Significance
The plant gibberellin receptor GID1 shows sequence similarity to carboxylesterase, suggesting that it is derived from an enzyme. However, how GID1 evolved and was modified is unclear. We identified two amino acids that are essential for GID1 activity, and we found that adjustment of these residues caused GID1 to recognize novel GAs carrying 13-OH as active GAs and to strictly refuse inactive GAs. Phylogenetic analysis of 169 GID1s revealed seven subtypes, and the B-type in core eudicots showed unique characteristics. In fact, certain B-type GID1s showed a higher nonsynonymous-to-synonymous divergence ratio in the region determining GA affinity. Such B-type GID1s with higher affinity were preferentially expressed in the roots in some core eudicot plants and conferred adaptive growth under stress.
Keywords: gibberellin, receptor, evolution, diversification, adaptation
Abstract
The plant gibberellin (GA) receptor GID1 shows sequence similarity to carboxylesterase (CXE). Here, we report the molecular evolution of GID1 from establishment to functionally diverse forms in eudicots. By introducing 18 mutagenized rice GID1s into a rice gid1 null mutant, we identified the amino acids crucial for GID1 activity in planta. We focused on two amino acids facing the C2/C3 positions of ent-gibberellane, not shared by lycophytes and euphyllophytes, and found that adjustment of these residues resulted in increased GID1 affinity toward GA4, new acceptance of GA1 and GA3 carrying C13-OH as bioactive ligands, and elimination of inactive GAs. These residues rendered the GA perception system more sophisticated. We conducted phylogenetic analysis of 169 GID1s from 66 plant species and found that, unlike other taxa, nearly all eudicots contain two types of GID1, named A- and B-type. Certain B-type GID1s showed a unique evolutionary characteristic of significantly higher nonsynonymous-to-synonymous divergence in the region determining GA4 affinity. Furthermore, these B-type GID1s were preferentially expressed in the roots of Arabidopsis, soybean, and lettuce and might be involved in root elongation without shoot elongation for adaptive growth under low-temperature stress. Based on these observations, we discuss the establishment and adaption of GID1s during plant evolution.
Gibberellins (GAs) are a large family of tetracyclic diterpenoid plant hormones that have diverse biological roles in plant growth and development, including stem elongation, seed germination, and floral induction (1). Although numerous GAs have been identified, only a few, including GA4, GA1, and GA3, are functionally active in plants (2). These bioactive GAs have structural commonalities, including a carboxyl group at the C6 position (C6-COOH), a hydroxyl group at the C3 position (C3-OH) of the ent-gibberellane skeleton, a γ-lactone ring, and a nonhydroxyl group at the C2 position (shown in yellow in Fig. 1A) (2), which indicates that GA receptors strictly distinguish active from inactive GAs on the basis of these features.
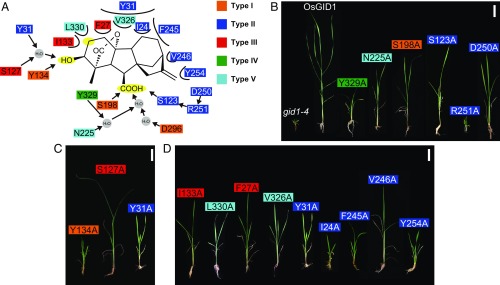
Fig. 1.
Effects of Ala substitution of GA4-interacting amino acids of OsGID1 on its activity in planta. (A) Amino acids are categorized by their commonality among GID1s and GID1-like CXEs in SI Appendix, Fig. S2: such as type I (orange), shared by all GID1s and GID1-like CXEs; type II (dark blue), shared by all GID1s but not CXEs; type III (red), shared by seed plants and ferns but not Selaginella; type IV (green), shared by seed plants but not nonseed plants; and type V (sky blue), not conserved among seed plants. Polar and nonpolar interactions are indicated by arrows and half circles, respectively. The C6-COOH, C3-OH, and C2 positions of GA4 are marked in yellow. (B–D) Rescued phenotypes by transformed mGID1s carrying mutated amino acids interacting with C6-COOH (B) and C3-OH (C), and involved in nonpolar interaction (D). (Scale bars: 5 cm.)
The GA receptor GID1 is structurally similar to carboxylesterases (CXEs), enzymes hydrolyzing short-chain fatty-acid esters, with the GA-binding site of GID1 corresponding to the catalytic site of CXEs and the movable lid at the N-terminal portion functioning to cover the GA molecule, resulting in stabilization at the binding site (3, 4). The N-terminal lid is also involved in the GA-dependent interaction of GID1 with DELLA proteins, which function as GA signaling repressors (3, 4). The structural similarity suggests that GID1 might have been derived from CXE in the process of plant evolution. However, when and how GID1 was established from CXE remains an open question. Previous studies have indicated that GID1 appeared after the divergence of vascular plants from the moss lineage as no GID1 homologs are found in Physcomitrella mosses or the liverwort Marchantia polymorpha (5–7). Furthermore, Hirano et al. (5) reported that GID1s in the lycophyte Selaginella moellendorffii (SmGID1s) have unique properties in comparison with angiosperm GID1s: namely, lower affinity to bioactive GAs and higher affinity to inactive GAs (lower specificity). This suggests that GID1 gained higher affinity and specificity to active GAs after its establishment.
In this study, we aimed to unravel the evolutionary process of GID1 from establishment to functional diversification in eudicots. First, we focused on two important amino acids in terms of GID1 evolution that are not shared by SmGID1s and euphyllophyte GID1s, and we quantitatively evaluated the effects of the differences on GA-binding affinity and elimination activity toward inactive GAs. In addition, we conducted a comprehensive phylogenetic analysis of GID1s in various plants species, and we found that important gene duplication occurred at the establishment of eudicots, which evolved to A- and B-type GID1s. Subsequently, certain eudicot plants evolved a novel hypersensitive B-type GID1, which was involved in achieving adaptive growth under inadequate conditions. Based on these observations, we propose a global evolutionary history of GID1 in the process of plant evolution. Our study provides insights into the molecular events during coevolution of a receptor and its ligands.
Results
Establishment of GID1 from CXEs.
