Significance
The conversion of extracellular chemical signals into electrical current across the cell membrane is a defining characteristic of the nervous system. This is mediated by proteins, such as acid-sensing ion channels (ASICs), membrane-bound receptors whose activation by decreased extracellular pH opens an intrinsic membrane-spanning sodium channel. Curiously, ASICs had only been reported in vertebrates, despite the homology of many other ion channels in vertebrates and invertebrates. Using molecular phylogenetics and electrophysiological recordings, we discover ASICs from tunicates, lancelets, sea urchins, starfish, and acorn worms. This shows that ASICs evolved much earlier than previously thought and suggests that their role in the nervous system is conserved across numerous animal phyla.
Keywords: ASIC evolution, ligand recognition, invertebrate, nervous system, proton sensing
Abstract
Acid-sensing ion channels (ASICs) are proton-gated ion channels broadly expressed in the vertebrate nervous system, converting decreased extracellular pH into excitatory sodium current. ASICs were previously thought to be a vertebrate-specific branch of the DEG/ENaC family, a broadly conserved but functionally diverse family of channels. Here, we provide phylogenetic and experimental evidence that ASICs are conserved throughout deuterostome animals, showing that ASICs evolved over 600 million years ago. We also provide evidence of ASIC expression in the central nervous system of the tunicate, Oikopleura dioica. Furthermore, by comparing broadly related ASICs, we identify key molecular determinants of proton sensitivity and establish that proton sensitivity of the ASIC4 isoform was lost in the mammalian lineage. Taken together, these results suggest that contributions of ASICs to neuronal function may also be conserved broadly in numerous animal phyla.
Neurons are set apart from other cells by their rapid conduction and exchange of information. This is in large part due to ligand-gated ion channels, membrane-bound receptors that convert chemical information, such as neurotransmitter release, into electrical current (1). Not surprisingly, the evolution of nervous systems is intricately linked to the emergence and expansion of ligand-gated ion channels in the genome (2). For example, certain neurotransmitter receptor families seem to have expanded uniquely in those basal animals with nervous systems (3), and these include the glutamate receptors typically associated with fast synaptic transmission and plasticity (4, 5).
It has recently emerged that protons act as neurotransmitters at certain glutamatergic synapses in the mammalian brain, where excitatory current through acid-sensing ion channels (ASICs) is required for synaptic plasticity (6, 7). This follows from several experiments implicating hippocampal and amygdala ASICs in learning and memory, fear, and depression (8). Members of the ASIC family have been cloned from simpler chordates, including the tunicate Ciona intestinalis and the jawless fish Lampetra fluviatilis, but these did not respond to protons (9, 10). This raises the possibility that proton sensitivity of ASICs emerged within the jawed vertebrates, well after this lineage had split from other, simpler deuterostome animals (9, 10). However, ASICs are widely distributed in the vertebrate nervous system and contribute to behaviors that are conserved across most animals, including nociception, chemosensation, and mechanosensation (11, 12). Furthermore, ASICs derive from the degenerin/epithelial Na+ channel (DEG/ENaC) family, genes of which are conserved throughout all major animal lineages (3), and some of which are expressed in invertebrate postsynaptic membranes (13). These facts are difficult to reconcile with ASICs being limited to the jawed vertebrates.
DEG/ENaC subunits form trimeric channels of diverse function. Examples include mechanically gated channels in roundworms (14), peptide-gated channels in snails and hydra (15, 16), and constitutively active ENaCs in vertebrates (17). It seems plausible that proton-gated channels could also contribute to physiology in more diverse animal lineages, and that we should therefore find ASICs in nonvertebrate genomes. However, previous studies suggest that ASICs are not readily identifiable through sequence analyses alone (9). This is likely due to our poor understanding of the molecular determinants of proton sensitivity, notwithstanding the finding that the mutation of several protonatable side chains in the extracellular domain decreases proton sensitivity of ASICs (18–22).
Here, we sought to illuminate the evolutionary emergence of ASICs and the molecular basis of proton sensing. Combining recent genetic data with molecular phylogenetics and electrophysiological recordings, we identified functional ASICs from a more representative sample of animals than previously possible. Through various mutagenesis experiments, we established determinants of proton-gated currents that underlie proton sensitivity throughout the ASIC family. Our results show that ASICs evolved much earlier than previously thought, suggest that proton-gated currents may contribute to neuronal signaling throughout deuterostome animals, and point toward a mechanism of proton sensing involving several parts of the channel.
Results
Proton-Gated ASICs Are Conserved Throughout the Deuterostomes.
The phylum Chordata includes vertebrates, tunicates (sister taxon to vertebrates), and cephalochordates (23). Chordates, together with echinoderms and hemichordates, comprise the deuterostomes, which are distinct from protostomes (e.g., mollusks, arthropods, and roundworms) and from the xenacoelomorphs (24). Together, deuterostomes, protostomes, and xenacoelomorphs comprise the Bilateria, one of the five major lineages of animals [the others being Cnidaria (e.g., hydra), Placazoa, Ctenophora (comb jellies), and Porifera (sponges)]. Previous investigation of ASIC evolution was limited by a vertebrate-centric literature, although an ASIC-like protein from the tunicate C. intestinalis was recombinantly expressed, yielding no proton-gated currents (9). To date, proton-activated currents have not been observed in protostome DEG/ENaC channels. To address the occurrence of ASICs in close relatives of the vertebrates, we assembled DEG/ENaC amino acid sequences from one vertebrate, one tunicate, two cephalochordates (both Branchiostoma spp.), two echinoderms, and two hemichordates, and assessed the phylogenetic relationship of 102 nonredundant sequences with maximum-likelihood methods (SI Appendix, Fig. S1 A and B). In the resulting tree, several basal branches were poorly supported in bootstrap resampling, but several well-supported clades were identified that include vertebrate channel subunits of established functional properties. One of these clades includes the vertebrate bile acid-sensing ion channel (BASIC), which is not activated by protons (25); another includes vertebrate ENaCs, which are constitutively active (16); and another includes vertebrate ASICs (SI Appendix, Fig. S1A). The ASIC clade includes sequences from each lineage investigated, suggesting that ASICs could be conserved throughout the deuterostomes.
