Significance
l-asparaginase (ASNase) is a critical component of treatment protocols for acute lymphoblastic leukemia (ALL). Although the cure rates have dramatically improved, the prognosis for patients with recurrent ALL remains poor. General control nonderepressible 2 (GCN2) plays a major role in cellular response to amino acid limitation. As inhibitors targeting GCN2 have been lacking, the potential of GCN2 inhibitors as cancer therapeutic agents remains unclear. Here we report potent GCN2 inhibitors that exhibit synergistic antiproliferative effects with ASNase in asparagine synthetase-low cancer. Our findings enhance the molecular understanding of the disrupted amino acid response caused by GCN2 inhibition under limited asparagine availability. Combined treatment with GCN2 inhibitors and ASNase shows promise for achieving improved outcomes in ALL and other types of cancer.
Keywords: GCN2, asparaginase, ASNS, ALL, pancreatic cancer
Abstract
General control nonderepressible 2 (GCN2) plays a major role in the cellular response to amino acid limitation. Although maintenance of amino acid homeostasis is critical for tumor growth, the contribution of GCN2 to cancer cell survival and proliferation is poorly understood. In this study, we generated GCN2 inhibitors and demonstrated that inhibition of GCN2 sensitizes cancer cells with low basal-level expression of asparagine synthetase (ASNS) to the antileukemic agent l-asparaginase (ASNase) in vitro and in vivo. We first tested acute lymphoblastic leukemia (ALL) cells and showed that treatment with GCN2 inhibitors rendered ALL cells sensitive to ASNase by preventing the induction of ASNS, resulting in reduced levels of de novo protein synthesis. Comprehensive gene-expression profiling revealed that combined treatment with ASNase and GCN2 inhibitors induced the stress-activated MAPK pathway, thereby triggering apoptosis. By using cell-panel analyses, we also showed that acute myelogenous leukemia and pancreatic cancer cells were highly sensitive to the combined treatment. Notably, basal ASNS expression at protein levels was significantly correlated with sensitivity to combined treatment. These results provide mechanistic insights into the role of GCN2 in the amino acid response and a rationale for further investigation of GCN2 inhibitors for the treatment of cancer.
The integrated stress response (ISR) is essential for maintaining cellular homeostasis under a wide range of stressors (1). ISR is regulated by four distinct kinases—heme-regulated initiation factor-2α kinase, dsRNA-activated protein kinase (PKR), PKR-like endoplasmic reticulum kinase (PERK), and general control nonderepressible 2 (GCN2) —all of which phosphorylate eukaryotic initiation factor 2α subunit (eIF2α) (1). The phosphorylation of eIF2α in turn leads to inhibition of global protein synthesis and active translation of specific mRNAs, such as those of activating transcription factor 4 (ATF4) (1). ATF4 functions as a transcriptional activator of genes encoding proteins involved in oxidative stress, nutrient uptake, and metabolism (2, 3).
GCN2, which is directly activated by uncharged tRNA resulting from amino acid deficiency, serves as a master regulator of the amino acid response (AAR) (4, 5). Amino acids constitute vital cellular building blocks for molecules such as protein, lipids, and nucleic acids, thereby serving as essential nutrients for rapidly proliferating cancer cells. The maintenance of amino acid homeostasis plays a significant role in tumor growth to sustain cancer cell survival under nutrient-limited tumor microenvironments. Notably, a previous study reported the activation of the GCN2 pathway in human primary tumors and described the essential role of this protein in xenograft tumor growth in mice (6). However, development of potent and selective inhibitors targeting the GCN2 pathway has been limited, and the potential of GCN2 inhibitors as cancer therapeutic agents has not been explored.
Recent advances in our knowledge of cancer metabolism suggest that targeting amino acid metabolism represents a promising strategy for the development of novel therapeutic agents (7). A traditional treatment that targets amino acids is l-asparaginase (ASNase). Escherichia coli-derived ASNase is a critical component of acute lymphoblastic leukemia (ALL) treatment (8, 9). Its mechanism of action involves catalyzing the hydrolysis of asparagine to aspartic acid and ammonia, with glutamine as a lower-affinity ASNase substrate. The antileukemic effects of ASNase result from the decreased expression of asparagine synthetase (ASNS) in ALL cells (10–12). ALL cells expressing low levels of ASNS are dependent on exogenous asparagine, making them particularly sensitive to ASNase treatment. Clinical resistance to ASNase may develop from ASNase inactivation by an anti-ASNase antibody and/or from the emergence of ASNase-insensitive tumor cells (13–15); however, the mechanisms underlying the insensitivity of ALL cells to ASNase remain controversial. Several studies have suggested that elevated ASNS expression leads to ASNase resistance, whereas other studies have reported different resistance mechanisms (16).
In this preclinical study, we generated potent and selective small-molecule inhibitors of GCN2 to investigate the contribution of GCN2 to ASNase sensitivity in ALL cells. By using gene-expression profiling, we performed mechanistic characterization of GCN2 inhibitors in combination with ASNase. Additionally, we explored the therapeutic potential of this combination in various types of cancer cells. Our findings establish a preclinical rationale for targeting GCN2 activity by using small-molecule inhibitors in combination with ASNase for the treatment of cancer.
Results
Reversal of ASNase Resistance by Inhibition of the GCN2/ASNS Pathway in ALL Cells.
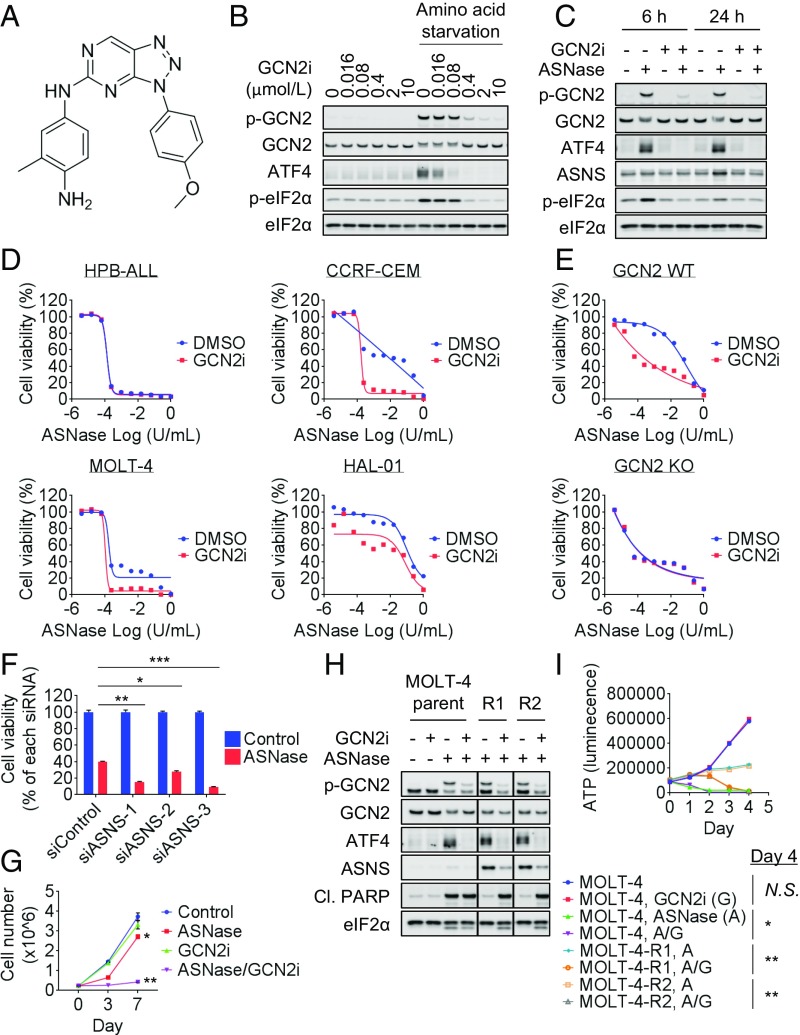
To explore the function of GCN2, we generated a potent kinase inhibitor of this protein (GCN2iA; Fig. 1A). Kinase assay revealed that this compound showed an IC50 value of 4.0 nmol/L for GCN2 (SI Appendix, Fig. S1A). In a focused panel of 27 kinases covering the major kinase families, GCN2iA at 1 µmol/L showed inhibition of most kinases by <50%, and only one kinase (GSK3β) showed >75% inhibition (SI Appendix, Fig. S1B). In U2OS cells, treatment with GCN2iA suppressed the phosphorylation of GCN2 and eIF2α as well as ATF4 expression in the absence of amino acids in the medium (Fig. 1B).
Fig. 1.