First, we conducted a phylogenetic analysis of CXEs and GID1s of Arabidopsis thaliana, Oryza sativa, S. moellendorffii, and Physcomitrella patens based on amino acid sequence alignment (SI Appendix, Fig. S1 and Dataset S1). The results showed that all GID1s were categorized into one subclade (shown in red) of the larger clade IV (SI Appendix, Fig. S1), confirming that GID1 was derived from one specific CXE group. Next, we aimed to identify the amino acids important for GA binding. Based on the structure of rice GID1 binding GA4 (4), we identified 18 amino acids involved in the direct interaction with GA4 (Fig. 1A), which were categorized into five types (I–V) on the basis of their conservation among GID1s and GID1-like CXEs (SI Appendix, Fig. S2). We introduced Ala-substituted mutant GID1s (mOsGID1s) into a rice gid1 null mutant. Introduction of WT-OsGID1 completely rescued the gid1 dwarfism while the mOsGID1s incompletely restored dwarfism to varying levels (Fig. 1 B–D and SI Appendix, Fig. S3). The effect of Ala substitution did not always correspond to the conservation state of the residue. For example, although S198 was categorized as type I (indicated in orange in SI Appendix, Fig. S2), shared by all GID1s and CXEs, S198A did not cause severe dwarfism (41% relative to WT-OsGID1) (Fig. 1B and SI Appendix, Fig. S3). We classified the amino acids by interaction type: polar interaction with C6-COOH or C3-OH of GA4, or nonpolar interaction (Fig. 1A), and representative rescued phenotypes are shown in Fig. 1 B–D. Regarding the interaction with C6-COOH, R251A caused the most severe defect (Fig. 1B). R251, shared by GID1s but not CXEs (type II), is involved in the establishment of a hydrogen bond network (Fig. 1A), indicating that this hydrogen bond network was important for GID1 establishment. Concerning the C3-OH interaction, Y134A caused the most severe effect (Fig. 1C). Although Y134 is replaced with Phe in certain CXEs (in yellow in Dataset S1), it is shared by all GID1-like CXEs (SI Appendix, Fig. S2, type I), suggesting that a Tyr-carrying member of GID1-like CXEs was selected for GID1 establishment. S127A caused intermediate defect in GID1 activity (Fig. 1C) while this residue is replaced with Met in SmGID1-2 (type III, in red), demonstrating that it was not essential for GID1 establishment (see below). Nonpolar interaction is also important for GA4–GID1 interaction (Fig. 1D), and Ala substitution of the conserved amino acid residues among GID1s (type II), such as I24, F245, and Y254, caused severe dwarfism. In contrast, I133A and L330A, which face the C2 position (Fig. 1A) and are diversified among GID1s (type III and V), had intermediate effects. As the C2 position is a target of hydroxylation by GA 2-oxidase (GA2ox) to inactivate active GAs, such nonpolar interaction could be important for eliminating inactive GAs (see below). Besides binding to GA, GID1 interacts with DELLA proteins through its so-called N-terminal “lid” domain (8). Six nonpolar residues in the lid (Leu18, Trp21, Leu29, Ile33, Leu45, and Tyr48, in red in Dataset S1) are involved in DELLA–GID1 interaction (4). Ala or Ser substitution of these six amino acids did not rescue gid1 dwarfism at all (SI Appendix, Fig. S4) although Ala substitution in mOsGID1 does not affect GA-binding activity in vitro (4). All six residues are shared by GID1s, but not GID1-like CXEs (Dataset S1), indicating that adjustment of the lid structure was also a prerequisite for GID1 establishment.
Adjustment of Amino Acids Facing the C2 and C3 Positions of GAs Enhances GID1 Function.
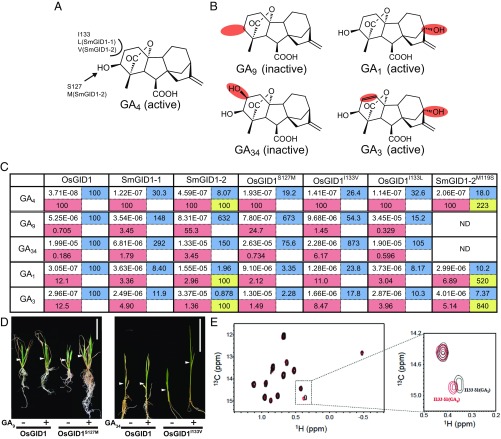
Two amino acids facing the C2 and C3 positions of the ent-gibberellane skeleton, I133 and S127, differed between euphyllophyte and S. moellendorffii GID1s (Fig. 2A). To address the effects of these amino acid differences, we examined the binding affinity of GID1s to GA4 (bioactive), GA9 (inactive by lacking C3-OH), and GA34 (inactive by the presence of C2-OH) (Fig. 2B). To estimate the binding affinity (KD) of GAs to various GID1s (SI Appendix, Fig. S5), we measured the DELLA–GID1 interaction at various GA concentrations under excess GID1 and DELLA by surface plasmon resonance (SPR) (Methods and SI Appendix, Fig. S6A) because DELLA can interact with GID1 carrying GAs, but not free GID1, and stabilize the GA–GID1 interaction. We performed three independent experiments for each GA–GID1 combination (SI Appendix, Figs. S7–S13), and the median KD values are presented in Fig. 2C. The KD of GA4–OsGID1 (3.07E−8 M) in the presence of SLR1 (rice DELLA protein) was 6.9 times lower than that in the absence of SLR1 (2.12E−7 M) (SI Appendix, Fig. S6 B and C). Further, GA1 hardly bound to SmGID1-1 and not at all to SmGID1-2 in the absence of DELLA (SI Appendix, Fig. S6 D and E). These results clearly demonstrate that the presence of SLR1 is essential for exact estimation of the GA–GID1 interaction affinity. The KD of GA4–OsGID1 was estimated as 3.71E−08 whereas that of GA9–OsGID1 and GA34–OsGID1 was less than 1% of GA4, 0.705% (highlighted in red in Fig. 2C) and 0.186%, respectively. The KD of GA4–SmGID1-1 and GA4–SmGID1-2 was 30.3% of GA4–OsGID1 (highlighted in blue in Fig. 2C) and 8.07%, respectively, confirming that SmGID1s have lower affinity for GA4 than OsGID1. The KD of SmGID1-1 to GA9 and GA34 was more than 1% relative to GA4 (3.45% and 1.79%, respectively) whereas that of SmGID1-2 was 55.3% and 3.45%, showing that the elimination ability of SmGID1s toward these inactive GAs is inferior to that of OsGID1. Replacement of S127 or I133 of OsGID1 with Met (S127M) or Val/Leu (I133V or I133L), the corresponding residues of SmGID1s, diminished OsGID1 binding affinity to GA4 and elimination ability toward GA9 or GA34. Indeed, the KD of GA4–OsGID1S127M was 19.2% of GA4–OsGID1 (highlighted in blue in Fig. 2C) while the KD of GA9–OsGID1S127M was 673% to GA9–OsGID1. In contrast, the elimination ability of OsGID1 toward GA34 was not changed by this replacement (0.734% of OsGID1S127M vs. 0.186% of Os-GID1). The KD of GA4–OsGID1I133V was 26.4% of GA4–OsGID1 while that of GA34–OsGID1I133V was 873% of GA34–OsGID1, suggesting that this replacement dramatically diminished the elimination ability of OsGID1 toward GA34. On the other hand, the replacement of I133 with Leu, the corresponding residue of SmGID1-1, did not significantly change the OsGID1 elimination ability toward GA9 (0.329% vs. 0.705%) or GA34 (0.596% vs. 0.186%).
Fig. 2.
