Significance
Light–oxygen–voltage (LOV) domain photoreceptors are found ubiquitously in nature and possess highly diverse signaling roles and mechanisms. Here, we show that a class of fungal LOV proteins dynamically associates with anionic plasma membrane phospholipids by a blue light-switched electrostatic interaction. This reversible association is rapidly triggered by blue light and ceases within seconds when illumination ceases. Within the native host, we predict that these proteins regulate G-protein signaling by the controlled recruitment of fused regulator of G-protein signaling (RGS) domains; in applied contexts, we anticipate that engineered chimeric versions of such proteins will be useful for rapid optogenetic membrane localization of fused proteins through direct interaction with the membrane itself, without requiring additional components to direct subcellular localization.
Keywords: light–oxygen–voltage domain, LOV, photobiology, optogenetics
Abstract
We report natural light–oxygen–voltage (LOV) photoreceptors with a blue light-switched, high-affinity (KD ∼ 10−7 M), and direct electrostatic interaction with anionic phospholipids. Membrane localization of one such photoreceptor, BcLOV4 from Botrytis cinerea, is directly coupled to its flavin photocycle, and is mediated by a polybasic amphipathic helix in the linker region between the LOV sensor and its C-terminal domain of unknown function (DUF), as revealed through a combination of bioinformatics, computational protein modeling, structure–function studies, and optogenetic assays in yeast and mammalian cell line expression systems. In model systems, BcLOV4 rapidly translocates from the cytosol to plasma membrane (∼1 second). The reversible electrostatic interaction is nonselective among anionic phospholipids, exhibiting binding strengths dependent on the total anionic content of the membrane without preference for a specific headgroup. The in vitro and cellular responses were also observed with a BcLOV4 homolog and thus are likely to be general across the dikarya LOV class, whose members are associated with regulator of G-protein signaling (RGS) domains. Natural photoreceptors are not previously known to directly associate with membrane phospholipids in a light-dependent manner, and thus this work establishes both a photosensory signal transmission mode and a single-component optogenetic tool with rapid membrane localization kinetics that approaches the diffusion limit.
Photoreceptors are complex protein machines that transduce optical sensory inputs into diverse outputs in signaling and energy conversion. Establishing the structure–function relationships and signaling modes of novel photosensory proteins enhances understanding of organismal adaptation to a universal cue of light. Among the known photoreceptors, light–oxygen–voltage (LOV) proteins (1–3) comprise the most ubiquitous and topologically diverse class. LOV photosensory signaling, which relies on a flavin chromophore bound within a Per–Arnt–Sim (PAS)-type sensory domain, is initiated by blue light-induced formation of a reversible covalent cysteinyl-flavin photoadduct. The subsequent structural changes in the surrounding protein alter the activity of effector domains and peptides fused up/downstream to the sensor domain itself, controlling biological function (1–6).
One way the diversity of LOV-regulated biochemical activities has been revealed is through the use of large-scale genomic analyses, including a recent study of ours that cataloged over 6,700 LOV domains with over 100 combinatorial sensor–effector arrangements (3). This topological flexibility confers great functional diversity in the ways signals are transmitted as well as the kinds of biological responses that are controlled by these photoreceptors (2–5, 7). Beyond their fundamental importance in photobiology, novel LOV proteins provide valuable components and molecular engineering principles for creating optogenetic tools to perturb the physiology of targeted cells. For example, bacterial LOV proteins have been adapted for light-regulated transcriptional activation (8) and adenylate cyclase activity (9) in eukaryotes. Likewise, truncated LOV sensor domains with established signaling structure–function offer numerous approaches to engineering chimeric proteins to confer optically inducible functions to a wide range of fusion partners (10–13).
Although the breadth of LOV function can be inferred from bioinformatics, reported protein-level biochemical characterization and structure–function analyses of novel-effector LOVs are still rare. As part of a broad survey of LOV proteins (3), we previously identified one such new class of dikarya fungal LOVs associated with regulator of G-protein signaling (RGS) domains (Fig. 1 A and B and SI Appendix, Fig. S1). RGS proteins are the primary fast terminators of G-protein–coupled receptor (GPCR) signaling, serving as GTPase accelerating proteins (GAPs) on activated Gα-protein partners (14–16). While other bioinformatics and cell biology studies have predicted the presence of RGS-LOV proteins (17–21), direct experimental evidence of photosensory function has not been shown. Their genetic deletion does not beget any pronounced phenotypic change (17), and their transcriptional levels are not under light-dependent transcriptional regulation (22). Thus, protein-level photochemical characterization is required to confirm bona fide photosensory activity for this class of LOV proteins.
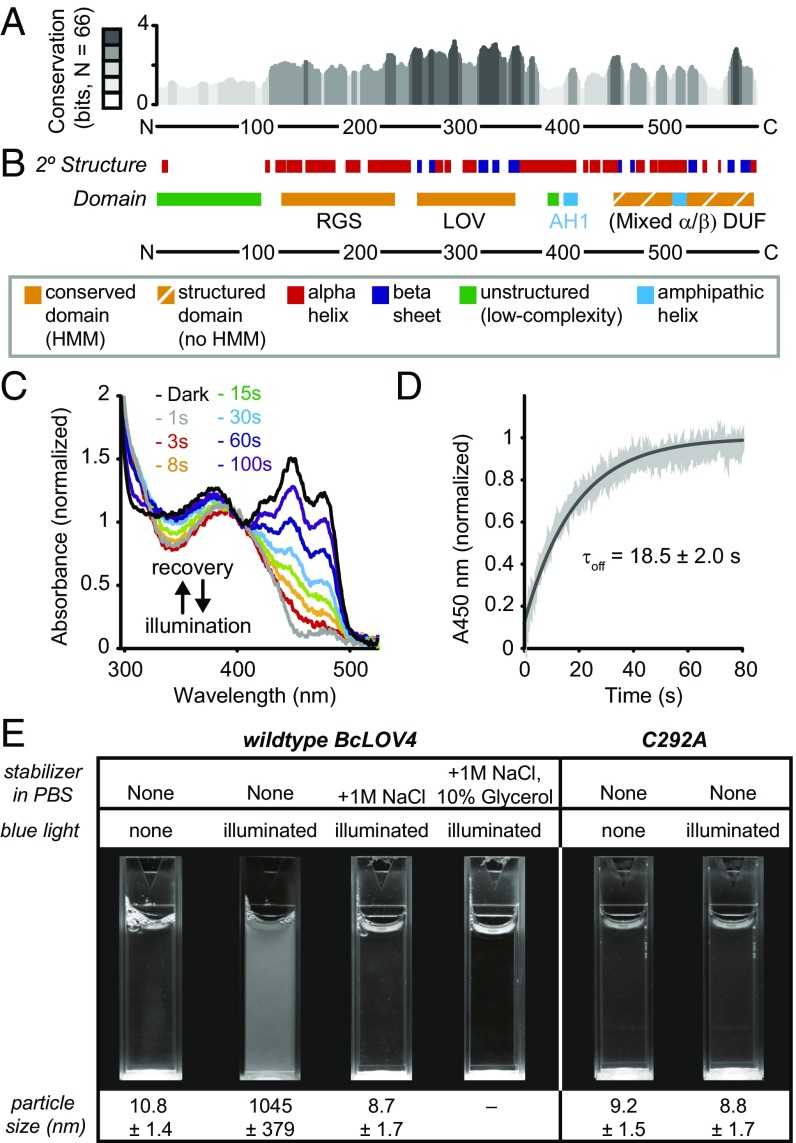
Fig. 1.