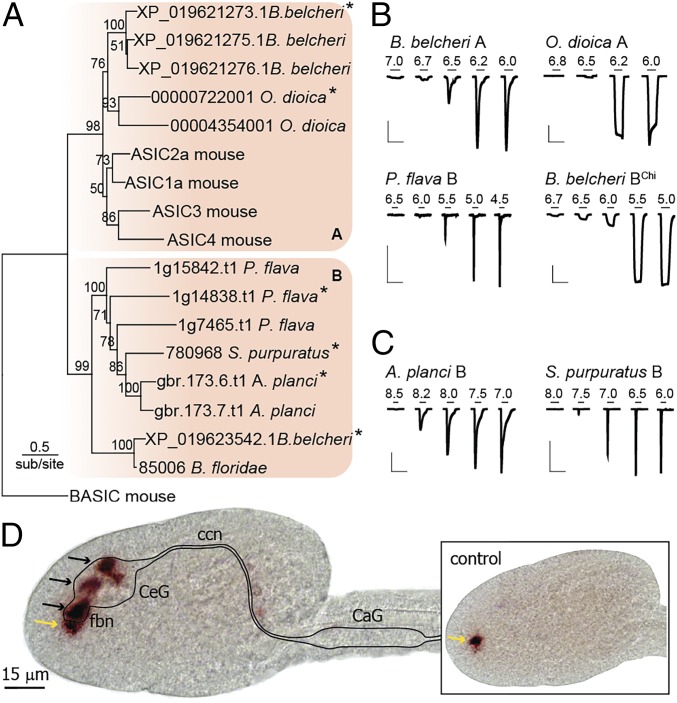
A maximum-likelihood tree of the deuterostome ASIC-like sequences, rooted to the outgroup of vertebrate BASIC, is shown in Fig. 1A, recapitulating the relationships of the overarching tree in SI Appendix, Fig. S1, including two well-supported clades within the ASICs (hereafter “group A” and “group B”). Proton-gated currents through channels formed by these subunits would confirm the conservation of ASICs throughout deuterostomes. We therefore synthesized cDNAs (de novo) of at least one subunit from each species addressed (asterisks in Fig. 1A), injected cRNAs into Xenopus oocytes, and measured current in response to decreased pH. For most channels, rapid application of acidic pH from a resting pH of 7.5 yielded robust proton-gated currents (Fig. 1B and Table 1). Acanthaster planci and Strongylocentrotus purpuratus group B ASICs showed constitutive and no proton-gated current, respectively, with a resting pH of 7.5, but when a resting pH of 9.0 was used, both channels were robustly activated by lower pH (Fig. 1C and Table 1). These experiments confirm that ASICs are conserved in each deuterostome lineage, suggesting that ASICs evolved or were already present in ancestral deuterostomes. Phylogenetic analysis including mollusk and arthropod DEG/ENaC sequences returned the very same clade of deuterostome-specific ASIC sequences (SI Appendix, Fig. S1E), suggesting that ASICs emerged after the protostome/deuterostome split.
Fig. 1.
Deuterostome ASICs identified through phylogenetic and functional analyses. (A) Maximum-likelihood tree inferred from alignment of 17 ASIC-like subunits from one vertebrate, one tunicate, two cephalochordates, one hemichordate, and two echinoderms, rooted to mouse BASIC. (B and C) pH-gated currents in oocytes expressing indicated cRNAs. Resting pH 7.5 (B) or 9.0 (C). [Scale bars: x 20 s; y 0.1 μA (B. belcheri A and P. flava B) or 3 μA (all others).] B. belcheri BChi is a chimera (SI Appendix, Supplementary Text). (D) In situ hybridization targeting O. dioica group A ASIC (black arrows). Illustrated CNS landmarks: CaG, caudal ganglion; ccn, cerebrocaudal nerve; CeG, cerebral ganglion; fbn, first brain nerve. (Inset) Control experiment using complementary sense probe showing nonspecific staining in the pharynx (yellow arrows in both sense and antisense experiments).
Table 1.
Characteristics of deuterostome ASICs (mean ± SD)
| Proton sensitivity | Permeability | ||||
| Deuterosome | pH50 | nH | n | PNa+/PK+ | n |
| Mouse ASIC1a | 6.8 ± 0.06 | 7.6 ± 4.4 | 6 | 8.7 ± 1.0 | 5 |
| O. doica A | 6.3 ± 0.02 | 9.9 ± 0.8 | 5 | 10.4 ± 1.9 | 6 |
| B. belcheri A | 6.5 ± 0.04 | 4.9 ± 1.2 | 5 | 12.2 ± 2.7 | 4 |
| B. belcheri B | 5.8 ± 0.03 | 4.5 ± 0.9 | 6 | 1.7 ± 0.2 | 5 |
| P. flava B | 5.5 ± 0.08 | 2.5 ± 0.9 | 6 | 1.9 ± 0.9 | 6 |
| S. purpuratus B | 7.0 ± 0.09 | 2.4 ± 0.5 | 5 | 1.6 ± 0.3 | 5 |
| A. planci B | 8.1 ± 0.05 | 5.1 ± 2.0 | 6 | 3.1 ± 0.4 | 5 |
Most DEG/ENaC channels selectively conduct sodium ions, with relative permeability over potassium (PNa+/PK+) ranging from ∼10 to over 100 (26). We measured PNa+/PK+ for newly identified ASICs (SI Appendix, Fig. S2A) and found that group A ASICs from Oikopleura dioica and Branchiostoma belcheri showed similar ion selectivity to mouse ASIC1a (also from group A) (Table 1). Surprisingly, group B ASICs showed relatively weak ion selectivity, with PNa+/PK+ values closer to unity (Table 1). Because a highly conserved glutamate residue in the channel pore determines ion selectivity in mouse ASIC1a (27), the presence of a neutral glutamine residue at the equivalent position in Ptychodera flava and S. purpuratus group B ASICs could underlie low selectivity in those channels (SI Appendix, Fig. S2B). The basis for low selectivity in B. belcheri and A. planci group B channels is unclear, however, as both possess this glutamate residue, although the A. planci channel lacks an aspartate residue near the top of the channel pore that contributes to ion conduction in human ASIC1a (28) (SI Appendix, Fig. S2B). Nonselective cation current through ASICs is still likely to be excitatory (29), and these results are therefore consistent with a role for ASICs in excitatory signaling throughout deuterostome animals.