Interaction affinities of GID1s to GAs. (A) Structure of GA4 with the amino acids featured in Fig. 1. I133 and S127 of OsGID1 interacting with the C3-OH and C2 positions of GA4 were replaced with the corresponding residues of SmGID1s. (B) Structures of GA9, GA1, GA34, and GA3, with sites distinct from those in GA4 marked in red. (C) Interaction affinity between indicated GID1s and five GAs as measured by SPR. All values are represented as molar concentration (M). The relative affinities of various GID1s relative to WT-OsGID1 are presented in the right, blue cell, whereas those of various GAs to GA4 are in the lower, red cell. The affinity of SmGID1-2M119S relative to WT-SmGID1-2 is presented in the lower right, yellow cell. ND, no data. (D) Effects of GA9 and GA34 on shoot elongation in rice plants overproducing GID1S127M and GID1I133V, respectively. (Scale bars: 5 cm.) (E) Comparison of the HSQC spectrum of [13Cδ1H3]-Ile–labeled OsGID1 carrying GA4 (red) or GA1 (black) with that of interaction with SLR1. The dotted square in the Left is enlarged in the Right.
The above observations suggested that the recognition of active versus inactive GAs mainly depends on I133 and S127. Indeed, gid1 plants overproducing OsGID1S127M or OsGID1I133V had elongated second-leaf sheaths when exposed to 10−6 M GA9 or GA34, respectively, while plants overproducing WT-OsGID1 did not (Fig. 2D and SI Appendix, Fig. S14). These results indicated that these mutated OsGID1s can accept GA9 or GA34 as active GAs while WT-OsGID1 cannot.
Additionally, we measured GID1 binding to GA1 or GA3, bioactive GAs carrying C13-OH (Fig. 2B). The presence of C13-OH decreased the binding affinity of OsGID1, with a KD of 3.05E−07 for GA1 (12.1% of GA4) and of 2.96E−07 for GA3 (12.5% of GA4) (Fig. 2C). The inhibitory effect of C13-OH was substantially greater for SmGID1s, with the affinities for GA1 and GA3 being less than 5% of that for GA4 in every combination, indicating that SmGID1s cannot perceive C13-OH-type GAs as active ones although the amino acids surrounding the C13 position are conserved between OsGID1 and SmGID1s (Fig. 1A). Interestingly, the affinities of GA1– and GA3–OsGID1S127M were substantially lower than that for binding to OsGID1 (2.12% vs. 12.1% for GA1 and 1.49% vs. 12.5% for GA3). In contrast, replacement of the corresponding M119 amino acid in SmGID1-2 with Ser (SmGID1-2M119S) increased its affinity to GA1 by 5.2 times and that to GA3 by 8.4 times whereas this replacement increased the binding affinity to GA4 by 2.2 times (shown in yellow in Fig. 2C). Further, I133L diminished the binding affinity of OsGID1 to GA1 and GA3 (3.04% vs. 12.1% and 3.96% vs. 12.5%, respectively), indicating that this residue also contributes to the low affinity of SmGID1-1 for GA1 and GA3. Together, these results indicate that the amino acids recognizing the C2 and C3 positions are also important to perceive C13-OH GAs as bioactive ones. To confirm this hypothesis, we compared the signal of methyl groups in Ile, Leu, and Val of OsGID1 interacting with GA4 or GA1 by NMR analysis, and we found no difference between these, with one exception of the δ1 signal of I133 (Fig. 2E), which demonstrates that C13-OH affects the hydrophobic interaction between GID1 and GA at the C2 position, which is located opposite of C13.
Diversification of GID1 in Angiosperms.
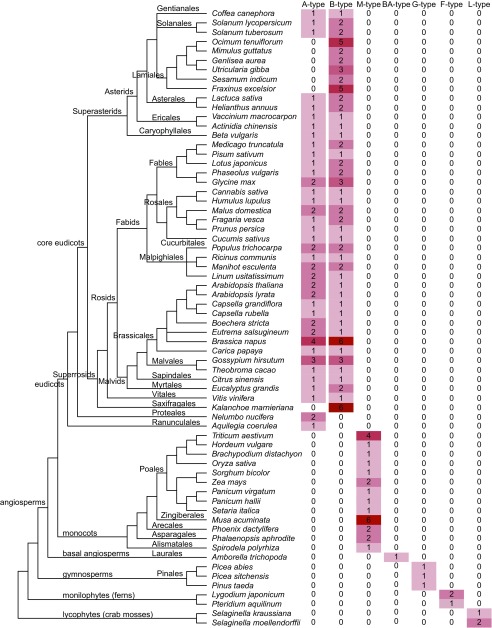
To investigate GID1 evolution, we conducted a phylogenetic analysis of 169 GID1 sequences from 59 angiosperms, three gymnosperms, two ferns, and two lycophytes (Fig. 3, SI Appendix, Fig. S15, and Datasets S2 and S3). In nonseed vascular plants, such as S. moellendorffii, Lygodium japonicum, and Pteridium aquilinum, GID1 is encoded by small, multicopy genes whereas all gymnosperms studied and Amborella trichopoda, the earliest angiosperm, have one gene copy (Fig. 3). In monocots, GID1 is also encoded by one copy with some exceptions (monocot (M)-type), which might be caused by recent genome duplication (9). Thus, the default copy number in the early stage of seed plants might have been one. Eudicot GID1s were divided into two clades, one of which, including AtGID1a and 1c, was referred to as “A-type,” and the other, including AtGID1b, as “B-type” (Fig. 3 and SI Appendix, Fig. S15). Nearly all eudicots, with a few exceptions, contained both types. Two basal eudicot species, Aquilegia coerulea and Nelumbo nucifera, contain a single GID1 type. We classified these GID1s as A-type because A. coerulea GID1 is included in the clade (SI Appendix, Fig. S15). However, all other core eudicots studied have B-type GID1, suggesting that B-type GID1 might have occurred just before or after the establishment of core eudicots. In contrast, we found no A-type, but multiple copies of B-type GID1, in Kalanchoe laxiflora and all Lamiales analyzed, indicating that these plants might have lost A-type GID1.
Fig. 3.
Copy number of seven types of GID1s in various plant species. Phylogenetic relationships among angiosperms are based on Angiosperm Phylogeny Group (APG) IV (39). We classified GID1s into seven different types: that is, A and B of eudicots, M of monocots, BA of basal angiosperms, G of gymnosperms, F of ferns, and L of lycophytes, based on the phylogenetic analysis. Detailed information is presented in Datasets S2 and S3 (list of 169 GID1s from 66 plant species and alignment) and SI Appendix, Fig. S15 (phylogenetic tree).