Bioinformatics annotation and photochemical competence of BcLOV4. (A) Secondary-structure conservation across 66 candidate RGS-LOV-DUF proteins, where height represents information content at a given position, in bits. Grayscale, bit score in fifths. (B) Consensus secondary-structure prediction and domain architecture of BcLOV4, from JPred, phyre2, PSIPRED, and i-TASSER (secondary structures), IUPRED (disorder), Heliquest (amphipathic helices), and Pfam hidden Markov models database (domains, HMM = match in database). (C) Representative flavin photocycling of BcLOV4 stabilized by 1 M NaCl and 10% glycerol to prevent photoinduced aggregation, measured by absorbance spectroscopy. Illumination, 15 mW/cm2; λ = 455 nm. Time indicates postillumination recovery period. (D) Recovery kinetics monitored at λ = 450-nm absorbance (A450). Black, exponential fit. Gray, mean ± SD (n = 3). (E) In vitro aggregration of BcLOV4 in direct response to blue light. The C292A mutant is unable to form a covalent cysteinyl-flavin photoadduct and is thus photochemically inactive. Illuminated samples become turbid but can be stabilized by high-salinity and/or molecular crowding agents. Illumination, 15 mW/cm2; λ = 455 nm. Particle size by DLS (mean ± SD).
Here, we report that these cytosolic LOV proteins dynamically and reversibly associate with the plasma membrane by directly light-regulated and high-affinity binding to anionic phospholipids, as revealed through a combination of bioinformatics, computational protein modeling in Rosetta, in vitro structure–function studies with purified recombinantly expressed protein, and optogenetic assays in multiple eukaryotic heterologous expression systems. The photosensory phenomenon was found to be directly coupled to flavin photocycling and is likely general across RGS-LOVs. This study establishes a significant signaling mechanism relevant to natural photoreceptors, and broadly applicable to single-component optogenetic tools for dynamic membrane localization.
Results
Domain Topology and in Vitro Photophysical Characterization.
Conserved domain analyses by us and others (3, 17–20) report a consensus RGS-LOV architecture with a low-complexity region and RGS domain located N-terminal to a single LOV domain. Secondary-structure predictions and structural modeling here indicate that there is an additional C-terminal domain of unidentified function (DUF) with mixed α-helix/β-sheet content as well (Fig. 1 A and B). The LOV and DUF domains are connected via a predicted LOV Jα-helix linker, which extends into a polybasic amphipathic helix (AH) and is known to mediate LOV signaling elsewhere (4–6). RGS-LOV candidates from five organisms were chosen for either their previously hypothesized functional roles by others (17, 18) or their short length, and were assessed for solubility as full-length proteins in bacterial expression systems (SI Appendix, Fig. S1). One of these, BcLOV4 (also named BcRGS1) from the noble rot fungus Botrytis cinerea (18, 21) (GenBank accession number CCD53251.1), could be produced in good yield as a dark-adapted oligomer and was used for further analyses (SI Appendix, Fig. S2).
BcLOV4 had an optical absorbance peak at λmax = 450 nm with triplet-peak fine structure (Fig. 1C) indicative of a LOV-bound flavin mononucleotide (FMN) cofactor (see SI Appendix, Fig. S2, for cofactor isolation). BcLOV4 photocycled with rapid thermal reversion kinetics (τoff = 18.5 s) (Fig. 1D). However, stable photocycle measurements required in vitro stabilization by high salinity alone (0.5–1 M NaCl) or in combination with glycerol (10%), or immobilization on solid-phase supports (see SI Appendix, Fig. S3, for photocycling summary). In the absence of such stabilization, illuminated BcLOV4 quickly aggregated into turbid solutions of micrometer-scale colloids as measured by dynamic light scattering (DLS) (Fig. 1E). This in vitro photoaggregration was preventable with high stabilization (e.g., 1 M NaCl), reversible with intermediate stabilization (e.g., 0.5 M NaCl), and irreversible in normal-salinity PBS, eventually precipitating from solution (SI Appendix, Fig. S4). This phenomenon was dependent on flavin photocycling, since it was abolished for a photochemically inactive C292A BcLOV4 mutant. This mutant is still a holoprotein, but the C292A mutation prevents the formation of the critical cysteinyl-flavin photoadduct (23) that initiates canonical LOV signaling, such that it mimics a permanently dark-adapted protein even in the presence of blue light.
Rapid Membrane Localization in Cells in Response to Blue Light.
Light-activated aggregation has not previously been reported for LOV proteins, although oligomerization into photobodies is known among natural phytochromes, cryptochromes, and their engineered optogenetic variants (24, 25). Thus, to functionally probe whether BcLOV4 forms photobodies in cells, BcLOV4 was visualized by fluorescence microscopy when heterologously expressed in mammalian cells (Fig. 2), which we used because B. cinerea is pathogenic and less genetically tractable than HEK cells. Cells expressing 3×-FLAG-tagged BcLOV4 were fixed in blue light or the dark, and then stained with fluorescent dye-labeled anti-FLAG monoclonal antibody. To our surprise, BcLOV4 did not primarily form photobodies in cells but instead localized to the plasma membrane in a blue light-dependent manner (Fig. 2A). Both the dynamic membrane localization in cells and in vitro photoaggregation were also observed with the homolog from the black yeast Cyphellophora europaea (hereon called CeRGS) (GenBank accession number ETN36999.1) (SI Appendix, Fig. S5). Thus, importantly, the in vitro and cellular phenomena are likely general to the class of RGS-LOV proteins. Due to low purified recombinant protein yield and heterologous expression levels of CeRGS in cellular assays, data reported hereafter focus on BcLOV4.
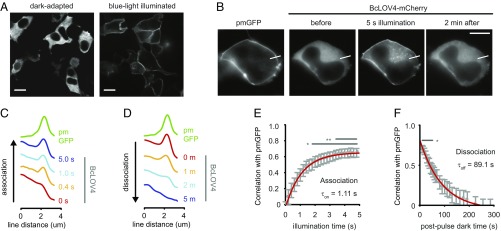
Fig. 2.
Light-activated membrane localization in HEK cells. (A) Spinning-disk confocal fluorescence micrographs of BcLOV4 show it is cytosolic in the dark and translocates to the plasma membrane in blue light. Cells were fixed with paraformaldehyde in the dark or under blue light, and stained with Alexa 488-conjugated anti-3×FLAG antibody. (Scale bar: 10 μm.) (B) Example single cell for quantitative membrane localization analysis. pm-GFP, isoprenylated GFP marker. Line section for C. (Scale bar: 10 μm.) (C) Line section profiles of pm-GFP and BcLOV4-mCherry from cell in B. Association, during 5-s illumination. (D) Same as C for dissociation (dissociation, dark after 5-s illumination). (E) Population analysis of translocation kinetics. Time constants were statistically determined by correlation analysis between the membrane marker and BcLOV line section profiles, for similarity (τon = 1.11 s; 95% CI, 1.05–1.18 s). n = 30 cells, *P < 0.05, **P < 0.01. (F) Same as E, for membrane dissociation. Time constants were also statistically determined by correlation analysis, for dissimilarity (τoff = 89.1 s; 95% CI, 83.0–96.3 s). (B–F) Blue light pulses: λ = 440/20 nm, 5 s, and 15 mW/cm2. GFP imaging (λex = 470/24 nm; λem = 525/50 nm). mCherry imaging (λex = 550/15 nm; λem = 630/75 nm).
To determine the dynamics of this translocation process, mCherry-tagged BcLOV4 variants were directly visualized, using cotransfected isoprenylated GFP as a plasma membrane marker in HEK cells. The functional kinetics of membrane association and undocking measured by live-cell imaging were fast (HEK τon = 1.11 s, τoff = 89.1 s) (Fig. 2 B–F). The membrane association was on the timescale of diffusion to the inner leaflet (Fig. 2 C and E) (∼0.7–1.6 s; see Materials and Methods for timescale estimate determination), and such kinetics is indicative of a high-affinity interaction between BcLOV4 and its membrane target. We hypothesized that such a light-switched interaction could occur directly between BcLOV4 and membrane lipids, because the photosensory signal-transmitting Jα-helix linker is fused to a polybasic amphipathic helix (AH1 in Fig. 1A, from residues 403–416) similar to those involved in membrane association in other systems (26–28).
