] [PubMed] [Google Scholar]
- 3.Oliveira-Garcia E, Valent B. How eukaryotic filamentous pathogens evade plant recognition. Curr Opin Microbiol. 2015;26:92–101. doi: 10.1016/j.mib.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 5.Whisson SC, Boevink PC, Wang S, Birch PR. The cell biology of late blight disease. Curr Opin Microbiol. 2016;34:127–135. doi: 10.1016/j.mib.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Schie CCN, Takken FLW. Susceptibility genes 101: How to be a good host. Annu Rev Phytopathol. 2014;52:551–581. doi: 10.1146/annurev-phyto-102313-045854. [DOI] [PubMed] [Google Scholar]
- 7.Boevink PC, et al. Oomycetes seek help from the plant: Phytophthora infestans effectors target host susceptibility factors. Mol Plant. 2016;9:636–638. doi: 10.1016/j.molp.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Kamoun S, et al. The top 10 oomycete pathogens in molecular plant pathology. Mol Plant Pathol. 2015;16:413–434. doi: 10.1111/mpp.12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fry WE, et al. Five reasons to consider Phytophthora infestans a reemerging pathogen. Phytopathology. 2015;105:966–981. doi: 10.1094/PHYTO-01-15-0005-FI. [DOI] [PubMed] [Google Scholar]
- 10.Wang S, et al. Delivery of cytoplasmic and apoplastic effectors from Phytophthora infestans haustoria by distinct secretion pathways. New Phytol. 2017;216:205–215. doi: 10.1111/nph.14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, et al. A host KH RNA binding protein is a susceptibility factor targeted by an RXLR effector to promote late blight disease. Mol Plant. 2015;8:1385–1395. doi: 10.1016/j.molp.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boevink PC, et al. A Phytophthora infestans RXLR effector targets plant PP1c isoforms that promote late blight disease. Nat Commun. 2016;7:10311. doi: 10.1038/ncomms10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbull D, et al. RXLR effector AVR2 up-regulates a brassinosteroid-responsive bHLH transcription factor to suppress immunity. Plant Physiol. 2017;174:356–369. doi: 10.1104/pp.16.01804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen LQ, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Römer P, et al. Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytol. 2010;187:1048–1057. doi: 10.1111/j.1469-8137.2010.03217.x. [DOI] [PubMed] [Google Scholar]
- 16.Streubel J, et al. Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 2013;200:808–819. doi: 10.1111/nph.12411. [DOI] [PubMed] [Google Scholar]
- 17.Lewis JD, et al. Quantitative Interactor Screening with next-generation Sequencing (QIS-Seq) identifies Arabidopsis thaliana MLO2 as a target of the Pseudomonas syringae type III effector HopZ2. BMC Genomics. 2012;13:8. doi: 10.1186/1471-2164-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui H, et al. Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe. 2010;7:164–175. doi: 10.1016/j.chom.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desveaux D, et al. Type III effector activation via nucleotide binding, phosphorylation, and host target interaction. PLoS Pathog. 2007;3:e48. doi: 10.1371/journal.ppat.0030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewezi T, Baum TJ. Manipulation of plant cells by cyst and root-knot nematode effectors. Mol Plant Microbe Interact. 2013;26:9–16. doi: 10.1094/MPMI-05-12-0106-FI. [DOI] [PubMed] [Google Scholar]
- 21.Hewezi T, et al. Cellulose binding protein from the parasitic nematode Heterodera schachtii interacts with Arabidopsis pectin methylesterase: Cooperative cell wall modification during parasitism. Plant Cell. 2008;20:3080–3093. doi: 10.1105/tpc.108.063065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang L, et al. Potato NPH3/RPT2-like protein StNRL1, targeted by a Phytophthora infestans RXLR effector, is a susceptibility factor. Plant Physiol. 2016;171:645–657. doi: 10.1104/pp.16.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bos JIB, et al. Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc Natl Acad Sci USA. 2010;107:9909–9914. doi: 10.1073/pnas.0914408107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi K, et al. SWAP70 functions as a Rac/Rop guanine nucleotide-exchange factor in rice. Plant J. 2012;70:389–397. doi: 10.1111/j.1365-313X.2011.04874.x. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi K, Kawasaki T. Function of Arabidopsis SWAP70 GEF in immune response. Plant Signal Behav. 2012;7:465–468. doi: 10.4161/psb.19562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krek W. BTB proteins as henchmen of Cul3-based ubiquitin ligases. Nat Cell Biol. 2003;5:950–951. doi: 10.1038/ncb1103-950. [DOI] [PubMed] [Google Scholar]
- 27.Pintard L, et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 2003;425:311–316. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]
- 28.Kotzer AM, et al. AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J Cell Sci. 2004;117:6377–6389. doi: 10.1242/jcs.01564. [DOI] [PubMed] [Google Scholar]
- 29.Richter S, Voss U, Jürgens G. Post-Golgi traffic in plants. Traffic. 2009;10:819–828. doi: 10.1111/j.1600-0854.2009.00916.x. [DOI] [PubMed] [Google Scholar]
- 30.Ebine K, et al. A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat Cell Biol. 2011;13:853–859. doi: 10.1038/ncb2270. [DOI] [PubMed] [Google Scholar]