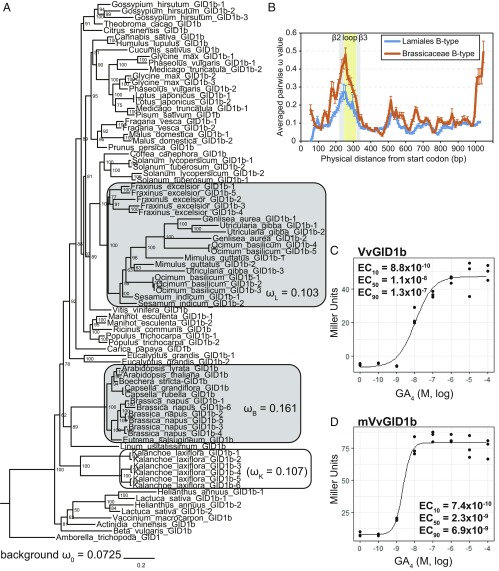
Because the phylogenetic data led us to speculate that the B-type GID1s in plants that lack A-type GID1s may have evolved more rapidly, we examined the ratio of nonsynonymous-to-synonymous divergence (dN/dS = ω), which allows estimating the balance between neutral mutations, purifying selection, and beneficial mutations, between these and other plants (Fig. 4A and SI Appendix, Tables S1 and S2). The ω value for Lamiales (ωL = 0.103) was significantly higher than the background (ω0 = 0.0725). For Kalanchoe, too, the ω value (ωK = 0.107) was higher than the background although statistical support was lacking (P = 0.472), presumably due to the lack of allele numbers, as the clade consisted of GID1 from one genus. These results indicated that the B-type GID1s in Lamiales are under relaxed purifying selection. To identify which region(s) of the B-type GID1s in Lamiales contribute to relaxation of the purifying selection, we conducted sliding-window analysis of their ω values. The ω values for the loop region located between β2 and β3 were markedly higher than those for other regions (Fig. 4B), suggesting that certain substitutions in this region could act for neofunctionalization of the B-type GID1 (see below). To confirm this hypothesis, we performed the same analysis using the B-type GID1s from Brassicaceae, for which 12 alleles were categorized in one clade (Fig. 4A). Similar to Lamiales, Brassicaceae significantly exhibited relaxed purifying selection in comparison with the background (0.161 (ωB) vs. 0.0725 (ω0)), which had clearly involved the loop region (Fig. 4 A and B).
Fig. 4.
Diversification of GID1s. (A) Phylogenetic analysis of B-type GID1s based on the alignment presented in Dataset S4. The ω values (dN/dS) calculated by using the codeml branch model (26) for background, Lamiales, Brassicaceae, and K. laxiflora, according to model Three (background, Lamiales, Brassicaceae) and model Two′ (K. laxiflora) (SI Appendix, Tables S1 and S2). Branch nodes show posterior probability. Horizontal branch lengths are proportional to the estimated number of amino acid substitutions per residue. A. trichopoda GID1 was used as an out-group. (B) The ω sliding window analysis of B-type GID1s in Brassicaceae and Lamiales using windows of 100 nucleotides with 10-bp step size. Error bars indicate SE. (C and D) Quantitative β-galactosidase assay for GA4 dose-dependence of the interactions of VvGID1 or mVvGID1b with A. thaliana GAI in Y2H. mVvGID1b. The loop region of VvGID1b (normal) was replaced with that of GmGID1b-2 (hypersensitive). β-Galactosidase activity was quantified in terms of Miller units by liquid assay. The 10%, 50%, and 90% of the maximum effective concentration (M) of GA4 (EC10, EC50, EC90) are shown in the graph. n = 3.
To examine the difference in biological function between A- and B-type GID1s, we compared the GID1 expression pattern in Lactuca sativa, representing Asterids, and Glycine max, representing Rosids. The B-type was preferentially expressed in the roots in both species (SI Appendix, Fig. S16), similar to findings in A. thaliana (10). We also examined the affinity of B-type GID1s from various species for GA4 by yeast two-hybrid assay (Y2H) (SI Appendix, Fig. S17). The interaction of AtGID1b and GAI with GA4 started around 10−10 M, and the 50% of maximum effective concentration (EC50) was 3.2 × 10−9 M, whereas that of AtGID1a was 1.8 × 10−7 M, indicating that AtGID1b has about 60 times higher affinity for GA4 than AtGID1a (SI Appendix, Fig. S17 A and B). Next, we examined the affinities of Vitis vinifera (Vv), Brassica napus (Bn), G. max (Gm), Gossypium hirsutum (Gh), and L. sativa (Ls) GID1s to GA4 (Fig. 4C and SI Appendix, Fig. S17 C–H). Some of these were hypersensitive like AtGID1b (EC50 = 1 to 3 × 10−9 M) while others showed normal sensitivity, similar to AtGID1a (10−7 to 10−8 M). Such hypersensitive GID1s were not grouped into one clade but scattered over the phylogenetic tree (blue and red arrows for normal and hypersensitive, respectively, in SI Appendix, Fig. S15), indicating that GA hypersensitivity might have been gained independently in each family or genus.
To study the mechanism of GA hypersensitivity, we focused on the loop located between β2 and β3 (Fig. 4B). The alignment showed that the hypersensitive type tended to preferentially contain basic amino acids (Arg and/or His) in the most variable region (boxed in SI Appendix, Fig. S18) while the normal type did not, with the exception of LsGID1b-2 (blue and pink for normal and hypersensitive, respectively, in SI Appendix, Fig. S18). Replacement of the loop of VvGID1b (normal) with that of GmGID1b-2 (hypersensitive) increased the sensitivity to GA4 by 4.7 times (Fig. 4 C and D), clearly indicating that the hypersensitivity of certain B-type GID1s depends on this hypervariable loop sequence.
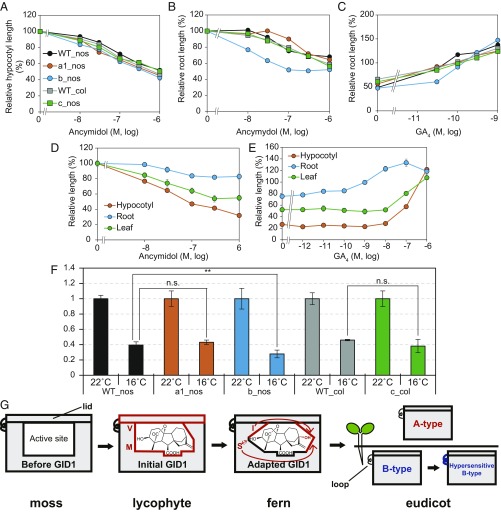
The observation by Tanimoto (11) that GA-dependent root elongation in rosette plants occurs at lower concentrations than that required for shoot elongation led us to speculate that root hypersensitivity to GA depends on hypersensitive B-type GID1s. We examined the effect of GA4 on root elongation in A. thaliana but found no response. We speculated that the endogenous level of active GA for root elongation might be saturated because the interaction of AtGID1b with GA4 is saturated at a very low level, around 5 × 10−8 M (SI Appendix, Fig. S17A). Thus, we examined the effect of ancymidol, a GA synthesis inhibitor, on hypocotyl and root elongation in A. thaliana gid1 mutants gid1a, gid1b, and gid1c. All mutants showed a response similar to that of WT in hypocotyl elongation while only gid1b was more sensitive than WT in view of root elongation (Fig. 5 A and B). When we investigated GA4-dependent recovery of roots stunted by ancymidol, gid1b was less responsive than WT and other mutants (Fig. 5C). Similar results were observed in lettuce; the inhibitory effect of ancymidol on root elongation was substantially lower than that on hypocotyl and leaf elongation while the rescue effect of GA4 on root elongation was considerably stronger than that on the aboveground organs (Fig. 5 D and E). Finally, we examined the effect of low temperature (16 °C) on the root growth of the three A. thaliana gid1 mutants, and we found that the gid1b mutant was affected more significantly than the others (Fig. 5F and SI Appendix, Fig. S19). All these observations strongly suggested that these species might use hypersensitive B-type GID1s for root elongation to achieve adaptive growth and that the divergence of GID1 genes in eudicots may be substantially conducive to survival under inadequate conditions (Discussion).