Directly Light-Regulated and High-Affinity Interaction with Anionic Phospholipids.
Initial protein–lipid overlay screening assays suggested that BcLOV4 bound anionic lipids but not zwitterionic ones, but this assay tests for headgroup interactions without recapitulating a membrane interface. Thus, to further test for a direct protein–lipid interaction with a more realistic membrane target (Fig. 3), we created droplets of water-in-oil (w/o) emulsions (29) containing purified recombinant BcLOV4-mCherry in the dispersed/aqueous phase, and phospholipid monolayers at the droplet interface to emulate the plasma membrane inner leaflet (Fig. 3A). Artificial membranes were composed of the zwitterionic phosphatidylcholine (PC) mixed with anionic phospholipids of varying concentration and headgroup charge density.
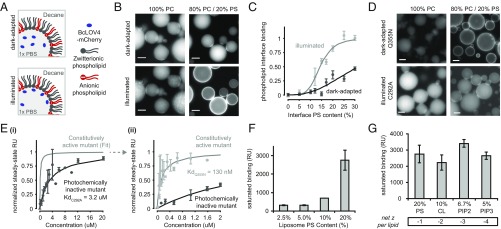
Fig. 3.
In vitro binding to anionic membrane lipids. (A) Schematic of BcLOV4 in lipid-stabilized w/o emulsions. (B) Fluorescence micrographs of wild-type BcLOV4 fused to mCherry. Translocation to the inner leaflet-like interface is observed with increasing anionic PS content, but not with purely zwitterionic PC interfaces. (C) Phospholipid interface binding curves, calculated as the membrane interface:dispersed phase ratio (normalized) of BcLOV4 in the light and dark. n = 20–75 droplets; error, SEM. (D) Constitutively active BcLOV4 Q355N structurally mimics the photoactivated signaling state, is localized to the interface in the dark, and retains its preference for net anionic phospholipids over zwitterionic ones. The photochemically inactive C292A mutant cannot form a covalent cysteinyl-flavin photoadduct and remains in the aqueous dispersed phase even upon illumination. (B–D) Blue light pulses: λ = 440/20 nm, 5 s, 15 mW/cm2. mCherry imaging: λex = 550/15 nm, λem = 630/75 nm. (Scale bar: 25 μm.) (E) Affinity measures by SPR to 80% PC/20% PS mixed liposomal bilayers. The interaction with constitutively active BcLOV4 is high affinity (KdQ355N = 130 ± 75 nM) and >20-fold enhanced over the photochemically inactive mutant (KdC292A = 3.2 ± 1.2 μM). (i) The 0–20 μM range, with fit only for constitutively active mutant for clarity, and (ii) 0–2 μM range. n = 2–7; error, SEM. (F) SPR measures of constitutively active mutant binding to mixed PC/PS liposomes of varying total anionic charge density. n = 3; error, SD. (G) SPR binding assessments of constitutively active mutant to lipids of different headgroup charge density, in liposomes of matching total anionic charge density of 20% (n = 3; error, SD). (F and G) CL, cardiolipin; PC, phosphatidylcholine; PIP2, phosphatidylinositol-(4,5)-biphosphate; PIP3, phosphatidylinositol-(3,4,5)-triphosphate; PS, phosphatidylserine.
These droplet assays allowed for complete control over illumination conditions and membrane compositions without complications introduced by the presence of other proteins. The facile customization and ability to multiplex on an automated fluorescence microscope made the system highly useful for screening and cross-validating other methods for establishing binding interactions, like surface plasmon resonance (SPR). As seen in 20% phosphatidylserine (PS)-containing emulsions of similar PS composition to mammalian membranes (Fig. 3 B and C), illuminated BcLOV4-mCherry primarily localized to the phospholipid interface, instead of aggregating as observed in lipid-free bulk solution experiments. Conversely, BcLOV4 formed colloids in lieu of binding pure zwitterionic PC interfaces with only positively charged headgroups. Both light-activated localization and aggregation within the aqueous compartment diminished as salinity increased (SI Appendix, Fig. S6), suggesting an electrostatic basis for these phenomena.
Localization was diminished in blue light for the photochemically inactive C292A mutant (23), confirming direct coupling of the phenomenon to signaling initiation by flavin photocycling as opposed to an unknown blue light interaction (Fig. 3D). Conversely, localization was persistent in the absence of illumination with a constitutively active Q355N mutant that structurally mimics the signal-transducing conformation of the LOV Jα-helix linker region as if it were in a permanently lit or active signaling state, even in the absence of illumination (30–32) (Fig. 3D). The constitutively active mutant also retained the BcLOV4 binding preference of light-activated wild-type BcLOV4 for net anionic lipids over purely zwitterionic PC interfaces. Thus, optical activation of the interaction with membrane phospholipid is consistent with known structure–function determinants of LOV signaling with respect to flavin photocycling and signal transmission via the Jα-helix.
Dissociation constants for BcLOV4 binding to immobilized liposomal bilayers were next measured by SPR. The measurements were made with the photochemically inactive C292A and constitutively active Q355N mutants, since controlled illumination within the instrument was not possible. BcLOV4-mCherry variants were used for SPR assays both to maintain consistency with droplet assays, and for improved solubility and protein yield. The BcLOV4 constitutively active mutant affinity for 20% PS liposomal bilayers was KdQ355N = 130 nM, or >20-fold enhanced vs. the photochemically inactive mutant KdC292A = 3.2 μM (Fig. 3E). Thus, consistent with biophysical inferences from cellular kinetics, BcLOV4 indeed possesses a high-affinity light-switched interaction with anionic phospholipids. Binding increased with total anionic content (with PS, Fig. 3F), but there were minimal differences between phospholipids of different headgroup charge density under conditions of matching total charge (Fig. 3G). Thus, BcLOV4 membrane binding is charge dependent but nonspecific to headgroup identity, unlike the well-established preference of pleckstrin homology (PH) domains for certain phosphatidylinositol phosphates (PIPs) (33), or similar lactadherin-C2 domain-specificity for PS (34).
Key Structure–Function of the Dynamic Protein–Lipid Interaction.
Having identified the light-switched interaction partner as an anionic phospholipid, we next sought to determine the protein binding site (Fig. 4) and focused on the polybasic amphipathic helix in the linker region that is largely conserved among the fungal homologs (AH1) (Fig. 4A). In BcLOV4, this helix possesses a conserved “FKK” motif (residues 412–414) found in membrane-interacting amphipathic helices of Bcl-2–associated death promoter (BAD) (35), kinase suppressor of RAS (KSR) (36), and cecropin anti-microbial peptides (37), and a “FFK” sequence (residues 408–410) found at the membrane interface of the M2 proton channel of influenza A [Protein Data Bank ID code 2rlf]. In such motifs, aromatic side chains putatively insert into the phospholipid bilayer, while the proximal lysine side chains electrostatically bind anionic lipids enriched in the inner leaflet without great headgroup specificity (27) (Fig. 4B).
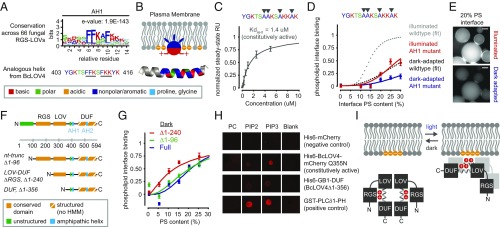
Fig. 4.
