Significance
Smyd1 is a muscle-specific histone methyltransferase, and its role in the regulation of growth and differentiation in skeletal and cardiac muscle is well established. However, despite the persistent expression of Smyd1 in postnatal cardiomyocytes, the role of Smyd1 in the adult heart is largely unknown. We show that Smyd1 regulates energy metabolism in the heart. Cardiac-specific ablation of Smyd1 in the mouse adult heart resulted in global downregulation of mitochondrial proteins involved in oxidative phosphorylation, concurrent with reduced mitochondrial respiration capacity. We further demonstrate that the regulation of Smyd1 in metabolism is through transcriptional control of PGC-1α, a key regulator of mitochondrial energetics. Thus, our data reveal a role for Smyd1 as a master regulator of cardiac energetics.
Keywords: heart, Smyd1, PGC-1a, metabolism, systems biology
Abstract
Smyd1, a muscle-specific histone methyltransferase, has established roles in skeletal and cardiac muscle development, but its role in the adult heart remains poorly understood. Our prior work demonstrated that cardiac-specific deletion of Smyd1 in adult mice (Smyd1-KO) leads to hypertrophy and heart failure. Here we show that down-regulation of mitochondrial energetics is an early event in these Smyd1-KO mice preceding the onset of structural abnormalities. This early impairment of mitochondrial energetics in Smyd1-KO mice is associated with a significant reduction in gene and protein expression of PGC-1α, PPARα, and RXRα, the master regulators of cardiac energetics. The effect of Smyd1 on PGC-1α was recapitulated in primary cultured rat ventricular myocytes, in which acute siRNA-mediated silencing of Smyd1 resulted in a greater than twofold decrease in PGC-1α expression without affecting that of PPARα or RXRα. In addition, enrichment of histone H3 lysine 4 trimethylation (a mark of gene activation) at the PGC-1α locus was markedly reduced in Smyd1-KO mice, and Smyd1-induced transcriptional activation of PGC-1α was confirmed by luciferase reporter assays. Functional confirmation of Smyd1’s involvement showed an increase in mitochondrial respiration capacity induced by overexpression of Smyd1, which was abolished by siRNA-mediated PGC-1α knockdown. Conversely, overexpression of PGC-1α rescued transcript expression and mitochondrial respiration caused by silencing Smyd1 in cardiomyocytes. These findings provide functional evidence for a role of Smyd1, or any member of the Smyd family, in regulating cardiac energetics in the adult heart, which is mediated, at least in part, via modulating PGC-1α.
Smyd1 is a striated muscle-specific histone methyltransferase (1, 2). It was identified initially as an essential regulator of cardiac differentiation and morphogenesis in systemic Smyd1-knockout (Smyd1-KO) mice, which die in utero due to cardiac defects (3). Recently, we found that Smyd1 expression is differentially regulated during pressure overload cardiac hypertrophy and failure in mice (4), consistent with its expression in the failing human heart (5). In addition, we showed that loss of Smyd1 in the adult mouse heart was sufficient to induce hypertrophic growth, which progressed to fulminant heart failure (4). Analysis of the cardiac transcriptome in these Smyd1-KO mice revealed dysregulation of pathways responsible for development, muscle growth, and metabolism (4). However, it remained unclear whether metabolic remodeling in Smyd1-KO mice is purely a consequence of hypertrophic cell growth or is an independent factor in the progression toward heart failure in this model.
Here, we applied untargeted metabolomics and proteomics approaches combined with targeted gene-expression analysis and mitochondrial respirometry assays to determine how Smyd1 regulates metabolic networks in the adult heart. We demonstrate that loss of Smyd1 leads to down-regulation of mitochondrial energetics prior to the onset of cardiac dysfunction. Furthermore, loss- and gain-of-function studies in primary cultured cardiomyocytes and ChIP assays provide further evidence that Smyd1 primarily regulates mitochondrial energetics through the peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), a master regulator of mitochondrial energetics. Overall, these results provide a heretofore unrecognized function for Smyd1 as a potent positive regulator of mitochondrial metabolism in the heart.
Results
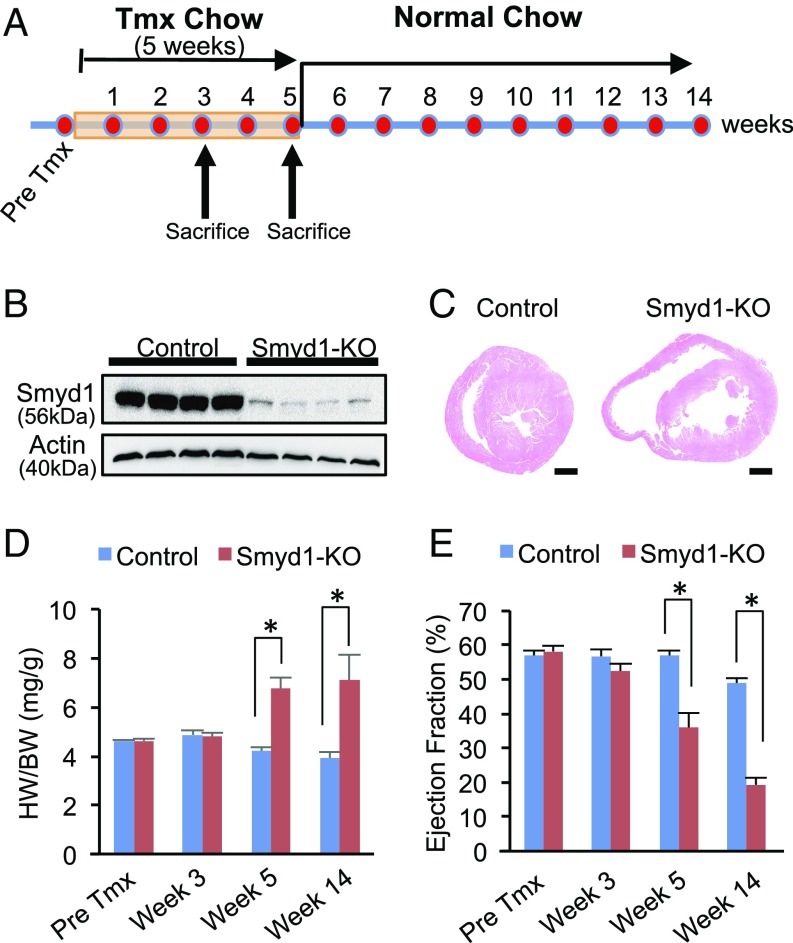
Our initial analysis of inducible, cardiac-specific Smyd1-KO revealed down-regulation of numerous genes involved in metabolism. However, these transcriptomic changes were accompanied by significant structural remodeling of the heart and severe heart failure (4). Thus, it remained unclear whether metabolic remodeling was primary or secondary to heart disease. To resolve this dilemma, we first sought to determine the time point at which Smyd1 expression is significantly reduced in the heart but cardiac function remains normal. Smyd1flox/flox Cre−/− and Smyd1flox/flox Cre+/− mice were fed tamoxifen-containing chow for 5 wk, followed by a normal chow diet (Fig. 1A). Prominent reduction of Smyd1 protein expression in Smyd1flox/flox Cre+/− mice was observed after 3 wk of tamoxifen treatment (Fig. 1B), whereas the heart weight to body weight (HW/BW) ratios and ejection fraction remained unchanged at this time point (Fig. 1 D and E). After 5 wk of tamoxifen treatment, cardiac disease was manifested as a significant increase in HW/BW ratios, a decrease in left ventricular ejection fraction, and subsequent chamber dilation in the heart of Smyd1flox/flox Cre+/− mice (Fig. 1 C–E). We therefore chose to determine whether metabolic remodeling is detectable in Smyd1-KO mice after 3 wk of tamoxifen treatment, the time point at which a significant reduction in Smyd1 expression has already occurred but the heart remains grossly normal.
Fig. 1.
Cardiac-specific deletion of the histone methyltransferase Smyd1 leads to cardiac dysfunction. (A) Smyd1flox/flox Cre−/− (control) and Smyd1flox/flox Cre+/− (Smyd1-KO) mice were fed tamoxifen (Tmx)-containing chow for up to 5 wk, followed by a normal chow diet. For molecular analysis, mice were euthanized after 3 wk or 5 wk of tamoxifen diet. (B) A robust reduction in the protein level of Smyd1 was observed after 3 wk of tamoxifen administration in the hearts from Smyd1flox/flox Cre+/− mice compared with Smyd1flox/flox Cre−/− mice. (C) H&E staining shows marked chamber dilation in Smyd1-deficient mice 8–10 wk after the return to normal chow. (D) The HW (in milligrams)/BW (in grams) ratios in control and Smyd1-KO mice show that cardiac hypertrophy developed in Smyd1-KO mice after 5 wk of tamoxifen diet. (E) The ejection fraction of Smyd1-KO mice remained normal through 3 wk of tamoxifen diet. The development of heart failure (ejection fraction < 40%) was apparent after 5 wk of tamoxifen diet. *P < 0.05.