Fig. 5.
Comparison of GA sensitivity in roots and above-ground tissues of A. thaliana and lettuce, low temperature tolerance of root elongation, and a model of GID1 evolution. (A and B) Inhibitory effect of ancymidol on hypocotyl (A) or root (B) elongation in A. thaliana gid1 mutants, gid1a-1 [background ecotype: Nossen (nos)], gid1b (nos), and gid1c [Columbia (col)]. (C) Recovery of Arabidopsis gid1 roots stunted by 3 × 10−7 M ancymidol by GA4. (D) Inhibitory effect of ancymidol on the elongation of root, hypocotyl, and leaf in lettuce. (E) Recovery of lettuce root stunted by 3 × 10−6 M ancymidol by GA4. (F) Effect of low temperature on A. thaliana gid1 root elongation. The root length of each genotype at 22 °C was set to 1. Error bars indicate SD. n ≥ 4. **P < 0.01 based on two-sided Student’s t test. n.s., not significant, P > 0.05. Actual root lengths are shown in SI Appendix, Fig. S19. (G) Model for the establishment and improvement of the GID1 receptor during plant evolution (for details, see text).
Discussion
Here, we studied the molecular mechanism of the establishment and evolution of the GA receptor GID1 in the process of plant evolution (Fig. 5G). Some CXEs show a monophyletic relationship to the GID1 subclade (SI Appendix, Fig. S1), suggesting that (a) member(s) of clade IV, GID1-like CXEs, are good candidates for GID1 ancestors (“Before GID1” in Fig. 5G). Even though these GID1-like CXEs display significant similarity with GID1s, various amino acids differ between these two proteins. For example, Y31 was recruited to establish a hydrogen bond for recognizing C3-OH whereas the corresponding residue in GID1-like CXEs is inconsistent. For recognizing C6-COOH, the development of a hydrogen bond by R251 was important. The adaptation of amino acids involved in nonpolar interaction was also essential for GID1 establishment.
Lycophyte SmGID1s can accept GA4 as GA receptors, but their affinity toward bioactive GAs and inactive GA elimination ability are inferior to those of GID1s in seed plants (“Initial GID1” in Fig. 5G). Their unique properties mainly depend on two amino acids facing the C2- and C3-positions of GA4 (highlighted in red in Fig. 5G). Indeed, a substitution of S127M in OsGID1 reduced its GA4-binding affinity and GA9-elimination ability (Fig. 2 B and C). SmGID1-1 and SmGID1-2 carry Leu and Val at the 133 position of OsGID1, both of which reduce the GA4-binding affinity, whereas Val also dramatically reduces the inactive GA34-elimination ability (Fig. 2 B and C). It was also confirmed that OsGID1S127M or OsGID1I133V can recognize GA34 or GA9 as active GA in planta, respectively (Fig. 3D). Thus, the adjustment of these two amino acids enhanced GID1 affinity to GA4 and capacity to eliminate inactive GAs.
Unexpectedly, these adjustments also allowed the perception of C13-OH GAs, GA1 and GA3, as active GAs (“Adapted GID1” in Fig. 5G), even though C13 is located at the opposite site of the adapted sites. The NMR results clearly demonstrated that C13-OH affects the Van der Waals interaction between Ile133 of GID1 and GA (Fig. 2E). Thus, the amino acid changes that increased the GA–GID1 interaction stability provided a leeway to accept unsuitable GAs carrying C13-OH. This raises the question why plants developed and continued to use GA1 as active GA although GA1 was not completely adaptable, even to the angiosperm GID1s, such as OsGID1 (12.1% for GA1 relative to GA4). As C13-OH GAs are more hydrophilic than the non-C13-OH GA4, their cell-to-cell movement is more restricted. Actually, when isotope-labeled GA20 was applied to pea leaves, radioactive GA1 was localized in the growing portions of the shoot (12), suggesting that GA1 is formed within the growing region and remains there. In contrast, in rice, GA4 produced specifically in the flowers could be transported to the stem to induce elongation (13) even though rice mainly uses GA1 as a bioactive GA in the vegetative stage. These findings led us to speculate that plants developed a new system that can properly use mobile and nonmobile GAs, depending on the situation, by gaining perception of C13-OH GAs as bioactive GAs.
Core eudicots, with a few exceptions, generally contain a set of A- and B-type GIDs, indicating that diversification of GID1 into A- and B-types occurred just before or after the establishment of core eudicots. We investigated why core eudicots developed two types of GID1 and found some differences in their properties, such as expression pattern and affinity to GA4 (Fig. 4 C and D and SI Appendix, Figs. S16 and S17). In addition, root growth was significantly attenuated at low temperature in gid1b mutants (Fig. 5F). Plants allocate biomass to the organs depending on the environmental conditions. Numerous studies have demonstrated that environmental factors, such as temperature, nutrition, and water availability, significantly affect organ development in various species (14). Tanimoto discussed that preferential root growth in eudicot plants, including A. thaliana and L. sativa, may depend on a difference in GA sensitivity between roots and shoots under low GA condition (11). We observed that root growth in the A. thaliana gid1b mutant was specifically attenuated under limited GA4 and low temperature compared with that in the other gid1 mutants (Fig. 5F), which led us to speculate that preferential root growth in these plants might depend on the unique properties of B-type GID1s, preferential root expression, and higher GA4 affinity, although it cannot be ruled out that other factors, such as higher GA penetration into roots, higher and lower levels of GID1 and DELLA proteins, respectively, and different sets of transcription factors for GA action might be involved in higher GA sensitivity of roots. Given the previous and our present results, the subfunctionalization of some B-type GID1s might have expanded the growing habitat in the process of eudicot evolution by the expression divergence of the two types of GID1 and neofunctionalization conferring higher sensitivity to GA4. In this context, the present results suggest that the loop region located between β2 and β3 might be a target for neofunctionalization as replacement of this loop of the normal B-type GID1 of VvGID1b with that of GmGID1b-2 (hypersensitive) increased the GA4 sensitivity by 4.7 times (Fig. 4 C and D). Consequently, one explanation is that the achievement of hypersensitive GID1 allowed for easier regulation of the body plan of the plants to adapt to inadequate conditions (“eudicot” in Fig. 5G).