If ASICs mediate excitatory neuronal signals in invertebrate deuterostomes, one might expect to find them in the central nervous system (CNS), as is largely the case in rodents and humans (30). To test this, we performed in situ hybridization on the tunicate O. dioica, using two antisense probes of different lengths against the O. dioica group A ASIC described above. The pattern of ASIC transcript signal was the same for both probes, but varied at different developmental stages. Here we report the temporally most consistent signal, which was localized to the cerebral ganglion, the main hub of the O. dioica CNS and the first brain nerves, which convey sensory information from the mouth region (Fig. 1D) (31). Control experiments using a complementary sense probe did not yield this signal, but rather nonspecific staining within the pharynx (Fig. 1D, control). The CNS localization of an invertebrate ASIC transcript provides additional support for the notion that ASICs contribute to neuronal signaling in invertebrate deuterostomes.
A Broader ASIC Comparison Establishes Crucial Determinants of Proton Sensitivity.
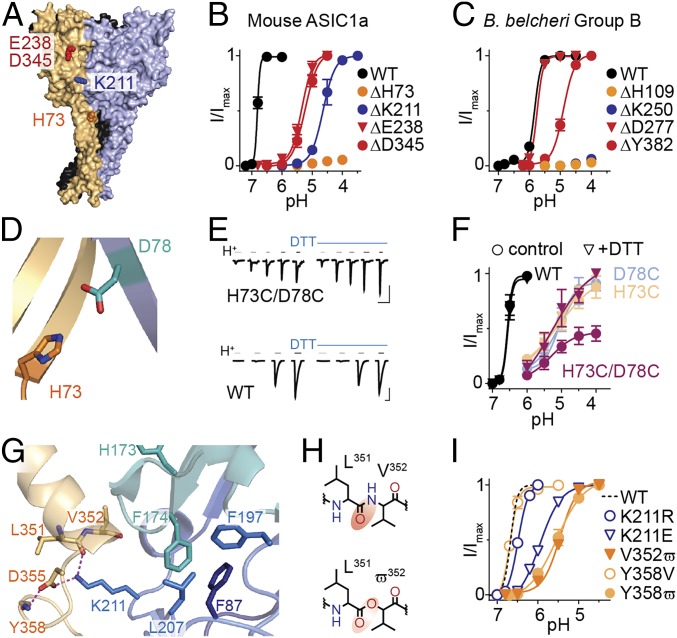
To distinguish between conserved, fundamental determinants of proton sensing and lineage-specific features, we compared the effects of mutating candidate proton sensors in two distantly related ASICs, mouse ASIC1a (group A) and B. belcheri group BChi (Fig. 1A). We focused on four extracellular positions that have emerged as potential proton sensors in structural and functional ASIC studies (18–21), including residues from the β1 strand (H73 in mouse ASIC1a), the β5/β6 loop (K211), the β6/β7 loop (E238), and the α5 helix (D345) (Fig. 2A and SI Appendix, Fig. S3A). The latter two both contribute to the “acidic pocket” that contains several acidic side chains (18). We measured proton sensitivity in mutants where residues were either deleted or substituted (Fig. 2 B and C and SI Appendix, Fig. S3B), as it is arguable whether typical charge-neutralizing substitutions, such as D345N, impair (32) or mimic (22) protonation.
Fig. 2.
Determinants of proton sensitivity in broadly related ASICs. (A) Chicken ASIC1 structure (PDB ID code 2QTS), different subunits colored orange, blue, and black. Selected side chains are shown in red (acidic pocket), blue (β5/β6 loop from adjacent subunit), and orange (β1 strand). All numbers in this figure refer to equivalent residues in mouse ASIC1a. (B and C) Normalized peak current responses to decreasing pH for mouse ASIC1a or B. belcheri group BChi ASIC mutants carrying indicated deletions (mean ± SEM, n = 5–7). (D) Magnified view of A showing H73 and D78 from adjacent subunits. (E) Example current responses to acidic pH (gray/black bars) from resting pH of 7.5, alone or with 2 mM DTT. pH for H73C/D87C 6.0/5.5/5.0/4.5/4.0; WT 7.0/6.8/6.5/6.0. [Scale bars: x 20 s; y 100 nA (H73C/D78C) and 3 µA (WT).] (F) Normalized responses to acidic pH with/without 2 mM DTT for WT and indicated mouse ASIC1a mutants (mean ± SEM, n = 4). (G) Magnified view of A showing K211 and selected vicinal side-chain/main-chain atoms. Dashed lines: distance = 2.9 Å. (H) Valine (V) and 2-hydroxy-3-methylbutanoic acid (ϖ), red ellipse highlighting decreased electron density. (I) Normalized responses to acidic pH for regular substitutions and noncanonical substitutions (mean ± SEM, n = 5–7).
Both approaches identified H73 as a crucial determinant of proton sensitivity in distantly related ASICs. In both mouse ASIC1a and in B. belcheri group BChi, the deletion of acidic pocket residues caused significant but relatively moderate decreases in proton sensitivity, whereas the β5/β6 lysine and, especially, the β1 histidine deletions were severely detrimental to proton-gated currents (Fig. 2 B and C and SI Appendix, Fig. S3A). In mouse ASIC1a, even the ΔE235-E238 deletion, removing most of this loop from the finger domain in the acidic pocket, did not decrease potency to the extent of ΔH73 or ΔK211 deletions (SI Appendix, Fig. S3A). The amino acid substitutions revealed a similar pattern, where β1 histidine mutations were by far the most detrimental (SI Appendix, Fig. S3B). Combined with the high conservation of this residue uniquely in ASICs and the lesser conservation of the β5/β6 lysine and acidic pocket residues, this suggests that the appearance of the histidine residue could underlie the emergence of proton-gated currents (SI Appendix, Fig. S3 C and D).
These results suggest intimate roles for the β1 strand and the β5/β6 loop in ASIC activation. Because recent structural and earlier mutagenesis studies implicate β1/β1 rearrangements in activation by protons (20, 33), we substituted H73 and D78 residues for cysteine in mouse ASIC1a and looked for changes in proton-gated currents in the presence of the reducing agent DTT, which would be indicative of a disulfide cross-link between adjacent β1 domains (34). As expected, activation of mutant H73C/D78C channels required much higher proton concentrations than WT channels (Fig. 2 E and F). In the presence of DTT, peak current amplitude was approximately doubled in H73C/D78C channels (and not in WT or single-mutant H73C or D78C channels), suggesting that a link between introduced cysteine residues forms spontaneously and is broken upon reduction. There was no significant difference in pH50 without (5.1 ± 0.1, n = 4) and with DTT (5.0 ± 0.4, n = 4; P = 0.959), suggesting that the β1-β1 cross-link causes smaller currents by precluding activation of many channels, rather than shifting the gating equilibrium of all channels. We also observed that application of DTT immediately preceding acidic pH increased current amplitudes, whereas coapplication of DTT during acidic pH had no effect on current amplitude (SI Appendix, Fig. S4A). These results confirm that β1/β1 interactions affect channel activation by protons and suggest that enforced 73/78 proximity in resting channels stabilizes a state from which activation proceeds poorly.