Deletion of Smyd1 Alters Substrate Metabolism in the Heart.
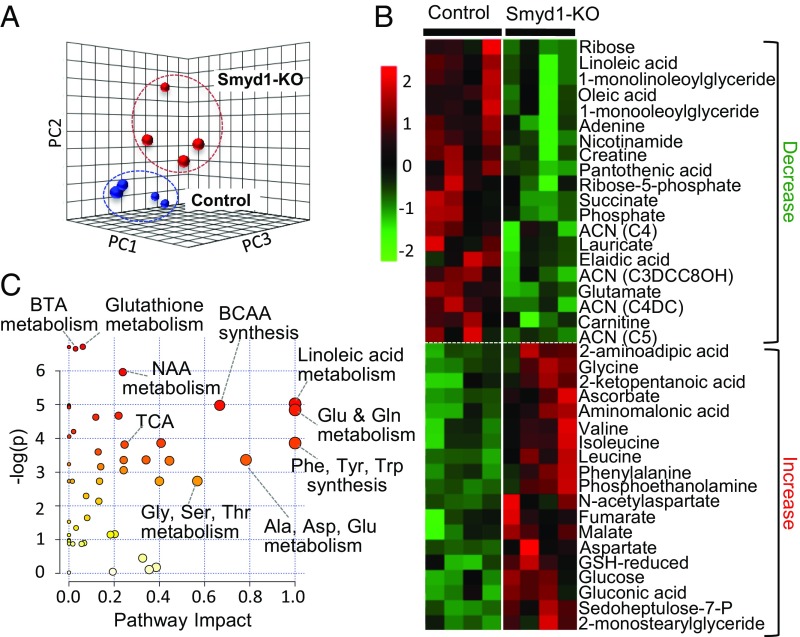
Untargeted metabolomic analyses by GC-MS and direct infusion MS/MS (detailed in Methods) of Smyd1-KO mice and age-matched controls at week 3 identified a total of 147 metabolites (Dataset S1). Principal component analysis (PCA) of these metabolites clearly distinguished between Smyd1-KO and control mice (Fig. 2A), suggesting that loss of Smyd1 in the adult mouse heart leads to significant alterations in the metabolomic profile in the absence of cardiac structural abnormalities. In total, 39 metabolites were significantly altered in the KO mice (20 decreased, 19 increased; P < 0.05) (Fig. 2B). Integrated pathway enrichment and pathway topology analysis (MetaboAnalyst 3.0) (6) identified multiple metabolic pathways affected by Smyd1 deletion (Fig. 2C). The pathways with the largest enrichment and/or impact included butanoate, glutathione, and nicotinamide metabolism as well as multiple interconnected amino acid metabolic pathways (Fig. 2C). The broad metabolic screening was supplemented by targeted quantitative analyses of acylcarnitines and organic acids, which allowed assessment of altered pool sizes of intermediates relevant to fatty acid oxidation (FAO), glycolysis, and the tricarboxylic acid (TCA) cycle. We found a 73.0% increase in the total pool of TCA cycle intermediates and a twofold increase in the lactate/pyruvate ratio (SI Appendix, Fig. S1 A and B), suggesting a bottleneck downstream of the TCA cycle leading to a diversion of glycolytic flux to lactate. A striking (more than 10-fold) increase in the level of 3-hydroxybutyric acid (SI Appendix, Fig. S1C) and a 32.6% reduction in the total pool of acylcarnitines (SI Appendix, Fig. S1D) further indicated dysregulation of ketone body metabolism and FAO in the Smyd1-KO heart. Overall, the results of these metabolic analyses clearly confirm alterations in multiple catabolic pathways converging on oxidative phosphorylation (OXPHOS) in the Smyd1-KO heart.
Fig. 2.
Metabolomic profile of Smyd1-KO mice at week 3 of tamoxifen diet. Metabolomic analysis was performed on left ventricle tissue from Smyd1-KO mice and age-matched control mice at week 3 of tamoxifen diet (n = 4 for Smyd1-KO mice and n = 4 for control mice), using GC/MS and MS/MS. (A) PCA of all 147 metabolites clearly separates the profiles of Smyd1-KO (red) and control (blue) mice. (B) The heat map represents all metabolites significantly altered in Smyd1-KO mice (20 decreased; 19 increased; P < 0.05). (C) Impact Pathway Analysis contains all the matched pathways (the metabolome) arranged by P values (from pathway enrichment analysis) on the y axis and pathway impact values (from pathway topology analysis) on the x axis. The node color is based on its P value, and the node radius is determined based on their pathway impact values. ACN, acylcarnitine; BCAA, branched-chain amino acid; BTA, butyric acid; GSH, glutathione; NAA, N-acetyl-l-aspartate; Sedoheptulose-7-P, sedoheptulose-7-phosphate; TCA, tricarboxylic acid cycle.
Proteomics and Mitochondrial Respirometry Reveal Down-Regulation of Mitochondrial Metabolism in Smyd1-KO Mice.
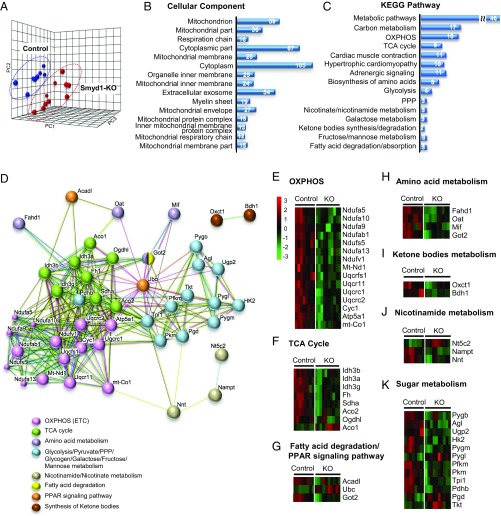
To complement our metabolomic analysis and further elucidate the role of Smyd1 in regulating metabolism in the heart, we performed LC-MS/MS–based label-free proteomic analysis of myocardial samples from Smyd1-KO mice and control mice at week 3. We detected a total of 1,429 proteins in myocardial left ventricular tissue but focused our analysis on the proteins that were detected in at least 60% of samples from each group (1,215 proteins; Dataset S2). A PCA plot generated from these data detected clear differences between control and Smyd1-KO mice (Fig. 3A). A heat map was generated from proteins differentially expressed in Smyd1-KO mice which included a total of 115 down-regulated (green) and 97 up-regulated (red) proteins (SI Appendix, Fig. S2A and Dataset S3). Using the STRING Database web-based tools (7), the 15 most significantly enriched gene ontology (GO) terms for cellular component and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Datasets S4 and S5) are shown in Fig. 3 B and C. At the top of the list for cellular component terms is mitochondrion, and the two most enriched KEGG pathways are the wide, encompassing pathway terms of metabolic pathways and carbon metabolism. More specifically, the two most enriched terms are OXPHOS and the TCA cycle, two key components of mitochondrial energetics, both of which were down-regulated in the Smyd1-KO heart (SI Appendix, Fig. S2C). Fig. 3D provides an interaction network view of the 46 significantly altered proteins within the metabolic pathways KEGG term. Note the highly interconnected clusters that include proteins involved in the electron transport chain (ETC) (pink) and the TCA cycle (green) as well as a prominent cluster of proteins involved in carbohydrate metabolism (light blue). The comparative analysis of those 46 proteins is depicted in heat maps (Fig. 3 E–K) displaying the direction of differential expression compared with the control group. This analysis in the subset of proteins involved in energy metabolism highlights the global down-regulation of proteins involved in the TCA cycle and OXPHOS (Fig. 3 E and F), which could explain, at least in part, the altered metabolic profile described above.
Fig. 3.
Proteomic profile of Smyd1-KO mice at week 3. Proteomic analysis was performed on left ventricle tissue from Smyd1-KO mice and age-matched control mice at week 3 of tamoxifen diet (n = 5 Smyd1-KO mice; n = 4 control mice) with two technical replicates (total of 1,215 proteins). (A) PCA of all detected proteins clearly separates the proteomic profiles of Smyd1-KO (red) and control (blue) mice. (B and C) Enrichment analyses of significantly regulated proteins for the GO terms cellular components (B) and KEGG pathways (C) reveal that loss of Smyd1 preferentially affects mitochondrial proteins involved in respiration and energetics. PPP, pentose phosphate pathway. (D) Network map of the 46 significantly changed proteins which belong to the collective KEGG term metabolic pathways in C. (E–K) Heat maps of the 46 proteins comprising the network map presented in C.