Structure–function determinants of the BcLOV4–lipid interaction. (A) Sequence logo of amphipathic helix (AH1) conserved among 66 RGS-LOV homologs (Top) and the specific sequence for BcLOV4, which includes known lipid binding motifs (underlined) (Bottom). (B) Schematized membrane insertion mechanism of the AH1, where hydrophobic residues embed into the hydrophobic bilayer and basic residues electrostatically bind anionic phospholipids. (C) SPR-determined affinity of AH1 mutant for 20% PS bilayers, with hydrophobic residues mutated to alanine, is reduced ∼10-fold from wild-type BcLOV4. SPR data are of constitutively active Q355N mutant (n = 2; error, SD). (D) Phospholipid interface binding curves, calculated as the membrane interface/dispersed phase ratio (normalized) of the AH1 mutant (no Q355N) when dark-adapted or illuminated with blue light in w/o emulsions, normalized to wild-type saturation level under illumination (n = 20–200 droplets; error, SEM). Dotted fits for perspective derived from Fig. 3. (E) Representative fluorescence micrographs showing that the AH1 mutant primarily remains in the aqueous dispersed phase upon illumination. (Scale bar: 25 μm.) (F) Truncations created to probe domain contributions to light-switched membrane association. HMM, existing hidden Markov model. (G) Phospholipid interface binding curves, calculated as the membrane interface/dispersed phase ratio (normalized) of BcLOV4, nt-truncated protein, and RGS-truncated LOV-DUF, in w/o emulsions and in the absence of illumination. Increased binding by deletion of the RGS suggests that the RGS domain inhibits the membrane interaction in the absence of illumination. Normalized to wild-type saturation level under illumination. n = 30–125 droplets; error, SEM. (H) Lipid-blot assays for DUF–lipid interaction (3-nmol lipid per spot). Visualized with IRDye680-conjugated anti-mouse IgG secondary antibody against primary mouse anti-His6 or anti-GST antibodies. The DUF bound negatively charged phosphoinositides, but not zwitterionic PC. (I) Schematized proposed signal transmission mode of BcLOV4, with the oligomer drawn as a dimer for clarity.
Thus, candidate phenylalanine and tyrosine residues within this region were mutated to alanines (Fig. 4 C–E). The BcLOV4-AH1 mutant (amphipathic helix mutant) photocycled similarly to wild-type protein (SI Appendix, Fig. S3). In SPR assays, the BcLOV4-AH1 constitutively active mutant showed a 10-fold reduction in affinity, KdAH1-Q355N = 1.4 μM, for 20% PS liposomal bilayers (Fig. 4C), providing evidence that light-induced exposure of the specific lipid-binding motifs drives membrane association. BcLOV4-AH1 also showed reduced binding to anionic phospholipids in droplets of w/o emulsions, and largely remained in the aqueous compartment/dispersed phase when illuminated with blue light (Fig. 4 D and E). The downward shift in the droplet-based phospholipid interface binding curves of the AH1 mutant from wild-type levels confirmed the direct lipid-binding roles of the aromatic side chains in the FKK and FFK motifs.
In vitro truncation analyses were performed to establish the relative contributions of the N-terminal and C-terminal domains in signal transmission from the LOV blue light sensor to the lipid-binding regions (Fig. 4 F–H). mCherry-fused RGS-truncated BcLOV4∆1–240 or “LOV-DUF,” aggregated in the absence of lipids in the dark (32 ± 43 nm by DLS) and exhibited an upward shift in the phospholipid interface binding curve vs. full-length (both as photochemically inactive mutants; Fig. 4G). Deletion of the unstructured N terminus alone, BcLOV4∆1–96, had no such effects. These data suggest that, first, the RGS domain serves an inhibitory role when dark-adapted and, second, that the LOV-DUF alone is sufficient for membrane association. While C-terminal truncations of BcLOV4 (i.e., ∆DUF) were insoluble, the isolated DUF region, which included the putative lipid-interacting AH1, was soluble as an in vitro refolded product (SI Appendix, Fig. S7). The DUF bound anionic phospholipids in protein–lipid overlay assays (Fig. 4H), further implicating the region C-terminal to the LOV sensor in lipid binding. In totality, these findings suggest that BcLOV4 signal transmission is mediated by light-induced structural rearrangements that expose a critical polybasic amphipathic helix at the LOV-DUF linker that is inhibited by the RGS domain in the dark (Fig. 4I).
Blue Light-Dependent Membrane Localization in Fungus.
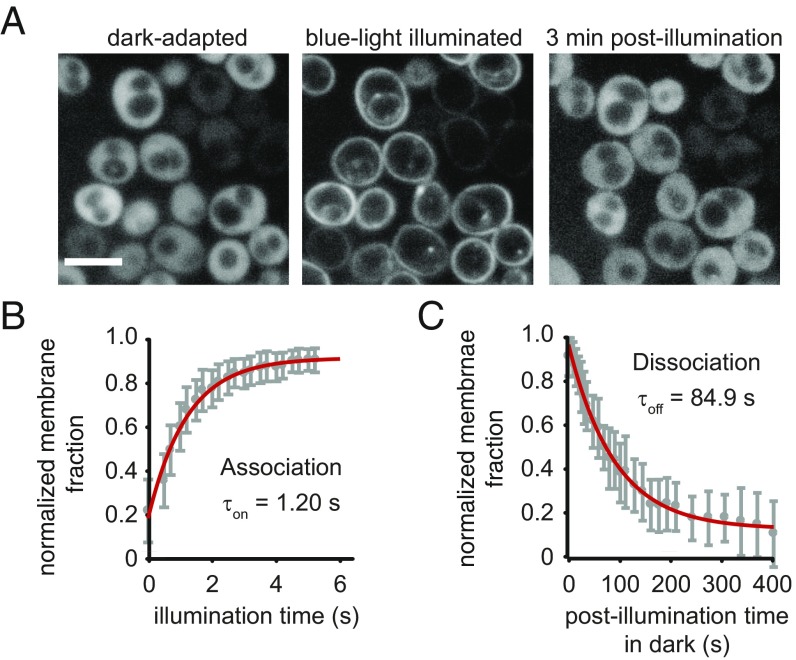
To determine whether the fungal-derived BcLOV4 associates with membranes in a blue light-inducible manner in fungus, dynamic localization assays were performed in Saccharomyces cerevisiae yeast (Fig. 5). Such confirmation of the photosensory response in fungal cells is important because they possess high cytoplasmic salinity and anionic membrane lipid content (38–40) that may influence the electrostatic interaction based on the binding studies here (Fig. 3F and SI Appendix, Fig. S6).
Fig. 5.
Light-regulated membrane localization in yeast (S. cerevisiae). (A) Spinning-disk confocal fluorescence micrographs of BcLOV4-mCherry in transformed yeast show reversible membrane localization in response to blue light. Cells were immobilized on agar pads immediately before imaging. (Scale bar: 5 μm.) (B) Population analysis of membrane association kinetics. Time constants determined from relative fraction of membrane-bound BcLOV4 from cellular line sections (mean ± SEM; n = 31 cells; τon = 1.20 s; 95% CI, 1.07–1.38 s). (C) Population analysis of membrane dissociation kinetics (τoff = 84.9 s; 95% CI, 83.3–93 s).
BcLOV4-mCherry in yeast indeed translocated from cytoplasm to plasma membrane (Fig. 5A) in response to blue light and in a dark-reversible manner in confocal microscopy analysis of agar-immobilized transformed cells. The measured kinetics in yeast (yeast τon = 1.20 s, τoff = 84.9 s) (Fig. 5 B and C) were similar to those measured in mammalian cells, although one should note that the similarity in dissociation timescales between the two eukaryotic expression systems here may be purely coincidental, given the electrostatic differences in cellular milieu. Membrane association kinetics on the timescale of intracellular diffusion to membrane (∼0.5–1.0 s; see Materials and Methods for determination of estimates) further confirmed the high-affinity membrane–lipid interaction. Thus, RGS-LOV photosensory signal transmission was consistent across all contexts studied, from pure in vitro systems to fungal and mammalian expression systems.