Sensitization of ALL cells to ASNase via GCN2 inhibition. (A) Compound structure and potency of GCN2iA. (B) U2OS cells were treated with amino acid-free medium and/or GCN2iA as indicated for 4 h. Cell lysates were analyzed by Western blot. (C) CCRF-CEM cells were treated with 1 mU/mL ASNase and/or 1 µmol/L GCN2iA as indicated. Cell lysates were analyzed by Western blot. (D) ALL cells were treated with ASNase and/or 1 µmol/L GCN2iA as indicated for 72 h, and cell viability was measured (mean; n = 2). (E) MEF cells (GCN2-WT or KO) were treated with ASNase and/or 1 µmol/L GCN2iA as indicated for 72 h, and cell viability was measured (mean; n = 2). (F) CCRF-CEM cells were transfected with siRNA as indicated; 24 h after transfection, cells were treated with 1 mU/mL ASNase for 72 h. Cell viability was measured and normalized against each siRNA-treated cell (mean with SD; n = 3; *P < 0.001, **P < 0.00001, and ***P < 0.000001). (G) CCRF-CEM cells were treated with 1 mU/mL ASNase and/or 1 µmol/L GCN2iA as indicated. Cell number was counted at days 0, 3, and 7 (mean with SD; n = 3; *P < 0.01 and **P < 0.00001). (H) MOLT-4, MOLT-4-R1, or MOLT-4-R2 cells were treated with 1 U/mL ASNase and/or 1 µmol/L GCN2iA as indicated for 24 h. Cell lysates were analyzed by Western blot. Individual blots with dividing lines were combined from a single electrophoresis gel. (I) MOLT-4, MOLT-4-R1, or MOLT-4-R2 cells were treated with 1 U/mL ASNase (marked as “A”) and/or 1 µmol/L GCN2iA (“G”) as indicated. Cell viability (i.e., ATP) was measured at the indicated time points (mean with SD; n = 3). Statistical analyses were performed at day 4 (*P < 0.0001 and **P < 0.00001; Cl.PARP, cleaved PARP; N.S., not significant, i.e., P > 0.05).
To examine the contribution of GCN2 to ASNase sensitivity in ALL cells, we first characterized four ALL cell types with different levels of sensitivity to ASNase: HPB-ALL cells are hypersensitive, MOLT-4 and CCRF-CEM cells are intermediately insensitive, and HAL-01 cells are hyperinsensitive (SI Appendix, Table S1). Western blot analysis revealed that HPB-ALL cells showed no detectable ASNS expression regardless of ASNase treatment (SI Appendix, Fig. S2A), whereas MOLT-4 and CCRF-CEM cells exhibited weak basal expression of ASNS protein, which was induced by ASNase treatment (0.001–1 U/mL; SI Appendix, Fig. S2A). In HAL-01 cells, basal levels of ASNS expression were relatively high and did not change following ASNase treatment (SI Appendix, Fig. S2A). Although there was no clear increase in eIF2α phosphorylation at 24 h after ASNase treatment, the up-regulation of eIF2α phosphorylation was observed within a shorter time period in CCRF-CEM cells (4 h; SI Appendix, Fig. S2B). The results were putatively attributable to feedback regulation by eIF2α-phosphatase–targeted growth arrest and DNA damage-inducible protein 34 (GADD34; SI Appendix, Fig. S2B). Importantly, GCN2 phosphorylation and ATF4 expression were clearly increased in HPB-ALL, MOLT-4, and CCRF-CEM cells after ASNase treatment (0.001–1 U/mL; SI Appendix, Fig. S2A). These results indicate that AAR was activated by treatment even with low concentrations of ASNase in the cell lines. On the contrary, in HAL-01 cells, the GCN2/ATF4 pathway was activated only following treatment with high concentrations of ASNase (1 U/mL), suggesting that high basal-level expression of ASNS mediated de novo asparagine synthesis and prevented GCN2 pathway activation. As ASNase is known to catalyze glutamine hydrolysis at high concentrations, we also measured extracellular asparagine/glutamine content by gas chromatography/MS (GC-MS). Asparagine and glutamine were depleted following treatment with high concentrations of ASNase (1 U/mL; SI Appendix, Fig. S2C). Glutamine depletion was considered to affect ASNS induction in HAL-01 cells (SI Appendix, Fig. S2A). Thus, we decided to use lower concentrations of ASNase in the subsequent mechanistic study to evaluate the effects of asparagine limitation without glutamine depletion.
We then tested GCN2iA in the presence of ASNase by using the ALL cell lines. ASNase-induced GCN2 phosphorylation, ATF4 expression, and ASNS expression were inhibited by GCN2iA in CCRF-CEM cells and MOLT-4 cells (Fig. 1C and SI Appendix, Fig. S2D). Notably, GCN2iA treatment rendered the intermediately insensitive ALL cells (CCRF-CEM and MOLT-4) highly sensitive to ASNase; in addition, GCN2iA treatment alone did not cause any single-agent effects on cell viability (Fig. 1D). In ASNase-hypersensitive HPB-ALL and hyperinsensitive HAL-01 cells, GCN2iA treatment did not extensively modulate ASNase sensitivity (Fig. 1D). Given that GCN2iA inhibited kinases other than GCN2, albeit with lower potency (SI Appendix, Fig. S1B), we next examined whether its cellular activities were on target. Importantly, the moderate antiproliferative effects achieved by combining ASNase and GCN2iA treatment were observed in GCN2- WT MEF cells, but not in GCN2-KO MEF cells, ruling out a significant impact from off-target effects (Fig. 1E). In MEF cells, we confirmed that ASNase treatment induced ATF4/ASNS expression in a GCN2-dependent manner (SI Appendix, Fig. S2E). Furthermore, siRNA-mediated depletion of ASNS sensitized the CCRF-CEM cells to ASNase (Fig. 1F and SI Appendix, Fig. S2F). Together, these results indicate the involvement of the GCN2/ASNS pathway in ASNase resistance.
We also performed a long-term proliferation assay to assess whether GCN2 inhibition prevents the development of resistance to ASNase (Fig. 1G). ASNase reduced cell proliferation of CCRF-CEM cells by day 3, but the cells started to grow, as did the control cells, from day 3 to day 7. Notably, the combined treatment of ASNase with GCN2 inhibitors persistently suppressed the cell growth over 7 d. Next, we established ASNase-resistant MOLT-4-R (R1 and R2) cells by incubating them with increasing concentrations of ASNase (0.00001–1 U/mL). Consistent with a previous report, MOLT-4-R cell lines showed higher levels of ASNS expression than the parental cells (Fig. 1H) (17). Additionally, the MOLT-4-R cell lines were capable of proliferation even when treated with high concentrations of ASNase (1 U/mL), although they showed slower growth than untreated parent cells (Fig. 1I). We observed that GCN2iA treatment in the presence of ASNase decreased ASNS expression and increased apoptosis marker PARP cleavage in these cell lines (Fig. 1H). Consistently, combined treatment with ASNase and GCN2iA decreased MOLT-4-R cell viability (Fig. 1I). These findings indicate that GCN2 inhibition has the potential to prevent and reverse ASNase resistance.
Correlation Between ASNS Expression and the Combined Effects of ASNase Treatment and GCN2 Inhibition.