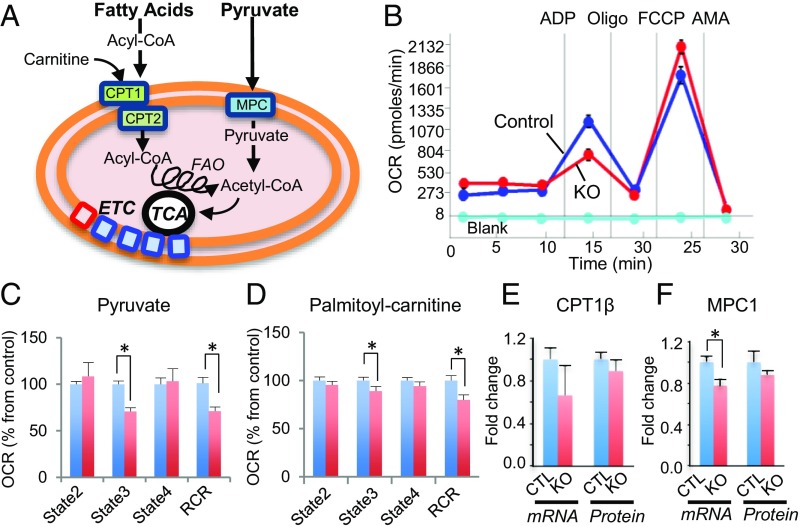
To determine if the observed changes in protein expression translate into altered mitochondrial function, we performed mitochondrial respirometry. We measured the O2 consumption rates (OCR) in isolated cardiac ventricular mitochondria from control and Smyd1-KO mice, using pyruvate and palmitoyl-carnitine as substrates for glucose and fatty acid β-oxidation, respectively (Fig. 4A). We found that ADP-stimulated (state 3) respiration was significantly diminished with both substrates in the Smyd1-KO group compared with the control group (P < 0.05) (Fig. 4 B–D). This indicated a reduced mitochondrial capacity for both FAO and glucose oxidation. In contrast, there was no difference between the two groups in state 4 respiration in the absence of ADP. This resulted in a significant reduction in the mitochondrial respiratory control ratios (RCRs, state 3/state 4) within the Smyd1-KO group (Fig. 4 C and D).
Fig. 4.
Smyd1 deletion leads to reduced mitochondrial respiration capacity. (A) Schematic of catabolic pathways of fatty acids and glucose in mitochondria. Fatty acyl-CoA enters the mitochondria through carnitine palmitoyltransferase 1 (CPT1) where carnitine is attached to form acylcarnitines. CPT2 converts acylcarnitines back to acyl-CoA, a substrate of FAO, yielding acetyl-CoA, which is oxidized in the TCA cycle, and ETC for ATP production. Pyruvate enters through the mitochondrial pyruvate carrier (MPC) and is converted to acetyl-CoA by the pyruvate dehydrogenase (PDH) complex. (B) Representative traces of OCRs in the control group (blue line), the Smyd1-KO group (red line), and baseline (aqua, no mitochondria) when pyruvate was used as a substrate. OCR was measured continuously throughout the experimental period at baseline followed by the addition of ADP, FCCP, and inhibitors. AMA, antimycin A. (C and D) Quantitative analysis of mitochondrial OCR from control and Smyd1-KO mice when pyruvate (C) and palmitoyl-carnitine (D) were used as a substrate. State 2, basal respiration in the absence of ADP; state 3, ADP-stimulated respiration; state 4, respiration in the absence of ADP. (E) No significant difference was found in the expression of CTP1β at mRNA levels and protein levels between control and KO. (F) The gene expression, but not the protein expression, of MPC was significantly decreased in Smyd1-KO mice. *P < 0.05.
To elucidate whether the reduced state respiration within Smyd1-KO hearts was associated with the reduced availability of mitochondrial uptake transporters, we assessed gene and protein expression of the mitochondrial pyruvate carrier 1 (MPC1) and carnitine-palmitoyl transferase 1 (CPT1), enzymes responsible for mitochondrial uptake of pyruvate and fatty acids, respectively (Fig. 4A). As shown in Fig. 4 E and F, only the mRNA levels of MPC1 were significantly reduced in Smyd-1KO mice compared with controls (P < 0.05). Thus, a decreased capacity for both glucose oxidation and FAO in Smyd1-KO mice likely results from derangements of the mitochondrial proteome involved in the TCA cycle and ETC (Fig. 3).
The effect of Smyd1 knockdown on mitochondrial respiration capacity was further confirmed by the Cell Mito Stress Test in H9c2 cardiomyocytes (8). We observed that siRNA-mediated Smyd1 knockdown led to a significant reduction in maximum respiration capacity when glucose or palmitate was used as a major respiration substrate (SI Appendix, Fig. S2). Overall, our data suggest that Smyd1 regulates mitochondrial oxidative capacity.
Smyd1 Modulates the Expression of Transcriptional Regulators of Energy Metabolism.
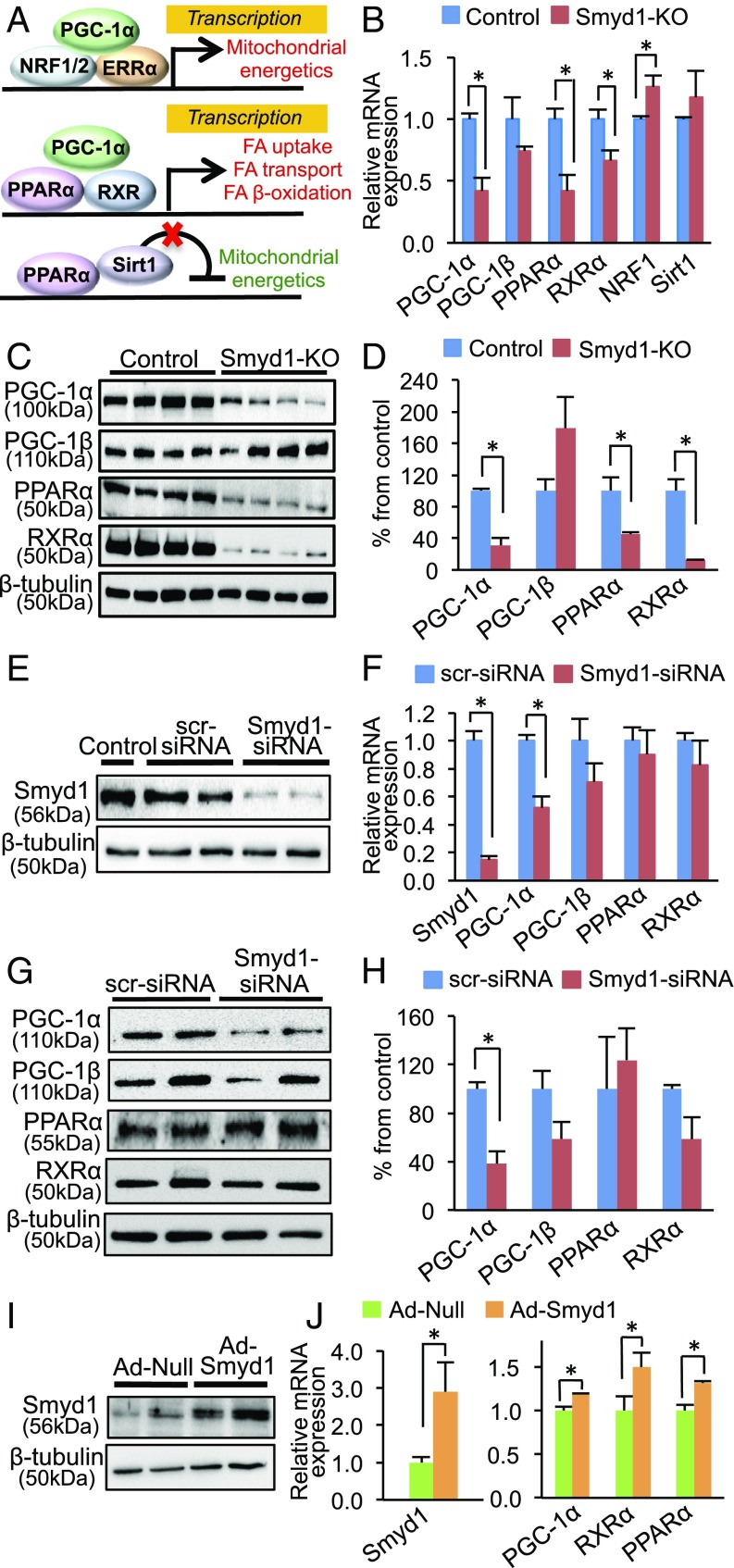
It is well established that cardiac energetics is regulated by PGC-1α. PGC-1α interacts with NRF1/2 and ERRα, leading to transcriptional activation of OXPHOS genes (9). PGC-1α also interacts with the peroxisome proliferator-activated receptor alpha (PPARα) and retinoid X receptor alpha (RXRα) to up-regulate FAO genes (Fig. 5A) (10, 11). Recent studies (12, 13) showed that PPARα also can transcriptionally repress genes involved in mitochondrial energetics by interacting with Sirtuin 1 (Sirt1) (Fig. 5A).
Fig. 5.