Discussion
Our cumulative findings suggest a photosensory signal transmission mode by RGS-LOV of rapidly blue light-inducible, and reversible, membrane association mediated by electrostatic interactions with anionic phospholipids. While other membrane-binding proteins contain PAS domain sensors (41, 42) related to LOV domains, such as PhoQ, Aer, and LuxQ (43, 44), these are ligand-regulated transmembrane proteins unlike the cytoplasmic photoreceptors reported here. We underscore that this signaling mode was not anticipated from hidden Markov model-based bioinformatics searches for conserved domains, which found nothing C-terminal to the LOV sensor. De novo secondary-structure and Rosetta (45) structural predictions suggest that the DUF in this region may adopt a PAS-like fold with antiparallel β-sheets (SI Appendix, Fig. S7D), and thus it is possible that the LOV–DUF interaction is an evolutionarily conserved PAS/PAS interaction as observed in other systems. It should be noted that other lipid interaction sites may exist beyond the critical amphipathic helix between the LOV and DUF domains. A future high-resolution structure of the lipid-bound state will greatly inform the proposed biophysical model, as well as conclusively determine whether the DUF is indeed a PAS domain.
Membrane localization is a known prerequisite for the canonical GAP activity of RGS proteins (46, 47). Therefore, it is plausible that the signal transmission mode proposed in Fig. 4I—and the photosensory response that was consistently observed across in vitro and cellular systems (including in yeast) and across different RGS-LOV proteins (BcLOV4 and CeRGS)—serves to regulate interactions with cognate Gα proteins at the membrane in-host. Little is known about the photobiological role of BcLOV4 (18), its interactions with the three Gα proteins of Botrytis (48), and the physiological roles of the latter. However, the isolated (or truncated) RGS domain of MoRGS5 (GenBank accession number EHA46884.1), the RGS-LOV of Magnaporthe oryzae (rice blast fungus), does interact in yeast two-hybrid assays with its cognate Gαi protein, MagB (17), which is involved with hydrophobic sensing and plant infectivity (49, 50). Genetic knockout of MoRGS5 (ΔMoRGS5) results in increased intracellular cAMP levels, further implicating a role for RGS-LOV proteins in cell signaling (17, 18). ΔMoRGS5 strains, however, exhibit no pronounced organism-level phenotypic difference from wild-type strains. Thus, while a conclusive photobiological role has yet to be established for RGS-LOV proteins (51), which of note have not been shown to be photochemically active to date, the findings here provide a potential biophysical mechanism by which they may affect fungal physiology: through light-regulated and reversible membrane association of a Gα-interacting photoreceptor to fine-tune Gα-dependent cAMP signaling (18).
In an applied context, BcLOV4 also contributes a useful single-component optogenetic system for photoinducible membrane localization that is compatible in yeast and mammalian expression systems. Its translocation kinetics was apparently limited by diffusion in a cellular context and thus approaches a practical limit for rapid optogenetic membrane localization. Unlike the indirect membrane binding of optogenetic tools that rely on heterodimerization between cytosolic and membrane-bound partners (52, 53), BcLOV4 as a single-component system is insensitive to heterogeneity in relative expression level tuning of two components, and is more facile in transgene delivery.
A common goal in optogenetics, and a common motivation for establishing fundamental structure–function that may beget new molecular engineering principles for creating better protein tools, is the identification of photoreceptor mutations that confer beneficial kinetic properties. For example, lengthening the photocycle of the sensor may extend the active signaling duration across the whole protein in some cases, thereby decreasing the stimulation fluence/duration required for sustained activity (54, 55). LOV sensor engineering to tune the photocycle, however, does not guarantee concomitant tuning of functional signaling outputs (56), such as membrane localization for BcLOV4. Screening mutations known to alter the photocycle in other LOV proteins (30, 55, 57–61) revealed that a BcLOV4 C258I mutation in the Aβ-sheet [analogous to I74 of Neurospora crassa VVD (21)] prolonged membrane residence in HEK cellular assays (HEK τoff = 622.7 s) (SI Appendix, Fig. S8), as well as the in vitro photocycle of the salt-stabilized mutant (τoff = 586.5 s). Similar to wild-type protein, membrane localization in this mutant persists longer than its photocycle duration. Given that the membrane undocking time of BcLOV4 is longer than its in vitro photocycle, thermal reversion of the photoexcited LOV may not disrupt all lipid interactions of the membrane-bound state across the whole multidomain protein, resulting in the overall longer time constant for undocking than thermal revision alone. This C258 residue is an interesting candidate for further structure–function studies on how flavin photochemistry couples to the signaling state in diverse LOV. More broadly, the direct readout of membrane localization assays makes RGS-LOV an interesting LOV class for such structure–function studies at large.
In summary, the myriad results presented here establish a photosensory signaling mode by RGS-LOV through a directly light-regulated, reversible, and high-affinity electrostatic interaction between anionic plasma membrane phospholipids and a polybasic amphipathic helix at the LOV interface with its C-terminal DUF. This work highlights the utility of convergent approaches that link bioinformatics, in vitro structure–function, and functional assays in live cells to define a mechanism by which photoreceptors dynamically regulate cellular physiology in response to sensory cues.
Materials and Methods
Genetic Constructs and Protein Expression.
Bacterial genetic constructs.
For protein expression, genes fragments encoding for BcLOV4 (GenBank accession number CCD53251.1), Cyphellophora europea LOV (ETN36999.1), Marsonnina brunnea LOV (EKD19672.1), Magnaporthe oryzae LOV (EHA46884.1), and Exophilia dermatitis LOV (EHY60539.1) were ordered from Integrated DNA Technologies as gBlocks and assembled by Gibson cloning or PCR assembly. Transgenes were cloned into a pET21/28-derived bacterial expression vector. C-terminal mCherry fusions with a (GGGS)2 linker were generated by Gibson cloning. Genetic constructs were transformed into competent Escherichia coli (C2984H; NEB Turbo). Mutants were generated by QuikChange site-directed mutagenesis. All sequences were verified by Sanger sequencing.
Mammalian genetic constructs.
DNA sequence of BcLOV4 was human codon-optimized (Genscript). The C-terminal mCherry fusion was created as described above. The mCherry-free variant with a C-terminal “3×FLAG” tag (Sigma Aldrich) had a GGGS linker. Transgenes were cloned into the pcDNA3.1 mammalian expression vector (Invitrogen).
Yeast genetic constructs.
BcLOV4-mCherry was cloned into a pRSII326 yeast expression vector with uracil auxotrophic marker (plasmid #35469; Addgene) (62), and transformed into S. cerevisiae (ATCC 201388 strain BY4741) competent cells prepared using a Zymo Research Frozen-EZ Yeast Transformation II Kit. Cells were cultured in uracil dropout medium (Sigma-Aldrich).
Recombinant protein expression, isolation, and purification.
Recombinant proteins were expressed in E. coli BL21(DE3). Cells were shaken (250 rpm) postinduction for 18–22 h at 18 °C in complete darkness, harvested by centrifugation, and dissolved in 50 mL of lysis buffer (50 mM sodium phosphate, 500 mM NaCl, 0.5% Triton X-100, pH 6.5) per liter of harvested culture. Samples were homogenized through a 21-gauge needle, sonicated, and clarified by centrifugation, all at ≤4 °C. His6-tagged protein was affinity-purified by fast protein liquid chromatography (FPLC) (AKTA Basic) on Ni-NTA (GE HisTrap FF) columns in darkness, using a stringent column wash (20–200 mM imidazole linear gradient). Protein was eluted with 500 mM imidazole and buffer exchanged into 1× PBS using PD-10 desalting columns and centrifuged to pellet insoluble debris.
In vitro refolding.