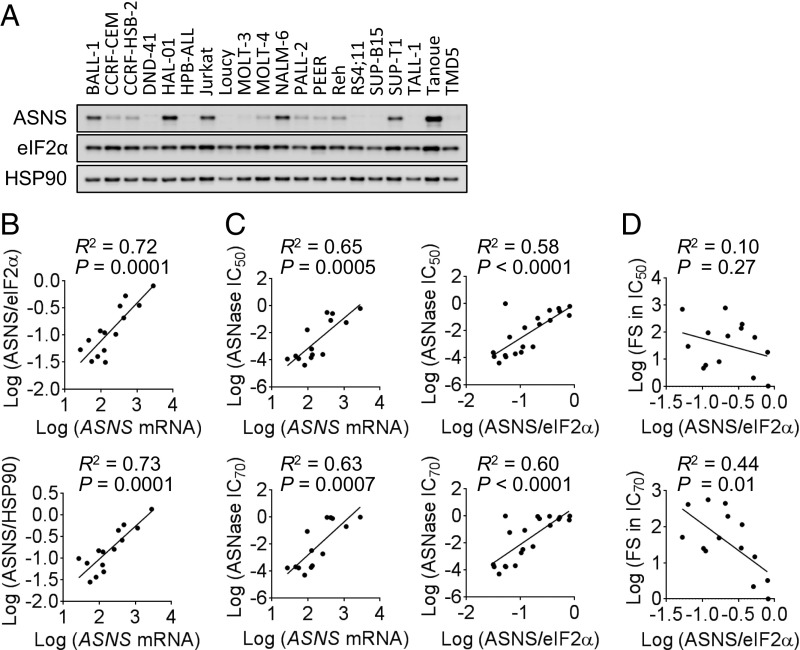
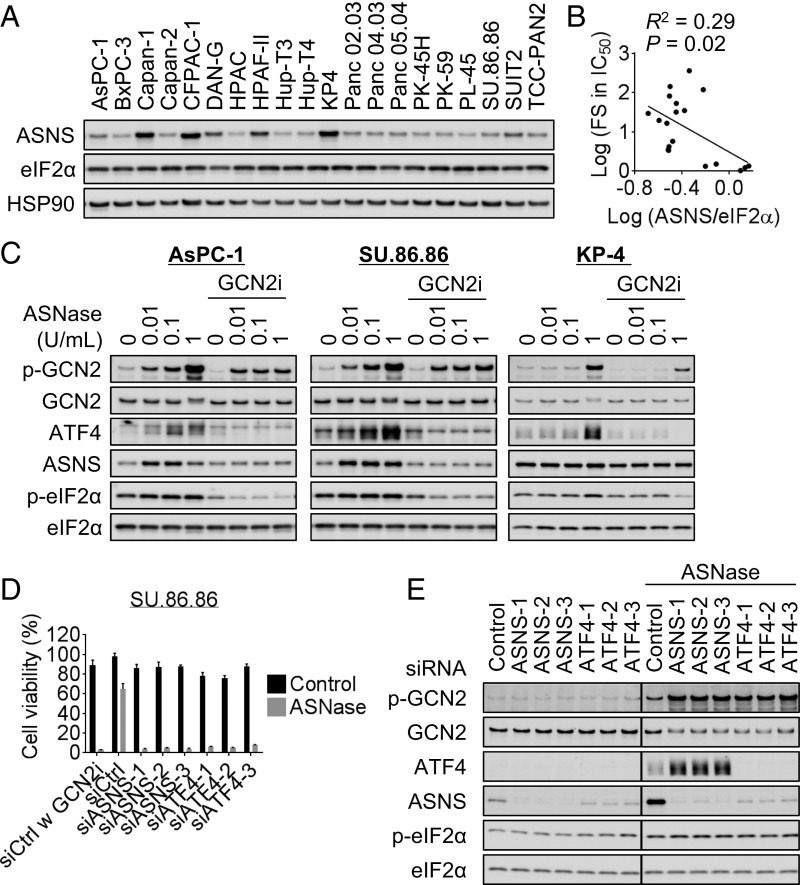
To determine whether there is a correlation between baseline ASNS expression and sensitivity to ASNase-GCN2iA combination treatment, particularly in ALL cells, we examined ASNS expression at the protein levels in 20 ALL cell lines, including the 4 previously mentioned cell lines (Fig. 2A and SI Appendix, Table S1). To analyze the mRNA expression of ASNS, we used information available from public databases (SI Appendix). Our analyses showed that the mRNA expression levels of ASNS were correlated with the ASNS protein levels, regardless of the protein used for normalization (eIF2α or HSP90; Fig. 2B). Consistent with the previous study (18), there was a significant correlation between ASNase sensitivity and baseline ASNS levels, even when we used different evaluation parameters (ASNase sensitivity, IC50 or IC70 value; ASNS expression, mRNA or protein level; Fig. 2C). To assess the combined effects of ASNase treatment and GCN2 inhibition, we measured the fold change in the IC50 and IC70 values. As six ALL cell lines were highly sensitive to ASNase alone (i.e., hypersensitive; SI Appendix, Table S1), we excluded them from subsequent correlation analyses. We found that the fold changes in the IC70 value, but not in the IC50 value, were associated with ASNS protein levels (Fig. 2D). Considering the dose–response curve in ALL cells with intermediate ASNase sensitivity (e.g., MOLT-4 cells in Fig. 1D), the fold change in IC70 value appeared to be a better parameter than IC50 value for assessing the effects of the combined treatment. Thus, our results indicate that ASNS protein levels are associated with sensitivity to the combined effects of ASNase treatment and GCN2 inhibition in ASNase-insensitive ALL cells.
Fig. 2.
Correlation between ASNS expression and the combined effect of ASNase treatment with GCN2 inhibition in ALL cells. (A) Western blot analysis of 20 ALL cell lines. (B) Correlation between ASNS mRNA levels (in 14 cell lines) and protein levels (normalized against eIF2α or HSP90 levels) was analyzed. (C) Correlation between IC50 or IC70 value of ASNase and ASNS mRNA or protein levels (normalized against eIF2α levels) was analyzed. (D) Correlation between the combined effects of ASNase treatment with GCN2 inhibition (FS, fold shift in IC50 or IC70 values) and ASNS protein levels (normalized against eIF2α) was analyzed. Linear regression analysis was performed, and R and P values determined by Pearson’s correlation are shown.
Our findings in CCRF-CEM and MOLT-4 cells (Fig. 1C and SI Appendix, Fig. S2D) indicate that GCN2 mediates the induction of ASNS following ASNase treatment in ALL cells, which show intermediate sensitivity to ASNase. To further support the hypothesis, we tested five ALL cell lines with intermediate sensitivity to ASNase and three ASNase-hypersensitive ALL cell lines. In the cell lines that were intermediately insensitive to ASNase, ASNS expression was induced by ASNase treatment, and GCN2iA treatment prevented this induction (SI Appendix, Fig. S2G). In contrast, ASNase-hypersensitive cells showed no induction of ASNS expression in response to ASNase treatment (SI Appendix, Fig. S2H).
Suppression of Protein Translation by GCN2 Inhibition in the Presence of ASNase.
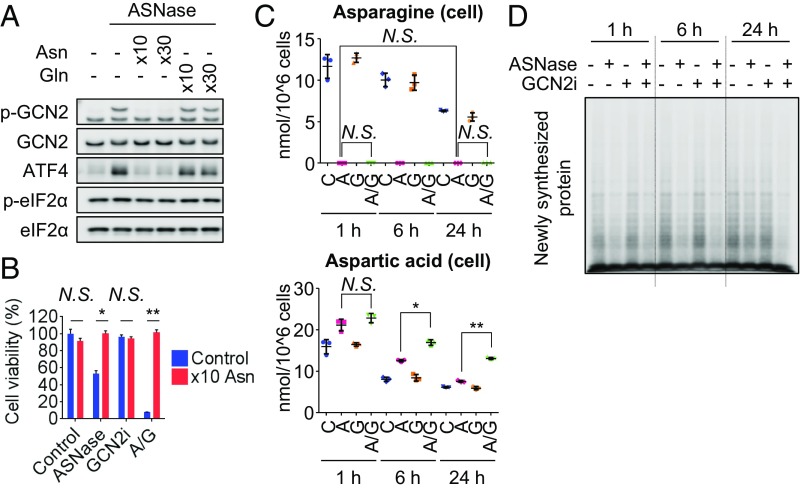
We then performed rescue experiments by using asparagine in CCRF-CEM cells. Supraphysiological concentrations of asparagine, but not those of glutamine, prevented GCN2 pathway activation following ASNase treatment and rescued the antiproliferative effects of the ASNase-GCN2iA combined treatment (Fig. 3 A and B). To further investigate the mechanism underlying the antiproliferative effect exerted by the combination of ASNase treatment and GCN2 inhibition, we measured the extracellular/intracellular amino acid levels. Our initial hypothesis was that ASNase treatment activates the GCN2/ASNS pathway, thereby recovering cellular asparagine concentration by up-regulating the expression of ASNS. Contrary to our expectations, the extracellular/intracellular asparagine levels were depleted 1 h after ASNase treatment and remained undetectable even at 24 h after ASNase treatment, although ASNS expression was increased (Figs. 1C and 3C). Further, the combined treatment with ASNase and GCN2iA caused the accumulation of cellular aspartic acid levels (Fig. 3C). Such accumulation was not observed in other amino acids tested, excluding glutamic acids, which showed modest accumulation following combined treatment (SI Appendix, Fig. S3B). As ASNS catalyzes the conversion of aspartic acid to asparagine, the increase in aspartic acid levels likely results from suppression of ASNS expression by GCN2 inhibition. Based on these results, we speculate that newly synthesized asparagine by ASNS was rapidly consumed, thereby precluding the observation of recovered asparagine levels.
Fig. 3.
Suppression of protein translation by inhibiting GCN2 in the presence of ASNase. (A) CCRF-CEM cells were treated with 1 mU/mL ASNase and supraphysiological levels of asparagine (10×, 4.3 mmol/L; 30×, 12.9 mmol/L) or glutamine (10×, 20 mmol/L; 30×, 60 mmol/L) as indicated for 24 h. Cell lysates were analyzed by Western blot. (B) CCRF-CEM cells were treated with 1 mU/mL ASNase (marked as “A”) and/or 1 µmol/L GCN2iA (“G”) along with supraphysiological levels of asparagine (10×, 4.3 mmol/L) as indicated for 72 h. Cell viability was measured (mean with SD; n = 3; *P < 0.0001 and **P < 0.000001). (C) CCRF-CEM cells were treated with 1 mU/mL ASNase (“A”) and/or 1 µmol/L GCN2iA (“G”) as indicated. Asparagine and aspartic acid levels in the cells were measured by GC-MS. Plots represent values from three biological replicates with SD (*P < 0.001 and **P < 0.00001). (D) CCRF-CEM cells were treated with 1 mU/mL ASNase and/or 1 µmol/L GCN2iA as indicated. Cells were then treated with 10 µmol/L puromycin for 10 min before being harvested. Cell lysates were analyzed by Western blot using an anti-puromycin antibody to detect newly synthesized proteins (N.S., not significant, i.e., P > 0.05).