Smyd1 regulates transcription factors involved in cardiac energetics. (A) Schematic of transcriptional control mediated by PPARα and PGC-1α with various binding partners. (Top) PGC-1α recruits nuclear respiratory factor (NRF) 1 and/or 2 and nuclear receptor estrogen-related receptor (ERR) alpha, which act as a transcriptional activator of mitochondrial energetics (56). (Middle) PPARα with its coactivator PGC-1α and its functional partner RXR acts as a transcriptional activator of genes involved in fatty acid (FA) uptake and β-oxidation (57). (Bottom) PPARα with the deacetylase Sirt1 acts as a transcriptional repressor of genes involved in the TCA cycle and the ETC (12). (B) Transcript levels of PPARα, PGC-1α, and RXRα were significantly reduced in Smyd1-KO mice (week 3), while the gene-expression levels of PGC-1β and Sirt1 were not changed, and NRF1 was up-regulated compared with control. (C and D) Western blotting analysis shows that protein levels of PPARα, PGC-1α, and RXRα were also significantly reduced in Smyd1-KO mice at week 3 of tamoxifen treatment (n = 4 per group). (E and F) Cultured NRVMs were transfected with either scrambled-siRNA (scr-siRNA) or Smyd1-siRNA for 24 h. qRT-PCR shows that siRNA-mediated knockdown of Smyd1 in NRVMs led to the down-regulation of PGC-1α (n = 4 in control, n = 5 in KD), whereas there was no significant change in gene expression of PGC-1β, PPARα, and RXR-α (n = 3 per group). (G and H) Western blotting analysis shows that PGC-1α was also down-regulated at the protein level in NRVMs that were transfected with Smyd1-siRNA for 48 h, whereas there was no significant change in the protein expression of PGC-1β, PPARα, and RXR-α in Smyd1-KD NRVMs. (I and J) Overexpression of Smyd1 in NRVMs by adenovirus infection for 24 h resulted in a significant increase in gene expression of PPARα, PGC-1α, and RXR-α. *P < 0.05.
Based on our bioinformatic analysis of Fig. 3, we reasoned that Smyd1 might regulate metabolism by modulating the gene expression of these key transcriptional metabolic regulators. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analyses indicated that mRNA levels of PPARα, PGC-1α, and RXRα were significantly reduced in Smyd1-KO mice (P < 0.05 each), whereas there was no significant change in the mRNA levels of PGC-1β and Sirt1, and NRF1 was slightly up-regulated (P = 0.01) (Fig. 5B). Immunoblotting showed that protein levels of PPARα, PGC-1α, and RXRα were also decreased in the Smyd1-KO heart (Fig. 5 C and D). Consistent with the down-regulation of PPARα, PPARα-targeted genes, such as CD36, MCAD, and ACOX1, were markedly reduced in Smyd1-KO hearts compared with controls (SI Appendix, Fig. S4).
Next, we assessed the effects of gain or loss of function of Smyd1 on metabolism in neonatal rat ventricular myocytes (NRVMs). Knockdown of Smyd1 (Smyd1-KD) with siRNAs for 24 h led to a 48% reduction in PGC-1α expression (P < 0.05) without significant changes in gene expression of PPARα and RXRα (Fig. 5 E and F). Consistently, PGC-1α was down-regulated in Smyd1-KD NRVMs at the protein level, whereas expression levels of PGC-1β, PPARα, and RXRα were not significantly different between control (scramble-siRNA) and Smyd1-KD cardiomyocytes (Fig. 5 G and H). An untargeted proteomic analysis of siRNA-mediated Smyd1-KD in NRVMs for 48 h showed that OXPHOS was the most perturbed metabolic pathway (SI Appendix, Fig. S5 and Datasets S6 and S7), as was consistent with the enrichment analysis from the Smyd1-KO heart (Fig. 3C). Adenovirus-mediated overexpression of Smyd1 in NRVMs, on the other hand, led to up-regulation of PGC-1α, PPARα, and RXRα (P < 0.05) (Fig. 5 I and J). Thus, gene expression of PGC-1α varied in parallel with Smyd1 expression, whereas loss of Smyd1 was less predictable for the other transcriptional regulators of metabolism we tested.
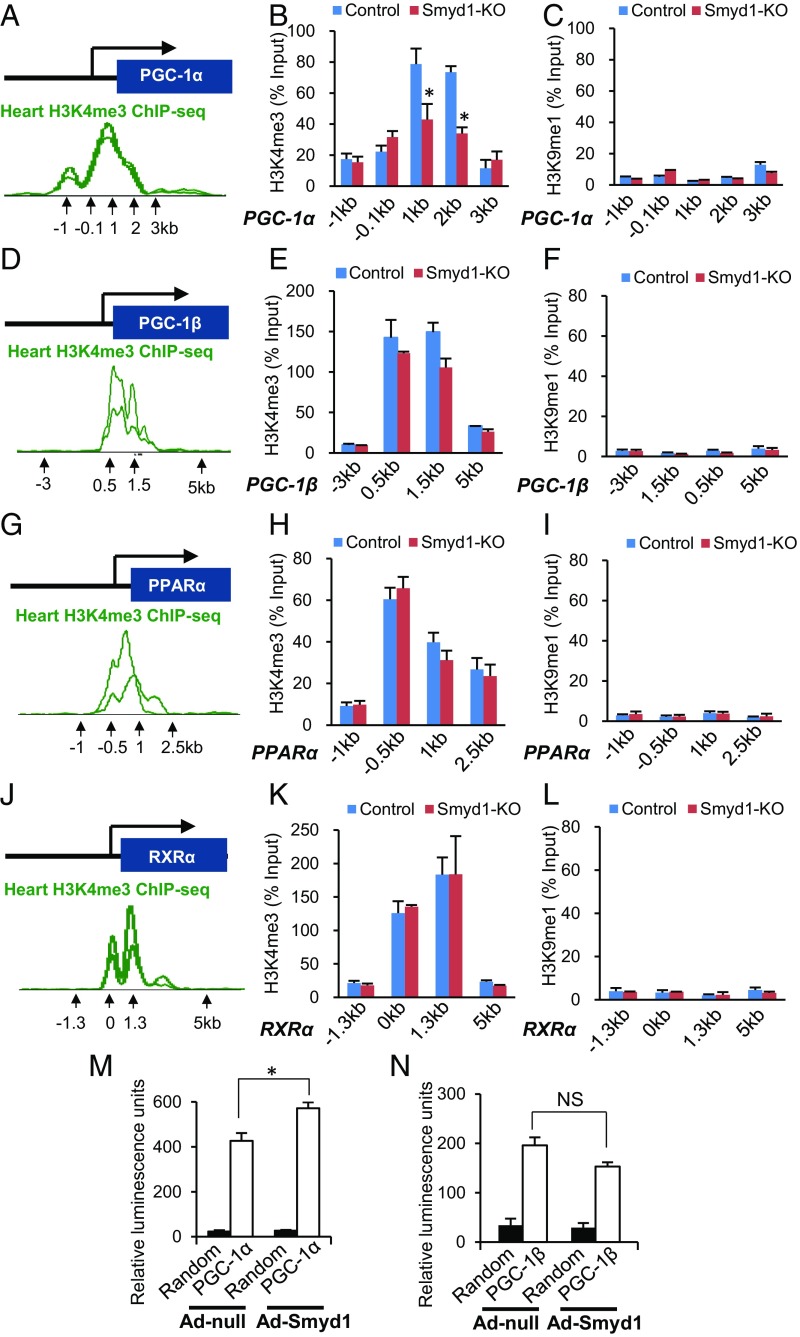
Smyd1 Acts as a Transcriptional Activator of PGC-1α Through Histone H3 Lysine 4 Trimethylation.
Several Smyd family members have been reported to methylate histone H3 lysine 4 (H3K4) (14–18). Trimethylation of H3K4 (H3K4me3) has been observed primarily at promoters of actively transcribed genes (19). We observed a consistent reduction of PGC-1α transcript expression in Smyd1-KO mice (Fig. 5B) and in Smyd1-KD NRVMs (Fig. 5F). If Smyd1 transcriptionally regulates PGC-1α, one might expect that the H3K4me3 profile of PGC-1α also may be altered in Smyd1-KO mice. Previous ChIP sequencing (ChIP-seq) studies have established enrichment of H3K4me3 within the PGC-1α locus (WashU EpiGenome database, epigenomegateway.wustl.edu) (Fig. 6A). Thus, we performed ChIP from cardiac tissue of Smyd1-KO and control mice using an H3K4me3-specific antibody. Consistent with previous observations, enrichment of H3K4me3 found within the PGC-1α locus in control mice was significantly reduced in Smyd1-KO mice (P < 0.05) (Fig. 6B). There was also a trend toward reduction of the H3K4me3 levels within the PGC-1β locus, but, it did not reach statistical significance (P = 0.095 at 1.5 kb) (Fig. 6E). In contrast, loss of Smyd1 did not affect the levels of H3K4me3 in the PPARα and RXRα loci (Fig. 6 G, H, J, and K). The H3K9me1 levels within the promoters of those genes, in which the H3K4me3 peaks were identified with the same primers, were negligible for both control and Smyd1-KO mice, confirming the specificity of ChIP assays in terms of pulldown for H3K4me3 (Fig. 6 C, F, I, and L). These results suggest that Smyd1 regulates PGC-1α gene expression through modulating chromatin accessibility at its promoter.