His6-Gb1–tagged BcLOV4 DUF (∆1–356) was expressed in BL21(DE3) E. coli. After lysing cells with a French Pressure Cell (Avestin EmulsiFlex-C5) and centrifuging, the pellet was resuspended in protein solubilization buffer [50 mM Tris⋅HCl (pH 8), 500 mM NaCl, 0.5% Triton X-100, 0.5 mM DTT, 6 M guanidine HCl, 2 mM EDTA]. After denaturation at 4 °C for 5–10 min, supernatant was added drop-by-drop to 500 mL of dilution buffer [50 mM Tris⋅HCl (pH 8), 500 mM NaCl] over a 2-h period to refold the protein. Protein was concentrated via Amicon stirred cell and FPLC-purified on a Superdex 75 or 200 size exclusion column.
Eukaryotic Cellular Assays.
Mammalian cell culture and transduction.
HEK293T cells were cultured in D10 media and maintained at 37 °C in a 5% CO2 incubator. Cells were seeded onto collagen-treated or poly-d-lysine–treated glass bottom dishes or into 24-well glass-bottom plates, and transfected at ∼20–30% confluence using the TransIT-293 transfection reagent. Cells were imaged 24–48 h posttransfection.
Yeast sample preparation.
Yeast strains were immobilized on agarose pads before imaging, as reported by others (63).
Optical microscopy.
Fluorescence microscopy was performed on an automated Leica DMI6000B fluorescence microscope as described previously (64, 65). When needed, isoprenylated GFP (66) was cotransfected as a plasma membrane marker. After a 5-s-long blue light pulse (15 mW/cm2), BcLOV4-mCherry images were collected every 200 ms (membrane association) or 5 s (membrane dissociation). Localization kinetics was measured for single cells by line section analysis in ImageJ and MATLAB. Spinning-disk confocal microscopy was performed as described previously (65).
HEK cell fixation and immunocytochemistry.
Cells were fixed in 4% paraformaldehyde in PBS for 15 min at room temperature, under dim red light (dark-adapted) or strobed illumination (Mightex; λ = 455 nm, ≥15 mW/cm2, 5-s on/25-s off). Immunocytochemistry analysis of 3×FLAG-tagged protein was performed by standard methods with Alexa 488-conjugated anti-3×FLAG (#5407; Cell Signaling Technology) or anti-3×FLAG antibody (#8146; Cell Signaling Technology) followed by an Alexa 488-conjugated secondary antibody (#4408; Cell Signaling Technology).
Diffusion estimates.
The intracellular diffusion constant (Dintra) of BcLOV4-mCherry was calculated as 8.8E-8 cm2/s, assuming an in vitro dark-adapted hydrodynamic radius of 10 nm measured by DLS and an intracellular viscosity of 2.5 centipoise in mammalian cells (67). The timescale for diffusion to the plasma membrane was considered the time to travel a length of a cell radius (rcell) in two dimensions [t ∼ rcell2/(4Dintra)] (68), assuming a 5- to 7.5-μm radius for HEK cells. The diffusion timescale in HEK was estimated as ∼0.7–1.6 s. Diffusion timescales in yeast (estimated as ∼0.5–1.0 s) were calculated similarly assuming a 2- to 3-μm radius and cytoplasmic viscosity of 10 centipoise (69).
In Vitro Protein Analysis: Nonlipid Interactions.
Absorbance spectroscopy and photocycling measurements.
Absorbance scans were measured on an Ocean optics USB2000+ spectrophotometer. Photocycle kinetics was measured by monitoring the absorbance at 450 nm (A450); after 15 s of baseline measurements, samples were stimulated with a collimated LED (Mightex; 10 s, λ = 455 nm, 15 mW/cm2), and recovery was monitored in the dark. For solid-phase photocycling measurements, 40 ng of His6-tagged protein was nutated with 0.5 mg of magnetic Ni-NTA beads (resin 88221; Thermo Fisher) in 400 μL of PBS for 1 h, washed, and resuspended in 200 μL of PBS; flavin fluorescence scans were then made on a Tecan Infinite M200 plate reader (λex = 450; λem = 505), similar to absorbance scans.
Protein quantification and flavin incorporation determination.
Flavin and holoprotein concentration were determined by A450 measurements (εFMN-450 = 12,500 M−1⋅cm−1). To estimate protein concentration from A280 measurements, the optical density loss contributions of flavin, mCherry, and photoaggregates were subtracted. A450-derived flavin concentration was converted to A280FMN (εFMN-280 = 20,300 M−1⋅cm−1), using the A450/A587 ratio for purified His6-mCherry control protein (∼0.01) to account for A450mCherry. Scattering contributions were accounted for as reported by others (70). Apoprotein extinction coefficients were calculated using ExPASy-ProtParam (71). Reported concentration is for holoprotein.
DLS and turbidity imaging.
Particle size analysis was performed using a Zetasizer Nano Series (Malvern Instruments; λ = 633 nm) for 5 µM protein in PBS. After establishing baseline values in the dark, samples were illuminated by a collimated LED (Mightex; 5 s, λ = 455 nm, 15 mW/cm2), and then returned to the dark DLS chamber.
TLC/cofactor identification.
Cofactor was isolated as described by others (72). TLC was performed on glass silica gel plates with n-butanol/acetic acid/water [3:1:1 (vol/vol)]. Plates were dried and imaged on a UV transilluminator. Rf values were as follows: BcLOV4 cofactor (0.26), FMN (0.26), FAD (0.14), and riboflavin (0.61).
Size exclusion chromatography with multiangle light scattering.
Protein in size exclusion chromatography–multiangle light scattering (MALS)-compatible buffer (50 mM sodium phosphate, 500 mM sodium chloride) was analyzed using an in-line HPLC (1200; Agilent Technologies), and MALS system (Wyatt DAWN HELEOS II and OPTILAB T-rEX, with Astra analysis software). Protein was loaded onto a Superdex 75 column (100 µL at 0.4 mg/mL) and was injected at a flow rate of 0.5 mL/min, over a 53-min-long profile.
Denaturing gel analysis and Western blots.
SDS/PAGE gels (4–12% Bis-Tris NuPAGE) were prepared by standard methods and visualized with InstantBlue Coomassie stain. Western blots were created by standard methods using mouse primary antibodies and IRDye 680RD goat anti-mouse IgG (LI-COR; 925-68070) secondary antibody for visualization on an Odyssey CLx Infrared Imaging System.
In Vitro Protein Analysis: Protein–Lipid Interactions.
SPR.
SPR measurements were made on a Biacore T200. Small unilamellar vesicles (SUVs) were generated by hydrating 1.5 mM total phospholipids in HBS-N buffer (25 mM Hepes, 150 mM NaCl, pH 7.4), sonicating, performing eight freeze/thaw cycles in a dry ice/ethanol bath, and passing the sample through an Avanti extruder (0.05-μm membrane, 15 times). SUVs were immobilized on a Sensor chip L1 at 2 μL/min for 30 min [∼4,000–10,000 resonance units (RU)]. His6-BcLOV4-mCherry proteins in HBS-N were passed over the chip surface (30 μL/min for 10 min). The 100 mM NaOH was injected (50 μL/min for 1 min) to regenerate the SUV-coated chip after each binding experiment. Steady-state equilibrium values were analyzed in MATLAB.
Water-in-oil emulsions.
Droplets were formed by vigorously pipetting 30 μL of 20 mM lipids (total in decane) with 1.28 μL of purified mCherry-tagged protein in PBS. Twenty microliters of the mixture were transferred to microwells and imaged with a 20× objective. Automated MATLAB scripts were used to segment the interface vs. dispersed phase, and to calculate fluorescence over these regions (area-normalized) and their ratios. All ratios were normalized to the max ratio for illuminated wild-type protein.
Protein–lipid overlay assay.
Blots were created based on methods reported by others (73), using 1 μL of 3 mM phospholipid per spot, and probed with mouse anti-His6 primary antibody (2366; Cell Signaling Technology) and IRDye 680RD goat (polyclonal) anti-mouse IgG (925–68070; LI-COR). Processed blots were imaged on an Odyssey Infrared Imaging System.