In addition to its function as a substrate for protein synthesis (19), asparagine has been shown to be important for protection against apoptosis under limited glutamine availability (20). Asparagine also functions as an amino acid exchange factor and regulates mTORC1 signaling (21). In CCRF-CEM cells treated with ASNase and/or GCN2iA, the extracellular and intracellular glutamine levels were not reduced compared with those in control cells, precluding the possibility of glutamine limitation (SI Appendix, Fig. S3). Meanwhile, ASNase and/or GCN2iA treatment did not decrease the levels of extracellular amino acids, other than those of asparagine. Besides aspartic acid and glutamic acid, only alanine levels showed a significant increase after ASNase treatment for 24 h (SI Appendix, Fig. S3A). The data suggest that the depletion of asparagine by ASNase does not have much impact on the import of other amino acids in CCRF-CEM cells. To further explore the potential use of asparagine, we evaluated de novo protein synthesis. Puromycin-incorporation assays revealed that the decrease in protein synthesis caused by ASNase was enhanced upon cotreatment with GCN2iA (24 h; Fig. 3D and SI Appendix, Fig. S4A). As a positive control for this assay, we confirmed that PERK inhibition recovered protein translation when combined with endoplasmic reticulum stressor tunicamycin (SI Appendix, Fig. S4B). Importantly, supraphysiological levels of asparagine caused the recovery of protein synthesis activity in CCRF-CEM cells treated with ASNase alone or with GCN2iA and ASNase, whereas supraphysiological levels of glutamine did not (SI Appendix, Fig. S4C). As mTORC1, which is activated upon amino acid stimulation, positively regulates protein translation, we additionally examined whether the reduction in protein synthesis following combined treatment with ASNase and GCN2iA is caused by inactivation of mTORC1 signaling. Phosphorylation of S6K was decreased by ASNase treatment; however, GCN2 inhibition reversed the decrease in S6K phosphorylation (SI Appendix, Fig. S4D), in accordance with a previous report showing that GCN2 suppresses mTORC1 signaling through up-regulation of Sestrin2 (22). We verified that Sestrin2 expression was induced by amino acid starvation in a GCN2-dependent manner in CCRF-CEM cells and MEF cells (SI Appendix, Fig. S4 D and E). Taken together, our findings indicate that GCN2 inhibition prevents ASNS expression and thereby augments the inhibition of protein translation in the presence of ASNase, independent of mTORC1 signaling.
Activation of MAPK Pathways by GCN2 Inhibition in Combination with ASNase Treatment.
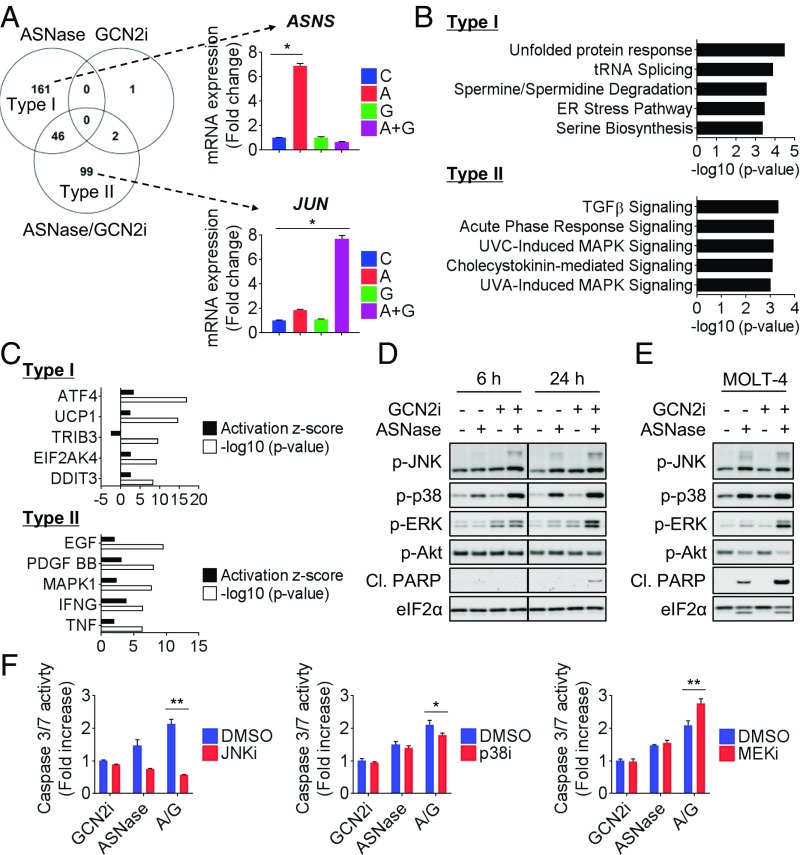
To examine the mechanism of action in an unbiased manner, we performed microarray analysis in CCRF-CEM cells: 207, 3, or 145 genes were differentially expressed following treatment with ASNase, GCN2iA, or ASNase-GCN2iA combination, respectively. The limited number of differentially expressed genes following GCN2iA treatment supported the idea that GCN2 serves as a regulator of amino acid homeostasis through its kinase activity, particularly under amino acid limitation. Among the 207 genes, 161 were modulated by ASNase treatment but not by GCN2iA or the combined treatment (type I; Fig. 4A). This finding indicates that most of the ASNase-mediated gene expression changes (∼80%) are dependent on GCN2. Interestingly, among genes differentially expressed following combined treatment with ASNase and GCN2iA, ∼70% (99 of 147) were not robustly modulated in response to treatment with ASNase or GCN2iA alone (type II; Fig. 4A). A comprehensive evaluation of transcriptional changes using Ingenuity Pathway Analysis (IPA) revealed that type I genes were enriched for multiple processes related to ISR or amino acid metabolism (Fig. 4B), in accordance with the known role of GCN2. We also performed upstream analysis and identified GCN2 and other ISR regulators as upstream factors for type I genes (Fig. 4C). Notably, the top-ranked biological processes and upstream factors enriched with type II genes suggested the induction of growth factor or stress-activated MAPK pathway (Fig. 4 B and C). Consistent with these results, Western blot analysis revealed that the phosphorylation of JNK, p38 MAPK, and ERK was robustly induced by combined treatment of CCRF-CEM cells with ASNase and GCN2iA (Fig. 4D). Similar results were observed in MOLT-4 cells (Fig. 4E). Considering that mitogen/stress-activated MAPK pathway regulates cell death and survival, we then examined whether the activation of the pathway contributes to cell death and survival induced by the combined treatment with ASNase and GCN2iA. In CCRF-CEM cells, the combined treatment increased PARP cleavage and caspase 3/7 activity, indicative of apoptosis induction (Fig. 4 D and F). Importantly, the increase in caspase 3/7 activity following combined treatment with ASNase and GCN2iA was reversed by treatment with JNK inhibitor SP600125 or p38 inhibitor SB203580. Conversely, the MEK inhibitor PD0325901 slightly potentiated the induction of apoptosis (Fig. 4F).
Fig. 4.
Transcriptional profiling of CCRF-CEM cells following combined treatment with ASNase and GCN2 inhibition. (A) CCRF-CEM cells were treated with 1 mU/mL ASNase (marked as “A”) and/or 1 µmol/L GCN2iA (“G”) as indicated for 6 h. Gene expression level was measured by microarray (mean with SD; n = 3). Venn diagram shows the number of genes altered (\fold change\ > 3), categorized as unique to ASNase treatment (type I) or unique to the combined treatment (type II). ASNS or JUN is shown as a representative of type I or type II genes, respectively (*P < 0.00001). (B) Pathway analysis of type I/II genes in CCRF-CEM cells by IPA. (C) Upstream analysis of type I/II genes in CCRF-CEM cells by IPA. (D) CCRF-CEM cells were treated with 1 mU/mL ASNase and/or 1 µmol/L GCN2iA as indicated. Cell lysates were analyzed by Western blot. (E) MOLT-4 cells were treated with 1 mU/mL ASNase and/or 1 µmol/L GCN2iA as indicated for 24 h. Cell lysates were analyzed by Western blot. (F) CCRF-CEM cells were treated with ASNase (“A”) and/or 1 µmol/L GCN2iA (“G”) along with 12.5 µmol/L JNKi (SP600125), 12.5 µmol/L p38i (SB203580), or 100 nmol/L MEKi (PD0325901) as indicated for 24 h. Caspase 3/7 activity was measured (mean; n = 3). *P < 0.05 and **P < 0.005; Cl.PARP, cleaved PARP.
In Vitro Antiproliferative Effects of Combined ASNase Treatment and GCN2 Inhibition on Various Types of Cancer Cells.