Fig. 6.
Smyd1 regulates H3K4me3 at the PGC-1α locus in mouse hearts. ChIP-qPCR measured H3K4me3 at the transcriptional start sites of the PGC-1α, PGC-1β, PPARα, and RXR-α loci in control and Smyd1-KO mice. (A, D, G, and J) Previous ChIP-seq studies have established enrichment of H3K4me3 within the promoter regions of those genes (WashU EpiGenome Database), which were targeted for qPCR reactions. (B, E, H, and K) Enrichment of H3K4me3 was found in the predicted promoter regions of PGC-1α, PGC-1β, PPARα, and RXR-α in control mice (blue bars). The level of H3K4me3 was remarkably reduced within the PGC-1α locus in Smyd1-KO mice (red bars), but no significant change was observed in the enrichment of H3K4me3 within the PGC-1β, PPARα, and RXR-α loci. (C, F, I, and L) Amplifications of the same loci regions with H3K9-monomethylation (H3K9me1) were used as negative controls. Values were normalized as a percentage of total input. Data represent the average of four control hearts and four Smyd1-KO hearts, ± SEM. (M and N) Luciferase reporter assays using the PGC-1α (M) and PGC-1β (N) promoters show that Smyd1 acts as a transcriptional activator of PGC-1α but does not directly regulate PGC-1β (n = 6 per group in PGC-1α; n = 4 per group in PGC-1β). *P < 0.05; NS, not significant.
Additionally, luciferase analyses confirmed that Smyd1 binds directly to the promoter region of PGC-1α and enhances the transcription of PGC-1α (Fig. 6M). Specifically, transactivation of the human 1-kb PGC-1α promoter was significantly increased by adenovirus-mediated overexpression of Smyd1 (Ad-Smyd1) compared with that in control (Ad-null) H9c2 cardiomyocytes. In contrast, the promoter activity of PGC-1β was not induced by Smyd1 (Fig. 6N). Thus, these data suggest that Smyd1 regulates the transcriptional activity of PGC-1α but not that of PGC-1β.
Regulation of Mitochondrial Respiration by Smyd1 Is Mediated by PGC-1α.
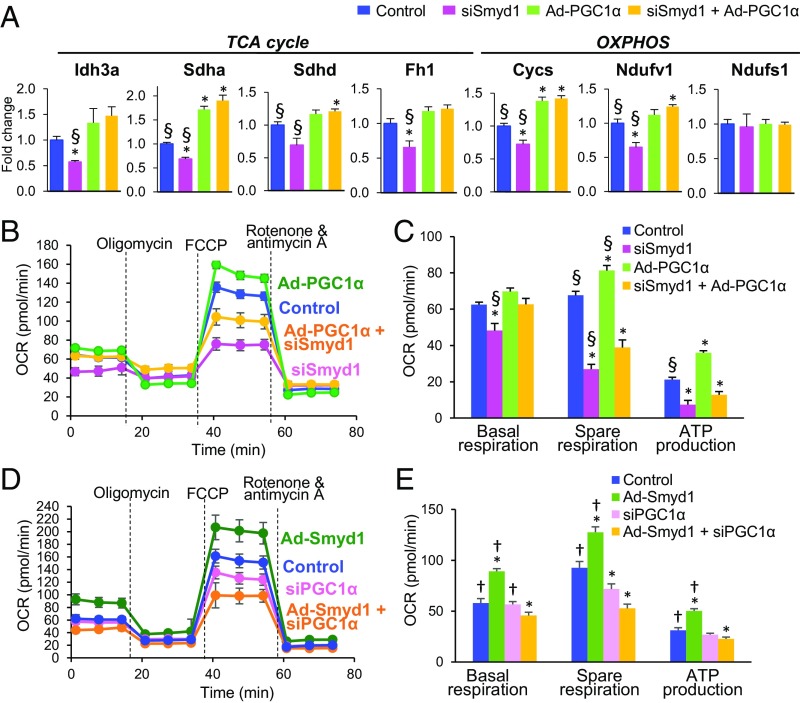
To affirm that Smyd1 regulates energetics, at least in part, through transcriptional control of PGC-1α, we tested whether PGC-1α overexpression can rescue mitochondrial gene expression and respiratory capacity in Smyd1-KD cells. Consistent with proteomic analysis of the Smyd1-KO heart, qRT-PCR revealed that siRNA-mediated Smyd1 knockdown (siSmyd1) in NRVMs led to the down-regulation of genes involved in the TCA cycle and OXPHOS (Idh3a, Sdha, Sdhd, Fh1, Cycs, and Ndufv1) (Fig. 7A). Adenovirus-mediated overexpression of PGC-1α not only fully abolished this down-regulation (yellow vs. pink in Fig. 7A) but for some relevant genes brought the levels of expression above that of the control (yellow vs. blue in Fig. 7A). Of note, neither PGC-1α overexpression (Ad-PGC-1α) nor Smyd1-KD (siSmyd1) affected the mRNA level of Ndufs1, which was also unchanged in the Smyd1-KO heart (Dataset S2). To analyze the mitochondrial respiratory capacity, we conducted the Cell Mito Stress Test in H9c2 cardiomyocytes. Consistent with previous findings (20), we observed increased spare respiration and ATP production in Ad-PGC-1α cells (green vs. blue in Fig. 7 B and C). The efficacy of adenovirus-mediated overexpression of PGC-1α in H9c2 cardiomyocytes (Ad-PGC-1α cells) was confirmed by qRT-PCR (SI Appendix, Fig. S6A). As expected (SI Appendix, Fig. S3 A and B), siRNA-mediated Smyd1 knockdown led to reduced respiration capacity (pink vs. blue in Fig. 7 B and C). Most importantly, overexpression of PGC-1α in Smyd1-KD cells (Ad-PGC-1α + siSmyd1) led to a complete rescue of basal respiration and a partial rescue of spare respiration (yellow vs. pink in Fig. 7 B and C).
Fig. 7.
Overexpression of PGC-1α partially rescues the down-regulation of cellular respiration caused by silencing of Smyd1, whereas overexpression of Smyd1 fails to increase respiration capacity in the absence of PGC-1α. (A) Overexpression of PGC-1α rescues the expression of genes involved in the TCA cycle and OXPHOS which were down-regulated by silencing Smyd1 in NRVMs. (B–E) A cell stress test was conducted using a Seahorse Bioscience XFe96 analyzer by injecting 1 µM oligomycin, 5 µM FCCP, and 1 µM rotenone + antimycin A, sequentially (see Methods for details). (B and C) H9c2 cardiomyocytes were transduced with Ad-PGC-1α or transfected with siRNA-Smyd or with both (Ad-PGC-1α + siSmyd1). (D and E) H9c2 cardiomyocytes were transduced with Ad-Smyd1 or transfected with siRNA-PGC-1α or with both (Ad-Smyd1 + siPGC-1α). The OCR values were normalized by the intensity of nuclear staining (Methods and ref. 55). Groups were compared using one-way ANOVA. Bonferroni (A) or Newman–Keuls test (C and E) were used for individual pairwise comparisons. *P < 0.05 vs. control; †P < 0.05 vs. Ad-Smyd1 + siPGC-1α; §P < 0.05 vs. siSmyd1 + Ad-PGC-1α (n = 5–6 per group in A; n = 6–8 per group in B–E).
To determine whether Smyd1 overexpression is able to increase respiration capacity in the absence of PGC-1α, we measured OCRs in PGC-1α-KD cells that were transduced with Ad-Smyd1 (siPGC-1α + Ad-Smyd1). Overexpression of Smyd1 significantly increased all respiration parameters (green vs. blue in Fig. 7 D and E), whereas knockdown of PGC-1α significantly reduced spare respiration (pink vs. blue in Fig. 7 D and E). However, overexpression of Smyd1 failed to increase mitochondrial respiration in the absence of PGC-1α (Ad-Smyd1 + siPGC-1α; yellow vs. pink in Fig. 7 D and E). This finding likely reflects a profound disturbance of mitochondrial metabolism caused by severe dysregulation of the Smyd1-PGC1α control circuit. Of note, we found that silencing of the PGC-1α gene in H9c2 cells causes down-regulation of Smyd1 (SI Appendix, Fig. S6B), which raises the possibility of a feedback loop between the two regulatory proteins. In summary, our data from mitochondrial stress tests suggest that the regulation of mitochondrial respiration capacity by Smyd1 is mediated, at least in part, by PGC-1α, although the mode of regulation may be more complex than a unidirectional signaling from Smyd1 to PGC1α.
Metabolic Remodeling in Smyd1-KO Hearts Expands with Time.