As phytohormones have definitive roles in various developmental processes throughout the plant kingdom, the coevolution between receptors and ligand chemicals in various circumstances throughout plant evolution poses an interesting research topic. Recently, two studies investigated the molecular evolution of the receptors of strigolactone (SL) and abscisic acid (ABA). Parasitic plants in Lamiales have developed specialized receptors to detect various kinds of SLs exuded by host plants (15). ABA receptors have undergone multiple duplications and sequence divergence, resulting in some of them attaining the capacity to recognize ABA catabolites, thus expanding adaptive plasticity (16). These studies revealed that, during evolution, plants have gained sophisticated adaptive traits through neofunctionalization of phytohormone receptors. In line with these findings, the present study revealed that GID1 has undergone molecular modifications in the process of plant evolution, contributing to the acquisition of novel properties that allowed adaptive growth under adverse conditions. This evolution of GID1 in coordination with the evolution of its ligand has rendered the GA perception system more sophisticated and, consequently, expanded its involvement in various biological events, as illustrated by the GA hypersensitivity of the roots of A. thaliana and lettuce owing to neofunctionalization of B-type GID1. The present study provided insights into the molecular events during coevolution of a receptor and its ligands, which will aid in studying the evolution of other ligand–receptor systems.
Methods
Collection of CXE and GID1 Sequences from Databases.
CXE sequences of P. patens, S. moellendorffii, O. sativa, and A. thaliana have been reported by Marshall et al. (17). Bacterial CXEs, WP_061301181.1 (Escherichia coli CXE1; EcCXE1) and WP_060616723.1 (EcCXE2), were collected by best-BLAST match searches in the National Center for Biotechnology Information (NCBI) database (18). GID1 sequences of various plant species were collected by reciprocal best-BLAST match searches in the following databases: Phytozome (19), NCBI, and the genome database of each species (Dataset S2). In some cases, partial sequences were concatenated to produce the entire gene-coding sequence.
Phylogenetic Analyses.
CXE amino acid sequences were aligned using ClustalW (Gonnet protein weight matrix). Bayesian estimation of phylogenetic topology was conducted with MrBayes (version 3.2.6) (20), using the WAG + gamma (G) + proportion of invariable sites (I) model, which was selected in ProtTest 3.4.2 (21). Bayesian Markov chain Monte Carlo (MCMC) analyses used flat priors and were run for 3,000,000 generations and four Markov chains (using default heating values) and were sampled every 1,000 generations. We inferred that the chains converged on a stable set of parameters by calculating the potential scale reduction factor using MrBayes. The initial 750,000 generations were discarded as burn-in. For amino acid sequence alignments of GID1s, MAFFT version 7 with the l-INS-i model (22) was used. Protein sequence alignment was converted to in-frame nucleotide sequence alignments using PAL2NAL (23). In all alignments, we removed unnecessary long gap sequences disturbing proper alignment. Bayesian estimation of phylogenetic topology was conducted using the general-time reversible (GTR) + gamma (G) + proportion of invariable sites (I) model, which was selected in jModeltest (24, 25), for 15 million generations. The initial 3.75 million generations were discarded as burn-in.
Identification of Selective Pressure Patterns in B-type GID1s.
For the phylogenetic tree used in the calculation of selective pressure, the B-type GID1 protein sequences alignment was converted to in-codon frame nucleotide alignment, as described above. The phylogenetic tree was constructed using MrBayes as described above, and PAML 4.9e (abacus.gene.ucl.ac.uk/software/paml.html) (26) was applied for in-frame alignment and constructing the phylogenetic tree. The ω values were computed across the gene sequence for designated portions of each phylogeny, with ω values closer to zero indicating stronger purifying selection. Likelihood ratio tests (LRTs) were conducted to compare the goodness of fit of the hypothesis models. To compare region-specific transitions of selective pressures (ω; dN/dS ratio) in B-type GID1s of Brassicaceae and Lamiales, informative single nucleotide polymorphisms (SNPs) in these alleles were analyzed with DnaSP 6.10.01 (27), according to Akagi et al. (28). Window-average ω values were calculated from the start codon (ATG) in a 100-bp window with a 10-bp step size, until the walking window reached the stop codon. B-type GID1b from Linum usitatissimum was excluded from the alignment of the loop regions in the B-type GID1s (SI Appendix, Fig. S18) because of low accuracy of the alignment.
Plasmid Construction.
To construct transgenic rice plants producing various mutated GID1s, site-directed mutagenesis was conducted using the primers listed in SI Appendix, Table S3. The products were cloned into pActNos/Hm2 at the SmaI target site as described previously (29). To construct transgenic rice plants producing 6Ala and 6Ser mutated GID1s, PCR was performed using mLid as template as described previously (4), and the product was cloned into the same target site.
For Y2H, AtGID1a, AtGID1b, GmGID1b-1, GmGID1b-2, VvGID1b, and BnGID1b in pGBKT7 were prepared as described previously (30). GhGID1b-1 was kindly provided by Randy D. Allen, Oklahoma State University, Ardmore, OK (31). LsGID1b-1 and LsGID1b-2 were constructed by RT-PCR using L. sativa mRNA and primers listed in SI Appendix, Table S3, and the constructs were cloned into the pGBKT7 vector. VvGID1b (loop GmGID1b-2) was constructed as previously described, using two sets of primers in SI Appendix, Table S3. To construct LsDELLA1 and LsDELLA2, RT-PCR was carried out using L. sativa mRNA and primers listed in SI Appendix, Table S3, and the constructs were cloned into pGADT7.
The construction of Trx-His-OsGID1, mutated Trx-His-OsGID1s, and Trx-His-SmGID1s using pET32a vector and of GST-SLR1 and GST-SmDELLA1 using pGEX-6P-1 vector have been described elsewhere (4, 5, 29). Mutated Trx-His-SmGID1s were produced using primers listed in SI Appendix, Table S3. All constructs were verified by sequencing.
Plant Material and Growth Condition.
gid1-4 mutant rice plants (29) were used to evaluate the effects of different GID1 mutations. Transgenic gid1-4 rice plants were produced by Agrobacterium-mediated transformation and were grown in the greenhouse as described previously (32).
A. thaliana gid1a-1 and gid1b in the Nossen background and gid1c in the Columbia background were kindly provided by Masatoshi Nakajima, The University of Tokyo, Tokyo (33). Sterilized A. thaliana seeds were sown on vertical agar plates containing 1% agar, 0.5× Murashige and Skoog salt, and 0.5% sucrose, with or without ancymidol and/or GA4. After 3 d of cold treatment (6 °C in the dark), seedlings were grown at 23 °C under long-day (16 h) light regimen for 10 to 14 d. For each treatment, root, hypocotyl, and leaf lengths of 15 seedlings were measured using a ruler. For leaf length, the largest cotyledon or the largest first leaf was selected from each seedling. To examine the effect of temperature, after cold treatment and 3 d of incubation at 22 °C, seedlings were transferred to 16 °C or 22 °C under continuous light for 4 d.