Structural data place the K211 side-chain nitrogen atom <3 Å from both the main chain carbonyl oxygen of L351 and the side chain carboxylate of D355 (Fig. 2G), and D355 mutations were previously shown to impair ASIC1a activation (20, 21). Here we questioned the role of a potential hydrogen bond (H bond) between K211 and L351 by replacing V352 with 2-hydroxy-3-methylbutanoic acid (ϖ). This withdraws electron density from the backbone carbonyl oxygen of L351 (Fig. 2H), making the latter a weaker H-bond acceptor (27). V352ϖ channels showed substantially reduced proton sensitivity compared with WT (Fig. 2I and SI Appendix, Fig. S4C) (pH50 = 5.47 ± 0.05, n = 7, P < 0.001 compare with WT), providing evidence that a K211/L351 H bond is important for ASIC1a activation. This is supported by our observation that the K211R substitution, retaining a strong H-bond donor, is well tolerated compared with the K211E substitution, which removes a strong H-bond donor (Fig. 2I). Another potential H bond in this domain appears between the carboxylate side chain of D355 and the main-chain nitrogen of Y358 (Fig. 2G), which we probed by measuring proton sensitivity of Y358ϖ channels, in which the main-chain H-bond donor (NH) is replaced by an acceptor (CO). The Y358ϖ substitution (pH50 = 5.51 ± 0.14, n = 7, P < 0.001 compared with WT) caused a similar decrease in proton sensitivity as the V352ϖ substitution, whereas the Y358V substitution did not differ from WT (Fig. 2I and SI Appendix, Fig. S4C). This provides evidence for an additional H bond in this domain contributing to proton sensitivity, and we conclude that in ASIC1a, K211 bridges thumb and palm domains of adjacent subunits via an H-bond network.
Unlike several other loops in the ASIC extracellular domain, the β5/β6 loop containing K211 is not stabilized by endogenous disulfides. We therefore questioned how K211 rearrangements could be stably coupled to conformational changes “downstream,” toward the channel domain. We noticed that two hydrophobic side chains of the β5/β6 loop interact closely with hydrophobic side chains from the β3/β4 and β1/β2 loops of the same subunit in crystal structures (Fig. 2G and SI Appendix, Fig. S5A). Hypothesizing that these interactions couple β5/β6 activity to channel activity, we tested the role of these large hydrophobic side chains by substituting them with alanine and recording proton-gated currents in mouse ASIC1a. β3/β4 F174A, β5/β6 F197A, and L207A, and especially β1/β2 F87A substitutions substantially reduced proton sensitivity compared with WT (SI Appendix, Fig. S5 B and C). Notably, the H173A mutation, adjacent to F174 but oriented away from this hub, had no effect on proton sensitivity (SI Appendix, Fig. S5). This suggests that a hub of hydrophobic side chains may provide a stable link between two crucial domains: the interface of adjacent β1 strands and the more extracellular interface of thumb (α5) and palm (β5/β6 loop) domains.
Loss of Proton Sensitivity in ASIC4 in Mammals.
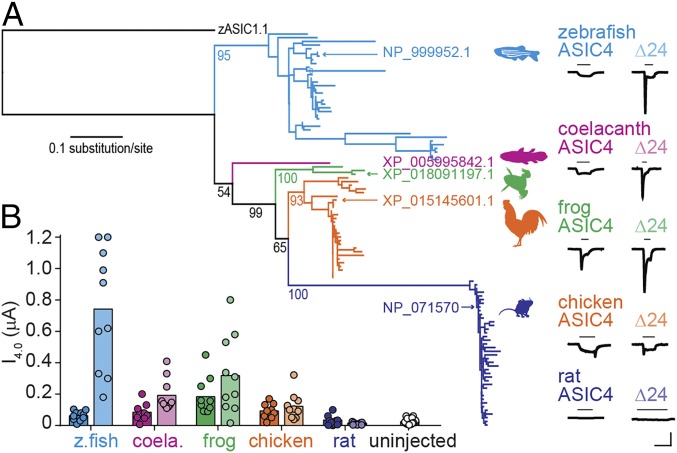
We next questioned why rat ASIC4 is apparently insensitive to protons, despite proton-gated currents in zebrafish ASIC4.1 and evidence for ASIC4 transcripts in the rat brain (35–37). Assessing the phylogenetic relationship of ASIC4 sequences showed broad conservation of ASIC4 throughout the vertebrates, with a maximum-likelihood tree of vertebrate ASIC4 sequences generally reflecting the overarching evolution of vertebrate animals (Fig. 3A). To establish where in the vertebrate lineage proton sensitivity of ASIC4 was lost, we tested ASIC4 channels from three major lineages between bony fishes and mammals: coelacanth (lobe-finned fish), frog, and chicken. Each of these ASICs showed proton-gated currents, although current amplitude was small and variable (Fig. 3). We therefore tested a mutated version of each channel, carrying a 24-amino acid deletion in the N terminal (hereafter “Δ24”), which has been shown to enhance zebrafish ASIC4 expression (38). Coelacanth, frog, and chicken ASIC4Δ24 channels showed larger currents in response to acidic pH, whereas rat ASIC4Δ24 remained unresponsive (Fig. 3). Thus, as ASIC4 in all vertebrate lineages except mammalian shows some proton-gated current, we conclude that ASIC4 was proton-sensitive in the ancestor of reptilian and mammalian lineages and lost proton sensitivity in the latter.
Fig. 3.
Proton sensitivity in vertebrate ASIC4. (A) Maximum-likelihood tree of ASIC4 amino acid sequences, rooted to zebrafish ASIC1.1. Bootstrap support (per 100) shown for selected branches. Example pH 4.0-gated currents for indicated ASICs shown on right. (Scale bars: x 10 s; y 0.1 µA.) “Δ24” constructs lack 24 N-terminal amino acids following the starting methionine. (B) Average (column) and individual (dots) pH 4.0-gated peak current amplitudes (color as in A). n = 10, except for uninjected, n = 25.