Bioinformatics.
Maximum-likelihood phylogenetic tree construction.
The tree was constructed by aligning all candidate sequences with MUSCLE, building a phylogenetic tree with PhyML, and rendering a tree with TreeDyn through the phylogeny.fr webserver (www.phylogeny.fr) (74). Taxonomic class assignments were made with the Interactive Tree of Life (iTOL) server (itol.embl.de) (75).
Secondary-structure modeling and consensus annotation.
Candidate amino acid sequences were submitted individually to iTASSER (76), Jpred (77), Phyre (78), and PSIPRED (79). A consensus secondary-structure prediction was generated by equally weighting α-helix and β-sheet predictions from the four algorithms at every amino acid residue and requiring two of four programs to agree on any given structural element. Amphipathic helices were predicted with the HeliQuest web server (80).
De novo energy minimization modeling in Rosetta.
De novo structural predictions were made with Rosetta, version 3.8, on 100 Intel E5-2665 2.4-GHz Xeon processors using the Abinitio Relax protocol. The consensus secondary-structure prediction was used throughout the process to filter out trajectories that were unlikely to converge to the supplied secondary structure. Near-native topologies were identified by determining the most frequently sampled conformations using clustering with rmsd as the distance metric. The lowest energy trajectory of the largest cluster was hypothesized to be the closest approximation of the native structure.
Supplementary Material
Acknowledgments
We thank Ivan Kuznetsov for computational structure predictions in Rosetta, the Dan Hammer laboratory for FPLC access, the Matt Good laboratory for technical assistance with emulsion studies, the Youhai Chen laboratory and the Wistar Institute molecular screening facility for technical assistance with liposome preparation and SPR, and Casey Sniffin for technical assistance. We also thank Ranganath Parthasarathy, Julia Schumacher, and Richard Neubig for helpful discussion. B.Y.C. acknowledges the support of National Science Foundation (NSF) Systems and Synthetic Biology (MCB 1652003), NSF Biophotonics (CBET 126497), NIH/National Institute on Drug Abuse (R21 DA040434), Penn Medicine Neuroscience Center, W. W. Smith Charitable Trust for the Heart, NIH/National Institute of Neurological Disorders and Stroke (NINDS) (R01 NS101106), and Defense Advanced Research Projects Agency (Living Foundries 5710003185). K.H.G. acknowledges support from NIH/National Institute of General Medical Sciences (R01 GM106239). S.T.G. acknowledges the fellowship support of the NSF Graduate Research Fellowship Program and the Penn Center for Neuroengineering and Therapeutics Training Grant (NIH/NINDS; T32 NS091006). B.S.S. acknowledges support from an NIH postdoctoral fellowship (F32GM119430).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The plasmid for mammalian codon-optimized BcLOV4 fused to mCherry and the plasmid for native sequence BcLOV4 fused to mCherry were deposited with AddGene (https://www.addgene.org) (accession nos. 114595 and 114596).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802832115/-/DCSupplemental.
References
- 1.Losi A, Gärtner W. Solving blue light riddles: New lessons from flavin-binding LOV photoreceptors. Photochem Photobiol. 2017;93:141–158. doi: 10.1111/php.12674. [DOI] [PubMed] [Google Scholar]
- 2.Herrou J, Crosson S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat Rev Microbiol. 2011;9:713–723. doi: 10.1038/nrmicro2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glantz ST, et al. Functional and topological diversity of LOV domain photoreceptors. Proc Natl Acad Sci USA. 2016;113:E1442–E1451. doi: 10.1073/pnas.1509428113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 5.Möglich A, Ayers RA, Moffat K. Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 7.Conrad KS, Bilwes AM, Crane BR. Light-induced subunit dissociation by a light-oxygen-voltage domain photoreceptor from Rhodobacter sphaeroides. Biochemistry. 2013;52:378–391. doi: 10.1021/bi3015373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motta-Mena LB, et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stierl M, et al. Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J Biol Chem. 2011;286:1181–1188. doi: 10.1074/jbc.M110.185496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie JM, Gawthorne J, Young G, Fraser NJ, Roe AJ. LOV to BLUF: Flavoprotein contributions to the optogenetic toolkit. Mol Plant. 2012;5:533–544. doi: 10.1093/mp/sss020. [DOI] [PubMed] [Google Scholar]
- 11.Toettcher JE, Voigt CA, Weiner OD, Lim WA. The promise of optogenetics in cell biology: Interrogating molecular circuits in space and time. Nat Methods. 2011;8:35–38. doi: 10.1038/nmeth.f.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pudasaini A, El-Arab KK, Zoltowski BD. LOV-based optogenetic devices: Light-driven modules to impart photoregulated control of cellular signaling. Front Mol Biosci. 2015;2:18. doi: 10.3389/fmolb.2015.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziegler T, Möglich A. Photoreceptor engineering. Front Mol Biosci. 2015;2:30. doi: 10.3389/fmolb.2015.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollinger S, Hepler JR. Cellular regulation of RGS proteins: Modulators and integrators of G protein signaling. Pharmacol Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- 15.Berman DM, Gilman AG. Mammalian RGS proteins: Barbarians at the gate. J Biol Chem. 1998;273:1269–1272. doi: 10.1074/jbc.273.3.1269. [DOI] [PubMed] [Google Scholar]
- 16.Urano D, et al. Endocytosis of the seven-transmembrane RGS1 protein activates G-protein-coupled signalling in Arabidopsis. Nat Cell Biol. 2012;14:1079–1088. doi: 10.1038/ncb2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, et al. Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae. PLoS Pathog. 2011;7:e1002450. doi: 10.1371/journal.ppat.1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumacher J. How light affects the life of Botrytis. Fungal Genet Biol. 2017;106:26–41. doi: 10.1016/j.fgb.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Krauss U, et al. Distribution and phylogeny of light-oxygen-voltage-blue-light-signaling proteins in the three kingdoms of life. J Bacteriol. 2009;191:7234–7242. doi: 10.1128/JB.00923-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu G, et al. Ubiquitous distribution of helmchrome in phototactic swarmers of the stramenopiles. Protoplasma. 2016;253:929–941. doi: 10.1007/s00709-015-0857-7. [DOI] [PubMed] [Google Scholar]
- 21.Foley BJ, et al. Characterization of a vivid homolog in Botrytis cinerea. Photochem Photobiol. April 23, 2018 doi: 10.1111/php.12927. [DOI] [PubMed] [Google Scholar]
- 22.Schumacher J, Simon A, Cohrs KC, Viaud M, Tudzynski P. The transcription factor BcLTF1 regulates virulence and light responses in the necrotrophic plant pathogen Botrytis cinerea. PLoS Genet. 2014;10:e1004040. doi: 10.1371/journal.pgen.1004040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christie JM, Swartz TE, Bogomolni RA, Briggs WR. Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 2002;32:205–219. doi: 10.1046/j.1365-313x.2002.01415.x. [DOI] [PubMed] [Google Scholar]
- 24.Zuo Z-C, et al. A study of the blue-light-dependent phosphorylation, degradation, and photobody formation of Arabidopsis CRY2. Mol Plant. 2012;5:726–733. doi: 10.1093/mp/sss007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods. 2013;10:249–252. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- 26.Heo WD, et al. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584:1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 28.Bernstein LS, Grillo AA, Loranger SS, Linder ME. RGS4 binds to membranes through an amphipathic alpha -helix. J Biol Chem. 2000;275:18520–18526. doi: 10.1074/jbc.M000618200. [DOI] [PubMed] [Google Scholar]