- 31.Saint-Jean B, Seveno-Carpentier E, Alcon C, Neuhaus JM, Paris N. The cytosolic tail dipeptide Ile-Met of the pea receptor BP80 is required for recycling from the prevacuole and for endocytosis. Plant Cell. 2010;22:2825–2837. doi: 10.1105/tpc.109.072215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liscum E, et al. Phototropism: Growing towards an understanding of plant movement. Plant Cell. 2014;26:38–55. doi: 10.1105/tpc.113.119727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts D, et al. Modulation of phototropic responsiveness in Arabidopsis through ubiquitination of phototropin 1 by the CUL3-Ring E3 ubiquitin ligase CRL3(NPH3) Plant Cell. 2011;23:3627–3640. doi: 10.1105/tpc.111.087999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melnick A, et al. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol Cell Biol. 2002;22:1804–1818. doi: 10.1128/MCB.22.6.1804-1818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genschik P, Sumara I, Lechner E. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): Cellular functions and disease implications. EMBO J. 2013;32:2307–2320. doi: 10.1038/emboj.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orosa B, et al. BTB-BACK domain protein POB1 suppresses immune cell death by targeting ubiquitin E3 ligase PUB17 for degradation. PLoS Genet. 2017;13:e1006540. doi: 10.1371/journal.pgen.1006540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullivan S, Thomson CE, Kaiserli E, Christie JM. Interaction specificity of Arabidopsis 14-3-3 proteins with phototropin receptor kinases. FEBS Lett. 2009;583:2187–2193. doi: 10.1016/j.febslet.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 38.Tseng TS, Whippo C, Hangarter RP, Briggs WR. The role of a 14-3-3 protein in stomatal opening mediated by PHOT2 in Arabidopsis. Plant Cell. 2012;24:1114–1126. doi: 10.1105/tpc.111.092130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tseng TS, Briggs WR. The Arabidopsis rcn1-1 mutation impairs dephosphorylation of Phot2, resulting in enhanced blue light responses. Plant Cell. 2010;22:392–402. doi: 10.1105/tpc.109.066423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wen F, Wang J, Xing D. A protein phosphatase 2A catalytic subunit modulates blue light-induced chloroplast avoidance movements through regulating actin cytoskeleton in Arabidopsis. Plant Cell Physiol. 2012;53:1366–1379. doi: 10.1093/pcp/pcs081. [DOI] [PubMed] [Google Scholar]
- 41.Suetsugu N, et al. RPT2/NCH1 subfamily of NPH3-like proteins is essential for the chloroplast accumulation response in land plants. Proc Natl Acad Sci USA. 2016;113:10424–10429. doi: 10.1073/pnas.1602151113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park CH, et al. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell. 2012;24:4748–4762. doi: 10.1105/tpc.112.105429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park CH, et al. The E3 ligase APIP10 connects the effector AvrPiz-t to the NLR receptor Piz-t in rice. PLoS Pathog. 2016;12:e1005529. doi: 10.1371/journal.ppat.1005529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacLean AM, et al. Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol. 2014;12:e1001835. doi: 10.1371/journal.pbio.1001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawano Y, Kaneko-Kawano T, Shimamoto K. Rho family GTPase-dependent immunity in plants and animals. Front Plant Sci. 2014;5:522. doi: 10.3389/fpls.2014.00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akamatsu A, et al. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe. 2013;13:465–476. doi: 10.1016/j.chom.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 47.Mbengue M, et al. Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc Natl Acad Sci USA. 2016;113:11034–11039. doi: 10.1073/pnas.1606004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nomura K, et al. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- 49.Nomura K, et al. Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:10774–10779. doi: 10.1073/pnas.1103338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy F, et al. The potato MAP3K StVIK is required for the Phytophthora infestans RXLR effector Pi17316 to promote disease. Plant Physiol. 2018;177:398–410. doi: 10.1104/pp.18.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He Q, et al. U-box E3 ubiquitin ligase PUB17 acts in the nucleus to promote specific immune pathways triggered by Phytophthora infestans. J Exp Bot. 2015;66:3189–3199. doi: 10.1093/jxb/erv128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J. 2002;31:777–786. doi: 10.1046/j.1365-313x.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- 53.McLellan H, et al. An RxLR effector from Phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathog. 2013;9:e1003670. doi: 10.1371/journal.ppat.1003670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gilroy EM, et al. Involvement of cathepsin B in the plant disease resistance hypersensitive response. Plant J. 2007;52:1–13. doi: 10.1111/j.1365-313X.2007.03226.x. [DOI] [PubMed] [Google Scholar]
- 55.Bos JIB, et al. The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 2006;48:165–176. doi: 10.1111/j.1365-313X.2006.02866.x. [DOI] [PubMed] [Google Scholar]
- 56.Gilroy EM, et al. CMPG1-dependent cell death follows perception of diverse pathogen elicitors at the host plasma membrane and is suppressed by Phytophthora infestans RXLR effector AVR3a. New Phytol. 2011;190:653–666. doi: 10.1111/j.1469-8137.2011.03643.x. [DOI] [PubMed] [Google Scholar]
- 57.Engelhardt S, et al. Relocalization of late blight resistance protein R3a to endosomal compartments is associated with effector recognition and required for the immune response. Plant Cell. 2012;24:5142–5158. doi: 10.1105/tpc.112.104992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cikos S, Bukovská A, Koppel J. Relative quantification of mRNA: Comparison of methods currently used for real-time PCR data analysis. BMC Mol Biol. 2007;8:113. doi: 10.1186/1471-2199-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.