According to the American Cancer Society, 266,120 women in the United States will be diagnosed with breast cancer and 40,920 will die in 2018 (1). Approximately 70% of all these breast cancers express estrogen receptor α (ERα) and are initially treated with endocrine therapies that target this nuclear hormone receptor (2). ERα plays a major role in the development of breast tumorigenesis, and its activity is mediated by binding of ligands, such as its naturally occurring major ligand, 17-β estradiol (E2). Ligand binding leads to dimerization and the subsequent binding of ERα to specific DNA sequences, followed by the recruitment of coactivators (CoActs) needed to transactivate ERα target genes (3). Three main US Food and Drug Administration (FDA)-approved endocrine therapies for inhibiting ERα signaling in patients who have breast cancer are as follows: aromatase inhibitors (AIs) [e.g., exemestane (4)] that decrease the E2 level in tumors, selective ERα modulators [e.g., tamoxifen (5)] that increase corepressors (and decrease CoActs) bound to ERα, and selective ERα down-regulators (SERDs) [e.g., fulvestrant (6)] that increase proteasome-mediated ERα protein turnover. However, roughly 50% of ERα-positive tumors acquire resistance to these therapies after an initial favorable response (7). This acquired resistance is often coupled to breast cancer metastasis [especially to bone (8)] that results in death. Thus, a much better understanding of the mechanisms driving endocrine therapy resistance and metastasis is critical. Previous studies have suggested multiple possible mechanisms of acquired resistance, including increased ERα and CoAct expression; mutations in the gene encoding ERα (ESR1) that give rise to estrogen-independent, constitutively active receptors (9, 10); and aberrant growth factor signaling (e.g., EGFR, HER2, IGF1R pathways) and other kinase pathways (e.g., mTOR, CDK4/6) (11, 12). While pharmacological targeting of EGFR and IGF1R did not improve patient survival over endocrine therapy alone, patients in clinical trials cotargeting other kinase pathways (e.g., mTOR, CDK4/6) in combination with an AI demonstrated increased survival (12, 13). However, additional mechanisms (including other unanticipated kinases) that may limit the effectiveness of endocrine therapy in the clinic still remain unclear. To begin to address this “knowledge gap,” Xiao et al. (14) conducted genome-wide, unbiased knockout screens on two ERα-expressing breast cancer cell lines to identify key mediators of endocrine resistance.
CRISPR/Cas9 Screens Identify Key Genes Driving Endocrine Resistance and Synthetic Lethal Vulnerabilities
The use of CRISPR-associated nuclease Cas9 loss-of-function screens employing guide RNAs targeting the human protein-encoding genome has revealed essential proteins that confer drug resistance in cancer cell lines in vitro and tumor metastasis in vivo [reviewed by Shalem et al. (15) and reported by Chen et al. (16)]. However, no such screens have been performed in the context of ERα-positive breast cancer until the study of Xiao et al. (14). Results from their CRISPR/Cas9 screen identified key genes essential for growth in two widely used ERα-expressing breast cancer cell lines, MCF7 and T47D, in both estrogen-deprived and E2-treated conditions. They found known ERα-positive breast cancer “driver” genes, such as the transcription factors ESR1, FOXA1, GATA3, and MYC, as well as the CoAct NCOA3, which, when knocked out, reduced growth in both conditions. Importantly, when assaying for genes whose disruption increased growth under estrogen-deprived conditions (mimicking AI treatment) versus E2, the authors found known tumor suppressor genes, including PTEN, TSC1/2, and NF1. However, the top candidate from this loss-of-function screen was the gene encoding C-terminal SRC kinase (CSK). Subsequent experiments revealed that the loss of CSK conferred enhanced growth in the absence of E2 through increased activation of SRC family tyrosine kinases (SFKs) as assayed by phosphorylation at Tyrosine 419 (Y419). CSK encodes a tumor suppressor kinase known to inhibit the action of oncogenic SFKs by phosphorylation at Y530 (17).
“Synthetic lethality” is invoked when a defect or loss in one gene does not impact cell viability, but is only lethal when combined with a defect or loss in another gene. It has been pursued as a therapeutic strategy by using pharmacological inhibitors as well as with RNA interference, and, more recently, in CRISPR screens [reviewed by O’Neil et al. (18)]. Xiao et al. (14) exploited this concept by performing a second CRISPR screen in T47D CSK-null cells to find synthetic lethal vulnerabilities to overcome E2-independent growth promoted by loss of CSK. Importantly, the authors found several candidate genes, such as EPHB2, PIK3R2, protooncogene CRK, and the p21-activated protein kinase 2 (PAK2). The authors focused on PAK2, as it is highly expressed in ERα-positive breast cancer cells and pharmacological inhibitors exist for this kinase. Of note, loss of PAK1, a highly related PAK family member, did not confer any synthetic lethality. Importantly, by using inhibitors against SFKs, activation of PAK2 as assayed by phosphorylation at Serine 141 (S141) was reduced. Altogether, the authors clearly show that loss of CSK enhances the activity of SFKs, and thus PAK2 activity, thereby promoting endocrine therapy-resistant cell growth (Fig. 1).