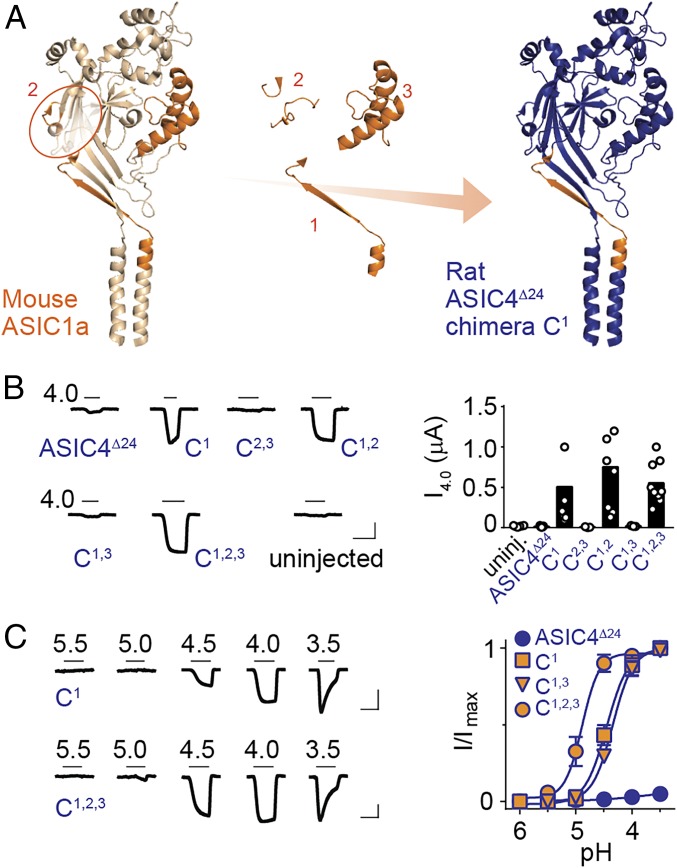
We aligned rat ASIC4 with proton-sensitive ASIC sequences, looking for amino acid sequence differences that might underlie the loss of proton sensitivity, but crucial residues were largely conserved, including β1 H73 and D78 and β1/β2 F87 (SI Appendix, Fig. S6A). We therefore turned to a different approach and generated chimeric channels based on proton-insensitive rat ASIC4Δ24 and containing one to three segments of highly proton-sensitive mouse ASIC1a sequence (Fig. 4A and SI Appendix, Fig. S6A). Segment 1 (25 residues) consisted primarily of the β1 strand containing H73/D78 and was chosen based on our results (Fig. 2) and on previous work showing that this segment is crucial for proton-gated currents (10, 38). Segment 2 (17 residues) included parts of β3/β4 and β5/β6 (including K211) loops, and segment 3 (54 residues) consisted primarily of the thumb domain α4 and α5 helices. These were chosen based on findings that interfacial interactions between thumb and β3/β4 and β5/β6 loops contribute to proton sensitivity (Fig. 2) (20, 39). We hypothesized that alone, segment 1 of ASIC1a might confer proton-gated currents on rat ASIC4Δ24 and the additional combination of segments 2 and 3 might render rat ASIC4Δ24 more sensitive to protons by introducing more favorable interactions between adjacent subunits.
Fig. 4.
ASIC1a β1 segment confers proton sensitivity on ASIC4. (A) Cartoon illustrating ASIC1a segments that were substituted into ASIC4 (segment 2 circled for clarity). (B) Example currents (Left) and mean peak current amplitude (black columns, Right) in response to pH 4.0 at indicated constructs. (C) Example currents (Left) and mean (±SEM, n = 5–6), normalized responses (Right) to decreasing pH. (Scale bars in B and C: x 5 s, y 0.1 μA.)
Remarkably, chimera C1, containing only segment 1 from mouse ASIC1a, showed robust proton-gated currents, with a pH50 of 4.5 ± 0.2 (n = 6) (Fig. 4 B and C), confirming that this segment is crucial for proton-gated currents. In contrast, chimera C2,3, containing only segments 2 and 3 and thus a putative ASIC1a-like intersubunit interface around K211, showed no responses to pH 4 (Fig. 4B). Combining either segment 2 or 3 with segment 1 (in chimeras C1,2 and C1,3) had no effect on proton sensitivity relative to chimera C1 or abolished proton-gated currents, respectively (Fig. 4 B and C). However, adding both extracellular segments to chimera C1, yielding chimera C1,2,3, caused a significant increase in proton sensitivity relative to chimera C1 (Fig. 4C), reflected in the higher pH50 value of 4.9 ± 0.2 for chimera C1,2,3 (n = 6, P = 0.001). Thus, the ASIC1a β1 segment renders ASIC4 a proton-gated channel, and adding ASIC1a-like thumb domain/palm domain interfaces increases proton sensitivity. Considering which β1 amino acid differences make mammal ASIC4 proton-insensitive, we noticed an insertion in mammal ASIC4 preceding the conserved β1/β2 phenylalanine residue (SI Appendix, Fig. S6A). Inserting this residue into proton-sensitive chimera C1 (rat ASIC4Δ24 with ASIC1a β1) abolished proton-gated currents (SI Appendix, Fig. S6B), suggesting that such a mutation could have contributed to the loss of proton sensitivity in mammal ASIC4. Deleting the equivalent residue from ASIC4 β1 failed to confer proton-gated currents on rat ASIC4Δ24, unlike replacing the entire ASIC4 β1 with that of ASIC1a (SI Appendix, Fig. S6B). Taken together, these results suggest that proton insensitivity in mammal ASIC4 is due to several differences from other ASICs in the β1 segment, including a different number of residues in this segment, and that low proton sensitivity in ASIC4 also derives from divergence in the intersubunit interface around K211.
Discussion
Gain of Proton Sensing and Potential Roles for ASICs in Invertebrate Neurophysiology.