L. sativa cultivar Cisco (Takii Seed Company) were grown on vertical filter papers dipped in culture medium consisting of 0.2× Hoagland’s No. 2 Solution (Sigma) supplemented with or without GA4 and/or ancymidol, as described previously (34, 35). For each treatment, root, hypocotyl, and cotyledon lengths of 15 seedlings were measured using a ruler after 4 to 5 d of incubation at 23 °C under continuous light.
Recombinant Protein Production.
Escherichia coli BL21 (DE3) cells were used for recombinant protein production. For affinity and kinetics studies, recombinant Trx·His-GID1s were produced from cells cultured in 500 mL of Terrific Broth (LB) medium, with addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for induction. Cells were suspended in sonication buffer containing 20 mM Tris⋅HCl, pH 7.5, 200 mM NaCl, and 15 mM n-octyl-β-d-glucoside (buffer A), and were sonicated (20 kHz, 30 × 5 s). The lysate was affinity-purified using 3 mL of IMAC Ni-Charged Resin and further purified by Superdex-200 gel filtration chromatography (GE Healthcare).
For the production of recombinant full-length GST-SLR1 and GST-SmDELLA1, cell culture was performed in the same way as for Trx·His-GIDs, except that 0.4 mM IPTG was used for induction. Cells were suspended in buffer A containing 1 mM DTT and sonicated (20 kHz, 30 × 5 s). The lysate was affinity-purified using 2.5 mL of Glutathione Sepharose 4B beads (GE Healthcare) and further purified by Superdex-200 gel filtration chromatography.
For sample preparation for NMR, methyl-labeled OsGID1 was produced using the E. coli expression system. E. coli BL21 (DE3) cells transformed with an expression vector encoding OsGID1 were cultured at 37 °C in M9 medium containing 0.1 mM GA4 [or GA1] and 50 μg/mL ampicillin. For selective methyl labeling, 35 mg of [methyl-13C; 3,3-2H2] α-ketobutyric acid sodium salt (for Ile δ 1) or 60 mg of [3-methyl-13C; 3,4,4,4-2H4] α-ketoisovaleric acid sodium salt (for Leu and Val methyls) was added to 500 mL of M9 medium when the OD600 reached ∼0.6. When the OD600 reached 0.9, the culture was stored on ice for 10 min, and then 0.5 mM IPTG was added. After induction, the culture was held at 25 °C for 18 h, and the cells were harvested by centrifugation. For sequence-specific assignment of Ile-133 δ1 methyl signal, Ile-133 was replaced with Leu (I133L). The procedure for the production of I133L OsGID1 was identical to that described for WT OsGID1. For the cellular expression of OsSLR1 (4E125R) protein, E. coli Rosetta (DE3) pLysS cells (Novagen) were transformed with pGEX6P1 encoding GST-tagged OsSLR1 (4E125R) and cultured at 37 °C in 500 mL of LB medium containing 50 μg/mL ampicillin. When the OD600 reached ∼0.6, the culture was stored on ice for 10 min, induced with 0.4 mM IPTG, and incubated at 16 °C for 18 h, and then the cells were collected by centrifugation.
Purification of the OsGID1–SLR1 protein complex for NMR analysis was carried out as follows. Cell pellets obtained from methyl-labeled OsGID1 and OsSLR1 (4E125R) cultures were suspended in sonication buffer [10 mM Na phosphate, pH 7.5, 150 mM NaCl, 0.5 mM DTT, 1 mM GA4 (or GA1) and 0.1× complete EDTA-free buffer (Roche)] and disrupted by sonication (20 kHz, 30 × 5 s). The lysate was purified with Glutathione Sepharose 4B resin (GE Healthcare), and then PreScission protease (GE Healthcare) was added to remove the GST-tag. The OsGID1–SLR1 protein complex was further purified using IMAC resin (Bio-Rad). The eluted protein was loaded onto a PD-10 column (GE Healthcare) that had been equilibrated with PD-10 buffer [10 mM Hepes-NaOH, pH 7.1, 1 mM EDTA, 0.5 mM Tris(2-carboxyethyl)phosphine hydrochloride, 2 mM GA4 (or GA1)]. The flow-through fraction, containing the purified protein complex, was concentrated by using an Amicon Ultra-10 filter (Millipore).
Affinity and Kinetic Studies.
SPR assays using a biosensor instrument (Biacore T100; GE Healthcare) were performed as described previously (30), with some modifications. DELLA proteins (entire SLR1 for OsGID1 or mutated OsGID1s, or entire SmDELLA1 for SmGID1-1 or -2, or mutated SmGID1-1 or -2) tagged with GST were immobilized to the sensor chip at a level of ∼2,000 resonance units of the ligand. GID1s (10−7 M; excess amount of GID1 over the mobilized DELLA protein) with various concentrations of GAs were used as the analyte.
Western Blot Analysis.
Western blot analysis was performed as described elsewhere (8).
NMR.
NMR experiments were performed at 37 °C, using an Avance900 spectrometer equipped with TCI CryoProbe (Bruker Biospin). 1H-13C heteronuclear single quantum correlation (HSQC) spectroscopy experiments on Leu, Val-[13CH3, 12CD3]-labeled OsGID1/SLR1 protein complexes with GA1 and GA4 were recorded using [13CH3, 12CD3] Leu/Val-labeled samples. The data size and spectral widths were 256 (t1) × 2,048 (t2) and 4,500 Hz (ω1, 13C) × 14,400 Hz (ω2, 1H), respectively, and the carrier frequency on 13C was set to 20 ppm. When observing Ile methyl signals using [13Cδ1H3]–Ile-labeled OsGID1/SLR1 and OsGID1(I133L)/SLR1, the data size and spectral widths were 256 (t1) × 2,048 (t2) and 3,400 Hz (ω1, 13C) × 14,400 Hz (ω2, 1H), respectively, and the 13C carrier frequency was set to 12 ppm. The repetition time was 2 s. The number of scan/free induction decay was 128. All NMR spectra were processed with the Topspin software (Bruker Biospin).
Y2H Assay.
The Y2H assay was carried out as described previously (8, 30). β-Gal activity was determined through a liquid assay with yeast (Y187) transformants. The drc package available in the software package R was used to model the dose–response curves and to estimate the effective concentrations (ECs) by a four-parameter log-logistic function as described by Ritz et al. (36).
RNA Extraction and RT-PCR.
Total RNA was extracted using an RNeasy Plant Mini kit (Qiagen). One microgram of total RNA was used to synthesize first-strand cDNA using the Omniscript RT Kit (Qiagen) and oligo(dT) primers. Real-time PCR was performed using SsoAdvanced SYBR Green Supermix (Bio-Rad) in a CFX96 Real-Time System (Bio-Rad). Ubiquitin and 60S ribosomal protein L30 (GmRPL30) genes were used as a control for lettuce and soybean, respectively (37, 38). Primers used in this study are listed in SI Appendix, Table S3.