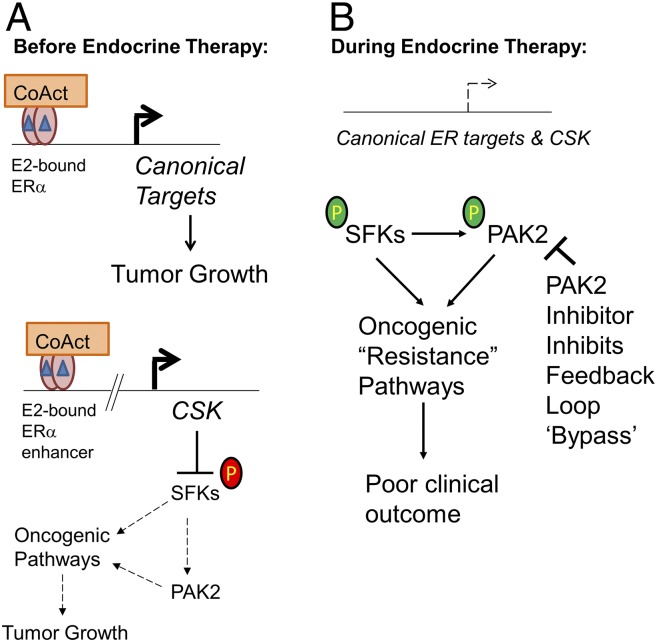
Fig. 1.
Development of endocrine therapy resistance in ERα-expressing breast cancers and a new proposed treatment paradigm. (A) E2 (blue triangles) promotes breast tumorigenesis by binding ERα and recruiting CoActs to stimulate canonical ERα target genes that promote growth. Xiao et al. (14) show that E2-liganded ERα activates expression of CSK to create a negative feedback loop to suppress oncogenesis mediated by the SFKs/PAK2 axis by inhibitory phosphorylation of SFKs (red circled “P”). Bent arrows indicate gene transcription. (B) Upon endocrine therapy targeting E2-bound ERα (main text), the expression of canonical ERα target genes, as well as the CSK-mediated negative feedback loop, is inhibited. The loss of CSK allows for more active SFKs/PAK2 kinase signaling (observed by activating phosphorylation of SFKs and PAK2, green circled “P”) and, with time, induces resistance to endocrine therapy by “bypassing” the negative feedback loop. Importantly, Xiao et al. (14) show that a PAK2 inhibitor effectively suppresses tumor growth in immunodeficient mice transplanted with CSK knockout cells.
Loss of an E2/ERα-Regulated Negative Feedback Loop Drives Endocrine Resistance
As CSK loss enhances growth in estrogen-deprived cells but not in E2-treated cells, Xiao et al. (14) investigated whether E2/ERα may directly regulate CSK expression. Indeed, the authors found that E2 induced (while tamoxifen and fulvestrant reduced) expression of the CSK gene. In addition, ERα bound to an enhancer 10 kb upstream of the CSK transcription start site, and subsequent CRIPSR-mediated knockout of the enhancer impaired E2 induction. Upon endocrine therapy treatment that inhibits ERα signaling (tamoxifen or fulvestrant), the SFKs/PAK2 kinase pathway became “hyper”-activated as a result of reduced CSK expression. RNA-sequencing experiments additionally revealed elevated expression of 292 CSK target genes normally repressed under nonperturbed conditions. Importantly, when patient outcomes following tamoxifen treatment were analyzed with gene expression data, the 292 CSK-suppressed gene signature score was correlated with poorer survival, and high PAK2 mRNA was also correlated with a worse outcome (Fig. 1).
Clinical Implications for Endocrine Therapy-Resistant ERα-Positive Breast Cancers
Since the loss of CSK with endocrine therapy correlated with higher PAK2 activity and worse patient outcome, Xiao et al. (14) tested whether a small-molecule inhibitor (FRAX597) that targets group 1 PAKs (PAK1, PAK2, and PAK3) would have preclinical efficacy in tumor reduction in female ovariectomized mice transplanted with MCF7 CSK-null cells. FRAX597 was chosen since a PAK2-selective inhibitor does not yet exist. FRAX597 monotherapy significantly inhibited tumor growth only in E2-deprived, but not E2-treated, animals, while fulvestrant inhibited tumor growth in both treatment groups. Importantly, the combination of a group 1 PAK inhibitor and a SERD synergistically inhibited tumor growth. The authors observed similar findings in an ERα-expressing, HER2-negative breast cancer patient-derived xenograft model as well.