Previous work identified ASIC subunits from the tunicate C. intestinalis, the jawless fish L. fluviatilis, and the cartilaginous fish Squalus acanthias, which formed membrane proteins that were not activated by protons (9, 10). With 34–66% identity with rat ASIC1a, these sequences certainly fit into the ASIC clade we now describe, but their proton insensitivity and the absence of data on other deuterostome ASICs at the time led to the logical conclusion that DEG/ENaC sequences developed proton-gated currents after the bony fishes split from the cartilaginous fishes (10), ∼420 Mya, well after chordates split from other deuterostomes (40). We sampled recent genetic data from a broad selection of deuterostome animals, to assess the phylogenetic relationships of ASIC sequences and measure the function of recombinantly expressed channels. Our phylogenetic and functional analyses identify ASICs from each deuterostome lineage that form a well-supported clade within the overarching DEG/ENaC family. Within the ASIC clade are two well-supported groups, and although ASICs of vertebrates and tunicates appear in only group A and ASICs of echinoderms and hemichordates appear only in group B, there is good support for cephalochordate ASICs in both clades (SI Appendix, Fig. S1C). This suggests that an ancestral chordate possessed genes from both groups, and group B ASICs were lost in Olfactores (tunicates + vertebrates), although this tentative hypothesis would benefit from broader sampling in future. In any case, the presence of ASICs in ancestral deuterostomes means that ASICs emerged at least 600 Mya, well before the Cambrian explosion (41). Tentative phylogenetic evidence suggests that ASICs evolved after the protostome/deuterostome split.
We find that in the tunicate O. dioica, transcripts of a functional ASIC are consistently found within the cerebral ganglion, a structure that is homologous to the vertebrate brain, integrates input from a variety of sensory nerves, and dynamically regulates more caudal CNS structures (42–44). This CNS localization accords with several roles for ASICs in the vertebrate CNS, including stress responses (45) and synaptic plasticity at central glutamatergic synapses (6, 7). Because protons are coreleased with glutamate (46), and glutamatergic signaling underlies synaptic plasticity in both deuterostomes and protostomes (4, 5), we suggest that ASICs could also contribute to synaptic plasticity in invertebrates.
Loss of Proton Sensing in ASIC4.
Vertebrates possess four ASIC genes, the products of which form homo- or hetero-trimeric channels of varying proton sensitivity and expression patterns (26). Vertebrate ASIC4 is curious in that transcripts are expressed in various loci of human, rat, mouse, and rabbit brain (30), yet when expressed recombinantly it does not form functional channels, despite cell surface localization (35, 47, 48). Certain studies suggest that ASIC4 may coassemble with other ASIC subunits and reduce their function through intracellular compartmentalization or degradation (47, 49). We report proton-gated currents through ASIC4 channels from fish, frog, and chicken and find that this property was completely lost in the mammalian lineage. It is conceivable that coassembly/internalization functions of ASIC4 evolved before it lost proton sensitivity, such that proton sensitivity became redundant in mammal ASIC4 and numerous substitutions detrimental to proton sensitivity could then accumulate. Nonetheless, our results with ASIC1a/ASIC4 chimeras and ASIC4 mutants indicate that inactivity of mammal ASIC4 is determined by a 24-amino acid segment just external to the channel domain, which includes a functionally detrimental single amino acid insertion. Certain other ASICs also appear to have lost proton sensitivity, including one from C. intestinalis (9) and another from O. dioica (SI Appendix, Fig. S1A), which contains a similar amino acid insertion as mammal ASIC4 (SI Appendix, Fig. S6C).
Molecular Determinants of Proton Sensitivity.
Our experiments with diverse proton-sensitive ASICs reiterate the key role of the β1 segment, in particular a histidine residue conserved in all ASICs we characterized, suggesting it was present in the ancestral deuterosome ASIC. Furthermore, we show that linking adjacent β1 strands in resting channels precludes channel activation. This is reflected in the dynamic role of β1 emerging from the assessment of resting and open-state chicken ASIC1 X-ray structures, where the base of β1 appears to move away from that of adjacent β1 strands, pulling the channel open (33). Others proposed that protonation of H73 facilitates a H73/D78 interaction (20), although H73/D78 Cα/Cα distances differ little in resting and open structures (9.3 Å in PDB ID code 5WKU; compare with 10.1 Å in PDB ID code 4NTW). However, the resting-state C73/C78 cross-link formed spontaneously, despite ∼6.3-Å separation of these sulfhydryls in 5WKU (as measured by H73/D78 Cγ atoms), implying flexibility in this domain even at rest.
H73 could, together with D78, act as both a proton sensor and part of a transduction pathway between more external proton sensors and the channel domain. Its position seems suited to this and is similar to that of key proton sensors in proton-gated channels from other protein families (50). Because H73 is conserved throughout the extended ASIC family (SI Appendix, Fig. S3C), it was likely present in the earliest deuterostome ASIC, and we speculate that it was the key to the evolution of proton-gated currents. We suppose that over time—before and after the emergence of proton-gated currents—ASICs have undergone other lineage-specific mutations that alter proton sensitivity. Such elaborations may be sufficient for activation even in the absence of H73 [e.g., in zebrafish ASIC1.1 (37)] and may include K211 and the acidic pocket, which is poorly developed in group B ASICs but more extensive in group A ASICs, such as vertebrate ASIC1a (18) (SI Appendix, Fig. S3C).
K211 contributes to proton sensitivity by bridging palm and thumb domains of adjacent subunits, consistent with a decrease in K211 Cα/L351 CO distance of 1.3 Å from resting to open channel structures (33, 51). Whether K211 is uncharged at neutral pH and is protonated by low pH remains unclear, but we note that pKa values for buried lysine side chains, especially in close proximity to carboxylate or carbonyl atoms, can be lower than 6.0 (52). Tentative support for the notion that K211 is uncharged in resting and charged in activated/desensitized channels comes from the absence of negatively charged chloride ions near K211 in the pH ∼ 8.0 resting-state structure (33) and close proximity of chloride ions to the K211 amine in pH 5.0–6.0 structures (18, 51). We identify a hydrophobic hub ideally poised to couple motions around β5/β6 K211 to those around β1 H73. In line with this hypothesis, in the hydrophobic hub, the Cα/Cα distance between β5/β6 L207 and β1/β2 F87 remains 5.5 Å in both the resting- and open-state structures, in contrast to the β5/β6 and β1 domains themselves, which undergo more noticeable rearrangements.
Summary.
This work shifts the estimate of the origin of ASICs from after to before the Cambrian explosion, and it shows that the loss of proton sensitivity in ASIC4 occurred after vertebrates moved onto land. The occurrence of ASICs in invertebrates points toward additional homology between invertebrate and vertebrate nervous systems regarding excitatory neurotransmission.