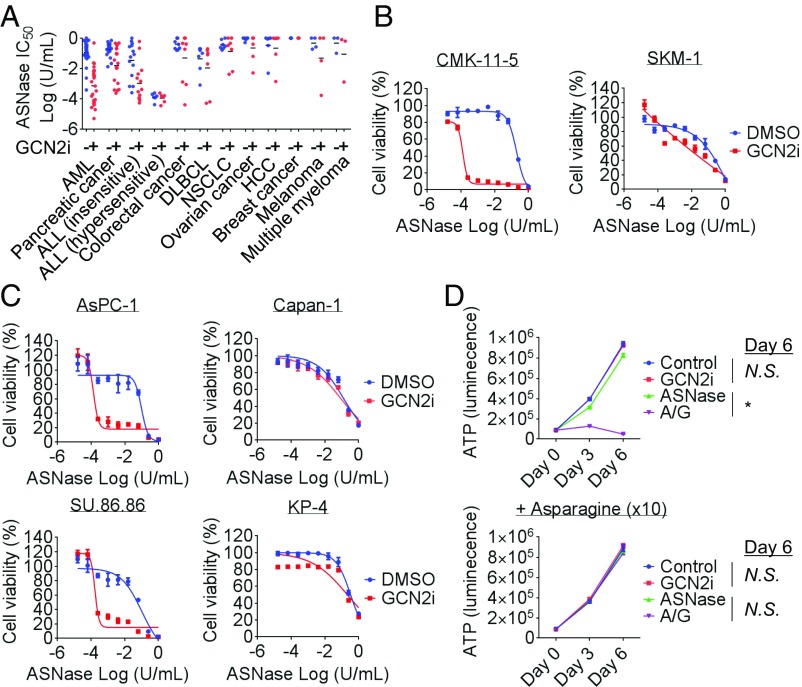
Preclinical and clinical studies have shown ASNase-related antitumor activities in various types of cancer (23). To identify the types of cancer that are particularly sensitive to the combination of GCN2 inhibition and ASNase treatment, we performed a cell-panel study with >100 cell lines, including ALL, acute myelogenous leukemia (AML), pancreatic cancer, colorectal cancer, diffuse large B-cell lymphoma, non–small-cell lung cancer, ovarian cancer, hepatocellular carcinoma, breast cancer, melanoma, and multiple myeloma cells (Fig. 5A and SI Appendix, Fig. S5). Similar to the observations in ALL cells, GCN2iA treatment alone did not exert an antiproliferative effect on these cells. Except for ASNase-hypersensitive ALL cells, most cell lines did not show extreme sensitivity to ASNase treatment (Fig. 5A and SI Appendix, Fig. S5). Notably, we found that the IC50 values of ASNase, when used in combination with GCN2 inhibition, were significantly lower against AML and pancreatic cancer cells (Fig. 5 A–C and SI Appendix, Fig. S5). Several cancer cell lines in other tumor types were also highly sensitive to ASNase-GCN2iA combination treatment; however, we did not observe statistically significant differences (Fig. 5A and SI Appendix, Fig. S5). Importantly, similar to the results observed in ALL cells, the antiproliferative effects of ASNase-GCN2iA combination treatment on pancreatic cancer cells were attenuated by treatment with supraphysiological levels of asparagine (Fig. 5D), indicating an identical mechanism of action.
Fig. 5.
In vitro antiproliferative effects of ASNase-GCN2iA combination in various types of cancer cells. (A) Cell lines of the indicated cancer types were treated with ASNase (0.00001–1 U/mL) and/or 1 µmol/L GCN2iA for 72 h. Cell viability was measured, and the IC50 value of ASNase was calculated. (B and C) AML (CMK-11–5 or SKM-1) and pancreatic cancer (AsPC-1, Capan-1, KP-4, or SU.86.86) cells were treated with ASNase and/or 1 µmol/L GCN2iA as indicated for 72 h. Cell viability was measured (mean with SD; n = 3). (D) SU.86.86 cells were treated with 1 mU/mL ASNase (marked as “A”) and/or 1 µmol/L GCN2iA (“G”) along with supraphysiological levels of asparagine (10×, 4.3 mmol/L) as indicated. Cell viability (i.e., ATP) was measured at the indicated time points (mean with SD; n = 3). Statistical analyses were performed at day 6 (*P < 0.000001; N.S., not significant, i.e., P > 0.05); DLBCL, diffuse large-cell B-cell lymphoma; HCC, hepatocellular carcinoma; NSCLC, non–small-cell lung cancer.
Previous studies have reported that 50–80% of pancreatic adenocarcinomas express null or low levels of ASNS compared with normal pancreatic tissues (24, 25). An in vitro study showed that pancreatic cancer cells expressing low levels of ASNS were sensitive to ASNase treatment, although only a limited number of cell lines were tested (25). Therefore, we investigated the correlation between baseline ASNS expression and sensitivity to ASNase or ASNase-GCN2iA combination treatment in pancreatic cancer cells. Unlike that in ALL cells, we observed no significant correlation between protein and mRNA levels of ASNS (Fig. 6A and SI Appendix, Fig. S6A); further, the baseline mRNA or protein expression of ASNS was not associated with ASNase sensitivity (SI Appendix, Fig. S6B and Table S2). However, we found that the combined effect of ASNase and GCN2iA treatment (measured by fold change in IC50 value) was associated with ASNS protein levels, but not mRNA levels (Fig. 6D and SI Appendix, Fig. S6C and Table S2). We did not use the IC70 value in the analysis of pancreatic cancer cells because of their intrinsic lower sensitivity to ASNase compared with ALL cells (Fig. 5A and SI Appendix, Tables S1 and S2). Consistent with the significant correlation between ASNS protein levels and sensitivity to the combined treatment, we found that ASNase treatment induced ASNS expression in a GCN2-dependent manner in pancreatic cancer SU86.86 and AsPC-1 cells (Fig. 6C), which were highly sensitive to ASNase-GCN2iA combination treatment (Fig. 5C). However, ASNS expression remained unchanged following the treatment of pancreatic cancer KP-4 cells with ASNase (Fig. 6C), in which GCN2 inhibition did not enhance sensitivity to ASNase (Fig. 5C).
Fig. 6.
Correlation between ASNS expression and the combined effects of ASNase treatment with GCN2 inhibition in pancreatic cancer cells. (A) Western blot analysis of 20 pancreatic cancer cell lines. (B) Correlation between the combined effects of ASNase treatment with GCN2iA inhibition (FS, fold shift in IC50 value) and ASNS protein levels (normalized against eIF2α level) was analyzed. Linear regression analysis was performed, and R and P values determined by Pearson’s correlation are indicated. PL-45 cells were excluded from the analysis because of their slow growth during the 72-h culture for the cell viability assay. (C) AsPC-1, SU.86.86, or KP-4 cells were treated with ASNase and/or 1 µmol/L GCN2iA as indicated for 24 h. Cell lysates were analyzed by Western blot. (D) SU.86.86 cells were transfected with siRNA as indicated; 24 h after transfection, cells were treated with 1 mU/mL ASNase and/or 1 µmol/L GCN2iA as indicated for 72 h. Cell viability was measured (mean with SD; n = 3). (E) SU.86.86 cells were transfected with siRNA as indicated; 24 h after transfection, cells were treated with 1 mU/mL ASNase for 24 h. Cell lysates were analyzed by Western blot. Individual blots with dividing lines were combined from a single electrophoresis gel.
We additionally performed siRNA experiments to validate our findings in pancreatic cancer cells. Depletion of ASNS or ATF4 phenocopied the antiproliferative effects of GCN2 inhibition on SU.86.86 cells in the presence of ASNase (Fig. 6D). Notably, GCN2 phosphorylation was elevated on depletion of ASNS or ATF4 only in the presence of ASNase (Fig. 6E), suggesting that suppression of ASNS-mediated asparagine synthesis caused an increase in the ratio of uncharged asparaginyl tRNA, thereby stimulating GCN2 autophosphorylation. Together, our data indicate that the GCN2/ASNS pathway plays a pivotal role in conferring ASNase insensitivity to pancreatic cancer cells.
In Vivo Antitumor Effects of GCN2 Inhibition Combined with ASNase Treatment.
We next examined the in vivo antitumor activity of combined GCN2 inhibition and ASNase treatment in mouse xenograft models. Before the combination study, we sought to optimize the dosing and schedule of ASNase administration by using ALL xenograft models. We observed that plasma asparagine levels were substantially depleted following a single treatment with ASNase at 4, 8, or 24 h (300–3,000 U/kg; SI Appendix, Fig. S7A) and after repeated ASNase treatment (1,000–10,000 U/kg; SI Appendix, Fig. S7B). Higher doses of ASNase (>1,000 U/kg) also depleted plasma glutamine levels (SI Appendix, Fig. S7B). Importantly, asparagine levels in CCRF-CEM xenograft tumors were found to be decreased at 4, 8, and 24 h after ASNase administration (300–3,000 U/kg; SI Appendix, Fig. S7A). The depletion of asparagine in the tumors appeared to be saturated at 1,000 U/kg ASNase. Consistent with these results, antitumor activities of ASNase in an ASNase-hypersensitive HPB-ALL xenograft model reached a plateau at 1,000 U/kg [for 300, 1,000, or 3,000 U/kg ASNase, treatment over control (T/C) values of 7%, −8%, or −9% (P < 0.0001), respectively; SI Appendix, Fig. S7C]. Therefore, we used a once-daily dose of 1,000 U/kg ASNase for the subsequent experiment.