As mentioned, a variety of suggested mechanisms may account for endocrine-resistant ERα-positive breast cancers. Since targeting receptor tyrosine kinases, such as EGFR or IGF1R, has not improved survival in patients with advanced ER-positive, HER2-negative breast cancer (12, 13), the work of Xiao et al. (14) holds promise for these patients. Other FDA-approved therapies to treat ER-positive metastatic breast cancer have resulted in positive clinical trials, such as the use of CDK4/6 (e.g., palbociclib) and mTOR (e.g., everolimus) inhibitors. As breast cancers resistant to these inhibitors will undoubtedly arise, a PAK2 inhibitor may be of additional benefit in this setting, although more studies will be needed as proof of principle. Additionally, patients with metastatic disease who experience disease progression during AI treatment often have mutations in the ESR1 gene [up to 39% (19)]. It will be of interest to know if endocrine therapy-resistant growth of ESR1 mutant cancer cells is sensitive to PAK2 inhibition. Of note, CSK gene expression is up-regulated in at least two ESR1 mutant (D538G and Y537N)-expressing MCF7 cell lines (20). Finally, whether PAK2 inhibition has any efficacy in reducing metastasis of ER-positive preclinical models should be tested. The timely and interesting study of Xiao et al. (14) provides new mechanistic insight (loss of CSK and SFK/PAK2 activation) to address an old and lethal problem of endocrine therapy resistance.
Acknowledgments
My research is supported by the Adrienne Helis Malvin Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with the Baylor College of Medicine Cancer Center.
Footnotes
The author declares no conflict of interest.
See companion article on page 7869 in issue 31 of volume 115.
References
- 1.American Cancer Society . Cancer Facts & Figures 2018. American Cancer Society; Atlanta: 2018. [Google Scholar]
- 2.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 3.Foulds CE, et al. Proteomic analysis of coregulators bound to ERα on DNA and nucleosomes reveals coregulator dynamics. Mol Cell. 2013;51:185–199. doi: 10.1016/j.molcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Asten K, Neven P, Lintermans A, Wildiers H, Paridaens R. Aromatase inhibitors in the breast cancer clinic: Focus on exemestane. Endocr Relat Cancer. 2014;21:R31–R49. doi: 10.1530/ERC-13-0269. [DOI] [PubMed] [Google Scholar]
- 5.Massarweh S, Schiff R. Unraveling the mechanisms of endocrine resistance in breast cancer: New therapeutic opportunities. Clin Cancer Res. 2007;13:1950–1954. doi: 10.1158/1078-0432.CCR-06-2540. [DOI] [PubMed] [Google Scholar]
- 6.Ciruelos E, et al. The therapeutic role of fulvestrant in the management of patients with hormone receptor-positive breast cancer. Breast. 2014;23:201–208. doi: 10.1016/j.breast.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Clarke R, Tyson JJ, Dixon JM. Endocrine resistance in breast cancer–An overview and update. Mol Cell Endocrinol. 2015;418:220–234. doi: 10.1016/j.mce.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang XH, Giuliano M, Trivedi MV, Schiff R, Osborne CK. Metastasis dormancy in estrogen receptor-positive breast cancer. Clin Cancer Res. 2013;19:6389–6397. doi: 10.1158/1078-0432.CCR-13-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang QX, Borg A, Wolf DM, Oesterreich S, Fuqua SA. An estrogen receptor mutant with strong hormone-independent activity from a metastatic breast cancer. Cancer Res. 1997;57:1244–1249. [PubMed] [Google Scholar]
- 10.Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations—A mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12:573–583. doi: 10.1038/nrclinonc.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med. 2011;62:233–247. doi: 10.1146/annurev-med-070909-182917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hart CD, et al. Challenges in the management of advanced, ER-positive, HER2-negative breast cancer. Nat Rev Clin Oncol. 2015;12:541–552. doi: 10.1038/nrclinonc.2015.99. [DOI] [PubMed] [Google Scholar]
- 13.Johnston SR. Enhancing endocrine therapy for hormone receptor-positive advanced breast cancer: Cotargeting signaling pathways. J Natl Cancer Inst. 2015;107:djv212. doi: 10.1093/jnci/djv212. [DOI] [PubMed] [Google Scholar]
- 14.Xiao T, et al. Estrogen-regulated feedback loop limits the efficacy of estrogen receptor–targeted breast cancer therapy. Proc Natl Acad Sci USA. 2018;115:7869–7878. doi: 10.1073/pnas.1722617115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okada M. Regulation of the SRC family kinases by Csk. Int J Biol Sci. 2012;8:1385–1397. doi: 10.7150/ijbs.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Neil NJ, Bailey ML, Hieter P. Synthetic lethality and cancer. Nat Rev Genet. 2017;18:613–623. doi: 10.1038/nrg.2017.47. [DOI] [PubMed] [Google Scholar]
- 19.Fribbens C, et al. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J Clin Oncol. 2016;34:2961–2968. doi: 10.1200/JCO.2016.67.3061. [DOI] [PubMed] [Google Scholar]
- 20.Jeselsohn R, et al. Allele-specific chromatin recruitment and therapeutic vulnerabilities of ESR1 activating mutations. Cancer Cell. 2018;33:173–186.e5. doi: 10.1016/j.ccell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]