Methods
ASICs were identified by alignment and phylogenetics of 102 DEG/ENaC amino acid sequences from deuterostomes. One sequence from each lineage (asterisks in Fig. 1A) was selected for commercial cDNA synthesis, cRNAs were injected into Xenopus laevis oocytes, and proton-gated currents were measured with two-electrode voltage clamp (TEVC). Whole-mount in situ hybridization (53) used 8-h postfertilization O. dioica and RNA probes targeting group A ASIC from O. dioica. Mouse ASIC1a and B. belcheri group B ASIC mutants (Fig. 2) were generated with site-directed mutagenesis and tested with TEVC. We used 108 vertebrate ASIC4 sequences to establish phylogenetic relationships of ASIC4 (Fig. 3). Three were selected for cDNA synthesis and TEVC, as were chimeric constructs from Fig. 4. See SI Appendix, Supplementary Text for detailed description of materials and methods.
Supplementary Material
Acknowledgments
This work was funded by the Danish Council for Independent Research (T.L.), the Lundbeck Foundation (T.L. and S.A.P.), the Carlsberg Foundation (S.A.P.), the Novo Nordisk Foundation (S.A.P.), and the Norwegian Research Council (J.C.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the Dryad Digital Data repository, doi: 10.5061/dryad.46320g1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806614115/-/DCSupplemental.
References
- 1.Smart TG, Paoletti P. Synaptic neurotransmitter-gated receptors. Cold Spring Harb Perspect Biol. 2012;4:a009662. doi: 10.1101/cshperspect.a009662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liebeskind BJ, Hillis DM, Zakon HH. Convergence of ion channel genome content in early animal evolution. Proc Natl Acad Sci USA. 2015;112:E846–E851. doi: 10.1073/pnas.1501195112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moroz LL, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510:109–114. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dale N, Kandel ER. L-glutamate may be the fast excitatory transmitter of Aplysia sensory neurons. Proc Natl Acad Sci USA. 1993;90:7163–7167. doi: 10.1073/pnas.90.15.7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicoll RA. A brief history of long-term potentiation. Neuron. 2017;93:281–290. doi: 10.1016/j.neuron.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 6.Du J, et al. Protons are a neurotransmitter that regulates synaptic plasticity in the lateral amygdala. Proc Natl Acad Sci USA. 2014;111:8961–8966. doi: 10.1073/pnas.1407018111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreple CJ, et al. Acid-sensing ion channels contribute to synaptic transmission and inhibit cocaine-evoked plasticity. Nat Neurosci. 2014;17:1083–1091. doi: 10.1038/nn.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wemmie JA, Taugher RJ, Kreple CJ. Acid-sensing ion channels in pain and disease. Nat Rev Neurosci. 2013;14:461–471. doi: 10.1038/nrn3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coric T, Passamaneck YJ, Zhang P, Di Gregorio A, Canessa CM. Simple chordates exhibit a proton-independent function of acid-sensing ion channels. FASEB J. 2008;22:1914–1923. doi: 10.1096/fj.07-100313. [DOI] [PubMed] [Google Scholar]
- 10.Coric T, Zheng D, Gerstein M, Canessa CM. Proton sensitivity of ASIC1 appeared with the rise of fishes by changes of residues in the region that follows TM1 in the ectodomain of the channel. J Physiol. 2005;568:725–735. doi: 10.1113/jphysiol.2005.087734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dymowska AK, Schultz AG, Blair SD, Chamot D, Goss GG. Acid-sensing ion channels are involved in epithelial Na+ uptake in the rainbow trout Oncorhynchus mykiss. Am J Physiol Cell Physiol. 2014;307:C255–C265. doi: 10.1152/ajpcell.00398.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price MP, et al. The DRASIC cation channel contributes to the detection of cutaneous touch and acid stimuli in mice. Neuron. 2001;32:1071–1083. doi: 10.1016/s0896-6273(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 13.Hill A, et al. The Drosophila postsynaptic DEG/ENaC channel ppk29 contributes to excitatory neurotransmission. J Neurosci. 2017;37:3171–3180. doi: 10.1523/JNEUROSCI.3850-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodman MB, et al. MEC-2 regulates C. elegans DEG/ENaC channels needed for mechanosensation. Nature. 2002;415:1039–1042. doi: 10.1038/4151039a. [DOI] [PubMed] [Google Scholar]
- 15.Golubovic A, et al. A peptide-gated ion channel from the freshwater polyp Hydra. J Biol Chem. 2007;282:35098–35103. doi: 10.1074/jbc.M706849200. [DOI] [PubMed] [Google Scholar]
- 16.Lingueglia E, Champigny G, Lazdunski M, Barbry P. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature. 1995;378:730–733. doi: 10.1038/378730a0. [DOI] [PubMed] [Google Scholar]
- 17.Canessa CM, Horisberger JD, Rossier BC. Epithelial sodium channel related to proteins involved in neurodegeneration. Nature. 1993;361:467–470. doi: 10.1038/361467a0. [DOI] [PubMed] [Google Scholar]
- 18.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 19.Li T, Yang Y, Canessa CM. Interaction of the aromatics Tyr-72/Trp-288 in the interface of the extracellular and transmembrane domains is essential for proton gating of acid-sensing ion channels. J Biol Chem. 2009;284:4689–4694. doi: 10.1074/jbc.M805302200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paukert M, Chen X, Polleichtner G, Schindelin H, Gründer S. Candidate amino acids involved in H+ gating of acid-sensing ion channel 1a. J Biol Chem. 2008;283:572–581. doi: 10.1074/jbc.M706811200. [DOI] [PubMed] [Google Scholar]
- 21.Sherwood T, et al. Identification of protein domains that control proton and calcium sensitivity of ASIC1a. J Biol Chem. 2009;284:27899–27907. doi: 10.1074/jbc.M109.029009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vullo S, et al. Conformational dynamics and role of the acidic pocket in ASIC pH-dependent gating. Proc Natl Acad Sci USA. 2017;114:3768–3773. doi: 10.1073/pnas.1620560114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 24.Cannon JT, et al. Xenacoelomorpha is the sister group to Nephrozoa. Nature. 2016;530:89–93. doi: 10.1038/nature16520. [DOI] [PubMed] [Google Scholar]
- 25.Wiemuth D, Assmann M, Gründer S. The bile acid-sensitive ion channel (BASIC), the ignored cousin of ASICs and ENaC. Channels (Austin) 2014;8:29–34. doi: 10.4161/chan.27493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellenberger S, Schild L. International Union of Basic and Clinical Pharmacology. XCI. Structure, function, and pharmacology of acid-sensing ion channels and the epithelial Na+ channel. Pharmacol Rev. 2015;67:1–35. doi: 10.1124/pr.114.009225. [DOI] [PubMed] [Google Scholar]