As shown in Fig. 1, between weeks 3 and 5 of tamoxifen treatment a major transition occurs in Smyd1-KO hearts from normal structure/function to overt signs of hypertrophy and hemodynamic decompensation. To assess the overall direction of metabolic remodeling during this transition, we compared the metabolic profiles from cardiac tissue after Smyd1-KO at these time points (note: the total number of significantly affected metabolites doubled at week 5 compared with week 3 of tamoxifen treatment: 87 versus 39, respectively; Dataset S8 for Smyd1-KO week 5). Changes in other metabolites were concordant, except for adenosine, hypoxanthine, and xanthine (the adenine nucleotide degradation pathway), which were reduced at week 3 but increased at week 5 of tamoxifen treatment (SI Appendix, Fig. S3A). At the proteomic level, the total number of significantly affected proteins belonging to the collective KEGG term metabolic pathways increased from 46 to 67 between week 3 and week 5 of tamoxifen treatment (Datasets S9–S11). Perturbations occurring at week 5 of tamoxifen treatment included further down-regulation of the ETC and the TCA (already evident at week 3) together with strong down-regulation of multiple catabolic pathways, including FAO and branched-chain amino acid (BCAA) oxidation and pyruvate, amino acid, and ketone body metabolism (SI Appendix, Figs. S3B, S4, and S5).
Discussion
Smyd1 is a striated muscle-specific histone methyltransferase (1, 2), which has been established as an essential regulator of cardiac differentiation and morphogenesis (3). Smyd1, as with other Smyd family members, contains the highly conserved Myeloid-Nervy-DEAF1 (MYND) and SET domains, which have been shown to regulate transcription by mediating distinct chromatin modifications (21, 22). Transcriptional control of Smyd1 by factors essential for muscle development, such as Hand2, has been demonstrated (3, 23). However, despite the persistent expression of Smyd1 in postnatal cardiomyocytes (3), its role in the regulation of the adult heart remains poorly understood.
Our previous study showed that Smyd1 is remarkably up-regulated in a mouse model of pressure overload-induced hypertrophy and failure (4). This differential regulation is consistent with Smyd1 (BOP) expression in human heart failure patients (5, 24). Additionally, our prior work revealed that conditional, cardiac-specific deletion of Smyd1 in the adult mouse resulted in cardiac hypertrophy and failure, suggesting that constitutive Smyd1 activity is essential for normal heart function (4). Here we provide strong evidence that an essential role of Smyd1 in the adult heart is to regulate mitochondrial metabolism. It is widely accepted that energy deficiency is a hallmark of the failing myocardium, irrespective of the etiology of heart failure (25). A large number of genetic manipulations associated with metabolism in mice produce heart failure (26–29), suggesting that impaired myocardial energetics plays an important role in the pathophysiology of heart failure. However, due to the close links among metabolism, cardiac function, and structure, it can be difficult to differentiate between the initial insult and secondary effects resulting from the developing heart disease. In this study, we demonstrate that loss of Smyd1 results in significant down-regulation of mitochondrial metabolism in the absence of structural remodeling. This occurs in concordance with the down-regulation of OXPHOS proteins and PGC-1α following silencing of Smyd1 in cultured primary cardiomyocytes. These initial metabolic perturbations expand over time, eventually leading to massive metabolic remodeling in a manner remarkably similar to that observed in mouse models of pressure overload-induced heart failure (30, 31), despite the absence of hemodynamic stress in the Smyd1-deficient model. This, when combined with our previous finding that Smyd1 inhibits the expression of genes controlling cell growth (4), supports the intriguing possibility that Smyd1 coordinates metabolic and structural remodeling of the myocardium in response to chronic hemodynamic stress. This link between cell growth and metabolism predicts that Smyd1 may also regulate myocardial remodeling during the development of physiological cardiac hypertrophy, diabetic cardiomyopathy, and other cardiac diseases.
Intensive studies over the past two decades have begun to uncover the staggering complexity of metabolic regulation directed by a plethora of cross-activating transcription factors, including nuclear receptor family members (12, 13, 32–34). Dynamic interactions between these factors orchestrate metabolic remodeling in response to various physiological and pathophysiological stimuli (35–40). Prominent among these factors is the transcriptional coactivator PGC-1α, which acts as a master regulator of mitochondrial oxidative metabolism by coordinating the capacity of each step required for ATP synthesis (20). Several studies suggest that PGC-1α is highly inducible in response to physiological conditions, such as short-term fasting (20), athletic training (37), and cold exposure (41), to send signals to increase myocardial ATP production. Our current study suggests that Smyd1 is a direct, upstream regulator of PGC-1α. Either gain or loss of Smyd1 function led to parallel changes in gene expression of PGC-1α. Overexpression of PGC-1α partially rescued cellular respiration in Smyd1-KD cardiomyocytes, (consistent with the normalization of genes involved in the TCA cycle and OXPHOS), whereas the increased respiration capacity due to Smyd1 overexpression was abolished and even reversed by PGC-1α knockdown. These data strongly suggest that Smyd1 regulates mitochondrial respiration in a PGC-1α–dependent manner.
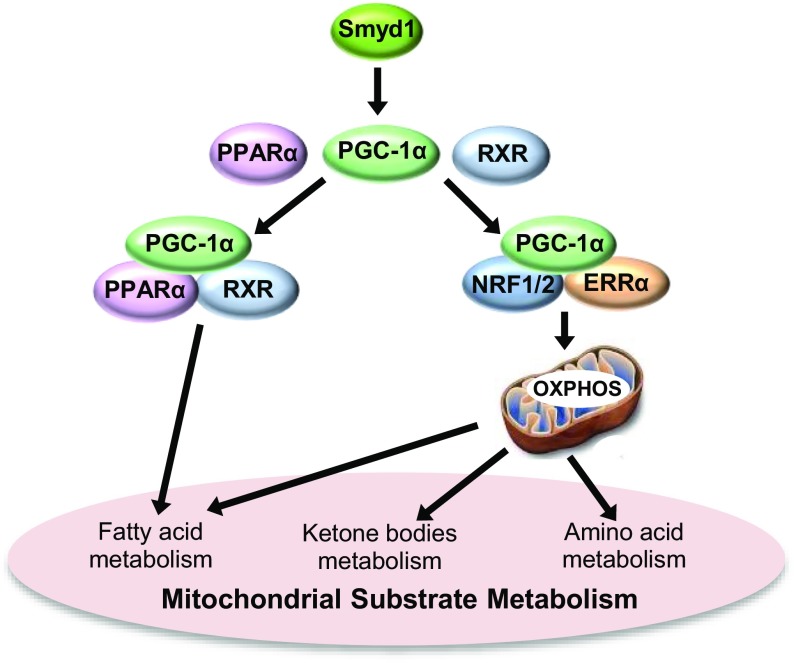
Although gene-expression analysis of Smyd1-KO mice at week 3 showed that PPARα and RXRα, the nuclear receptors forming a heterodimer that regulates FAO genes (42), as well as several well-established PPARα target genes, were down-regulated in Smyd1-KO mice, siRNA-mediated silencing of Smyd1 in NRVMs had no effect on expression of PPARα and RXRα. Furthermore, our ChIP data revealed a marked reduction in the enrichment of H3K4me3 (a marker of gene activation) within the PGC-1α locus, whereas the H3K4me3 levels were not changed in the promoters of PPARα and RXRα. This suggests that Smyd1 regulates cardiac metabolism primarily through transactivation of PGC-1α, whereas changes in PPARα and RXRα expression are mediated by PGC-1α signaling and/or attendant changes in the metabolic milieu. This conjecture is supported by the fact that Smyd1-KO mice exhibited down-regulation of OXPHOS at the earliest stage of metabolic remodeling (week 3), whereas significant down-regulation of FAO occurred later (week 5). The progressive nature of metabolic remodeling following the loss of Smyd1 was also evident in Smyd1-KD NRVMs, in which significant alterations in the metabolic proteome were detectable after 48 h, but not after 24 h, of Smyd1 gene silencing. In both cases, PGC-1α expression was significantly reduced before the earliest time point analyzed. We speculate that the metabolic remodeling caused by loss of Smyd1 is a sequential process in which down-regulation of PGC-1α is followed by down-regulation of other metabolic transcription factors and, finally, by a decrease in protein levels of metabolic enzymes (Fig. 8).
Fig. 8.
Smyd1 regulates cardiac metabolic networks via PGC-1α. Smyd1 globally regulates metabolic pathways and mitochondrial energetics through transcription activation of key regulatory genes. Our data suggest that in cardiomyocytes Smyd1 primarily regulates PGC-1α, which interacts with various transcriptional factors involved in OXPHOS and mitochondrial substrate metabolism, including fatty acid metabolism, amino acid metabolism, and ketone metabolism.