Supplementary Material
Acknowledgments
We thank Yasushi Yukawa, Shin-ichiro Kidou (Nagoya City University), and Masaki Itoh (Nagoya University) for allowing us to use their facilities for growth experiments, and Hiroyuki Tsuji (Yokohama City University) for providing computer equipment. We also thank Mitsuyasu Hasebe (National Institute for Basic Biology) and Takayuki Kohchi (Kyoto University) for critical reading of the manuscript and for providing insightful comments. This work was supported by Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology, Japan. This work was partially supported by Grants-in-Aid for Scientific Research on Innovative Areas [Grants 16H06464 (to M.U.-T.) and 16H06468 (to M.U.-T. and M.M.)], a Grant-in-Aid for Scientific Research (B) [Grant 16H04907 (to M.U.-T.)], and Japan Society for the Promotion of Science (JSPS) Grant 17J09723 (to H.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806040115/-/DCSupplemental.
References
- 1.Olszewski N, Sun TP, Gubler F. Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell. 2002;14(Suppl):S61–S80. doi: 10.1105/tpc.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacMillan J. Occurrence of gibberellins in vascular plants, fungi, and bacteria. J Plant Growth Regul. 2001;20:387–442. doi: 10.1007/s003440010038. [DOI] [PubMed] [Google Scholar]
- 3.Murase K, Hirano Y, Sun TP, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- 4.Shimada A, et al. Structural basis for gibberellin recognition by its receptor GID1. Nature. 2008;456:520–523. doi: 10.1038/nature07546. [DOI] [PubMed] [Google Scholar]
- 5.Hirano K, et al. The GID1-mediated gibberellin perception mechanism is conserved in the Lycophyte Selaginella moellendorffii but not in the Bryophyte Physcomitrella patens. Plant Cell. 2007;19:3058–3079. doi: 10.1105/tpc.107.051524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yasumura Y, Crumpton-Taylor M, Fuentes S, Harberd NP. Step-by-step acquisition of the gibberellin-DELLA growth-regulatory mechanism during land-plant evolution. Curr Biol. 2007;17:1225–1230. doi: 10.1016/j.cub.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 7.Bowman JL, et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 2017;171:287–304.e15. doi: 10.1016/j.cell.2017.09.030. [DOI] [PubMed] [Google Scholar]
- 8.Ueguchi-Tanaka M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- 9.Schnable J, Lyons E. 2015 Plant paleopolyploidy. Figshare. Available at https://figshare.com/articles/Plant_Paleopolyploidy/1538627/1. Accessed July 23, 2018.
- 10.Griffiths J, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanimoto E. Tall or short? Slender or thick? A plant strategy for regulating elongation growth of roots by low concentrations of gibberellin. Ann Bot. 2012;110:373–381. doi: 10.1093/aob/mcs049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingram TJ, et al. Internode length in Pisum : The Le gene controls the 3β-hydroxylation of gibberellin A20 to gibberellin A 1. Planta. 1984;160:455–463. doi: 10.1007/BF00429763. [DOI] [PubMed] [Google Scholar]
- 13.Itoh H, et al. A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol. 2004;54:533–547. doi: 10.1023/B:PLAN.0000038261.21060.47. [DOI] [PubMed] [Google Scholar]
- 14.Poorter H, et al. Biomass allocation to leaves, stems and roots: Meta-analyses of interspecific variation and environmental control. New Phytol. 2012;193:30–50. doi: 10.1111/j.1469-8137.2011.03952.x. [DOI] [PubMed] [Google Scholar]
- 15.Conn CE, et al. Plant Evolution. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science. 2015;349:540–543. doi: 10.1126/science.aab1140. [DOI] [PubMed] [Google Scholar]
- 16.Weng JK, Ye M, Li B, Noel JP. Co-evolution of hormone metabolism and signaling networks expands plant adaptive plasticity. Cell. 2016;166:881–893. doi: 10.1016/j.cell.2016.06.027. [DOI] [PubMed] [Google Scholar]
- 17.Marshall SD, Putterill JJ, Plummer KM, Newcomb RD. The carboxylesterase gene family from Arabidopsis thaliana. J Mol Evol. 2003;57:487–500. doi: 10.1007/s00239-003-2492-8. [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Goodstein DM, et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suyama M, Torrents D, Bork P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 27.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 28.Akagi T, Henry IM, Morimoto T, Tao R. Insights into the Prunus-specific S-RNase-based self-incompatibility system from a genome-wide analysis of the evolutionary radiation of S locus-related F-box genes. Plant Cell Physiol. 2016;57:1281–1294. doi: 10.1093/pcp/pcw077. [DOI] [PubMed] [Google Scholar]
- 29.Ueguchi-Tanaka M, et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell. 2007;19:2140–2155. doi: 10.1105/tpc.106.043729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto Y, et al. A rice gid1 suppressor mutant reveals that gibberellin is not always required for interaction between its receptor, GID1, and DELLA proteins. Plant Cell. 2010;22:3589–3602. doi: 10.1105/tpc.110.074542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aleman L, et al. Functional analysis of cotton orthologs of GA signal transduction factors GID1 and SLR1. Plant Mol Biol. 2008;68:1–16. doi: 10.1007/s11103-008-9347-z. [DOI] [PubMed] [Google Scholar]
- 32.Shimada A, et al. The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 2006;48:390–402. doi: 10.1111/j.1365-313X.2006.02875.x. [DOI] [PubMed] [Google Scholar]
- 33.Iuchi S, et al. Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J. 2007;50:958–966. doi: 10.1111/j.1365-313X.2007.03098.x. [DOI] [PubMed] [Google Scholar]
- 34.Tanimoto E. Gibberellin-dependent root elongation in Lactuca sativa: Recovery from growth retardant-suppressed elongation with thickening by low concentration of GA3. Plant Cell Physiol. 1987;28:963–973. [Google Scholar]
- 35.Tanimoto E, Watanabe J. Automated recording of lettuce root elongation as affected by indole-3-acetic acid and acid pH by a new rhizometer with minimum mechanical contact to root. Plant Cell Physiol. 1986;27:1475–1487. [Google Scholar]
- 36.Ritz C, Baty F, Streibig JC, Gerhard D. Dose-response analysis using R. PLoS One. 2015;10:e0146021. doi: 10.1371/journal.pone.0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Argyris J, Dahal P, Hayashi E, Still DW, Bradford KJ. Genetic variation for lettuce seed thermoinhibition is associated with temperature-sensitive expression of abscisic acid, gibberellin, and ethylene biosynthesis, metabolism, and response genes. Plant Physiol. 2008;148:926–947. doi: 10.1104/pp.108.125807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bansal R, et al. Recommended reference genes for quantitative PCR analysis in soybean have variable stabilities during diverse biotic stresses. PLoS One. 2015;10:e0134890. doi: 10.1371/journal.pone.0134890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chase MW, et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 2016;181:1–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.