- 27.Lynagh T, et al. A selectivity filter at the intracellular end of the acid-sensing ion channel pore. eLife. 2017;6:e24630. doi: 10.7554/eLife.24630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L, Palmer LG. Ion conduction and selectivity in acid-sensing ion channel 1. J Gen Physiol. 2014;144:245–255. doi: 10.1085/jgp.201411220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wollmuth LP. Ion permeation in ionotropic glutamate receptors: Still dynamic after all these years. Curr Opin Physiol. 2018;2:36–41. doi: 10.1016/j.cophys.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deval E, Lingueglia E. Acid-sensing ion channels and nociception in the peripheral and central nervous systems. Neuropharmacology. 2015;94:49–57. doi: 10.1016/j.neuropharm.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Søviknes AM, Glover JC. Spatiotemporal patterns of neurogenesis in the appendicularian Oikopleura dioica. Dev Biol. 2007;311:264–275. doi: 10.1016/j.ydbio.2007.08.058. [DOI] [PubMed] [Google Scholar]
- 32.Yang H, et al. Inherent dynamics of the acid-sensing ion channel 1 correlates with the gating mechanism. PLoS Biol. 2009;7:e1000151. doi: 10.1371/journal.pbio.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoder N, Yoshioka C, Gouaux E. Gating mechanisms of acid-sensing ion channels. Nature. 2018;555:397–401. doi: 10.1038/nature25782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akabas MH. Cysteine modification: Probing channel structure, function and conformational change. In: Ahern C, Pless S, editors. Novel Chemical Tools to Study Ion Channel Biology. Springer; New York: 2015. pp. 25–54. [DOI] [PubMed] [Google Scholar]
- 35.Akopian AN, Chen CC, Ding Y, Cesare P, Wood JN. A new member of the acid-sensing ion channel family. Neuroreport. 2000;11:2217–2222. doi: 10.1097/00001756-200007140-00031. [DOI] [PubMed] [Google Scholar]
- 36.Gründer S, Geissler HS, Bässler EL, Ruppersberg JP. A new member of acid-sensing ion channels from pituitary gland. Neuroreport. 2000;11:1607–1611. doi: 10.1097/00001756-200006050-00003. [DOI] [PubMed] [Google Scholar]
- 37.Paukert M, et al. A family of acid-sensing ion channels from the zebrafish: Widespread expression in the central nervous system suggests a conserved role in neuronal communication. J Biol Chem. 2004;279:18783–18791. doi: 10.1074/jbc.M401477200. [DOI] [PubMed] [Google Scholar]
- 38.Chen X, Polleichtner G, Kadurin I, Gründer S. Zebrafish acid-sensing ion channel (ASIC) 4, characterization of homo- and heteromeric channels, and identification of regions important for activation by H+ J Biol Chem. 2007;282:30406–30413. doi: 10.1074/jbc.M702229200. [DOI] [PubMed] [Google Scholar]
- 39.Gwiazda K, Bonifacio G, Vullo S, Kellenberger S. Extracellular subunit interactions control transitions between functional states of acid-sensing ion channel 1a. J Biol Chem. 2015;290:17956–17966. doi: 10.1074/jbc.M115.641688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brazeau MD, Friedman M. The origin and early phylogenetic history of jawed vertebrates. Nature. 2015;520:490–497. doi: 10.1038/nature14438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swalla BJ, Smith AB. Deciphering deuterostome phylogeny: Molecular, morphological and palaeontological perspectives. Philos Trans R Soc Lond B Biol Sci. 2008;363:1557–1568. doi: 10.1098/rstb.2007.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bollner T, Storm-Mathisen J, Ottersen OP. GABA-like immunoreactivity in the nervous system of Oikopleura dioica (Appendicularia) Biol Bull. 1991;180:119–124. doi: 10.2307/1542435. [DOI] [PubMed] [Google Scholar]
- 43.Kreneisz O, Glover JC. Developmental characterization of tail movements in the appendicularian urochordate Oikopleura dioica. Brain Behav Evol. 2015;86:191–209. doi: 10.1159/000439517. [DOI] [PubMed] [Google Scholar]
- 44.Søviknes AM, Chourrout D, Glover JC. Development of putative GABAergic neurons in the appendicularian urochordate Oikopleura dioica. J Comp Neurol. 2005;490:12–28. doi: 10.1002/cne.20629. [DOI] [PubMed] [Google Scholar]
- 45.Coryell MW, et al. Acid-sensing ion channel-1a in the amygdala, a novel therapeutic target in depression-related behavior. J Neurosci. 2009;29:5381–5388. doi: 10.1523/JNEUROSCI.0360-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown JT, et al. Vesicular release of glutamate utilizes the proton gradient between the vesicle and synaptic cleft. Front Synaptic Neurosci. 2010;2:15. doi: 10.3389/fnsyn.2010.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donier E, Rugiero F, Jacob C, Wood JN. Regulation of ASIC activity by ASIC4—New insights into ASIC channel function revealed by a yeast two-hybrid assay. Eur J Neurosci. 2008;28:74–86. doi: 10.1111/j.1460-9568.2008.06282.x. [DOI] [PubMed] [Google Scholar]
- 48.Gründer S, Pusch M. Biophysical properties of acid-sensing ion channels (ASICs) Neuropharmacology. 2015;94:9–18. doi: 10.1016/j.neuropharm.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 49.Schwartz V, Friedrich K, Polleichtner G, Gründer S. Acid-sensing ion channel (ASIC) 4 predominantly localizes to an early endosome-related organelle upon heterologous expression. Sci Rep. 2015;5:18242. doi: 10.1038/srep18242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nemecz Á, et al. Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel. PLoS Biol. 2017;15:e2004470. doi: 10.1371/journal.pbio.2004470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baconguis I, Bohlen CJ, Goehring A, Julius D, Gouaux E. X-ray structure of acid-sensing ion channel 1-snake toxin complex reveals open state of a Na(+)-selective channel. Cell. 2014;156:717–729. doi: 10.1016/j.cell.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Isom DG, Castañeda CA, Cannon BR, García-Moreno B. Large shifts in pKa values of lysine residues buried inside a protein. Proc Natl Acad Sci USA. 2011;108:5260–5265. doi: 10.1073/pnas.1010750108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mikhaleva Y, Kreneisz O, Olsen LC, Glover JC, Chourrout D. Modification of the larval swimming behavior in Oikopleura dioica, a chordate with a miniaturized central nervous system by dsRNA injection into fertilized eggs. J Exp Zool B Mol Dev Evol. 2015;324:114–127. doi: 10.1002/jez.b.22607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.