A previous study demonstrated that PGC-1α and PGC1-β have overlapping roles in mitochondrial bioenergetics (43). We examined whether Smyd1 also regulates PGC1-β. Although we observed a trend for reduction in PGC1-β gene expression and the H3K4me3 levels in its promoter in the Smyd1-KO heart, neither reached statistical significance. In addition, luciferase reporter gene assays confirmed the absence of transcriptional activation of Smyd1 in the PGC-1β promoter. Given that siRNA-mediated PGC-1α reduced the mRNA levels of PGC1-β in H9c2 cells (P > 0.05) (SI Appendix, Fig. S6B), it is plausible that the reduced mRNA levels of PGC1-β in the Smyd1-KO heart and Smyd1-KD cardiomyocytes (both P > 0.05) (Fig. 5 B and F) were secondary to the down-regulation of PGC-1α.
Smyd1-KO mice develop signs of heart failure following week 5 of tamoxifen treatment, which coincides with massive down-regulation of energy metabolism. It is of interest that, at this stage, the metabolic profile strongly resembles that observed in pressure overload-induced heart failure (achieved by transverse aortic constriction, TAC). Specifically, both cases exhibit coordinated down-regulation of FAO and BCAA oxidation (30). In previous work (4), we showed that Smyd1 is a suppressor of genes involved in pathological cardiomyocyte growth. In the current study, we observed that Smyd1 exerts positive control over energy metabolism, which likely is independent of the regulation of cell growth. Given these different functions of Smyd1, the development of heart failure in Smyd1-KO mice could result from an inability to restrict growth or the down-regulation of energy metabolism, or both. In any event, deficiencies in mitochondrial energetics observed early in Smyd1-KO mice are likely to become a major contributing factor of eventual heart failure. The convergent pattern of metabolic remodeling observed in Smyd1-KO and TAC models points to the possibility of a common pathophysiological pathway originating in mitochondria. Indeed, there is significant evidence suggesting that deficiencies in mitochondrial energetics are causally linked to the development of heart failure (44, 45).
In summary, this study revealed a role for Smyd1 as a positive regulator of cardiac metabolism. We suggest that its regulation of PGC-1α, which interacts with various transcription factors, leads to the amplification of Smyd1 signaling to OXPHOS and eventually spreads into mitochondrial substrate metabolism, including FAO and amino acid and ketone body metabolism (Fig. 8). In addition, Smyd1 might directly regulate other key nodal enzymes, from which the perturbation spreads across the metabolic networks due to dynamic interactions between coupled metabolic reactions.
Methods
Animals and Tissue Harvest.
All animal experiments were approved by the University of Utah Institutional Animal Care and Use Committee and were conducted according to the Guide for the Care and Use of Laboratory Animals (46). Inducible, cardiac-specific Smyd1-KO mice (Smyd1flox/flox Cre+/−) and control mice (Smyd1flox/flox Cre−/−) were developed as described previously (4). These animals at the age of 8–10 wk were fed a diet containing tamoxifen, 0.4 mg/g of chow diet (catalog no. TD.07262; Harlan). Hearts were harvested from mice after 3 wk and 5 wk on a tamoxifen diet. Tissue samples from ventricles were immediately frozen in liquid nitrogen and stored at −80 °C until they were used for analysis.
Metabolomic Analysis.
Ventricular tissue samples harvested at week 3 of tamoxifen treatment from Smyd1-KO mice and control animals were analyzed using GC/MS and direct infusion MS/MS at the University of Utah Metabolomics Core Facility and Associated Regional and University Pathologists, Inc. (ARUP) Laboratories, as described previously (47). Quantitative measurements of organic acids and acylcarnitines were performed at ARUP Laboratories following established diagnostic test protocols. Ventricular samples from week 5 of tamoxifen treatment were analyzed using two metabolomic platforms [GC-TOF MS and hydrophilic interaction chromatography (HILIC)-quad-time of flight (QTOF)/MS] for untargeted metabolomic screening, which was performed at the NIH West Coast Metabolomics Center at the University of California, Davis, as described previously (48, 49). The PCA, heat maps, and pathway impact analysis of the metabolomic data were carried out using MetaboAnalyst 3.0 software (50–52). Differences in the abundance of individual metabolites were determined using a Student’s t test (two-tailed, unpaired comparison). A value of P < 0.05 was considered statistically significant. Data are given as mean ± SEM.
Proteomic Analysis.
Proteomic analyses of ventricular tissue collected from Smyd1-KO and control mice at week 3 and week 5 of tamoxifen treatment were conducted using label-free quantitative LC-MS/MS, as described in our previous publication (47), with some modification. Label-free quantitation of protein expression was accomplished using the PEAKS Studio 7.5 software (Bioinformatics Solutions, Inc.). The significance of changes in proteins was determined by a Student’s t test of control and KO areas. Proteins identified by at least one unique peptide and two or more total peptides, with a P value less than 0.01, were considered significant. Enrichment analyses in biological processes, cellular components, and KEGG pathways were performed using the STRING database (53).
Mitochondrial Respirometry Analysis.
Mitochondria were isolated from ventricle tissue of Smyd1-KO and control mice by differential centrifugation as described previously (54), with minor modifications. The OCRs of isolated mitochondria were measured using a Seahorse XF24 analyzer (Agilent).
Cell Mito Stress Test.
OCRs in H9c2 cardiomyocytes were measured using a Seahorse XFe96 Analyzer (Agilent) as described previously (8), with some modifications. Briefly, the cells were plated in 96-well Seahorse analyzer plates (2 × 104 cells per well), which were transduced with adenovirus and/or transfected with siRNA. After 48 h, OCR measurements were conducted periodically every 6.5 min over 74 min. Three readings were taken after each injection of oligomycin, carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP), and rotenone + antimycin A. The OCR values were normalized by the intensity of nuclear staining as described previously (55).
ChIP-qPCR.
Ventricular tissue from control and Smyd1-KO mice was fixed in 1% formaldehyde. Lysed samples were sonicated to fragment DNA. ChIP was performed using a commercially available ChIP kit (53040; Active Motif) according to the manufacturer’s instructions. DNA-bound proteins were immunoprecipitated using anti-H3K4me3 (ab8580; Abcam) and anti-H3K9me1 (ab9045; Abcam). The DNA recovered was analyzed by qRT-PCR using primer sets that amplified the promoter regions of PGC-1α, PGC-1β, PPARα, and RXRα (see SI Appendix for primer sequences). qPCR was performed in duplicate with equal amounts of specific antibody-immunoprecipitated samples and input. Values were normalized to input measurements, and enrichment was calculated using the ΔΔCt method.
Luciferase Reporter Assay.
H9c2 cells were cultured in a 96-well plate to ∼70% confluency and were transduced with either Ad-CMV–null (empty virus) or Ad-Smyd1a-Flag adenovirus at a multiplicity of infection (MOI) of 300 in medium. After 48 h, the cells were transfected with 50 ng per well of human PGC-1α, PGC-1β, negative control (scrambled), or positive control (ACTB) luciferase reporter constructs (product nos. S722424, S705203, S790001, and S717678, respectively; SwitchGear Genomics), using 8 ng per well of a pGL4 13 [luc2 SV40] vector (catalog no. E668A; Promega Corporation) and were incubated for 24 h. Luciferase activity was assayed using the LightSwitch luciferase assay reagent (catalog no. E1910; SwitchGear Genomics) according to the manufacturer’s instructions and was measured using a Veritas microplate luminometer (Turner Biosystems, Inc.).
Additional details about methods are given in SI Appendix.
Supplementary Material
Acknowledgments
We thank Chris Hunter, Wenqing Yu, Alicja Booth, Jayne Davis, Nancy Allen, and June Harriss for excellent technical assistance. This study was supported by NIH Grants R01HL130424-01 (to S.F.), 1S10OD021505-01 (to J.E.C.), Diabetes Research Center at Washington University Grant 5 P30 DK020579 (to J.S.W.), Nora Eccles Treadwell Foundation Research Grants (to A.V.Z., S.F., and J.S.W.), American Heart Association Grant-in-Aid 17GRNT33440031 (to S.-i.O.), New Jersey Health Foundation Research Grant Programs PC56-16 and PC80-17 (to S.-i.O.), NIH Grant R01CA31534 (to H.O.T.), Cancer Prevention Research Institute of Texas Grant RP120459 (to H.O.T.), the Marie Betzner Morrow Centennial Endowment (H.O.T.), the Undergraduate Research Opportunity Program of the University of Utah (A.D.S. and K.M.C.), and an NIH Medical Research Scholars Program Fellowship (D.W.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800680115/-/DCSupplemental.