- 29.Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R. Cytoplasmic volume modulates spindle size during embryogenesis. Science. 2013;342:856–860. doi: 10.1126/science.1243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nash AI, Ko W-H, Harper SM, Gardner KH. A conserved glutamine plays a central role in LOV domain signal transmission and its duration. Biochemistry. 2008;47:13842–13849. doi: 10.1021/bi801430e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganguly A, Thiel W, Crane BR. Glutamine amide flip elicits long distance allosteric responses in the LOV protein Vivid. J Am Chem Soc. 2017;139:2972–2980. doi: 10.1021/jacs.6b10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gleichmann T, Diensthuber RP, Möglich A. Charting the signal trajectory in a light-oxygen-voltage photoreceptor by random mutagenesis and covariance analysis. J Biol Chem. 2013;288:29345–29355. doi: 10.1074/jbc.M113.506139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kavran JM, et al. Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J Biol Chem. 1998;273:30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- 34.Yeung T, et al. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 35.Hekman M, et al. Reversible membrane interaction of BAD requires two C-terminal lipid binding domains in conjunction with 14-3-3 protein binding. J Biol Chem. 2006;281:17321–17336. doi: 10.1074/jbc.M600292200. [DOI] [PubMed] [Google Scholar]
- 36.Andresen BT, Rizzo MA, Shome K, Romero G. The role of phosphatidic acid in the regulation of the Ras/MEK/Erk signaling cascade. FEBS Lett. 2002;531:65–68. doi: 10.1016/s0014-5793(02)03483-x. [DOI] [PubMed] [Google Scholar]
- 37.Sato H, Feix JB. Peptide-membrane interactions and mechanisms of membrane destruction by amphipathic α-helical antimicrobial peptides. Biochim Biophys Acta. 2006;1758:1245–1256. doi: 10.1016/j.bbamem.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 38.Ariño J, Ramos J, Sychrová H. Alkali metal cation transport and homeostasis in yeasts. Microbiol Mol Biol Rev. 2010;74:95–120. doi: 10.1128/MMBR.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suomalainen H, Nurminen T. The lipid composition of cell wall and plasma membrane of baker’s yeast. Chem Phys Lipids. 1970;4:247–256. [Google Scholar]
- 40.Zinser E, et al. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Möglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henry JT, Crosson S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol. 2011;65:261–286. doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheung J, Bingman CA, Reyngold M, Hendrickson WA, Waldburger CD. Crystal structure of a functional dimer of the PhoQ sensor domain. J Biol Chem. 2008;283:13762–13770. doi: 10.1074/jbc.M710592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neiditch MB, et al. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 46.Srinivasa SP, Bernstein LS, Blumer KJ, Linder ME. Plasma membrane localization is required for RGS4 function in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:5584–5589. doi: 10.1073/pnas.95.10.5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy AA, Lemberg KE, Chidiac P. Recruitment of RGS2 and RGS4 to the plasma membrane by G proteins and receptors reflects functional interactions. Mol Pharmacol. 2003;64:587–593. doi: 10.1124/mol.64.3.587. [DOI] [PubMed] [Google Scholar]
- 48.Amselem J, et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011;7:e1002230. doi: 10.1371/journal.pgen.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bosch DE, et al. A P-loop mutation in Gα subunits prevents transition to the active state: Implications for G-protein signaling in fungal pathogenesis. PLoS Pathog. 2012;8:e1002553. doi: 10.1371/journal.ppat.1002553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu S, Dean RA. G protein α subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol Plant Microbe Interact. 1997;10:1075–1086. doi: 10.1094/MPMI.1997.10.9.1075. [DOI] [PubMed] [Google Scholar]
- 51.Idnurm A, Verma S, Corrochano LM. A glimpse into the basis of vision in the kingdom Mycota. Fungal Genet Biol. 2010;47:881–892. doi: 10.1016/j.fgb.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tischer D, Weiner OD. Illuminating cell signalling with optogenetic tools. Nat Rev Mol Cell Biol. 2014;15:551–558. doi: 10.1038/nrm3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hallett RA, Zimmerman SP, Yumerefendi H, Bear JE, Kuhlman B. Correlating in vitro and in vivo activities of light-inducible dimers: A cellular optogenetics guide. ACS Synth Biol. 2016;5:53–64. doi: 10.1021/acssynbio.5b00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 55.Zoltowski BD, Vaccaro B, Crane BR. Mechanism-based tuning of a LOV domain photoreceptor. Nat Chem Biol. 2009;5:827–834. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diensthuber RP, et al. Biophysical, mutational, and functional investigation of the chromophore-binding pocket of light-oxygen-voltage photoreceptors. ACS Synth Biol. 2014;3:811–819. doi: 10.1021/sb400205x. [DOI] [PubMed] [Google Scholar]
- 57.Zayner JP, Antoniou C, French AR, Hause RJ, Jr, Sosnick TR. Investigating models of protein function and allostery with a widespread mutational analysis of a light-activated protein. Biophys J. 2013;105:1027–1036. doi: 10.1016/j.bpj.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Diensthuber RP, Bommer M, Gleichmann T, Möglich A. Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure. 2013;21:1127–1136. doi: 10.1016/j.str.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 59.Kawano F, Aono Y, Suzuki H, Sato M. Fluorescence imaging-based high-throughput screening of fast- and slow-cycling LOV proteins. PLoS One. 2013;8:e82693. doi: 10.1371/journal.pone.0082693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zoltowski BD, Motta-Mena LB, Gardner KH. Blue light-induced dimerization of a bacterial LOV-HTH DNA-binding protein. Biochemistry. 2013;52:6653–6661. doi: 10.1021/bi401040m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song S-H, et al. Modulating LOV domain photodynamics with a residue alteration outside the chromophore binding site. Biochemistry. 2011;50:2411–2423. doi: 10.1021/bi200198x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chee MK, Haase SB. New and redesigned pRS plasmid shuttle vectors for genetic manipulation of Saccharomyces cerevisiae. G3 (Bethesda) 2012;2:515–526. doi: 10.1534/g3.111.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okada S, Lee ME, Bi E, Park HO. Probing Cdc42 polarization dynamics in budding yeast using a biosensor. Methods Enzymol. 2017;589:171–190. doi: 10.1016/bs.mie.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hannanta-Anan P, Chow BY. Optogenetic control of calcium oscillation waveform defines NFAT as an integrator of calcium load. Cell Syst. 2016;2:283–288. doi: 10.1016/j.cels.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hannanta-Anan P, Chow BY. Optogenetic inhibition of Gαq protein signaling reduces calcium oscillation stochasticity. ACS Synth Biol. 2018;7:1488–1495. doi: 10.1021/acssynbio.8b00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kennedy MJ, et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mastro AM, Babich MA, Taylor WD, Keith AD. Diffusion of a small molecule in the cytoplasm of mammalian cells. Proc Natl Acad Sci USA. 1984;81:3414–3418. doi: 10.1073/pnas.81.11.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lauffenburger DA, Linderman J. Receptors: Models for Binding, Trafficking, and Signaling. Oxford Univ Press; Oxford: 1993. [Google Scholar]
- 69.Burns VW. Measurement of viscosity in living cells by a fluorescence method. Biochem Biophys Res Commun. 1969;37:1008–1014. doi: 10.1016/0006-291x(69)90232-0. [DOI] [PubMed] [Google Scholar]
- 70.Birdsall B, et al. Correction for light absorption in fluorescence studies of protein-ligand interactions. Anal Biochem. 1983;132:353–361. doi: 10.1016/0003-2697(83)90020-9. [DOI] [PubMed] [Google Scholar]
- 71.Artimo P, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): Binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci USA. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fayngerts SA, et al. TIPE3 is the transfer protein of lipid second messengers that promote cancer. Cancer Cell. 2014;26:465–478. doi: 10.1016/j.ccr.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dereeper A, et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Letunic I, Bork P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang J, et al. The I-TASSER suite: Protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 80.Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: A web server to screen sequences with specific alpha-helical properties. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.