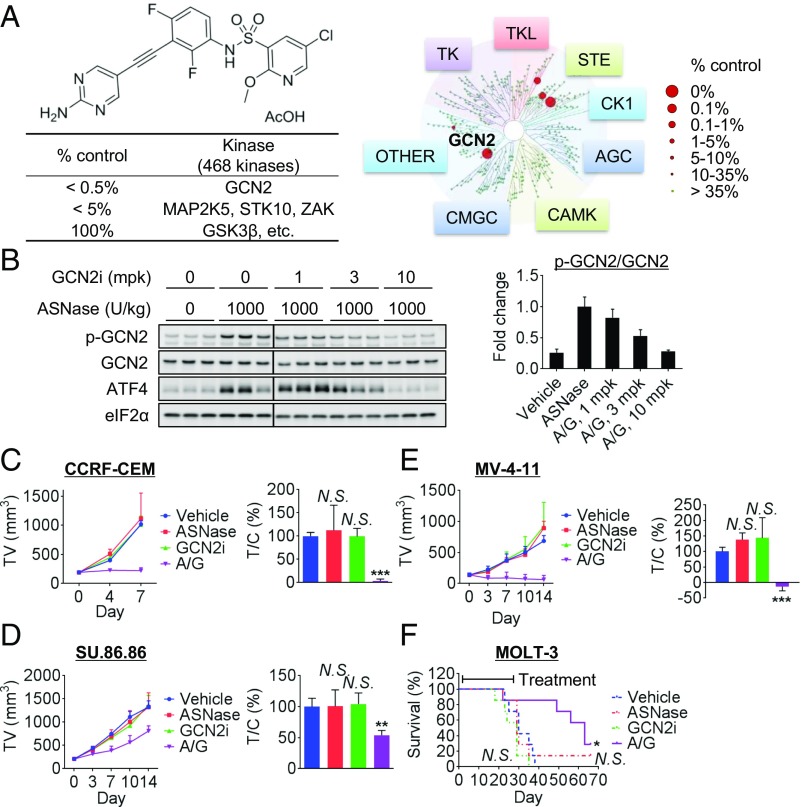
Because GCN2iA was not suitable for in vivo use because of its poor pharmacokinetic profile, we generated GCN2iB (Fig. 7A), an ATP-competitive GCN2 inhibitory compound with a better pharmacokinetic profile. The compound showed an IC50 value of 2.4 nmol/L for GCN2 (Fig. 7A) and potent cellular activity (SI Appendix, Fig. S8A). In a panel of 468 kinases, only GCN2 showed >99.5% inhibition, and three kinases (MAP2K5, STK10, and ZAK) showed >95% inhibition at 1 µmol/L GCN2iB (Fig. 7A), demonstrating high kinase selectivity. Unlike GCN2iA, GCN2iB did not inhibit GSK3β (0% inhibition; Fig. 7A). We verified that GCN2iB exhibited in vitro antiproliferative effects in combination with ASNase against GCN2-WT MEF cells, but not against GCN2-KO MEF cells (SI Appendix, Fig. S8B), indicating that the combined effects were indeed mediated by the GCN2-inhibitory activity of the compound. As observed in the case of GCN2iA, the robust in vitro antiproliferative effects were also elicited following combinatorial treatment with ASNase and GCN2iB in ALL (CCRF-CEM), AML (MV-4-11), and pancreatic cancer (SU.86.86) cells (SI Appendix, Fig. S8 C and D). Next, we performed the combination study in xenograft models of these cells. With respect to in vivo target engagement, GCN2iB suppressed GCN2 pathway activation when cotreated with ASNase in the CCRF-CEM xenograft model (Fig. 7B). In the antitumor activity study of the CCRF-CEM xenografts, ASNase or GCN2iB alone did not significantly affect tumor growth (P = 0.84 and P = 0.99, respectively; Fig. 7C). Notably, a combination of ASNase and GCN2iB elicited potent antitumor activity (P = 0.0002; Fig. 7C) with synergistic effects (main effect of ASNase, P = 0.0053; main effect of GCN2iB, P = 0.0006; interaction effect of ASNase and GCN2iB, P = 0.0007). In MV-4–11 and SU.86.86 xenografts, robust antitumor activity of the combination of GCN2iB and ASNase was observed (P = 0.0003 and P = 0.0038; Fig. 7 D and E) with synergistic effect (main effect of ASNase, P = 0.0019 or P = 0.0045; main effect of GCN2iB, P = 0.00038 or P = 0.022; interaction effect of ASNase and GCN2iB, P < 0.0001 or P = 0.0079), respectively. For MV-4–11 xenografts, we measured tumor volume until 1 wk after drug cessation. As shown in SI Appendix, Fig. S9A, ASNase/GCN2i-treated tumors did not show significant growth even after drug cessation. To evaluate the effects on long-term survival, we tested a disseminated ALL xenograft model (MOLT-3). The combination of ASNase and GCN2iB yielded survival advantage compared with the vehicle-treated control (P = 0.011; Fig. 7F) with synergistic effect (main effect of ASNase, P = 0.56; main effect of GCN2iB, P = 0.10; interaction effect of ASNase and GCN2iB, P = 0.046). Severe body weight loss was not induced by the treatments throughout the study period, although the SU.86.86 model showed a modest cachectic body weight reduction (SI Appendix, Fig. S9 B–E). Thus, our data demonstrate that the combination of ASNase treatment with GCN2 inhibition synergistically blocks in vivo tumor growth in ALL, AML, and pancreatic xenograft models.
Fig. 7.
In vivo antitumor activity of ASNase treatment combined with GCN2 inhibition. (A) Compound structure, potency, and kinase selectivity of GCN2iB. (B) Mice bearing CCRF-CEM xenografts were pretreated with 1,000 U/kg ASNase for 24 h and then with GCN2iB and/or ASNase as indicated. Tumor xenografts were harvested 8 h after treatment and analyzed by Western blot. (Left) Individual blots with dividing lines are combined from a single electrophoresis gel. (Right) Bars represent densitometric analysis of phospho-GCN2 divided by total GCN2, normalized to ASNase-treated control (mean with SD; n = 3). (C) Mice bearing CCRF-CEM xenografts were treated daily with GCN2iB [marked as “G,” 10 mg/kg (mpk) twice daily] and daily with ASNase (“A,” 1,000 U/kg once daily) as indicated for 7 d; (Left) tumor volume (TV) and (Right) T/C values on day 7. (D and E) Mice bearing MV-4–11 or SU.86.86 xenografts were first treated 3 d per week with GCN2iB (“G,” 10 mpk twice daily) and/or ASNase (“A,” 1,000 U/kg once per day) for 14 d as indicated; (Left) tumor volume (TV) and (Right) T/C values on day 14. Data are mean tumor volume or T/C values with SD (n = 5). (F) MOLT-3 cells were inoculated into SCID mice via the tail vein. Mice were first treated 3 d per week with GCN2iB (“G,” 10 mpk twice daily) and/or ASNase (“A,” 1,000 U/kg once daily) as indicated for 28 d. Data indicate survival rates (n = 7). Day 1 is the beginning of treatment (*P < 0.05, **P < 0.005, and ***P < 0.0005; N.S., not significant, i.e., P > 0.05 vs. vehicle-treated control).
Discussion
The development of pharmacological tools that selectively modulate a target molecule has been facilitated by strategies enabling the evaluation of biological function and therapeutic potential of such agents. ASNS contributes to cancer cell survival and metastasis (6, 26), and ASNS inhibitors have been developed previously (27, 28); however, information on small-molecule inhibitors of GCN2 has been limited (29). In this study, we developed two potent and structurally different GCN2 inhibitory compounds. Although comparable results were obtained with the use of these compounds, we cannot completely deny the possibility of off-target effects. For example, the concentrations of GCN2iA that do not affect eIF2α phosphorylation are sufficient to reduce ATF4 expression (Fig. 1B), whereas such effects were not observed in the case of GCN2iB (SI Appendix, Fig. S8A). However, both compounds suppressed ASNase-mediated activation of the GCN2 pathway and showed comparable antiproliferative activity when combined with ASNase in multiple cell lines tested. Consistently, the GCN2 inhibitors phenocopied the effect of GCN2 KO in the proliferation of MEF cells. In addition, treatment of GCN2iA as a single agent did not have much impact on cell viability (in various cell lines) and on global gene expression (in CCRF-CEM). These results suggest that the potential off-target effect of GCN2iA may not hamper our conclusion. To the best of our knowledge, the compound described here is the first small-molecule inhibitor of GCN2 with high potency and kinase selectivity and a good pharmacokinetic profile to be reported. The compound will be an important tool for investigating the function of GCN2 in vitro and in vivo.
Despite its long history as a critical component of ALL treatment, new formulations of ASNase are still being developed to improve clinical outcomes. An E. coli-derived ASNase encapsulated in red blood cells was shown to have a high response rate and an ability to significantly reduce the incidence of allergic reactions and other toxic effects such as coagulopathy. ERY-ASP has also been tested in clinical trials for patients with AML or pancreatic cancer (24) (ClinicalTrials.gov identifier NCT01810705). These trials are supported by previous findings that indicated higher sensitivity of these tumors to ASNase treatment (23, 25). In our cell-panel study, ALL, AML, and pancreatic cancer cells were particularly sensitive to combined treatment with ASNase and GCN2 inhibitors. One caveat for this interpretation is that the panel includes a larger number of these cell lines than other cell lines; therefore, it remains possible that other types of cancer cells show sensitivity to the combined treatment. In the in vivo setting, our data demonstrated robust antitumor activities of combined treatment with ASNase and GCN2 inhibitors in ALL, AML, and pancreatic cancer cells compared with the results of single-agent ASNase or GCN2 inhibitor treatment. Thus, GCN2 inhibitors may represent ideal sensitizing agents to ASNase used for treating these tumors. Furthermore, considering the function of GCN2 as a regulator of AAR, further testing of whether GCN2 inhibitors synergize with other agents targeting amino acid metabolism is warranted.