References
- 1.Wu J, et al. Biochemical characterization of human SET and MYND domain-containing protein 2 methyltransferase. Biochemistry. 2011;50:6488–6497. doi: 10.1021/bi200725p. [DOI] [PubMed] [Google Scholar]
- 2.Spellmon N, Holcomb J, Trescott L, Sirinupong N, Yang Z. Structure and function of SET and MYND domain-containing proteins. Int J Mol Sci. 2015;16:1406–1428. doi: 10.3390/ijms16011406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gottlieb PD, et al. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- 4.Franklin S, et al. The chromatin binding protein Smyd1 restricts adult mammalian heart growth. Am J Physiol Heart Circ Physiol. 2016;311:H1234–H1247. doi: 10.1152/ajpheart.00235.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borlak J, Thum T. Hallmarks of ion channel gene expression in end-stage heart failure. FASEB J. 2003;17:1592–1608. doi: 10.1096/fj.02-0889com. [DOI] [PubMed] [Google Scholar]
- 6.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0–Making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szklarczyk D, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung KG, et al. Sirtuin-3 (SIRT3) protein attenuates doxorubicin-induced oxidative stress and improves mitochondrial respiration in H9c2 cardiomyocytes. J Biol Chem. 2015;290:10981–10993. doi: 10.1074/jbc.M114.607960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mudd JO, Kass DA. Tackling heart failure in the twenty-first century. Nature. 2008;451:919–928. doi: 10.1038/nature06798. [DOI] [PubMed] [Google Scholar]
- 10.Schoonjans K, Staels B, Auwerx J. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J Lipid Res. 1996;37:907–925. [PubMed] [Google Scholar]
- 11.van Bilsen M, van der Vusse GJ, Gilde AJ, Lindhout M, van der Lee KA. Peroxisome proliferator-activated receptors: Lipid binding proteins controling gene expression. Mol Cell Biochem. 2002;239:131–138. [PubMed] [Google Scholar]
- 12.Oka S, et al. PPARα-Sirt1 complex mediates cardiac hypertrophy and failure through suppression of the ERR transcriptional pathway. Cell Metab. 2011;14:598–611. doi: 10.1016/j.cmet.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oka S, et al. Peroxisome proliferator activated receptor-α association with silent information regulator 1 suppresses cardiac fatty acid metabolism in the failing heart. Circ Heart Fail. 2015;8:1123–1132. doi: 10.1161/CIRCHEARTFAILURE.115.002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamamoto R, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 15.Berkholz J, Orgeur M, Stricker S, Munz B. skNAC and Smyd1 in transcriptional control. Exp Cell Res. 2015;336:182–191. doi: 10.1016/j.yexcr.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 16.Tan X, Rotllant J, Li H, De Deyne P, Du SJ. SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc Natl Acad Sci USA. 2006;103:2713–2718, erratum (2006) 103:7935. doi: 10.1073/pnas.0509503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abu-Farha M, et al. The tale of two domains: Proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol Cell Proteomics. 2008;7:560–572. doi: 10.1074/mcp.M700271-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Tracy C, et al. The Smyd family of methyltransferases: Role in cardiac and skeletal muscle physiology and pathology. Curr Opin Physiol. 2018;1:140–152. doi: 10.1016/j.cophys.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 20.Lehman JJ, et al. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 22.Jenuwein T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 2001;11:266–273. doi: 10.1016/s0962-8924(01)02001-3. [DOI] [PubMed] [Google Scholar]
- 23.Phan D, et al. BOP, a regulator of right ventricular heart development, is a direct transcriptional target of MEF2C in the developing heart. Development. 2005;132:2669–2678. doi: 10.1242/dev.01849. [DOI] [PubMed] [Google Scholar]
- 24.Abaci N, et al. The variations of BOP gene in hypertrophic cardiomyopathy. Anadolu Kardiyol Derg. 2010;10:303–309. doi: 10.5152/akd.2010.087. [DOI] [PubMed] [Google Scholar]
- 25.Ingwall JS. Energy metabolism in heart failure and remodelling. Cardiovasc Res. 2009;81:412–419. doi: 10.1093/cvr/cvn301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finck BN, et al. The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus. J Clin Invest. 2002;109:121–130. doi: 10.1172/JCI14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe K, et al. Constitutive regulation of cardiac fatty acid metabolism through peroxisome proliferator-activated receptor alpha associated with age-dependent cardiac toxicity. J Biol Chem. 2000;275:22293–22299. doi: 10.1074/jbc.M000248200. [DOI] [PubMed] [Google Scholar]
- 28.Karamanlidis G, et al. Mitochondrial complex I deficiency increases protein acetylation and accelerates heart failure. Cell Metab. 2013;18:239–250. doi: 10.1016/j.cmet.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips D, et al. Mice over-expressing the myocardial creatine transporter develop progressive heart failure and show decreased glycolytic capacity. J Mol Cell Cardiol. 2010;48:582–590. doi: 10.1016/j.yjmcc.2009.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai L, et al. Energy metabolic reprogramming in the hypertrophied and early stage failing heart: A multisystems approach. Circ Heart Fail. 2014;7:1022–1031. doi: 10.1161/CIRCHEARTFAILURE.114.001469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sansbury BE, et al. Metabolomic analysis of pressure-overloaded and infarcted mouse hearts. Circ Heart Fail. 2014;7:634–642. doi: 10.1161/CIRCHEARTFAILURE.114.001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puigserver P, et al. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 34.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1alpha. J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 35.Kaimoto S, et al. Activation of PPARalpha in the early stage of heart failure maintained myocardial function and energetics in pressure overload heart failure. Am J Physiol Heart Circ Physiol. 2016;312:H305–H313. doi: 10.1152/ajpheart.00553.2016. [DOI] [PubMed] [Google Scholar]
- 36.Smeets PJ, et al. Transcriptomic analysis of PPARalpha-dependent alterations during cardiac hypertrophy. Physiol Genomics. 2008;36:15–23. doi: 10.1152/physiolgenomics.90296.2008. [DOI] [PubMed] [Google Scholar]
- 37.Jeong TS, et al. Acute simulated soccer-specific training increases PGC-1α mRNA expression in human skeletal muscle. J Sports Sci. 2015;33:1493–1503. doi: 10.1080/02640414.2014.992937. [DOI] [PubMed] [Google Scholar]
- 38.Rowe GC, Jiang A, Arany Z. PGC-1 coactivators in cardiac development and disease. Circ Res. 2010;107:825–838. doi: 10.1161/CIRCRESAHA.110.223818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huss JM, Kelly DP. Nuclear receptor signaling and cardiac energetics. Circ Res. 2004;95:568–578. doi: 10.1161/01.RES.0000141774.29937.e3. [DOI] [PubMed] [Google Scholar]
- 40.Madrazo JA, Kelly DP. The PPAR trio: Regulators of myocardial energy metabolism in health and disease. J Mol Cell Cardiol. 2008;44:968–975. doi: 10.1016/j.yjmcc.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 41.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 42.Finck BN, Kelly DP. Peroxisome proliferator-activated receptor alpha (PPARalpha) signaling in the gene regulatory control of energy metabolism in the normal and diseased heart. J Mol Cell Cardiol. 2002;34:1249–1257. doi: 10.1006/jmcc.2002.2061. [DOI] [PubMed] [Google Scholar]
- 43.Lai L, et al. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22:1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horton JL, et al. Mitochondrial protein hyperacetylation in the failing heart. JCI Insight. 2016;2:e84897. doi: 10.1172/jci.insight.84897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jain-Ghai S, et al. Complex II deficiency–A case report and review of the literature. Am J Med Genet A. 2013;161A:285–294. doi: 10.1002/ajmg.a.35714. [DOI] [PubMed] [Google Scholar]
- 46.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies; Washington, DC: 2011. [Google Scholar]
- 47.Shibayama J, et al. Metabolic remodeling in moderate synchronous versus dyssynchronous pacing-induced heart failure: Integrated metabolomics and proteomics study. PLoS One. 2015;10:e0118974. doi: 10.1371/journal.pone.0118974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meissen JK, et al. Induced pluripotent stem cells show metabolomic differences to embryonic stem cells in polyunsaturated phosphatidylcholines and primary metabolism. PLoS One. 2012;7:e46770. doi: 10.1371/journal.pone.0046770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cajka TFO. Increasing lipidomic coverage by selecting optimal mobile-phase modifiers in LC–MS of blood plasma. Metabolomics. 2016;12:1–11. [Google Scholar]
- 50.Ghobakhlou A, et al. Metabolomic analysis of cold acclimation of Arctic Mesorhizobium sp. strain N33. PLoS One. 2013;8:e84801. doi: 10.1371/journal.pone.0084801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0–a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012;40:W127–W133. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franceschini A, et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boudina S, et al. Contribution of impaired myocardial insulin signaling to mitochondrial dysfunction and oxidative stress in the heart. Circulation. 2009;119:1272–1283. doi: 10.1161/CIRCULATIONAHA.108.792101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kam YJN, Clayton J, Held P, Dranka BP. 2017. Normalization of Agilent Seahorse XF Data by In-Situ Cell Counting Using a BioTek Cytation 5, Application Note (Agilent Technologies, Santa Clara, CA)
- 56.Ruiz M, et al. A cardiac-specific robotized cellular assay identified families of human ligands as inducers of PGC-1α expression and mitochondrial biogenesis. PLoS One. 2012;7:e46753. doi: 10.1371/journal.pone.0046753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rakhshandehroo M, et al. Comprehensive analysis of PPARalpha-dependent regulation of hepatic lipid metabolism by expression profiling. PPAR Res. 2007;2007:26839. doi: 10.1155/2007/26839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.