In addition to ASNS, other proteins involved in amino acid metabolism (ADSL, ASS1, and DARS), amino acid transport (SLC1A4 and SLC38A2), apoptosis (BCL2L13), folate metabolism (DHFR and MTHFD1/2), and transcription (ATF5), as well as ribosomal proteins (RPL3–6 and RPL11), have been implicated in ASNase sensitivity (18, 30–36). Microarray analysis revealed that ASNase treatment induced changes in the expression of some of these genes in a GCN2-dependent manner (ASS1, SLC1A4, SLC38A2, MTHFD1/2, and ATF5; SI Appendix, Fig. S10). Therefore, the possibility that these factors might affect ASNase sensitivity synergistically, with each other or with ASNS, cannot be ignored. Notably, mRNA levels of the ribosomal proteins (RPL4–6 or RPL11) decreased significantly following ASNase treatment with GCN2 inhibition (SI Appendix, Fig. S10), supporting the idea that the combined treatment may attenuate the protein translation machinery.
Phosphorylation of eIF2α disrupts the formation of a ternary complex comprising Met-tRNAi and GTP-bound eIF2α, resulting in the disassembly of the 43S preinitiation complex and the suppression of protein translation initiation (1). Analysis of GCN2-KO cells has revealed GCN2/eIF2α-dependent inhibition of translation under UV radiation or essential amino acid starvation (37, 38). Our results demonstrated that GCN2 inhibition prevented eIF2α phosphorylation after ASNase treatment but did not elicit the recovery of protein synthesis. In addition, GCN2 inhibition reversed S6K phosphorylation in the presence of ASNase, which indicates recovered mTORC1 activity. We speculate that the suppression of ASNS expression induced by GCN2 inhibition caused a decrease in protein translation through the attenuation of asparagine synthesis, even at reduced levels of eIF2α phosphorylation and recovered levels of S6K phosphorylation. More detailed analysis (e.g., via ribosome profiling) is required to improve our understanding of how GCN2 inhibition affects protein translation machinery.
GC-MS analysis revealed that cellular amino acid levels (excluding those of asparagine) were up-regulated by ASNase treatment in CCRF-CEM cells and that GCN2 inhibition partially reversed the up-regulation. This is not surprising, as the expression of various enzymes involved in amino acid metabolism is controlled by the GCN2/ATF4 pathway. Considering that ASNase suppresses protein synthesis, the up-regulation of cellular amino acid levels may also be influenced by reduced consumption of amino acids in de novo protein synthesis.
JNK and p38 have important roles in the signaling cascades that orchestrate cellular responses to various types of stresses (39). In response to amino acid limitation, JNK modulates the transcription of amino acid-regulated genes by phosphorylation of transcription factors such as ATF2 and c-Jun (40). The formation of homo- and/or heterodimers between the ATF and JUN families of proteins forms an integrated transcription factor network that determines the initiation, magnitude, and duration of the cellular response to amino acid limitation (41). Activation of JNK and p38 may also impact apoptosis induction in response to cellular stresses. Meanwhile, the MEK/ERK pathway is essential for GCN2 pathway activation following amino acid limitation in hepatocellular carcinoma cells (42). Thus, the GCN2/ATF4 pathway is considered to regulate AAR in cooperation with these mitogen/stress-activated MAPK pathways. Our results indicate that GCN2 inhibition combined with asparagine depletion (i.e., ASNase treatment) aberrantly activates the MAPK pathways at the kinase signaling level and leads to cell death resulting from the mixed signal for apoptosis and cell survival (Fig. 4 and SI Appendix, Fig. S10). The balance between the signaling pathways triggered by AAR may be critical for maintaining cellular homeostasis under amino acid-limited condition.
Gene expression is controlled at the translation level by upstream ORFs (uORFs). Given that ASNS, similar to ATF4, contains 5′-uORFs in its transcript (43), ASNS expression may be regulated not only at the transcription level, but also at the translation level. Here, we observed no correlation between the mRNA and protein levels of ASNS in pancreatic cancer cells, suggesting that these cells possess specific mechanisms for ASNS regulation. Although ASNS expression has been reported to be lower in pancreatic cancer tissues than in normal tissues (24, 25), the mechanisms underlying this difference, as well as its implications, remain unknown. Further investigation should allow us to better understand the regulatory mechanisms underlying ASNS expression and its implications for pancreatic cancer.
Our xenograft studies demonstrated that combined treatment with ASNase and GCN2 inhibitors exerted synergistic antitumor activity without causing severe body weight loss. Previous studies have shown that GCN2-KO mice are viable, fertile, and display no gross phenotypic abnormalities unless fed on diets lacking an essential amino acid (44). Additionally, GCN2-KO mice are more sensitive to ASNase treatment, which promotes liver and/or pancreas dysfunction (45, 46). In our mouse xenograft study, blood test and histopathological examination revealed that ASNase-related toxicity in the liver and pancreas of mice was not exacerbated by combining ASNase treatment with a GCN2 inhibitor. It is possible that the difference in ASNase dosage affected these results, as previous studies had used 3,000 U/kg ASNase (45, 46), whereas we used 1,000 U/kg ASNase. In our analysis, 3,000 U/kg ASNase, but not 1,000 U/kg ASNase, caused a depletion of plasma glutamine levels after repeated administration. Given that such substantial glutamine depletion after ASNase administration has not been observed in rationally designed clinical studies (47, 48), 1,000 U/kg ASNase appeared to be an appropriate dose for use, at least in our model.
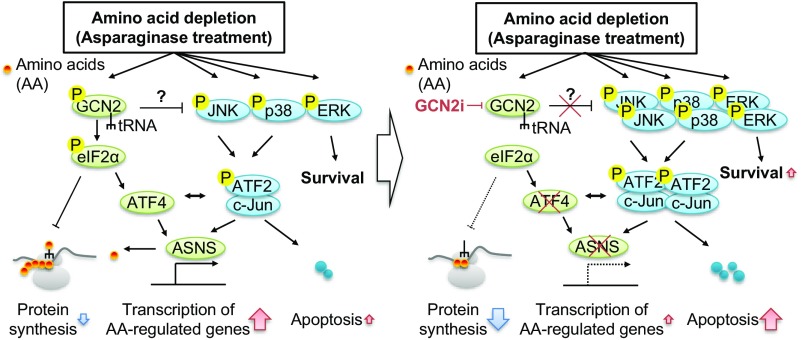
In summary, our preclinical findings demonstrate that GCN2 inhibition enhances the sensitivity to ASNase treatment by preventing ASNS induction in cancer cells with low ASNS expression at basal levels. Mechanistically, the inhibition of GCN2 activity in the presence of ASNase led to sustained suppression of protein translation and induction of apoptosis via stress-activated MAPK signaling (Fig. 8). Our findings offer valuable insights into the importance of GCN2 in determining ASNase sensitivity in tumors for which ASNase treatment has been clinically used or is under evaluation through clinical trials. Future study using more clinically relevant models (e.g., patient-derived xenograft models) should demonstrate the clinical translatability of our findings.
Fig. 8.
Schematic model of GCN2 inhibition-mediated disruption in AAR. (Left) Activation of GCN2 and MAPKs in response to amino acid limitation (i.e., ASNase treatment). (Right) Disruption of AAR by GCN2 inhibition. Detailed information is provided in the Discussion.
Materials and Methods
Detailed protocols regarding compound synthesis, kinase assay, kinase panel, transfection, Western blot, amino acid measurement, microarray, cell viability assay, caspase 3/7 assay, animal study, and statistical analysis, as well as information regarding siRNA, antibody, and cell lines, is provided in SI Appendix.
GCN2 inhibitors and a PERK inhibitor were synthesized at Takeda Pharmaceutical Company Limited (49). ASNase, asparagine, glutamine, and puromycin were purchased from Sigma-Aldrich. SB203580, SP600125, and PD0325901 were purchased from Selleck Chemicals. For in vivo use of ASNase, Leunase was purchased from Kyowa Hakko Kirin.
Supplementary Material
Acknowledgments
We thank Akihiro Ohashi, Kazuhide Nakamura, and Hiroshi Miyake for their helpful discussions; Yusa Tadashi, Satoshi Okanishi, Yuki Kurahashi, Yoji Sagiya, Masahiro Yaguchi, Kenichi Iwai, Ryo Dairiki, and Hitoshi Miyashita for providing technical support for the pharmacological experiments and/or bioinformatics; Tsuyoshi Ishii for performing enzymatic analyses; Junpei Takeda and Katsuhiko Yamamoto for performing analyses of physical properties; Yasuyuki Debori and Maki Miyamoto for performing pharmacokinetic analyses; and Yuuichi Takai for performing toxicological evaluations.
Footnotes
Conflict of interest statement: The authors are employees of Takeda Pharmaceutical Company Limited.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805523115/-/DCSupplemental.
References
- 1.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 2.Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 3.Siu F, Bain PJ, LeBlanc-Chaffin R, Chen H, Kilberg MS. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J Biol Chem. 2002;277:24120–24127. doi: 10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- 4.Wek RC, Ramirez M, Jackson BM, Hinnebusch AG. Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol Cell Biol. 1990;10:2820–2831. doi: 10.1128/mcb.10.6.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15:4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye J, et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lukey MJ, Katt WP, Cerione RA. Targeting amino acid metabolism for cancer therapy. Drug Discov Today. 2017;22:796–804. doi: 10.1016/j.drudis.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ali U, et al. L-asparaginase as a critical component to combat acute lymphoblastic leukaemia (ALL): A novel approach to target ALL. Eur J Pharmacol. 2016;771:199–210. doi: 10.1016/j.ejphar.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 9.Oettgen HF, et al. Inhibition of leukemias in man by L-asparaginase. Cancer Res. 1967;27:2619–2631. [PubMed] [Google Scholar]
- 10.Broome JD. Evidence that the L-asparaginase of guinea pig serum is responsible for its antilymphoma effects. II. Lymphoma 6C3HED cells cultured in a medium devoid of L-asparagine lose their susceptibility to the effects of guinea pig serum in vivo. J Exp Med. 1963;118:121–148. doi: 10.1084/jem.118.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broome JD. Evidence that the L-asparaginase of guinea pig serum is responsible for its antilymphoma effects. I. Properties of the L-asparaginase of guinea pig serum in relation to those of the antilymphoma substance. J Exp Med. 1963;118:99–120. doi: 10.1084/jem.118.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haskell CM, Canellos GP. l-asparaginase resistance in human leukemia–Asparagine synthetase. Biochem Pharmacol. 1969;18:2578–2580. doi: 10.1016/0006-2952(69)90375-x. [DOI] [PubMed] [Google Scholar]
- 13.Asselin BL, et al. Prognostic significance of early response to a single dose of asparaginase in childhood acute lymphoblastic leukemia. J Pediatr Hematol Oncol. 1999;21:6–12. doi: 10.1097/00043426-199901000-00003. [DOI] [PubMed] [Google Scholar]
- 14.Burke MJ. How to manage asparaginase hypersensitivity in acute lymphoblastic leukemia. Future Oncol. 2014;10:2615–2627. doi: 10.2217/fon.14.138. [DOI] [PubMed] [Google Scholar]
- 15.Kaspers GJ, et al. In vitro cellular drug resistance and prognosis in newly diagnosed childhood acute lymphoblastic leukemia. Blood. 1997;90:2723–2729. [PubMed] [Google Scholar]
- 16.Chen SH. Asparaginase therapy in pediatric acute lymphoblastic leukemia: A focus on the mode of drug resistance. Pediatr Neonatol. 2015;56:287–293. doi: 10.1016/j.pedneo.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Hutson RG, et al. Amino acid control of asparagine synthetase: Relation to asparaginase resistance in human leukemia cells. Am J Physiol. 1997;272:C1691–C1699. doi: 10.1152/ajpcell.1997.272.5.C1691. [DOI] [PubMed] [Google Scholar]
- 18.Fine BM, Kaspers GJ, Ho M, Loonen AH, Boxer LM. A genome-wide view of the in vitro response to l-asparaginase in acute lymphoblastic leukemia. Cancer Res. 2005;65:291–299. [PubMed] [Google Scholar]
- 19.Ubuka T, Meister A. Studies on the utilization of asparagine by mouse leukemia cells. J Natl Cancer Inst. 1971;46:291–298. [PubMed] [Google Scholar]
- 20.Zhang J, et al. Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion. Mol Cell. 2014;56:205–218. doi: 10.1016/j.molcel.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krall AS, Xu S, Graeber TG, Braas D, Christofk HR. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat Commun. 2016;7:11457. doi: 10.1038/ncomms11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye J, et al. GCN2 sustains mTORC1 suppression upon amino acid deprivation by inducing Sestrin2. Genes Dev. 2015;29:2331–2336. doi: 10.1101/gad.269324.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emadi A, Zokaee H, Sausville EA. Asparaginase in the treatment of non-ALL hematologic malignancies. Cancer Chemother Pharmacol. 2014;73:875–883. doi: 10.1007/s00280-014-2402-3. [DOI] [PubMed] [Google Scholar]
- 24.Bachet JB, et al. Asparagine synthetase expression and phase I study with L-asparaginase encapsulated in red blood cells in patients with pancreatic adenocarcinoma. Pancreas. 2015;44:1141–1147. doi: 10.1097/MPA.0000000000000394. [DOI] [PubMed] [Google Scholar]
- 25.Dufour E, et al. Pancreatic tumor sensitivity to plasma L-asparagine starvation. Pancreas. 2012;41:940–948. doi: 10.1097/MPA.0b013e318247d903. [DOI] [PubMed] [Google Scholar]
- 26.Knott SRV, et al. Asparagine bioavailability governs metastasis in a model of breast cancer. Nature. 2018;554:378–381. doi: 10.1038/nature25465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutierrez JA, et al. An inhibitor of human asparagine synthetase suppresses proliferation of an L-asparaginase-resistant leukemia cell line. Chem Biol. 2006;13:1339–1347. doi: 10.1016/j.chembiol.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikeuchi H, et al. A sulfoximine-based inhibitor of human asparagine synthetase kills L-asparaginase-resistant leukemia cells. Bioorg Med Chem. 2012;20:5915–5927. doi: 10.1016/j.bmc.2012.07.047. [DOI] [PubMed] [Google Scholar]
- 29.Brazeau JF, Rosse G. Triazolo[4,5-d]pyrimidine derivatives as inhibitors of GCN2. ACS Med Chem Lett. 2014;5:282–283. doi: 10.1021/ml500052f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aslanian AM, Kilberg MS. Multiple adaptive mechanisms affect asparagine synthetase substrate availability in asparaginase-resistant MOLT-4 human leukaemia cells. Biochem J. 2001;358:59–67. doi: 10.1042/0264-6021:3580059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen SH, et al. A genome-wide approach identifies that the aspartate metabolism pathway contributes to asparaginase sensitivity. Leukemia. 2011;25:66–74. doi: 10.1038/leu.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Estes DA, Lovato DM, Khawaja HM, Winter SS, Larson RS. Genetic alterations determine chemotherapy resistance in childhood T-ALL: Modelling in stage-specific cell lines and correlation with diagnostic patient samples. Br J Haematol. 2007;139:20–30. doi: 10.1111/j.1365-2141.2007.06763.x. [DOI] [PubMed] [Google Scholar]
- 33.Holleman A, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351:533–542. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 34.Holleman A, et al. The expression of 70 apoptosis genes in relation to lineage, genetic subtype, cellular drug resistance, and outcome in childhood acute lymphoblastic leukemia. Blood. 2006;107:769–776. doi: 10.1182/blood-2005-07-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rousseau J, et al. ATF5 polymorphisms influence ATF function and response to treatment in children with childhood acute lymphoblastic leukemia. Blood. 2011;118:5883–5890. doi: 10.1182/blood-2011-05-355560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scherf U, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 37.Deng J, et al. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr Biol. 2002;12:1279–1286. doi: 10.1016/s0960-9822(02)01037-0. [DOI] [PubMed] [Google Scholar]
- 38.Harding HP, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 39.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 40.Chaveroux C, et al. Identification of a novel amino acid response pathway triggering ATF2 phosphorylation in mammals. Mol Cell Biol. 2009;29:6515–6526. doi: 10.1128/MCB.00489-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kilberg MS, Balasubramanian M, Fu L, Shan J. The transcription factor network associated with the amino acid response in mammalian cells. Adv Nutr. 2012;3:295–306. doi: 10.3945/an.112.001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thiaville MM, et al. MEK signaling is required for phosphorylation of eIF2alpha following amino acid limitation of HepG2 human hepatoma cells. J Biol Chem. 2008;283:10848–10857. doi: 10.1074/jbc.M708320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 44.Anthony TG, et al. Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J Biol Chem. 2004;279:36553–36561. doi: 10.1074/jbc.M404559200. [DOI] [PubMed] [Google Scholar]
- 45.Wilson GJ, Bunpo P, Cundiff JK, Wek RC, Anthony TG. The eukaryotic initiation factor 2 kinase GCN2 protects against hepatotoxicity during asparaginase treatment. Am J Physiol Endocrinol Metab. 2013;305:E1124–E1133. doi: 10.1152/ajpendo.00080.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phillipson-Weiner L, et al. General control nonderepressible 2 deletion predisposes to asparaginase-associated pancreatitis in mice. Am J Physiol Gastrointest Liver Physiol. 2016;310:G1061–G1070. doi: 10.1152/ajpgi.00052.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pieters R, et al. Pharmacokinetics, pharmacodynamics, efficacy, and safety of a new recombinant asparaginase preparation in children with previously untreated acute lymphoblastic leukemia: A randomized phase 2 clinical trial. Blood. 2008;112:4832–4838. doi: 10.1182/blood-2008-04-149443. [DOI] [PubMed] [Google Scholar]
- 48.Tong WH, et al. A prospective study on drug monitoring of PEGasparaginase and Erwinia asparaginase and asparaginase antibodies in pediatric acute lymphoblastic leukemia. Blood. 2014;123:2026–2033. doi: 10.1182/blood-2013-10-534347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atkins C, et al. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res. 2013;73:1993–2002. doi: 10.1158/0008-5472.CAN-12-3109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.